Abstract
Mammals express four highly conserved TEAD/TEF transcription factors that bind the same DNA sequence, but serve different functions during development. TEAD-2/TEF-4 protein purified from mouse cells was associated predominantly with a novel TEAD-binding domain at the amino terminus of YAP65, a powerful transcriptional coactivator. YAP65 interacted specifically with the carboxyl terminus of all four TEAD proteins. Both this interaction and sequence-specific DNA binding by TEAD were required for transcriptional activation in mouse cells. Expression of YAP in lymphocytic cells that normally do not support TEAD-dependent transcription (e.g., MPC11) resulted in up to 300-fold induction of TEAD activity. Conversely, TEAD overexpression squelched YAP activity. Therefore, the carboxy-terminal acidic activation domain in YAP is the transcriptional activation domain for TEAD transcription factors. However, whereas TEAD was concentrated in the nucleus, excess YAP65 accumulated in the cytoplasm as a complex with the cytoplasmic localization protein, 14-3-3. Because TEAD-dependent transcription was limited by YAP65, and YAP65 also binds Src/Yes protein tyrosine kinases, we propose that YAP65 regulates TEAD-dependent transcription in response to mitogenic signals.
Keywords: TEAD, TEF, YAP, 14-3-3, transcription, Src
Transcription enhancer factor-1 (TEF-1) was first cloned in human cells as an activator of the simian virus 40 (SV40) enhancer (Davidson et al. 1988), and later recognized as a family of four ubiquitous and highly conserved transcription factors found in mammals, birds, fungi, flies, and yeast (Kaneko and DePamphilis 1998; Jacquemin et al. 1998). Because these proteins share in common the TEA DNA binding domain, we refer to them as TEA domain (TEAD) proteins to avoid ambiguity with acronyms for other proteins. At least one TEAD gene is expressed in most adult tissues, and all four are abundantly expressed in some tissues, such as lung (Yockey et al. 1996; Kaneko et al. 1997). However, each protein exhibits different expression patterns with respect to tissue and developmental stage specificity (Kaneko et al. 1997; Jacquemin et al. 1998), suggesting that each protein has a unique function. Moreover, disruption of the TEAD-1/TEF-1 gene prevents heart biogenesis during mouse development, arguing that TEAD gene functions are not redundant (Chen et al. 1994). Although TEAD-1/TEF-1 is required for gene expression in cardiac muscle (Chen et al. 1994; Gupta et al. 1997; Butler and Ordahl 1999; Ueyama et al. 2000), TEAD-4/TEF-3 appears to play a specific role in activating skeletal muscle genes (Jacquemin et al. 1996; Yockey et al. 1996). TEAD-3/TEF-5 is expressed primarily in the placenta (Jacquemin et al. 1998; Jiang et al. 1999) and in cardiac muscle (Azakie et al. 1996). TEAD-2/TEF-4 is the only TEAD gene expressed in mouse embryos immediately after fertilization and during the first seven days of development (Kaneko et al. 1997; Wang and Latham 2000). Thus, TEAD-2/TEF-4 is one of the first transcription factors expressed at the onset of zygotic gene expression where it presumably plays a role in activating transcription of other genes during preimplantation development.
All four TEAD proteins have virtually identical DNA binding domains that recognize, with essentially the same affinity (Kaneko and DePamphilis 1998), a canonical M-CAT motif (5′-TCATTCCT-3′) found in promoters specific for transcription in muscle (Larkin et al. 1996; Jiang et al. 2000). This motif is also found within the enhancers of either wild-type SV40 or host range mutants of polyomavirus (PyV) that can replicate in undifferentiated mouse embryonal carcinoma and embryonic stem cells (Xiao et al. 1991). These PyV mutant enhancers, as well as one or more tandem copies of the M-CAT motif, can stimulate promoter activity in early mouse embryos up to 600-fold (Martinez-Salas et al. 1989; Melin et al. 1993), identifying TEAD-dependent enhancers and promoters as important transcriptional control elements at the beginning of mammalian development.
TEAD proteins appear to require a transcriptional coactivator, because ectopic expression of TEAD-1 in cells that do not express TEAD proteins does not elicit TEAD-dependent transcription, and overexpression of TEAD-1 in cells that do express TEAD proteins results in repression of transcription, consistent with titrating out (squelching) of a coactivator activity (Xiao et al. 1991; Jiang and Eberhardt 1996). Several candidates for this coactivator have been reported. TEAD-1 can bind TATA-box binding protein (TBP) (Jiang and Eberhardt 1996), and TEAD-2 can bind a nuclear receptor coactivator protein (Belandia and Parker 2000), but the effects of these proteins on TEAD-dependent transcription is only 2- to 3-fold. Similarly, MAX, a nuclear phosphoprotein that forms a heterodimer with MYC, and poly(ADP-ribose) polymerase, a chromatin bound protein, can also bind TEAD-1 and stimulate expression of cardiac muscle specific genes 3- to 5-fold (Gupta et al. 1997; Butler and Ordahl 1999). However, neither protein has a transcriptional activation domain, and neither alone is sufficient to activate TEAD-dependent transcription or to relieve squelching. Perhaps the most promising candidate is TONDU, a mammalian homolog of the Drosophila Vestigial (Vg) gene that interacts specifically with all four TEAD proteins. Vg is a transcriptional coactivator of Scalloped (Sd), a Drosophila protein that contains the TEA DNA-binding domain and is required for wing formation. TONDU can substitute for Vg in Drosophila, suggesting that TONDU is a specific transcriptional coactivator of TEAD proteins in tissues where it is expressed (Vaudin et al. 1999), although this has not been demonstrated. Thus, although it is clear that the action of TEAD proteins can be modified by a variety of cellular proteins, none of the ones identified so far fill the role of a the putative, general TEAD transcriptional coactivator.
In an effort to identify mammalian proteins that modify TEAD-2 activity, we expressed a tagged-mouse TEAD-2 protein in mouse cells and then purified TEAD-2 protein complexes by affinity chromatography. The advantage of this strategy is that it reveals the relative abundance of each protein within a complex that has been assembled in vivo under native conditions, and identifies all proteins that associate with the tagged protein, regardless of whether they bind independently or in concert with other proteins. The results revealed that TEAD-dependent transcription by any one of the four TEAD proteins required a transcriptional coactivator identified as YAP65 (abbreviated YAP), a protein whose carboxyl terminus contains an acidic transcriptional activation domain similar to that found in the herpesvirus VP16 protein (Yagi et al. 1999). Furthermore, TEAD activity was limited by the intracellular level of YAP, accounting for previous reports of squelching by TEAD proteins. Surprisingly, although TEAD was concentrated in the nucleus, YAP was concentrated in the cytoplasm where most of it was associated with a 14-3-3, a protein involved in transporting its binding partner from the nucleus to the cytoplasm (Fu et al. 2000). Because YAP also binds membrane-associated PDZ domain proteins and Src/Yes protein tyrosine kinases (Sudol 1994) that are components of signal transduction pathways, we propose that the activity of TEAD transcription factors is regulated by release of its cytoplasmic coactivator protein in response to mitogenic signals.
Results
TEAD-2 protein complexes assembled in vivo
TEAD protein complexes were purified from mouse 3T3 cells by expressing an N-FLAG-hemaglutinin–[mouse TEAD-2] fusion protein (FH–TEAD-2) in vivo, and then purifying the FH–TEAD-2 protein by double immuno-affinity chromatography from cell extracts prepared in 100 mM KCl. First, anti-FLAG monoclonal antibody attached to agarose beads was used to isolate FLAG-tagged proteins by eluting the beads with FLAG peptide under nondenaturing conditions. Next the eluted proteins were further purified by immunoprecipitation using an anti-hemaglutinin peptide (anti-HA) monoclonal antibody attached to sepharose beads, and then eluted from the beads with the HA peptide under nondenaturing conditions. The proteins present at each step were fractionated by SDS-PAGE, and visualized by silver staining. The FH–TEAD-2 protein band was identified by its molecular weight and by immuno-blotting with anti-FLAG and anti-mTEAD-2 antibodies.
