Abstract
Pax6 is a key transcription factor in eye development, particularly in lens development, but its molecular action has not been clarified. We demonstrate that Pax6 initiates lens development by forming a molecular complex with SOX2 on the lens-specific enhancer elements, e.g., the δ-crystallin minimal enhancer DC5. DC5 shows a limited similarity to the binding consensus sequence of Pax6 and is bound poorly by Pax6 alone. However, Pax6 binds cooperatively with SOX2 to the DC5 sequence, resulting in formation of a high-mobility form of ternary complex in vitro, which correlates with the enhancer activation in vivo. We observed Pax6 and SOX2-interdependent factor occupancy of DC5 in a chromatin environment in vivo, providing the molecular basis of synergistic activation by Pax6 and SOX2. Subtle alterations of the Pax6-binding-site sequence of DC5 or of the inter-binding-sites distance diminished the cooperative binding and caused formation of a non-functional low-mobility form complex, suggesting DNA sequence-guided and protein interaction-induced conformation change of the Pax6 protein. When ectopically expressed in embryo ectoderm, Pax6 and SOX2 in combination activate δ-crystallin gene and elicit lens placode development, indicating that the complex of Pax6 and SOX2 formed on specific DNA sequences is the genetic switch for initiation of lens differentiation.
Keywords: Sox2, Pax6, lens induction, lens placode, transcriptional synergy, expression cloning
The embryonic lens of vertebrate has been used as a model system to study mechanisms of cell differentiation from tissue induction to maturation (Grainger 1992; Jean et al. 1998; Kondoh 1999; Ogino and Yasuda 2000). The primitive head ectoderm is the origin of the lens, which is supposed to be biased toward lens formation compared with the ectoderm of other regions. Differentiation of the lens placode (a group of thickened columnar cells) from the head ectoderm is initiated by an inductive influence of the closely apposed optic vesicle (the retina primordium). The lens placode invaginates to form a cup and is finally pinched off from the rest of the ectoderm to form the lens vesicle. Cells of the posterior side of the vesicle facing the retina differentiate into primary lens fiber cells. These morphological alterations are accompanied by specific activation of crystallin genes. In the chicken, expression of δ-crystallin is initiated as early as the placode stage and hence provides a good probe for transcriptional regulations that govern early processes of lens differentiation.
Lens-specific expression of the chicken δ-crystallin gene is dependent on the 1-kb-long intronic enhancer, in which the 30-bp-long minimal enhancer DC5 is responsible for determination of the lens-specific action of the enhancer (Hayashi et al. 1987; Goto et al. 1990; Kamachi and Kondoh 1993). The DC5 by itself has lens-specific enhancer activity that is dependent on the binding by Group B1 SOX proteins, SOX1, SOX2, or SOX3, which are similar in amino acid sequences (Kamachi et al. 1995, 1998). It is remarkable that expression of Sox2 and/or Sox3 (Sox2 and Sox3 in chicken; Sox2 in mouse; Sox3 in frog and fish) in the head ectoderm is up-regulated prior to placode formation in response to inductive signals of the optic vesicle (Furuta and Hogan 1998; Kamachi et al. 1998; Zygar et al. 1998); this suggests that SOX2 and SOX3 play a role in initiation of lens differentiation rather than only in activation of δ-crystallin. Sox1 starts to be expressed in the lens pit and appears to function in differentiation of lens fiber cells (Kamachi et al. 1998; Nishiguchi et al. 1998).
It has been shown that SOX1/SOX2/SOX3 bind to the 5′ half of the DC5 sequence, and that binding of one of these SOX proteins is essential for the DC5 enhancer activity. Our previous work has also indicated that DC5 enhancer is activated by cooperative action of one of SOX1/SOX2/SOX3 and their partner factor δEF3, which interacts with 3′ half of the DC5 sequence and is present in the lens but absent in fibroblasts (Kamachi et al. 1995; Kamachi et al. 1998; Kamachi et al. 1999). Mutations of the DC5 sequence that alter either the SOX-binding site or the putative δEF3-binding site abolish the enhancer activity, indicating that synergy of SOX1/SOX2/SOX3 and δEF3 is essential for the enhancer action. Consistently, overexpression of SOX1/SOX2/SOX3 and even the SOX2-VP16 fusion protein fail to activate the DC5 enhancer in non-lens cells lacking δEF3, whereas these increase the enhancer activity in lens cells having δEF3 (Kamachi et al. 1995, 1998, 1999).
It is suggested that many SOX proteins including SOX1/SOX2/SOX3 regulate differentiation of various cell types through cooperation with partner factors unique to the cell type; e.g., SOX1/SOX2/SOX3 are also expressed in the nervous system and appear to play a role in neural determination by regulating a different set of target genes employing a partner factor distinct from δEF3 (Pevny et al. 1998; Kamachi et al. 1999, 2000; Wegner 1999). Molecular identification of δEF3, the partner factor for SOX1/SOX2/SOX3 in the lens, is of critical importance in understanding lens differentiation, and, more generally, how SOX proteins regulate cell differentiation through partnering with other transcription factors.
Another key transcription factor in lens development is Pax6 (Jean et al. 1998; Gehring and Ikeo 1999; Kondoh 1999; Ogino and Yasuda 2000). Pax6 was originally identified as a member of the Pax protein family having paired DNA-binding domains, and has been recognized as a key regulator of eye development. Mouse small eye phenotype and human Aniridia result from heterozygous mutations in Pax6 (Jean et al. 1998), and homozygotes fail to develop eyes (Hogan et al. 1986). In addition to eye development, Pax6 is also necessary for normal development of the nose, pancreas, and the central nervous system (Hogan et al. 1986; Glaser et al. 1994; Grindley et al. 1995; St-Onge et al. 1997). Although Pax6 is expressed both in the optic vesicle and in the prospective lens ectoderm, the following evidence indicates the importance of cell-autonomous function of Pax6 in the head ectoderm of the prospective lens region: (1) Pax6 is first expressed in a broad region of the head ectoderm, then the expression becomes restricted to the presumptive lens placode, and is maintained in the differentiating lens (Li et al. 1994; Grindley et al. 1995; Zygar et al. 1998); (2) tissue recombination experiments have demonstrated that the Pax6−/− ectoderm displays no response to the wild-type optic cup, whereas the Pax6−/− optic cup can induce a lens in the wild-type ectoderm (Fujiwara et al. 1994); (3) Pax6 chimera analysis has shown no contribution of Pax6−/− cells in the lens placode (Collinson et al. 2000); and (4) Cre/loxP-mediated Pax6 inactivation in the eye surface ectoderm (Le-mutant) has demonstrated that Pax6 activity is required for lens placode development (Ashery-Padan et al. 2000). Although these studies clearly indicate the importance of Pax6 for initiation of lens differentiation, the molecular action of Pax6 has not been determined.
Here, we report that Pax6 functions as a co-DNA-binding partner factor of SOX2 to regulate their downstream target genes during early lens development. In a search for the SOX1/SOX2/SOX3 partner factor δEF3, we performed a functional expression screen using yeast cells, and identified Pax6 as δEF3. Pax6 and SOX2 interact directly with each other and bind cooperatively to the DC5 sequence, forming a ternary complex recognized as a high-mobility form complex in a gel mobility shift assay. Formation of the high-mobility form complex in vitro correlates with synergistic activation by Pax6 and SOX2 in cultured cells. Consistent with this, chromatin immunoprecipitation analysis indicates that Pax6 and SOX2 interdependently associate with the DC5 sequence in vivo. Finally, we show that exogenous SOX2 and Pax6 in the head ectoderm can induce ectopic lens placode development. Thus, partnership between Pax6 and SOX2 is crucial for initiation of lens development. The identification of Pax6 as an interactive partner of SOX2 may imply a general requirement of partner factors for transcriptional regulation by the members of SOX and Pax protein families and possible SOX-Pax partnerships in other developmental contexts.
Results
Identification of Pax6 as a partner factor of SOX1/SOX2/SOX3
SOX1/SOX2/SOX3 require cooperative action of their partner factor δEF3 to activate the DC5 enhancer. Although our previous data suggest that SOX1/SOX2/ SOX3 and δEF3 bind to the DC5 sequence side by side, conventional cDNA cloning strategies such as southwestern screen and yeast one-hybrid screen failed to isolate cDNA of δEF3. We presumed that synergistic activation of the DC5 enhancer by SOX1/SOX2/SOX3 and δEF3 largely result from cooperative binding of the two factors (Kamachi et al. 1999), and developed a functional screening strategy based on cooperative action of the two factors on a DNA sequence.
The developed method is based on a functional cDNA screen using yeast cells and can be regarded as a modified one-hybrid method, where the DC5 enhancer-dependent reporter genes will be activated only when δEF3 is expressed together with SOX1 or SOX2 (Fig. 1A, section a). Octamerized DC5 sequence was placed upstream of the reporter genes yeast HIS3 or Escherichia coli lacZ, and the reporter genes were integrated into the yeast genome (Fig. 1A, sections a,b). Expression of SOX1 or SOX2 alone did not activate the reporter genes, as anticipated from the nature of the DC5 enhancer. A GAL4 activation domain (GAL4AD)-fused cDNA library derived from embryonic chicken lenses was then introduced into SOX1- or SOX2-expressing yeast cells, and transformants were screened for histidine prototrophy and β-galactosidase expression. Two double-positive clones were identified among 35 His+ clones. These clones had cDNAs coding for Pax6, one full length and the other lacking C-terminal 29 aa. Thus, Pax6 became a strong candidate molecule for δEF3.
