Abstract
The Tap protein mediates the sequence-specific nuclear export of mRNAs bearing the retroviral constitutive transport element (CTE) and also plays a critical role in the sequence nonspecific export of cellular mRNAs. Previously, we have demonstrated that CTE function displays species specificity, that is, the CTE functions in human but not quail cells. Here, we demonstrate that quail Tap fails to support CTE function because it cannot bind the CTE. However, changing a single residue in quail Tap, glutamine 246, to arginine, the residue found in human Tap, rescues both CTE function and CTE binding. This residue, which is located on the exterior of a recently reported molecular structure of Tap, defines a surface on Tap that is critical for CTE binding. These data emphasize the potential importance of cross-species genetic complementation in the identification and characterization of cellular factors that are critical for different aspects of viral replication.
Keywords: Gene regulation, nuclear export, retrovirus, RNA binding
Retroviral replication requires the nuclear export and cytoplasmic translation of both unspliced and spliced forms of the initial, genome-length retroviral transcript (for review, see Cullen 2000). However, cells have developed surveillance mechanisms that prevent the inappropriate export of incompletely spliced cellular mRNAs, that is, pre-mRNAs, and these same mechanisms also have the potential to block the nuclear export of intron-containing retroviral mRNAs. Retroviruses have developed at least two mechanisms to deal with this problem. Thus, human immunodeficiency virus type 1 (HIV-1) encodes a regulatory protein, termed Rev, that recruits the cellular nuclear export factor Crm1 to a cis-acting RNA target present in all incompletely spliced viral mRNAs (Malim et al. 1989b; Fornerod et al. 1997; Neville et al. 1997; Stade et al. 1997). Surprisingly, Crm1 is not believed to play a direct role in cellular mRNA export (Fischer et al. 1995; Bogerd et al. 1998; Neville and Rosbash 1999) and, instead, normally acts to export snRNAs and a range of proteins from the nucleus, in the latter case by binding to a short leucine-rich consensus sequence that also functions as a nuclear export signal (NES) (Malim et al. 1991; Fischer et al. 1995).
Although simple retroviruses, by definition, do not encode regulatory proteins equivalent to Rev they face the same problem of how to export incompletely spliced viral mRNAs out of the nucleus in the face of cellular pre-mRNA retention mechanisms. In the case of the simian type D viruses, this problem has been solved by evolution of a constitutive transport element (CTE), a cis-acting RNA sequence that activates unspliced viral RNA export independently of any viral gene product (Bray et al. 1994). The cellular cofactor for the CTE, termed Tap or NXF1, was subsequently identified based on its ability to bind the CTE found in the retrovirus Mason-Pfizer Monkey Virus (MPMV) with high affinity in vitro (Grüter et al. 1998).
Two lines of evidence suggest that Tap also plays a key role in the sequence-nonspecific export of most, or possibly all, mRNA molecules from the eukaryotic nucleus. First, microinjection of excess levels of the MPMV CTE into Xenopus oocyte nuclei specifically blocks the nuclear export of not only CTE-containing RNAs but also of coinjected cellular mRNAs. In contrast, nuclear export of other classes of RNA, such as snRNA and tRNA, remains unaffected (Pasquinelli et al. 1997; Saavedra et al. 1997). Importantly, nuclear export of both CTE-containing and cellular mRNAs could be rescued by injection of recombinant Tap protein, implying that Tap plays a key role in this process (Grüter et al. 1998). A second line of evidence supporting a critical role for Tap in nuclear mRNA export comes from the finding that the yeast homolog of Tap, termed Mex67p, is essential for global poly(A)+ RNA export in yeast cells (Segref et al. 1997). Remarkably, the viability of Mex67p-deficient yeast cells could be rescued by expression of human Tap (hTap) together with a proposed hTap cofactor termed p15–1 or NXT-1 (Katahira et al. 1999). These data imply that the critical importance of Tap in mediating nuclear mRNA export has been conserved through much of eukaryotic evolution.
An interesting aspect of the MPMV CTE is that this RNA element fails to support nuclear RNA export in quail cells (Kang and Cullen 1999). However, CTE function can be rescued by expression of hTap in trans. This observation, together with in vitro experiments performed in parallel, has greatly facilitated the identification and characterization of functional domains within hTap. Specifically, the 619-amino-acid hTap protein is now known to contain a domain that mediates CTE binding (approximately amino acids 96–372), a central domain that recruits the p15–1 cofactor (∼370–490), and an essential carboxy-terminal domain that directly interacts with several nucleoporins and that also functions as an NES (∼550–619) (Braun et al. 1999; Kang and Cullen 1999; Katahira et al. 1999). Recently, an X-ray crystallographic analysis of the molecular structure of the CTE-binding domain of hTap has been presented (Liker et al. 2000). This region of hTap was shown to consist of a noncanonical ribonucleoprotein (RNP) domain linked to an adjacent leucine-rich repeat (LRR) domain, both of which are required for CTE binding.
In this paper, we have defined the molecular basis for the lack of MPMV CTE function in quail cells. Using a series of in vivo and in vitro assays, we demonstrate that the MPMV CTE fails to function in quail cells because quail Tap (qTap) cannot bind to the CTE. However, both CTE binding by qTap and CTE function in quail cells can be completely rescued by changing a single residue in qTap, glutamine 246, to arginine, the residue found at the equivalent position in hTap. Although this arginine residue is located on the surface of the Tap LRR domain structure described recently by Liker et al. (2000), its location is inconsistent with their proposal that CTE binding by Tap is structurally comparable to U2 snRNA binding by the U2B"–U2A‘ heterodimer. Instead, this natural sequence variant defines an RNA binding surface on Tap that predicts a novel structure for the hTap:CTE complex.
Results
Molecular cloning of a cDNA encoding quail Tap
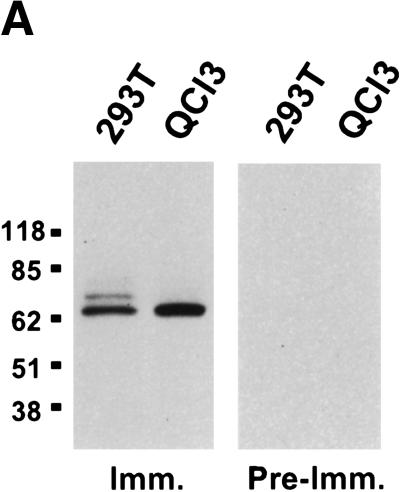
Homologs of the hTap nuclear RNA export factor have been reported in a range of species, including fruit flies, nematodes, and fungi (Katahira et al. 1999; Kang et al. 2000; Tan et al. 2000). Although it seemed improbable that the inability of the MPMV CTE to function in quail cells would result from the lack of a Tap homolog in these cells, we nevertheless wished to confirm that quail cells do express a protein related to hTap. Using a rabbit polyclonal antiserum directed against residues 61–372 of hTap, we performed a Western blot analysis of proteins extracted from an equivalent number of human (293T) or quail (QCl-3) cells. As shown in Figure 1A, this anti-hTap antiserum detected a single protein band in the QCl-3 cells that comigrated with the major band detected in human cells. We therefore conclude that quail cells express a protein that is closely related in size and probably also amino acid sequence to hTap.
Figure 1.
