Abstract
Acid phosphatases of budding yeast have been studied for more than forty years. This paper covers biochemical characteristics of acid phosphatases and different aspects in expression regulation of eukaryotic genes, which were researched using acid phosphatases model. A special focus is devoted to cyclin-dependent kinase Pho85p, a negative transcriptional regulator, and its role in maintaining mitochondrial genome stability and to pleiotropic effects of pho85 mutations.
1. Introduction
Reactions of protein phosphorylation and dephosphorylation play a significant regulatory role in cell processes. Activity of many proteins, such as regulatory proteins, histones, permeases of various compounds, and plenty of enzymes, depends on working of protein kinases and phosphatases.
Phosphorus is known to be one of the most necessary macroelements, used in biosynthesis of the most important cell macromolecules, such as nucleic acids, proteins, and lipids. Reducing the level of inorganic phosphate (Pi) in medium leads to changes of cell physiology, influences on ATP synthesis, DNA replication, and other key processes in the cell [1]. In case of phosphate shortage, the cell can replace it by different phosphate-containing organic compounds, which undergo the cleavage of ether bonds by the phosphatases that results in release of Pi and its subsequent importation into the cell. Most of the genes encoding phosphatases are activated and repressed concordantly depending on Pi concentration in the medium. The special positive and negative regulators in and out of cell transduce the signals about that [2].
The budding yeast Saccharomyces cerevisiae appeared to be the most convenient object for researching structure and functions of the phosphatases. Family of nonspecific acid phosphatases is one of the most popular molecular models in yeast genetics that gives unique opportunities for investigating different functions of phosphatases and protein kinases.
2. Phosphate Metabolism
In living organisms, the phosphate is present basically in the form of orthophosphate (HPO4 2−). In the yeast, Pi can be found as free ion, but the most part of it is bound in phospholipids, nucleotides, phosphoproteins, and phosphorylated hydrocarbons. An excess of Pi is accumulated in the form of polyphosphates, which represent linear polymers of orthophosphoric acid. The polyphosphate comprises phosphate atoms linked by anhydride bonds that results in its capability of storing energy and releasing it by the bonds' hydrolysis [3]. Also Pi has an important role in the intracellular pH maintenance. Beside this, Pi acts as a substrate and an effector for many enzymes (for example, phosphofructokinase) and regulates many metabolic pathways. Reaction speed alterations of any process leading to Pi release or its consumption affect the level of intracellular Pi. Decrease of Pi quantity in the medium and in the cell can be compensated by using the intracellular phosphate resources as ATP, phosphoenolpyruvate, sugar phosphates, and polyphosphates [4]. Analysis of almost 6200 yeast genes revealed 22 genes whose expression is sharply increased for the lack of phosphate. This gene group was designated as PHO-regulon. Also Pi concentration influences on the expression of many other genes, which may not participate directly in phosphate metabolism, but their transcription is activated or repressed for the lack of phosphate in the medium [5].
The enzymes of phosphate metabolism include the following: isozymes of the nonspecific acid phosphatase (AP), which provide detachment of phosphate group from the phosphate-containing organic compounds in medium; transport proteins, that is, permeases with different phosphate affinity; alkaline phosphatases; polyphosphatases; polyphosphate kinases; also enzymes with phytase activity [6]. APs of S. cerevisiae are of special interest because they being extracellular enzymes are localized in cell wall and periplasmic space that significantly simplifies qualitative and quantitative methods of enzyme activity testing and also procedure of their extraction. Studying AP isozymes of yeast strain GRF18 (Dr. Hinnen, USA) and the strains from Peterhoff Genetic Collection (PGK), St. Petersburg, Russia revealed that yeast S. cerevisiae synthesize three isozymes of acid phosphatases designated as AP1, AP2, and AP3 [7]. AP1 is synthesized constitutively, while others are repressed by high phosphate concentration. Fractionating on Sephadex G-200 columns showed that all three AP isozymes (AP1, AP2, and AP3) represent glycoproteins with molecular masses more than 200 kDa (Figure 1) [8, 9]. Molecular masses of the polypeptides after deglycosylation were shown to be 57 kDa for AP1, 58 kDa for AP2, and 55 kDa for AP3 [9]. These results agree with the data of Bostian and coworkers obtained in translation product analysis of the total yeast polyA-RNA in cell-free system [10]. Studying properties of the APs showed that all of them have optimum pH value in acid range and that AP1 has lower optimum pH value near 3,7–3,8 than AP2 and AP3 (pH 4,6) (Figure 2). This feature provides the ability of hydrolyzing extracellular substrates in periplasmic space.
Figure 1.
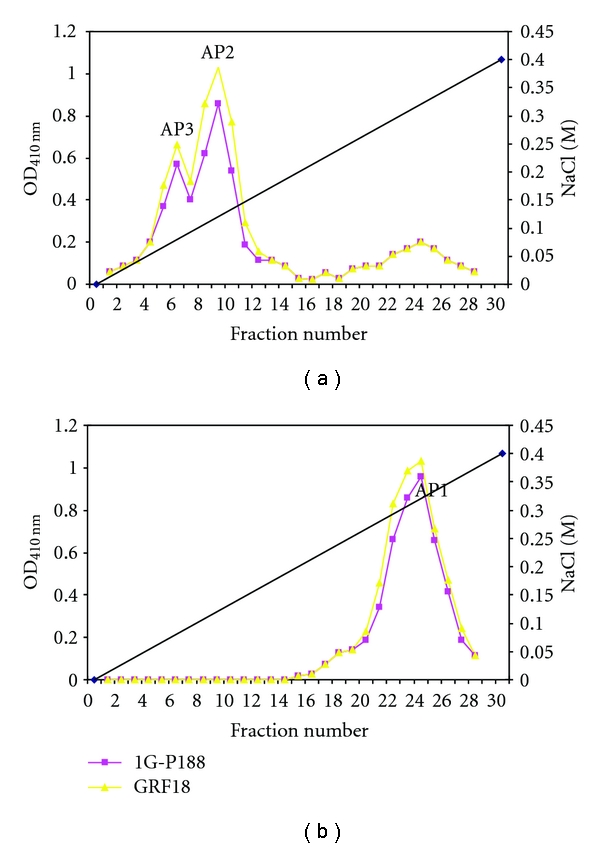
Gel-filtration of APs: (a) yeast strains 1G-P188 (PGK) and GRF18 (USA), cultivated in low-Pi medium; (b) strains 1G-P188 (PGK) and GRF18 (USA), cultivated in high-Pi medium.
Figure 2.
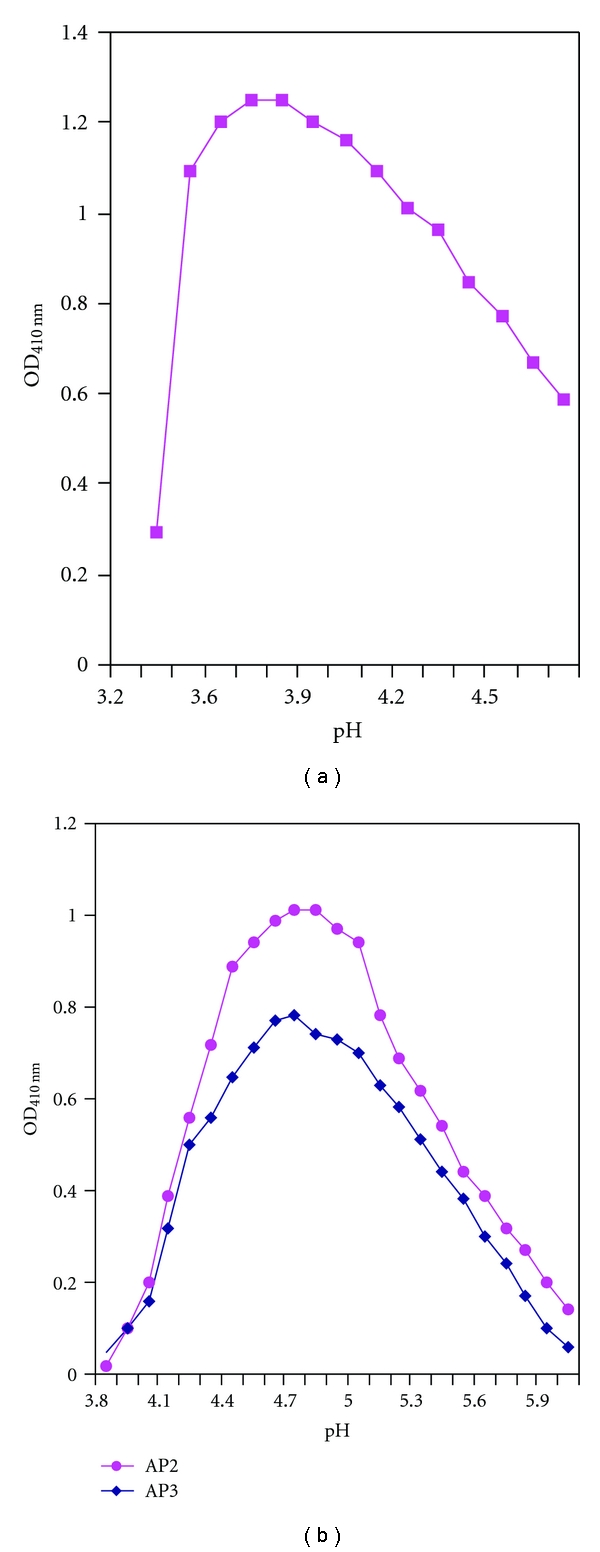
The pH dependence of enzyme activity: (a) AP1; (b) AP2 and AP3.
Constitutive AP1 is more thermostable than repressible APs (Figure 3). AP1 shows full activity at 40°C, while AP2 maintains 40% of its activity during 15 minutes, and AP3 is almost inactivated at 40°C [8, 9].
Figure 3.
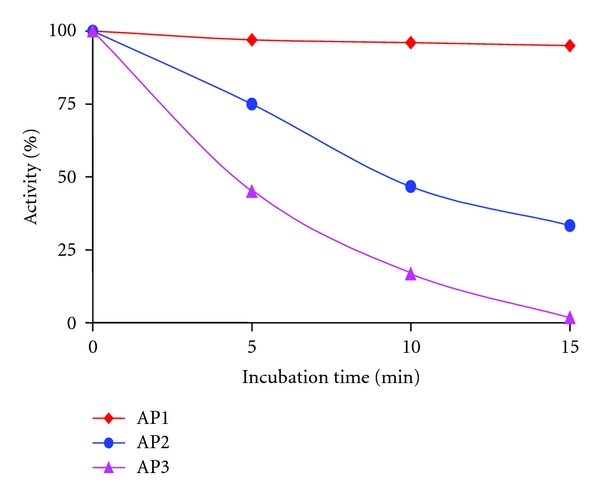
Thermostability of AP's at 40°C.
Exploration of the conditions for repressible APs accumulation in cultural medium revealed that AP3 activity inhibition occurs at the lower concentration of KH2PO4 than that for AP2 [9] (Figure 4).
Figure 4.
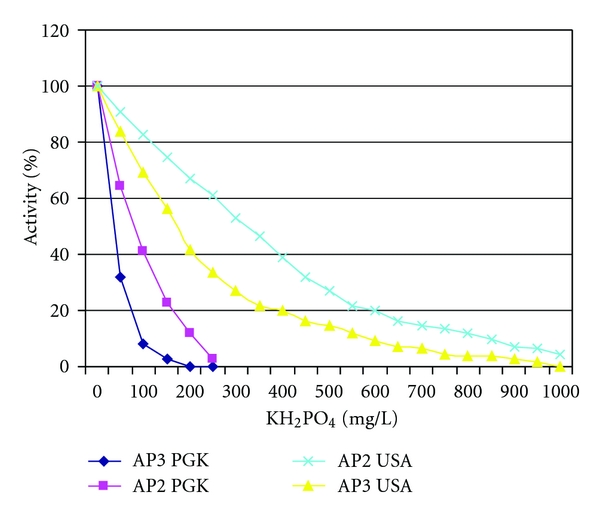
AP2 and AP3 enzyme activity at different KH2PO4 medium concentration. APs from yeast strains of PGK (St. Petersburg, Russia) and GRF18 (USA) were analyzed.
The constitutive AP1 is encoded by gene PHO3 of S. Cerevisiae; it hydrolyses different phosphate-containing substrates in periplasmic space (thiamine pyrophosphates, in particular). The increase of thiamine concentration in medium leads to PHO3 transcription blockage [11].