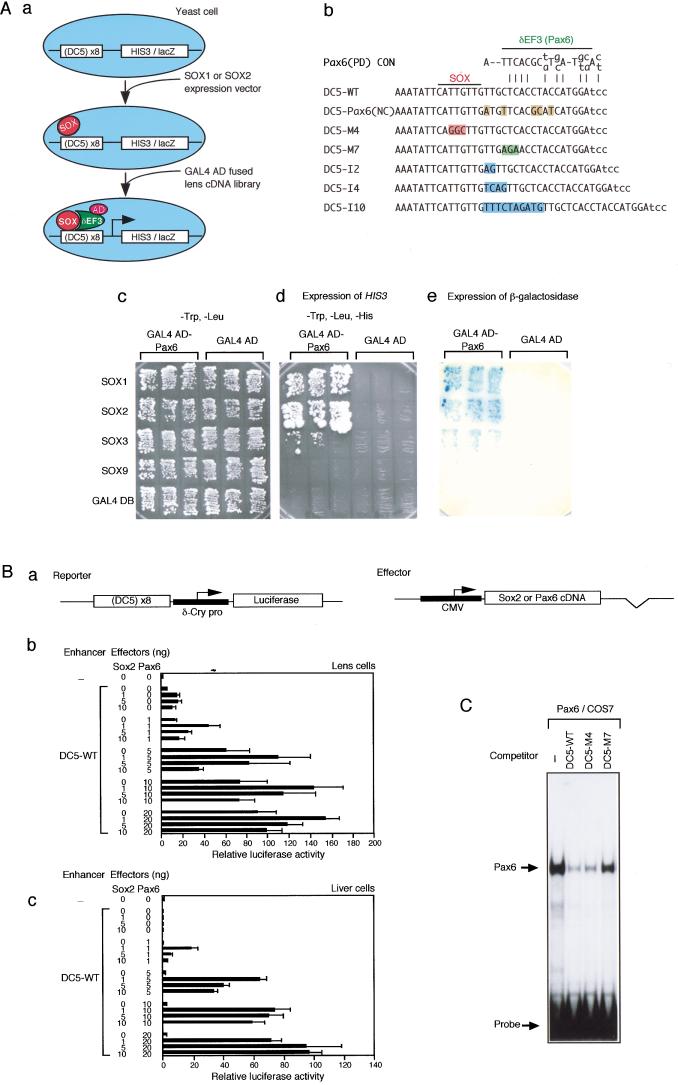
Figure 1.
Identification of Pax6 as SOX1/SOX2/SOX3-partner factor δEF3 by functional expression screen. (A) Functional expression screen using yeast cells. (a) The scheme of the screening procedure. Yeast cells carrying the DC5 enhancer-dependent HIS3 and lacZ reporters were first transformed with a SOX1 or SOX2 expression vector. GAL4 activation domain-fused lens cDNA library was then introduced. Yeast cells that harbor both SOX1/SOX2 vector and a δEF3 clone from the library are expected to express HIS3 and lacZ genes. (b) Wild-type (WT) and mutant sequences of the DC5 enhancer used in this study. Altered or inserted nucleotides are shaded. SOX and δEF3 (Pax6) binding regions are shown at the top. (c–e) The growth and β-galactosidase expression of the HIS3/lacZ reporter-containing yeast cells, which harbor expression plasmids for various Sox and Pax6 cDNAs. (c,d) The growth of three clones in the presence (c) and in the absence (d) of histidine; (e) expression of β-galactosidase. (B) Cell type-independent activation of the DC5 enhancer by SOX2 and Pax6. (a) Structures of the luciferase reporter and effector plasmids. (b,c) Varying amounts of effector plasmids to express SOX2 or Pax6 and the reporter plasmids were transfected into lens cells (b) and liver cells (c). The luciferase activity generated by the enhancer-less reporter was taken as 1. (C) Binding of Pax6 to the DC5 sequence and sequence-specific competition. The probe was DC5-WT sequence. 50 ng of poly dA-dT was included in the reaction to achieve a stringent binding condition.
In lens cells, the DC5 enhancer is preferentially activated by SOX1, SOX2 and SOX3 but not by SOX9 or SOX11, suggesting that δEF3 selectively cooperates with SOX1, SOX2 and SOX3 (Kamachi et al. 1999). Thus, we re-introduced GAL4AD-Pax6 in combination with various SOX proteins or the GAL4 DNA-binding domain (GAL4BD) as an unrelated protein into the reporter-containing yeast cells, and examined HIS3 and lacZ expression. The GAL4AD-Pax6 fusion increased expression of reporter genes only in the presence of SOX1, SOX2, and SOX3 (Fig. 1A, sections c–e). This indicates that GAL4AD-Pax6 efficiently activates the DC5 enhancer together with SOX1, SOX2 and SOX3 in yeast cells, but not by itself or with other SOX proteins, as expected for δEF3.
We inquired whether synergism of SOX2 and Pax6 is sufficient for activation of the DC5 enhancer. To test this model, the DC5 enhancer-bearing luciferase reporter vector was cotransfected with effector vectors expressing SOX2 or Pax6 into lens cells and liver cells prepared from chicken embryos (Fig. 1B). In lens cells, exogenous expression of either SOX2 or Pax6 augmented DC5 enhancer activity (Fig. 1B, section b), reflecting endogenously expressed SOX1/SOX2/SOX3 and Pax6. In these assays, exogenous expression of Pax6 was more effective than SOX2, indicating that endogenous Pax6 protein is limited in cultured lens cells. Higher-level activation of DC5 enhancer was observed when SOX2 and Pax6 were overexpressed together. In liver cells, however, no substantial activation of DC5 occurred when SOX2 or Pax6 was singly expressed, while their simultaneous expression resulted in DC5-dependent activation to levels similar to those achieved in lens cells (Fig. 1B, section c). It was noted that activation levels were dependent not only on the amount of transfected vectors, but also on the ratio of the two effectors. Data analogous to Figure 1B were also obtained when we used chick fibroblasts and murine 10T1/2 cells (data not shown), indicating that cooperative action of SOX2 and Pax6 is cell independent and sufficient for DC5 activation. This accounts for the lens-specific activity of the enhancer, since the lens is one of the few cell types coexpressing SOX2 and Pax6. SOX1 and SOX3 also cooperated with Pax6 in activating the DC5 enhancer similar to SOX2 (data not shown), indicating that Pax6 cooperates efficiently with all of the subgroup B1 SOX proteins.
Pax6 is a transcription factor containing the paired domain and homeodomain. The binding consensus sequences of individual domains have been determined (Epstein et al. 1994a; Czerny and Busslinger 1995). The δEF3 site of the DC5 enhancer shows similarity to the Pax6 paired domain-consensus sequences, although having a number of mismatches (Fig. 1A, section b). Thus, we tested binding of the Pax6 protein to the DC5 probe using a gel mobility shift assay. Pax6 protein expressed in COS7 cells gave rise to a complex with the probe, which was sensitive to competition by excess DNA of DC5-WT and DC5-M4 (SOX site mutant), but not by DC5-M7 (δEF3 site mutant) (Kamachi et al. 1995, 1999) (Fig. 1A, section b), indicating that Pax6 binds to the δEF3 site of DC5 in a sequence-specific manner (Fig. 1C). Taken together, these data in Figure 1 demonstrate that the Pax6 gene codes for the authentic δEF3 protein.
SOX2 and Pax6 cooperatively bind to the DC5 sequence in vitro
The data presented above (Fig. 1) all indicate that Pax6 activates the DC5 enhancer in cooperation with SOX1/SOX2/SOX3 but there are previous reports that Pax6 alone can activate transcription through binding to the artificial Pax6 paired domain-consensus sequence (Epstein et al. 1994b; Czerny and Busslinger 1995; Tang et al. 1998). We thus compared binding of Pax6 to the consensus binding sequence and to the DC5 sequence. The paired domain of Pax6 tagged with 6xHis (Fig. 2A), Pax6(169), was synthesized in E. coli, purified, and used for gel mobility shift assays. By changing the amount of Pax6(169) in the assay, we found that its binding affinity to the DC5 sequence was approximately 25-fold lower than that to the consensus sequence (Fig. 2B).
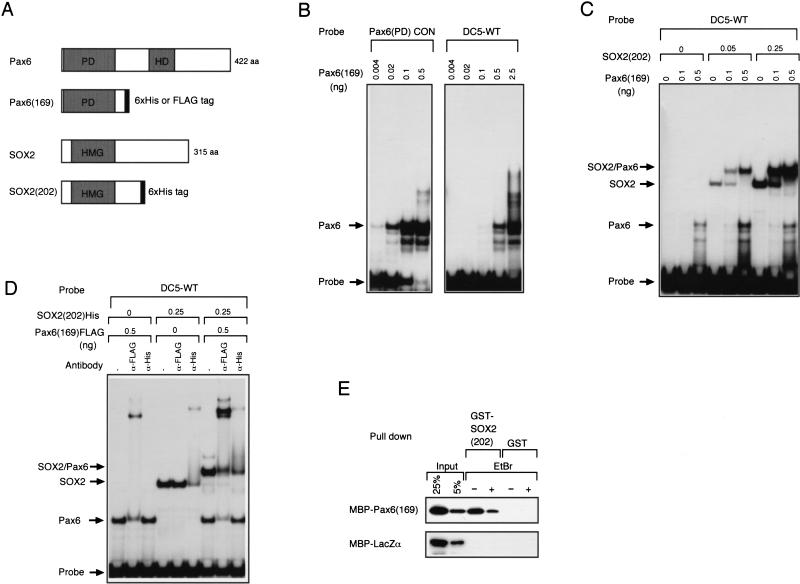
Figure 2.