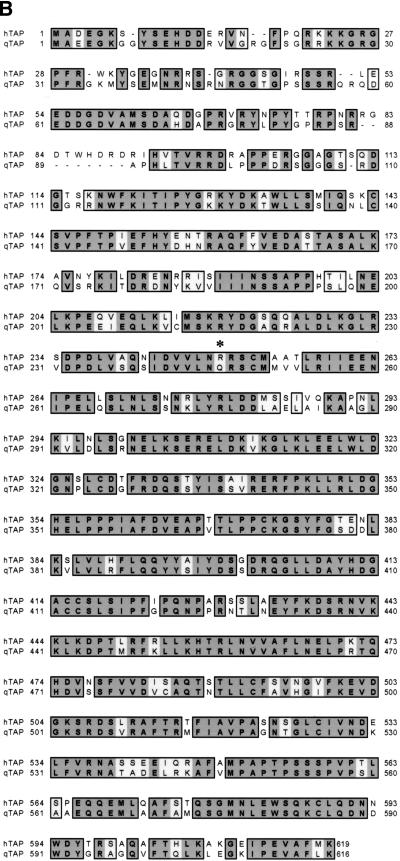
Identification of the qTap gene product. (A) Total cellular proteins, prepared from human 293T or quail QCl-3 cells, were resolved on a 4%–15% acrylamide/SDS gel, and blotted to nitrocellulose. Membranes were probed with pre-immune sera (Pre-Imm.) or with polyclonal sera raised in the same rabbit against bacterially expressed GST–hTap (61–372) (Imm.). Reactive proteins were visualized by enhanced chemiluminescence. (B) Amino acid sequence comparison of hTap and qTap. Identical and similar amino acids are indicated. A crucial arginine residue (R249) required for CTE function is denoted by an asterisk. The sequence of qTap has been deposited into the DDBJ/EMBL/GenBank database (accession no. AF343749).
To molecularly clone qTap we prepared a phage λ cDNA library using poly(A)+ mRNA derived from the QCl-3 cell line and then recovered part of the qTap cDNA sequence by PCR amplification using primers complementary to tap sequences conserved in humans and fruit flies. A 32P-labeled form of the resultant PCR generated qTap DNA fragment was then used to probe the quail cDNA library, resulting in the recovery of several full-length or partial cDNA copies of the qTap gene. In Figure 1B, the predicted amino acid sequence of the qTap protein encoded by the complete qTap cDNA sequence is aligned with the hTap protein. The qTap protein is predicted to be 75% identical and 85% similar to hTap and is only three amino acids shorter than the 619-amino-acid hTap sequence. The greatest sequence divergence, including several small gaps and insertions, was found within the amino-terminal ∼90 amino acids of Tap, which are known to be largely dispensable for hTap-mediated export of mRNAs bearing the MPMV CTE (Braun et al. 1999; Kang and Cullen 1999). Known functional domains in hTap, including the MPMV CTE-binding domain (∼96–372), the p15–1 cofactor-binding domain (∼370–490), and the NES/nucleoporin-binding sequence (∼550–619) are in contrast well conserved (Kang and Cullen 1999; Katahira et al. 1999; Bachi et al. 2000; Kang et al. 2000; Liker et al. 2000).
The quail Tap protein can support sequence nonspecific nuclear mRNA export
The hTap protein is believed to play a critical role in not only the sequence-specific nuclear export of incompletely spliced MPMV mRNAs bearing the viral CTE but also in the sequence-nonspecific export of global poly(A)+ mRNAs (Pasquinelli et al. 1997; Saavedra et al. 1997; Katahira et al. 1999). Although the importance of hTap in the former export pathway can be readily demonstrated by functional complementation in quail cells or frog oocytes (Grüter et al. 1998; Kang and Cullen 1999), demonstration of the latter activity of Tap is difficult in metazoan cells. However, it has been demonstrated that hTap, together with its cofactor p15–1, can rescue the viability of yeast Saccharomyces cerevisiae cells lacking the essential yeast Tap homolog Mex67p and/or the proposed yeast p15–1 homolog Mtr2p, presumably by rescuing nuclear mRNA export (Katahira et al. 1999). We therefore wished to test whether qTap would also support nuclear mRNA export in Mex67p- and/or Mtr2p-deficient yeast cells.
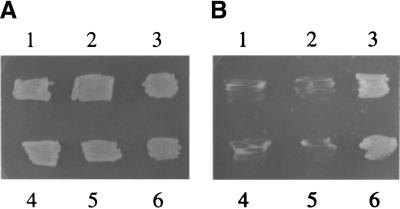
The experiment shown in Figure 2, A and B, uses a previously reported yeast strain (Katahira et al. 1999) that lacks a functional genomic mex67 gene but is complemented by wild-type mex67 on a plasmid bearing the ura marker. This marker also allows growth on uracil-deficient media but is toxic to yeast on media containing 5-fluoroorotic acid (5-FOA). Complementation by metazoan tap and/or p15–1 genes can be tested by introducing one or both of these genes on plasmids that encode the selectable markers leu or trp, followed by growth on plates containing 5-FOA, that is, under conditions that select for loss of the complementing ura plasmid encoding Mex67p.
Figure 2.
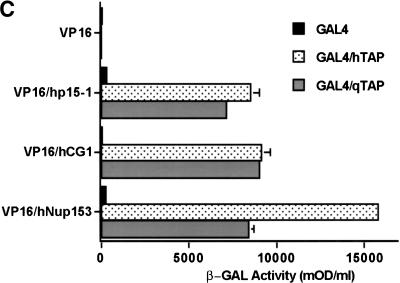
The qTap protein can support sequence nonspecific nuclear mRNA export. (A,B) Complementation of a yeast mex67− null strain by coexpression of hTap(61–619) or qTap and human p15–1. The yeast strain mex67::HIS3, complemented with plasmid pURA3–MEX67, was cotransformed with the following plasmids: (1) pPGK–TRP/hTap(61–619) and pPGK–LEU; (2) pPGK–TRP and pPGK–LEU/p15–1; (3) pPGK–TRP/hTap(61–619) and pPGK–LEU/p15–1; (4) pPGK–TRP and pPGK–LEU; (5) pPGK–TRP/qTap and pPGK–LEU; (6) pPGK–TRP/qTap and pPGK–LEU/p15–1. Transformants were grown on SDC-leu-trp-ura for 3 days and then transferred to SDC-trp-leu plates (A) or SDC-trp-leu plates containing 5-FOA (B). (C) The ability of qTap to bind to human p15–1 and to the FG-repeat regions of two human nucleoporins, CG1 and Nup153, was measured by yeast two-hybrid analysis. hTap and qTap proteins were expressed as GAL4 DNA-binding domain fusions, while p15–1, hCG1 and Nup153 were expressed fused to the VP16 transcription activation domain. After selection for transformants, induced β-gal activity was measured as described (Bogerd et al. 1993). Results shown are the average of 3 experiments and are expressed as mOD (milli-optical-density units)/mL.