Repressible AP2 and AP3, encoded by genes PHO5 and PHO10,11, respectively, are synthesized in conditions of low phosphate only. Therefore, the regulation system of repressible APs is a convenient model for comparable genetic analysis of enzymes biosynthesis mechanisms [12]; In derepression conditions, the APs' fraction is mainly composed of AP2 (Pho5p), and the minor fraction consists of Pho10p and Pho11p, which have 87% amino acid sequence identity with Pho5p. Genes PHO3 and PHO5 are linked in II chromosome of S. cerevisiae [13]. The coding regions of both genes have high rate of homology in nucleotide (82%) and amino acid (87%) sequences. This rate decreases to 65% towards flanking noncoding regions and becomes very low in promoter regions of these genes [14]. Genes of acid phosphatases family PHO3, PHO5, PHO10, and PHO11 are possibly the result of ancestral gene duplication [15].
2.1. Phosphate Transport
Transmembrane transportation of Pi is the first and obligatory step of its assimilation by cell. Uncombined phosphate is transported into cell by special permeases. There are three transport systems, which depend on concentrations of Pi and also H+ and Na+ ions. The presence of different transport systems with high and low phosphate affinity provides flexibility for yeast phosphate metabolism in continuously changing environmental conditions. Mutations affecting Pi transport occur in a set of genes: PHO84, PHO86, PHO87, PHO88, PHO89, and GTR1 and still unknown gene encoding low-affinity transporter. Such mutants are characterized by constitutive PHO5 expression. High-affinity transport system includes two phosphate permeases Pho84p and Pho89p with dissociation constant Km~8 μM [16, 17]. Expression of genes PHO84 and PHO89 is repressed by phosphate abundance [18].
The main phosphate permease of this system is Pho84p [16]. It has Km value ranging from 1 to 15 μM and works at pH 4,5. Pho84p is a symporter of H+ and symports 2-3 ions with one phosphate. Furthermore, Pho84p was shown to transport selenium [19] and manganese [20]. Pho84p activity is inhibited by arsenate that allows to get selectively mutations pho84 by picking yeast clones resistant to arsenate [21]. The quantity of Pho84p decreases in case of phosphate concentration increase or carbon source depletion in the medium [22]. Pho84p combines into complexes with some other proteins participating in Pi transport: Pho86p, Pho87p, Pho88p, and Gtr1p. The last one plays a special role: N-terminal domain of Gtrp1p (35,8 kDa) is similar to yeast protein Ypt1p and comprises a region for binding GTP. Gtr1p is a member of ras family of GTP binding proteins. Gene GTR1 is localized in yeast chromosome XIII near gene PHO84. The disruption pho84 leads to constitutive synthesis of APs and affects Pi assimilation by the cell. The complex Gtr1p-Pho84p is supposed to have a regulatory function and to be a receptor of Pi concentration signals [23]. Permease Pho84p is packed to COP II-coated vesicles and transported to the cell surface with the help of Pho86p [24, 25].
Another Pi transporter with high affinity is encoded by gene PHO89. It has Km value near 0,5 μM and performs Na+-associated Pi cotransportation and works at pH 9,5 [26].
Low-affinity phosphate transport system includes three permeases: Pho87p, Pho90p, and Pho91p with dissociation constant Km ~ 770 μM [16].
Expression of genes PHO84, PHO89, PHO87, PHO90, and PHO91 is regulated according to the models of positive and negative feedback loops [18]. Low-affinity permeases' activity is regulated with the participation of protein Spl2p [27]. The expression of genes PHO84 and SPL2 is controlled by transcription factor Pho4p whose activity depends on intracellular Pi concentration.
2.2. Genetic Control of AP1 Synthesis
Transcription rate of gene PHO3 is regulated by thiamine (vitamin B1), which is a necessary component for enzymatic reactions in Krebs cycle and pentose-phosphate pathway of carbohydrate oxidation. The yeast is able to receive extracellular thiamine from a medium using the special membrane transporter, encoded by gene THI7, and also to create its own intracellular thiamine using the enzyme, encoded by gene THI4 [28]. There are some recessive mutations revealed in PHO3, which lead to blocking AP1 activity and changing its properties [7, 29, 30]. Unlike wild-type yeast cells, pho3 mutants are characterized by significantly reduced intensity of thiamine pyro- and monophosphates transport. It means that AP1 is responsible for hydrolyzing thiamine phosphates in periplasmic space and is specialized on this class of phosphate-containing compounds [31]. Thiamine addition results in repressing Pho3p activity and thiamine biosynthesis enzymes [32]. PHO3 expression was shown also to depend on the type of nitrogen source in cultural medium [33]. It was proved that PHO3 expression level is decreased while yeast cells use poor nitrogen sources like urea [34].
2.3. Genetic Control of Repressible APs Synthesis
The regulation of repressible APs genes' expression is one of the most well-studied genetic systems in yeast S. cerevisiae [2]. The regulation of PHO5 gene, encoding major repressible AP, differs from PHO10 and PHO11 by higher rate of expression during phosphate absence and stringent repression during phosphate abundance [14]. Genes PHO10 and PHO11 are the result of duplication [35]. Promotor regions of nonlinked genes PHO5 and PHO11 have a strong homology and are regulated in coordination [36]. Because of its accurate regulating mechanism, gene PHO5 promotor is widely used in biotechnology for industrial production of heterologous proteins in the yeast [37].
There are some recessive mutations revealed in gene PHO5, which affect AP2 activity; its deletion leads to growth speed decrease in complete and mineral media [12, 38]. Mutations pho5 occurred in signal peptide damage a secretion level of the protein that was synthesized in cells successfully [39]. Intracellular AP2 represents a dimer [40]; in periplasmic space, APs form an oligomerous enzyme comprising Pho5p, Pho11p, and Pho12p [41].
Nowadays there are multiple levels of gene PHO5 expression regulation discovered that displays an importance of this gene for the yeast cell [42–44].
The expression of the genes encoding phosphate metabolism enzymes (PHO-regulon) is regulated by transcriptional activators Pho4p and Pho2p, cyclin-CDK complex Pho85p-Pho80p, and inhibitor of cyclin-CDK complex Pho81p [25, 45].
Transcription induction of gene PHO5 is associated with chromatin reorganization in promotor region [46, 47]. While in repression conditions (when phosphate is abundant), promotor of PHO5 gene is packaged into four nucleosomes and only 70 bp region remains unoccupied and nuclease sensitive. Two nucleosomes flank the UAS1 region (5′-AATTAGCACGTTTTCGCCATA-3′) that is unoccupied, while UAS2 (5′-GCACTCACACGTGGGA-3′) and TATA-box are packed into the nucleosomes-2 and -1, respectively [48]. When phosphate concentration is low, activator Pho4p is imported in the nucleus and docks UAS1 and UAS2 regions of promotor. This leads to changing the chromatin structure and nucleosome removal [49]. It was shown that mutations in UAS1 or UAS2 lead to tenfold promotor activity decrease, and double mutations in both regulatory sequences are followed by lack of PHO5 induction [50].
When extracellular phosphate concentration is high, cyclin-CDK complex Pho85p-Pho80p hyperphosphorylates the transcriptional activator Pho4p that leads to its localization in cytoplasm and inactivation [51]. Free of TFs gene PHO5 promotor is a target for nucleosomal chaperon Spt6p that restores nucleosome structures in the promotor. Spt6p inhibition results in that PHO5 promotor stays free of nucleosomes and transcription occurs in vivo even in the absence of the activator. Therefore, Pho4p function may be to maintain promotor in nucleosome-depleted state [52]. It was proposed earlier that chromatin remodeling takes place in the absence of DNA replication [53] and requires the presence of Pho4p activating domain [54]. This domain can interact with the transcription apparatus and assist in forming the initiation complex on the promoter only after nucleosomes' removal [55]. It is remarkable that Spt6p absence and transcription-prone chromatin can be followed by DNA replication [56].
Also SAGA protein complex contributes to PHO5 expression activation. The attraction of this complex depends on Pho4p activity. Histone acetyltransferase Gcn5p is one of the complex subunits. Deletion gcn5 leads to nucleosome rearrangements in PHO5 promotor and drastic reduction of transcription rate [57]. Another member of the SAGA complex is Spt3p. Its functions differ from Gcn5p: deletion spt3 leads to the reduction of PHO5 transcription affecting TBP interaction with TATA-box but not touching the chromatin structures. Double mutant gcn5spt3 displays the same phenotype as mutants spt7 or spt20 [58]. They are not able to form the SAGA complex at all. Another subunit is Ada2p, whose absence results in chromatin remodeling delay like in case of mutation gcn5. Factor Ada2p controls Gcn5p activity via SANT domain. Thus, the SAGA complex appears to be coactivator of PHO5 promotor along with Pho4p [59].
2.4. Phosphate Concentration Signals Transduction Pathways
The AP2 and AP3 synthesis is repressed in response to high concentration of Pi. To research transduction pathways for this signal, the mutants were selected characterized by constitutive synthesis of APs. As a result, some more genes were identified in addition to known PHO2, PHO4, PHO80, PHO85, and PHO81. Gene PMA1 encodes ATPase of plasma membrane. Gene ACC1 is involved in fatty acid synthesis. Also a novel, nonessential gene PHO23 was identified. The mutants pho84, pho86, and pma1 are defective in high-affinity phosphate uptake, whereas acc1 and pho23 are not. Hence, the products of these genes control different steps of signal transduction about high phosphate concentration to the regulatory factors of PHO-regulon expression [60].
Pho84p was shown to be not only high-affinity permease but also a signal transmitter of phosphate and glycerol-3-phosphate quantities. A novel term was proposed for the molecules like, “transceptor” [61].
2.5. Cyclin-Dependent Protein Kinase Pho85p
Yeast S. cerevisiae is the most well-studied eukaryotic model object for signal transmission and adaptive response molecular mechanisms studies. The main components of signalling networks are well conserved from yeast to higher eukaryotes. The central role in the regulation of signalling cascades in yeast is executed by nutrients, while in higher eukaryotic cells, the same role is played by hormones and growth factors. In yeast S. Cerevisiae, several nutrient-controlled pathways which adapt cell growth and proliferation, metabolism, and stress resistance, have been estimated. These pathways form general signalling network which guarantees that yeast cells enter resting phase G0 in order to pass through the period of nutrient depletion and, on the other hand, to be capable of cell proliferation fast resumption, when nutrient conditions become favorable. Key roles in this signalling network play nutrient-sensory protein kinases PKA, Snf1p, Tor1p, Tor2p, Sch9p, and Pho85p. Pho85 is a cyclin-dependent protein kinase with a wide range of substrates, which is a functional homologue of Cdk5p [62]. Protein kinase Pho85p takes part in phosphate and glycogen metabolism regulation, autophagy regulation, stress adaptation, proline utilization, regulates proteolysis, and cell polarity.
Cyclin-dependent protein kinases (CDKs) are the group of serine-threonine kinases and have multiple regulatory functions of great importance in such global processes as cell cycle, cell differentiation, and cell response to environmental changes. S. cerevisiae CDK family comprises Cdc28p, Ssn3p, Kin28p, Pho85p, and Cak1p [63]. Binding CDK with cyclin protein followed by phosphorylation leads to the activation of catalytic CDK subunits [64]. Cyclins define CDK substrate specificity. Nowadays, ten cyclins Pcl1–10 and at least nineteen substrates (Pho4p, Sic1p, Rvs167p, etc.,) of Pho85p are known [63, 65].
Gene analysis indicated that PHO85 encodes protein kinase highly homologous to Cdc28p (51% amino acid identity), the key cell cycle regulator in the yeast [62, 66]. But still the overexpression of one CDK does not compensate mutations in the other one. In view of the inviability of strains with cdc28 mutations, Cdc28p is thought to be the main cell cycle kinase [67]. The presence of Pho85p is not necessary for viability, but this protein kinase assures normal metabolic and cell cycle dynamics reversion after stress [63].
Mutations pho85 were firstly identified as recessive mutations of repressible APs' constitutive synthesis in the sake yeast strains (original gene name—PHO U) and in the strains of Peterhoff Genetic Collection (originally—ACP82). Studying epistatic interactions between different pho mutations and mutations in gene PHO85 revealed a negative role of CDK in PHO5 expression regulation [12, 68, 69].