Cooperative binding of SOX2 and Pax6 to the DC5 sequence in vitro. (A) SOX2 and Pax6 proteins and their variants with oligopeptide tags. DNA binding domains are shaded: PD, paired domain; HD, homeodomain; HMG, HMG domain. (B) Difference of affinity of the Pax6 paired domain in binding to the consensus sequence [Pax6(PD)CON] and to the DC5 sequence indicated by gel mobility shift assays. (C) Cooperative binding of the Pax6 paired domain and the SOX2 HMG domain to the DC5 sequence in vitro. (D) FLAG-tagged Pax6(169) and His-tagged SOX2(202) were used to make a complex on DC5 probe, and effect of anti-tag antibodies on the complex was examined. (E) Direct interaction of the Pax6 paired domain and the SOX2 HMG domain without intervention of DNA. MBP-tagged proteins were incubated with GST-SOX2 or GST and precipitated with glutathione sepharose. Effect of Escherichia coli DNA in the protein preparations was quenched by EtBr.
We then examined how Pax6 interacts with the DC5 sequence in the presence of SOX2. SOX2(202) protein tagged with 6xHis (Fig. 2A) was produced by a baculovirus expression system, purified, and used with Pax6(169). When both Pax6(169) and SOX2(202) were included in binding reactions, a slowly migrating band, probably representing the ternary complex containing Pax6(169), SOX2(202), and the probe, appeared even at a low concentration of Pax6(169) (Fig. 2C). This ternary complex band was efficiently formed at 0.1 ng of Pax6(169) if SOX2(202) was present, although 0.1 ng of Pax6(169) alone failed to show a band with the DC5 probe. In addition, the amount of the SOX2/Pax6/DC5 complex in the presence of 0.25 ng of SOX2(202) and 0.5 ng of Pax6(169) was greater than those formed with the individual protein of the same amount (Fig. 2C), indicating that Pax6(169) and SOX2(202) bind cooperatively to the DC5 sequence.
To confirm the presence of both Pax6(169) and SOX2(202) in the slowly migrating complex, we performed supershift assays utilizing epitope tagging and anti-tag antibodies; in this experiment, Pax6(169) was tagged with the FLAG sequence (Fig. 2A). Inclusion of either anti-FLAG or anti-His antibody disrupted and/or supershifted the slowly migrating complex band (Fig. 2D), demonstrating that this band represents a ternary complex formed among Pax6(169)-FLAG, SOX2(202)-6xHis, and the DC5 DNA.
Cooperative binding of SOX2(202) and Pax6(169) to the DC5 sequence suggests that they may interact with each other. We therefore synthesized glutathione S-transferase (GST)-fused SOX2(202) and maltose binding protein (MBP)-fused Pax6(169) in E. coli, and examined their direct interaction in vitro. When MBP-Pax6(169) was incubated with GST-SOX2(202), approximately 20% of the input MBP-Pax6(169) was coprecipitated as a complex with GST-SOX2(202) bound-glutathione resin (Fig. 2E). Some fraction of the complex was disrupted by ethidium bromide (EtBr), an inhibitor of DNA binding, but a significant level of the interaction was resistant to EtBr. This indicates that SOX2(202) and Pax6(169) directly interact with each other, but that the interaction is stabilized by residual E. coli DNA present in the preparation (Lai and Herr 1992), and that the ternary complex involves both DNA–protein and protein–protein interactions.
SOX2 and Pax6 cooperatively associate with the DC5 enhancer in vivo
The results of in vitro binding reaction (Fig. 2) indicate that factor occupancy on the DC5 enhancer is highly dependent on cooperative binding of SOX2 and Pax6. We presumed that the cooperativity may be more pronounced in a chromosomal environment in vivo, considering the strict requirement of both factors for activation of DC5 enhancer (Fig. 1; see discussion). Thus, the in vivo factor binding was analyzed in yeast cells using a chromatin immunoprecipitation procedure (ChIP) (Strahl-Bolsinger et al. 1997; Orlando 2000). Yeast cells were used because they do not have a SOX- or a Pax-like activity. After formaldehyde fixation of yeast cells expressing epitope-tagged SOX2 and/or Pax6 and carrying the DC5 enhancer-dependent lacZ reporter, SOX2-DC5 and Pax6-DC5 complexes were purified using anti-HA antibody, and the DC5 sequence in the immunoprecipitate was assessed by polymerase chain reaction (Fig. 3). When SOX2 and Pax6 were individually expressed, no signal above the background was observed (Fig. 3B, cf. lanes 2,3 with lane 1). However, when SOX2 and Pax6 were coexpressed, significant extents of factor occupancy on the DC5 enhancer were demonstrated (Fig. 3B, lanes 4,5). Only a weak background signal was detected for the lacZ fragment (Fig. 3C), ensuring specificity of the assay. Thus, the in vivo condition more clearly demonstrates the cooperative binding of SOX2 and Pax6 to the DC5 sequence, and that one factor alone does not associate with the enhancer in the chromatin. Moreover, the observed factor occupancy, i.e., the amounts of immunoprecipitated DC5 DNA, well correlated with lacZ reporter expression (Fig. 3D). These indicate that cooperative binding of the two factors is the most critical process in the synergistic activation of the DC5 enhancer by SOX2 and Pax6 in vivo.
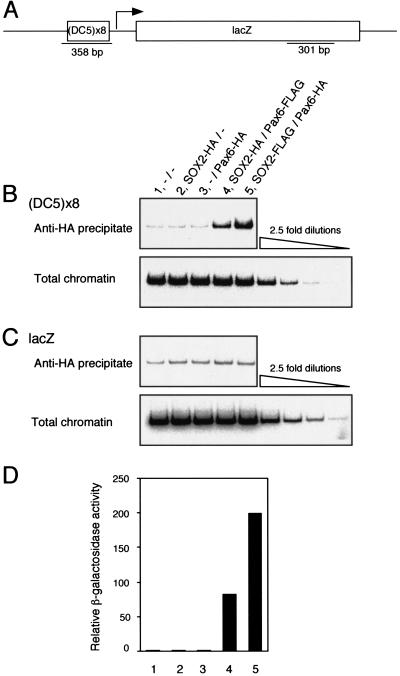
Figure 3.
Interdependent association of SOX2 and Pax6 with the DC5 enhancer in a chromatin environment in vivo. (A) The DC5-lacZ reporter construct introduced into yeast genome and the location of the sequences amplified by PCR. (B,C) Chromatin immunoprecipitation analysis using yeast cells carrying various combinations of SOX2, Pax6, and empty expression vectors as indicated: –, empty vectors; SOX2-HA and SOX2-FLAG, C-terminally 3xHA- and 3xFLAG-tagged SOX2 expression vectors, respectively, containing the TRP1 selection marker; GAL4AD-Pax6-HA and GAL4AD-Pax6-FLAG, C-terminally 3xHA- and 3xFLAG-tagged GAL4AD-Pax6 expression vectors, respectively, containing the LEU2 selection marker. Immunoprecipitation was performed with anti-HA antibody and PCR was done on the DC5 sequence (B) and the lacZ gene (C). PCR products from 2.5-fold serial dilutions of total chromatin demonstrate the linear response of the PCR. (D) Expression of the reporter lacZ gene in yeast cells having the combinations of SOX2/Pax6 effectors and used for chromatin immunoprecipitation.
Activation-competent ternary complex is formed only on the wild-type DC5 sequence
The data presented above indicate that SOX2 and Pax6 form the stable ternary complex through DNA–protein and protein–protein interactions. Thus, we examined the contribution of each interaction and the nature of the ternary complex. In addressing these questions, we first tested whether ternary complex formation requires both SOX2 and Pax6 binding sites by using the DC5 probes carrying mutations in either the SOX2 (M4) or Pax6 (M7) binding sites (Fig. 1A, section b) in gel mobility shift assays. Mutation M4 diminished the binding of SOX2 to the probe and abolished formation of the ternary complex (Fig. 4A). Thus the protein–protein interaction between SOX2 and Pax6 is not sufficient to form the ternary complex. Mutation M7 still allowed binding of Pax6 to the DC5 probe under the low stringent condition used (i.e., without nonspecific competitor DNA), but the mutation significantly decreased the production of the band containing SOX2(202) and Pax6(169) (Fig. 4A). Interestingly, the SOX2(202)/Pax6(169)-containing complex formed on the DC5-M7 probe migrated more slowly than that with the DC5-WT probe (SOX2/Pax6/DC5-WT complex is marked by “H” for high-mobility complex; SOX2/Pax6/DC5-M7 complex is marked by “L” for low-mobility complex. Fig. 4A). Difference in the gel mobility suggests that conformation of the ternary complex made on the wild-type sequence might be in a more compact form than that with the M7 mutant sequence, which appears to be more relaxed.
