Abstract
In liver, the synthesis of cholesterol and fatty acids increases in response to cholesterol deprivation and insulin elevation, respectively. This regulatory mechanism underlies the adaptation to cholesterol synthesis inhibitors (statins) and high calorie diets (insulin). In nonhepatic cells, lipid synthesis is controlled by sterol regulatory element-binding proteins (SREBPs), membrane-bound transcription factors whose active domains are released proteolytically to enter the nucleus and activate genes involved in the synthesis and uptake of cholesterol and fatty acids. SCAP (SREBP cleavage-activating protein) is a sterol-regulated escort protein that transports SREBPs from their site of synthesis in the endoplasmic reticulum to their site of cleavage in the Golgi. Here, we produced a conditional deficiency of SCAP in mouse liver by genomic recombination mediated by inducible Cre recombinase. SCAP-deficient mice showed an 80% reduction in basal rates of cholesterol and fatty acid synthesis in liver, owing to decreases in mRNAs encoding multiple biosynthetic enzymes. Moreover, these mRNAs failed to increase normally in response to cholesterol deprivation produced by a cholesterol synthesis inhibitor and to insulin elevation produced by a fasting–refeeding protocol. These data provide in vivo evidence that SCAP and the SREBPs are required for hepatic lipid synthesis under basal and adaptive conditions.
Keywords: Cholesterol, fatty acids, insulin, SREBP, SCAP, liver-specific gene targeting
In multicellular organisms, embryonic development requires precise control of the lipid composition of the membranes that separate cells from their environment. Inherited defects in lipid metabolism cause developmental defects in the brain, including hydrocephalus and lissencephaly (Farese and Herz 1998). Lipids also control the development of adipose tissue by interacting with transcriptional activators such as peroxisome proliferator-activated receptor-γ (PPAR-γ) (Rosen and Spiegelman 2000). In liver, disordered regulation of lipid metabolism causes fetal biliary atresia (Schwarz et al 2000). Postnatally, disordered hepatic lipid metabolism leads to fatty liver (Shimano et al. 1996) as well as hyperlipidemia and atherosclerosis (Brown and Goldstein 1986).
Control of lipid synthesis is especially important in the liver, which synthesizes lipids not only for its own uses but also for export into the plasma as lipoproteins. Levels of plasma lipoprotein cholesterol are lowered by treatment with drugs called statins, which inhibit 3-hydroxy-3-methylglutaryl CoA reductase (HMG CoA reductase), the rate-controlling enzyme of cholesterol synthesis in liver (Witztum 1996). The response to statins is limited because the liver responds to cholesterol depletion by massive up-regulation of the mRNAs for HMG CoA reductase and other cholesterologenic enzymes, thereby rendering the competitive inhibitor less efficient (Brown and Goldstein 1986). Hepatic fatty acid synthesis is elevated when plasma insulin rises, as in states of obesity and noninsulin-dependent diabetes mellitus. The fatty acids are exported from the liver in lipoproteins, and they reach extrahepatic organs in which they increase insulin resistance and worsen the diabetic state (McGarry 1998; Shimomura et al. 2000).
The mechanisms that regulate lipid synthesis have come into focus recently with the discovery of sterol regulatory element-binding proteins (SREBPs), which are membrane-bound transcription factors that control the rates of lipid synthesis in animal cells (Wang et al. 1994; Kim and Spiegelman 1996). To activate lipid biosynthesis, SREBPs must be transported from the endoplasmic reticulum (ER) to the Golgi apparatus, in which they are cleaved sequentially by two proteases that release the cytosolic NH2-terminal transcription factor domains (Brown and Goldstein 1997, 1999). These fragments enter the nucleus, where they bind to sterol regulatory elements in the enhancer regions of more than a score of genes whose products mediate the synthesis of cholesterol and unsaturated fatty acids (Horton and Shimomura 1999; Edwards et al. 2000).
The proteolytic processing of SREBPs is negatively regulated by sterols, which thereby repress their own synthesis. The key to sterol regulation is SREBP cleavage-activating protein (SCAP), a polytopic membrane protein that forms a complex with SREBPs in the ER (Brown and Goldstein 1999). In cultured fibroblasts, SCAP is absolutely required to stabilize the SREBPs and to transport them to the Golgi apparatus. Mutant cells that lack SCAP have low levels of SREBP precursors, apparently because these proteins are unstable in the absence of SCAP. Even when SREBP precursors are elevated in these cells by cDNA transfection, they are not transported to the Golgi for cleavage. As a result of these abnormalities, SCAP-deficient CHO cells cannot synthesize cholesterol, and they require external sources of cholesterol for growth (Rawson et al. 1999). SCAP is also the cholesterol sensor. When sterols build up in cells, SCAP no longer transports SREBPs, and proteolytic processing is abolished (DeBose-Boyd et al. 1999; Nohturfft et al. 1999, 2000). The sterol-sensing function is mediated by the polytopic membrane domain of SCAP, which contains a motif that is shared with other proteins that are postulated to interact with sterols (Brown and Goldstein 1999). Point mutations in this sterol-sensing motif prevent sterol repression of SREBP cleavage and lead to unregulated overproduction of cholesterol (Hua et al. 1996; Nohturfft et al. 1996).
The SREBP/SCAP system has been studied most extensively in cultured fibroblast-like cell lines. Less is known about the proteolytic processing of SREBPs in liver. Liver produces all three isoforms of SREBP, designated SREBP-1a, SREBP-1c, and SREBP-2 (Shimomura et al. 1997). Transgenic mice that overexpress the soluble transcription-activating domain of SREBP-1a develop massive fatty liver, owing primarily to the overproduction of fatty acids and secondarily to overproduction of cholesterol (Shimano et al. 1996). Overexpression of the soluble transcription-activating domain of SREBP-2 also produces a fatty liver, but in this case cholesterol is the predominant lipid (Horton et al. 1998b). Overexpression of SREBP-1c (also known as ADD-1) (Kim and Spiegelman 1996) causes less total lipid accumulation, and all of it is derived from fatty acids (Shimano et al. 1997a).
Under normal conditions, the liver does not produce soluble truncated forms of SREBPs. Rather, it produces full-length SREBP-1a, SREBP-1c, and SREBP-2 precursors that must be processed proteolytically to activate transcription. The liver expresses SCAP and the two proteases, Site-1 protease (S1P) and Site-2 protease (S2P), that process SREBPs in nonhepatic cells. Evidence that SCAP plays a regulatory role in liver came from experiments in which transgenic mice overexpressed a mutant SCAP that is sterol resistant, owing to a point mutation in the sterol-sensing domain (Korn et al. 1998). Livers from these mice overproduced cholesterol, and they failed to show normal feedback inhibition when cholesterol was fed.
Correlative evidence suggests that SREBPs are involved in the regulatory response to hepatic cholesterol deprivation. When hepatic cholesterol levels are lowered by treatment with statins, the level of nuclear SREBP-2 rises, and this is correlated with an increase in the mRNAs for target genes, including HMG CoA synthase, HMG CoA reductase, and the low-density lipoprotein (LDL) receptor (Sheng et al. 1995). Whether SCAP is required for this regulatory response is unknown.
In addition to activating sterol synthesis, SREBPs activate fatty acid synthesis in response to insulin, which increases the mRNA and protein levels for one isoform, SREBP-1c, in liver (Foretz et al. 1999; Shimomura et al. 1999b, 2000). It is likely that insulin-induced SREBP-1c must be processed proteolytically for insulin to stimulate fatty acid synthesis, and this processing may require SCAP.