As shown in Figure 2B, and as reported previously (Katahira et al. 1999), yeast cells lacking Mex67p were not viable (B4) and this defect could not be complemented by expression of hTap (B1) or human p15–1 (B2) alone. However, expression of both hTap and p15–1 rescued viability and growth (B3). Similarly, expression of only qTap did not rescue the viability of Mex67p-deficient yeast (B5), but expression of full-length qTap together with human p15–1 did rescue viability (B6). The lack of growth of four of the transformants analyzed in Figure 2B did not result from an intrinsic loss of viability, as all transformants were viable on plates that selected for trp, leu, and ura (Fig. 2A). We therefore conclude that qTap can rescue the mRNA export defect present in Mex67p-deficient yeast cells but only when coexpressed with human p15–1. Exactly the same pattern of complementation was observed upon transformation of Mtr2p deficient yeast (data not shown).
The data presented in Figure 2B imply that qTap must be able to functionally interact with human p15–1 and also suggest that qTap can target yeast mRNA molecules to the nuclear pore complex, that is, that qTap, like hTap, should be able to interact with specific nucleoporins. To test this hypothesis, we performed a yeast two-hybrid analysis (Fields and Song 1989) comparing the ability of hTap and qTap to bind to human p15–1 and to the human nucleoporins CG1 and Nup153, both known targets for hTap (Bachi et al. 1999; Katahira et al. 1999). As shown in Figure 2C, qTap and hTap were both found to bind p15–1, CG1 and Nup153 specifically in this assay.
A single amino acid change in qTap rescues CTE-dependent nuclear mRNA export
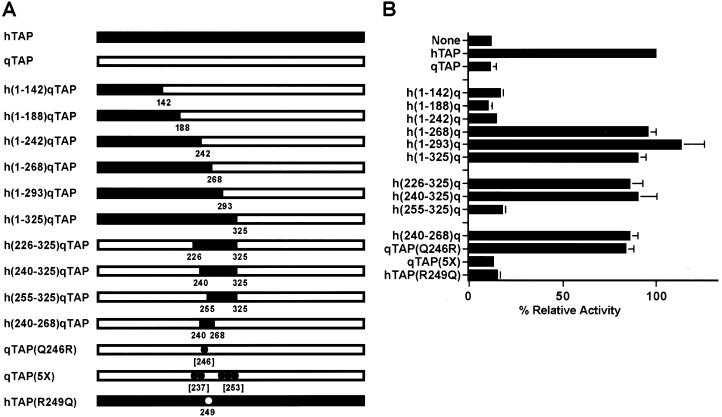
The extensive sequence similarity between hTap and qTap (Fig. 1B), together with evidence suggesting that qTap can support sequence nonspecific nuclear RNA export (Fig. 2), suggested that the inability of qTap to support MPMV CTE function might be due to only one or a small number of sequence differences with hTap. To define the sequences within qTap that are responsible for this phenotypic difference with hTap, we constructed a set of hTap/qTap chimeras, some of which are presented in Figure 3A and then measured their ability to rescue MPMV CTE function in transfected quail cells (Fig. 3B). The assay used to measure CTE function utilizes the previously described indicator plasmid pDM128/CTE (Hope et al. 1990; Malim et al. 1991; McDonald et al. 1992; Bogerd et al. 1998; Kang and Cullen 1999). This plasmid contains a cat gene and the MPMV CTE, flanked by 5′ and 3′ splice sites, under the transcriptional control of the cytomegalovirus immediate early promoter. Expression of the cat indicator gene requires the nuclear export of an unspliced cat mRNA and this only occurs effectively when the CTE is functional.
Figure 3.
A single amino acid change in qTap rescues CTE function. (A) Schematic diagram of the various human/quail chimeras and mutant Tap constructs used in this study. hTap sequences are in black; qTap sequences are in white. All coordinates refer to the hTap sequence except for those in brackets, which refer to qTap. (B) QCl-3 cells (35-mm cultures) were transfected with 25 ng of the pDM128/CTE reporter plasmid, 50 ng of the pBC12/CMV/β-gal internal control plasmid and 200 ng of the indicated effector plasmid. The parental plasmid pBC12/CMV served as a negative control. Induced CAT activities are expressed relative to the culture transfected with the pHA–hTap expression plasmid. Average of three transfection experiments with standard deviation indicated. (C) To confirm that all Tap variants were expressed, a Western analysis of transfected QCl-3 cells was performed using a monoclonal antibody directed towards an amino-terminal HA epitope tag. Mock transfected cells served as the negative control.
As described previously (Kang and Cullen 1999; Kang et al. 2000), transfection of QCl-3 cells with the pDM128/CTE indicator plasmid induces only a low level of CAT enzyme activity (Fig. 3B). However, cotransfection of an hTap expression plasmid results in an ∼10-fold increase in CAT expression because of the activation of MPMV CTE-dependent export of unspliced cat mRNA from the quail cell nucleus. In contrast, cotransfection of a similar qTap expression plasmid has no detectable effect on the level of CAT expression (Fig. 3B).
We first constructed a nested set of full-length hTap/qTap chimeras containing hTap amino-terminal sequences substituted in place of the equivalent qTap residues (Fig. 3A). Chimeras containing hTap residues 1–142, 1–188, or 1–242 failed to rescue CTE function in quail cells, whereas chimeras containing hTap residues 1–268, 1–293, or 1–325 were as active as wild-type hTap (Fig. 3B). We therefore conclude that the carboxy-terminal border of the hTap sequence that permits rescue of CTE function in quail cells lies between hTap residues 242 and 268.
We next attempted to map the amino-terminal border of these critical hTap sequences. As shown in Figure 3, substitution of hTap residues 226–325 or 240–325 into qTap also rescued CTE function, whereas residues 255–325 had no beneficial effect. These data suggested that substitution of only 29 hTap residues, located between 240 and 268, should suffice to rescue the ability of qTap to support CTE function, as indeed proved to be the case (plasmid h[240–268]q Tap, Fig. 3B).
Comparison of residues 240–268 in hTap with the equivalent qTap sequence (Fig. 1B) revealed seven amino acid differences. We particularly noted the substitution of Arg 249 in hTap with a Gln in qTap, as basic residues can play a key role in nucleic acid binding. We therefore constructed a point mutant in qTap in which this Gln (residue 246 in qTap) was substituted with Arg to give qTap(Q246R). As shown in Figure 3B, this single amino acid substitution fully rescued the ability of qTap to support MPMV CTE function in transfected quail cells. In contrast, mutation of five other divergent residues in the qTap 240–265 sequence to their human equivalent failed to enhance the ability of qTap to support MPMV CTE function (plasmid qTap[5X], Fig. 3B). We also mutated residue Arg 249 in hTap to Gln (hTap[R249Q]) to make the exact converse of the qTap(Q246R) mutant. As shown in Figure 3B, this single amino acid mutation blocked the ability of hTap to support MPMV CTE function. Importantly, the inactive hTap(R249Q) mutant, as well as all other Tap mutants functionally analyzed in Figure 3B, were found to be expressed at comparable levels as determined by Western blot (Fig. 3C). We therefore conclude that a single glutamine residue, which is substituted by an arginine in hTap, is responsible for the inability of qTap to support MPMV CTE function.
qTap fails to support MPMV CTE function because it cannot bind the CTE
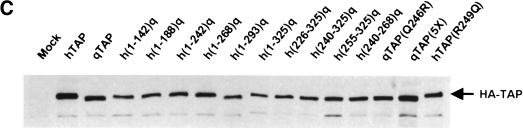
The apparent ability of qTap to support sequence nonspecific nuclear mRNA export (Fig. 2) but not CTE-dependent mRNA export (Fig. 3) suggested that qTap might be defective in binding to the MPMV CTE. To address this issue, we used in vivo and in vitro assays to analyze the ability of hTap, qTap, and selected qTap mutants and chimeras to bind to the MPMV CTE (Fig. 4).