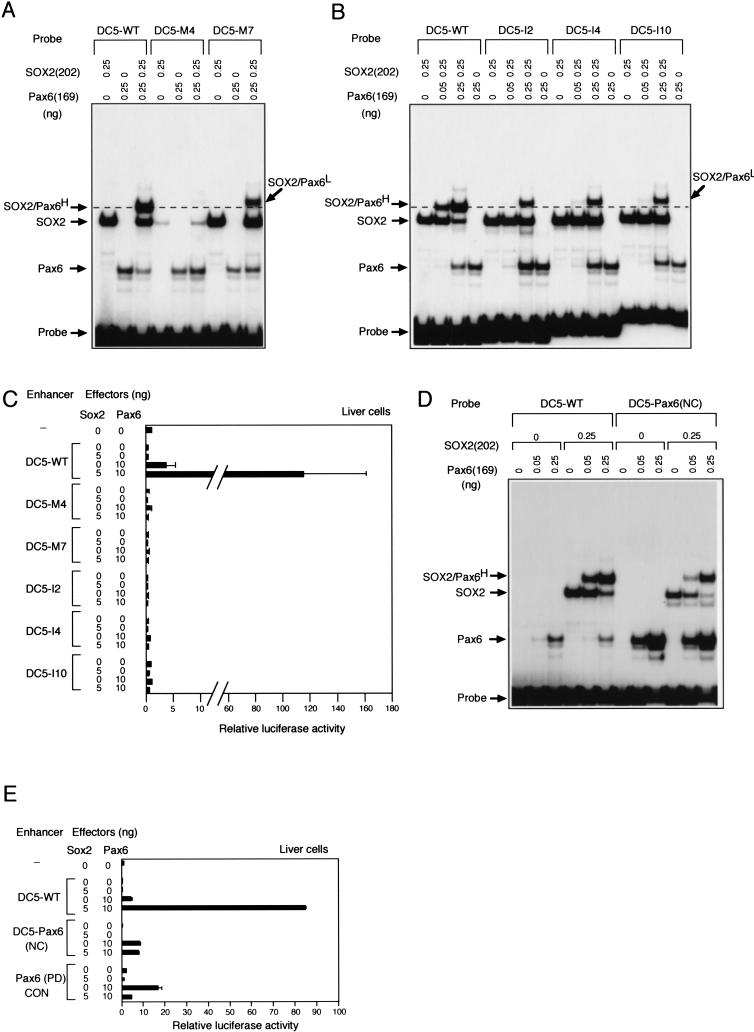
Figure 4.
Formation of the high-mobility ternary complex in vitro that correlates with synergistic activation of the DC5 enhancer by SOX2 and Pax6 in cultured cells. (A,B) Differences in ternary complex formation on wild type and mutant sequences of DC5. (A) Nucleotide replacement mutants (M4 and M7); (B) nucleotide insertion mutants (I2, I4, and I10) (Fig. 1). High-mobility and low-mobility forms of the ternary complex are indicated by superscripts H and L, respectively. (C) Wild type and mutant DC5 enhancers in (A) and (B) were tested for activation by exogenous SOX2 and Pax6 in liver cells. (D) Alteration of the Pax6 binding sequence of DC5 closer to the consensus binding sequence [DC5-Pax6(NC)] increases the binding of Pax6(169) itself, but disrupts cooperative binding of SOX2(202) and Pax6(169). (E) Loss of the cooperative binding on the DC5-Pax6(NC) correlates with loss of synergistic activation by SOX2 over activation by Pax6 itself.
Cooperative binding and direct interaction of SOX2 and Pax6 suggest that ternary complex formation requires a particular spatial organization of these factors on the DC5 sequence. To address this possibility, we inserted spacer sequences of 2 bp, 4 bp, or 10 bp in DC5 between the binding sites of SOX and Pax6 (Fig. 1A, section b), and examined their effects on the complex formation. As shown in Figure 4B, these insertion sequences significantly decreased the efficiency of ternary complex formation. With wild-type DC5 sequence, 0.05 ng of Pax6(169) in the presence of SOX2(202) was sufficient to create a strong band of the ternary complex, but with insertion mutants of DC5, 0.25 ng of Pax6(169) protein was required to give rise to an analogous band intensity of the ternary complex. Moreover, the complex was the low-mobility form similar to that observed using the DC5-M7 probe. Thus, the strict spatial arrangement of the factor binding sites within the DC5 enhancer is essential for the assembly of the high-mobility ternary complex, suggesting that direct protein–protein interaction facilitates the assembly.
We examined whether the form of the ternary complex observed in vitro correlates with activation of the DC5 enhancer by SOX2 and Pax6 in vivo. Luciferase reporters bearing these DC5 mutants used in Figure 4 panels A and B were transfected with effector vectors expressing SOX2 or Pax6 into cultured liver cells. Under the same condition where the DC5-WT enhancer was highly activated by coexpression of SOX2 and Pax6, none of these DC5 mutant enhancers was activated (Fig. 4C). This result clearly indicates that the enhancer activation in vivo requires the wild-type DC5 DNA sequence, to which SOX2 and Pax6 cooperatively bind and form the activation-competent high-mobility ternary complex in vitro.
These results indicate that binding sites of SOX2 and Pax6 must be arranged in correct spacing on their target DNA sequences. Are correctly spaced binding sites sufficient for the functional ternary complex? We altered the Pax6 binding sequence of the DC5 enhancer to resemble the Pax6 consensus sequence [DC5-Pax6 (NC)] (Fig. 1A, section b), and tested the effect on cooperative binding of Pax6(169) and SOX2(202), and on the synergistic activation of the enhancer by Pax6 and SOX2. As shown in Figure 4D, Pax6(169) bound to the DC5-Pax6(NC) probe with an affinity comparable to the Pax6(PD)CON probe (Fig. 3A). However, formation of the ternary complex was less efficient than using the DC5 wild-type sequence, indicating that the Pax6 consensus sequence is not compatible with cooperative binding with SOX2. In transfection assays, the DC5-Pax6(NC) mutant was activated by Pax6 comparable to Pax6(PD)CON element, but it was not activated any further by coexpressed SOX2 (Fig. 4E). This indicates that correctly spaced binding sites for SOX2 and Pax6 are not sufficient, and that the native DC5 sequence, which by itself is a poor Pax6 site, is essential for organizing the activation-competent functional complex. Perhaps interaction with the DC5 sequence and with SOX2 induces a conformational change in Pax6 required for formation of the high-mobility ternary complex, but interaction with the Pax6(NC) sequence inhibits this change.
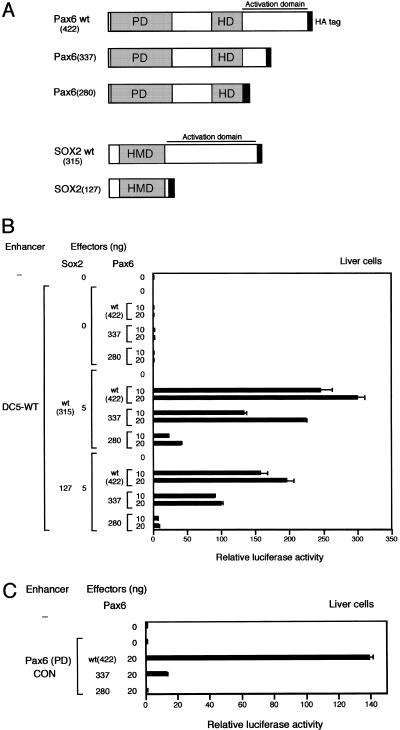
Transactivation by the SOX2/Pax6 complex largely relies on the activation domain of Pax6
Since the activation level of the DC5 enhancer attained by synergism of SOX2 and Pax6 is extremely high, we speculate that the synergism also involves interaction of the activation domains. We examined this possibility using deletion mutants of the activation domains of Pax6 and SOX2. Activation domain of SOX2 is assigned to the C-terminal half of the protein that is actually composed of multiple subdomains contributing various degrees to the overall activation level (Kamachi et al. 1998; Ambrosetti et al. 2000; Nowling et al. 2000). Activation by Pax6 also depends on subdomains distributed in its C-terminus PST region (Glaser et al. 1994; Czerny and Busslinger 1995; Tang et al. 1998). These C-terminal regions of SOX2 and Pax6 also function as transactivation domains when fused to the GAL4 DNA-binding domain. We made two Pax6 mutant forms with C-terminal deletions, Pax6(337) and Pax6(280), and tested their activity by transfecting their expression vectors with or without those for wild-type SOX2 and SOX2(127) that lacks the whole SOX2 activation domain (Fig. 5A). In the absence of SOX2 protein of any form, neither wild type nor mutant forms of Pax6 activated the DC5-bearing luciferase gene (Fig. 5B). In combination with wild-type SOX2, Pax6(337), which lacks the distal half of the activation domain, displayed >50% activity compared with the case using wild-type Pax6 (Fig. 5B), while Pax6(280) lacking the entire Pax6 activation domain under the same condition showed only ∼15% level of wild-type Pax6. The low activation level observed by combination of wild-type SOX2 and Pax6(280) may represent the contribution of the SOX2 activation domain in the overall activity of the complex. On the other hand, combination of SOX2(127) and wild-type Pax6 caused only moderate reduction of the activation of the DC5 enhancer, and ∼60% of the activity was retained compared with the level with wild-type SOX2 and Pax6 (Fig. 5B). No substantial activation of the DC5 enhancer was observed when SOX2(127) and Pax6(280), totally lacking the activation domains, were combined. These findings indicate that activation by the SOX2/Pax6 complex relies more on the Pax6 activation domain than on the SOX2 activation domain, although synergy between the two factors is also notable. In fact, appreciable degrees of synergistic activation were observed in any combinations tested except SOX2(127)/Pax6(280). The mechanism of synergy may be primarily in the cooperative binding of SOX2 and Pax6 to the DC5 sequence, but ternary interaction-dependent alteration of activation potential is also implied. For instance, Pax6(337) shows a very limited activation when bound singly to Pax6(PD)CON sequence (Fig. 5C), while the same Pax6(337) showed more than half the level of wild-type Pax6 when bound to DC5 sequence in cooperation with SOX2(wt) or SOX2(127) (Fig. 5B). One can speculate that there is conformational change of the proteins including the activation domains in the ternary complex.
