Abstract
POP2 protein of Saccharomyces cerevisiae is a component of a protein complex that regulates the transcription of many genes. We found that the 97th threonine residue (Thr 97) of Pop2p was phosphorylated upon glucose limitation. The Thr 97 phosphorylation occurred within 2 min after removing glucose and was reversed within 1 min after the readdition of glucose. The effects of hexokinase mutations and glucose analogs indicate that this phosphorylation is dependent on glucose phosphorylating activity. We purified a protein kinase that phosphorylates a peptide containing Thr 97 of Pop2p and identified it as Yak1p, a DYRK family kinase. Phosphorylation of Pop2p was barely detectable in a yak1Δ strain. We found that Yak1p interacted with Bmh1p and Bmh2p only in the presence of glucose. A GFP-Yak1p fusion protein shuttled rapidly between the nucleus and the cytoplasm in response to glucose. A strain with alanine substituted for Thr 97 in Pop2p showed overgrowth in the postdiauxic transition and failed to stop the cell cycle at G1 phase in response to glucose deprivation. Thus, Yak1p and Pop2p are part of a novel glucose-sensing system in yeast that is involved in growth control in response to glucose availability.
Keywords: POP2, YAK1, glucose sensing, 14-3-3 protein
Glucose is the favored carbon source for most cells. Depending on the availability of glucose, cells dramatically change expression of many genes involved in processes such as metabolism, growth, and proliferation. In the yeast Saccharomyces cerevisiae, glucose represses or induces the expression of many genes, including those encoding proteins in the respiratory pathway, enzymes for the use of alternative carbon sources, and glucose transporters. These phenomena are well known as glucose repression and glucose induction, respectively. The Snf1 kinase plays a central role in the glucose repression mechanism (Carlson 1999). Activation of Snf1 kinase activity in response to glucose limitation is caused by conformational changes in the kinase complex triggered by its phosphorylation (Jiang and Carlson 1996). A major role of the Snf1 kinase is to inhibit the repression of glucose-repressible genes through the phosphorylation of Mig1p, a zinc finger protein that mediates the glucose repression of several genes. Phosphorylation of Mig1p is necessary for targeting it to the nucleus, as well as for its repressor activity (De Vit and Johnston 1999). In another glucose-sensing system, Snf3p and Rgt2p function as glucose sensors for the glucose-induced transcription of HXT (glucose transporter) genes (Ozcan et al. 1998; Johnston 1999). In the absence of glucose, another zinc finger protein, Rgt1p, binds to the HXT promoters and represses their transcription (Ozcan et al. 1996). Glucose generates a signal and causes the SCFGrr1 complex to inactivate the Rgt1p repressor, thereby derepressing HXT gene expression (Li and Johnston 1997). Glucose also triggers a rapid elevation of the intracellular level of cAMP and then activates protein kinase A (PKA) (Thevelein and de Winde 1999), whose downstream targets are metabolic enzymes involved in intermediary carbon metabolism. Other targets of this process are Msn2p and Msn4p, transcription factors required for the expression of STRE (stress response element)-controlled genes (Gorner et al. 1998). For glucose-activated cAMP synthesis, a G-protein-coupled receptor system, consisting of a putative glucose receptor, Gpr1p, and its G-protein Gpa2, is required (Yun et al. 1998; Kraakman et al. 1999).
The POP2 gene is required for expression of glucose-repressed genes in S. cerevisiae (Sakai et al. 1992). Cells carrying a pop2 mutation show many defects, such as an inability to derepress glucose-repressed genes; temperature-sensitive growth; reduced level of reserved carbohydrate, isocitrate lyase, and invertase activities; and abnormal entry into stationary phase (Sakai et al. 1992; Hata et al. 1998). Pop2p is ubiquitous, and a mouse homolog can suppress the yeast pop2 null mutation (Draper et al. 1995; Shimizu-Yoshida et al. 1999). Pop2p, Ccr4p (required for transcription of glucose-repressible ADH2), and Dhh1p (RNA helicase) form a protein complex (CCR4-NOT complex) that regulates gene expression both positively and negatively (Draper et al. 1995; Hata et al. 1998; Bai et al. 1999).
In this paper, we show that Pop2p is a phosphoprotein, and the phosphorylation is regulated rapidly by glucose. We also present evidence showing that Yak1 kinase, a DYRK (dual specificity Yak1-related kinase) family kinase, is the major Pop2p kinase in vivo. The intracellular localization of Yak1p is regulated by glucose, probably through the binding of Bmh1p and Bmh2p. Phosphorylation of Pop2p by Yak1p is required for proper cell cycle arrest upon glucose limitation. We discuss the role of Yak1p-Pop2p in glucose sensing in yeast.
Results
Pop2p is phosphorylated at its 97th Thr residue
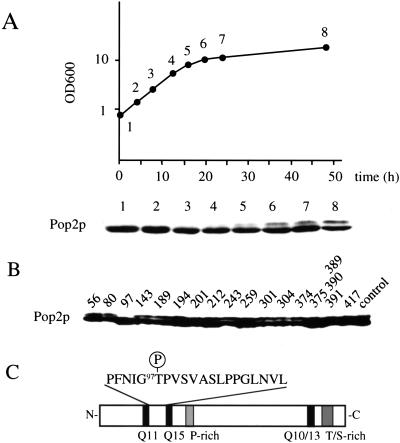
Because the POP2 gene is required for entry into stationary phase (Sakai et al. 1992), we analyzed Pop2p in cells in various growth phases using SDS-PAGE followed by Western blotting. Pop2p was identified as a single 50-kD band in exponentially growing cells. A band with slower migration appeared where the cell density reached its maximum (defined in this article as post-log phase; Fig. 1A), indicating a posttranslational modification. Alkaline phosphatase treatment of immunoprecipitated Pop2p from post-log phase cells eliminated the slowly migrating form, indicating that it is the phosphorylated form of Pop2p. To confirm this, Pop2p 32P-labeled in vivo was prepared from post-log phase cells by immunoprecipitation and analyzed. We discovered that (1) both the 50-kD form and the slower migration form were phosphorylated, and (2) two-dimensional thin-layer chromatography and phosphoamino acid analysis of the tryptic peptides derived from both forms of protein revealed that the 50-kD form is phosphorylated at a serine residue(s), and the slowly migrating form is phosphorylated at serine and threonine residues (data not shown). We therefore concluded that Pop2p is phosphorylated at a serine residue(s) in exponentially growing cells and an additional phosphorylation occurred at a threonine residue(s) in post-log phase cells, as well as in response to glucose deprivation (see below). To identify the threonine(s) that is phosphorylated, we constructed 16 Pop2p mutants in which one threonine residue in each was replaced with alanine. Each mutant gene was introduced into the pop2 null mutant cells, and the mobility shift of Pop2p in post-log phase cells was examined. As shown in Figure 1B, the mobility shift was detected in all of the mutant Pop2p, except for the residue 97 mutation (POP2-T97A). The position of Thr 97 in Pop2p is presented in Figure 1C. This result indicates strongly that a serine/threonine kinase(s) phosphorylates Thr 97 of Pop2p in post-log phase.
Figure 1.
(A) Appearance of a slower migration Pop2p in post-log phase. Yeast strain D24 (wild-type diploid) was inoculated into YPD media and cultivated at 24°C. At each culture time indicated, protein sample was prepared from the cells and Pop2p was detected by Western blot analysis. (B) The 97th threonine is responsible for the mobility shift. Sixteen mutant Pop2p, harboring threonine to alanine substitutions (one had a triple replacement) at the indicated positions, were introduced into the pop2Δ-3 (A2280) cells. The cells were grown to post-log phase and Pop2p was detected by Western blotting. (C) Schematic illustration of Pop2p including the amino acid sequence around the 97th threonine residue.