Figure 4.
CTE-binding activity of hTap/qTap chimeras. (A) The indicated Tap variants were expressed fused to the HIV-1 Tat protein and their ability to bind to either the MPMV CTE or the M1-AB CTE mutant then assayed in vivo. Human 293T cells (35-mm cultures) were transfected with 100 ng of either pCTE/CAT or of the pCTE(M1-AB)/CAT negative control plasmid, 50 ng of pBC12/CMV/β-gal and 500 ng of the indicated effector plasmid. Induced CAT activities are expressed relative to the culture transfected with the pTAT–hTAP expression plasmid. Average of three experiments. (B) Binding to the MPMV CTE in vitro was analyzed by electrophoretic mobility shift assay using a 32P-labeled CTE RNA probe. The fusion proteins used consisted of GST linked to wild-type or mutant forms of the RNA-binding domain of hTap (amino acids 61–372) or the equivalent sequence in qTap (amino acids 68–369), as indicated. (FP) Free probe; (C), bound probe.
We have described previously (Kang and Cullen 1999) an assay for the detection of sequence-specific RNA binding in the mammalian cell nucleus that uses the unusual Tat transcriptional activator encoded by HIV-1. Tat normally activates transcription from the HIV-1 long terminal repeat (LTR) promoter after binding to a promoter-proximal viral RNA target termed TAR (for review, see Cullen 1998). However, Tat can also effectively activate the HIV-1 LTR promoter when recruited to a heterologous RNA target, substituted in place of TAR, when expressed as a fusion with a protein displaying the appropriate RNA-binding specificity. In this context, Tat serves as a transcription activation domain whereas the fused protein serves exclusively as an RNA-binding domain.
As demonstrated previously (Kang and Cullen 1999), a fusion protein consisting of Tat fused to hTap activates the expression of a cat indicator gene linked to an HIV-1 LTR bearing the MPMV CTE in place of TAR (Fig. 4A). This activation is specific, as point mutations that block CTE function also block the ability of the CTE to support transcriptional activation by the Tat–hTap fusion. As shown in Figure 4A, a Tat–qTap fusion protein was, in contrast, unable to support transcriptional activation of the HIV-1 LTR bearing the MPMV CTE. Analysis of a set of hTap/qTap chimeras or mutants demonstrated that all of the Tap proteins that failed to support CTE function in quail cells (Fig. 3B) were also unable to support recruitment of the fused Tat activation domain to an HIV-1 LTR containing the MPMV CTE in cis (Fig. 4A). Importantly, substitution of Gln 246 with Arg entirely rescued in vivo binding of the Tat–qTap fusion protein to the MPMV CTE, whereas the converse mutation, that is, substitution of Arg 249 with Gln in hTap, blocked recruitment of the Tat-hTap fusion protein to the CTE (Fig. 4A). All Tat fusions were expressed at comparable levels, as documented by Western blot analysis (data not shown). Closely comparable data confirming that qTap differs from hTap in being unable to bind to the MPMV CTE in vivo were also obtained using the yeast three-hybrid assay (SenGupta et al. 1996) (data not shown).
We wished to demonstrate that the inability of qTap to effectively bind to the MPMV CTE was not due to the presence or absence of some cellular factor but was instead an intrinsic property of qTap. We therefore used a previously described gel retardation assay (Kang and Cullen 1999) to analyze the ability of recombinant proteins consisting of the hTap CTE-binding domain (amino acids 61–372), or the equivalent sequence in qTap (amino acids 68–369), fused to glutathione-S-transferase (GST) to bind the CTE in vitro. As shown in Figure 4B, both hTap and the qTap(Q246R) mutant proved able to bind to a 32P-labeled CTE RNA probe whereas both qTap and the hTap(R249Q) mutant failed to bind detectably. These in vivo (Fig. 4A) and in vitro (Fig. 4B) data therefore clearly demonstrate that substitution of Gln 246 in qTap with arginine, the residue found in hTap, rescues not only the ability of qTap to support CTE function (Fig. 3B) but also CTE binding (Fig. 4).
Recruitment of hTap or qTap to a heterologous RNA target activates nuclear RNA export
The data presented thus far imply that qTap is a fully functional nuclear mRNA export factor. They also indicate that the inability of qTap to support the sequence-specific nuclear export of mRNAs bearing the CTE encoded by the primate retrovirus MPMV results entirely from the inability of qTap to bind to this heterologous RNA target. These data therefore predict that qTap should be as active as hTap in mediating nuclear mRNA export in human cells if it could be effectively recruited to an mRNA target.
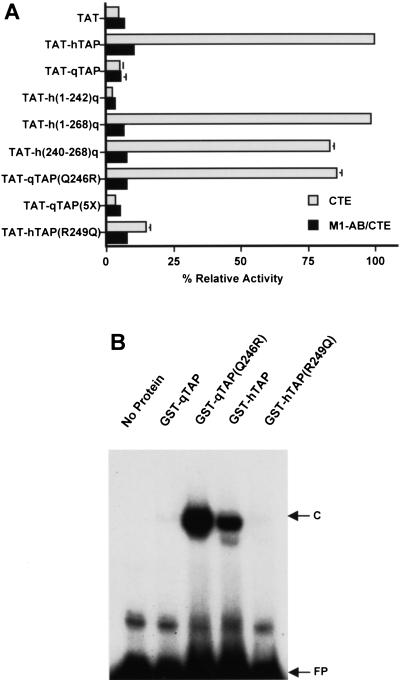
Previously, it has been reported that the Rev nuclear mRNA export factor encoded by HIV-1 is able to induce the nuclear export of RNAs bearing the bacteriophage MS2 translational operator RNA target when expressed as a fusion to the MS2 coat protein (McDonald et al. 1992). This activity required a minimum of two MS2 RNA-binding sites but was most robust when four MS2 sites were present in cis and was specific in that RNA export was blocked by mutation of the critical NES present in Rev. We therefore asked whether the Tap protein would also induce RNA export when recruited to a heterologous RNA-binding site. The indicator construct used to address this question, termed pDM128/4XMS2, is similar to the pDM128/CTE plasmid used in Figure 3B except that it bears four tandem MS2 RNA-binding sites within the intron instead of a copy of the MPMV CTE. The parental plasmid for both pDM128/CTE and pDM128/4XMS2, termed pDM128/PL, bears an intronic polylinker rather than an inserted RNA target and was used as a negative control.
As shown in Figure 5, nuclear export of the unspliced cat mRNA bearing the four MS2-binding sites was activated by an MS2–Rev fusion protein but not by expression of an MS2 fusion to the M10 mutant of Rev, which lacks a functional NES (Malim et al. 1989a). A fusion protein consisting of MS2 linked to full-length hTap was also able to potently activate expression of the cat mRNA encoded by pDM128/4XMS2 but did not affect cat expression from the pDM128/PL negative control plasmid. This activation was again specific, as it was blocked by introduction of the A17 mutation into hTap (residues 593-NWD-595 to alanine) that we have shown previously inhibits both the NES function and the nucleoporin-binding activity of the essential hTap carboxy-terminal domain (Kang and Cullen 1999; Kang et al. 2000). We therefore conclude that hTap is indeed fully competent to induce the nuclear export of an mRNA when tethered to that RNA via a heterologous RNA-binding site.