Figure 5.
Requirement of activation domains of SOX2 and Pax6 for synergistic activation of the DC5 enhancer. (A) Schemes of the deletion mutants of SOX2 and Pax6, which have the 3xHA tag at their C-termini. (B) Various combinations of effector vectors to express SOX2 and Pax6 in wild-type or mutant forms were transfected into chicken liver cells with the DC5 enhancer-containing luciferase reporter plasmid. (C) Effector vectors for the wild-type and mutant forms of Pax6 were transfected into chicken liver cells with the Pax6(PD)CON sequence-containing luciferase reporter plasmid.
Exogenous SOX2 and Pax6 induces ectopic δ-crystallin expression and lens differentiation
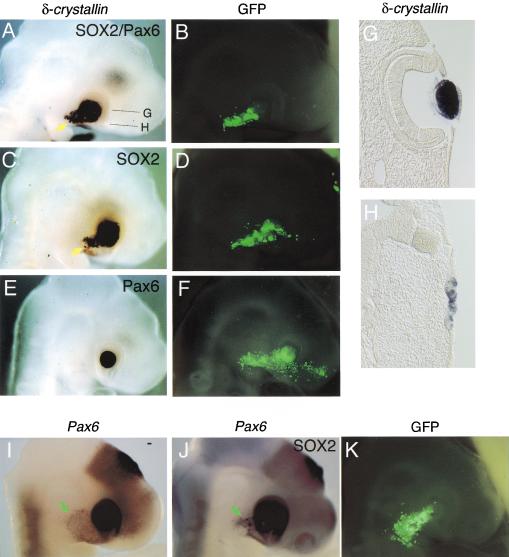
In chicken embryos, Pax6 is expressed in a wide region of the head ectoderm covering the presumptive lens area earlier than lens placode induction (Li et al. 1994; Kamachi et al. 1998). Then in the ectodermal region, Sox2/Sox3 expression is activated in the area that was in contact with the optic vesicle, immediately followed by lens placode development and δ-crystallin expression (Kamachi et al. 1998). Thus, it appears that, in lens placode cells, SOX2/SOX3 and Pax6 form a ternary complex with the DC5 enhancer of the δ-crystallin gene, and possibly with other analogous target sequences. Because the DC5 enhancer constitutes the core of the intronic enhancer in the gene, we asked whether exogenous SOX2 and Pax6 could induce expression of the endogenous δ-crystallin gene in non-lens cells. When vectors to express SOX2 and Pax6 were introduced into head ectoderm of chicken embryos at stage 10 by in ovo electroporation, we observed ectopic expression of the δ-crystallin gene outside of the eyes in a population of electroporated ectodermal cells (Fig. 6A), which were marked by GFP expression (Fig. 6B). SOX1 and SOX3 also gave essentially the same result as SOX2 (data not shown), consistent with the similar characteristics of Group B1 SOX proteins indicated by transfection assays. Pax6 alone did not induce the δ-crystallin gene (Fig. 6E,F). In this series of experiments, exogenous SOX2 expression was sufficient to activate δ-crystallin (Fig. 6C). This is explained by the wide expression of endogenous Pax6 in the surface ectoderm at the time of electroporation (Li et al. 1994; Kamachi et al. 1998), and persistent low expression still observed even after the lens is formed (Fig. 6I). In addition, exogenous SOX1/SOX2/SOX3 expression in the head ectoderm up-regulates endogenous Pax6 expression (Fig. 4J for SOX2; data not shown). This up-regulated endogenous Pax6 must have cooperated with exogenous SOX1/SOX2/SOX3. The data also suggests that Pax6 expression is maintained, at least in part, by SOX1/SOX2/SOX3 in normal processes of lens development.
Figure 6.
Induction of ectopic δ-crystallin expression by exogenous SOX2 and Pax6 in chicken embryo ectoderm. (A–F) Chicken embryos were electroporated in ovo at stage 10 with vectors expressing SOX2 and Pax6 (A,B), SOX2 alone (C,D), or Pax6 alone (E,F), and then analyzed at stage 17 for expression of the δ-crystallin gene by whole mount in situ hybridization (A,C,E). Ectopic δ-crystallin expression is indicated by yellow arrows. Electroporated cells were marked by GFP expression (B,D,F). (G,H) Sections of the embryo expressing ectopic δ-crystallin shown in (A); section showing endogenous δ-crystallin expression in the lens (G), and section showing ectopic δ-crystallin expression in the ectodermal cells with the morphology of thickened placode (H). (I) Low level expression of Pax6 in the head ectoderm of stage 17− normal embryo (green arrow). (J,K) Up-regulation of Pax6 expression in the head ectoderm by exogenous SOX2 in a stage 17+ embryo (J, green arrow). The low level Pax6 expression observed in (I) is extinct by this stage. Electroporated cells were marked by GFP expression (K).
In addition to the ectopic δ-crystallin expression, the Sox2/Pax6 electroporated cells underwent morphological changes characteristic of the lens placode (Fig. 6H), suggesting that the SOX2/Pax6 complex also regulates other target genes responsible for these changes. Thus, activation of SOX2/SOX3 in the Pax6-expressing ectoderm is the critical step in the initiation of δ-crystallin expression and in lens development.
Discussion
Here, we report that Pax6 acts as a co-DNA-binding partner factor of SOX2 in initiating lens differentiation. Pax6 alone binds poorly to the δ-crystallin gene enhancer but binds cooperatively with SOX2 to form an organized ternary complex. The action of Pax6 with SOX2 is totally dependent on the non-canonical binding sequence, and alteration of the sequence to the high affinity Pax6 consensus deteriorates the partnership with SOX2, suggesting a DNA sequence-guided conformational change of the Pax6 protein. Moreover, we successfully correlated synergistic activation by Pax6 and SOX2 with interdependent factor occupancy in the chromatin in vivo. Our findings shed light on the molecular mechanism of early processes of lens development involving Pax6 and SOX2.
Partnership of SOX2 and Pax6 in synergistic transactivation
We demonstrate here that SOX2 and Pax6 cooperatively bind to the DC5 enhancer sequence in both in vitro and in vivo assays, which is correlated with synergistic transcriptional activation. This confirms our previous notion that SOX1/SOX2/SOX3 and possibly other SOX proteins as well, participate in transcriptional regulation through interaction with co-DNA-binding partner proteins. Our ChIP analysis indicates that synergistic activation of the DC5 enhancer by SOX2 and Pax6 results from highly cooperative and interdependent factor occupancy on the enhancer in vivo (Fig. 3). To our knowledge, our ChIP data are the first demonstration of cooperative binding of two transcription factors in an in vivo reconstituted system. Requirement of partner factors for binding of SOX proteins to enhancers in vivo accounts for the observation that most SOX proteins are not able to substantially activate transcription by themselves (Kamachi et al. 2000). An analogous example is a partnership between SOX2 and Oct3 in activation of Fgf4 and UTF1 enhancers in the early processes of mouse development. In these cases also, SOX2 and Oct3, under an in vitro condition, bind cooperatively to these enhancer sequences, and synergistically activate the enhancers in vivo (Ambrosetti et al. 1997, 2000; Nishimoto et al. 1999). We have demonstrated here that in an in vivo situation where DNA is in a chromatin structure, SOX binding may not be sufficient to form a stable protein–DNA complex, although its DNA binding is demonstrable under a particular in vitro condition. Our data shown in Figure 3 indicate that requirement of cooperative binding of transcription factors for transactivation is more pronounced in vivo and that importance of cooperative binding may have been underestimated in many cases.
Similarly, Pax proteins may usually employ partner factors (e.g., SOX proteins) in specifically regulating the target genes, since most of the natural Pax binding sequences deviate from the Pax consensus binding sequences, and binding of Pax proteins to these sequences presumably occurs with a considerably lower affinity than to the consensus sequences. For instance, Pax6 binding sites have been mapped in the regulatory sequences of several crystallin genes (Cvekl et al. 1995; Cvekl and Piatigorsky 1996). Those in the δ-crystallin enhancer reported by Cvekl et al. (1995) are distinct from the Pax6 site in the DC5 sequence. Our data suggest that since these Pax6 binding sequences deviate from the consensus sequence like the site in the DC5, significance of these potential sites also should be considered in terms of a factor complex with SOX2 or other partner factors if they exist. It is also known that Pax5 protein forms a functional complex with Ets proteins on the non-canonical binding sequences in the B-cell–specific mb-1 gene promoter (Fitzsimmons et al. 1996). Furthermore, possible partnership between Pax3 and SOX10 is suggested in the regulation of MITF and c-Ret genes during melanocyte differentiation (Bondurand et al. 2000; Lang et al. 2000; Potterf et al. 2000; Smit et al. 2000). This implies that coregulation of gene expression by SOX and Pax family proteins might be a common mechanism of cell differentiation, since there are many tissues such as the central nervous system that express SOX and Pax proteins together.