The current studies were designed to test the hypothesis that SCAP is required for SREBP processing in liver and that it plays a role in the liver's response to cholesterol deprivation and to insulin signaling. We did not attempt a germ-like knockout of the SCAP gene because prior experiments showed that a homozygous knockout of a single SREBP isoform (SREBP-2) results in embryonic lethality, and we anticipated that a SCAP knockout would have the same effect. We therefore targeted the SCAP gene by flanking exon 1 with loxP sites, which are targets for bacteriophage P1 Cre recombinase. We produced mice that are homozygous for the loxP-tagged SCAP allele and also carry a transgene encoding Cre under control of the interferon-inducible MX1 promoter. When these mice are treated with double-stranded RNA (polyinosinic acid-polycytidylic acid, pIpC), interferon is induced. This activates the Cre recombinase in liver (Kuhn et al. 1995; Rohlmann et al. 1998), and it leads to the near-total inactivation of the SCAP gene. The SCAP-deficient animals exhibit a decline in the precursor and nuclear forms of SREBP-1c and SREBP-2. As a result, they manifest a major decrease in synthesis of cholesterol and fatty acids, and they do not show the normal increase in these processes in response to cholesterol deprivation or insulin.
Results
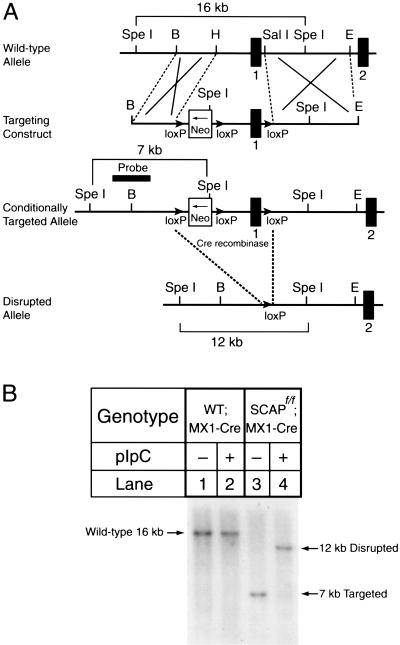
Figure 1A shows the vector that was designed to target mouse SCAP conditionally. A bacterial neomycin resistance gene (neo) flanked by loxP sites were inserted into the 5′-flanking region of the SCAP gene, and an additional loxP site was inserted into the first intron. After introducing the vector into mouse ES cells, we used G418 resistance to select a clone that had incorporated the neo cassette into genomic DNA by homologous recombination. The ES cells were injected into mouse blastocysts, and we bred the resultant chimeric males to establish a line of mice that is homozygous for the floxed allele, designated SCAPf/f. The mice were bred with a line of transgenic mice that express the Cre recombinase under the control of the MX1 promoter, which responds to pIpC. These mice are designated SCAPf/f;MX1-Cre.
Figure 1.
Generation and characterization of a floxed SCAP allele. (A) Schematic of the sequence-replacement gene-targeting strategy. The map of the wild-type allele spans the SCAP promoter region (the sequence upstream of exon 1), exon 1, intron 1, and exon 2. The targeting vector containing the neo gene driven by the pgk-neo-pA cassette (phosphoglyceride kinase promoter followed by the 3′-untranslated region of the bovine growth hormone gene containing the polyadenylation signal) (Soriano et al. 1991) is flanked on each side by one copy of the loxP sequence as denoted by the solid arrows (Kuhn et al. 1995). The transcriptional direction of the neo gene is shown by the arrow in its box. The entire cassette was inserted 3 kb upstream of the SCAP gene. The downstream loxP sequence was inserted within intron 1. Excision of the sequences between the loxP sites by the Cre recombinase deletes 3 kb of upstream sequence and exon 1, which includes the initiator methionine and residues encoding the first transmembrane domain of SCAP. The location of the probe used for Southern analysis (2-kb EcoRI–HindIII fragment) is denoted by a solid box. (B) Representative Southern blot analysis of SpeI-digested genomic DNA from the livers of wild-type;MX1-Cre transgenic mice and SCAPf/f;MX1-Cre transgenic mice that were treated with three intraperitoneal injections of pIpC (250 μg/injection) as described in Materials and Methods. Genomic DNA from liver was prepared 14 d after the last injection. The positions of migration of the fragments derived from wild-type, targeted, and disrupted alleles are indicated.
Figure 1B shows mouse genomic DNA that was digested with SpeI and blotted with the indicated probe from the SCAP gene (shown in Fig. 1A). A 16-kb fragment was visualized in DNA from wild-type mice that were transgenic for MX1-Cre. The band was not affected when these mice were injected with pIpC, which induces interferon and induces the Cre recombinase. In the SCAPf/f;MX1-Cre mice, the SpeI digest produced a 7-kb band that corresponds to the targeted allele. Injection of pIpC led to a nearly complete loss of the 7-kb band and the appearance of a 12-kb band, which represents recombination between the 5′ and 3′ loxP sites. This recombination eliminates the promoter and the first exon of the SCAP gene.
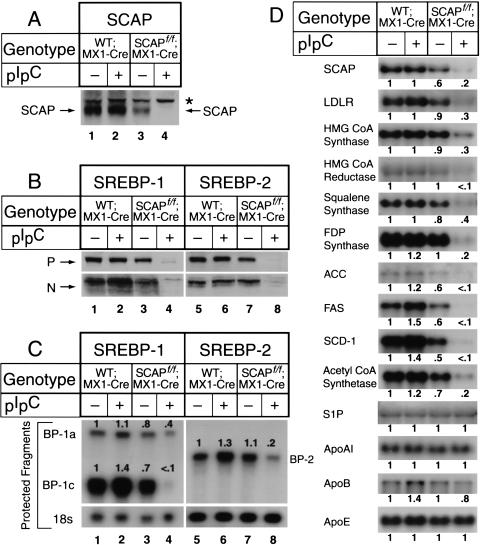
When wild-type;MX1-Cre mice were injected with pIpC, there was no change in the amount of SCAP protein that was visualized by immunoblotting in liver extracts (Fig. 2A, lanes 1,2). Even without pIpC, the livers from the SCAPf/f;MX1-Cre mice expressed a reduced amount of SCAP protein, perhaps owing to transcription interference by the neo cassette (Fig. 2A, lane 3). When these animals were injected with pIpC, the amount of SCAP declined dramatically (Fig. 2A, lane 4). We estimated that the reduction was 90%–95%.
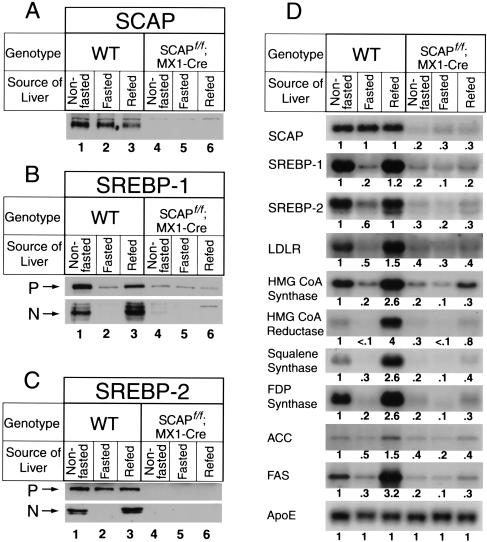
Figure 2.