The results revealed that FH–TEAD-2 was associated tightly with 12 other polypeptides designated T1 to T12. These polypeptides were associated with FH–TEAD-2 protein recovered from cells expressing FH-TEAD-2 (Fig. 1A, lane 6), but not from control cells (Fig. 1A, lane 3), and they were associated with FH–TEAD-2 throughout both affinity purification steps (compare lanes 4, 5 and 6 in Fig. 1A). The anti-HA step efficiently eliminated most proteins that were not specifically retained by the anti-FLAG resin. Only those proteins found exclusively in the HA peptide eluate were considered specifically associated with FH–TEAD-2. For example, actin (42 kD) was found mostly in the flow-through fraction (Fig. 1A, lane 4), and it appeared in the HA peptide eluate from control cells (Fig. 1A, lane 3), suggesting that actin associated nonspecifically with the resins. The same complexes were isolated in 300 mM and 500 mM KCl extraction buffer, revealing that this collection of proteins were stably bound to FH-TEAD-2.
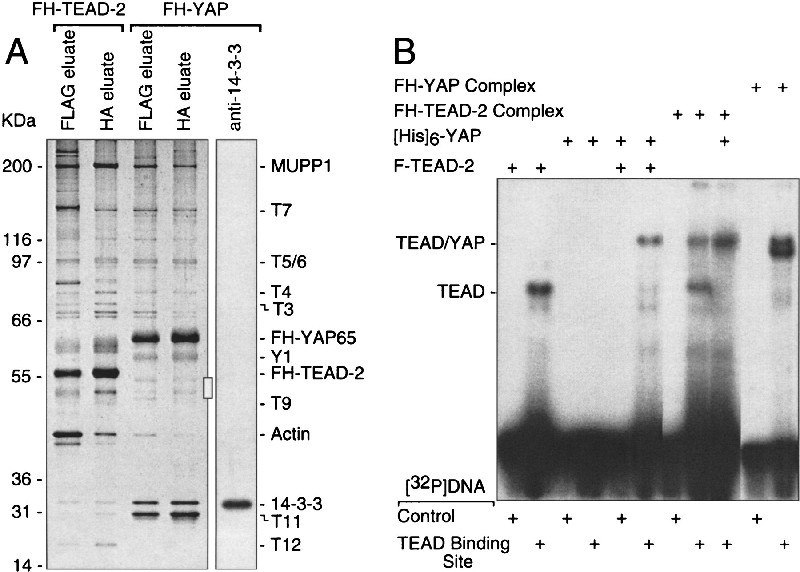
Figure 1.
Purification of TEAD-2 protein complexes from mouse cells. (A) A TEAD-2 protein complex was purified either from 3T3 cells (lanes 1–3) or from 3T3 cells expressing FH-TEAD-2 protein (lanes 4–6). FLAG tagged proteins in the extracts were bound to anti-FLAG resin, washed with buffer, and then eluted with FLAG peptide (lanes 2, 5). The FLAG eluate was then bound to an anti-HA resin, unbound proteins removed with buffer (lanes 1, 4), and the bound proteins eluted with HA peptide (lanes 3, 6). Aliquots of each fraction were subjected to SDS-PAGE, and the proteins were stained with silver. A set of standard proteins (Invitrogen) were fractionated in parallel on each gel to determine molecular weights (kD). Twelve proteins (T1-T12) were associated specifically with FH-TEAD-2 (lane 6), because they were absent under the same conditions when nonexpressing cells were used (lane 3). Three proteins were identified by mass spectroscopy as MUPP1 (T8), YAP65 (abbreviated YAP) (T1/2), and actin. TEAD-2 was identified by its reactivity with anti-FLAG and anti-TEAD-2 peptide antibodies, and by its molecular weight. (B) The HA eluate (see A, lane 6) was fractionated by sedimentation in a glycerol gradient, and individual fractions were subjected to SDS-PAGE and then stained with silver. (C) Aliquots of each fraction in B were fractionated by SDS-PAGE and subjected to immunoblotting with anti-FLAG antibody to quantify the amount of FH-TEAD-2 in each fraction.
To determine whether or not these proteins existed as more than one complex, the purified FH–TEAD-2 complex preparation was fractionated by sedimentation through a glycerol gradient (Fig. 1B). These results revealed the presence of three forms of FH–TEAD-2 protein. About 20% of the FH–TEAD-2 identified by anti-mTEAD-2 antibody (Fig. 1C) sedimented as monomeric TEAD protein (Fig. 1C, lanes 1,2) that was not apparently associated with other proteins. About 5% sedimented as a multiprotein complex containing all of the proteins in the initial FH–TEAD-2 complex preparation in approximately equimolar amounts (Fig. 1C, lanes 6–8). About 75% of the FH–TEAD-2 sedimented as a complex with proteins T1 and T2 (Fig. 1C, lanes 3,4), which were shown later to be product of a single gene. Thus, T1/T2 appeared to be the major partner of mTEAD-2 in vivo.
TEAD-2 associates in vivo with the transcriptional coactivator YAP
The identity of four TEAD-2-associated proteins was obtained by using mass spectrometry to determine the molecular weights of parent peptide and their fragments and then comparing them with those in the NCBI nonredundancy protein and EST sequence databases. T8 was identified as the multiple-PDZ-domain protein MUPP1, and the 42 kD band was identified as actin on the basis of one MS/MS spectrum and the MALDI/TOF spectrum for each protein. One MS/MS spectrum from T1 and one from T2 corresponded to the YES kinase-associated protein, YAP, a protein recently reported to act as a transcriptional coactivator (Yagi et al. 1999). The MALDI/TOF spectrums obtained for T1 and T2 were virtually indistinguishable, and more than 10 tryptic peptides from each band could be assigned to the same protein within a mass accuracy of 20 ppm. This combination of peptide chromatography and mass spectroscopic analysis ensured a high degree of confidence in the identification (Fenyo et al. 1998). Thus, T2 appears to be a posttranslationally modified form of T1, most likely phosphorylated (Sudol 1994). Clear matches for the other nine TEAD-2 associated proteins were not found, presumably because of the absence of data in the available mouse sequence data bases.
The carboxy-terminal half of TEAD-2 is required to bind YAP
To determine which domain of TEAD-2 is required to bind YAP, the ability of various TEAD-2 deletion mutants to bind native YAP protein was determined in vitro. The glutathione-S-transferase (GST) gene was fused to the amino terminus of full length YAP, expressed in Escherichia coli and purified by immobilization on glutathione-sepharose. [35S]TEAD-2 proteins were synthesized in the presence of [35S]methionine using a coupled in vitro transcription/translation system. Similar amounts of each [35S]TEAD-2 protein (Fig. 2B, input) were analyzed for their ability to bind full-length GST–YAP protein in vitro, and fraction bound (Fig. 2B, bound) normalized against the binding of the full-length [35S]TEAD protein (Fig. 2A, % YAP binding). Deletions at virtually any site between amino acids 224 and 445 resulted in essentially complete loss of YAP binding. These deletions ranged from 16 to 139 amino acids. In contrast, deletion of 113 amino acids from the amino terminus (protein A) had no effect on binding, and deletion of amino acids 115 to 223 reduced binding by 43% (protein B). These data revealed that the carboxy-terminal 75% portion of TEAD protein (aa 115 to 445) was required to efficiently bind YAP protein.
Figure 2.
Identification of the YAP protein binding domain in TEAD-2 protein. (A) Each TEAD-2 protein used in B and C is shown, together with the map number of their terminal amino acid and the efficiency with which they bound full-length YAP protein. Each data point is the mean of 3–5 independent determinations (sem was 6% to 11% of the mean value). The DNA binding domain (aa 40–112) (Kaneko and DePamphilis 1998), the total YAP binding domain (aa 115–445), and the essential YAP binding domain (aa 224–445) are indicated by shaded blocks. The transcriptional activation domain mapped in human TEAD-1 (Hwang et al. 1993) is indicated by a solid bar. (B) To identify the YAP binding domain in TEAD-2 protein, full-length GST–YAP protein was attached to beads and incubated either with full-length [35S]TEAD-2 protein (TEAD–2) or with the indicated TEAD-2 deletion mutant (A to O). The amount of full-length [35S]TEAD–2 bound to full-length GST–YAP was taken as “100% YAP binding.” YAP binding was corrected for variation in the amount of [35S]TEAD-2 added to each assay ([35S]TEAD-2 input). Binding of [35S]TEAD–2 to GST alone was not detected (GST). (C) To identify fragments of TEAD-2 protein that can bind YAP, full-length GST–TEAD-2 protein or protein fragment (GST-P to S) were attached to beads and incubated with full-length [35S]YAP protein.