The Thr 97 phosphorylation of Pop2p is regulated by glucose
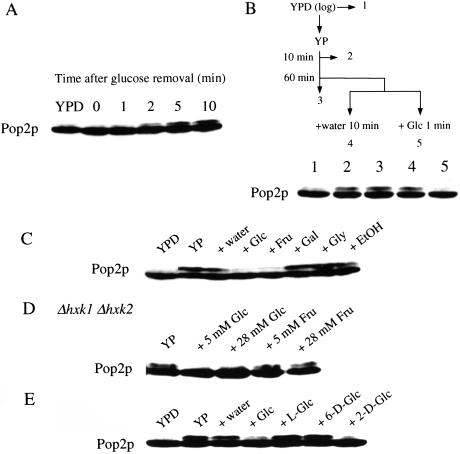
The post-log phase in which Pop2p was phosphorylated is known as the “diauxic phase,” when glucose is exhausted. Therefore, the slower migration form of Pop2p could be induced by limitation of glucose in the media. As shown in Figure 2A, when exponentially growing cells were transferred to glucose-deprived media, phosphorylated Pop2p appeared within 2 min and increased for 2–3 h, but the maximum amount never exceeded 50% of the total Pop2p. When glucose was added to the glucose-starved culture, the phosphorylated form disappeared within 1 min (Fig. 2B, lane 5). Addition of 100 μg/mL of cycloheximide did not affect either the phosphorylation or the dephosphorylation of the Pop2p, indicating that the quick and reversible phosphorylation was independent of protein synthesis.
Figure 2.
(A) Phosphorylation of Pop2p. Exponentially growing D24 cells in YPD medium were collected by centrifugation, washed once with YP medium, and resuspended in YP medium. At the times indicated, Pop2p from the cells was detected by Western blot analysis. (B) Dephosphorylation of Pop2p. Exponentially growing D24 cells in YPD medium were transferred to YP medium, and glucose was added to the culture to a final concentration of 20 mM. Pop2p from the cells was detected by Western blotting. Effects of various carbon sources (C), glucose analogs (D), and mutations of hexokinase genes (E) on Pop2p phosphorylation. Exponentially growing D24 cells (in A and B) or A1643 (hxk1Δ hxk2Δ mutant, in C) in YPD medium (YPD) were transferred to YP medium for 60 min (YP), and indicated carbon sources were added to the culture to a final concentration of 20 mM, unless otherwise stated. After 10 min, protein samples were prepared from the cells and Pop2p was detected by Western blotting. (Glc) Glucose; (Frc) fructose; (Gal) galactose; (Gly) glycerol; (EtOH) ethanol; (L-Glc) L-glucose; (6-D-Glc) 6-deoxyglucose; (2-D-Glc) 2-deoxyglucose.
The dephosphorylation of Thr 97 is coupled to glucose phosphorylating activity
To determine whether dephosphorylation of Pop2p was specific to glucose, we tested the effects of several carbon sources on the dephosphorylation. Figure 2C shows that glucose and fructose caused dephosphorylation of Pop2p whereas galactose, glycerol, and ethanol did not. Because both glucose and fructose are phosphorylated by hexokinase, we investigated the effects on Pop2p phosphorylation of mutations in the three genes encoding hexokinases (Herrero et al. 1995). The addition of glucose to a hxk1 hxk2 glk1 triple mutant failed to eliminate the phosphorylated form (data not shown). Cells carrying hxk1 hxk2 mutations, which block phosphorylation of fructose but not glucose (Gancedo et al. 1977), failed to dephosphorylate Pop2p in response to fructose (Fig. 2D). These results indicate that either ATP production by glycolysis or phosphorylation of glucose might be necessary for dephosphorylation of the Thr 97 phosphorylation of Pop2p.
To clarify this, we examined the effects of various glucose analogs on dephosphorylation of Pop2p. Pop2p was dephosphorylated by 2-deoxyglucose, which is incorporated into cells and phosphorylated by hexokinases but not metabolized further, but not L-glucose, which is not incorporated into cells, or 6-deoxyglucose, which is incorporated into cells but not phosphorylated by hexokinases (Fig. 2E), indicating that dephosphorylation of Pop2p was not dependent on ATP production but instead was related to hexokinase activity.
Purification of a protein kinase that phosphorylates Thr 97 of Pop2p
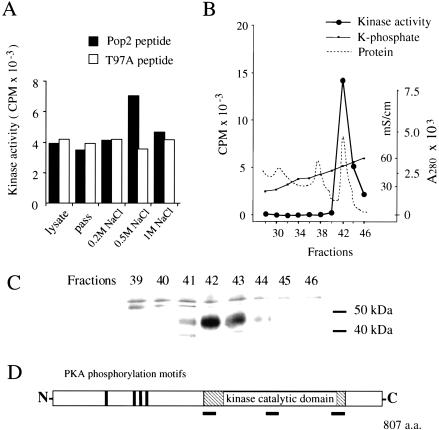
Next, we tried to identify a protein kinase that phosphorylates Thr 97 of Pop2p. Pop2p was phosphorylated normally in all the protein kinase mutant strains we tested (snf1, pkc1, bck1, mpk1, hog1, tpk1, tpk2, tpk3, and tpk1 tpk2 tpk3w). To identify the protein kinase that phosphorylates Thr 97 of Pop2p, we developed a kinase assay system using a 22-mer peptide containing Thr 97 and flanking amino acids of Pop2p (Pop2 peptide) and the T97A peptide, another 22-mer peptide that has a threonine to alanine substitution. Because phosphorylation of Thr 97 was observed in stationary-phase cells, we attempted to detect Pop2 peptide kinase activity from stationary-phase yeast cells. As shown in Figure 3A, Pop2 peptide-specific kinase activity was detected in a 0.2–0.5 M NaCl elution fraction from a phosphocellulose column. Using six chromatographic steps (see Materials and Methods), we succeeded in isolating Pop2 peptide kinase close to homogeneity (Table 1). The elution profile of the last column (ceramic hydroxyapatite column) is shown in Figure 3B. The peak fractions of the peptide kinase activity contained mainly a 45-kD protein (Fig. 3C). The partial amino acid sequences of this 45-kD protein (SRTEYLTQSITEAK, LAPEESSSSTQ, and QEFTGEWFPPGSSLPG) are identical to those of Yak1p of S. cerevisiae (positions 402 to 415, 607 to 617, and 707 to 722, respectively [Fig. 3D]). Yak1p is a 97-kD protein kinase and a negative growth regulator that antagonizes the PKA pathway (Garrett and Broach 1989; Garrett et al. 1991). Judging from its amino acid sequence and molecular weight, the 45-kD protein we purified was the C-terminal half of Yak1p, which contains the catalytic domain of the protein kinase (Fig. 3D). Endogenous protease digestion must have occurred during the purification.
Figure 3.
Detection, purification, and identification of the Pop2p peptide kinase. (A) Detection of Pop2 peptide-specific phosphorylation activity in phosphocellulose column fraction. Lysate of stationary-phase yeast cells was loaded on a phosphocellulose (P11) column. Protein was eluted with the NaCl concentrations indicated. The Pop2 peptide and T97A peptide kinase activities of each fraction were determined. (B) Elution profile of the final step of the Pop2 peptide kinase purification (ceramic hydroxyapatite chromatography). Pop2 peptide kinase activity (Kinase activity, CPM), K-phosphate concentration (K-phosphate, mS/mL), and protein concentration (Protein, A280) of each fraction are shown. (C) Silver staining image of the SDS-PAGE-separated elution fractions of the column indicated in panel B. The molecular weights indicated on the left side were estimated by SDS-PAGE standards (Bench-Mark protein ladder, GIBCO BRL). (D) The purified Pop2 peptide kinase was Yak1p. Amino acid sequences of three peptides derived from purified 45-kD Pop2p peptide kinase were determined and analyzed as described in the Materials and Methods. The positions of the peptides determined are shown as lines under the structure of Yak1p.