Expression of mRNAs and proteins in livers of wild-type;MX1-Cre and SCAPf/f;MX1-Cre. The mice used in these experiments are described in Table 1. (A,B) Immunoblot analysis of SCAP, SREBP-1, and SREBP-2 in hepatic membranes and nuclear extracts. After treatment of mice with or without pIpC, the livers from the four groups in Table 1 were separately pooled, and aliquots of the membrane fraction (50 μg protein) (A and top gel in B) and nuclear extract fraction (30 μg) (bottom gel in B) were subjected to 6% SDS-PAGE for membranes and 8% SDS-PAGE for nuclear extracts. Immunoblot analysis was performed with 5 μg/mL of rabbit anti-hamster SCAP IgG, rabbit anti-mouse SREBP-1 IgG, or rabbit anti-mouse SREBP-2 IgG as the primary antibody and 0.25 μg/mL horseradish peroxidase-coupled donkey anti-rabbit IgG as the secondary antibody. Filters were exposed to Reflection NEF496 film 15–30 sec at room temperature. (*) A non-specific band. (P and N) The precursor and cleaved nuclear forms of SREBPs, respectively. (C) Amounts of mRNAs for SREBP-1a, SREBP-1c, and SREBP-2, as measured by RNase protection assay. The cRNA probe for SREBP-1 generates protected fragments of 262 bp for SREBP-1a and 168 bp for SREBP-1c. Total RNA isolated from livers of mice (Table 1) was pooled, and 15-μg aliquots were subjected to the RNase protection assay as described in Materials and Methods. After RNase digestion, the protected fragments were separated by gel electrophoresis and exposed to film for 16 h at −80°C, and then quantified. The intensity of each band relative to lane 1 is denoted above the band. (D) Amounts of various mRNAs in livers, as measured by Northern blot hybridization. Total RNA isolated from livers of mice (Table 1) was pooled, and 20-μg aliquots were subjected to electrophoresis and blot hybridization with the indicated 32P-labeled cDNA probe. Filters were exposed to film with intensifying screens at −80°C for 8–24 h, and then quantified. The intensity of each band relative to lane 1 is denoted below the band.
Livers of wild-type;MX1-Cre mice expressed both the precursor (P) and processed nuclear (N) forms of SREBP-1 and SREBP-2 (Fig. 2B, lanes 1,5), and expression was not affected by pIpC injection (Fig. 2B, lanes 2,6). In the absence of pIpC, the livers of the SCAPf/f;MX1-Cre mice expressed normal amounts of SREBP-1 and SREBP-2 (Fig. 2B, lanes 3,7). Injection of pIpC caused a severe reduction in the amounts of both the precursor and the nuclear forms of SREBP-1 and SREBP-2 (Fig. 2B, lanes 4,8). There was also a reduction in the amounts of mRNA encoding SREBP-1c and SREBP-2 as determined by an RNase protection assay (Fig. 2C, lanes 4,8).
Figure 2D shows the mRNAs for a variety of SREBP target genes in liver as measured by Northern blotting. None of these mRNAs was altered when wild-type;MX1-Cre mice were injected with pIpC. When the SCAPf/f; MX1-Cre mice were injected with pIpC, the SCAP mRNA declined by 80%, and there was a severe decline in mRNAs for all of the measured SREBP target genes, including a gene for lipid uptake (LDL receptor), four genes in the cholesterol biosynthetic pathway (encoding HMG CoA synthase, HMG CoA reductase, squalene synthase, and farnesyl diphosphate synthase); three genes required for synthesis of unsaturated fatty acids (encoding acetyl CoA carboxylase, fatty acid synthase, and stearoyl CoA desaturase-1), and a gene required for acetate activation (encoding acetyl CoA synthetase). In contrast, there was no change in four genes that are not regulated by SREBPs, including Site-1 protease (S1P), which processes SREBPs, and three apoproteins of the lipoprotein transport system (apo AI, apo B, and apo E).
Table 1 shows the effects of the pIpC injection on various properties of Cre-expressing wild-type and SCAPf/f mice. pIpC did not change the measured parameters in wild-type;MX1-Cre mice with the exception of a slight increase in plasma triglycerides. In the SCAPf/f;MX1-Cre mice, pIpC produced a significant decrease in the content of cholesterol and triglycerides in liver and in plasma. Free fatty acids also declined in plasma. There was no change in plasma glucose or insulin.
Table 1.
Comparison of wild-type;MX1-Cre mice and SCAPf/f;MX1-Cre mice treated with or without pIpC
| Parameter
|
Wild-type;MX1-Cre
|
SCAPf/f;MX1-Cre
|
||
|---|---|---|---|---|
| PBS
|
pIpC
|
PBS
|
pIpC
|
|
| Number of mice | 3 | 3 | 3 | 3 |
| Body weight (g) | 25 ± 0.9 | 24 ± 2.9 | 25 ± 1.1 | 24 ± 0.7 |
| Liver weight (g) | 1.3 ± 0.04 | 1.3 ± 0.12 | 1.3 ± 0.12 | 1.4 ± 0.11 |
| Liver cholesterol content (mg/g) | 2.4 ± 0.16 | 2.2 ± 0.19 | 2.1 ± 0.09 | 1.7 ± 0.04* |
| Liver triglyceride content (mg/g) | 11 ± 0.88 | 8.2 ± 2.0 | 9.8 ± 2.7 | 3.4 ± 0.1* |
| Total plasma cholesterol (mg/dL) | 86 ± 6 | 73 ± 6 | 79 ± 3 | 60 ± 3* |
| Total plasma triglyceride (mg/dL) | 108 ± 8 | 136 ± 12 | 102 ± 15 | 48 ± 2** |
| Plasma free fatty acid (mM) | 1.4 ± 0.02 | 1.5 ± 0.9 | 1.2 ± 0.05 | 0.77 ± 0.03** |
| Plasma insulin (ng/mL) | 0.5 ± 0.02 | 0.5 ± 0.02 | 0.6 ± 0.08 | 0.5 ± 0.03 |
| Plasma glucose (mg/dL) | 184 ± 13 | 176 ± 14 | 200 ± 16 | 206 ± 11 |
Male mice (7–9 weeks of age) were injected three times intraperitoneally with either PBS or pIpC (250 μg/injection) as described in Materials and Methods. On day 14 after the last injection, the mice were killed, and blood and tissues were obtained for analysis. Each value represents the mean ± sem of three animals. Wild-type mice were littermates of SCAPf/f mice. Asterisks denote the level of statistical significance (student's t-test). *P < 0.05 and **P < 0.01.
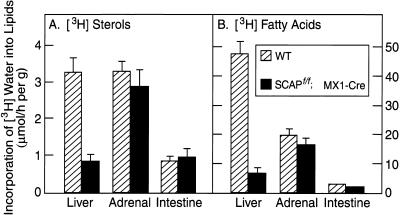
To measure the rates of lipid synthesis, we injected mice with 3H-labeled water intraperitoneally and determined the tissue content of 3H-labeled sterols and 3H-labeled fatty acids 1 h later (Fig. 3). This assay has been shown to reflect the rates of lipid synthesis in various tissues (Dietschy and Spady 1984). Injection of pIpC caused a 71% reduction in the synthesis of cholesterol (Fig. 3A) and an 84% reduction in the synthesis of fatty acids (Fig. 3B) in the liver. There was no change in these synthetic rates in adrenal or intestine. In experiments not shown, we found that pIpC did not decrease SCAP mRNA in the adrenal, kidney, testis, white adipose tissue, or heart.
Figure 3.
Synthesis of cholesterol and fatty acids in liver, adrenal gland, and small intestine in wild-type mice and SCAPf/f; MX1-Cre transgenic mice. Mice (five females per group; 7–9 wk of age) were injected three times intraperitoneally with pIpC (250 μg/injection) as described in Materials and Methods. Fourteen days after the last injection, each mouse was injected intraperitoneally with 3H-labeled water (50 mCi in 0.25 mL of isotonic saline), and 1 h later the indicated tissue was removed for measurement of its content of 3H-labeled fatty acids and digitonin-precipitable sterols as described in Materials and Methods. Bars, mean ± sem of the values from five mice.