To determine whether or not this conclusion was an artifact of the experimental protocol used, the protocol was reversed: Amino-terminal GST-tagged TEAD-2 fragments were tested for their ability to bind full-length [35S]YAP (Fig. 2C). The results confirmed the deletion analysis. The carboxy-terminal half of TEAD-2 bound YAP, but the amino-terminal half did not (Fig. 2A,C, proteins R and S). In contrast, smaller fragments from the carboxy-terminal half had no affinity for YAP (proteins Q and P). The remarkable sensitivity of YAP:TEAD-2 binding for an intact TEAD-2 structure argues strongly for a highly specific TEAD:YAP interaction.
YAP binds all four mouse TEAD transcription factors
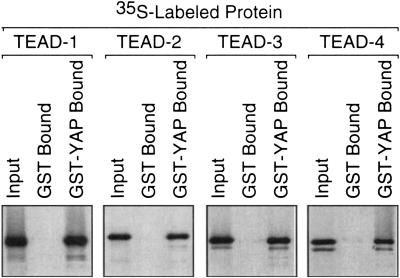
The YAP binding domain in mouse TEAD-2 is 79%–91% similar in sequence to the corresponding regions of the other three TEAD proteins, suggesting that YAP should bind specifically to all four TEAD proteins. To test this hypothesis, the same protein binding assay was repeated using 35S-labeled mouse TEAD proteins and full-length mouse GST–YAP. TEAD-1, -2, -3, and -4 each bound YAP with essentially the same efficiency (Fig. 3).
Figure 3.
YAP binds to all four TEAD proteins. To determine whether or not full-length TEAD-1,-2, -3, and -4 proteins can bind YAP with similar efficiency, full-length GST–YAP or GST alone was attached to beads as in Figure 2 and incubated with the indicated full-length [35S]TEAD protein.
YAP contains a novel TEAD-specific protein binding domain
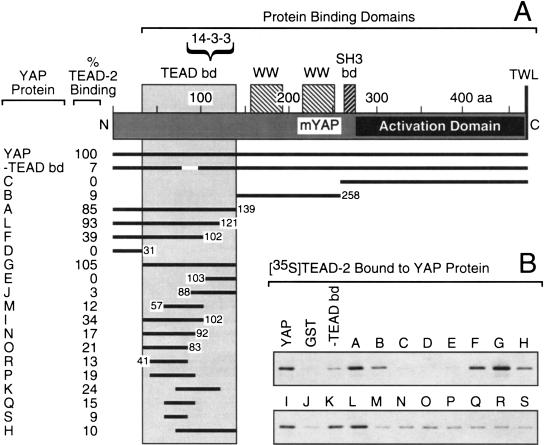
To identify the protein domain in YAP that is responsible for binding TEAD-2, the ability of various YAP protein fragments to bind native TEAD-2 protein was determined in vitro as described above. Various GST–YAP protein fragments were analyzed for their ability to bind full-length [35S]TEAD-2 protein in vitro (Fig. 4B). The results revealed that amino acids 32–139 near the amino terminus of YAP were required for binding TEAD-2 (Fig. 4A). YAP fragments B, C, and D lacking this region failed to bind TEAD-2, whereas YAP fragments A, L, and G containing all or most of this region did bind TEAD-2. Several fragments containing only a portion of this region bound poorly, but reproducibly, whereas fragments F and I had higher binding activity. A 20 amino acid deletion in the center of this region (−TEAD bd) eliminated binding activity. These data revealed that TEAD-2 binds specifically to a novel YAP protein domain at the opposite end from the YAP transcription activation domain, and that this interaction likely involves spatial relationships between specific amino acid residues.
Figure 4.
Identification of the TEAD protein binding domain in YAP. (A) Each YAP protein used in B is shown, together with the map number of their terminal amino acid and the efficiency with which they bound full-length TEAD–2 protein. The TEAD binding domain (aa 32–139), the 14-3-3 binding domain (Kanai et al. 2000), protein binding domains WW, SH3, and TWL (shaded bars), and the YAP transcriptional activation domain (solid bar) are indicated (Yagi et al. 1999). (B) The indicated full-length GST–YAP protein (YAP), YAP(–TEAD bd), a deletion of amino acids 77 to 96, and YAP protein fragments (A to S) were attached to beads and then incubated with full-length [35S]TEAD-2 protein to determine their binding efficiency (“% TEAD-2 binding”), as described in Figure 2.
TEAD-dependent transcription in vivo requires YAP
To determine whether or not YAP is a transcriptional coactivator of TEAD in vivo, four different mouse cell lineages were cotransfected with a mixture of plasmids that expressed the firefly luciferase gene driven by a TEAD-dependent promoter (pGT4Tluc), the indicated HA-tagged mouse TEAD gene [pCI(H-TEAD)], the mouse YAP gene [pSI(YAP)], and the E. coli β–galactosidase gene [pRI(βgal)]. The level of luciferase enzyme activity was determined by the amounts of TEAD and its putative coactivator, YAP, that were provided either by the cell or by the corresponding expression vector. pRI(βgal) was included to correct for variation in transfection efficiency. The promoters for each gene were selected to minimize competition for the same set of transcription factors.
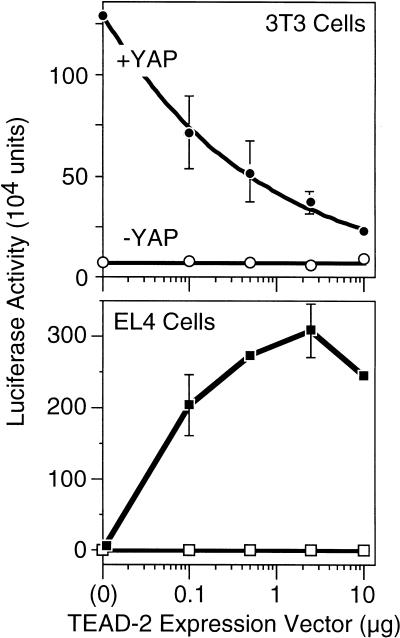
Blotting-hybridization with appropriate [32P]DNA probes confirmed that 3T3 embryonic fibroblasts contain mRNA from YAP and all four TEAD genes (data not shown). When 3T3 cells were transfected with pGT4Tluc in the absence of pCI(H–TEAD-2), the presence of pSI(YAP) stimulated luciferase production ⋝100-fold, demonstrating that YAP can stimulate the activity of endogenous TEAD transcription factor activity in 3T3 cells. Moreover, addition of increasing amounts of pCI(H–TEAD-2) resulted in decreasing levels of TEAD-dependant transcription in the presence of pSI(YAP) (Fig. 5), presumably because excess TEAD-2 protein competed with TEAD:YAP complexes for binding to the TEAD-dependent promoter. Therefore, the concentration of YAP in 3T3 cells appeared to be the rate-limiting factor in determining the amount of TEAD transcription factor activity.
Figure 5.
TEAD-dependent transcription requires YAP. Mouse 3T3 embryonic fibroblasts (which express YAP and TEAD-1, -2, -3, and -4 genes) or EL-4 T-lymphocytes (which express neither YAP nor TEAD proteins) were transfected with a mixture of three plasmids: pGT4Tluc, which expresses firefly luciferase only in the presence of a TEAD transcription factor; an increasing amount of pCI[H–TEAD–2], which expresses mouse H–TEAD–2 to activate transcription of the luciferase gene in pGT4Tluc; and pRI(βgal), which expresses Escherichia coli β–galactosidase to measure transfection efficiency. The amount of luciferase activity (measured as light units) was normalized to the amount of β–galactosidase activity in each assay. Where indicated (solid symbols), a fixed amount of pSI(FH-mYAP) was included to produce mouse YAP protein to determine whether or not YAP was required for TEAD-dependent transcription. The mean values (±sem) from three independent experiments are given.
This hypothesis was confirmed by carrying out the same experiment in EL4 T-lymphocytes that lacked YAP and TEAD mRNAs (data not shown). When EL4 cells were transfected with pGT4Tluc in the absence of pCI(H–TEAD-2), the presence of pSI(YAP) did not stimulate luciferase production. Therefore, YAP alone could not stimulate transcription from a TEAD-dependent promoter. However, addition of increasing amounts of pCI(H–TEAD-2) increased TEAD-dependant transcription up to 300-fold, but only in the presence of pSI(YAP) (Fig. 5). Therefore, YAP can function as a transcriptional coactivator for TEAD, even in cells that do not express either protein. Eventually, further addition of pCI(H–TEAD-2) began to reduce TEAD-dependant transcription, as the amount of intracellular TEAD presumably exceeded the amount of intracellular YAP.