The role of protein kinase is not restricted by transcription regulation. A great number of pho85 phenotypic effects were described [70, 71]. Yeast mutants pho85 generally do not grow at 37°C, have abnormal cell morphology [72], are incapable of growing on nonfermentable carbon substrates (ethanol and glycerol), accumulate glycogen [73], are sensitive to aminoglycoside antibiotics [74], and cannot use proline as a sole nitrogen source [75]. Disruption of gene PHO85 leads to forming phenotype of nuclear thermosensitivity (ts) and respiratory incompetence [rho-] affecting mitochondrial DNA [70]. The diploids heterozygous for pho85 are characterized by defects of sporulation [76], that makes it difficult to analyze these features of the mutants. The pho85 phenotypes' research is of special interest because of homology between the yeast kinase Pho85p and the main mammalian neurogenesis protein kinase Cdk5p [77]. Mitochondria dysfunction is typical for many neurodegenerative diseases.
PHO85 transcription does not depend on cell cycle stage, phosphate concentration, and α-factor. PHO85 expression increases during sporulation, nitrogen depletion, and after heat shock. It is remarkable that the quantity of kinase monomers is constantly high in the cell.
Ogawa and coworkers showed that CDK Pho85p has a significant role in gene expression regulation [5]. They used flavopiridol, a specific inhibitor of kinases Cdc28p and Pho85p, to analyze its influence on genome expression. This confirmed Pho85p participation in transcription control of PHO-regulon, stress response genes (CTT1, HSP12, and UBI4) and other genes working in stationary phase [78].
2.6. CDK Structure
Cyclin-dependent protein kinase structure is extremely conservative. Take space structure of human Cdk2p as an example (http://pdbwww1.sdsc.edu/pdb/explore.do?structureId=1B38). Cdk2 consists of 11 subdomains, which are packed in two lobes with an active site cleft among. The N-terminus of Cdk2p is important for ATP binding and correct orientation of the molecule to the catalytic and protein-binding sites. The C-terminus determines binding of protein substrates. Amino acid residues which are localized in three short β-sheet sites between two lobes are other important players for Cdk2p catalysis [79]. The conservative PSTAIRE motive is necessary for cyclin binding in all CDKs, and Cdk2p is not a singularity [80].
The functional analysis of PHO85 gene mutations was done in two laboratories [70, 81]. It is rather interesting to mention that CDK Pho85p needs no phosphorylation by other kinase in order to be active [82].
2.7. Pho85p Regulators
2.7.1. Cyclins
The expression of cyclin genes is coordinated according to cell cycle phases. Cyclins play a double role in kinase activity regulation. On the one hand, programmed cyclin synthesis and degradation through the cell cycle leads to the well-timed kinase activation; on the other, cyclins determine substrate specific binding of kinase [83]. Cyclins are identified in many species, and it is shown that despite the specificity of cyclin-kinase interaction, CDK can be activated by different cyclins, and cyclins in its turn can activate different kinases. An exception to this rule applies yeast S. cerevisiae, and the principle of “one cyclin activates one kinase” was formed above them. All cyclins have conservative domain—cyclin box, which consists of 5 α-helixes [84]. It should be mentioned that the role of cyclins is not limited by cell cycle control.
The fact, gene PHO85 encodes CDK, and its mutations lead to different pleiotropic effects, engaged Pho85p cyclins, and substrates searches. Due to two-hybrid screening it is known that ten cyclins Pcl1p-Pcl10p can interact with Pho85p. PCL1–PCL10-deleted strains were constructed and characterized, on which ground Pcl1–Pcl10 cyclins were divided into two subfamilies: Pho80 subfamily (Pho80p, Pcl6p, Pcl7p, Pcl8p, and Pcl10p) and Pcl1, 2 subfamily (Pcl1p, Pcl2p, Pcl5p, Pcl9p, and Clg1) [85].
The Subfamily of Pho80p Cyclins —
Cyclins of this family take part in metabolism control, such as stress response, temperature shock, nitrogen starvation, and stationary stage of growth.
Pho80p —
Pho80p was one of the first identified Pho85 cyclins. Unlike the other cyclins, PHO80 expression is not changed through the cell cycle progression [86]. PHO80 mutations have recessive inheritance and lead to constitutive synthesis of repressible acid phosphatases [87]. In pho80 PGC yeast strains, two types of mutations were identified: recessive with constitutive synthesis of repressible acid phosphatases and semidominant mutations, which lead to the lack of repressible acid phosphatase expression phenotype [88]. PHO80 expression is not governed by phosphate concentration, but still it does depend on Pho85p functionality. There are some contradictory data concerning Pho80p self-transcription regulation [88, 89]. PHO80 transcription level is decreased during heat shock or nitrogen starvation, but during the stationary stage of growth, PHO80 mRNA level is increased several times [33]. PHO80 disruption has no lethal effect. There are some pleiotropic effects of pho80 mutation, just to name few: permeability deoxythymidine monophosphate increase [90], vacuoles segregation defects [29, 30, 91, 92], decreased growth on nonfermentable carbon source containing medium [93], and methyl-methane sulphonate sensibility [94]. PHO80 mutations lead to aminoglycosidic antibiotics resistance [33, 74]; pho80 strains are characterized by ions K+, Mg2+, Zn2+, Ca2+, and Mn2+ sensibility increase [34, 95]. One of the most possible Pho80p partners is kinase Pho85p in all the above-mentioned processes and phenotypes. PHO85 gene overdose compensates the effects of pho80 deletion partially and shows allele-specific effect [93].
Pho80p-Pho85p complex phosphorylates transcriptional factor bHLH-Pho4p [96, 97], as a result Pho4p stays in cytoplasm and is not capable of PHO-regulon genes activation on the medium with phosphate abundance [98]. PHO80 overexpression leads to PHO5 transcription inhibition restoration in pho85 strains, which means that cyclin abundance can lead to nonspecific interactions with other kinases [98].
Pcl6p and Pcl7p —
Pcl6p and Pcl7p cyclins have 46% identity. PCL6 mRNA level is constant during all stages of cell cycle, but is increased during heat shock and nitrogen starvation and decreased in stationary phase of growth significantly, while PCL7 mRNA level is increased by sixfold. PCL6 insertional mutants have complete medium growth defects [33]. Pcl6p-Pho85p complex is able to phosphorylate Pho4p in vitro, but in vivo it is still unknown whether Pho4p is Pcl6p-Pho85p complex's substrate or not. Pcl6p and Pcl7p cyclin's role in metabolism control is rather contradictory. Some researchers have shown that Pcl6p-Pho85p complex regulates transcription of genes PUT1 and PUT2, which encode proline utilization components [5, 75]. PCL6 and PCL7 mRNA increase during nitrogen starvation suits this hypothesis. But in another research work, it has been shown that Pcl6p-Pho85p and Pcl7p-Pho85p complexes control glycogen accumulation [99]. PCL7 expression varies during cell cycle progression unlike other cyclin's genes of Pho80p subfamily. PCL7 mRNA level is increased in the middle of S-phase of growth. Pcl7p-Pho85p complex activity is inhibited by Pho81p during phosphate starvation. Authors have considered a role of Pcl7p-Pho85p complex in cell cycle regulation [100]. Since cyclins Pcl7 and Pcl6 interact with common substrate, it is fair to assume that this substrate is Pho4p as in PUT1,2 gene expression regulation and so in glycogen accumulation process.
Pcl8p and Pcl10p —
Other members of Pho80p cyclin subfamily are Pcl8p and Pcl10p, which also form a complex with kinase Pho85p and phosphorylate glycogen synthase Gsy2p as in vitro so in vivo. Gsy2p phosphorylation prevents glycogen accumulation on phosphate-abundant medium [101]. As a conclusion of Pho80p subfamily cyclins description, it should be mentioned that protein interaction analysis by HMS-PCI (high-throughput mass spectrometric protein complex identification) method showed Pho85p interaction only with cyclins Pcl6p, Pcl7p, Pcl8p, and Pcl10p [102].
Pcl1/2p Subfamily Cyclins —
Pcl1/2p subfamily cyclins are characterized by expression fluctuation during cell cycle progression. PCL1/PCL2 expression pattern research has shown mRNA concentration decrease during heat shock, nitrogen starvation, and stationary stage of growth. Therefore, cyclins, that provide metabolic processes, are of key importance at stress conditions, rather than cyclins, which are necessary for cell cycle progression.
PCL1, PCL2, and PCL9 have maximum expression at early G1 phase PCL5 mRNA level is constant during cell cycle progression but increases during nitrogen starvation, thermo shock, or stationary phase of growth [33]. Pcl1/pcl1, pcl2/pcl2/, clg1/clg1, pcl5/pcl5, pcl9/pcl9 diploids, and pho85-deleted strains have common phenotype of abnormal cell morphology. Haploid cells of Δpho85 strains are characterized by large round cells with giant vacuoles. Homozygous pho85/pho85 diploids also have some morphological defects: wide bud neck, granular cytoplasm, and elongated cells, but this phenotype is more noticeable on complete medium rather than on mineral [103]. One of the most interesting Pcl2p-Pho85p and Pcl9p-Pho85p substrates is protein Rvs167, which is homologous to mammalian amphiphysin-1. Rvs167p takes part in actin cytoskeleton regulation and cell survival after starvation [104]. PCL9 transcription is activated by transcriptional factor Swi5p. Pcl9p-Pho85p complex is able to phosphorylate Pho4p in vitro and plays an important role in M/G1 checkpoint transition [105]. PCL2 transcription activation also takes place after α-factor treatment and PCL1; PCL9 mRNA level is decreased on the contrary. α-factor treatment leads to G1 stage arrest as far as CLN1, CLN2 genes are low expressed and Cdc28p is inactive. Cell cycle progression is blocked, and Pcl2p-Pho85p is still functionally active.
Clg1p is another member of Pcl1/2p subfamily with 27% homology to Pcl1p. CLG1-mutated strains have no visual phenotypic effects, and CLG1 expression is constant during cell cycle progression [65]. CLG1 mRNA level fluctuations take place during environment changes: it is decreased during amino acid and nitrogen starvation and is increased at stationary phase four times [33]. It is possible that Clg1p is the cyclin, which is responsible for the regulation of stationary phase genes by protein kinase Pho85p.
Cyclins compete with each other for Pho85p kinase binding, so that cyclins expression fluctuations make the delicate regulation different depending on Pho85p processes possible.
Pho85p-Pho80p Substrates —
Cyclins determine the kinase substrate specificity. 19 Pho85p substrates were identified [62, 63]. Bound with different cyclins, protein kinase Pho85p takes part in the regulation of G1/S progression, cell polarity, PHO-regulon genes expression, and signal transduction [106]. Strains with pho85 deletion are characterized by different phenotypic effects, such as constitutive synthesis of repressible acid phosphatases, slow growth with G1 arrest, glycogen accumulation, cell morphology changes, sporulation and cell polarization defects, irregular cell divisions, actin depolarization, endocytosis defects, and high frequency of thermosensitive and respiratory-deficient cells genesis [107].
Respiratory-deficient phenotype is associated with mitochondrion functions defects. Mitochondria participation in different vital cellular processes dictates the necessity of delicate organelle function regulation existence according to the environmental changes. In cells of higher eukaryotic organisms, kea role in numerous stress response signals is played by transcription, factor p53 which blocks cell cycle progression and triggers apoptosis in many cell types, including neurons. Phosphorylated by Cdk5 transcription factor p53 is accumulated in nucleus, where it induces pro-apoptotic genes and mitochondria-mediated apoptosis in response to genotoxic or oxidative stress. Cdk5p prevents p53 ubiquitylation and downregulation [108]. Cdk5 is implicated in both development and disease of the mammalian central nervous system. Protein kinase Pho85p is a functional homologue of Cdk5p. The loss of Cdk5 in mice is perinatal lethal, and overactive Cdk5 induces apoptosis, meanwhile pho85 mutations have no lethality effect. The existence of high conservativity in mitochondria functions regulation mechanisms permits to project revealed in yeast mechanisms to higher eukaryotes.