Figure 5.
Recruitment of Tap to a heterologous RNA target activates nuclear RNA export. Fusion proteins consisting of the MS2 coat protein linked to hTap, qTap, and the indicated Tap mutants and chimeras were assayed in 293T cells for their ability to induce the nuclear export of an unspliced cat mRNA, encoded by pDM128/4XMS2, that contains four copies of the MS2 operator RNA. The parental pDM128/PL plasmid served as the negative control. Transfections and CAT assays were performed as described in Fig. 3 and are expressed relative to the culture transfected with pMS2–hTAP.
A fusion protein consisting of MS2 fused to full-length qTap was also able to effectively induce the nuclear export and expression of an unspliced cat mRNA bearing MS2-binding sites, when expressed in human 293T cells (Fig. 5). Introduction of the A17 mutation into qTap again blocked this activity. Importantly, all of the various hTap/qTap chimeras and mutants analyzed in Figure 3 proved fully capable of supporting the nuclear export of an unspliced cat mRNA bearing MS2 target sites, when expressed as MS2 coat protein fusions. For example, the hTap(R249Q) point mutant that had lost the ability to support CTE-dependent nuclear mRNA export (Fig. 3B) proved entirely effective at inducing nuclear export of a similar cat mRNA when tethered via the MS2 RNA-binding domain (Fig. 5). We therefore conclude that qTap can indeed function as an effective sequence-specific nuclear mRNA export factor when recruitment to the relevant RNA target is efficient.
Discussion
Many critical steps in the replication cycle of viruses are dependent on the interaction of cellular proteins with specific target sites on virally encoded proteins and nucleic acids. Because these binding events do not necessarily recapitulate interactions that are required by the host cell itself, they are not invariably conserved across species barriers. As a result, most viruses show a clear species tropism.
A good example of this generalization is seen in HIV-1, which fails to replicate in rodent cells because of their inability to support several critical steps in the HIV-1 replication. Known defects include the inability of the rodent homologs of the human CD4 and CCR5 receptors to support HIV-1 infection (Maddon et al. 1986; Bieniasz et al. 1997), a rodent-specific defect in HIV-1 virion morphogenesis (Bieniasz and Cullen 2000; Mariani et al. 2000), and the failure of the critical Tat transcriptional activator to function in rodent cells (Bieniasz et al. 1998; Garber et al. 1998). These defects have all been found to be recessive, that is, they can be rescued by fusion of rodent cells to permissive human cells and, where known, by expression of the relevant human protein in trans. In the case of Tat, function in rodent cells can also be rescued by mutation of a single residue in the murine homolog of human cyclin T1, a critical Tat cofactor, from tyrosine to cysteine (Bieniasz et al. 1998; Garber et al. 1998). This single change rescues Tat function because it permits the specific recruitment of the Tat:Cyclin T1 complex to the viral TAR RNA target in rodent cells.
Because species-specific defects in critical steps in viral replication cycles can frequently be complemented by introduction of a single gene product from a permissive species, species tropism can provide an invaluable, but perhaps underappreciated, system for the identification and genetic analysis of key cellular cofactors for viral replication. This is the case for the MPMV CTE, which fails to function in quail cells but can be fully rescued by expression of the hTap protein in trans (Kang and Cullen 1999). Although this observation has faciliated the mutational definition of domains in hTap that play a role in CTE-dependent nuclear mRNA export, there is always the concern that inactivating mutations might have pleotropic effects that make their interpretation impossible. In contrast, natural sequence variation in a particular cellular factor is unlikely to compromise the role of that factor in the normal life of the cell. Therefore, the identification of the species-specific protein sequence variations that control viral tropism can shed unique light on the molecular mechanisms underlying key steps in a viral replication cycle.
In this paper, we have examined the molecular basis for the inability of qTap to support MPMV CTE function. We demonstrate that changing a single glutamine residue in the 616-amino-acid qTap protein to arginine (Q246R), the residue found in hTap, fully rescues the ability of qTap to support CTE function (Fig. 3). We further demonstrate that the inability of wild-type qTap to support MPMV CTE function results entirely from the inability of qTap to bind to the CTE. Introduction of the Q246R mutation into qTap rescues CTE binding both in vivo and in vitro (Fig. 4) whereas introduction of the converse mutation into hTap, R249Q, blocks the ability of hTap to support CTE function (Fig. 3) and to bind the CTE (Fig. 4). Consistent with the hypothesis that qTap is fully competent to support the sequence-nonspecific export of cellular poly(A)+ mRNA from the nucleus, we demonstrate that qTap, like hTap, can rescue the viability of yeast lacking Mex67p, the yeast Tap homolog, when coexpressed together with the human p15–1 cofactor (Fig. 2). In support of the hypothesis that the inability of the MPMV CTE to recruit qTap fully accounts for the failure of the CTE to function in quail cells, we also demonstrate that both hTap and qTap can activate the nuclear export of an mRNA bearing the bacteriophage MS2 operator RNA target, when expressed as MS2 coat protein fusions (Fig. 5). Parenthetically, this latter result strongly argues that the sole role of the MPMV CTE is to function as a binding site for Tap and therefore implies that other reported CTE-binding proteins (Tang et al. 1997) are unlikely to play a significant role in mediating CTE-dependent mRNA export. The demonstration that Tap can induce the nuclear export of an RNA to which it is tethered via a heterologous RNA-binding domain also suggests that it should be possible to define the minimal Tap sequences that are required for the export of, as opposed to recruitment to, target RNAs.
Recently, Herold et al. (2000) described two human proteins closely related to Tap, termed NXF2 and NXF3, and proposed orthologs of Tap have also been described in mice, fruit flies, and nematodes (Bear et al. 1999; Kang et al. 2000; Tan et al. 2000). Among these proteins, the critical R249 residue is conserved in murine Tap, Drosophila Tap, and in human NXF2 but has been substituted in both C. elegans Tap and in human NXF3. Although murine Tap retains the ability to both bind the CTE and rescue CTE function in quail cells, neither NXF2 nor NXF3 display this ability (Herold et al. 2000; data not shown). Therefore, R249, although critical for CTE binding, is obviously not sufficient.