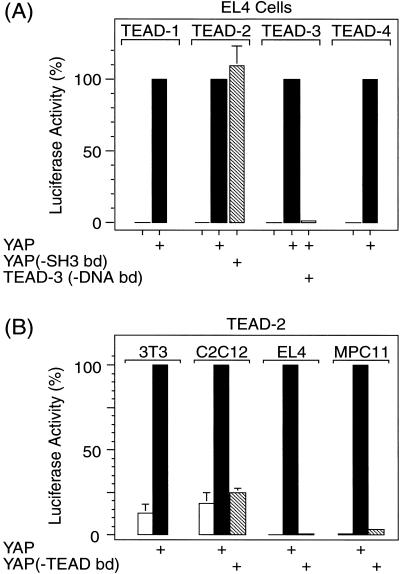
To determine whether or not YAP was a transcriptional coactivator for all four TEAD proteins, EL4 cells were transfected with a plasmid expressing one of these genes, in the presence or absence of pSI(YAP). In each case, YAP was required for TEAD-dependent expression of the luciferase gene, and the extent of transcription among the four TEAD proteins was similar (Fig. 6A). As expected, sequence specific DNA binding of TEAD protein also was required for TEAD-dependent transcription, because a mutant TEAD-3 protein with a single amino acid change in its DNA-binding domain that eliminated its ability to recognize its DNA binding site in vitro (Kaneko and DePamphilis 1998) prevented luciferase expression (Fig. 6A). In addition, two YAP deletion mutants were tested for their ability to stimulate luciferase production in EL4 cells. Deletion of amino acids 77 to 96 from YAP, which eliminated binding to TEAD-2 (Fig. 4), also failed to stimulate transcription by TEAD-2 (Fig. 6B). In contrast, mutation of amino acids 267 to 269, which disrupted the SH3 binding domain in YAP, diminished neither binding to TEAD-2 protein nor stimulation of transcription by TEAD-2 (Fig. 6A).
Figure 6.
TEAD-dependent transcription by all four TEAD transcription factors requires YAP with a functional TEAD binding domain and TEAD with a functional DNA binding domain. (A) Conditions for comparing TEAD-1, -2, -3, and -4 proteins in EL4 cells were the same as in Figure 5, except that each assay contained 0.1 μg pCI[H-TEAD1, -2, -3, or -4], and 10 μg pSI[YAP] or pSI when pSI[YAP] was omitted. YAP(-SH3 bd) is a mutation in the SH3 binding domain that has three proline residues at positions 267–269 mutated to glycines destroying the Src/Yes kinase SH3 domain consensus binding sequence (Sparks et al. 1996). TEAD(-DNA bd) contains a G to D amino acid substitution at position 74 in the DNA binding domain (Kaneko and DePamphilis 1998). To facilitate comparison, luciferase activity observed for each TEAD protein in the presence of YAP was taken as 100%. Luciferase activity (104 units) was 29 (TEAD-1), 29 (TEAD-2), 30 (TEAD-3) and 12 (TEAD-4). (B) Conditions for evaluating YAP activity in 3T3 embryonic fibroblasts, C2C12 muscle fibroblasts, EL4 T-lymphocytes, and MPC-11 B-lymphocytes were the same as in Figure 5, except that 0.1 μg pCI[H-TEAD-2] was used to produce H-TEAD-2, and 10 μg of either pSI[YAP] or pSI[YAP(-TEAD bd) was used to produce either YAP or a deletion mutant of YAP that failed to bind TEAD protein (Fig. 4). The mean values (±sem) from four independent experiments are given. Luciferase activity observed in the presence of YAP was taken as 100% for each cell line.
Previous studies have shown that TEAD-1 activates muscle-specific genes in C2C12 muscle fibroblasts (Yockey et al. 1996). These cells behaved like 3T3 cells in that both cell lines exhibited endogenous TEAD activity and both cell lines required a YAP protein with a functional TEAD binding domain to stimulate TEAD-dependent transcription (Fig. 6B). Thus, YAP is utilized for TEAD-dependent transcription in muscle as well as embryonic fibroblasts.
Previous studies also have shown that MPC-11 cells, which do not express any of the TEAD genes (data not shown), do not support TEAD-dependent transcription even when TEAD-1 (Xiao et al. 1991), -2, -3, or -4 (data not shown) are expressed ectopically, suggesting that MPC-11 cells lack a necessary TEAD cofactor. As with EL4 T-lymphocytes, MPC-11 B-lymphocytes required ectopic expression of both TEAD and a YAP protein with a functional TEAD binding domain to elicit TEAD-dependent transcription (Fig. 6B). Thus, YAP is capable of providing the coactivator activity that is absent in MPC-11 cells.
The results described above showed that TEAD proteins interact specifically with YAP both physically and functionally. The proteins used here were active in vivo, and TEAD-dependent transcription in vivo required YAP protein with a functional TEAD binding domain as well as a TEAD protein with a functional DNA binding domain.
YAP protein complexes assembled in vivo contain TEAD/YAP dimers that bind TEAD-specific DNA sequence
To confirm the existence of TEAD/YAP complexes in vivo, FH-YAP was expressed in 3T3 cells and purified, as described for FH–TEAD-2. As expected, FH-YAP and FH-TEAD-2 were associated with the same proteins, although the relative amounts of some of these proteins differed (Fig. 7A, cf. lanes 2 and 4). T12 was absent from the YAP complex, and at least two new bands were present in the YAP complex (Fig. 7A, Y1 and  ). The mobility of the
). The mobility of the  bands was consistent with TEAD proteins, but they were present in much lower amounts than FH-YAP, and TEAD-2 was not detected by immuno-blotting. Nevertheless, TEAD proteins could be detected in the FH-YAP complex by their ability to bind specifically [32P]DNA containing the GT-IIC sequence (Fig. 7B, [32P]DNA TEAD-binding site). The GT-IIC sequence has been shown to bind TEAD proteins specifically and to bestow TEAD-dependent transcription to promoters and enhancers (Melin et al. 1993; Larkin et al. 1996; Jiang et al. 2000).
bands was consistent with TEAD proteins, but they were present in much lower amounts than FH-YAP, and TEAD-2 was not detected by immuno-blotting. Nevertheless, TEAD proteins could be detected in the FH-YAP complex by their ability to bind specifically [32P]DNA containing the GT-IIC sequence (Fig. 7B, [32P]DNA TEAD-binding site). The GT-IIC sequence has been shown to bind TEAD proteins specifically and to bestow TEAD-dependent transcription to promoters and enhancers (Melin et al. 1993; Larkin et al. 1996; Jiang et al. 2000).
Figure 7.
Comparison of YAP protein complexes with TEAD-2 protein complexes assembled in mouse cells. (A) FLAG-HA-tagged protein complexes were purified from 3T3 cells expressing either FH–TEAD–2 protein (lanes 1, 2) or FH-YAP protein (lanes 3, 4) by sequential affinity chromatography, as described in Figure 1, fractionated in parallel by SDS-PAGE, and then stained with silver to compare peptide bands. A duplicate of lane 4 was immunoblotted with anti-14-3-3 serum (Upstate Biotechnology). (B) Formation of a YAP/TEAD/DNA complex. Purified FH-YAP and FH–TEAD–2 complexes, or the indicated purified proteins were mixed with [32P]DNA containing either a wild-type PyV enhancer sequence ([32P]DNA control) that does not bind TEAD proteins, or a single base pair change in this PyV sequence that converts it into a GT-IIC sequence that does bind TEAD proteins ([32P]DNA TEAD binding site), and then fractionated by gel electrophoresis. The positions of [32P]DNA/TEAD complexes and [32P]DNA/TEAD/YAP complexes are indicated. All samples were run on the same gel. Some lanes were deleted for simplicity, and a lighter exposure of the two “FH–YAP complex” lanes is shown to see the presence of two bands.
Affinity purified F-TEAD-2 monomer bound [32P]DNA containing the TEAD-specific DNA binding sequence, but not [32P]DNA containing the same sequence with a single base pair change that eliminates TEAD binding. In contrast, affinity purified [His]6-YAP monomer bound neither DNA fragment. When these proteins were mixed together, the GT-IIC sequence was bound specifically by a higher molecular weight complex, and this complex was present in both the FH-TEAD-2 and FH-YAP complexes. In the FH-TEAD-2 complex, both a [32P]DNA/TEAD band and a [32P]DNA/TEAD/YAP band were present in equal amounts, consistent with the distribution of FH-TEAD-2 protein between monomeric and dimeric forms (Fig. 1B). Addition of purified YAP to the FH-TEAD-2 complex shifted all of the FH-TEAD-2 into the dimeric complex, confirming the formation of a TEAD/YAP complex in vitro. In the FH-YAP complex isolated from 3T3 cells, two [32P]DNA/protein complexes were detected, one corresponding to [32P]DNA/TEAD-2 and a smaller molecular complex that presumably represents [32P]DNA/TEAD-1, -3, or -4 proteins. Therefore, the FH-YAP complex contained TEAD proteins.