Respiratory, carbon, and amino acid metabolisms take part in the regulation of mitochondrial functions in yeast [109]. An important role in mitochondria functions plays phosphate, but the consequences of phosphate metabolism defects on mitochondria functions in yeast are deficiently studied. Respiratory-deficient phenotype of pho85 strains was thought to be a pleiotropic effect of the mutation [81]. Genetic analysis has shown that respiratory deficiency of pho85 strains is estimated by nonnuclear determinant [110]. Respiratory deficiency of pho85 strains can be the consequence of different reasons:
point mutations or deletions of mitochondria DNA (mtDNA), as it was shown for abf2, msh1, mip1, and rim1 mutants [111];
mtDNA synthesis defects in the consequence of nucleotide pool changes (adk1, sml1) [112, 113];
changes of protein quantity, which take part in mtDNA synthesis and compactization, because of nuclear genes expression level changes (ILV5, ACO1) or organelle protein transport defects (tom20, pam17, and tom6) [114, 115];
mitochondria transport defects, which are determined by changes in mitochondria morphology (mje1, pam18, ssc1, tim17, tim23, tim44, tom20, tom40, tom7, and tom70) or by actin cytoskeleton structure or regulation changes (act1, trp1, bni4, rvs167, sac6, sla1, prk1, and myo3) [115, 116];
mtDNA destabilization by mitochondrial mitochore structure changes or by mtDNA transport defects (puf3, mdm31, mdm34, mmm1, mdm10, and mdm12) [117];
mitochondria membrane potential changes, as a consequence of membrane permeability defects (crd1, atp10, atp11, atp12, atp25, fmc1, oxa1, and mir1) [118].
Respiratory-deficient cells of pho85 strains appear on the medium with glucose when mitochondria functions are repressed. Mitochondria GFP marking has shown that respiratory-deficient phenotype of pho85 strains is not a consequence of mitochondria morphology or transport defects [119, 120]. It was shown that pho85 strains are characterized by high nuclear DNA mutation rate [121].
mtDNA mutations frequency of active dividing cells depends neither on the moment of mutation appearance during cell divisions nor on replication regulation factors, mtDNA recombination, and segregation. These processes determine mutation fixation rate in multicopy pool of mtDNA. It was shown that pho85 mutations lead to changes in expression pattern of genes ABF2, ACO1, CCE1, MSH1-6, PIF1, MDM12, MMM1, MMM2, KGD1, and RPO41, which encode mtDNA replication, reparation, and segregation proteins [5]. We have shown that [rho0] appearance in pho85 strains is not a consequence of mutation accumulation, but is a result of mtDNA loss. Nucleoids loss of pho85 strains is taking place on the medium with glucose and high concentration of phosphate. It was shown that in such environment respiratory-deficient cells have selective advantage [120].
The loss of mtDNA is a dynamic process, and respiratory-deficient cell population consists of cells with full mtDNA loss [rho0] so of respiratory-deficient cells with low mtDNA copy number, which is not enough for respiration [122]. [rho0] colonies would not be able to revert on glycerol medium, but cells with low mtDNA copy number could reproduce normal mtDNA quantity, respiration, and growth on glycerol. We selected revertants, which had restored pho85 strains respiration capability. Phenotype analysis and test for allelism revealed that respiration restoration of pho85 strains is a consequence of pho4, pho81, pho84, and pho87 mutations.
In pho85 strains, transcription factor Pho4p is always localized in nucleus, where it activates the expression of PHO-regulon genes, which encode phosphate permeases Pho84p and Pho87p, for example. As a result, an intracellular phosphate level is uncontrollably growing. Under such conditions, pho85 cells with secondary mutations in genes encoding permeases Pho84p or Pho87p, in regulatory genes PHO4 or PHO81 or unidentified genes, get a selective advantage [120].
Increase of intracellular phosphate concentration can lead to some negative effects, like plasmalemma or mitochondria membrane potential changes [123], PKA signal pathway activation [124], and so forth. Mutations preventing uncontrolled phosphate transport apparently normalize its cytoplasm concentration. Therefore, in the course of Saccharomyces evolution, regulatory networks which modify mitochondrion functions according to the changes not only of carbon or nitrogen source but also of changes in phosphate concentration has formed [120].
Pho85p and Cyclin's Chaperones —
Chaperones take part in protein kinases space structure organization. Yeast protein kinase Cdc28p form complexes with cyclins through some extra factors. Hsp90p's cochaperone Cdc37p is one of them. It was shown that Cdc37p is an important factor for Cdc28p-Cln2p and Cdc28p-Clb2p activation in vitro. Cdc37p has genetic interactions with 4 protein kinases: Cdc5p, Cdc7, Cdc15p, and Cak1p [125]. It is assumed that protein kinases stabilize inactive Cdc28p preparing it for cyclin binding [126]. Cdc37p and Cdc28p also have a genetic interaction, which was shown in synthetic lethality and suppressions analysis, but there are no evidences of Cdc37p and Cdc28p physical interactions. Two-hybrid analysis has shown physical interaction of N-terminus Cdc28p with C-terminus of Cdc37p, but still full-size Cdc28p protein did not interact with Cdc37p [125].
Pho85p's chaperones have not been identified yet. We have selected different pho85 missense mutants, which led to molecule conformation changes. One of these mutants had F82L substitution, which had led to Pho85p folding defects and constitutive synthesis of repressible acid phosphatases. Extragenic suppressions search of strains, which restore normal repressible acid phosphatases synthesis, have revealed a few genes. One of these suppressor genes is localized to a 2 cM interval from PHO85, like what EGD1 gene does. EGD1 encodes chaperone of Gal4p and of some ribosomal proteins [70]. It is fair enough to assume that Egd1p can regulate Pho85p conformation, but extra experiments are necessary for an exact conclusion.
2.7.2. Pho85p Inhibitors
Inhibitors and cyclins extend opposite effects. The majority of CDK's activity is controlled by cyclin binding. CDK inhibitors play a role of tumor suppressors and development regulators in mammalian cells so search of mechanisms it acts with is of great importance. Nowadays, three CDK inhibitors in yeast S. Cerevisiae are known:
Far1p, which is activated by phosphorylation during alpha factor treatment and leads to cell cycle arrest at G1 stage by Cdc28p-Cln1p and Cdc28p-Cln2 inhibition [127];
Sic1p, which inhibits Clb5p-Cdc28p and Clb6p-Cdc28p during G1 and Pcl2p-Pho85p; it also prevents DNA synthesis as long as cell passes through G1/S boundary [128];
Pho81p is the only one identified Pho85p cyclin. PHO81 mutations are characterized by two types of phenotype: one of them is recessive and leads to the lack of PHO5 derepression, and other one is dominant and is characterized by constitutive synthesis of PHO5 [5]. PHO81 gene has been cloned [129].
PHO81 promotor has Pho4p, Pho2p binding sites and negative regulatory sequence (NRS). NRS-localized deletions lead to PHO81 expression increase at fourfold [130]. Main PHO-regulon regulatory proteins take part in PHO81 transcription regulation. Pho81p contains 6 ankyrin repeats, that in mammalian homologues are used for cyclin-kinase complex attaching [131]. In case of Pho81p, it was shown that, ankyrin repeats do not influence the inhibitor-cyclin binding so inhibition itself. However, neither N-terminus nor C-terminus of the protein has an influence on interaction of cyclin-kinase complex with Pho81p. The 80 amino acid (645–724 aa) sequence was identified which is sufficient for cyclin-kinase complex interaction.
Activity regulation of a great number of proteins is determined by its localization. There are conformable proteins activators and Cln1p, Cln2p cyclins. Pho81p is predominantly localized in nucleus, but still it is can be situated in cytoplasm and endoplasmic membranes. Pho81p-Pho80p-Pho85p complex is constitutive, but kinase inhibition by inhibitor takes place only under phosphate starvation conditions. It is supposed that Pho81p inhibits kinase by means of cyclin binding, which leads to changes in 3(R121), 5(E154) helixes of cyclin, and kinase's PSTAIRE domain interaction [132].
Pho81p is also able to inhibit Pcl7p-Pho85p complex functioning during phosphate starvation. PCL7 mRNA level is maximally increased during S-phase, so it is fair to assume that replication arrest is taking part according to the phosphate level [100].
So that Pho81p receives signals of phosphate concentration changes and transmits it to other proteins of signal network. Pho81p also interacts with adenylate cyclase Cdc35p. Adenylate cyclase controls PKA producing cAMP and is a part of RAS signal pathway, which in its turn takes part in glucose starvation signal pathway. Pho81p-Cdc37p interaction can lead to cAMP concentration changes in response to phosphate availability and transmit signal to PKA taking part in control of growth, proliferation, and other vital processes.
Pho81p-Cdc35p complex is localized in cytoplasmic membrane. It was shown that Pho81 can interact with a great number of other proteins, like mitotic growth component Emg1p or cortical cytoskeleton component Cof1p, but these complexes have cytoplasm localization [102]. It is possible that Pho81p influences Pho85p-cyclin complex functions according to phosphate concentration changes. Another Pho80p-Pho85p complex inhibitor is Spl2p. Spl2p and Pho81p have 27% identity [133].
Spl2p controls phosphorylation of other substrates except Pho4p, but SPL2 transcription is regulated by Pho4p. SPL2 mutations were identified as supressors of plc1 mutants. PLC1 gene encodes phosphoinositide-specific phospholipase C involved in regulating many cellular processes.
Proteolytic degradation also plays a significant role in kinase inhibitors regulation. Far1 and Sic1 target Cdc28p for degradation. Cdc28p inactivation leads to cell accumulation of inhibitors.
2.8. Transcription Regulation of PHO and ADE Genes by Metabolic Intermediates
Recent studies have revealed that several key metabolic intermediates take part in phosphate and purine metabolism [134, 135]. A 5-phosphoribosyl-5-amino-4-imidazolecarboxamide (AICAR) and succinyl-AICAR belong to these regulatory molecules [136]. TF Pho2p is involved in the regulation of AICAR-dependent genes. During AICAR accumulation, the affinity of Pho2p to promotors of phosphate and purine metabolism genes also raises. It was shown that AICAR in vitro interacts with TF Pho2p and Pho4p in purine starvation conditions. In vivo Pinson and coauthors have revealed that AICAR can stimulate the formation of Pho2p-Pho4p and Pho2p-Bas1p complexes [44]. Thus, under the purine starvation conditions, TF Bas1p and Pho4p compete for Pho2p binding that provides the coordinated regulation of nucleotides biosynthesis and Pi uptake in cell.
It was shown that SAICAR affects only the formation of Pho2p-Bas1p complex and correspondingly the regulation of purine metabolism. Supposably the physiological reason of different cell response on the accumulation of AICAR and SAICAR appears to be the result of the adenylosuccinate lyase (ASL) function which metabolizes SAICAR in the inosin-5′-monophospate (IMP) and AMP biosynthesis de novo [137].
Another interesting example that illustrates cross-regulation of purine biosynthesis and phosphate metabolism is the constitutive transcription of PHO5 on the background of ado1 and adk1, encoding adenosine and adenylate kinases. In both cases, Pho4p is localized in nucleus even if the phosphate concentration is high [138].
Gauthier and coauthors have shown that Ado1p and Adk1p work upstream Pho81p, independently of phosphate concentration, but in response to concentration changes of ATP as the target product and AICAR as intermediate product [139].
Beside the regulation of PHO-genes by Pi and purine metabolism intermediates, some other mechanisms were also identified. There are models illustrating that PHO-genes expression also depends on the concentration of another Pi sources in the cell such as polyphosphates and inositol pyrophosphates.
2.9. Polyphosphates Metabolism
Polyphosphates are commonly used throughout almost all organisms as the reserve pool of Pi [3]. Polyphosphates are the linear polymers of phosphoric acid where the phosphate residues are linked by phosphoanhydride bonds as in ATP. Thus, polyphosphates are not only the reserve pool of phosphates but also play role as high-energy molecules. The number of phosphate residues can range from 3 to 1000. In yeast, 37% of all phosphate is accumulated as polyphosphate and 90% of polyphosphates, are localized in vacuoles.
It is known that the polymerization of phosphates is catalyzed by polyphosphates kinase in Escherichia coli (E. coli). The reverse reaction is the result of exopolyphosphatase function [1]. The major exopolyphosphatase in yeast S. cerevisiae is encoded by gene PPX1 [140]. Ppx1p is localized in mitochondrial matrix, cytosol, and cell membrane. Ppx1p has restricted substrate specificity and can not cleave pyrophosphates, ATP, and cyclic forms of tripolyphosphates. Other enzymes were also discovered: exopolyphosphatases similar to Ppx1p localized in cell membrane and vacuoles (Ppx2p, Ppx3p) and the vacuolar endopolyphosphatase Ppn1p [140–143]. Many of the genes encoding enzymes of polyphosphates metabolism were discovered by the DNA Chip technology while searching the genes regulated by PHO-system.