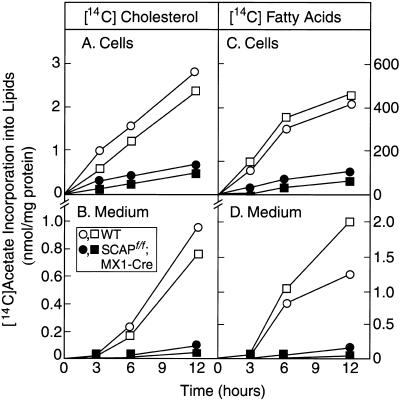
The decrease in cholesterol and fatty acid synthesis in livers of the pIpC-treated SCAPf/f;MX1-Cre mice was also apparent when fresh primary hepatocytes were incubated with 14C-labeled acetate (Fig. 4). The rates of 14C-labeled acetate incorporation into 14C-labeled cholesterol and ;14C-labeled fatty acids were reduced by 84% and 87%, respectively (Fig. 4A,C). There was also a profound reduction in the rates of secretion of these lipids into the culture medium (decreases of 90% and 93%, respectively) (Fig. 4B,D).
Figure 4.
Rates of lipid synthesis and secretion by primary hepatocytes from wild-type mice (□, ○) and SCAPf/f;MX1-Cre transgenic mice (▪, ●). Male mice (6 weeks of age) were injected intraperitoneally with pIpC four times (300 μg/injection) as described in Materials and Methods. Fourteen days after the last injection, primary hepatocytes were prepared as described in Materials and Methods. After a 2-h attachment period, the hepatocytes were incubated with 0.5 mM sodium 14C-labeled acetate (50 dpm/pmole) in DMEM supplemented with 5% human lipoprotein-deficient serum. At the indicated time, the medium was removed, the monolayers were washed, and the cells were harvested. The content of 14C-labeled cholesterol and fatty acids in cells and medium was quantified as described in Material and Methods. Each value is the average of duplicate incubations. Similar results were obtained in three other experiments.
We next carried out studies to determine whether the mRNAs encoding enzymes of lipid synthesis could be up-regulated in response to insulin or cholesterol depletion in livers of SCAP-deficient mice. To study insulin-mediated regulation, we injected mice with pIpC and then subjected them to a 24-h fast (which lowered insulin by 25%–50%), followed by a 12-h period of food intake (which increased insulin by threefold over the fasted level in wild-type;MX1-Cre mice and SCAPf/f; MX1-Cre mice) (Table 2). The liver weight declined with fasting in both mouse lines. The hepatic content of cholesterol and triglyceride was lower in the SCAPf/f;MX1-Cre mice. These lipids increased with fasting in both lines, presumably owing to mobilization from adipose tissue. Plasma cholesterol and triglyceride were low in the SCAPf/f;MX1-Cre mice, but they rose with fasting so that the difference between the wild-type;MX1-Cre mice and SCAPf/f;MX1-Cre mice was minimized in the fasting state. This normalization may be attributable to the mobilization of cholesterol and fatty acids from adipose tissue to liver upon fasting, which partially relieves the lipid deficiency in the livers of the SCAPf/f;MX1-Cre mice.
Table 2.
Effect of fasting and refeeding on wild-type and SCAPf/f;MX1-Cre mice treated with pIpC
| Parameter
|
Wild-type
|
SCAPf/f;MX1-Cre
|
||||
|---|---|---|---|---|---|---|
| nonfasted
|
fasted
|
refed
|
nonfasted
|
fasted
|
refed
|
|
| Number of mice | 4 | 4 | 4 | 4 | 3a | 4 |
| Body weight (g) | 26 ± 0.6 | 23 ± 1.3* | 25 ± 0.6 | 26 ± 1.0 | 22 ± 0.8* | 26 ± 1.0 |
| Liver weight (g) | 1.5 ± 0.10 | 1.1 ± 0.04** | 1.5 ± 0.05 | 1.5 ± 0.07 | 1.0 ± 0.04** | 1.4 ± 0.10 |
| Liver weight/body weight (%) | 5.6 ± 0.3 | 4.8 ± 0.1* | 6.1 ± 0.3 | 5.9 ± 0.1 | 4.6 ± 0.1** | 5.6 ± 0.2 |
| Liver cholesterol content (mg/g) | 1.9 ± 0.08 | 2.9 ± 0.09** | 1.7 ± 0.05 | 1.5 ± 0.06 | 2.0 ± 0.2** | 1.3 ± 0.06 |
| Liver triglyceride content (mg/g) | 5.2 ± 0.4 | 83 ± 15.4** | 20 ± 6.9 | 1.7 ± 0.1 | 25 ± 9.4 | 4.6 ± 1.4 |
| Total plasma cholesterol (mg/dL) | 102 ± 9 | 101 ± 5 | 123 ± 13 | 53 ± 7 | 76 ± 9* | 75 ± 7* |
| Total plasma triglyceride (mg/dL) | 86 ± 7 | 99 ± 19 | 151 ± 15** | 51 ± 9 | 90 ± 17* | 57 ± 10 |
| Plasma insulin (ng/mL) | 0.6 ± 0.2 | 0.3 ± 0.01* | 1.0 ± 0.2** | 0.4 ± 0.02 | 0.3 ± 0.01 | 0.8 ± 0.04** |
| Plasma glucose (mg/dL) | 222 ± 16 | 84 ± 7** | 205 ± 8 | 158 ± 12 | 121 ± 23 | 179 ± 19 |
Male mice (8–10 weeks of age) were injected intraperitoneally three times with pIpC (250 μg/injection) as described in Materials and Methods. On day 14 after the last injection, the mice were subjected to fasting and refeeding as described in Materials and Methods. Each value represents the mean ± sem of 4 mice. Asterisks denote the level of statistical significance compared with the nonfasted group (student's t-test). *P < 0.05 and **P < 0.01.
One animal in this group showed virtually no Cre-mediated recombination of the SCAP gene by Southern blotting of liver genomic DNA. This animal was omitted from the analysis.
Fasting and refeeding did not affect the low levels of SCAP protein in livers of pIpC-injected SCAPf/f;MX1-Cre mice or the normal levels in wild-type;MX1-Cre mice (Fig. 5A). Fasting reduced the amounts of the precursor and nuclear forms of SREBP-1 (Fig. 5B), and the nuclear form of SREBP-2 in the wild-type;MX1-Cre mice (Fig. 5C). As reported previously (Horton et al. 1998a), refeeding caused an overshoot of nuclear SREBP-1 to super-normal levels, and it returned nuclear SREBP-2 to the same levels as in the nonfasted animals (Fig. 5B,C). The precursor and nuclear forms of both SREBPs were barely detectable in the SCAPf/f;MX1-Cre mice under all conditions.
Figure 5.
Expression of mRNAs and proteins in livers of wild-type mice and SCAPf/f;MX1-Cre subjected to fasting and refeeding. The mice used in these experiments are described in Table 2. (A–C) Immunoblot analysis of SCAP, SREBP-1, and SREBP-2 in hepatic membranes and nuclear extracts. After treatment with pIpC, the wild-type and SCAPf/f;MXI-Cre mice were each divided into three groups. The nonfasted group was maintained ad lib, the fasted group was fasted for 24 h, and the refed group was fasted for 24 h and then refed for 12 h prior to study. The treatments were staggered so that all animals could be killed at the same time. Livers from each group were separately pooled, and aliquots of the membrane pellet (50 μg protein) for SCAP, SREBP-1, and SREBP-2 and aliquots of the nuclear extract (30 μg) for SREBPs were subjected to SDS-PAGE and immunoblot analysis as described in the legend to Fig. 2. Filters were exposed to film for 15–30 sec at room temperature. (D) Amounts of various mRNAs in livers, as measured by Northern blot hybridization. Aliquots of pooled total RNA (20 μg) isolated from livers of the indicated mice were subjected to electrophoresis and blot hybridization with the indicated 32P-labeled cDNA probe. Filters were exposed to film with intensifying screens at −80°C for 8–24 h, and then quantified. The intensity of each band relative to lane 1 is shown below the band.