Definition of a surface on hTap that contacts the CTE
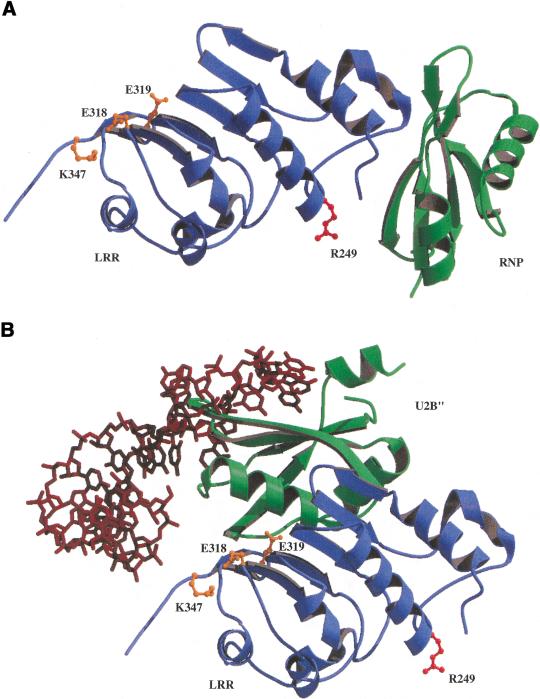
Recently, Liker et al. (2000) reported an X-ray crystallographic analysis of the structure of the CTE-binding domain of hTap (amino acids 102–372) that identified two structural domains, that is, an RNP domain extending from residue 119 to 198 and an LRR domain extending from residue 203 to 362. Consistent with the critical role in CTE-binding demonstrated in this paper, R249 is located on the surface of the LRR domain (Fig. 6A). Surprisingly, however, R249 is located precisely on the opposite side of the LRR from the surface that has been suggested previously to be critical for CTE binding (Fig. 6A). This proposal arose from the hypothesis (Liker et al. 2000) that the interaction of Tap with the MPMV CTE was similar to the interaction of the U2B"(RNP)–U2A‘(LRR) spliceosomal heterodimer with U2 snRNA (Price et al. 1998). However, as shown in Figure 6, superimposition of the hTap LRR domain on the structurally similar U2A‘ molecule demonstrates that the hTap RNP domain and the U2B" RNP domain are located in very different positions, relative to the LRR, in the spliceosomal complex (Fig. 6B) versus the hTap(102–372) structure (Fig. 6A). It therefore seems unlikely that binding by the U2B"–U2A‘ heterodimer to its target RNA (Fig. 6B) is in fact structurally comparable to hTap binding to the CTE.
Figure 6.
Model of the hTap RNA binding domain. (A) A ribbon rendering of the LRR and RNP domains of the hTap protein is shown with the LRR domain in blue and the RNP domain in green. LRR domain residues E318, E319, and K347 are highlighted in ball-and-stick models in orange and R249 in red. The residues shown in orange are located on the opposite side of the LRR domain from R249. (B) A ribbon rendering showing the position of the U2B" RNP domain in green relative to the LRR domain in blue (This is the LRR domain from Tap that has been superimposed on the similar spliceosomal U2A‘ LRR. In the spliceosomal complex U2A‘ (LRR) and U2B" (RNP) are bound to the U2 snRNA, shown as a brick red stick model. The LRR domain has been rotated slightly from the view shown in A to allow visualization of the U2 snRNA. The same residues as in A are shown highlighted in orange and red. The position of R249 suggests that interaction of Tap with the CTE requires the surface on the opposite side of the LRR to that proposed previously (Liker et al. 2000).
It might be argued that CTE RNA binding could produce a conformational shift in Tap that would allow Tap to adopt a conformation more comparable to the U2A‘–U2B" heterodimer. However, the short linker region located between the Tap RNP and LRR motifs, which was not visualized in the Tap structure reported by Liker et al. (2000), would only be ∼12 Å in length even when fully extended. In contrast, the amino terminus of the Tap LRR domain and the carboxyl terminus of the U2B" RNP are ∼40 Å apart in the complex proposed by Liker et al. (2000) and modeled in Figure 6B. Finally, we note that the U2 snRNA actually makes few direct contacts with the U2A‘ LRR domain in the spliceosomal complex and then only with residues located towards the carboxy-terminal end of the LRR (Fig. 6B) (Price et al. 1998). However, mutation of potentially equivalent charged residues on the surface of the hTap LRR domain, which might be predicted to be important if CTE binding by hTap indeed followed the spliceosomal paradigm (shown in orange in Fig. 6), failed to significantly inhibit CTE binding (Liker et al. 2000). In contrast, the substitution of R249 with Q, which affects a residue located approximately in the center of the opposite face of the LRR domain (shown in red in Fig. 6), entirely blocks CTE binding (Fig. 4). We note that the location of R249 on the surface of the LRR that faces the hTap RNP domain (Fig. 6A) is consistent with the finding that CTE binding by hTap requires that the LRR and RNP domains are located in cis, not in trans as seen in the U2B"–U2A‘ heterodimer (Liker et al. 2000). We therefore conclude that, although hTap and the U2B"–U2A‘ heterodimer do contain structurally similar LRR and RNP domains, their interaction with substrate RNAs is likely to be distinct. Although the structural model presented in Figure 6A makes predictions about which surfaces of the hTap LRR and RNP domains are likely to be directly involved in CTE binding that could be tested by mutational analysis, true resolution of this question must await the successful conclusion of ongoing efforts to solve the molecular structure of the hTap:CTE complex.
Materials and methods
Quail cDNA library construction and isolation of a cDNA encoding qTap
Total RNA from quail QCl-3 cells was isolated using the SV Total RNA Isolation system (Promega). Poly(A)+ mRNA was purified on an oligo(dT) column and cDNA synthesis and cloning into λ phage was performed using the ZAP-cDNA synthesis and cloning kit (Stratagene). Plating and screening of the λ phage cDNA library was carried out using standard methods. A randomly 32P-labeled DNA probe was generated from quail cDNA by PCR using oligonucleotide primers directed towards an hTap DNA sequence (5′ primer: 5′-CCCAAAAAAAAG CACGGAC-3′ and 3′ primer: 5′-AAGCACTTCTGGGAC CACTCGAGGTTCAT-3-‘). After screening ∼7.5 × 105 plaque forming units, 10 positive clones were subjected to successive rounds of plaque purification. Plasmid DNA was rescued from purified bacteriophage clones and cDNA inserts were sequenced on both strands. Sequence alignments with the hTap protein were performed using the ClustalW program.
Plasmid construction
All vertebrate expression plasmids are based on pBC12/CMV (Cullen 1986). The following expression plasmids have been described: pcTat, pBC12/CMV/β-gal (Tiley et al. 1992), pHA-hTap (Kang and Cullen 1999), pcTat-hTap (Kang et al. 1999); the indicator constructs pDM128/PL and pDM128/CTE (Bogerd et al. 1998); reporter plasmids containing the cat gene under the control of a modified HIV-1 LTR, where the TAR element has been replaced with the MPMV CTE or the inactive CTE double-loop mutant M1-AB (Kang and Cullen 1999).