By the analysis of genes expression levels on the medium with, high- and low-Pi concentrations, three genes were revealed in the wild-type and PHO-genes mutants: PHM2, PHM3, and PHM4, which had the expression levels similar to PHO5 and PHO84. The homology of PHM2, PHM3, PHM4, and PHM1 was also identified; PHM1 showed the expression level similar to PHO8 and PHO86. These genes encode integral proteins of vacuolar membrane, which form the heterotetrameric complex and regulate the activity of vacuolar H+-ATPase [144, 145]. Besides, genes encoding glycerol phosphatase Hor2p and ATP-phosphoribosyl transferase His1p are also regulated by the Pi concentration, and under starvation conditions their expression also increases along with PHO5, PHO11, PHO12, PHO8, and PHM5.
The AA sequences of Phm1p and Phm2p are identical for 58%. The N-end of the Phm1p, Phm2p, and Phm3p has a domain, which shows 33% homology with the N-end of Pho81p and Pho87p [18]. Strains Δphm1, Δphm2, Δphm3, Δphm4, and Δphm1Δphm2 are viable and have the same growth dynamics as the wild-type strain. Meanwhile, such mutant cells demonstrate different levels of polyphosphate accumulation: Δphm1 has low level of polyphosphate, and Δphm2 leads to the significant decrease of polyphosphate level and to the reduction of polyphosphate chain length. In strains with double deletion Δphm1Δphm2, none of the polyphosphates are detected, which indicates the dubbing functions of Phm1p and Phm2p. Strains Δphm3 and Δphm4 do not contain polyphosphate also. Thus, Phm1p, 2p, 3p, and 4p play an important role in polyphosphate accumulation.
In yeast, polyphosphates serve as emergency stores of phosphate. When polyphosphate synthesis critically decreases, unassociated phosphates are accumulated in cell. Accordingly to this situation, the phosphate transport by phosphate permeases is going to be locked by the gene expression decrease. Thus, metabolisms of phosphates and polyphosphates have mutual mechanisms of regulation.
Such a scheme of mutual regulation also exists in bacteria E. coli and Vibrio cholera. Bacterial promotors of ppk-ppx operons have so named pho-box [5, 146], while phosphate starvation regulatory factor phoB-phoR docks pho-box and activates the polyphosphatases transcription [146].
Thus, the regulation of polyphosphate metabolism genes by phosphate concentration is a conservative feature of yeast and bacteria.
2.10. Inositol pyrophosphates: The New Form of Intracellular Signaling
Inositol pyrophosphates are high-energy compounds involved in many biological functions of a cell. Inositol pyrophosphates take part in the regulation of such processes as vesicular transport, apoptosis, DNA reparation, telomere stability, and stress-motivated signaling, but exact mechanisms of this influence are still not clear [147]. Inositol pyrophosphates IP7 and IP8 are the most characterized molecules of this signaling system, which were found in all eukaryotic cells from amoeba to neurons of higher mammals. Pyrophosphorylation reactions constantly occur, so the concentration of IP7 and IP8 normally is high. A lot of substrates which are phosphorylated by IP7 were found in yeast and mammals [148]. The main advantage of phosphorylation by IP7 is the absence of necessity to special kinase functioning, like in ATP-dependent phosphorylation. IP7 phosphorylates substrates in direct way. The phosphorylated IP7 peptides are more resistant to phosphatases cleaving than peptides, which have been phosphorylated by means of ATP, because of another type of phosphate bond. By energy content, pyrophosphate bond of IP7 is equal to or even exceeds pyrophosphate bond of ATP, what indicates a great potential of these molecules in cell [149]. Pyrophosphorylation can be a new signaling system of eukaryotic cells [150].
The main players in inositol pyrophosphates biosynthesis are inositol hexakisphosphate kinases (IP6K) [151]. In yeast, S. cerevisiae IP6K are encoded by KCS1 and VIP1 genes. Strains Δkcs1 almost do not contain IP7 and IP8, but deletion of Ddp1p phosphatase gene (DDP1) on the Δkcs1 background suppresses this phenotype due to IP6K Vip1 [152]. IP6Ks Vip1 and Kcs1 synthesize different isomers of pyrophosphates. The existence of such differences in IP7 enantiomer biosynthesis might have a great physiological value. Thus, Lee with coauthors showed that IP7 which has been synthesized by kinase Vip1p represses Pho85p-Pho80p complex [153]. Meanwhile, the products of Kcs1p do not repress Pho85p-Pho80p. But the Kcs1p influence on PHO-regulon also exists as far as mutants kcs1 are characterized by constitutive expression of PHO5 and phosphate transport efficiency decrease [43]. Thus, the mechanisms of PHO-system regulation differ in the case of two IP7 enantiomers.
Despite the complicated phosphate cellular regulation, which includes the mechanisms of mutual control of orthophosphates, polyphosphates, and inositol pyrophosphates pathways, in order to regulate the cytoplasm phosphate concentration, Pho85p functions are of critical importance.
3. Conclusion
Recent researches have revealed a significant role that nonprotein factors play in gene expression regulation in eukaryotic and prokaryotic cell. Especially, lots of data were obtained about regulatory function of different RNA molecules in response to environmental signals. The yeast S. cerevisiae have some well-studied samples of gene regulation by noncoding RNAs [154–156].
Three well-known asRNAs in yeast relate to phosphate metabolism. First, there is RNA molecule transcribed in antisense orientation from gene PHO84 of the high-affinity permease of phosphates. This asRNA participates in the repression of its own gene in senescent cells [154]. Second, short asRNA is transcribed from gene KCS1 in the presence of activator Pho4p and low phosphate concentration in medium. KCS1 encodes structure of kinase involved in signal transduction about phosphate depletion. This asRNA blocks normal synthesis of Kcs1p that leads to the increase of Vip1p kinase activity. Vip1p competes with Kcs1p and intensifies derepression effect from phosphate depletion [157]. The third case is the one known sample of positive regulation by asRNA in the yeast. This asRNA (2400 n.) is transcribed in PHO5 locus in repressive conditions of high phosphate, exceeds the PHO5 ORF length (1400 n.), and binds the upstream region of gene PHO5. This binding leads to chromatin remodeling, histone removal from promotor DNA, and therefore PHO5 transcription activation as soon as phosphate concentration goes down in the medium [158].
The latest analysis of cDNA libraries and RNA polymerase II binding sites mapping in S. cerevisiae genome revealed much more antisense RNAs with unknown functions. The future of expression control researches in eukaryotes is connected with studying the RNA role in yeast gene regulation, and PHO-regulon genes stand first in a queue.
References
- 1.Kornberg A, Rao NN, Ault-Riché D. Inorganic polyphosphate: a molecule of many functions. Annual Review of Biochemistry. 1999;68:89–125. doi: 10.1146/annurev.biochem.68.1.89. [DOI] [PubMed] [Google Scholar]
- 2.Oshima Y. The phosphatase system in Saccharomyces cerevisiae. Genes and Genetic Systems. 1997;72(6):323–334. doi: 10.1266/ggs.72.323. [DOI] [PubMed] [Google Scholar]
- 3.Kulaev IS. Inorganic polyphosphates and its role in evolution. Educational Soros Journal. 1996;2:28–35. [Google Scholar]
- 4.Rouxel T, Danchin A, Henaut A. METALGEN.DB: metabolism linked to the genome of Escherichia coli, a graphics-oriented database. Computer Applications in the Biosciences. 1993;9(3):315–324. doi: 10.1093/bioinformatics/9.3.315. [DOI] [PubMed] [Google Scholar]
- 5.Ogawa N, DeRisi J, Brown PO. New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Molecular Biology of the Cell. 2000;11(12):4309–4321. doi: 10.1091/mbc.11.12.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veide J, Andlid T. Improved extracellular phytase activity in Saccharomyces cerevisiae by modifications in the PHO system. International Journal of Food Microbiology. 2006;108(1):60–67. doi: 10.1016/j.ijfoodmicro.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 7.Padkina MV, Krasnopevtceva NG, Petrashen MG, Kozhin SA, Smirnov MN. Genetical and biochemical research of acid phosphatases in yeast Saccharomyces cerevisiae. I. Acid phosphatases characteristics. Genetika. 1974;10:100–110. [Google Scholar]
- 8.Padkina MV. Nonspecific acid phosphatases of yeast Saccharomyces cerevisiae: regulation of biosynthesis. Vestnik of Saint-Petersburg State University. 1998;3(4):52–57. [Google Scholar]
- 9.Krasnopevtseva NG, Urazmanova NA, Padkina MV. Genetical and biochemical research of acid phosphatases in yeast Saccharomyces cerevisiae. XII. Yeast repressible acid phosphatases isolation and characteristics. Vestnik of Saint Petersburg University. Management Series. 1986;3:98–106. [Google Scholar]
- 10.Bostian KA, Lemire JM, Cannon LE, Halvorson HO. In vitro synthesis of repressible yeast acid phosphatase: identification of multiple mRNAs and products. Proceedings of the National Academy of Sciences of the United States of America. 1980;77(8):4504–4508. doi: 10.1073/pnas.77.8.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nosaka K, Nishimura H, Iwashima A. Effect of tunicamycin on thiamine transport in Saccharomyces cerevisiae. Biochimica et Biophysica Acta. 1986;858(2):309–311. doi: 10.1016/0005-2736(86)90336-6. [DOI] [PubMed] [Google Scholar]
- 12.Samsonova MG, Padkina MV, Krasnopevtseva NG. Genetical and biochemical research of acid phosphatases in yeast Saccharomyces cerevisiae. V. Genetic control of acid phosphatase II synthesis. Genetika. 1975;11:104–115. [PubMed] [Google Scholar]
- 13.Meyhack B, Bajwa W, Rudolph H, Hinnen A. Two yeast acid phosphatase structural genes are the result of a tandem duplication and show different degrees of homology in their promoter and coding sequences. EMBO Journal. 1982;1(6):675–680. doi: 10.1002/j.1460-2075.1982.tb01229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bajwa W, Meyhack B, Rudolph H, Schweingruber AM, Hinnen A. Structural analysis of the two tandemly repeated acid phosphatase genes in yeast. Nucleic Acids Research. 1984;12(20):7721–7739. doi: 10.1093/nar/12.20.7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansche PE. Gene duplication as a mechanism of genetic adaptation in Saccharomyces cerevisiae. Genetics. 1975;79(4):661–674. doi: 10.1093/genetics/79.4.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamai Y, Toh-e A, Oshima Y. Regulation of inorganic phosphate transport systems in Saccharomyces cerevisiae. Journal of Bacteriology. 1985;164(2):964–968. doi: 10.1128/jb.164.2.964-968.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Persson BL, Petersson J, Fristedt U, Weinander R, Berhe A, Pattison J. Phosphate permeases of Saccharomyces cerevisiae: structure, function and regulation. Biochimica et Biophysica Acta. 1999;1422(3):255–272. doi: 10.1016/s0304-4157(99)00010-6. [DOI] [PubMed] [Google Scholar]
- 18.Bun-Ya M, Nishimura M, Harashima S, Oshima Y. The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Molecular and Cellular Biology. 1991;11(6):3229–3238. doi: 10.1128/mcb.11.6.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazard M, Blanquet S, Fisicaro P, Labarraque G, Plateau P. Uptake of selenite by Saccharomyces cerevisiae involves the high and low affinity orthophosphate transporters. The Journal of Biological Chemistry. 2010;285(42):32029–32037. doi: 10.1074/jbc.M110.139865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen LT, Ajua-Alemanji M, Culotta VC. The Saccharomyces cerevisiae High Affinity Phosphate Transporter Encoded by PHO84 Also Functions in Manganese Homeostasis. The Journal of Biological Chemistry. 2003;278(43):42036–42040. doi: 10.1074/jbc.M307413200. [DOI] [PubMed] [Google Scholar]