Table 1.
Purification of Pop2 peptide kinase
| Fraction
|
Volume (mL)
|
Total protein (μg)
|
Total activity (cpm/min)
|
Specific activity (cpm/min/mg)
|
Enrichment (fold)
|
|---|---|---|---|---|---|
| P11 | 60 | 216,000 | 71,600 | 0.331 | 1 |
| Methyl-HIC | 12 | 13,400 | 408,000 | 30.4 | 92 |
| Bio-Gel HTP (NaCl) | 4.5 | 563 | 335,000 | 595 | 1800 |
| Bio-Gel HTP (Phosphate) | 4.5 | 59.6 | 219,000 | 3670 | 11,000 |
| Bio-Scale CHT2-I | 0.4 | [10]a | 38,200 | [3820]a | [11,500]a |
Quantity of total protein in Bio-Scale CHT2-1 peak fraction estimated by Coomassie Brilliant Blue staining on SDS-PAGE.
YAK1 kinase phosphorylates Pop2p in vitro and in vivo
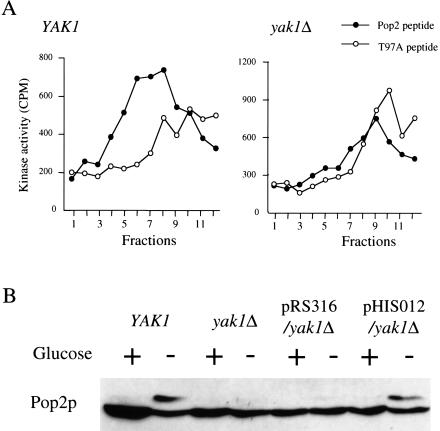
To confirm that Yak1p is the Pop2p kinase in vitro and in vivo, we constructed a yak1Δ mutant strain and determined its Pop2p peptide kinase activity. Pop2 peptide kinase activity was detected in the fractions from the phosphocellulose column (Fig. 4A, left) of wild type, but not the yak1Δ cell lysate (Fig. 4A, right). These results show clearly that YAK1 is the major Pop2 peptide kinase activity in the yeast cell extract. The band corresponding to the Thr 97 phosphorylated product of Pop2p was observed in wild-type cells when glucose was depleted, but not in the yak1Δ cells (Fig. 4B). Introduction into the yak1Δ cells of a plasmid harboring the YAK1 gene (pHIS012) restored the phosphorylated form of Pop2p. These results indicate strongly that Yak1p is the major Pop2p kinase in vivo. It should be noted that we were still able to observe a small amount of phosphorylated Pop2p after long exposure to enhanced chemiluminescence (ECL) on Western blotting (data not shown). Because this band is never observed in POP2-T97A cells, another Pop2p protein kinase seems to exist in yeast.
Figure 4.
Kinase activities in the yak1 null mutant strain. (A) In vitro peptide kinase activities of peptide kinase activity in the phosphocellulose (P11) column fractions in wild-type (YAK1; H213) and yak1Δ (H214) cell lysates. The stationary-phase cell lysate of each strain was loaded on a P11 column and then eluted with a 0 to 0.5 M NaCl gradient. Peptide kinase activity of each elution fraction was determined as described in the Materials and Methods. (B) In vivo phosphorylation of the Pop2p in wild type (YAK1; H213), yak1Δ (H214), H214 harboring pRS314, and H214 harboring YCpYAK1(pHIS012). Exponentially growing cells of each strain were washed by YP medium and resuspended in YPD medium (Glucose +) or YP medium (Glucose −). After 5 min, protein samples were prepared from the cells and Pop2p was detected by Western blotting.
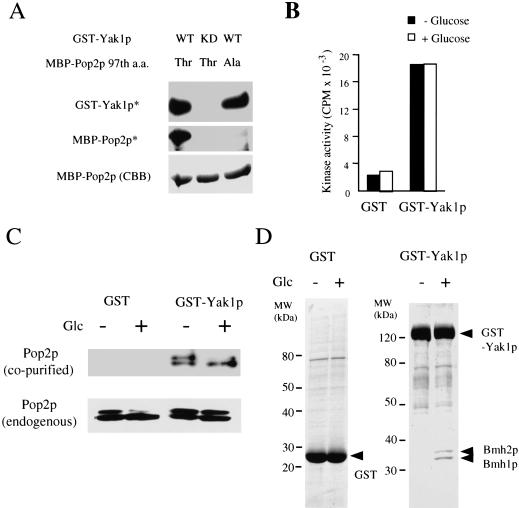
To determine if Yak1p could phosphorylate full-length Pop2p, we constructed a plasmid that expressed glutathione S-transferase (GST)-Yak1p fusion protein from the GAL1 promoter in yeast. In cells overexpressing GST-Yak1p, the Thr 97 phosphorylation of Pop2p was still observed 5 min after the addition of glucose (Fig. 5C). Full-length Pop2p and Pop2p-T97A were purified as maltose-binding protein (MBP) fusions expressed in Escherichia coli. GST-Yak1p could phosphorylate MBP-Pop2p, but not MBP-Pop2p-T97A (Fig. 5A). In addition, kinase-negative Yak1p (GST-Yak1p-K398R), which has a mutation in its ATP binding site, failed to phosphorylate the MBP-Pop2p. These results strongly support the idea that Yak1p is the major Pop2p kinase under glucose-limiting conditions.
Figure 5.
Analysis of Yak1p using GST-Yak1p fusion protein. (A) GST-Yak1p phosphorylates Thr 97 of full-length Pop2p. Phosphorylation assays were performed as described in the Materials and Methods. (WT) Wild-type Yak1p; (KD, kinase dead) K398R mutant of Yak1p; (Thr) wild-type Pop2p; (Ala) alanine substitution mutant for the 97th threonine of Pop2p; (GST-Yak1p*) autophosphorylation product of GST-Yak1p; (MBP-Pop2p*) phosphorylated MBP-Pop2p; (MBP-Pop2p [CBB]) CBB staining of MBP-Pop2p. (B) In vitro Pop2 peptide kinase activity of GST-Yak1p was not regulated by glucose. The Pop2 peptide phosphorylation activities of GST fusion proteins purified from yeast after treatment with the glucose conditions indicated were determined. (C) Pop2p associates with Yak1p. GST fusion proteins were purified from yeast cells after 10 min incubation in YPD medium (Glc +) or YP (Glc −) and separated by SDS-PAGE, and the proteins were stained with CBB. The Pop2p copurified with the GST fusion proteins was detected using anti-Pop2p specific antibody (Pop2p [co-purified]). The endogenous Pop2p in each yeast cell was detected by Western blotting (Pop2p [endogenous]). (D) Yak1p associates with Bmh1/2p in a glucose-dependent manner. SDS-PAGE image of GST and GST-Yak1p purified in panel C. The amino acid sequences of peptides derived from Yak1p-associated proteins were determined and analyzed as described in the Materials and Methods.
Regulation of Yak1p by glucose
To determine if Yak1p kinase activity is regulated by glucose, GST-Yak1p was purified from cells cultured transiently in YPD or YP medium, and Pop2 peptide kinase activity was determined. As shown in Figure 5B, we failed to detect any difference in Yak1p activity on Pop2 peptide or MBP-Pop2p between the two glucose conditions. We also purified a protein extract by the same method reported for Snf1 kinase (Wilson et al. 1996), but GST-Yak1p activity was not affected by the glucose condition. We obtained the same result using C-terminal hexahistidine-tagged Yak1p (Yak1p-His6) expressed from the YAK1 promoter, showing that these results are not due to addition of GST to the N terminus of Yak1p. Based on these observations, we conclude that the kinase activity of Yak1p is not regulated by glucose. It has been reported recently that deletion of the N-terminal region of Yak1p activates the kinase activity and that phosphorylation of the tyrosine residue is required for the activity of Yak1p (Kassis et al. 2000). However, we failed to find any evidence indicating the existence of a cleaved product of Yak1p or a difference in the Tyr phosphorylation on Yak1p between the two glucose conditions.