Molecular organization of the SOX2/Pax6/DNA ternary complex
We should also emphasize that the molecular organization of the high-mobility form of ternary complex is critical for the transactivation potential. To form this complex, Pax6 depends not only on co-DNA-binding of SOX2, but strictly on the particular DNA sequence of DC5 which by itself is a poor Pax6 binding site (Fig. 4). In gel mobility shift analysis, the ternary complex of SOX2/Pax6/wild-type DC5 shows faster mobility than that containing the mutant DC5 sequences, to which Pax6 and SOX2 bind in a less cooperative fashion (Fig. 4). These suggest that, in the ternary complex of SOX2/Pax6/DC5, the Pax6 protein takes a particular conformation to be compatible with interaction with SOX2 and the non-canonical binding sequence. Thus, DNA sequence-guided and protein interaction-induced conformational changes of Pax6 protein occur in the ternary complex. Consistent with this, it has been shown that the Pax6 paired domain is largely structureless in solution, but alters its conformation upon DNA binding to the consensus binding sequence (Epstein et al. 1994a). Conformation of Pax6 in the ternary complex of SOX2/Pax6/wild-type DC5 may be different from that in the binary complex of Pax6/consensus sequence, since alteration of the sequence to the high affinity Pax6 consensus deteriorates the partnership with SOX2. It is known that the SOX HMG domains induce a sharp bend to the DNA that they bind to. DNA bending induced by SOX2 might be involved in the formation of the ternary complex of SOX2/Pax6/DC5 by altering DNA conformation of the DC5 sequence and/or facilitating the protein interaction.
Partnership of SOX2 and Pax6 in lens development
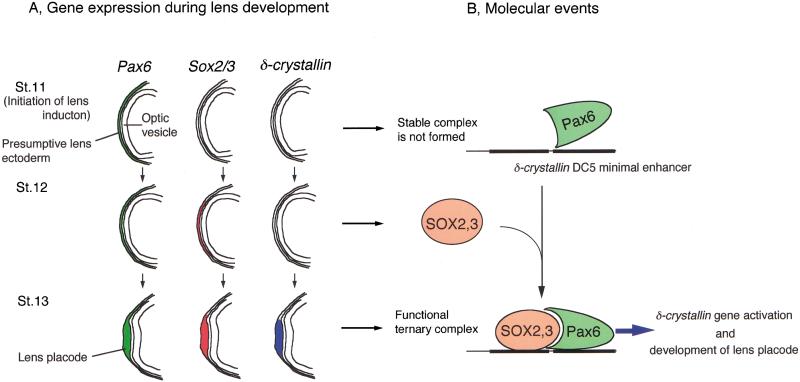
We show that SOX2 and Pax6 cooperatively bind to and form a ternary complex with the target DNA sequence, which is essential for synergistic transcriptional activation. We propose a model that, in the head ectoderm of chicken embryos, Pax6 and SOX2 are expressed in the sequence, and they cooperatively interact with the DC5 enhancer and possibly other target sequences, and initiate lens development (Fig. 7). Expression of Pax6 starts before placode formation in the eye surface ectoderm of most vertebrate species. Expression of Sox2 and/or Sox3 is activated by the inductive signals of the optic vesicle (Sox2 and Sox3 in chicken embryos; Sox2 in mouse embryos [Kamachi et al. 1998]; and Sox3 in frog embryos [Zygar et al. 1998]) within the Pax6-expressing head ectodermal cells. In vertebrates, therefore, Pax6 and Sox2/Sox3 (or one of them) are expressed together in the presumptive lens ectoderm, and then cells that express both genes undergo lens placode development (Fig. 7).
Figure 7.
A model of a genetic switch composed of SOX2/SOX3 and Pax6, which regulates initiation of lens development. (A) Expression of Pax6 (green), Sox2 and Sox3 (red), and δ-crystallin (blue) in the ectoderm during lens induction in chicken embryos. A very low level of Sox2 expression observed in the stage 11 head ectoderm is not illustrated. Expression of the genes in the optic vesicle is omitted. (B) Molecular events that occur on the δ-crystallin enhancer during early lens development. At stage 11, Pax6 protein is expressed in the head ectoderm, but does not make a stable complex with the target DNA sequences such as DC5. After Sox2/Sox3 expression commences at stage 12, Pax6 cooperatively binds with SOX2/SOX3 to the target DNA sequences, and activates genes for lens differentiation such as δ-crystallin. Ternary complex formation presumably involves conformational changes of Pax6 and SOX2/SOX3 as well as DNA.
In the chicken, expression of δ-crystallin is a good indication of the synergistic action of Pax6 and SOX2/SOX3. This notion is corroborated by our previous observation that low level expression of δ-crystallin in non-lens tissues such as Rathke's pouch and the developing spinal cord is always accompanied by Sox2/Sox3 and Pax6 expression (Kamachi et al. 1998), and our recent demonstration that the adenohypophysis primordium (Rathke's pouch in the chicken) can give rise to lens tissues in certain mutants of zebrafish and chicken (Kondoh et al. 2000). However, simultaneous expression of SOX2/SOX3 and Pax6 is not always sufficient for δ-crystallin expression in non-lens cells. For instance, the nasal placode in which endogenous SOX2/SOX3 and Pax6 are present does not express δ-crystallin. Not all cells expressing exogenous SOX2/SOX3 and Pax6 cause ectopic induction of δ-crystallin in the head ectoderm (Fig. 6). When exogenous SOX2 and Pax6 were introduced into trunk ectoderm of chicken embryos, we rarely observed this expression (data not shown). These observations suggest that additional transcription factors are required for full expression of the δ-crystallin gene. Good candidates for such factors are Maf proteins which activate the δ-crystallin gene in Sox2-expressing cells (Ogino and Yasuda 1998, 2000; Kondoh 1999).
Synergism of SOX2/SOX3 and Pax6 is probably a prerequisite for activation of δ-crystallin and other genes involved in early lens development, since induction of δ-crystallin by exogenous SOX1/SOX2/SOX3 and Pax6 is accompanied by morphological changes characteristic of the lens placode (Fig. 6). In normal lens development, changes of cell shape are associated with expression of the cell adhesion molecule N-cadherin. We have recently found that expression of N-cadherin is, in fact, induced in these ectopic lens placodes (M. Uchikawa, M. Matsumata, Y. Kamachi, and H. Kondoh, in prep.). A recent report showed that Pax6 is also essential for expression of N-cadherin in the lens placode (van Raamsdonk and Tilghman 2000). Therefore, it is possible that N-cadherin is also regulated by cooperative action of SOX2 and Pax6 in the lens. We speculate that partnership of SOX2/SOX3 and Pax6 plays an important role in regulation of other lens-specific genes. It is interesting to determine which lens-specific genes are controlled by the SOX2/SOX3-Pax6 system, although binding sequence-based prediction of target genes will not always be possible since binding sequences of the SOX2/SOX3–Pax6 complex may deviate from their individual consensus binding sequences.
Another important link between Sox2/Sox3 and Pax6 in lens development is that their expression is interdependent. We observed that Pax6 expression is up-regulated by exogenous SOX1/SOX2/SOX3 in the head ectoderm (Fig. 6). Köster et al. (2000) have reported similar results that exogenous SOX3 up-regulates Pax6 in medaka fish . In normal development, early Pax6 expression in a broad region of the surface ectoderm is down-regulated, but expression is specifically maintained in the developing lens placode, where Sox2/Sox3 expression has started. This suggests that SOX2/SOX3 proteins are required for maintenance of Pax6 expression in the lens. As Pax6 itself is also essential for maintenance of its own expression in the lens placode, (Grindly et al. 1995; Ashery-Padam et al. 2000) its maintenance might be under the control of the SOX2/Pax6 complex. On the other hand, up-regulation of Sox2 expression in the presumptive lens placode is dependent on Pax6. In normal development, strong expression of Sox2 starts just after the eye head ectoderm is apposed by the optic vesicle, while low level expression of Sox2 in a broad domain of the head ectoderm is seen before its up-regulation in the presumptive lens ectoderm (Furuta and Hogan 1998; Kamachi et al. 1998). This up-regulation is not observed in Pax6−/− mice (Furuta and Hogan 1998). These together suggest that reciprocal regulation by SOX2 and Pax6 restricts and maintains their expression in the lens placode. An interesting possibility is that the SOX2/Pax6 complex may be involved in this process.
Ectopic expression of Sox3 in medaka fish (Köster et al. 2000) and that of Pax6 in Xenopus laevis (Chow et al. 1999) have been reported to cause ectopic lens formation in limited regions of the head ectoderm. These processes probably involve the cross-regulation between SOX2/SOX3 and Pax6 as described above. Our results suggest that the ectopic lenses are derived from cells that possess SOX2/SOX3 and Pax6 proteins of either endogenous or exogenous origin. In medaka fish, the C-terminal deletion mutant of SOX3 phenocopies the effect of ectopic wild-type SOX3 expression (Köster et al. 2000). This might be explained by the fact that transactivation by the SOX2/Pax6 complex largely relies on the C-terminal activation domain of Pax6, and that the effect of the deletion of the SOX2 activation domain is mild (Fig. 5).