- 21.Sambuk EV, Alenin VV, Kozhin SA. Genetic and biochemical study of yeast acid phosphatases. XI. Gene ACP80 controls inorganic phosphate transport. Genetika. 1985;21(9):1449–1454. [PubMed] [Google Scholar]
- 22.Martinez P, Persson BL. Identification, cloning and characterization of a derepressible Na-coupled phosphate transporter in Saccharomyces cerevisiae. Molecular and General Genetics. 1998;258(6):628–638. doi: 10.1007/s004380050776. [DOI] [PubMed] [Google Scholar]
- 23.Bun-ya M, Harashima S, Oshima Y. Putative GTP-binding protein, Gtr1, associated with the function of the PHO84 inorganic phosphate transporter in Saccharomyces cerevisiae. Molecular and Cellular Biology. 1992;12(7):2958–2966. doi: 10.1128/mcb.12.7.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau WTW, Howson RW, Malkus P, Schekman R, O’Shea EK. Pho86p, an endoplasmic reticulum (ER) resident protein in Saccharomyces cerevisiae, is required for ER exit of the high-affinity phosphate transporter Pho84p. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(3):1107–1112. doi: 10.1073/pnas.97.3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wykoff DD, O’Shea EK. Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics. 2001;159(4):1491–1499. doi: 10.1093/genetics/159.4.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serrano R, Ruiz A, Bernal D, Chambers JR, Ariño J. The transcriptional response to alkaline pH in Saccharomyces cerevisiae: evidence for calcium-mediated signalling. Molecular Microbiology. 2002;46(5):1319–1333. doi: 10.1046/j.1365-2958.2002.03246.x. [DOI] [PubMed] [Google Scholar]
- 27.Wykoff DD, Rizvi AH, Raser JM, Margolin B, O’Shea EK. Positive Feedback Regulates Switching of Phosphate Transporters in S. cerevisiae. Molecular Cell. 2007;27(6):1005–1013. doi: 10.1016/j.molcel.2007.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singleton CK. Identification and characterization of the thiamine transporter gene of Saccharomyces cerevisiae. Gene. 1997;199(1-2):111–121. doi: 10.1016/s0378-1119(97)00354-5. [DOI] [PubMed] [Google Scholar]
- 29.To-E A, Ueda Y, Kakimoto SI, Oshima Y. Isolation and characterization of acid phosphatase mutants in Saccharomyces cerevisiae. Journal of Bacteriology. 1973;113(2):727–738. doi: 10.1128/jb.113.2.727-738.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ter-Avanesyan MD, Inge-Vechtomov SG, Petrashen MG. Genetical and biochemical research of acid phosphatases in yeast Saccharomyces cerevisiae. II. Research of mutations which influence acid phosphatase 1 activity. Genetika. 1974;10(12):101–109. [Google Scholar]
- 31.Nosaka K, Kaneko Y, Nishimura H, Iwashima A. A possible role for acid phosphatase with thiamin-binding activity encoded by PHO3 in yeast. FEMS Microbiology Letters. 1989;60(1):55–60. doi: 10.1016/0378-1097(89)90077-3. [DOI] [PubMed] [Google Scholar]
- 32.Nosaka K, Nishimura H, Iwashima A. Identity of soluble thiamine-binding protein with thiamine repressible acid phosphatase in Saccharomyces cerevisiae. Yeast. 1989;5:S447–451. [PubMed] [Google Scholar]
- 33.Gasch AP, Spellman PT, Kao CM, et al. Genomic expression programs in the response of yeast cells to environmental changes. Molecular Biology of the Cell. 2000;11(12):4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Savinov V, Rumyancev A, Fizikova A, Sambuk E. The regulators of nitrogen and phosphorus metabolisms affect gene PHO3 expression in yeast Saccharomyces cerevisiae. Yeast. 2009;24:p. 160. [Google Scholar]
- 35.Venter U, Hörz W. The acid phosphatase genes PHO10 and PHO11 in S. cerevisiae are located at the telomeres of chromosomes VIII and I. Nucleic Acids Research. 1989;17(4):1353–1369. doi: 10.1093/nar/17.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kramer RA, Anderson N. Isolation of yeast genes with mRNA levels controlled by phosphate concentration. Proceedings of the National Academy of Sciences of the United States of America. 1980;77(11):6541–6545. doi: 10.1073/pnas.77.11.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rudolph H, Hinnen A. The yeast PHO5 promoter: phosphate-control elements and sequences mediating mRNA start-site selection. Proceedings of the National Academy of Sciences of the United States of America. 1987;84(5):1340–1344. doi: 10.1073/pnas.84.5.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toh EA, Kakimoto S, Oshima Y. Genes coding for the structure of the acid phosphatases in Saccharomyces cerevisiae. Molecular and General Genetics. 1975;143(1):65–70. doi: 10.1007/BF00269421. [DOI] [PubMed] [Google Scholar]
- 39.Monod M, Haguenauer-Tsapis R, Rauseo-Koenig I, Hinnen A. Functional analysis of the signal-sequence processing site of yeast acid phosphatase. European Journal of Biochemistry. 1989;182(2):213–221. doi: 10.1111/j.1432-1033.1989.tb14820.x. [DOI] [PubMed] [Google Scholar]
- 40.Mizunaga T, Izawa M, Ikeda K, Maruyama Y. Secretion of an active nonglycosylated form of the repressible acid phosphatase of Saccharomyces cerevisiae in the presence of tunicamycin at low temperatures. Journal of Biochemistry. 1988;103(2):321–326. doi: 10.1093/oxfordjournals.jbchem.a122268. [DOI] [PubMed] [Google Scholar]
- 41.Shnyreva MG, Petrova EV, Egorov SN, Hinnen A. Biochemical properties and excretion behavior of repressible acid phosphatases with altered subunit composition. Microbiological Research. 1996;151(3):291–300. doi: 10.1016/S0944-5013(96)80027-9. [DOI] [PubMed] [Google Scholar]
- 42.Barbaric S, Luckenbach T, Schmid A, Blaschke D, Hörz W, Korber P. Redundancy of chromatin remodeling pathways for the induction of the yeast PHO5 promoter in vivo. The Journal of Biological Chemistry. 2007;282(38):27610–27621. doi: 10.1074/jbc.M700623200. [DOI] [PubMed] [Google Scholar]
- 43.Auesukaree C, Tochio H, Shirakawa M, Kaneko Y, Harashima S. Plc1p, Arg82p, and Kcs1p, enzymes involved in inositol pyrophosphate synthesis, are essential for phosphate regulation and polyphosphate accumulation in Saccharomyces cerevisiae. The Journal of Biological Chemistry. 2005;280(26):25127–25133. doi: 10.1074/jbc.M414579200. [DOI] [PubMed] [Google Scholar]
- 44.Pinson B, Vaur S, Sagot I, Coulpier F, Lemoine S, Daignan-Fornier B. Metabolic intermediates selectively stimulate transcription factor interaction and modulate phosphate and purine pathways. Genes and Development. 2009;23(12):1399–1407. doi: 10.1101/gad.521809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Auesukaree C, Homma T, Kaneko Y, Harashima S. Transcriptional regulation of phosphate-responsive genes in low-affinity phosphate-transporter-defective mutants in Saccharomyces cerevisiae. Biochemical and Biophysical Research Communications. 2003;306(4):843–850. doi: 10.1016/s0006-291x(03)01068-4. [DOI] [PubMed] [Google Scholar]
- 46.Almer A, Hörz W. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO Journal. 1986;5(10):2681–2687. doi: 10.1002/j.1460-2075.1986.tb04551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Almer A, Rudolph H, Hinnen A, Hörz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO Journal. 1986;5(10):2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vogel K, Horz W, Hinnen A. The two positively acting regulatory proteins PHO2 and PHO4 physically interact with PHO5 upstream activation regions. Molecular and Cellular Biology. 1989;9(5):2050–2057. doi: 10.1128/mcb.9.5.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Svaren J, Hörz W. Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends in Biochemical Sciences. 1997;22(3):93–97. doi: 10.1016/s0968-0004(97)01001-3. [DOI] [PubMed] [Google Scholar]
- 50.Barbarić S, Münsterkötter M, Svaren J, Hörz W. The homeodomain protein PHO2 and the basic-helix-loop-helix protein PHO4 bind DNA cooperatively at the yeast PHO5 promoter. Nucleic Acids Research. 1996;24(22):4479–4486. doi: 10.1093/nar/24.22.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnston M, Carlson M. Molecular and Cellular Biology: Gene Expression. New York, NY, USA: Cold Spring Harbor; 1992. Regulation of carbon and phosphate utilization of the yeast Saccharomyces cerevisiae; pp. 283–317. [Google Scholar]
- 52.Adkins MW, Tyler JK. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Molecular Cell. 2006;21(3):405–416. doi: 10.1016/j.molcel.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 53.Schmid A, Fascher KD, Horz W. Nucleosome disruption at the yeast PHO5 promoter upon PHO5 induction occurs in the absence of DNA replication. Cell. 1992;71(5):853–864. doi: 10.1016/0092-8674(92)90560-y. [DOI] [PubMed] [Google Scholar]
- 54.Svaren J, Schmitz J, Horz W. The transactivation domain of PHO4 is required for nucleosome disruption at the PHO5 promoter. EMBO Journal. 1994;13(20):4856–4862. doi: 10.1002/j.1460-2075.1994.tb06812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gaudreau L, Schmid A, Blaschke D, Ptashne M, Hörz W. RNA polymerase II holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter. Cell. 1997;89(1):55–62. doi: 10.1016/s0092-8674(00)80182-8. [DOI] [PubMed] [Google Scholar]
- 56.Ohsawa R, Adkins M, Tyler JK. Epigenetic inheritance of an inducibly nucleosome-depleted promoter and its associated transcriptional state in the apparent absence of transcriptional activators. Epigenetics Chromatin. 2009;2(1):p. 11. doi: 10.1186/1756-8935-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gregory PD, Schmid A, Zavari M, Liu L, Berger SL, Hörz W. Absence of Gcn5 HAT activity defines a novel state in the opening of chromatin at the PHO5 promoter in yeast. Molecular Cell. 1998;1(4):495–505. doi: 10.1016/s1097-2765(00)80050-7. [DOI] [PubMed] [Google Scholar]
- 58.Nishimura K, Yasumura K, Igarashi K, Harashima S, Kakinuma Y. Transcription of some PHO genes in Saccharomyces cerevisiae is regulated by Spt7p. Yeast. 1999;15(16):1711–1717. doi: 10.1002/(SICI)1097-0061(199912)15:16<1711::AID-YEA497>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 59.Barbaric S, Reinke H, Hörz W. Multiple mechanistically distinct functions of SAGA at the PHO5 promoter. Molecular and Cellular Biology. 2003;23(10):3468–3476. doi: 10.1128/MCB.23.10.3468-3476.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lau WTW, Schneider KR, O’Shea EK. A genetic study of signaling processes for repression of PHO5 transcription in Saccharomyces cerevisiae. Genetics. 1998;150(4):1349–1359. doi: 10.1093/genetics/150.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Popova Y, Thayumanavan P, Lonati E, Agrochão M, Thevelein JM. Transport and signaling through the phosphate-binding site of the yeast PHO84 phosphate transceptor. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(7):2890–2895. doi: 10.1073/pnas.0906546107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nishizawa M, Kanaya Y, Toh-e A. Mouse cyclin-dependent kinase (Cdk) 5 is a functional homologue of a yeast Cdk, PHO85 kinase. The Journal of Biological Chemistry. 1999;274(48):33859–33862. doi: 10.1074/jbc.274.48.33859. [DOI] [PubMed] [Google Scholar]
- 63.Huang K, Ferrin-O’Connell I, Zhang W, Leonard GA, O’Shea EK, Quiocho F. Structure of the PHO85-PHO80 CDK-Cyclin Complex of the Phosphate-Responsive Signal Transduction Pathway. Molecular Cell. 2007;28(4):614–623. doi: 10.1016/j.molcel.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilson WA, Mahrenholz AM, Roach PJ. Substrate targeting of the yeast cyclin-dependent kinase PHO85 p by the cyclin Pcl10p. Molecular and Cellular Biology. 1999;19(10):7020–7030. doi: 10.1128/mcb.19.10.7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Measday V, Moore L, Retnakaran R, et al. A family of cyclin-like proteins that interact with the PHO85 cyclin- dependent kinase. Molecular and Cellular Biology. 1997;17(3):1212–1223. doi: 10.1128/mcb.17.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Toh-e A, Tanaka K, Uesono Y, Wickner RB. PHO85 , a negative regulator of the PHO system, is a homolog of the protein kinase gene, CDC28, of Saccharomyces cerevisiae. MGG Molecular & General Genetics. 1988;214(1):162–164. doi: 10.1007/BF00340196. [DOI] [PubMed] [Google Scholar]