Northern blotting studies of liver mRNA from pIpC-injected wild-type mice showed that fasting reduced the levels of mRNA encoding SREBP-1 and SREBP-2 and all of the target genes (Fig. 5D). Refeeding caused an overshoot to supernormal levels in several of these target genes, most notably HMG CoA synthase, HMG CoA reductase, squalene synthase, and fatty acid synthase.
As expected from the previous experiments, the mRNAs for the SREBPs and all of the target genes were decreased in the ad lib-fed SCAP-deficient mice (Fig. 5D). These mRNAs decreased slightly during fasting and rose slightly during refeeding, but the maximal level was reduced by 3- to 12-fold when compared with the refed wild-type mice.
To test the liver's response to cholesterol deprivation, we performed two experiments (Fig. 6). In one experiment we injected mice with pIpC and then treated them for 10 d with a mixture of the HMG CoA reductase inhibitor lovastatin and the bile acid-binding resin colestipol in the diet (Fig. 6A–C). In wild-type;MX1-Cre mice, the lovastatin/colestipol treatment increased the amount of SCAP protein (Fig. 6A) and raised the nuclear content of SREBP-2 (Fig. 6B, lane 6). Cholesterol deprivation lowered the amount of nuclear SREBP-1 (Fig. 6B, lane 2), a phenomenon that has been attributed to the loss of an endogenous sterol ligand for LXR, a nuclear receptor that is required for high-level transcription of the SREBP-1c gene (Repa et al. 2000; DeBose-Boyd et al. 2001). In the pIpC-injected SCAPf/f;MX1-Cre mice, SCAP protein was not detectable (Fig. 6A, lanes 3,4), the amounts of precursor and nuclear SREBP-1 and SREBP-2 were reduced severely (Fig. 6B, lanes 1,3 and 5,7), and these proteins did not change in response to lovastatin/colestipol (Fig. 6B, lanes 2,4 and 6,8). The mRNAs for the cholesterol-related target genes increased markedly when the wild-type;MX1-Cre mice were treated with lovastatin/colestipol, and there was also a significant increase in the fatty acid synthase mRNA (Fig. 6C). The SCAP-deficient mice exhibited marked reductions in all of these mRNAs. All of them increased significantly with lovastatin/colestipol treatment, but the levels remained well below those seen in the wild-type;MX1-Cre mice.
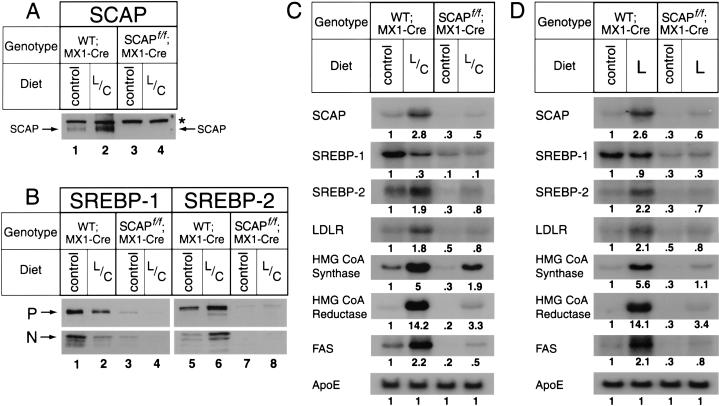
Figure 6.
Expression of mRNAs and proteins in wild-type;MX1-Cre mice and SCAP f/f;MX1-Cre mice fed a diet supplemented with 0.2% lovastatin plus 2% colestipol (L/C) for 10 d (A–C) or 0.2% lovastatin (L) alone for 4 d (D). Mice (9–10 wk of age) were injected intraperitoneally with pIpC four times (300 μg/injection). The dietary treatments were begun immediately after the fourth injection. (A,B) Immunoblot analysis. Livers from five mice in each group (2 males, 3 females) were pooled, and aliquots of the membrane fraction (50 μg protein for SCAP and SREBPs) and nuclear extracts (30 μg) were subjected to SDS-PAGE and immunoblot analysis as described in the legend to Fig. 2. Filters were exposed to film for 15–30 sec at room temperature. (*) A nonspecific band. (P and N) The precursor and cleaved nuclear forms of SREBPs. (C,D) Northern blot analysis. Livers from five mice in each group (2 males, 3 females) were pooled for preparation of total RNA, and 20-μg aliquots were subjected to electrophoresis and blot hybridization with the indicated 32P-labeled cDNA probes as described in Materials and Methods. Filters were exposed to film with intensifying screens at −80°C for 0.5–12 h, and then quantified. The intensity of each band relative to lane 1 is shown below the band.
In another cholesterol-deprivation experiment, mice were treated with lovastatin alone for 4 d (Fig. 6D). The levels of SCAP, SREBP-1, and SREBP-2 proteins were the same as we observed with the lovastatin/colestipol combination (data not shown). In each line of mice, the mRNAs for all of the target genes responded to lovastatin alone (Fig. 6D) in the same way that they responded to the lovastatin/colestipol mixture (Fig. 6C).
Discussion
The current results establish SCAP as a required element for high-level synthesis of cholesterol and fatty acids in mouse liver. Disruption of the SCAP gene in liver led to a profound reduction in the nuclear content of SREBP-1 and SREBP-2. This decrease led to a reduction in the basal levels of mRNAs encoding the crucial enzymes in cholesterol and fatty acid biosynthesis, and this reduction was associated with a decrease in lipid synthetic rates as measured both in vivo and in vitro. The SCAP-deficient livers showed a blunted response to insulin, as indicated by a failure of the lipogenic mRNAs to increase normally in response to a fasting–refeeding protocol. In the SCAP-deficient livers, the cholesterologenic mRNAs failed to show the normal increase in response to cholesterol deprivation as revealed by the experiments with lovastatin and colestipol.
The SCAP-deficient livers showed a decrease in the precursor forms of SREBP-1 and SREBP-2 as well as in the processed nuclear forms (Figs. 2, 5, 6). The decrease in SREBP precursors may have several causes. First, studies in cultured cells have suggested that these precursors are unstable when they cannot form complexes with SCAP (Rawson et al. 1999). Second, SCAP deficiency caused a reduction in the mRNAs encoding SREBP-1c and SREBP-2 (Figs. 2C, 5D, and 6C,D). The enhancer regions of the SREBP-1c and SREBP-2 genes have sterol regulatory element sequences (SREs) that are required for high-level transcription of these genes (Sato et al. 1996; Miserez et al. 1997; Amemiya-Kudo et al. 2000;). When SREBP processing is blocked by SCAP deficiency, nuclear SREBPs decline and the feed-forward stimulation of SREBP gene transcription is disrupted, leading to a fall in SREBP mRNAs. The decrease in SREBP mRNA levels appears to be more profound in SCAP-deficient livers than it is in SCAP-deficient CHO cells (Rawson et al. 1999), suggesting that the feed-forward control of transcription is more important in liver than it is in nonhepatic cells.
Although the lipogenic mRNAs fell 80% on average in the SCAP-deficient livers, they did not decline to zero. Moreover, although the responses to insulin and cholesterol depletion were blunted, there was still a detectable increase (Figs. 5, 6). We estimated that the pIpC treatment reduced SCAP mRNA and protein levels by 90%–95%, apparently owing to the presence of cells in which both copies of the SCAP gene were not disrupted by recombination. We attempted to increase the extent of recombination by giving up to four injections of pIpC at 48-h intervals, but we were never able to achieve total recombination. We cannot rule out the possibility that the residual levels of the lipogenic mRNAs and the blunted but detectable response to refeeding and cholesterol deprivation are attributable to other factors that increase these mRNAs, either by enhancing transcription or by mRNA stabilization.