TEAD and YAP are localized in different cellular compartments
TEAD proteins were present but underrepresented in purified YAP protein complexes, because YAP protein was concentrated in the cytoplasm, whereas TEAD proteins were concentrated in the nucleus. More than 85% of the FH-YAP protein was found in the cytoplasmic fraction when nuclei were isolated from 3T3 cells in 10 mM KCl buffer that did not contain a detergent (silver staining of PAGE analysis, data not shown). The remaining FH-YAP protein could be extracted from the nuclear pellet with 300 mM KCl and 0.1% NP-40. In contrast, only 25% of the FH–TEAD-2 protein was in the cytoplasmic fraction, whereas 75% was in the nuclear pellet.
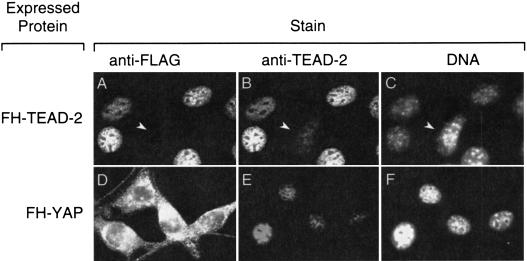
This distribution was confirmed by immuno-fluorescence analysis of 3T3 cells expressing either FH-TEAD-2 or FH-YAP. The cellular distribution of FH–TEAD-2 (Fig. 8A) was indistinguishable from that of native TEAD-2 protein (Fig. 8E); both proteins were localized in the nuclei of interphase cells in a speckled pattern that excluded nucleoli. Anti-FLAG antibody did not stain nuclei that did not express FH-TEAD-2 (Fig. 8, arrow in panels A–C), whereas anti-TEAD-2 antibody stained all 3T3 cell nuclei. In contrast, FH-YAP was concentrated in the cytoplasm (Fig. 8D), consistent with its appearance in the cytoplasmic fraction of cell extracts. Thus, ectopically expressed FH-TEAD-2 colocalized with endogenous TEAD-2 in the nucleus, while its transcriptional coactivator, YAP, is localized in the cytoplasm. Not all cells in the population expressed FH-TEAD-2 (Fig. 8B, arrowhead). The intensity of anti-TEAD-2 staining of the nuclei in these cells was ∼10-fold less, on average, than the nuclei in cells expressing FH-TEAD-2.
Figure 8.
Subcellular localization of TEAD-2 and YAP proteins. Cells in A–C were expressing FH-TEAD-2, whereas cells in D–F were expressing FH-YAP. Arrows in A–C identify a nucleus that expressed endogenous TEAD-2, but not FH-TEAD-2.
The cytoplasmic localization of YAP together with the equimolar presence of proteins of 30 to 32 kD in the purified FH-YAP complex (Fig. 7A, lane 4) suggested that 14-3-3 protein, which is involved in transporting proteins from the nucleus to the cytoplasm (Fu et al. 2000), may be a component of the FH-YAP complex. In fact, immuno-blotting revealed that T10 was 14-3-3 (Fig. 7A, lane 5), and fractionation of the purified FH-YAP protein complex by glycerol gradient sedimentation confirmed that most of the YAP protein existed as a complex with 14-3-3 and T11 (data not shown).
Discussion
YAP is a general transcriptional coactivator for TEAD transcription factors
One or more members of the TEAD/TEF family of transcription factors have been implicated in the activation of specific genes at different stages throughout animal development, and their action appears to involve one or more cofactors that may serve to modify their activity. However, identification of the putative general coactivator for all TEAD proteins that is absent from lymphocytic cells has proven elusive. Here we confirm that TEAD-dependent transcription in mammalian cells requires a transcriptional coactivator, and identify this coactivator as YAP, a ubiquitous, powerful transcriptional coactivator with an acidic activation domain.
YAP contains a carboxy-terminal acidic activation domain (pI = 3.79) that can activate transcription when tethered to a protein DNA-binding domain (Yagi et al. 1999). In this regard, YAP is equivalent to herpes simplex virus VP16, which contains a similar acidic activation domain (pI = 3.43), suggesting that YAP, like VP16, interacts directly with components of the transcriptional machinery including TFIIB, TBP, TFIIA, and TFIIH (Flint and Shenk 1997). Thus, sequence-specific DNA-binding proteins that bind YAP could utilize its activation domain to initiate transcription. One example is PEBP whose PY motif binds to one of the two WW domains (Fig. 4) in the amino-terminal half of YAP (Yagi et al. 1999). WW domains potentially can bind to the PY motif in other transcription factors such as c-Jun, AP-2, NF-E2, C/EBP alpha and PEBP2/CBF.
Here we show that the amino terminus of YAP also contains a novel 108 amino acid domain (Fig. 4) that binds all four members of the TEAD family of transcription factors in vitro (Fig. 3) and in vivo (Fig. 6). This domain is highly conserved; it is 91% identical to the same sequence in human and chicken YAP proteins. Conversely, TEAD proteins contain a conserved YAP binding domain that consists of the entire carboxy-terminal half (332 amino acids) of TEAD proteins (Fig. 2). Interaction between TEAD and YAP appears to be highly sensitive to the 3-dimensional structure of the TEAD protein, because all deletions within this region eliminated binding activity, although none of the smaller fragments from this region contained significant binding activity. These results are quite similar to those obtained in identifying the “transcriptional activation domain” (Fig. 2) for TEAD-1-dependent transcription in vivo (Hwang et al. 1993). Because human and mouse TEAD-1 proteins are 99% identical, the same transcriptional activation domain must exist in mTEAD-1. Because the carboxy-terminal halves of human or mouse TEAD-1 and TEAD-2 proteins are 71% identical, we conclude that the “transcriptional activation domain” identified in TEAD-1 is actually the YAP binding domain identified in TEAD-2 (Fig. 2). Thus, the real TEAD transcriptional activation domain is the carboxy-terminal acidic domain in YAP.
Several pieces of evidence support this conclusion. First, the tagged forms of TEAD and YAP proteins used in these experiments consistently reflected the behavior of their native counterparts. The tagged proteins were localized in the same cellular compartments as their native counterparts, stimulated transcription in vivo using the same domains identified in vitro, and bound the native form of their cognate protein in vivo, regardless of their levels of expression. Similar results to those in Figure 1 were visible at ∼5-fold less FH-TEAD-2 (data not shown). Second, TEAD proteins bound to a specific DNA sequence, while YAP protein neither interfered with TEAD binding to DNA nor exhibited any DNA binding of its own (Fig. 7B). Third, DNA/TEAD/YAP complexes of the same size were detected both in a mixture of purified TEAD and YAP proteins (Fig. 7B, lane 6), and in purified FH-TEAD and FH-YAP complexes purified from mouse cells (Fig. 7B, lanes 8, 11). Therefore, stable TEAD/YAP dimers exist in vivo that are capable of binding TEAD-specific DNA sequences, and TEAD can recruit YAP to specific promoters in vivo (Fig. 9). Fourth, formation of DNA/TEAD/YAP complexes was necessary for TEAD-dependent transcription in mouse cells, because TEAD-dependent transcription required both a TEAD protein with a functional DNA-binding domain, and YAP protein with a functional TEAD-binding domain (Figs. 5, 6). Thus, in vivo, TEAD and YAP proteins interacted functionally as well as physically. Whether or not other transcriptional coactivators can also serve this purpose remains to be determined.
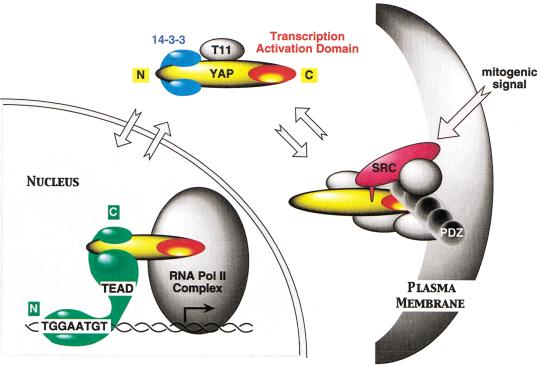
Figure 9.