We next analyzed proteins bound to Yak1p. Intrinsic Pop2p was copurified with GST-Yak1p (Fig. 5C), but the amount of Pop2p was indistinguishable under the two glucose conditions. Although the GST-Yak1p overexpression caused phosphorylation of endogenous Pop2p under both glucose-existing and nonexisting conditions, the copurified Pop2p with GST-Yak1p was dephosphorylated only under the glucose-existing condition (Fig. 5C). We believe that a Pop2p phosphatase could be activated under the glucose-existing condition and Pop2p was dephosphorylated during the purification (see Discussion and Fig. 8). We found that two proteins (∼30 kD) copurified with GST-Yak1p only in the glucose-treated conditions (Fig. 5D). Four peptide sequences derived from the larger protein (LAEQAERYEEMVE, SEHQVELI, IETELTK, and ATNSSLEAYK) indicated that it is Bmh2p (the corresponding positions were amino acids 14–27, 77–84, 91–97, and 146–155, respectively). Two peptide sequences derived from the smaller protein (VFYYK and ATNASLEAYK) showed that it is Bmh1p (the corresponding positions were amino acids 120–124 and 146–155, respectively). Bmh1p and Bmh2p are homologs of mammalian 14-3-3 protein (van Heusden et al. 1992, 1995). The 14-3-3 proteins are known to bind to the amino acid sequence motifs that contain a phospho-serine residue (Yaffe et al. 1997) and to function as nuclear export molecules (Kumagai and Dunphy 1999).
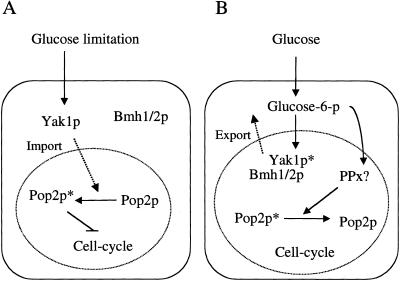
Figure 8.
Models of the Pop2p phosphorylation and Yak1p.
Yak1p localization is altered by glucose
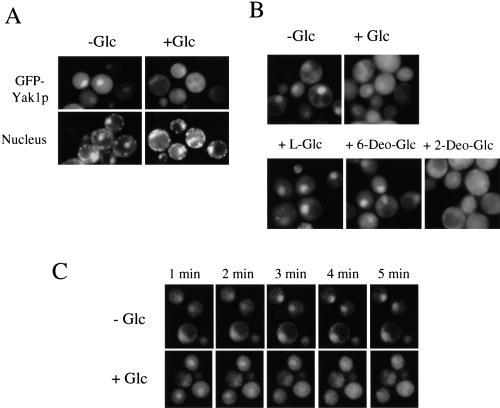
To observe the intracellular localization of Yak1p, we used green fluorescent protein (GFP)-Yak1p fusion protein expressed under the control of the YAK1 promoter on a single-copy plasmid. We confirmed that the GFP-YAK1 plasmid could complement the defect of the Pop2p phosphorylation of yak1Δ strain (data not shown). As shown in Figure 6A, in the presence of glucose, GFP-Yak1p did not show any specific localization. However, GFP-Yak1p accumulated in the nucleus when glucose was removed from the culture medium. When glucose was readded to glucose-starved cells, GFP-Yak1p spread reversibly throughout the entire cell. A deletion of the YAK1 C-terminal region (amino acids 189 to 807) from GFP-YAK1 resulted in the loss of the specific localization (data not shown), indicating that this localization pattern was dependent on Yak1p. Exclusion from the nucleus was also dependent on the hexose phosphorylation activity (Fig. 6B). This alteration in localization was a rapid phenomenon and was completed within 3 min (Fig. 6C).
Figure 6.
Intracellular localization of GFP-Yak1p. (A) Localization of GFP-Yak1p is regulated by glucose. The yak1Δ (H214) cells harboring the single-copy and GFP-Yak1p-expressing plasmid (pHIS063) grown in SCM-Ura were collected and resuspended in SCM-Ura (+Glc) or SCM-Ura minus glucose (−Glc). After 3 min, fluorescent images were taken. (B) Effect of the glucose analogs on the nuclear localization of GFP-Yak1p. The glucose analogs shown were added to pHIS063/H214 cells precultured in SCM-Ura minus glucose. (Glc) D-glucose; (L-Glc) L-glucose; (6-Deo-Glc) 6-deoxy-D-glucose; (2-Deo-Glc) 2-deoxy-D-glucose. (C) Time course of the translocation of GFP-Yak1p. Images were taken for the time periods indicated after the pHIS063/H214 cells were transferred to SCM-Ura from SCM-Ura minus glucose (+Glc), or SCM-Ura minus glucose from SCM-Ura (−Glc).
Physiological roles of the phosphorylation on Thr 97 of Pop2p
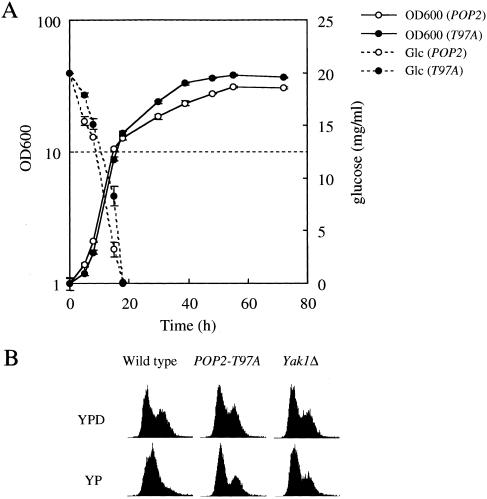
The POP2-T97A cells with pop2 null allele did not show any known phenotype of the pop2 null mutant cells, including temperature-sensitive growth, glucose-regulated expression of the SUC2 gene, caffeine sensitivity, accumulation of glycogen, STRE promoter activity, and survival in stationary phase (data not shown). Here, we found that the saturation density of POP2-T97A cells always reached 1.3 to 1.5 times higher than that of the wild-type cells. As shown in Figure 7A, after glucose in the culture medium was exhausted, POP2-T97A cells grew more rapidly than wild type, although the growth rate of them in log phase was indistinguishable. In contrast, the pop2 null mutant cells grow more slowly and show reduced saturation density in stationary phase (Sakai et al. 1992; Hata et al. 1998). The POP2-T97A is dominant because POP2-T97A with the POP2 wild-type allele also showed the same phenotype. We also found that the POP2-T97D and POP2-T97E mutations had the same overgrowth phenotypes as the POP2-T97A cells (data not shown). Yeast cells grow using ethanol as a carbon source after the diauxic transition, but the growth rate of POP2-T97A and wild type in YPethanol medium was unchanged. Thus, the overgrowth phenotype seems to require a carbon source transition. In the diauxic transition, the growth rate of yeast cells is reduced dramatically (cell cycle progression is retarded) compared with exponentially growing cells (Fig. 7A). We therefore tested the POP2-T97A cells in cell cycle regulation upon glucose limitation. As shown in Figure 7B, G1-phase cells accumulated in the wild-type cells after glucose removal, but the POP2-T97A cells failed to accumulate G1-phase cells 2 h after glucose deprivation. This indicates that the cell cycle is still progressing in the POP2-T97A cells 2 h after glucose deprivation, and this may be one of the reasons for the overgrowth after diauxic transition. As described previously, the yak1Δ cells did not show any growth defect (Garrett and Broach 1989). We failed to find any difference in the growth curve between the yak1Δ cells and the wild-type cells. Furthermore, the yak1 cells did not show the overgrowth phenotype (data not shown). However, we found that they showed a very similar defect in accumulating G1-phase cells upon glucose depletion as POP2-T97A cells. As described above, Pop2p was still phosphorylated in the yak1Δ cells at a very low level. This low-level phosphorylation could be enough to prevent the overgrowth phenotype but could not be enough for rapid cell cycle arrest upon glucose deprivation.
