Abstract
The development of compounds to regulate the activation of the complement system in non-primate species is of profound interest because it can provide models for human diseases. The peptide compstatin inhibits protein C3 in primate mammals and is a potential therapeutic agent against unregulated activation of complement in humans but is inactive against nonprimate species. Here, we elucidate this species specificity of compstatin by molecular dynamics simulations of complexes between the most potent natural compstatin analog and human or rat C3. The results are compared against an experimental conformation of the human complex, determined recently by X-ray diffraction at 2.4-Å resolution. The human complex simulations provide information on the relative contributions to stability of specific C3 and compstatin residues. In the rat simulations, the protein undergoes reproducible conformational changes, which eliminate or weaken specific interactions and reduce the complex stability. The simulation insights can be used to design improved compstatin-based inhibitors for human C3 and active inhibitors against lower mammals.
Keywords: molecular dynamics simulations, compstatin analogs, C3 inhibitors, complement system, innate immune response
INTRODUCTION
The complement system provides the first line of defense against foreign pathogens.1 The role of the component system in host defense involves opsonization, inflammation, and lysis and includes autoimmunity, clearance of immune complexes, debris removal, and response to tissue injury. Activation of the complement system proceeds through the convergence of three independently activated but related pathways (classical, alternative, and lectin) to a common pathway. The convergence point is the cleavage of protein C3 to C3b and C3a. Inappropriate activation of the complement system may cause or aggravate several pathological conditions, such as asthma, adult respiratory distress syndrome, hemolytic anemia, rheumatoid arthritis, rejection of xenotransplantation, stroke, and heart attack.2,3 Therefore, the development of drugs that can control the activation of complement is of profound interest. Protein C3 is essential in all pathways and represents a good target for complement inhibition.
The 13-residue cyclic peptide compstatin prevents the proteolytic activation of complement component C3 by binding to the C3 β-chain3–5 and constitutes a promising candidate for the therapeutic treatment of unregulated complement activation. Compstatin has the sequence Ile1-Cys2-Val3-Val4-Gln5-Asp6-Trp7-Gly8-His9-His10-Arg11-Cys12-Thr13-NH2 and is maintained in a cyclic conformation via the disulfide bridge Cys2-Cys12. It was first discovered by using a phage-displayed random peptide library for binding against C3b.6 Structural nuclear magnetic resonance (NMR), mutational, and computational studies have examined systematically the properties of compstatin and several mutant derivatives in solution.4,7–20 These studies have shown that native compstatin is highly flexible in solution and possesses a thermodynamically dominant conformer, with residues 5–8 in a Type I β-turn (probability 42–63%),7 and the terminal residues 1–4 and 12–13 in a hydrophobic cluster.10
Recent X-ray crystallography studies have determined a 2.4-Å resolution structure of the complex between the complement component C3c (the major proteolytic fragment of C3) and the N-terminal acetylated (Ac) double mutant Ac-Val4Trp/His9Ala (W4A9).3 W4A9 is more active than native compstatin by 45-fold (15-fold, compared with a native variant, acetylated at the N-terminal end).4,9,10,12,16,21,22 Its conformation in the C3c complex is significantly different from the thermodynamically dominant conformer of native compstatin in solution; the 5–8 β-turn becomes extended, and a new β-turn is formed by residues 8–11. The average backbone Cα atom root mean square difference between the conformation of W4A9 in the complex and the thermodynamically dominant conformation of native compstatin in solution is 3.7 Å.3
Mutational studies have shown that residues 5–8 and portion of the N-terminal hydrophobic cluster (residues 2, 3, and 12) are critical for activity,7,10 even though certain property-preserving point mutations can be tolerated (Val3Leu, Gln5Asn).10 The compstatin activity depends partly on specific intermolecular and intramolecular interactions, formed in the complex. In the case of W4A9, the critical for activity residues Val3 and Trp7 are buried in hydrophobic pockets and participate in hydrophobic interactions with C3c.3 The Gln5 and Trp7 side chains form specific hydrogen-bonding interactions with nearby residues.