One interesting aspect of these studies is the apparent good health of the SCAP-deficient mice. Despite marked reductions in hepatic lipogenesis, these animals appeared normal and gained weight normally during the 14-d period following the last pIpC injection. We have followed a small number of these mice for up to 3 mo after pIpC treatment. When the animals were killed, their organs appeared grossly normal. Northern blotting and immunoblotting studies revealed that the hepatic deficiency of SCAP and the nuclear SREBPs persisted, and the levels of the lipogenic mRNAs remained low. The apparent health of these mice is in marked contrast to the early embryonic lethality (approximately post-coitum day 7.5) that was observed when both copies of one SREBP gene (SREBP-2) were disrupted in the germ line by homologous recombination (Shimano et al. 1997a and unpubl. obs.). These findings may suggest that SREBPs are more essential for life in some tissue other than the liver. Alternatively, it is possible that the mechanisms that compensate for hepatic SREBP deficiency in the adult cannot compensate during embryonic life.
Materials and methods
We obtained polyinosinic acid-polycytidylic acid (pIpC, Cat. No P9582) from Sigma (St. Louis, Mo), lovastatin from Merck & Co., Inc., and colestipol from Pharmacia & Upjohn. The plasma concentration of cholesterol, triglycerides, insulin, glucose, and free fatty acids, and liver cholesterol and triglycerides were measured as described previously (Ishibashi et al. 1993; Shimomura et al. 1998; Yokode et al. 1990). Lipoprotein-deficient serum (density > 1.215 g/mL) was prepared by centrifugation (Goldstein et al. 1983). DNA manipulations were performed by standard molecular techniques (Sambrook and Russell 2001).
Cloning of mouse SCAP gene
A mouse genomic SCAP clone was obtained from a 129/SvJ mouse bacterial artificial chromosome library (BAC, Genome Systems Inc, St. Louis, Mo). The library was screened with a 123-bp probe produced by PCR using a 129/Sv mouse genomic DNA as template and the following primers from conserved regions between human and hamster SCAP sequences: 5′ primer, 5′-CCTGACTGAAAGGCTGCGTGAGAAGATATC 3′, and 3′ primer, 5′-GGGTAGCAGCAGGCTAAGATGCA-3′, corresponding to amino acids 2–12 and 36–43 of hamster SCAP, respectively (Hua et al. 1996). The mouse SCAP genomic clone was mapped by sequencing and restriction enzyme analysis.
Construction of targeting vector for conditional disruption of SCAP gene
The strategy for conditional disruption of the SCAP gene involved the insertion of a loxP-flanked pgk-neo-pA cassette (Soriano et al. 1991) 3 kb upstream of the initiator ATG codon of exon 1 of SCAP and the insertion of a downstream loxP sequence within intron 1 (Fig. 1A). Exon 1 encodes the first 41 amino acids of SCAP, including the initiator methionine and residues in the first transmembrane domain (Nohturfft et al. 1998). The conditional targeting vector was constructed in four steps as follows: (1) A 3.1-kb BglII–HindIII (blunt end) fragment, which is 3 kb upstream of the initiator ATG codon, and a 2.1-kb SalI–BamHI (blunt end) fragment of pFlox (Rohlmann et al. 1996) containing the pgk-neo-pA cassette were ligated into the SalI–BamHI site of pBluescript II KS (Stratagene, La Jolla, CA). The resulting plasmid was designated pBK-KO3. (2) A 3.4-kb SalI–HindIII (blunt end) fragment containing exon 1 from the mouse SCAP gene was ligated into the SalI–XbaI (blunt end) site of pBK-KO3. The resulting plasmid was designated pBK-KO4. (3) A pair of oppositely oriented oligonucleotides containing a ClaI site, loxP sequence, and BglII site, respectively (5′ primer, 5′-TCGACAGATCTATAACTTCGTATAGCATACATTATACG AAGTTATATCGATC-3′, and 3′ primer, 5′-TCGAGATCGA TATAACTTCGTATAATGTATGCTATACGAAGTTATAGA TCTG-3′) were annealed and ligated into the SalI site of pBK-KO4. The resulting plasmid was designated pBK-KO5. (4) A 9.7-kb SalI–EcoRI fragment, which contains intron 1 of mouse SCAP, was ligated into the SalI–EcoRI site of pBK-KO5 to generate the SCAP conditional targeting vector. The lengths of the short and long arm were 3.1 and 9.7 kb, respectively. The transcriptional orientation of the neo gene was opposite to that of the SCAP gene. Restriction analysis and DNA sequencing of all ligation junctions confirmed the integrity of all plasmids.
ES cell culture for disruption of SCAP gene
Low passage (P8) SM-1 ES cells, derived from 129Sv/Ev blastocysts (Shimano et al. 1997b), were cultured on leukemia inhibitory factor-producing STO feeder cells (Ishibashi et al. 1993). On day 0, a total of 2 × 107 cells were transfected by electroporation (275 V, 330 μF, low resistance; GIBCO BRL Electroporator; Life Technologies, Gaithersburg, MD) with 7 μg of NotI-linearized targeting vector and seeded onto irradiated STO feeder cells. On day 2, ES cells were subjected to selection with 250 μg/mL of G418 (Life Technologies). G418-resistant clones were isolated on day 8, and recombinant clones were identified by PCR and Southern blotting.
Generation of SCAPf/f mice
An ES clone containing a single SCAPflox allele was established and injected into C57BL/6J blastocysts, yielding chimeric males whose coat color (agouti) indicated a contribution of ES cells ranging from 25–100%. All 14 chimeric males whose coat color was > 90% agouti were fertile, and 3 produced offspring that carried the SCAPflox allele through the germ line.
Mice homozygous for the SCAPflox allele (referred to as SCAPf/f mice) were bred with MX1-Cre transgenic mice (Rohlmann et al. 1998) to produce SCAPf/f;MX1-Cre mice. Cre expression was induced by intraperitoneal injection of the interferon-inducer, polyinosinic-polycytidylic acid (pIpC) (Kuhn et al. 1995; Rohlmann et al, 1998). Each mouse received three or four injections of 250 or 300 μL of a 1 mg/mL solution of pIpC in water administered every 48 h. Mice were analyzed 10 or 14 d after the final injection of pIpC.
Diet studies
For the fasting and refeeding experiments, mice from each genotype were divided into three treatment groups. The nonfasted group was fed ad lib, the fasted group was fasted for 24 h, and the refed group was fasted for 24 h and then refed for 12 h prior to study. For the lovastatin/colestipol diet studies, mice of each genotype were fed for 10 d with a chow diet supplemented with 0.2% (wt/wt) lovastatin plus 2% (wt/wt) colestipol. Another group of mice received lovastatin alone (0.2% wt/wt) for 4 d as indicated in the legend to Fig. 6.
All mice were housed in colony cages and maintained on a 12-h light/12-h dark cycle and fed Teklad Mouse/Rat Diet #7002 from Harlan Teklad Premier Laboratory Diets (Madison, WI).
Immunoblotting
Nuclear extract and membrane fractions were prepared from mouse livers as described previously (Sheng et al. 1995). Immunoblot analyses of mouse SREBP-1 (Shimano et al. 1996), SREBP-2 (Shimano et al. 1997b), and SCAP (Rawson et al. 1999) were carried out as described in the indicated reference.