Figure 7.
Physiological roles of Pop2p phosphorylation by Yak1p. (A) Growth curves of the POP2-T97A cells (open circles) and wild-type cells (closed circles) and glucose concentration of each culture. Two independent experiments were performed, and the standard deviations of each duplicated experiment are indicated as error bars. (B) The phosphorylation of Pop2p by Yak1p is required for proper cell cycle arrest upon glucose deprivation. Exponentially growing wild-type (H213), POP2-T97A (A2079), and yak1Δ (H214) cells were transferred to YPD medium and YP medium. After 2 h culture time, cell cycle profiles of each sample were analyzed by FACS analysis.
Discussion
Glucose-regulated phosphorylation of Thr 97 of Pop2p and its role in growth control
Here, we report that Thr 97 of Pop2p is phosphorylated within 2 min after glucose removal and reversed within 1 min after glucose readdition (Fig. 2A,B). Dephosphorylation of Thr 97 of Pop2p is dependent on the first step of glucose metabolism (glucose phosphorylating activity) (Fig. 2C,D). We believe that the regulation of Thr 97 of Pop2p is one of the earliest events in glucose sensing in yeast. Although POP2-T97A cells did not show any phenotype observed in pop2 null mutant cells, they showed one interesting phenotype associated specifically with glucose limitation and growth: After the diauxic transition, the POP2-T97A cells grow more rapidly than the wild-type cells (Fig. 7A). In addition, the POP2-T97A cells failed to arrest the cell cycle after glucose deprivation (Fig. 7B).
Our current hypothesis is as follows: Pop2p regulates growth depending on the carbon source via phosphorylation of its Thr 97 residue. When glucose is present in the medium, Pop2p is dephosphorylated, and this form of Pop2p supports rapid growth (rapid cell cycle progression). When glucose is depleted from the medium, Thr 97 of Pop2p is phosphorylated, and this form of Pop2p is required for proper cell cycle arrest. POP2-T97A is a dominant-negative mutation that allows cells to receive the glucose signal continuously. This growth control does not seem to be performed through STRE, a major cis-element that functions in the induction of many genes during the diauxic transition (Boy-Marcotte et al. 1998). In the POP2-T97A cells, the expression of a reporter gene from STRE was not affected during diauxic transition (data not shown). Search for downstream targets of the growth control by the phosphorylation of Thr 97 of Pop2p will be required.
We also provided evidence that Yak1p is the major protein kinase that phosphorylates Pop2p upon glucose depletion (Fig. 4). Yak1p was identified as a protein kinase that functions as a negative growth regulator by antagonizing the PKA pathway (Garrett and Broach 1989). Our present results that Yak1p phosphorylates Pop2p, and that this phosphorylation is required for proper cell cycle arrest upon glucose limitation, supports the previous belief that Yak1p functions as a negative growth regulator. In addition, our findings add new insight into the function of YAK1 in glucose sensing through the phosphorylation of Thr 97 of Pop2p. We do not believe that Yak1p is the only protein kinase that phosphorylates Thr 97 of Pop2p in vivo, because weak phosphorylation is still observed in the yak1Δ cells and the yak1Δ cells did not show the overgrowth phenotype observed in the POP2-T97A cells (see Results). Because the POP2-T97A mutation could not rescue the growth defect of the PKA pathway, although the yak1Δ mutation could (data not shown), Pop2p does not seem to be the only target of Yak1p. We believe that another target for Yak1p to antagonize the PKA pathway exists in yeast cells.
Regulation of Yak1p by glucose
Transcription of the YAK1 gene and the kinase activity of Yak1p increase in stationary phase and correlate with cell cycle arrest (Garrett et al. 1991). Our results showed that Yak1p phosphorylated Pop2p in response to glucose removal, but the kinase activity of Yak1p was not changed by glucose (Fig. 5). Instead, we found another possible mode of regulation of Yak1p: Glucose regulates the association of Bmh1p and Bmh2p (14-3-3 proteins) and its translocation to the nucleus (Figs. 5,6). The 14-3-3 proteins bind to phospho-serine residues and function as nuclear export molecules (Yaffe et al. 1997; Kumagai and Dunphy 1999). Yak1p could be phosphorylated at its serine residues (not identified) and exported from the nucleus by the binding of Bmh1p and Bmh2p in the presence of glucose. As expected, GFP-Yak1p was shuttling rapidly from the cytoplasm to the nucleus and was affected by glucose. On the removal of glucose, GFP-Yak1p accumulated in the nucleus within minutes. Furthermore, with the readdition of glucose, GFP-Yak1p spread reversibly throughout the entire cell within minutes. This localization pattern correlates exactly with the association pattern of Bmh1/2p and supports their predicted function as nuclear export molecules. This change in localization is also dependent on glucose phosphorylating activity as in the case of the dephosphorylation of Pop2p (Fig. 6). Currently, the most likely upstream protein kinase is PKA, which is known to be active in the presence of glucose. Yak1p has five potential PKA phosphorylation sites and can be phosphorylated by PKA in vitro (Garrett et al. 1991). However, disruption of the GPR1 gene (see introduction) did not affect Yak1p localization, and phosphorylation of Yak1p by PKA in vitro did not enhance the binding of Bmh1p (data not shown).
Yak1p as a member of the DYRK family
Yak1p is a member of the DYRK family, which also includes DYRK1A/B/C, DYRK2, and DYRK3 (mammalian), minibrain (Drosophila), and YakA (Dictyostelium) (Becker and Joost 1999). These kinases share several sequence motifs in the catalytic domain that distinguish them from all other known protein kinases, and they are involved in the regulation of growth and development. DYRK1 has been suggested to be involved in Down's syndrome (Tejedor et al. 1995; Song et al. 1996), minibrain is required for proper neuroblast cell formation and maintenance, and YakA is essential for starvation-induced development and growth arrest (Souza et al. 1998). Despite their significant biological roles, however, the participation of DYRK family kinases in a particular signal transduction pathway and its native substrate have not been shown. The consensus amino acid sequences of targets for DYRK1A (Himpel et al. 2000) and Yak1p (Kassis et al. 2000) using artificial substrates (histoneH1 and myelin basic protein, respectively) have been determined and found potentially to be proline-directed kinases. The Thr 97 of Pop2p is also positioned before proline. To the best of our knowledge, our finding is the first report of a native substrate and particular pathway for a DYRK family kinase.
Conclusion: Yak1p-Pop2p system in the glucose-sensing system in yeast
Figure 8 shows our current model. When glucose is limited, Yak1p accumulates in the nucleus where it phosphorylates Pop2p, which is required for proper cell cycle arrest (Fig. 8A). In the presence of glucose, Yak1p is phosphorylated by an as yet unknown protein kinase at its serine residue(s) and associates with Bmh1p and Bmh2p, and is then exported from the nucleus to the cytoplasm. Glucose phosphorylation is required for this reaction (Fig. 8B). There clearly are unknown components in this model: a protein kinase that phosphorylates Yak1p and a protein phosphatase that dephosphorylates Pop2p in the presence of glucose. Further analysis is required to identify these components.