Summary of TEAD and YAP65 interactions (see Discussion).
The data also suggest that YAP is the predominant, if not exclusive, transcriptional activation domain for TEAD proteins. In the presence of excess FH-TEAD-2 protein, the TEAD/YAP dimer constituted the major TEAD complex in 3T3 cells (Fig. 1B). Moreover, YAP functioned as a transcriptional coactivator for all four TEAD proteins (Fig. 6A), and YAP supported TEAD-dependent transcription in different cell types, regardless of whether or not YAP was already produced in these cells (Fig. 6B). With the exception of lymphocytic cells, YAP is present in both mouse and human tissues and in most cell types, from 2-cell embryos (BLAST search of EST data bases), to adults (Sudol et al. 1995). This is consistent with the fact that TEAD-specific transcription factor activity is first detected at the 2-cell stage in mouse development, concomitant with expression of the TEAD-2 gene, and that one or more TEAD genes are expressed in most tissues (Kaneko and DePamphilis 1998; Wang and Latham 2000). One example is the requirement of TEAD-1 for heart muscle development in the mouse, and expression of muscle specific genes in mouse C2C12 muscle fibroblasts. The fact that YAP could bind to TEAD-1, stimulate TEAD-1 transcription activity, and function in C2C12 cells strongly suggests that TEAD-1-dependent activation of muscle-specific genes is mediated by YAP. In human cells that already express TEAD proteins, ectopic expression of TEADsquelches TEAD-dependent transcription, because the excess TEAD protein titrates out a required coactivator (Xiao et al. 1991; Jiang and Eberhardt 1996). This coactivator appears to be YAP, because ectopic expression of YAP in mouse cells that already express TEAD proteins strongly stimulates endogenous TEAD transcriptional activity, whereas ectopic expression of additional TEAD protein titrates out the YAP activity (Fig. 5, 3T3 cells). Taken together, these observations suggest that YAP mediates TEAD function in most, if not all, mammalian cells.
Regulation of TEAD activity
A surprising feature of TEAD/YAP-dependent transcription activity is that TEAD is localized in the nucleus, while its transcriptional coactivator is localized in the cytoplasm (Fig. 8). YAP has been shown previously to bind to the Src/Yes protein tyrosine kinase family via its SH3 binding domain (Sudol 1994), and to a PDZ domain protein (EBP50) that results in concentration of YAP at the apical membrane in human epithelial cells (Mohler et al. 1999). PDZ domains, consisting of ∼90 amino acid residues, have been found in several proteins involved in the assembly of protein signaling complexes on the membranes of synaptic junctions (Garner et al. 2000). Thus, the function of the PDZ protein MUPP1 in the multimeric TEAD-2 protein complex (Fig. 1B) may be to attach this complex to the plasma membrane. Because MUPP1 has 13 PDZ domains, it potentially can bind 13 different proteins and thereby serve as a scaffold on which PDZ binding proteins, such as YAP, can be organized. The amount of either FH-TEAD-2 or FH-YAP multimeric protein complex was too small to allow detection of Src protein. However, the fact that deletion of the SH3 binding domain in YAP did not affect its ability to activate TEAD transcription factors in vivo (Fig. 6A), means that YAP does not have to associate with this Src/Yes proteins to activate nuclear transcription factors such as TEAD.
Overexpression of YAP resulted in the accumulation of a YAP/14-3-3/T11 complex in the cytoplasm. The 14-3-3 family of proteins bind a multitude of functionally diverse signaling proteins, including kinases, phosphatases, and transmembrane receptors, and are involved in shuttling these proteins out of the nucleus and thereby localizing them in the cytoplasm (Fu et al. 2000). The 31-kD T11 protein may also be a member of this family, although it reacted only weakly with the antisera we tested. Thus, the YAP/14-3-3 complex appears to represent an intermediate between YAP/transcription factor complexes in the nucleus and YAP/signal transduction complexes associated with the plasma membrane. Moreover, accumulation of YAP/14-3-3 complexes in the cytoplasm demonstrated that mammalian cells regulate the concentration of YAP in their nucleus, and thereby regulate the activity of sequence specific transcription factors such as TEAD that require YAP as a transcriptional coactivator. Furthermore, the 14-3-3 binding domain in TAZ, a transcriptional coactivator that shares homology with YAP, requires phosphorylation of TAZ on a single serine residue (Kanai et al. 2000). This residue lies within the TEAD binding domain (Fig. 4), suggesting that 14-3-3 and TEAD would compete for YAP binding.
Taken together, these observations suggest a regulatory pathway for transcriptional activity of TEAD and other proteins that utilize YAP as their transcription activation domain (Fig. 9). YAP, which is normally the rate-limiting factor for TEAD-dependent transcription, is sequestered at the plasma membrane by association with a PDZ-protein such as MUPP1 and by association with Src/Yes protein tyrosine kinases. Mitogenic signals (or ectopic expression) release YAP into the cytoplasm where it forms a complex with 14-3-3. YAP can then be transported into and out of the nucleus, where it forms a transcriptionally active complex with TEAD.
Materials and methods
Cell culture
NIH/3T3 cells (ATCC CRL1658) were cultured in high glucose DMEM supplemented with 7.5% calf serum, 4 mM glutamine and 1 mM sodium pyruvate. EL4 (ATCC TIB39), MPC11 (ATTC CCL167), and C2C12 (ATTC CRL1772) cells were cultured in high glucose DMEM supplemented with 10% FCS, 4 mM glutamine, 1 mM sodium pyruvate, and nonessential amino acids. For EL4 and MPC11 cells, 0.05 mM β–mercaptoethanol was included.
Purification of tagged protein complexes assembled in vivo
A full-length mouse YAP open reading frame (ORF) was isolated from a plasmid DNA containing an EST (GenBank Accession No. AW211560). YAP was cloned into the multicloning site in pTIN, a modified version of the Moloney murine leukemia virus (MMLV)-derived retroviral vector pRetro-Off (Clontech). The puromycin resistance gene in pRetro-Off vector was replaced in pTIN with the IL-2Rα protein ORF to express IL-2Rα surface antigen constitutively under the control of 5′-LTR viral promoter. The multicloning site in pRetro-Off was altered to contain the sequence ACCATGgactacaaggacgacgatgacaagCTCG ATGGAGGAtacccctacgacgtgcccgactacgccGGAGGACTCGAG-Y AP. Translation began at a Kozak consensus sequence (underlined) followed by sequences for the FLAG and hemaglutinin epitopes (lowercase) separated by a linker. Amino-terminal- FH-YAP was expressed under the control of a tetracycline-repressible promoter. Cells that were transduced with pTIN were grown in the presence of 3 μg/ml tetracycline to suppress expression of the cloned protein, and expression of FH-YAP was induced by culturing cells in the absence of tetracycline for 24 h, according to the Clontech protocol.
Mouse TEAD-2 gene (Kaneko and DePamphilis 1998) was cloned into pOZ, a MMLV derived retroviral vector (B. Howard, NIH, Bethesda, MD), upstream of the IL-2Rα ORF so that both genes were expressed from a bi-cistronic mRNA. Translation of the cloned TEAD gene began at the Kozak consensus sequence [ACCATGgactacaaggacgacgatgacaagCTCGATGGAGGAtacccc tacgacgtgcccgactacgccGGAGGACTCGAG-TEAD-2], while translation of the IL-2Rα gene began at the IRIS sequence.
Several mouse cell lines were tested for their ability to stabile express mouse TEAD or YAP proteins. 3T3 cells were transduced with either pOZ or pTIN recombinant viruses using the Clontech protocol described for pRetro-Off vector. Transduced cells expressing IL-2Rα on their surface were collected by affinity cell sorting using anti-ILR2α antibodies (UPSTATE Biotechnology) and Dynabeads according to the manufacturer protocol (Dynal). Cells (107) expressing IL-2Rα were cultured to 90% confluence and then extracted for 30 min with 200 μl buffer B100 (20 mM Tris-HCl at pH 7.9, 100 mM KCl, 5 mM MgCl2, 10% glycerol, 0.1% Nonidet P-40, and 10 μM leupeptin, 1 μM pepstatin, 1 μM PMSF, 0.5 μg/ml aprotinin). All steps were carried out on ice or at 4°C. The extract was incubated with 2.5 μl M2 anti-FLAG agarose (Sigma) for 4 h with rotation. Beads were washed in B100 and then bound proteins were eluted by incubation for 1 h with 0.2 mg/ml FLAG peptide (Sigma) in B100 with rotation. The eluate was incubated for 2 h with 2.5 μl protein A sepharose (Pharmacia) that was coupled to 12CA5 anti-HA antibody (Roche) as described previously (Harlow and Lane 1999). The resin was washed with B100, and then eluted by incubation for 1 h with 0.5 mg/ml HA peptide (Roche) in buffer B100. The yield from 107 cells was 35 ng FH-TEAD-2 or 80 ng FH-YAP protein.