Despite the insights gained from the crystallographic structure, several questions still remain. The development of higher activity compstatin analogs against human C3 is an area of active research.4,10,12–16,23–25 Furthermore, experimental studies have demonstrated that compstatin is active against other primate C3 proteins but fails to inhibit the activation of proteins from lower mammalian species.6,26 This is an important question, because the development of effective inhibitors against nonprimate species can be used to test disease models in nonprimate animals. A sequence alignment of primate and nonprimate C3 proteins is shown in Figure 1. Experiments of compstatin analogs with a large number of nonprimate mammalian species have shown that the lack of compstatin activity against these proteins is not due to steric or electrostatic repulsion between the ligand and the protein.26 Thus, it is plausible that the lack of compstatin binding is due to the loss of specific interactions and/or due to structural rearrangements of the nonprimate proteins.
Figure 1.
Alignment of C3 primary sequences from primate and nonprimate species. The alignment was prepared with the program CLUSTAL W v. 2.0.12.53 The color code used is: red: hydrophobic, green: polar, blue: negatively charged, and purple: positively charged aminoacids. An asterisk denotes invariant amino acids, a colon strongly similar, and a period weakly similar amino acids. The regions interacting with compstatin are enclosed in black boxes.
Molecular dynamics (MD) simulations provide detailed information on the structural properties and stabilizing interactions of biomolecular systems.27,28 In this study, we employ multi-ns MD simulations to compare complexes of the double-mutant W4A9 with the human C3c protein and the rat C3c protein from Rattus norvegicus. The simulations of the human C3c:W4A9 complex are in agreement with the corresponding, recently determined crystallographic structure3 and reproduce well the interactions observed in the crystal complex. An analysis identifies important intermolecular interactions contributing to the stability of the complex and provides an interpretation of the above mutational results in terms of specific interactions.
The simulations of the rat C3c complex provide significant insights on the lack of compstatin activity against the C3 protein from rat and other nonprimate mammals. The rat C3 protein undergoes local conformational changes, which disrupt the interactions with the ligand and reduce the stability of the complex. These changes are reproducible in several independent runs. Furthermore, the behavior of the systems provides insights for the development of higher activity compstatin analogs against human C3c and active analogs against lower mammalian species.
METHODS
Simulation systems
Human and rat complexes
Compstatin
We simulated the double mutant Val4Trp/His9Ala (W4A9) with sequence COCH3-Ile1-Cys2-Val3-Trp4-Gln5-Asp6-Trp7-Gly8-Ala9-His10-Arg11-Cys12-Thr13-NH2. The crystallographic structure of W4A9 in complex with human C3c was recently determined.3 The disulfide bond Cys2-Cys12 was maintained by a disulfide patch of the CHARMM topology file. Titratable residues were assigned their most common ionization state at physiological pH (charged Asp6 and Arg10, neutral His9 and His10). All simulations were conducted with the molecular mechanics program CHARMM, version c35a1.29
C3c
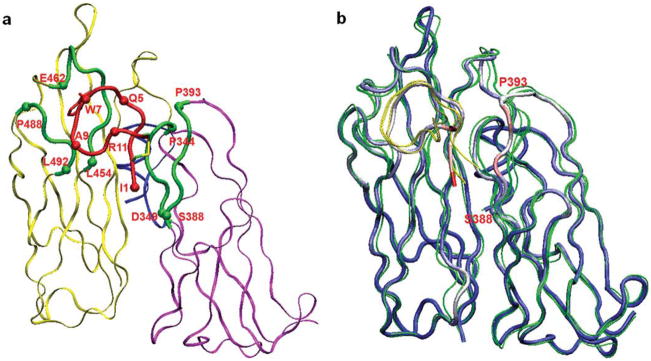
The C3c:W4A9 complex is too large to be entirely included in the simulation system (e.g., the first monomer in the asymmetric crystallographic unit of PDB entry 2QKI has 1120 residues).3 We simulated a truncated C3c region, containing: (i) The entire two domains MG4 (residues 329–424) and MG5 (residues 425–534) of the compstatin binding site and (ii) segment 607–620, which is proximal to MG4 and MG5. The resulting protein-ligand complex [shown in Fig. 2(a)] had dimensions 40 ×53 × 56 Å. Compstatin bound on the solvent-exposed side of the truncated model and was at least 21 Å away from any protein atom omitted in the simulation. The nearest charged residue in the omitted region (Asp26) was ~27 Å away from compstatin Asp6. Poisson calculations with an extended 60-Å protein sphere (centered on compstatin) confirmed that the electrostatic interactions between compstatin and any charged residues, present in the extended sphere and omitted in the simulation system, were small. For each of these residues, we computed its solution and vacuum electrostatic potential on compstatin, using the UHBD program,30 a zero ionic strength, and protein/ligand dielectric constants ε = 1/80 (solution) and 1/1 (vacuum). The resulting solution potentials were smaller by a factor of ~80 compared with the vacuum potentials, that is, they were sufficiently screened by solvent.