Materials and methods
Strains, media, and growth conditions
All S. cerevisiae strains used in this study were derived from S288C and are listed in Table 2. E. coli strain JM109 was used for DNA manipulations, and BL21(DE3) (Novagen) was used for the expression of the maltose binding protein (MBP) fusion of Pop2p. The transformation of yeast was performed by the LiOAc-method (Ito et al. 1983). The basic culture medium used for S. cerevisiae was YPD medium containing 1% Bacto-yeast extract, 2% Bacto-peptone, and 2% dextrose. For YP medium, the dextrose was removed from YPD medium. The synthetic medium was CSM medium containing 0.67% yeast nitrogen base without amino acids, 2% dextrose, and amino acids as required (Sherman et al. 1986). The media were solidified with 2% Bacto-agar for plates. Crossing, sporulation, and tetrad analysis were performed by standard genetic methods (Sherman et al. 1986). Unless otherwise specified, the permissive and restrictive temperatures were 24°C and 37°C, respectively.
Table 2.
Yeast strains used in this study
| Strain
|
Genotype
|
Source
|
|---|---|---|
| D24 | MATa/α HIS3/his3 trp1/trp1 ura3/ura3 LEU2/leu2 | A. Sakai |
| D51 | MATa/α his3/his3 trp1/trp1 ura3/ura3 LEU2/leu2 | A. Sakai |
| A1643 | MATa leu2 trp1 ura3 hxk1Δ::LEU2 hxk2Δ::URA3 | This study |
| A2260 | MATa his3 leu2 trp1 ura3 hxk1Δ::LEU2 hxk2Δ::URA3 glk1Δ::HIS3 | This study |
| A2085 | MATa his3 trp1 ura3 leu2 pep4Δ::TRP1 | A. Sakai |
| A2279 | MATa his3 leu2 trp1 ura3 | This study |
| A2280 | MATα his3 leu2 trp1 ura3 pop2-Δ3::LEU2 | This study |
| A2281 | MATa his3 leu2 trp1 ura3 URA3::POP2-T97A | This study |
| A2282 | MATα his3 leu2 trp1 ura3 pop2-Δ3::LEU2 URA3::POP2-T97A | This study |
| H213 | MATα his3 trp1 ura3 leu2 | This study |
| H214 | MATa his3 trp1 ura3 leu2 yak1Δ::TRP1 | This study |
Constructions of yeast mutant strains
To construct the yak1 deletion mutant, an EcoRV fragment of the YAK1 gene (bases +900 to +1760 from the start codon) of strain D51 was replaced with the TRP1 gene by one-step gene replacement (Rothstein 1983) using pYAK1Δ2 (a kind gift of Akira Matsuura, National Institute for Longevity Sciences, Aich, Japan). To construct the hxk1 deletion mutant, an EcoRV fragment of HXK1 gene (bases +736 to +1309 from the start codon) of strain D24 was replaced with the LEU2 gene. To construct the hxk2 deletion mutant, an EcoRV fragment of the HXK2 gene (bases +736 to +856 from the start codon) of strain D24 was replaced with the URA3 gene. To construct the glk1 deletion mutant, a SalI fragment of the GLK1 gene (bases +736 to +1309 from the start codon) of strain D51 was replaced with the HIS3 gene. The diploid transformant cells were sporulated, and haploid cells were recovered by a standard micromanipulation technique. Each disruption was confirmed by PCR and Southern hybridization. Haploid cells carrying hxk1Δ::LEU2 hxk2Δ::URA3 or hxk1Δ::LEU2 hxk2::URA3 glk1Δ::HIS3 were constructed by a standard genetic cross. The POP2 alanine replacement mutants were constructed as follows: Plasmids harboring each replacement mutant were constructed using the “splice by overlap extension” method (Horton et al. 1990) against pYUK220, which has the full-length POP2 on pRS316 (Shimizu-Yoshida et al. 1999). Each mutant plasmid was introduced into the pop2-Δ3 mutant (A2280) cells. Cells carrying POP2-T97A were constructed as follows: The POP-T97A DNA (A to G transition on base +289 from the start codon of the POP2 gene) constructed as above was subcloned into pRS306 (Sikorski and Hieter 1989) and introduced at the URA3 locus of pop2-Δ3 cells.
Plasmids used in this study
Standard DNA manipulations were performed as described previously (Sambrook et al. 1989). PCR was performed using KOD dash (Toyobo) DNA Polymerase according to the supplier's protocol. The YAK1 plasmid pHIS012 (YCpYAK1) was made by inserting a 3.5-kb YAK1 fragment (bases −663 to +2773 from the start codon, amplified by PCR from yeast genomic DNA) into pRS316 (Sikorski and Hieter 1989). The plasmid used for the expression of GST-Yak1p fusion protein pHIS020 was constructed by inserting a 2.4-kb YAK1 fragment (from the start codon to the stop codon, amplified by PCR from pHIS012) into pEG(KT) (Mitchell et al. 1993). pMAL-POP2 was made by inserting a 1.3-kb POP2 fragment (from the start codon to the stop codon) into an MBP fusion vector pPEPKmalE. pMAL-T97A was constructed by inserting a 1.3-kb POP2-T97A fragment (from the start codon to the stop codon) into pPEPKmalE. The plasmid used for the observation of GFP-Yak1p fusion protein pHIS063 was prepared by inserting a 0.7-kb GFP fragment (amplified by PCR from pQBI63 [Wako]) immediately after the start codon of YAK1 on pHIS012.
Detection of Pop2p by Western blot analysis
Yeast cells (OD600 = 1.0) of each strain in various conditions were collected by centrifugation. The cell extracts were prepared by boiling for 5 min in 20 μL of 2× SDS-PAGE loading buffer (125 mM Tris-HCl at pH 6.8, 4% SDS, 0.01% bromophenol blue, 14% glycerol, 10% [v/v] 2-mercaptoethanol). Ten microliters of each extract was separated on an 8% SDS–PAGE and then blotted onto a PVDF membrane (Fluorotrans W membrane, Pall) using a semidry electroblotting system (Model BE-300, Biocraft). The membrane was incubated with the affinity-purified antibody against Pop2p (Shimizu-Yoshida et al. 1999) at a 1:1000 dilution, followed by anti-mouse/-rabbit antibody conjugated to horseradish peroxidase (Histofine simple stain PO [multi], Nichirei). The antibody on the membrane was then detected with an ECL system (Supersignal west pico chemiluminescent substrate, Pierce).
Kinase assays
Peptide kinase assays were performed as follows: 80 mM K-phosphate (pH 7.5), 20 mM MgCl2, 100 μM ATP, 1 mM peptide (Pop2 peptide: RRFNIGTPVSVASLPPGL or T97A peptide: RRFNIGAPVSVASLPPGL), 1 μCi [γ-32P]ATP, and 5 μL protein sample were mixed in a 25 μL solution. This reaction mixture was incubated for 20 min at 30°C. The mixture was spotted on P81 phosphocellulose filters (Whatman), and the filters washed three times with 1% phosphate and then dried. The radioactivity of each filter was measured by scintillation counting. The MBP-Pop2p kinase assays and autophosphorylation assays were performed as follows: 80 mM K-phosphate (pH 7.5), 20 mM MgCl2, 100 μM ATP, 10 μCi [γ-32P] ATP, 1.2 M (NH4)2SO4, and 5 μL of kinase sample (purified Pop2p kinase or GST-Yak1p beads) and 5 μL of substrate (MBP-Pop2p or MPB-Pop2p-T97A) were mixed in a 25 μL solution. This reaction mix was incubated for 30 min at 30°C. The reaction products were separated by SDS-PAGE. After Coomassie Brilliant Blue (CBB) staining, the phosphorylated proteins were visualized using an imaging analyzer (FLA2000). Autophosphorylation assays were performed in the same way without substrate.