Implication of SOX-partner factor interaction for cell differentiation
The present work underscores the importance of the pairs of SOX proteins and their co-DNA-binding partner factors in eliciting cell differentiation. During lens development of chicken embryos, Sox2 and Sox3 are activated within the broader Pax6-expressing head ectodermal cells, and then cells that express both genes undergo lens placode development (Fig. 7). Our data of in vivo factor occupancy suggest that Pax6 protein can associate with the target enhancer sequences such as δ-crystallin DC5 only after SOX2/SOX3 proteins become available in the same cells. This enables the ectodermal cells to develop a sharp boundary between future lens cells and non-lens ectodermal cells. Thus, SOX2/SOX3 and Pax6 appear to organize a genetic switch that regulates initiation of lens development.
Partnering with co-DNA-binding factors accounts for how SOX proteins distinguish their regulatory targets and act in a cell type-specific fashion. SOX proteins including SRY recognize only six to seven base pair DNA sequences with allowance of considerable degeneracy. But they appear to regulate different sets of target genes depending on the cell type in which they are expressed. For instance, it is now apparent that SOX2 regulates at least two sets of target genes by pairing with Pax6 or Oct3 in distinct developmental contexts. Cooperative interactions between SOX proteins and their partner factors may allow stable association with their specific target enhancer sequences composed of binding sites for SOX and its partner factor, as shown by our ChIP analysis. It appears that distribution of partner factors is cell-type specific like Pax6, and the cell-type specificity of the action of SOX proteins may be ascribed to their interaction with the cell-specific partner factors. Thus, this mechanism will endow SOX proteins with highly specific target recognition and cell type-dependent regulatory functions.
Materials and methods
Functional screening of δEF3
Reporter vectors for one hybrid system (Clontech) were adopted for the functional screen in yeast cells. Octamerized DC5 enhancer sequence was cloned into pHisi and pLacZi(AUR1-C). pLacZi(AUR1-C) is a modified version of pLacZi, in which URA3 of pLacZi was replaced with AUR1-C for Aureobasidin A selection (Takara Shuzo). These reporter vectors were integrated into the genome of yeast YM4271. cDNAs of Sox1 and Sox2 were cloned into pBD-GAL4(Cam) (Stratagene) by replacing the sequence coding for the GAL4 DNA-binding domain, and expressed under the control of ADH1 promoter. A cDNA library was constructed with the HybriZAP vector system (Stratagene) using mRNA prepared from lenses of 7-d-old chicken embryos. The reporter-carrying yeast cells were sequentially transformed with the SOX expression vector that contains the TRP1 gene and the lens cDNA library that contains the LEU2 gene, and then grown on selectable plates lacking histidine, tryptophan, and leucine. His+ clones were further assessed for β-galactosidase expression by filter lift assay.
Protein expression and DNA binding assays
SOX2(202) tagged with 6xHis at its C-terminus was expressed by the Bac-to-Bac baculovirus expression system (Gibco BRL) and purified with TALON metal affinity resin (Clontech). Pax6(169) tagged with 6xHis at its C-terminus was first expressed as an N-terminal GST fusion protein, and then cleaved with PreScission protease (Amersham Pharmacia), followed by purification with His tag. Pax6(169) tagged with FLAG at its C-terminus was synthesized similarly. Probes used in gel mobility shift assays were 61-bp fragments carrying wild-type or mutant DC5 sequences excised from pUC19BEX vector (Kamachi and Kondoh 1993) with SalI and Asp718I and labeled with [α-32P]dCTP by Klenow fragment. Binding reactions were carried out in buffer containing 20 mM Hepes (pH 7.9), 45 mM KCl, 10 mM NaCl, 1 mM EDTA, 10% glycerol, 0.1% NP-40, 0.2 μg/μL BSA, 1 mM DTT, and subjected to gel electrophoresis as described in Kamachi and Kondoh (1993).
Protein interaction assay
Three micrograms of GST-SOX2(202) and 0.5 μg of MBP-Pax6(169) were incubated at 4°C for 2 h in 100 μL of the binding buffer containing 20 mM Hepes (pH 7.9), 100 mM KCl, 10% glycerol, 0.1% Triton X-100, 0.5 μg/μL BSA, 1 mM DTT. The complex was precipitated with a 20 μL bed volume of glutathione sepharose (Amersham Pharmacia) and beads were washed three times with the binding buffer without BSA. EtBr was included at 12.5 μg/mL in binding and wash buffers. Bound proteins were subjected to Western blot analysis using anti-MBP antibody (Santa Cruz).
Cell culture and transfection analysis
Lens cell cultures were prepared from 14-d-old chicken embryos as described in Kamachi et al. (1999), while liver cell cultures were prepared from 10-d-old chicken embryos by seeding at 1.8 × 105 cells per 35-mm-diameter dish in DMEM/Ham's F12 medium with 10% FCS two days before transfection. Transfections were carried out as described in Kamachi et al. (1999). Briefly, plasmid DNA containing 1.3 μg of luciferase reporter, 0.1 μg (total) of SOX2/Pax6/empty expression vector (pCMV/SV) mixture, and 0.1 μg of β-galactosidase reference vector (pMiwZ) was transfected by a calcium phosphate precipitation method.
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation was carried out as described (Strahl-Bolsinger et al. 1997) with modifications. Cell lysis and immunoprecipitation were performed in RIPA buffer (50 mM Tris-HCl [pH 7.5], 250 mM NaCl, 1 mM EDTA, 1% NP-40, 0.1% sodium deoxycholate, 0.1% SDS, Complete Protease Inhibitor [Roche]). Immunoprecipitation was done with anti-HA affinity matrix of 20 μL bed volume (Roche). One-fiftieth of the precipitated DNA and 1/50,000 of the total DNA were used in quantitative PCR analysis using the primer pairs shown in Figure 3. The PCR products labeled with [α-32P]dCTP were detected by a Fuji BAS2500 phosphorimage analyzer after separation in a 5% polyacrylamide gel.
in ovo electroporation
Introduction of plasmid vectors into chicken embryos was done by in ovo electroporation as described (Ogino and Yasuda 1998; Nakamura et al. 2000). Plasmid DNA solution at 6 μg/μL was delivered around the head ectoderm of stage 10 chicken embryos and an electric pulse at 5 V/mm for 50 msec was applied five times using a pair of platinum electrodes 4 mm apart.
Acknowledgments
We thank Drs. H. Sasaki, Y. Higashi, and members of H.K.'s laboratory for discussion, and Drs. H. Nakamura and J. Funahashi for electroporation technique. This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan to Y.K., M.U., and H.K. M.U. is the recipient of a fellowship from the Japan Society for the Promotion of Science.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.887101.