- 67.Mendenhall MD, Hodge AE. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiology and Molecular Biology Reviews. 1998;62(4):1191–1243. doi: 10.1128/mmbr.62.4.1191-1243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Uesono Y, Tanaka K, Toh-e A. Negative regulators of the PHO system in Saccharomyces cerevisiae: isolation and structural characterization of PHO85. Nucleic Acids Research. 1987;15(24):10299–10309. doi: 10.1093/nar/15.24.10299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sambuk EV, Kuchkartaev AI, Padkina MV, Smirnov MN. Genetic mapping of genes regulating synthesis of acid phosphatases in yeast Saccharomyces cerevisiae of Peterhoff yeast collection. Genetika. 1991;27(4):644–648. [PubMed] [Google Scholar]
- 70.Sambuk EV, Popova YG, Fizikova AY, Padkina MV. Genetic analysis of pleiotropic effects of PHO85 mutations in yeast Saccharomyces cerevisiae. Genetika. 2003;39(8):1039–1045. [PubMed] [Google Scholar]
- 71.Lenburg ME, O’Shea EK. Genetic evidence for a morphogenetic function of the Saccharomyces cerevisiae PHO85 cyclin-dependent kinase. Genetics. 2001;157(1):39–51. doi: 10.1093/genetics/157.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zou J, Friesen H, Larson J, et al. Regulation of cell polarity through phosphorylation of Bni4 by PHO85 G1 cyclin-dependent kinases in Saccharomyces cerevisiae. Molecular Biology of the Cell. 2009;20(14):3239–3250. doi: 10.1091/mbc.E08-12-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Timblin BK, Tatchell K, Bergman LW. Deletion of the gene encoding the cyclin-dependent protein kinase PHO85 alters glycogen metabolism in Saccharomyces cerevisiae. Genetics. 1996;143(1):57–66. doi: 10.1093/genetics/143.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wickert S, Finck M, Herz B, Ernst JF. A small protein (Ags1p) and the Pho80p-Pho85p kinase complex contribute to aminoglycoside antibiotic resistance of the yeast Saccharomyces cerevisiae. Journal of Bacteriology. 1998;180(7):1887–1894. doi: 10.1128/jb.180.7.1887-1894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Popova IuG, Padkina MV, Sambuk EV. Effect of mutations in PHO85 and PHO4 genes on utilization of proline in Saccharomyces cerevisiae yeasts. Genetika. 2000;36(12):1622–1628. [PubMed] [Google Scholar]
- 76.Friesen H, Murphy K, Breitkreutz A, Tyers M, Andrews B. Regulation of the yeast amphiphysin homologue Rvs167p by phosphorylation. Molecular Biology of the Cell. 2003;14(7):3027–3040. doi: 10.1091/mbc.E02-09-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang D, Patrick G, Moffat J, Tsai LH, Andrews B. Mammalian Cdk5 is a functional homologue of the budding yeast PHO85 cyclin-dependent protein kinase. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(25):14445–14450. doi: 10.1073/pnas.96.25.14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Timblin BK, Bergman LW. Elevated expression of stress response genes resulting front deletion of the PHO85 gene. Molecular Microbiology. 1997;26(5):981–990. doi: 10.1046/j.1365-2958.1997.6352004.x. [DOI] [PubMed] [Google Scholar]
- 79.Bourne Y, Watson MH, Hickey MJ, et al. Crystal structure and mutational analysis of the human CDK2 kinase complex with cell cycle-regulatory protein CksHs1. Cell. 1996;84(6):863–874. doi: 10.1016/s0092-8674(00)81065-x. [DOI] [PubMed] [Google Scholar]
- 80.Hanks SK, Hunter T. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB Journal. 1995;9(8):576–596. [PubMed] [Google Scholar]
- 81.Santos RC, Waters NC, Creasy CL, Bergman LW. Structure-function relationships of the yeast cyclin-dependent kinase PHO85. Molecular and Cellular Biology. 1995;15(10):5482–5491. doi: 10.1128/mcb.15.10.5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Espinoza FH, Farrell A, Nourse JL, Chamberlin HM, Gileadi O, Morgan DO. Cak1 is required for Kin28 phosphorylation and activation in vivo. Molecular and Cellular Biology. 1998;18(11):6365–6373. doi: 10.1128/mcb.18.11.6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nigg EA. Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. BioEssays. 1995;17(6):471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- 84.Andersen G, Busso D, Poterszman A, et al. The structure of cyclin H: common mode of kinase activation and specific features. EMBO Journal. 1997;16(5):958–967. doi: 10.1093/emboj/16.5.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Spellman PT, Sherlock G, Zhang MQ, et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Molecular Biology of the Cell. 1998;9(12):3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lange P, Hansche PE. Mapping of a centromere-linked gene responsible for constitutive acid phosphatase synthesis in yeast. Molecular and General Genetics. 1980;180(3):605–607. doi: 10.1007/BF00268067. [DOI] [PubMed] [Google Scholar]
- 87.Samsonova MG, Padkina MV, Krasnopevtseva NG. Genetic and biochemical research of acid phosphatases in yeast Saccharomyces cerevisiae. V. Genetic control of acid phosphatase II synthesis regulation. Genetika. 1975;11:104–115. [PubMed] [Google Scholar]
- 88.Gilliquet V, Legrain M, Berben G, Hilger F. Negative regulatory elements of the Saccharomyces cerevisiae PHO system: interaction between PHO80 and PHO85 proteins. Gene. 1990;96(2):181–188. doi: 10.1016/0378-1119(90)90251-l. [DOI] [PubMed] [Google Scholar]
- 89.Toh-e A, Shimauchi T. Cloning and sequencing of the PHO80 gene and CEN15 of Saccharomyces cerevisiae. Yeast. 1986;2(2):129–139. doi: 10.1002/yea.320020209. [DOI] [PubMed] [Google Scholar]
- 90.Bisson LF, Thorner J. Mutations in the PHO80 gene confer permeability to 5′-mononucleotides in Saccharomyces cerevisiae. Genetics. 1982;102(3):341–359. doi: 10.1093/genetics/102.3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nicolson TA, Weisman LS, Payne GS, Wickner WT. A truncated form of the PHO80 cyclin redirects the PHO85 kinase to disrupt vacuole inheritance in S. cerevisiae. Journal of Cell Biology. 1995;130(4):835–845. doi: 10.1083/jcb.130.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nicolson T, Conradt B, Wickner W. A truncated form of the PHO80 cyclin of Saccharomyces cerevisiae induces expression of a small cytosolic factor which inhibits vacuole inheritance. Journal of Bacteriology. 1996;178(14):4047–4051. doi: 10.1128/jb.178.14.4047-4051.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gilliquet V, Berben G. Positive and negative regulators of the Saccharomyces cerevisiae ‘PHO system’ participate in several cell functions. FEMS Microbiology Letters. 1993;108(3):333–339. doi: 10.1111/j.1574-6968.1993.tb06124.x. [DOI] [PubMed] [Google Scholar]
- 94.Bandyopadhyay S, Mehta M, Kuo D, et al. Rewiring of genetic networks in response to DNA damage. Science. 2010;330(6009):1385–1389. doi: 10.1126/science.1195618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mao XC, Xia YL, Hu YF, Lu CD. Involvement of PHO80 and PHO85 genes in Saccharomyces cerevisiae ion tolerance. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 2003;35(1):86–91. [PubMed] [Google Scholar]
- 96.O’Neill EM, Kaffman A, Jolly ER, O’Shea EK. Regulation of PHO4 nuclear localization by the PHO80-PH085 cyclin-CDK complex. Science. 1996;271(5246):209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- 97.Jayaraman PS, Hirst K, Goding CR. The activation domain of a basic helix-loop-helix protein is masked by repressor interaction with domains distinct from that required for transcription regulation. EMBO Journal. 1994;13(9):2192–2199. doi: 10.1002/j.1460-2075.1994.tb06496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Madden SL, Johnson DL, Bergman LW. Molecular and expression analysis of the negative regulators involved in the transcriptional regulation of acid phosphatase production in Saccharomyces cerevisiae. Molecular and Cellular Biology. 1990;10(11):5950–5957. doi: 10.1128/mcb.10.11.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang Z, Wilson WA, Fujino MA, Roach PJ. The yeast cyclins Pc16p and Pc17p are involved in the control of glycogen storage by the cyclin-dependent protein kinase PHO85 p. FEBS Letters. 2001;506(3):277–280. doi: 10.1016/s0014-5793(01)02914-3. [DOI] [PubMed] [Google Scholar]
- 100.Lee M, O’Regan S, Moreau JL, Johnson AL, Johnston LH, Goding CR. Regulation of the PcI7-PHO85 cyclin-cdk complex by Pho81. Molecular Microbiology. 2000;38(2):411–422. doi: 10.1046/j.1365-2958.2000.02140.x. [DOI] [PubMed] [Google Scholar]
- 101.Tan YSH, Morcos PA, Cannon JF. PHO85 phosphorylates the Glc7 protein phosphatase regulator Glc8 in vivo. The Journal of Biological Chemistry. 2003;278(1):147–153. doi: 10.1074/jbc.M208058200. [DOI] [PubMed] [Google Scholar]
- 102.Ho Y, Gruhler A, Heilbut A, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415(6868):180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 103.Huang D, Moffat J, Wilson WA, et al. Cyclin partners determine PHO85 protein kinase substrate specificity in vitro and in vivo: control of glycogen biosynthesis by Pcl8 and Pcl10. Molecular and Cellular Biology. 1998;18(6):3289–3299. doi: 10.1128/mcb.18.6.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee J, Colwill K, Aneliunas V, et al. Interaction of yeast Rvs167 and PHO85 cyclin-dependent kinase complexes may link the cell cycle to the actin cytoskeleton. Current Biology. 1998;8(24):1310–1321. doi: 10.1016/s0960-9822(07)00561-1. [DOI] [PubMed] [Google Scholar]
- 105.Tennyson CN, Lee J, Andrews BJ. A role for the Pcl9-PHO85 cyclin-cdk complex at the M/G boundary in Saccharomyces cerevisiae. Molecular Microbiology. 1998;28(1):69–79. doi: 10.1046/j.1365-2958.1998.00773.x. [DOI] [PubMed] [Google Scholar]
- 106.Carroll AS, O’Shea EK. PHO85 and signaling environmental conditions. Trends in Biochemical Sciences. 2002;27(2):87–93. doi: 10.1016/s0968-0004(01)02040-0. [DOI] [PubMed] [Google Scholar]
- 107.Wysocki R, Javaheri A, Kristjansdottir K, Sha F, Kron SJ. CDK PHO85 targets CDK inhibitor Sic1 to relieve yeast G1 checkpoint arrest after DNA damage. Nature Structural and Molecular Biology. 2006;13(10):908–914. doi: 10.1038/nsmb1139. [DOI] [PubMed] [Google Scholar]
- 108.Schmid G, Strosznajder JB, Wesierska-Gadek J. Interplay between the p53 tumor suppressor protein family and Cdk5: novel therapeutic approaches for the treatment of neurodegenerative diseases using selective Cdk inhibitors. Molecular Neurobiology. 2006;34(1):27–50. doi: 10.1385/mn:34:1:27. [DOI] [PubMed] [Google Scholar]
- 109.Kucej M, Kucejova B, Subramanian R, Chen XJ, Butow RA. Mitochondrial nucleoids undergo remodeling in response to metabolic cues. Journal of Cell Science. 2008;121(11):1861–1868. doi: 10.1242/jcs.028605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sambuk EV, Popova YG, Padkina MV. Genetic analysis of spontaneous suppressors of the PHO85 mutation in yeast Saccharomyces cerevisiae. Genetika. 2003;39(1):18–24. [PubMed] [Google Scholar]
- 111.Miyakawa I, Kanayama M, Fujita Y, Sato H. Morphology and protein composition of the mitochondrial nucleoids in yeast cells lacking Abf2p, a high mobility group protein. Journal of General and Applied Microbiology. 2010;56(6):455–464. doi: 10.2323/jgam.56.455. [DOI] [PubMed] [Google Scholar]