The DNA sequence encoding amino acids 493–616 of qTap was amplified by PCR from the qTap cDNA clone with Pfu Turbo (Stratagene). The PCR products were digested with NcoI and SalI and ligated into the NcoI and XhoI sites of the eukaryotic expression vector pBC12/CMV/HA. Two DNA fragments, an NcoI/BamHI fragment encompassing qTap residues 1–232 and a BamHI/XhoI fragment encompassing qTap residues 233–580, were generated by PCR or were excised from the qTap cDNA clone, respectively. Both fragments were ligated into the modified pBC12/CMV/HA vector that had been digested with NcoI and XhoI. The resultant plasmid was named pHA–qTap. Chimeric hTap/qTap variants were generated by recombinant PCR with oligonucleotides containing flanking NcoI sites. The human/quail alleles were subsequently ligated into NcoI-digested pHA–qTap. Additional amino acid changes were introduced into either pHA–qTap or pHA–hTap by Quick change site-directed mutagenesis (Stratagene) or by recombinant PCR. Plasmids that express qTap and its variants fused to the carboxyl terminus of the MS2 coat protein were constructed from the pBC12/CMV derivative containing an NcoI/SalI fragment encoding the carboxy-terminal 493–616 amino acids of qTap. Oligonucleotide primers were used to PCR-amplify a DNA fragment encoding the MS2 coat protein gene from genomic DNA isolated from the yeast indicator strain L40-coat (SenGupta et al. 1996). This fragment was digested with BspHI and NcoI and ligated into the NcoI site of the modified pBC12/CMV vector, destroying the upstream NcoI site. qTap and the hTap/qTap variants were transferred from pBC12/CMV/HA to this vector by digestion of pHA–qTap and its variants with NcoI followed by ligation into the NcoI site of the resultant plasmid. The indicator construct pDM128/4XMS2 was generated by PCR amplification of MS2 RNA from pIII/MS2–2 (SenGupta et al. 1996) followed by ligation into the BglII and ClaI sites of the polylinker present in pDM128/PL. Fusions of qTap and the hTap/qTap chimeras with HIV-1 Tat were generated by PCR. The fragments were digested with EcoRI and SalI and cloned in-frame and downstream of the tat gene in a modified version of pcTat (Tiley et al. 1992).
The following yeast expression plasmids have been described previously: pVP16 (Bogerd et al. 1993); pGAL4/hTap, which expresses hTap (61–619) fused to the GAL4 DNA-binding domain; and pVP16/p15–1 (Kang et al. 2000). Plasmids expressing the FG repeat domains of CG1 and Nup153 fused to a VP16 activation domain were obtained from a two-hybrid cDNA library using pGAL4/hTap(61–619) as a bait. A construct that expresses full-length qTap fused to the GAL4 DNA-binding domain, pGAL4/qTap, was constructed by insertion of the PCR-amplified qTap cDNA into the EcoRI and SalI sites of pGBT9 (Clontech). The expression plasmid pPGK–TRP is derived from pGBT9, but the existing GAL4 DNA-binding domain and the ADH promoter and terminator sequences have been replaced with those of the PGK gene. The hTap and qTap genes were inserted into this plasmid as EcoRI/XhoI fragments to generate pPGK–TRP/hTap (61–619) and pPGK-TRP/qTap, respectively. The plasmid pPGK–LEU is derived from pVP16 (Bogerd et al. 1993). cDNA sequences encoding the VP16 activation domain were removed and the human p15–1 gene was cloned into EcoRI and XhoI sites to generate pPGK-LEU/p15–1.
Sequences encoding residues 68–369 of qTap or qTap(Q246R) or residues 61–372 of hTap(R249Q) were PCR amplified and inserted into the EcoRI and XhoI sites present in the bacterial GST fusion protein expression plasmid pGEX-4T-1 to generate pGST–qTap(68–369), pGST–qTap(Q246R/68–369), and pGST–hTap(R249Q/61–372), respectively. The bacterial expression plasmid, pGST–hTap(61–372), and the half-CTE in vitro transcription vector, pT7–1/2CTE, have been described (Kang and Cullen 1999).
Cell culture, transfection, and Western blot analyses
Human 293T cells and quail QCl-3 cells were maintained as described previously (Cullen et al. 1983; Bogerd et al. 1998) and were transfected using Fugene-6 (Roche Molecular Biochemicals) or DEAE-Dextran (Cullen et al. 1983), respectively. All transfections used cells cultured in 35-mm plates. Levels of DNA used in each transfection experiment are denoted in the relevant figure legend, with pBC12/CMV/β-gal included as an internal control. In all transfection experiments, CAT enzyme levels were determined ∼48 h after transfection and normalized to the level of β-gal activity present in the cell lysate (Bogerd et al. 1998). Western blot analysis of protein expression levels in transfected QCl-3 cells were performed using a monoclonal mouse anti-HA tag antibody (Roche Molecular Biochemicals) essentially as described previously (Kang et al. 2000). Western analysis of nontransfected 293T and QCl-3 cells was performed using a rabbit polyclonal antiserum raised against bacterially expressed residues 61–372 of hTap fused to GST. The anti-Tap antiserum was used at a dilution of 1:500.
Yeast techniques
Yeast two-hybrid analyses were performed essentially as described (Kang and Cullen 2000). Plasmids encoding the appropriate GAL4(1–147) DNA-binding domain or VP16 activation domain fusions were transformed into the yeast indicator strain Y190 (Harper et al. 1993) by standard techniques. After 3 days of growth at 30°C on selective culture plates, double transformants were selected and grown overnight in liquid media. The harvested cells were then lysed and assayed for β-gal activity as described previously (Kang and Cullen 2000). The yeast strains mex67::HIS3/pURA3–MEX67 and mtr2::HIS3/pURA3–MTR2 were a gift of Dr. E. Hurt. Plasmid-shuffling experiments were performed essentially as described (Katahira et al. 1999). The yeast shuffle strains were cotransformed with the expression plasmid pPGK–LEU/p15–1 and either pPGK–TRP/hTap(61–619) or pPGK–TRP/qTap. After three days of growth at 30°C on selective media, SDC-leu-trp-ura, transformants were re-streaked on SDC-leu-trp selective plates, or on SDC-leu-trp plates containing 5-FOA, and incubated for an additional 15 days at 23°C.
Electrophoretic mobility shift analysis
GST fusion proteins, containing the Tap RNA-binding domain (amino acids 61–372 of hTap or 68–369 of qTap), were expressed in bacteria and purified on glutathione affinity resin as described previously (Kang and Cullen 1999). The half-CTE probe (Kang and Cullen 1999) was labeled with [32P]CTP using the Riboprobe transcription system (Promega). Total isotope incorporation was determined after gel purification. The binding reaction was carried out with ∼104 cpm (∼0.1 ng) of the probe and (∼50–100 ng) of GST fusion proteins in 20 μL of binding buffer (150 mM KCl, 10 mM HEPES-NaOH at pH 7.5, 0.5 mM EGTA, 10% glycerol, 2 mM MgCl2, 1 mM DTT) containing 1 μg bacterial rRNA and 0.5 μg of yeast tRNA. Binding was allowed to proceed for 20 min at 4°C, and free and bound forms of the probe were then resolved on a 5% native polyacrylamide gel and visualized by autoradiography.
Molecular modeling
Structural models of U2A‘ (la9n) and of the LRR domain (1ft8) from hTap were superimposed in O (Jones et al. 1991) using conserved secondary structural elements including residues (14–15, 23–25, 37–40, 92–94, 103–108, 117–119, 125–127, 131–138, 144–145, 148–150) from U2A‘ and residues (220–221, 226–228, 257–260, 295–297, 306–311, 319–321, 327–329, 337–344, 350–351, 354–356) from hTap. An overall rmsd of 2.8Å for 37 residues indicates that the structures are similar but not identical. Ribbon renderings of the structural models were generated using MOLSCRIPT (Kraulis 1991) and RASTER3D (Merritt and Bacon 1997).
Acknowledgments
The authors thank Dr. Ed Hurt for the gift of yeast strains. This research was supported by the Howard Hughes Medical Institute, by NIH grant R01 GM55026 (to M.M.G.), and by NIH training grant GM08319 (to D.N.H.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL culle002@mc.duke.edu; FAX (919) 681-8979.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.888201.