References
- Ambrosetti D-C, Basilico C, Dailey L. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol Cell Biol. 1997;17:6321–6329. doi: 10.1128/mcb.17.11.6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosetti D-C, Scholer HR, Basilico C, Dailey L. Modulation of the activity of multiple transcriptional activation domains by the DNA binding domains mediates the synergistic action of Sox2 and Oct-3 on the fibroblast growth factor-4 enhancer. J Biol Chem. 2000;275:23387–23397. doi: 10.1074/jbc.M000932200. [DOI] [PubMed] [Google Scholar]
- Ashery-Padan R, Marquardt T, Zhou X, Gruss P. Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes & Dev. 2000;14:2701–2711. doi: 10.1101/gad.184000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondurand N, Pingault V, Goerich DE, Lemort N, Sock E, Caignec CL, Wegner M, Goossens M. Interaction among SOX10, PAX3 and MITF, three genes altered in Waardenburg syndrome. Hum Mol Genet. 2000;9:1907–1917. doi: 10.1093/hmg/9.13.1907. [DOI] [PubMed] [Google Scholar]
- Chow RL, Altmann CR, Lang RA, Hemmati-Brivanlou A. Pax6 induces ectopic eyes in a vertebrate. Development. 1999;126:4213–4222. doi: 10.1242/dev.126.19.4213. [DOI] [PubMed] [Google Scholar]
- Collinson JM, Hill RE, West JD. Different roles for Pax6 in the optic vesicle and facial epithelium mediate early morphogenesis of the murine eye. Development. 2000;127:945–956. doi: 10.1242/dev.127.5.945. [DOI] [PubMed] [Google Scholar]
- Cvekl A, Piatigorsky J. Lens development and crystallin gene expression: Many roles for Pax-6. BioEssays. 1996;18:621–630. doi: 10.1002/bies.950180805. [DOI] [PubMed] [Google Scholar]
- Cvekl A, Sax CM, Li X, McDermott JB, Piatigorsky J. Pax-6 and lens-specific transcription of the chicken δ1-crystallin gene. Proc Natl Acad Sci. 1995;92:4681–4685. doi: 10.1073/pnas.92.10.4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerny T, Busslinger M. DNA-binding and transactivation properties of Pax-6: Three amino acids in the paired domain are responsible for the different sequence recognition of Pax-6 and BSAP (Pax-5) Mol Cell Biol. 1995;15:2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein J, Cai J, Glaser T, Jepeal L, Maas R. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J Biol Chem. 1994a;269:8355–8361. [PubMed] [Google Scholar]
- Epstein JA, Glaser T, Cai J, Jepeal L, Walton DS, Maas RL. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes & Dev. 1994b;8:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- Fitzsimmons D, Hodsdon W, Wheat W, Maira S-M, Wasylyk B, Hagman J. Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promoter. Genes & Dev. 1996;10:2198–2211. doi: 10.1101/gad.10.17.2198. [DOI] [PubMed] [Google Scholar]
- Fujiwara M, Uchida T, Osumi-Yamashita N, Eto K. Uchida rat (rSey): A new mutant rat with craniofacial abnormalities resembling those of the mouse Sey mutant. Differentiation. 1994;57:31–38. doi: 10.1046/j.1432-0436.1994.5710031.x. [DOI] [PubMed] [Google Scholar]
- Furuta Y, Hogan BLM. BMP4 is essential for lens induction in the mouse embryo. Genes & Dev. 1998;12:3764–3775. doi: 10.1101/gad.12.23.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring WJ, Ikeo K. Pax6: Mastering eye morphogenesis and eye evolution. Trends Genet. 1999;15:371–377. doi: 10.1016/s0168-9525(99)01776-x. [DOI] [PubMed] [Google Scholar]
- Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, Maas RL. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7:463–471. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- Goto K, Okada TS, Kondoh H. Functional cooperation of lens-specific and nonspecific elements in the δ1-crystallin enhancer. Mol Cell Biol. 1990;10:958–964. doi: 10.1128/mcb.10.3.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger RM. Embryonic lens induction: shedding light on vertebrate tissue determination. Trends Genet. 1992;8:349–355. doi: 10.1016/0168-9525(92)90280-h. [DOI] [PubMed] [Google Scholar]
- Grindley JC, Davidson DR, Hill RE. The role of Pax-6 in eye and nasal development. Development. 1995;121:1433–1442. doi: 10.1242/dev.121.5.1433. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Goto K, Okada TS, Kondoh H. Lens-specific enhancer in the third intron regulates expression of the chicken δ1-crystallin gene. Genes & Dev. 1987;1:818–828. doi: 10.1101/gad.1.8.818. [DOI] [PubMed] [Google Scholar]
- Hogan BLM, Horsburgh G, Cohen J, Hetherington CM, Fisher G, Lyon MF. Small eyes (Sey): A homozygous lethal mutation on chromosome 2 which affects the differentiation of both lens and nasal placodes in the mouse. J Embryol Exp Morphol. 1986;97:95–110. [PubMed] [Google Scholar]
- Jean D, Ewan K, Gruss P. Molecular regulators involved in vertebrate eye development. Mech Dev. 1998;76:3–18. doi: 10.1016/s0925-4773(98)00117-8. [DOI] [PubMed] [Google Scholar]
- Kamachi Y, Cheah KSE, Kondoh H. Mechanism of regulatory target selection by the SOX high-mobility-group domain proteins as revealed by comparison of SOX1/SOX2/SOX3 and SOX9. Mol Cell Biol. 1999;19:107–120. doi: 10.1128/mcb.19.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi Y, Kondoh H. Overlapping positive and negative regulatory elements determine lens-specific activity of the δ1-crystallin enhancer. Mol Cell Biol. 1993;13:5206–5215. doi: 10.1128/mcb.13.9.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi Y, Sockanathan S, Liu Q, Breitman M, Lovell-Badge R, Kondoh H. Involvement of SOX proteins in lens-specific activation of crystallin genes. EMBO J. 1995;14:3510–3519. doi: 10.1002/j.1460-2075.1995.tb07357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi Y, Uchikawa M, Collignon J, Lovell-Badge R, Kondoh H. Involvement of Sox1, 2 and 3 in the early and subsequent molecular events of lens induction. Development. 1998;125:2521–2532. doi: 10.1242/dev.125.13.2521. [DOI] [PubMed] [Google Scholar]
- Kamachi Y, Uchikawa M, Kondoh H. Pairing SOX off with partners in the regulation of embryonic development. Trends Genet. 2000;16:182–187. doi: 10.1016/s0168-9525(99)01955-1. [DOI] [PubMed] [Google Scholar]
- Kondoh H. Transcription factors for lens development assessed in vivo. Curr Opin Genet Dev. 1999;9:301–308. doi: 10.1016/s0959-437x(99)80045-8. [DOI] [PubMed] [Google Scholar]
- Kondoh H, Uchikawa M, Yoda H, Takeda H, Furutani-Seiki M, Karlstrom RO. Zebrafish mutations in Gli-mediated hedgehog signaling lead to lens transdifferentiation from the adenohypophysis anlage. Mech Dev. 2000;96:165–174. doi: 10.1016/s0925-4773(00)00387-7. [DOI] [PubMed] [Google Scholar]
- Köster RW, Kühnlein RP, Wittbrodt J. Ectopic Sox3 activity elicits sensory placode formation. Mech Dev. 2000;95:175–187. doi: 10.1016/s0925-4773(00)00356-7. [DOI] [PubMed] [Google Scholar]
- Lai J-S, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D, Chen F, Milewski R, Li J, Lu MM, Epstein JA. Pax3 is required for enteric ganglia formation and functions with Sox10 to modulate expression of c-ret. J Clin Invest. 2000;106:963–971. doi: 10.1172/JCI10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H-S, Yang J-M, Jacobson RD, Pasko D, Sundin O. Pax-6 is first expressed in a region of ectoderm anterior to the early neural plate: Implications for stepwise determination of the lens. Dev Biol. 1994;162:181–194. doi: 10.1006/dbio.1994.1077. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Watanabe Y, Funahashi J. Misexpression of genes in brain vesicles by in ovo electroporation. Dev Growth Differ. 2000;42:199–201. doi: 10.1046/j.1440-169x.2000.00501.x. [DOI] [PubMed] [Google Scholar]
- Nishiguchi S, Wood H, Sockanathan S, Kondoh H, Lovell-Badge R, Episkopou V. Sox1 directly regulates the γ-crystallin genes and is essential for lens development in mice. Genes Dev. 1998;12:776–781. doi: 10.1101/gad.12.6.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto M, Fukushima A, Okuda A, Muramatsu M. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol Cell Biol. 1999;19:5453–5465. doi: 10.1128/mcb.19.8.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowling TK, Johnson LR, Wiebe MS, Rizzino A. Identification of the transactivation domain of the transcription factor Sox-2 and an associated co-activator. J Biol Chem. 2000;275:3810–3818. doi: 10.1074/jbc.275.6.3810. [DOI] [PubMed] [Google Scholar]
- Ogino H, Yasuda K. Induction of lens differentiation by activation of bZIP transcription factor, L-Maf. Science. 1998;280:115–118. doi: 10.1126/science.280.5360.115. [DOI] [PubMed] [Google Scholar]
- ————— Sequential activation of transcription factors in lens induction. Dev Growth Differ. 2000;42:437–448. doi: 10.1046/j.1440-169x.2000.00532.x. [DOI] [PubMed] [Google Scholar]
- Orlando V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem Sci. 2000;25:99–104. doi: 10.1016/s0968-0004(99)01535-2. [DOI] [PubMed] [Google Scholar]
- Pevny LH, Sockanathan S, Placzek M, Lovell-Badge R. A role for SOX1 in neural determination. Development. 1998;125:1967–1978. doi: 10.1242/dev.125.10.1967. [DOI] [PubMed] [Google Scholar]
- Potterf SB, Furumura M, Dunn KJ, Arnheiter H, Pavan WJ. Transcription factor hierarchy in Waardenburg syndrome: Regulation of MITF expression by SOX10 and PAX3. Hum Genet. 2000;107:1–6. doi: 10.1007/s004390000328. [DOI] [PubMed] [Google Scholar]
- Smit DJ, Smith AG, Parsons PG, Muscat GE, Sturm RA. Domains of Brn-2 that mediate homodimerization and interaction with general and melanocytic transcription factors. Eur J Biochem. 2000;267:6413–6422. doi: 10.1046/j.1432-1327.2000.01737.x. [DOI] [PubMed] [Google Scholar]
- St-Onge L, Sosa-Pineda B, Chowdhury K, Mansouri A, Gruss P. Pax6 is required for differentiation of glucagon-producing α-cells in mouse pancreas. Nature. 1997;387:406–409. doi: 10.1038/387406a0. [DOI] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes & Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- Tang HK, Singh S, Saunders GF. Dissection of the transactivation function of the transcription factor encoded by the eye developmental gene PAX6. J Biol Chem. 1998;273:7210–7221. doi: 10.1074/jbc.273.13.7210. [DOI] [PubMed] [Google Scholar]
- van Raamsdonk CD, Tilghman SM. Dosage requirement and allelic expression of PAX6 during lens placode formation. Development. 2000;127:5439–5448. doi: 10.1242/dev.127.24.5439. [DOI] [PubMed] [Google Scholar]
- Wegner M. From head to toes: The multiple facets of Sox proteins. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zygar CA, Cook TLJ, Grainger RM. Gene activation during early stages of lens induction in Xenopus. Development. 1998;125:3509–3519. doi: 10.1242/dev.125.17.3509. [DOI] [PubMed] [Google Scholar]