Purification of Pop2 peptide kinase
All purification procedures were performed at 4°C. All chromatographic steps were performed using an Econo system (Bio-Rad), and each chromatographic matrix was packed in a suitably sized Econo Column (Bio-Rad) unless otherwise stated. All chromatographic columns except for the Methyl-HIC columns were equilibrated with KPG (15 mM K-phosphate at pH 7.5, 10% glycerol) before loading a sample. The conductivity of each elution fraction was determined using a B-173 (Horiba) conductivity meter. The protein concentration of each elution fraction was determined using a protein assay kit (Bio-Rad). Yeast cells (D24) were cultured for 4 d at 30°C in 3 L of YPD medium containing 4% glucose, harvested, and resuspended in 150 mL of KPG with one tablet of protease inhibitor mix (Complete, Roche) per 50 mL of solution. The cells were disrupted using a Bead-Beater with 0.5-mm diameter glass beads (Biospec Products). The lysate was clarified by centrifugation at 40,000g for 60 min, passed through glass wool, and then loaded onto a 40-mL (bed volume) DEAE-cellulose (DE-52, Whatman) column. The flow-through was then loaded onto a 30-mL (bed volume) phosphocellulose (P11, Whatman) column. Following extensive washing with KPG, the proteins were eluted by a single step with 60 mL of KPG containing 0.4 M NaCl. Solid (NH4)2SO4 was added to the P11 elution to a final concentration of 2.0 M and then loaded onto a 10-mL Methyl-HIC (Macro-Prep Methyl HIC, Bio-Rad) column equilibrated with 2.4 M (NH4)2SO4-KPG. Following extensive washing with 2.4 M (NH4)2SO4-KPG, a 40-mL linear gradient of 2.4 M to 0 M (NH4)2SO4 in KPG was used for elution. Fractions (2 mL) were collected and analyzed for Pop2 peptide kinase activity as described above. Fractions containing the kinase activity were pooled (about 12 mL), diluted with 2 volumes of KPG, and loaded onto a 3-mL (bed volume) hydroxyapatite (Bio-Gel HTP, Bio-Rad) column. Following extensive washing with KPG, a 30-mL linear gradient of 15 mM to 400 mM K-phosphate (pH 7.5) was used for elution. Fractions (1 mL) were collected and analyzed for activity. Fractions containing the kinase activity were pooled (about 4.5 mL), diluted with 3 volumes of KPG, and loaded onto a 0.5-mL (bed volume) Bio-Gel HTP column. After extensive washing with KPG, a 12.5-mL linear gradient of 0 to 1 M KCl in KPG was used for elution. Fractions (0.5 mL) were collected and the kinase activity determined. Fractions containing the kinase activity were pooled (about 4.5 mL), diluted with 2 volumes of KPG, and loaded onto a ceramic hydroxyapatite column (Bio-Scale CHT2-I, Bio-Rad). Following extensive washing with KPG, a 12-mL linear gradient of 0 mM to 400 mM K-phosphate (pH 7.5) was used for elution. Fractions (0.4 mL) were collected and the kinase activity determined. The fractions with kinase activity were also separated by SDS-PAGE followed by silver-staining and autophosphorylation assay.
Determination of partial amino acid sequence of Pop2 peptide kinase and Yak1p-associated proteins
Proteins separated by SDS-PAGE were blotted onto a polyvinylidene difluoride (PVDF) membrane (Problott membranes, Applied Biosystems) and stained with CBB. Protein spots corresponding to a target protein were excised and digested overnight with 0.2 μg of Lysyl-endopeptidase AP-1 (Wako) in 100 μL of 20 mM Tris-HCl (pH 8.5) with 8% acetonitrile at 37°C. The digested peptides were extracted with 8% acetonitrile and purified using an HPLC system (Model 172, Applied Biosystems) with a C8 column (RP-300, Applied Biosystems). The amino acid sequences of the recovered peptides were determined using a pulse liquid phase sequencer (Model 492 cLC, Applied Biosystems). Sequence homology was analyzed with a GENETYX computer software program (Software Development).
Affinity purification of GST-Yak1p
A2085 yeast cells harboring a plasmid (pHIS020) or a control (pEG[KT]) were cultured overnight in 10 mL of CSM medium minus uracil (CSM-Ura medium). The cells were transferred to 100 mL of YP-galactose (2%) medium and cultured overnight. After collecting the cells by centrifugation, they were resuspended in 10 mL of YPD medium or YP medium, incubated for 10 min at 24°C with shaking, and then harvested by centrifugation. The cells were resuspended in 0.5 mL of KYB (KPG containing 50 mM NaF, 5 mM Na-pyrophosphate, and one tablet of protease inhibitor mix [Complete, Roche] per 50 mL of buffer), and then broken by vortexing with glass beads (0.5-mm diameter, Biospec Products). The cell extract was clarified by centrifugation at 15,000 rpm for 10 min at 4°C, and 100 μL of glutathione-sepharose 4B beads (50% slurry, Pharmacia) was added to the extract. After incubation for 1 h at 4°C , the beads were washed three times with KYB and suspended in KYB. These GST-Yak1p beads were used for further analysis in a 50% slurry form.
Preparation of MBP-Pop2p
E. coli cells expressing MBP-Pop2p (from pMBP-POP2) or MBP-Pop2p-T97A (from pMBP-T97A) were harvested and disrupted by sonication in TSB (50 mM Tris-HCl at pH 7.5, 100 mM NaCl, 5 mM DTT). The clarified cell lysate was loaded onto an amylose resin column (New England Biolabs), and the MBP-fusion protein was eluted with TSB containing 10 mM maltose. The concentration of MBP-fusion protein was 0.5 mg/mL.
Microscopic observation
The log-phase yeast cells cultured in CSM-Ura medium were collected and washed with CSM-Ura minus glucose medium. For the glucose-minus condition, the cells were resuspended in CSM-Ura minus glucose medium. For the glucose-plus condition, glucose (or glucose analogs [Sigma]) was then added to a final concentration of 2%. DAPI was added to a final concentration of 2.5 μg/mL. The cells were observed using a fluorescent microscope (AX-80, Olympus Optical Co., Ltd.). A U-MWIBA/GFP cube was used for the GFP fluorescent observation, and a WU cube unit was used for the DAPI fluorescent observation. Images were taken using a SenSys CCD camera system (Photometrics) and IPLab Spectrum software (Signal Analytics) and cropped and processed with Adobe Photoshop 5.0 software.
FACS analysis
Log-phase cells of wild-type and mutant strains were transferred to glucose-deprived medium for 2 h and fixed with 70% ethanol. DNA was stained with propidium iodide, and FACS analysis was performed as described (Moriya and Isono 1999).
Acknowledgments
We thank Drs. M. Carlson, M. Inoue, K. Irie, D. Levin, J. Nikawa, K. Matsumoto, and T. Noda for providing yeast strains, K. Satoh for the peptide synthesis, A. Matsuura and T. Kohno for providing the plasmids, Y. Kamada for discussion, and Mr. Stephen McKay for his critical reading of the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sakai@libra.ls.m-kagaku.co.jp; FAX +81-42-724-6316.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.884001.