For glycerol gradient sedimentation, 50 μl purified protein complex was placed on top of a 3.7 ml 10%–35% linear glycerol gradient in buffer B100 and centrifuged in a Beckman SW60 rotor (55,000 rpm for 3.5 h at 4°C).
Plasmids and antibodies
Full-length mouse TEAD gene sequences cloned into pOZ (see above) were amplified together with their HA-tag using PCR and inserted into pCI (Promega). Full-length YAP, as well as the indicated fragments (Fig. 4), were generated by PCR and inserted into pBS-KSII+ (Stratagene), pTIN, pSI (Promega), or pGEX-6P-1 (Pharmacia) vectors. Deletions in the wild-type TEAD-2 and YAP sequences were generated by PCR-based mutagenesis (Ausubel et al. 1997). pGT4Tluc contains the firefly luciferase gene driven by four tandem GT-IIC sequences (Kaneko et al. 1997), the TEAD DNA binding site found in wild-type SV40, and PyV host range enhancers. PCI[H-TEAD-2] contains the mouse H-TEAD-2 gene driven by the cytomegalovirus intermediate-early promoter/enhancer. pSI(FH-mYAP) contains the mouse YAP gene driven by the SV40 early gene promoter and enhancer. pRI(βgal) contains the E. coli β–galactosidase gene driven by the Rous Sarcoma Virus long terminal repeat. All constructions were confirmed by DNA sequencing, and equivalent results were obtained with at least two independent clones. Rabbit anti-mTEAD-2 polyclonal antibody was generated against a synthetic peptide (aa 2 to 14, [Kaneko et al. 1997]).
Identification of proteins by mass spectrometry
The separated proteins in SDS-PAGE gel were visualized by the Colloidal Blue Staining (Novex). The bands of interest were excised, destained in 25 mM NH4HCO3 (pH 8) in methanol:water (1:1), cleared in water:acetonitrile:acetic acid (45:45:10, v/v/v)) for 20 h, swollen in water for 2 h, equilibrated with 20 μl 50 mM NH4HCO3, (pH 8) for 4 min, and then digested at 37 °C for 2 h with 0.2 μg trypsin (Roche sequencing grade). Tryptic peptides were extracted twice with 70% acetonitrile in water and concentrated in a speed vac. Each dried sample was dissolved in 6 μl of HPLC buffer A solution (water:acetonitrile:acetic acid, 97.5:2:0.5 [v/v/v]) for mass spectrometric (MS) analysis.
HPLC-MS/MS analysis was performed in a LCQ (Finnigan MAT) coupled on-line with a capillary HPLC system (Eldex MicroPro syringe pumping system). Two μl were loaded on capillary HPLC connected with a C18 column (5–10 cm length, 75 μm ID). Peptides eluted from the HPLC column were sprayed directly from the tip of the capillary column to LCQ mass spectrometer for MS analysis (Gatlin et al. 1998). LCQ was operated in a data-dependent mode where the machine measured intensity of all peptide ions in the mass range 400 to 1400 (mass-to-charge ratios) and isolated the peptide peak with the highest intensity for collision-induced dissociation. Thus, masses of both the parent peptide and its daughter ions were detected.
Molecular weights of tryptic peptides were also determined in a matrix-assisted laser desorption ionization time of flight (MALDI-TOF) mass spectrometer (Hillenkamp et al. 1991) equipped with delay ion extraction (Voyager STR, Perseptive Biosystems, Inc.). 0.8 μl of peptide mixture and 0.8 μl of matrix solution (6 times dilution of the saturated 2,5-dihydroxybensoic acid in ACN/H2O (1:1) solution was mixed on a sample plate and dried at room temperature prior to MS analysis. The best match between experimentally determined masses of tryptic peptides and their fragments and theoretically calculated masses for each tryptic peptide from proteins or ESTs in the database were determined using PepFrag (http://prowl1.rockefeller.edu/prowl/pepfragch.html) (Fenyo et al. 1998).
Identification of protein-binding domains in vitro
TEAD-2 proteins were radio-labeled with [35S]-methionine using an in vitro coupled transcription/translation system (TnT Quick, Promega). GST–YAP proteins were expressed in E. coli using pGEX-6P1 vector (Pharmacia). GST–YAP protein was bound to glutathione-CL4B Sepharose (Pharmacia) as follows. All steps were at 4°C. Bacteria were extracted for 30 min with B100 (1 ml/10 culture) by vortexing with 200 μl of 600 micron glass beads (SIGMA) per ml. After centrifuging at 12,000xg for 30 min, the supernatant was incubated with glutathione-CL4B Sepharose for 1 h with rotation. After washing the beads with B100, the amount of bound GST protein was measured by subjecting them to SDS-PAGE along with bovine serum albumin standards, staining the proteins with Coomassie brilliant blue R-250, and quantifying protein bands by densitometry. Beads (2 μl) containing 200 ng of GST–YAP were incubated with 15 μl completed TnT Quick translation reaction for 30 min at 4°C in 100 μl B100 (final volume) containing 100 μg/ml BSA. After washing the beads with B100, the bound proteins were released by boiling in SDS sample buffer and fractionated by SDS-PAGE (NuPAGE gradient gel, Invitrogen). Proteins were stained with Coomassie brilliant blue R-250. The gel was dried, and 35S-labeled proteins were visualized by autoradiography, and quantified by densitometry. The amount of bound [35S]-TEAD protein was normalized against the amount of Coomassie stained GST–YAP in each sample.
TEAD-dependent transcription in vivo
Electroporation, luciferase, and β–galactosidase assays were carried out as described previously (Kaneko and DePamphilis 2000). Cells were transfected with a mixture of plasmid DNAs consisting of 10 μg pGT4Tluc, 10 μg pRI(βgal), 0 to 10 μg pCI(H-TEAD), and where indicated, 10 μg pSI(YAP). The total amount of DNA was adjusted to 40 μg with salmon sperm DNA in 20 μl final volume of TE buffer. This DNA was mixed with 107 cells in 300 μl culture medium and electroporated at 1180 μF and either 200 V (3T3 cells) or 250 V (EL4 cells). Cells were then cultured for 48 h before lysing them in reporter lysis buffer (Promega) and quantifying the amounts of luciferase and β–galactosidase activities in suitable aliquots.
Immunofluorescence
3T3 cells were fixed at 50%–70% confluence with methanol (−20°C, 5 min) and incubated anti-FLAG M2 antibody (Sigma) and anti-TEAD-2 rabbit serum followed by fluorochrome-conjugated secondary antibodies (Jackson Immunoresearch) diluted in PBS containing 0.1 mg/ml Hoechst 33258 (Sigma) to visualize DNA. Images were taken using a digital camera (Princeton Instruments) attached to a Nikon E600 photomicroscope equipped with epifluorescence optics using a 100× objective.
Acknowledgments
We are indebted to Dr. Bruce Howard and members of our laboratory for providing the pOZ retroviral expression vector and for advising us on its application.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL depamphm@mail.nih.gov; FAX (301) 480-9354.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.888601.