References
- Bachi A, Braun IC, Rodriques JP, Panté N, Ribbeck K, von Kobbe C, Kutay U, Wilm M, Görlich D, Carmo-Fonseca M, Izaurralde E. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA. 2000;6:136–158. doi: 10.1017/s1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear J, Tan W, Zolotukhin AS, Tabernero C, Hudon EA, Felber BK. Identification of novel import and export signals of human TAP, the protein that binds to the constitutive transport element of the type D retrovirus mRNAs. Mol Cell Biol. 1999;19:6306–6317. doi: 10.1128/mcb.19.9.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz PD, Cullen BR. Multiple blocks to human immunodeficiency virus type 1 replication in rodent cells. J Virol. 2000;74:9868–9877. doi: 10.1128/jvi.74.21.9868-9877.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz PD, Fridell RA, Aramori I, Ferguson SSG, Caron MG, Cullen BR. HIV-1 induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR-5 co-receptor. EMBO J. 1997;16:2599–2609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz PD, Grdina TA, Bogerd HP, Cullen BR. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 1998;17:7056–7065. doi: 10.1093/emboj/17.23.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Fridell RA, Blair WS, Cullen BR. Genetic evidence that the Tat proteins of human immunodeficiency virus types 1 and 2 can multimerize in the eukaryotic cell nucleus. J Virol. 1993;67:5030–5034. doi: 10.1128/jvi.67.8.5030-5034.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Echarri A, Ross TM, Cullen BR. Inhibition of human immunodeficiency virus Rev and human T-cell leukemia virus Rex function, but not Mason-Pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to Crm1. J Virol. 1998;72:8627–8635. doi: 10.1128/jvi.72.11.8627-8635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun IC, Rohrbach E, Schmitt C, Izaurralde E. TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J. 1999;18:1953–1965. doi: 10.1093/emboj/18.7.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M, Prasad S, Dubay JW, Hunter E, Jeang K-T, Rekosh D, Hammarskjöld M-L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. Trans-activation of human immuodeficiency virus occurs via a bimodal mechanism. Cell. 1986;46:973–982. doi: 10.1016/0092-8674(86)90696-3. [DOI] [PubMed] [Google Scholar]
- ————— HIV-1 auxiliary proteins: Making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- ————— Nuclear RNA export pathways. Mol Cell Biol. 2000;20:4181–4187. doi: 10.1128/mcb.20.12.4181-4187.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR, Skalka AM, Ju G. Endogenous avian retroviruses contain deficient promoter and leader sequences. Proc Natl Acad Sci. 1983;80:2946–2950. doi: 10.1073/pnas.80.10.2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S, Song O-k. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Fischer U, Huber J, Boelens WC, Mattaj IW, Lührmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Garber ME, Wei P, KewalRamani VN, Mayall TP, Herrmann CH, Rice AP, Littman DR, Jones KA. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes & Dev. 1998;12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber BK, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Herold A, Suyama M, Rodrigues JP, Braun IC, Kutay U, Carmo-Fonseca M, Bork P, Izaurralde E. TAP (NXF1) belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Mol Cell Biol. 2000;20:8996–9008. doi: 10.1128/mcb.20.23.8996-9008.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope TJ, Huang X, McDonald D, Parslow TG. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: Mapping cryptic functions of the arginine-rich motif. Proc Natl Acad Sci. 1990;87:7787–7791. doi: 10.1073/pnas.87.19.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kang Y, Cullen BR. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes & Dev. 1999;13:1126–1139. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Bogerd HP, Cullen BR. Analysis of cellular factors that mediate nuclear export of RNAs bearing the Mason-Pfizer monkey virus constitutive transport element. J Virol. 2000;74:5863–5871. doi: 10.1128/jvi.74.13.5863-5871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J, Strasser K, Podtelejnikov A, Mann M, Jung JU, Hurt E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis PJ. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J Appl Cryst. 1991;24:946–950. [Google Scholar]
- Liker E, Fernandez E, Izaurralde E, Conti E. The structure of the mRNA export factor TAP reveals a cis arrangement of a non-canonical RNP domain and an LRR domain. EMBO J. 2000;19:5587–5598. doi: 10.1093/emboj/19.21.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddon PJ, Dalgleish AG, McDougal JS, Clapham PR, Weiss RA, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- Malim MH, Böhnlein S, Hauber J, Cullen BR. Functional dissection of the HIV-1 Rev trans-activator-Derivation of a trans-dominant repressor of Rev function. Cell. 1989a;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- Malim MH, Hauber J, Le S-Y, Maizel JV, Cullen BR. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989b;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- Malim MH, McCarn DF, Tiley LS, Cullen BR. Mutational definition of the human immunodeficiency virus type 1 Rev activation domain. J Virol. 1991;65:4248–4254. doi: 10.1128/jvi.65.8.4248-4254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani R, Rutter G, Harris ME, Hope TJ, Krausslich H-G, Landau NR. A block to human immunodeficiency virus type 1 assembly in murine cells. J Virol. 2000;74:3859–3870. doi: 10.1128/jvi.74.8.3859-3870.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Hope TJ, Parslow TG. Posttranscriptional regulation by the human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex proteins through a heterologous RNA binding site. J Virol. 1992;66:7232–7238. doi: 10.1128/jvi.66.12.7232-7238.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt EA, Bacon DJ. Raster3d: Photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- Neville M, Rosbash M. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 1999;18:3746–3756. doi: 10.1093/emboj/18.13.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville M, Stutz F, Lee L, Davis LI, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Ernst RK, Lund E, Grimm C, Zapp JL, Rekosh D, Hammarskjöld M-L, Dahlberg JE. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. EMBO J. 1997;16:7500–7510. doi: 10.1093/emboj/16.24.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price SR, Evans PR, Nagai K. Crystal structure of the spliceosomal U2B″–U2A‘ protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- Saavedra C, Felber B, Izaurralde E. The simian retrovirus-1 constitutive transport element, unlike the HIV-1 RRE, utilises factors required for the export of cellular mRNAs. Curr Biol. 1997;7:619–628. doi: 10.1016/s0960-9822(06)00288-0. [DOI] [PubMed] [Google Scholar]
- Segref A, Sharma K, Doye V, Hellwig A, Huber J, Lührmann R, Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SenGupta DJ, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Tan W, Zolotukhin AS, Bear J, Patenaude DJ, Felber BK. The mRNA export in Caenorhabditis elegans is mediated by Ce-NXF-1, an ortholog of human TAP/NXF and Saccharomyces cerevisiae Mex67p. RNA. 2000;6:1762–1772. doi: 10.1017/s1355838200000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Gaietta GM, Fischer WH, Ellisman MH, Wong-Staal F. A cellular cofactor for the constitutive transport element of type D retrovirus. Science. 1997;276:1412–1415. doi: 10.1126/science.276.5317.1412. [DOI] [PubMed] [Google Scholar]
- Tiley LS, Madore SJ, Malim MH, Cullen BR. The VP16 transcription activation domain is functional when targeted to a promoter-proximal RNA sequence. Genes & Dev. 1992;6:2077–2087. doi: 10.1101/gad.6.11.2077. [DOI] [PubMed] [Google Scholar]