References
- Bai Y, Salvadore C, Chiang YC, Collart MA, Liu HY, Denis CL. The CCR4 and CAF1 proteins of the CCR4-NOT complex are physically and functionally separated from NOT2, NOT4, and NOT5. Mol Cell Biol. 1999;19:6642–6651. doi: 10.1128/mcb.19.10.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker W, Joost HG. Structural and functional characteristics of Dyrk, a novel subfamily of protein kinases with dual specificity. Prog Nucleic Acid Res Mol Biol. 1999;62:1–17. doi: 10.1016/s0079-6603(08)60503-6. [DOI] [PubMed] [Google Scholar]
- Boy-Marcotte E, Perrot M, Bussereau F, Boucherie H, Jacquet M. Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae. J Bact. 1998;180:1044–1052. doi: 10.1128/jb.180.5.1044-1052.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M. Glucose repression in yeast. Curr Opin Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- De Vit MJ, Johnston M. The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr Biol. 1999;9:1231–1241. doi: 10.1016/s0960-9822(99)80503-x. [DOI] [PubMed] [Google Scholar]
- Draper MP, Salvadore C, Denis CL. Identification of a mouse protein whose homolog in Saccharomyces cerevisiae is a component of the CCR4 transcriptional regulatory complex. Mol Cell Biol. 1995;15:3487–3495. doi: 10.1128/mcb.15.7.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo JM, Clifton D, Fraenkel DG. Yeast hexokinase mutants. J Biol Chem. 1977;252:4443–4444. [PubMed] [Google Scholar]
- Garrett S, Broach J. Loss of Ras activity in Saccharomyces cerevisiae is suppressed by disruptions of a new kinase gene, YAKI, whose product may act downstream of the cAMP-dependent protein kinase. Genes & Dev. 1989;3:1336–1348. doi: 10.1101/gad.3.9.1336. [DOI] [PubMed] [Google Scholar]
- Garrett S, Menold MM, Broach JR. The Saccharomyces cerevisiae YAK1 gene encodes a protein kinase that is induced by arrest early in the cell cycle. Mol Cell Biol. 1991;11:4045–4052. doi: 10.1128/mcb.11.8.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorner W, Durchschlag E, Martinez-Pastor MT, Estruch F, Ammerer G, Hamilton B, Ruis H, Schuller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes & Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata H, Mitsui H, Liu H, Bai Y, Denis CL, Shimizu Y, Sakai A. Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics. 1998;148:571–579. doi: 10.1093/genetics/148.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero P, Galindez J, Ruiz N, Martinez-Campa C, Moreno F. Transcriptional regulation of the Saccharomyces cerevisiae HXK1, HXK2 and GLK1 genes. Yeast. 1995;11:137–144. doi: 10.1002/yea.320110205. [DOI] [PubMed] [Google Scholar]
- Himpel S, Tegge W, Frank R, Leder S, Joost HG, Becker W. Specificity determinants of substrate recognition by the protein kinase DYRK1A. J Biol Chem. 2000;275:2431–2438. doi: 10.1074/jbc.275.4.2431. [DOI] [PubMed] [Google Scholar]
- Horton RM, Cai ZS, Ho N, Pease LR. Gene splicing by overlap extension: Tailor-made genes using the polymerase chain reaction. BioTechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bact. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Carlson M. Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes & Dev. 1996;10:3105–3115. doi: 10.1101/gad.10.24.3105. [DOI] [PubMed] [Google Scholar]
- Johnston M. Feasting, fasting and fermenting. Glucose sensing in yeast and other cells. Trends Genet. 1999;15:29–33. doi: 10.1016/s0168-9525(98)01637-0. [DOI] [PubMed] [Google Scholar]
- Kassis S, Melhuish T, Annan RS, Chen SL, Lee JC, Livi GP, Creasy CL. Saccharomyces cerevisiae Yak1p protein kinase autophosphorylates on tyrosine residues and phosphorylates myelin basic protein on a C-terminal serine residue. Biochem J. 2000;348 Pt 2:263–272. [PMC free article] [PubMed] [Google Scholar]
- Kraakman L, Lemaire K, Ma P, Teunissen AW, Donaton MC, Van Dijck P, Winderickx J, de Winde JH, Thevelein JM. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol Microbiol. 1999;32:1002–1012. doi: 10.1046/j.1365-2958.1999.01413.x. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG. Binding of 14-3-3 proteins and nuclear export control the intracellular localization of the mitotic inducer Cdc25. Genes & Dev. 1999;13:1067–1072. doi: 10.1101/gad.13.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FN, Johnston M. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: Coupling glucose sensing to gene expression and the cell cycle. EMBO J. 1997;16:5629–5638. doi: 10.1093/emboj/16.18.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DA, Marshall TK, Deschenes RJ. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast. 1993;9:715–722. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- Moriya H, Isono K. Analysis of genetic interactions between DHH1, SSD1 and ELM1 indicates their involvement in cellular morphology determination in Saccharomyces cerevisiae. Yeast. 1999;15:481–496. doi: 10.1002/(SICI)1097-0061(199904)15:6<481::AID-YEA391>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Ozcan S, Leong T, Johnston M. Rgt1p of Saccharomyces cerevisiae, a key regulator of glucose-induced genes, is both an activator and a repressor of transcription. Mol Cell Biol. 1996;16:6419–6426. doi: 10.1128/mcb.16.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S, Dover J, Johnston M. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 1998;17:2566–2573. doi: 10.1093/emboj/17.9.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein RJ. One-step gene disruption in yeast. Meth Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sakai A, Chibazakura T, Shimizu Y, Hishinuma F. Molecular analysis of POP2 gene, a gene required for glucose-derepression of gene expression in Saccharomyces cerevisiae. Nucleic Acids Res. 1992;20:6227–6233. doi: 10.1093/nar/20.23.6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- Shimizu-Yoshida Y, Sasamoto M, Yoshida A, Yoshioka T, Matsumoto A, Sakai A. Mouse CAF1, a mouse homologue of the yeast POP2 gene, complements the yeast pop2 null mutation. Yeast. 1999;15:1357–1364. doi: 10.1002/(SICI)1097-0061(19990930)15:13<1357::AID-YEA465>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WJ, Sternberg LR, Kasten-Sportes C, Keuren ML, Chung SH, Slack AC, Miller DE, Glover TW, Chiang PW, Lou L, et al. Isolation of human and murine homologues of the Drosophila minibrain gene: Human homologue maps to 21q22.2 in the Down syndrome “critical region.”. Genomics. 1996;38:331–339. doi: 10.1006/geno.1996.0636. [DOI] [PubMed] [Google Scholar]
- Souza GM, Lu S, Kuspa A. YakA, a protein kinase required for the transition from growth to development in Dictyostelium. Development. 1998;125:2291–2302. doi: 10.1242/dev.125.12.2291. [DOI] [PubMed] [Google Scholar]
- Tejedor F, Zhu XR, Kaltenbach E, Ackermann A, Baumann A, Canal I, Heisenberg M, Fischbach KF, Pongs O. minibrain: A new protein kinase family involved in postembryonic neurogenesis in Drosophila. Neuron. 1995;14:287–301. doi: 10.1016/0896-6273(95)90286-4. [DOI] [PubMed] [Google Scholar]
- Thevelein JM, de Winde JH. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1999;33:904–918. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- van Heusden GP, Wenzel TJ, Lagendijk EL, de Steensma HY, van den Berg JA. Characterization of the yeast BMH1 gene encoding a putative protein homologous to mammalian protein kinase II activators and protein kinase C inhibitors. FEBS Lett. 1992;302:145–150. doi: 10.1016/0014-5793(92)80426-h. [DOI] [PubMed] [Google Scholar]
- van Heusden GP, Griffiths DJ, Ford JC, Chin AWTF, Schrader PA, Carr AM, Steensma HY. The 14-3-3 proteins encoded by the BMH1 and BMH2 genes are essential in the yeast Saccharomyces cerevisiae and can be replaced by a plant homologue. Eur J Biochem. 1995;229:45–53. [PubMed] [Google Scholar]
- Wilson WA, Hawley SA, Hardie DG. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr Biol. 1996;6:1426–1434. doi: 10.1016/s0960-9822(96)00747-6. [DOI] [PubMed] [Google Scholar]
- Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- Yun CW, Tamaki H, Nakayama R, Yamamoto K, Kumagai H. Gpr1p, a putative G-protein coupled receptor, regulates glucose-dependent cellular cAMP level in yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1998;252:29–33. doi: 10.1006/bbrc.1998.9600. [DOI] [PubMed] [Google Scholar]