Figure 2.
(a) The C3c-compstatin simulation segment system. Protein and ligand side chains and water are omitted for clarity. Compstatin is shown in red, segment MG4 (residues 329–424) in yellow, segment MG5 (residues 425–534) in magenta, and segment 607–620 is in blue. Four protein sectors in direct contact with compstatin (344–349, 388–393, 454–462, and 488–492) are shown in green. (b) Conformations of the rat complex at the end of the runs R1–R3 (C3c in green, compstatin in yellow). The crystal structure of the human complex is also shown as thick, multicolored tube; its residues are colored, based on the average residue-RMSD values of the final conformations in the rat simulations (blue indicates small and red-white large values). Restrained segment 607–620 is omitted for clarity. Sector 388–393 moves consistently toward the same direction in all three runs, away from the ligand. Compstatin has high RMSD values due to net displacements, which maintain the shape of the bound conformation (see text). The pictures were prepared with VMD version 1.8.7.54
The truncated complexes were immersed in a water box that was replicated in all directions by periodic boundary conditions. An analogous setup was used in Refs. 31,32 In our case, the water box had the shape of a 89-Å truncated octahedron. Overlapping water molecules were omitted, and five chloride anions were added (seven ions in the rat system), to neutralize the total charge. The final human complex had 35,751 atoms (3679 protein-ligand atoms); the rat complex had 35,763 atoms (3641 protein-ligand atoms).
In the human complex, the initial coordinates of the protein and peptide heavy atoms were taken from the crystallographic structure (PDB entry 2QKI).3 In the rat C3c:W4A9 complex, the initial positions of backbone heavy atoms (with the exception of loop 369–378 analyzed below) were also taken from the crystallographic structure of the human complex. With this choice, we avoided introducing any a priori structural differences between the human and rat complexes. Thus, our simulations investigated whether the rat C3c:W4A9 complex was able to maintain the conformation of the human C3c:W4A9 complex, or had the propensity to undergo conformational changes, with a concurrent decrease in compstatin affinity. This was indeed the case as shown in the Results section. Loop 369–378 contains a deletion in the rat protein (Fig. 1 shows an alignment of primate and nonprimate C3 proteins). The initial conformation of this loop was constructed with the program MODELLER33 and had a root mean square difference (RMSD) of 1.39 Å from the corresponding conformation in the human C3c. The heavy atoms of invariant side chains were initially placed on the corresponding coordinates of the human complex. The initial positions of mutated side chains were modeled with the SCWRL4 program.34 Hydrogens were positioned by the HBUILD algorithm of the CHARMM program.
Free C3c protein
The experimental conformation of the free human (FH) C3c fragment has also been determined by X-ray crystallography (PDB code 2A74).35 As discussed in Ref. 3, W4A9 binding conserves the orientation of domains MG4 and MG5 and induces minor, local structural rearrangements in the compstatin binding site (Fig. 3 of Ref. 3). In this study, we conducted a control run of the free human C3c fragment, starting from the conformation of the complex (2QKI). The protein model employed in this simulation was identical to the one of the human complex. The objective of this run was to test whether the compstatin-binding site of human C3c would tend to rearrange, in the absence of compstatin, toward the experimental conformation of the free protein. Our simulation model was indeed able to capture this tendency, as discussed in the Results section.
To compare the behavior of the rat protein in the presence and absence of compstatin, we conducted an additional simulation of the free rat C3c protein. The protein model employed in the simulation was identical to the one of the rat complex. The simulation started from the conformation of the human C3c complex, as was done for the rat C3c complex.
Force field specifications
The peptide atomic charges, van der Waals and stereo-chemical parameters, were taken from the CHARMM22 all-atom force field,36 including a Cross-term MAP (C-MAP) backbone ϕ/ψ energy correction37 and revised indole parameters.38 The water was represented by a modified TIP3P water model.39,40 Electrostatic interactions were calculated without truncation by the particle-mesh Ewald method,41 with a parameter κ = 0.33333 for the charge screening, and sixth-order splines for the mesh interpolations. The Lennard-Jones interactions between atom pairs were switched to zero at a cutoff distance of 14 Å. The temperature was kept at T = 300 K by a Nosé-Hoover thermostat 42,43 by using a mass of 1000 kcal ps−12 for the thermostat. The pressure was maintained at P = 1 atm with a Langevin piston,44 using a 500 amu mass and a 5 ps−1 collision frequency for the piston. The classical equations of motion were integrated by the Leap-Frog integrator, using a time step of 2 fs. Bond lengths to hydrogen atoms and the internal water geometry were constrained to standard values with the SHAKE algorithm,45 implemented into CHARMM.