Blot hybridization of RNA
All cDNA probes for Northern blot analysis have been described previously (Shimano et al. 1996; Shimomura et al., 1998, 1999a, 2000; Luong et al. 2000). The livers from three to five mice were pooled, and total RNA was prepared with an RNA STAT-60 kit (TEL-TEST&“B”. Inc., Friendswood, TX). Aliquots (20 μg) of total RNA were subjected to electrophoresis in a 1% formaldehyde agarose gel and transferred to Hybond N+ membranes (Amersham). cDNA probe preparation, hybridization conditions, and quantification of bands were carried out as described (Shimomura et al. 1998).
RNase protection assay
The RNase protection assay was performed as described (Shimomura et al. 1999b). Aliquots of total RNA (15 μg) from each pooled liver sample were subjected to the RNase protection assay with a HybSpeed RPA kit (Ambion, Inc.). Each assay tube contained a cRNA probe for the mRNA of SREBP-1a plus a cRNA probe for 18S RNA. In preparing the probes, we adjusted the specific activities of the [α-32P]CTP individually to give an 18S RNA signal comparable with the signal for SREBP mRNAs. After digestion with RNase A/T1, protected fragments were separated on 8 M urea/4.8% polyacrylamide gels, and the gels were dried and subjected to autoradiography by use of reflection film and intensifying screens. The protected fragments corresponding to mouse SREBP-1a (262 bp), and SREBP-1c (168 bp) were visualized on gels at −80°C and quantified as described previously (Shimomura et al. 1998).
Cholesterol and fatty acid synthesis in vivo
The amounts of cholesterol and fatty acid synthesized in liver, adrenal gland, and small intestine were measured 1 h after intraperitoneal injection of 3H-labeled water as described previously (Shimano et al. 1996). The rates of hepatic cholesterol and fatty acid synthesis were calculated as μM of 3H-labeled water incorporated into fatty acids or digitonin-precipitable sterols/h per gram of tissue.
Primary culture of mouse hepatocytes
Primary mouse hepatocytes were prepared as described previously for rat hepatocytes (Shimomura et al. 1999b, 2000) with several modifications. Mice were anesthetized, and livers were perfused with 30 mL of Liver Perfusion Medium (GIBCO BRL) and then with 30 mL of Liver Digestion Medium (GIBCO BRL) at a rate of 3 mL/min via the portal vein. The temperature of the perfusion was 37°C. The livers were removed, the hepatic capsule was peeled off, and hepatocytes were dispersed by shaking the digested liver in Liver Digestion Medium at 37°C, followed by filtration through gauze. Digestion was stopped by addition of an equal volume of ice-cold Dulbecco's modified Eagle medium (DMEM) containing 5% (v/v) newborn calf lipoprotein-deficient serum, 10 mM Hepes (pH 7.4), 100 units/mL penicillin G sodium, and 100 μg/mL streptomycin sulfate. The cells were washed twice and resuspended in the same medium. Cell viability, determined by trypan blue dye exclusion, was 90%–95%. The cells were plated on 60- or 35-mm dishes coated with mouse type IV collagen in the above medium at a density of 1.5 × 106 cells or 0.5 × 106 cells per dish, respectively. After incubation for 2 h (period of attachment), the cells were used for experiments.
Lipid synthesis and secretion in mouse primary hepatocytes
Lipid synthesis and secretion in hepatocytes were measured as described previously (Horton et al. 1999). After attachment, the hepatocytes were incubated with DMEM containing 5% newborn calf lipoprotein-deficient serum, 100 units/mL penicillin G sodium, 100 μg/mL streptomycin sulfate, and 0.5 mM sodium 14C-labeled acetate (50 dpm/pmole). After incubation for the indicated time, the medium was removed, and cells were harvested. After saponification of the cells and medium, the cholesterol and fatty acids were extracted and separated by thin-layer chromatography. The bands were excised, and 14C-labeled acetate incorporated into cholesterol and fatty acids were counted in a liquid scintillation counter. The results are expressed as nanomoles of 14C-labeled acetate incorporated into cholesterol and fatty acids per milligram of cell protein. Cell protein content was measured with a BCA Kit (Pierce).
Acknowledgments
We thank Judy Webber, Janet Johnstone, Scott Clark, Liz Lummus, and Richard Gibson for excellent technical assistance. We also thank Drs. Yuriy Bashmakov and Jiafu Ou for reagents and helpful suggestions and Dr. Stephen Turley for help with the 3H-labeled water studies. B.S.K. was supported by Medical Scientist Training Grant GM08014. R.K. is the recipient of a 2000 Banyu Fellowship Award in Cardiovascular Medicine from The Merck Co. Foundation. J.D.H. is a Pew Scholar in the Biomedical Sciences and is the recipient of an Established Investigator Grant from the American Heart Association. This work was supported by grants from the National Institutes of Health (HL20948), the Moss Heart Foundation, the Perot Family Foundation, and the W.M. Keck Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.891301.
References
- Amemiya-Kudo M, Shimano H, Yoshikawa T, Yahagi N, Hasty AH, Okazaki H, Tamura Y, Shionoiri F, Iizuka Y, Ohashi K, et al. Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. J Biol Chem. 2000;275:31078–31085. doi: 10.1074/jbc.M005353200. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- ————— The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- ————— A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBose-Boyd RA, Brown MS, Li W-P, Nohturfft A, Goldstein JL, Espenshade PJ. Transport-dependent proteolysis of SREBP: Relocation of Site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell. 1999;99:703–712. doi: 10.1016/s0092-8674(00)81668-2. [DOI] [PubMed] [Google Scholar]
- DeBose-Boyd RA, Ou J, Goldstein JL, Brown MS. Expression of sterol regulatory element-binding protein 1c (SREBP-1c) mRNA in rat hepatoma cells requires endogenous LXR ligands. Proc Natl Acad Sci. 2001;98:1477–1482. doi: 10.1073/pnas.98.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietschy JM, Spady DK. Measurement of rates of cholesterol synthesis using tritiated water. J Lipid Res. 1984;25:1469–1476. [PubMed] [Google Scholar]
- Edwards PA, Tabor D, Kast HR, Venkateswaran A. Regulation of gene expression by SREBP and SCAP. Biochim Biophys Acta. 2000;1529:103–113. doi: 10.1016/s1388-1981(00)00140-2. [DOI] [PubMed] [Google Scholar]
- Farese RV, Jr, Herz J. Cholesterol metabolism and embryogenesis. Trends Genet. 1998;14:115–120. doi: 10.1016/s0168-9525(97)01377-2. [DOI] [PubMed] [Google Scholar]
- Foretz M, Guichard C, Ferre P, Foufelle F. Sterol regulatory element binding protein-1c is a major mediator of insulin action on the hepatic expression of glucokinase and lipogenesis-related genes. Proc Natl Acad Sci. 1999;96:12737–12742. doi: 10.1073/pnas.96.22.12737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, Basu SK, Brown MS. Receptor-mediated endocytosis of LDL in cultured cells. Meth Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- Horton JD, Shimomura I. Sterol regulatory element-binding proteins: Activators of cholesterol and fatty acid biosynthesis. Curr Opin Lipidol. 1999;10:143–150. doi: 10.1097/00041433-199904000-00008. [DOI] [PubMed] [Google Scholar]