- 112.Schricker R, Magdolen V, Strobel G, Bogengruber E, Breitenbach M, Bandlow W. Strain-dependent occurrence of functional GTP:AMP phosphotransferase (AK3) in Saccharomyces cerevisiae. The Journal of Biological Chemistry. 1995;270(52):31103–31110. doi: 10.1074/jbc.270.52.31103. [DOI] [PubMed] [Google Scholar]
- 113.Zhao X, Muller EGD, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Molecular Cell. 1998;2(3):329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 114.Bateman JM, Iacovino M, Perlman PS, Butow RA. Mitochondrial DNA instability mutants of the bifunctional protein Ilv5p have altered organization in mitochondria and are targeted for degradation by Hsp78 and the Pim1p protease. The Journal of Biological Chemistry. 2002;277(49):47946–47953. doi: 10.1074/jbc.M209071200. [DOI] [PubMed] [Google Scholar]
- 115.Lithgow T, Junne T, Wachter C, Schatz G. Yeast mitochondria lacking the two import receptors Mas20p and Mas70p can efficiently and specifically import precursor proteins. The Journal of Biological Chemistry. 1994;269(21):15325–15330. [PubMed] [Google Scholar]
- 116.Steger HF, Söllner T, Kiebler M, et al. Import of ADP/ATP carrier into mitochondria: two receptors act in parallel. Journal of Cell Biology. 1990;111(6):2353–2363. doi: 10.1083/jcb.111.6.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dimmer KS, Fritz S, Fuchs F, et al. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Molecular Biology of the Cell. 2002;13(3):847–853. doi: 10.1091/mbc.01-12-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Contamine V, Picard M. Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast. Microbiology and Molecular Biology Reviews. 2000;64(2):281–315. doi: 10.1128/mmbr.64.2.281-315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sambuk EV, Fizikova AI, Zakharova KV, Smirnov AM, Padkina MV. The lack of cyclin-dependent phosphoprotein kinase PHO85 p leads to defects in mitochondrial nucleoid transmission in yeast Saccharomyces cerevisiae. Tsitologiia. 2005;47(10):917–924. [PubMed] [Google Scholar]
- 120.Fizikova AIu, Padkina MV, Sambuk EV. The absence of cyclin-dependent protein kinase PHO85 affects stability of mitochondrial DNA in yeast Saccharomyces cerevisiae. Genetika. 2009;45(6):745–752. [PubMed] [Google Scholar]
- 121.Smirnov AM, Sambuk EV. PHO-regulatory genes mutations influence on genetic material stability of yeast Saccharomyces cerevisia. Ecological Genetics. 2008;6:40–47. [Google Scholar]
- 122.Kaufman BA, Kolesar JE, Perlman PS, Butow RA. A function for the mitochondrial chaperonin Hsp60 in the structure and transmission of mitochondrial DNA nucleoids in Saccharomyces cerevisiae. Journal of Cell Biology. 2003;163(3):457–461. doi: 10.1083/jcb.200306132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fristedt U, Berhe A, Ensler K, Norling B, Persson BL. Isolation and characterization of membrane vesicles of Saccharomyces cerevisiae harboring the high-affinity phosphate transporter. Archives of Biochemistry and Biophysics. 1996;330(1):133–141. doi: 10.1006/abbi.1996.0235. [DOI] [PubMed] [Google Scholar]
- 124.Giots F, Donaton MCV, Thevelein JM. Inorganic phosphate is sensed by specific phosphate carriers and acts in concert with glucose as a nutrient signal for activation of the protein kinase A pathway in the yeast Saccharomyces cerevisiae. Molecular Microbiology. 2003;47(4):1163–1181. doi: 10.1046/j.1365-2958.2003.03365.x. [DOI] [PubMed] [Google Scholar]
- 125.Mort-Bontemps-Soret M, Facca C, Faye G. Physical interaction of Cdc28 with Cdc37 in Saccharomyces cerevisiae. Molecular Genetics and Genomics. 2002;267(4):447–458. doi: 10.1007/s00438-002-0676-3. [DOI] [PubMed] [Google Scholar]
- 126.Gerber MR, Farrell A, Deshaies RJ, Herskowitz I, Morgan DO. Cdc37 is required for association of the protein kinase Cdc28 with G and mitotic cyclins. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(10):4651–4655. doi: 10.1073/pnas.92.10.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Peter M, Herskowitz I. Direct inhibition of the yeast cyclin-dependent kinase Cdc28-Cln by Far1. Science. 1994;265(5176):1228–1231. doi: 10.1126/science.8066461. [DOI] [PubMed] [Google Scholar]
- 128.Mendenhall MD. An inhibitor of p34(CDC28) protein kinase activity from Saccharomyces cerevisiae. Science. 1993;259(5092):216–219. doi: 10.1126/science.8421781. [DOI] [PubMed] [Google Scholar]
- 129.Coche T, Prozzi D, Legrain M, Hilger F, Vandenhaute J. Nucleotide sequence of the PHO81 gene involved in the regulation of the repressible acid phosphatase gene in Saccharomyces cerevisiae. Nucleic Acids Research. 1990;18(8):p. 2176. doi: 10.1093/nar/18.8.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Creasy CL, Shao D, Bergman LW. Negative transcriptional regulation of PHO81 expression in Saccharomyces cerevisiae. Gene. 1996;168(1):23–29. doi: 10.1016/0378-1119(95)00737-7. [DOI] [PubMed] [Google Scholar]
- 131.Schneider KR, Smith RL, O’Shea EK. Phosphate-regulated inactivation of the kinase PHO80-PHO85 by the CDK inhibitor PHO81. Science. 1994;266(5182):122–126. doi: 10.1126/science.7939631. [DOI] [PubMed] [Google Scholar]
- 132.Ogawa N, Noguchi KI, Sawai H, Yamashita Y, Yompakdee C, Oshima Y. Functional domains of Pho81p, an inhibitor of PHO85 p protein kinase, in the transduction pathway of P(i) signals in Saccharomyces cerevisiae. Molecular and Cellular Biology. 1995;15(2):997–1004. doi: 10.1128/mcb.15.2.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Flick JS, Thorner J. An essential function of a phosphoinositide-specific phospholipase C is relieved by inhibition of a cyclin-dependent protein kinase in the yeast Saccharomyces cerevisiae. Genetics. 1998;148(1):33–47. doi: 10.1093/genetics/148.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shen X, Xiao H, Ranallo R, Wu WH, Wu C. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science. 2003;299(5603):112–114. doi: 10.1126/science.1078068. [DOI] [PubMed] [Google Scholar]
- 135.Sze IS-Y, McFarlan SC, Spormann A, Hogenkamp HPC, Follman H. A possible new class of ribonucleotide reductase from Methanobacterium thermoautotrophicum. Biochemical and Biophysical Research Communications. 1992;184(2):1101–1107. doi: 10.1016/0006-291x(92)90705-p. [DOI] [PubMed] [Google Scholar]
- 136.Rébora K, Desmoucelles C, Borne F, Pinson B, Daignan-Fornier B. Yeast AMP pathway genes respond to adenine through regulated synthesis of a metabolic intermediate. Molecular and Cellular Biology. 2001;21(23):7901–7912. doi: 10.1128/MCB.21.23.7901-7912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Guetsova ML, Lecoq K, Daignan-Fornier B. The isolation and characterization of Saccharomyces cerevisiae mutants that constitutively express purine biosynthetic genes. Genetics. 1997;147(2):383–397. doi: 10.1093/genetics/147.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Huang S, O’Shea EK. A systematic high-throughput screen of a yeast deletion collection for mutants defective in PHO5 regulation. Genetics. 2005;169(4):1859–1871. doi: 10.1534/genetics.104.038695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gauthier S, Coulpier F, Jourdren L, et al. Co-regulation of yeast purine and phosphate pathways in response to adenylic nucleotide variations. Molecular Microbiology. 2008;68(6):1583–1594. doi: 10.1111/j.1365-2958.2008.06261.x. [DOI] [PubMed] [Google Scholar]
- 140.Wurst H, Shiba T, Kornberg A. The gene for a major exopolyphosphatase of Saccharomyces cerevisiae. Journal of Bacteriology. 1995;177(4):898–906. doi: 10.1128/jb.177.4.898-906.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Andreeva AL, Andreev OA, Borejdo J. Structure of the 265-kilodalton complex formed upon EDC cross-linking of subfragment 1 to F-actin. Biochemistry. 1993;32(50):13956–13960. doi: 10.1021/bi00213a027. [DOI] [PubMed] [Google Scholar]
- 142.Kumble KD, Kornberg A. Endopolyphosphatases for long chain inorganic polyphosphate in yeast and mammals. The Journal of Biological Chemistry. 1996;271(43):27146–27151. doi: 10.1074/jbc.271.43.27146. [DOI] [PubMed] [Google Scholar]
- 143.Kulakovskaya TV, Lichko LP, Vagabov VM, Kulaev IS. Inorganic polyphosphates in mitochondria. Biochemistry. 2010;75(7):825–831. doi: 10.1134/s0006297910070035. [DOI] [PubMed] [Google Scholar]
- 144.Cohen A, Perzov N, Nelson H, Nelson N. A novel family of yeast chaperons involved in the distribution of V- ATPase and other membrane proteins. The Journal of Biological Chemistry. 1999;274(38):26885–26893. doi: 10.1074/jbc.274.38.26885. [DOI] [PubMed] [Google Scholar]
- 145.Müller O, Bayer MJ, Peters C, Andersen JS, Mann M, Mayer A. The Vtc proteins in vacuole fusion: coupling NSF activity to V trans-complex formation. EMBO Journal. 2002;21(3):259–269. doi: 10.1093/emboj/21.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Makino K, Shinagawa H, Nakata A. Regulation of the phosphate regulon of Escherichia coli K-12: regulation and role of the regulatory gene phoR. Journal of Molecular Biology. 1985;184(2):231–240. doi: 10.1016/0022-2836(85)90376-6. [DOI] [PubMed] [Google Scholar]
- 147.Bennett M, Onnebo SMN, Azevedo C, Saiardi A. Inositol pyrophosphates: metabolism and signaling. Cellular and Molecular Life Sciences. 2006;63(5):552–564. doi: 10.1007/s00018-005-5446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Saiardi A, Bhandari R, Resnick AC, Snowman AM, Snyder SH. Phosphorylation of proteins by inositol pyrophosphates. Science. 2004;306(5704):2101–2105. doi: 10.1126/science.1103344. [DOI] [PubMed] [Google Scholar]
- 149.Stephens L, Radenberg T, Thiel U, et al. The detection, purification, structural characterization, and metabolism of diphosphoinositol pentakisphosphate(s) and bisdiphosphoinositol tetrakisphosphate(s) The Journal of Biological Chemistry. 1993;268(6):4009–4015. [PubMed] [Google Scholar]
- 150.Bhandari R, Saiardi A, Ahmadibeni Y, et al. Protein pyrophosphorylation by inositol pyrophosphates is a posttranslational event. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(39):15305–15310. doi: 10.1073/pnas.0707338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Saiardi A, Erdjument-Bromage H, Snowman AM, Tempst P, Snyder SH. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Current Biology. 1999;9(22):1323–1326. doi: 10.1016/s0960-9822(00)80055-x. [DOI] [PubMed] [Google Scholar]
- 152.Mulugu S, Bai W, Fridy PC, et al. A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science. 2007;316(5821):106–109. doi: 10.1126/science.1139099. [DOI] [PubMed] [Google Scholar]
- 153.Lee YS, Mulugu S, York JD, O’Shea EK. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science. 2007;316(5821):109–112. doi: 10.1126/science.1139080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA Stabilization Induces Transcriptional Gene Silencing via Histone Deacetylation in S. cerevisiae. Cell. 2007;131(4):706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 155.Hongay CF, Grisafi PL, Galitski T, Fink GR. Antisense Transcription Controls Cell Fate in Saccharomyces cerevisiae. Cell. 2006;127(4):735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 156.Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429(6991):571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 157.Nishizawa M, Komai T, Katou Y, Shirahige K, Ito T, Toh-e A. Nutrient-regulated antisense and intragenic RNAs modulate a signal transduction pathway in yeast. PLoS Biology. 2008;6(12, article e326):2817–2830. doi: 10.1371/journal.pbio.0060326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Uhler JP, Hertel C, Svejstrup JQ. A role for noncoding transcription in activation of the yeast PHO5 gene. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(19):8011–8016. doi: 10.1073/pnas.0702431104. [DOI] [PMC free article] [PubMed] [Google Scholar]