References
- Ausubel F, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short protocols in molecular biology. New York: John Wiley & Sons, Inc.; 1997. [Google Scholar]
- Azakie A, Larkin SB, Farrance IK, Grenningloh G, Ordahl CP. DTEF-1, a novel member of the transcription enhancer factor-1 (TEF-1) multigene family. J Biol Chem. 1996;271:8260–8265. doi: 10.1074/jbc.271.14.8260. [DOI] [PubMed] [Google Scholar]
- Belandia B, Parker MG. Functional interaction between the p160 coactivator proteins and the transcriptional enhancer factor family of transcription factors. J Biol Chem. 2000;275:30801–30805. doi: 10.1074/jbc.C000484200. [DOI] [PubMed] [Google Scholar]
- Butler AJ, Ordahl CP. Poly(ADP-ribose) polymerase binds with transcription enhancer factor 1 to MCAT1 elements to regulate muscle-specific transcription. Mol Cell Biol. 1999;19:296–306. doi: 10.1128/mcb.19.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Friedrich GA, Soriano P. Transcriptional enhancer factor 1 disruption by a retroviral gene trap leads to heart defects and embryonic lethality in mice. Genes & Dev. 1994;8:2293–2301. doi: 10.1101/gad.8.19.2293. [DOI] [PubMed] [Google Scholar]
- Davidson I, Xiao JH, Rosales R, Staub A, Chambon P. The HeLa cell protein TEF-1 binds specifically and cooperatively to two SV40 enhancer motifs of unrelated sequence. Cell. 1988;54:931–942. doi: 10.1016/0092-8674(88)90108-0. [DOI] [PubMed] [Google Scholar]
- Fenyo D, Qin J, Chait BT. Protein identification using mass spectrometric information. Electrophoresis. 1998;19:998–1005. doi: 10.1002/elps.1150190615. [DOI] [PubMed] [Google Scholar]
- Flint J, Shenk T. Viral transactivating proteins. Annu Rev Genet. 1997;31:177–212. doi: 10.1146/annurev.genet.31.1.177. [DOI] [PubMed] [Google Scholar]
- Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: Structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- Garner CC, Nash J, Huganir RL. PDZ domains in synapse assembly and signalling. Trends Cell Biol. 2000;10:274–280. doi: 10.1016/s0962-8924(00)01783-9. [DOI] [PubMed] [Google Scholar]
- Gatlin CL, Kleemann GR, Hays LG, Link AJ, Yates JR., 3rd Protein identification at the low femtomole level from silver-stained gels using a new fritless electrospray interface for liquid chromatography-microspray and nanospray mass spectrometry. Anal Biochem. 1998;263:93–101. doi: 10.1006/abio.1998.2809. [DOI] [PubMed] [Google Scholar]
- Gupta MP, Amin CS, Gupta M, Hay N, Zak R. Transcription enhancer factor 1 interacts with a basic helix-loop-helix zipper protein, Max, for positive regulation of cardiac alpha-myosin heavy-chain gene expression. Mol Cell Biol. 1997;17:3924–3936. doi: 10.1128/mcb.17.7.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Using antibodies, a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- Hillenkamp F, Karas M, Beavis RC, Chait BT. Matrix-assisted laser desorption/ionization mass spectrometry of biopolymers. Anal Chem. 1991;63:1193A–1203A. doi: 10.1021/ac00024a002. [DOI] [PubMed] [Google Scholar]
- Hwang JJ, Chambon P, Davidson I. Characterization of the transcription activation function and the DNA binding domain of transcriptional enhancer factor-1. EMBO J. 1993;12:2337–2348. doi: 10.1002/j.1460-2075.1993.tb05888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemin P, Hwang JJ, Martial JA, Dolle P, Davidson I. A novel family of developmentally regulated mammalian transcription factors containing the TEA/ATTS DNA binding domain. J Biol Chem. 1996;271:21775–21785. doi: 10.1074/jbc.271.36.21775. [DOI] [PubMed] [Google Scholar]
- Jacquemin P, Sapin V, Alsat E, Evain-Brion D, Dolle P, Davidson I. Differential expression of the TEF family of transcription factors in the murine placenta and during differentiation of primary human trophoblasts in vitro. Dev Dyn. 1998;212:423–436. doi: 10.1002/(SICI)1097-0177(199807)212:3<423::AID-AJA10>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Jiang SW, Eberhardt NL. TEF-1 transrepression in BeWo cells is mediated through interactions with the TATA-binding protein, TBP. J Biol Chem. 1996;271:9510–9518. doi: 10.1074/jbc.271.16.9510. [DOI] [PubMed] [Google Scholar]
- Jiang SW, Wu K, Eberhardt NL. Human placental TEF-5 transactivates the human chorionic somatomammotropin gene enhancer. Mol Endocrinol. 1999;13:879–889. doi: 10.1210/mend.13.6.0288. [DOI] [PubMed] [Google Scholar]
- Jiang SW, Desai D, Khan S, Eberhardt NL. Cooperative binding of TEF-1 to repeated GGAATG-related consensus elements with restricted spatial separation and orientation. DNA Cell Biol. 2000;19:507–514. doi: 10.1089/10445490050128430. [DOI] [PubMed] [Google Scholar]
- Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, Hisaminato A, Fujiwara T, Ito Y, Cantley LC, et al. TAZ: A novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 2000;19:6778–6791. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko KJ, DePamphilis ML. Regulation of gene expression at the beginning of mammalian development and the TEAD family of transcription factors. Dev Genet. 1998;22:43–55. doi: 10.1002/(SICI)1520-6408(1998)22:1<43::AID-DVG5>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- ————— Soggy, a spermatocyte-specific gene, lies 3.8 kb upstream of and antipodal to TEAD-2, a transcription factor expressed at the beginning of mouse development. Nucleic Acids Res. 2000;28:3982–3990. doi: 10.1093/nar/28.20.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko KJ, Cullinan EB, Latham KE, DePamphilis ML. Transcription factor mTEAD-2 is selectively expressed at the beginning of zygotic gene expression in the mouse. Development. 1997;124:1963–1973. doi: 10.1242/dev.124.10.1963. [DOI] [PubMed] [Google Scholar]
- Larkin SB, Farrance IK, Ordahl CP. Flanking sequences modulate the cell specificity of M-CAT elements. Mol Cell Biol. 1996;16:3742–3755. doi: 10.1128/mcb.16.7.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Salas E, Linney E, Hassell J, DePamphilis ML. The need for enhancers in gene expression first appears during mouse development with formation of a zygotic nucleus. Genes & Dev. 1989;3:1493–1506. doi: 10.1101/gad.3.10.1493. [DOI] [PubMed] [Google Scholar]
- Melin F, Miranda M, Montreau N, DePamphilis ML, Blangy D. Transcription enhancer factor-1 (TEF-1) DNA binding sites can specifically enhance gene expression at the beginning of mouse development. EMBO J. 1993;12:4657–4666. doi: 10.1002/j.1460-2075.1993.tb06154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler PJ, Kreda SM, Boucher RC, Sudol M, Stutts MJ, Milgram SL. Yes-associated protein 65 localizes p62(c-Yes) to the apical compartment of airway epithelia by association with EBP50. J Cell Biol. 1999;147:879–890. doi: 10.1083/jcb.147.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks AB, Rider JE, Hoffman NG, Fowlkes DM, Quillam LA, Kay BK. Distinct ligand preferences of Src homology 3 domains from Src, Yes, Abl, Cortactin, p53bp2, PLCgamma, Crk, and Grb2. Proc Natl Acad Sci. 1996;93:1540–1544. doi: 10.1073/pnas.93.4.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudol M. Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product. Oncogene. 1994;9:2145–2152. [PubMed] [Google Scholar]
- Sudol M, Bork P, Einbond A, Kastury K, Druck T, Negrini M, Huebner K, Lehman D. Characterization of the mammalian YAP (Yes-associated protein) gene and its role in defining a novel protein module, the WW domain. J Biol Chem. 1995;270:14733–14741. doi: 10.1074/jbc.270.24.14733. [DOI] [PubMed] [Google Scholar]
- Ueyama T, Zhu C, Valenzuela YM, Suzow JG, Stewart AF. Identification of the functional domain in the transcription factor RTEF-1 that mediates alpha 1-adrenergic signaling in hypertrophied cardiac myocytes. J Biol Chem. 2000;275:17476–17480. doi: 10.1074/jbc.M001970200. [DOI] [PubMed] [Google Scholar]
- Vaudin P, Delanoue R, Davidson I, Silber J, Zider A. TONDU (TDU), a novel human protein related to the product of vestigial (vg) gene of Drosophila melanogaster interacts with vertebrate TEF factors and substitutes for Vg function in wing formation. Development. 1999;126:4807–4816. doi: 10.1242/dev.126.21.4807. [DOI] [PubMed] [Google Scholar]
- Wang Q, Latham KE. Translation of maternal messenger ribonucleic acids encoding transcription factors during genome activation in early mouse embryos. Biol Reprod. 2000;62:969–978. doi: 10.1095/biolreprod62.4.969. [DOI] [PubMed] [Google Scholar]
- Xiao JH, Davidson I, Matthes H, Garnier JM, Chambon P. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65:551–568. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yockey CE, Smith G, Izumo S, Shimizu N. cDNA cloning and characterization of murine transcriptional enhancer factor-1-related protein 1, a transcription factor that binds to the M-CAT motif. J Biol Chem. 1996;271:3727–3736. doi: 10.1074/jbc.271.7.3727. [DOI] [PubMed] [Google Scholar]