- Horton JD, Bashmakov Y, Shimomura I, Shimano H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc Natl Acad Sci. 1998a;95:5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Shimomura I, Brown MS, Hammer RE, Goldstein JL, Shimano H. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J Clin Invest. 1998b;101:2331–2339. doi: 10.1172/JCI2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Shimano H, Hamilton RL, Brown MS, Goldstein JL. Disruption of LDL receptor gene in transgenic SREBP-1a mice unmasks hyperlipidemia resulting from production of lipid-rich VLDL. J Clin Invest. 1999;103:1067–1076. doi: 10.1172/JCI6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Nohturfft A, Goldstein JL, Brown MS. Sterol resistance in CHO cells traced to point mutation in SREBP cleavage activating protein (SCAP) Cell. 1996;87:415–426. doi: 10.1016/s0092-8674(00)81362-8. [DOI] [PubMed] [Google Scholar]
- Ishibashi S, Brown MS, Goldstein JL, Gerard RD, Hammer RE, Herz J. Hypercholesterolemia in LDL receptor knockout mice and its reversal by adenovirus-mediated gene delivery. J Clin Invest. 1993;92:883–893. doi: 10.1172/JCI116663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Spiegelman BM. ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes & Dev. 1996;10:1096–1107. doi: 10.1101/gad.10.9.1096. [DOI] [PubMed] [Google Scholar]
- Korn BS, Shimomura I, Bashmakov Y, Hammer RE, Horton JD, Goldstein JL, Brown MS. Blunted feedback suppression of SREBP processing by dietary cholesterol in transgenic mice expressing sterol-resistant SCAP(D443N) J Clin Invest. 1998;102:2050–2060. doi: 10.1172/JCI5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Luong A, Hannah VC, Brown MS, Goldstein JL. Molecular characterization of human acetyl CoA synthetase, an enzyme regulated by SREBPs. J Biol Chem. 2000;275:26458–26466. doi: 10.1074/jbc.M004160200. [DOI] [PubMed] [Google Scholar]
- McGarry JD. Glucose-fatty acid interactions in health and disease. Am J Clin Nutr. 1998;67:500S–504S. doi: 10.1093/ajcn/67.3.500S. [DOI] [PubMed] [Google Scholar]
- Miserez AR, Cao G, Probst L, Hobbs HH. Structure of the human gene encoding sterol regulatory element binding protein 2 (SREBF2) Genomics. 1997;40:31–40. doi: 10.1006/geno.1996.4525. [DOI] [PubMed] [Google Scholar]
- Nohturfft A, Hua X, Brown MS, Goldstein JL. Recurrent G-to-A substitution in a single codon of SREBP cleavage-activating protein causes sterol resistance in three mutant CHO cell lines. Proc Natl Acad Sci. 1996;93:13709–13714. doi: 10.1073/pnas.93.24.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohturfft A, Brown MS, Goldstein JL. Topology of SREBP cleavage-activating protein, a polytopic membrane protein with a sterol-sensing domain. J Biol Chem. 1998;273:17243–17250. doi: 10.1074/jbc.273.27.17243. [DOI] [PubMed] [Google Scholar]
- Nohturfft A, DeBose-Boyd RA, Scheek S, Goldstein JL, Brown MS. Sterols regulate cycling of SREBP cleavage-activating protein (SCAP) between endoplasmic reticulum and Golgi. Proc Natl Acad Sci. 1999;96:11235–11240. doi: 10.1073/pnas.96.20.11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohturfft A, Yabe D, Goldstein JL, Brown MS, Espenshade PJ. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell. 2000;102:315–323. doi: 10.1016/s0092-8674(00)00037-4. [DOI] [PubMed] [Google Scholar]
- Rawson RB, DeBose-Boyd R, Goldstein JL, Brown MS. Failure to cleave sterol regulatory element-binding proteins (SREBPs) causes cholesterol auxotrophy in Chinese hamster ovary cells with genetic absence of SREBP cleavage-activating protein. J Biol Chem. 1999;274:28549–28556. doi: 10.1074/jbc.274.40.28549. [DOI] [PubMed] [Google Scholar]
- Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro J-MA, Shimomura I, Shan B, Brown MS, Goldstein JL, Mangelsdorf DJ. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRα and LXRβ. Genes & Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlmann A, Gotthardt M, Willnow TE, Hammer RE, Herz J. Sustained somatic gene inactivation by viral transfer of Cre recombinase. Nature Biotechnol. 1996;14:1562–1565. doi: 10.1038/nbt1196-1562. [DOI] [PubMed] [Google Scholar]
- Rohlmann A, Gotthardt M, Hammer RE, Herz J. Inducible inactivation of hepatic LRP gene by cre-mediated recombination confirms role of LRP in clearance of chylomicron remnants. J Clin Invest. 1998;101:689–695. doi: 10.1172/JCI1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, Spiegelman BM. Molecular regulation of adipogenesis. Annu Rev Cell Dev Biol. 2000;16:145–171. doi: 10.1146/annurev.cellbio.16.1.145. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sato R, Inoue J, Kawabe Y, Kodama T, Takano T, Maeda M. Sterol-dependent transcriptional regulation of sterol regulatory element-binding protein-2. J Biol Chem. 1996;271:26461–26464. doi: 10.1074/jbc.271.43.26461. [DOI] [PubMed] [Google Scholar]
- Schwarz M, Wright AC, Davis DL, Nazer H, Bjorkhem I, Russell DW. Expression cloning of 3β-hydroxy-Δ5-C27-steroid oxidoreductase gene of bile acid synthesis and its mutation in progressive intraheptic cholestasis. J Clin Invest. 2000;106:1175–1184. doi: 10.1172/JCI10902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Z, Otani H, Brown MS, Goldstein JL. Independent regulation of sterol regulatory element binding proteins 1 and 2 in hamster liver. Proc Natl Acad Sci. 1995;92:935–938. doi: 10.1073/pnas.92.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano H, Horton JD, Hammer RE, Shimomura I, Brown MS, Goldstein JL. Overproduction of cholesterol and fatty acids causes massive liver enlargement in transgenic mice expressing truncated SREBP-1a. J Clin Invest. 1996;98:1575–1584. doi: 10.1172/JCI118951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano H, Horton JD, Shimomura I, Hammer RE, Brown MS, Goldstein JL. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J Clin Invest. 1997a;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano H, Shimomura I, Hammer RE, Herz J, Goldstein JL, Brown MS, Horton JD. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Invest. 1997b;100:2115–2124. doi: 10.1172/JCI119746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura I, Shimano H, Horton JD, Goldstein JL, Brown MS. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J Clin Invest. 1997;99:838–845. doi: 10.1172/JCI119247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura I, Hammer RE, Richardson JA, Ikemoto S, Bashmakov Y, Goldstein JL, Brown MS. Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: Model for congenital generalized lipodystrophy. Genes & Dev. 1998;12:3182–3194. doi: 10.1101/gad.12.20.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura I, Bashmakov Y, Horton JD. Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J Biol Chem. 1999a;274:30028–30032. doi: 10.1074/jbc.274.42.30028. [DOI] [PubMed] [Google Scholar]
- Shimomura I, Bashmakov Y, Ikemoto S, Horton JD, Brown MS, Goldstein JL. Insulin selectively increases SREBP-1c mRNA in livers of rats with streptozotocin-induced diabetes. Proc Natl Acad Sci. 1999b;96:13656–13661. doi: 10.1073/pnas.96.24.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura I, Matsuda M, Hammer RE, Bashmakov Y, Brown MS, Goldstein JL. Decreased IRS-2 and increased SREBP-1c lead to mixed insulin resistance and sensitivity in livers of lipodystrophic and ob/ob mice. Mol Cell. 2000;6:77–86. [PubMed] [Google Scholar]
- Soriano P, Montgomery C, Geske R, Bradley A. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- Wang X, Sato R, Brown MS, Hua X, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- Witztum JL. Goodman & Gilman's the pharmacological basis of therapeutics. New York, NY: McGraw-Hill; 1996. Drugs used in the treatment of hyperlipoproteinemias; pp. 875–897. [Google Scholar]
- Yokode M, Hammer RE, Ishibashi S, Brown MS, Goldstein JL. Diet-induced hypercholesterolemia in mice: Prevention by overexpression of LDL receptors. Science. 1990;250:1273–1275. doi: 10.1126/science.2244210. [DOI] [PubMed] [Google Scholar]