Simulation protocols
To avoid structural deformations at the protein boundary due to the truncation, the main-chain heavy atoms of an external protein shell, with atoms at least 20 Å away from any atom of compstatin, were harmonically restrained to their initial crystallographic positions. Segments 373–377 of the reconstructed loop (373–376 in rat) were also harmonically restrained. The structure was initially optimized by 150 energy minimization steps with the steepest-descent and adopted-basis Newton-Raphson (ABNR) algorithms. This was followed by an equilibration run, consisting of: (i) 30 ps of dynamics, with all protein and ligand heavy atoms harmonically restrained by a force constant of 10 Kcal mol−1 Å −2 and (ii) five 50-ps segments, in which the harmonic force constants were gradually lowered to 1.5 kcal mol−1 Å −2 in the external shell, and to 0 kcal mol−1 Å −2 elsewhere. The systems were then simulated for 7 ns, retaining the harmonic restraints at the end of equilibration.
Side-chain contacts analysis
Probability-density maps of intermolecular side-chain contacts were computed with the WORDOM package.46 Two side chains were considered in contact if the distance of their geometric centers was smaller than 6.0 Å.
Computation of association free energies
To compare the compstatin affinity for the human and rat C3c protein, we estimated the corresponding association free energies (second column in Table III) by the relation
Table III.
Average Distance (% Hydrogen Bond Occupancy) of Important Intermolecular and Intramolecular (Ligand) Hydrogen-Bonding Atom Pairs
| Compstatin | C3c | X1 | X2 | Average distance (% Hydrogen-bond occupancy)
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Human
|
Rat
|
|||||||||
| H1 | H2 | Average occupancy | R1 | R2 | R3 | Average occupancy | ||||
| Intermolecular Atom Pairs | ||||||||||
| Ile1 OY | Asn390 OD1 | 2.9 | 3.0 | 5.1 (7) | 7.6 (<1) | 4 | 4.4 (18) | 6.2 (1) | 8.2 (4) | 8 |
| Ile1 N | Asn390 OD1 | 2.9 | 2.5 | 3.8 (57) | 6.0 (6) | 32 | 4.6 (30) | 5.0 (5) | 6.7 (16) | 17 |
| Cys2 N | Asn390 OD1 | 3.4 | 3.9 | 3.3 (79) | 4.0 (66) | 73 | 5.8 (9) | 4.3 (40) | 6.4 (1) | 17 |
| Cys2 O | Asn390 ND2 | 5.0 | 3.5 | 4.4 (37) | 3.4 (74) | 56 | 5.0 (27) | 5.5 (24) | 7.2 (1) | 17 |
| Trp4 N | Gly345 CO | 3.1 | 3.0 | 3.0 (97) | 2.9 (100) | 99 | 3.3 (80) | 3.1 (93) | 4.5 (40) | 71 |
| Trp4 NE1 | Thr391 CO | 3.4 | 3.3 | 3.7 (35) | 3.7 (29) | 32 | 7.4 (1) | 5.8 (9) | 7.7 (<1) | 3 |
| Trp4 O | Arg456 NE | 3.5 | 3.1 | 3.7 (33) | 2.9 (98) | 66 | 4.0 (4) | 3.0 (95) | 4.2 (41) | 47 |
| Trp4 O | Arg456 NH* | 3.0 | 3.6 | 3.7 (38) | 3.5 (47) | 43 | 3.0 (86) | 3.1 (88) | 4.3 (33) | 69 |
| Gln5 OE1 | Met/Thr457 N | 3.6 | 2.9 | 3.3 (77) | 3.0 (98) | 88 | 6.5 (5) | 3.2 (86) | 3.6 (60) | 50 |
| Gln5 NE2 | Leu455 CO | 3.6 | 2.7 | 3.6 (47) | 3.2 (89) | 68 | 5.4 (5) | 4.0 (25) | 3.9 (35) | 22 |
| Gln5 NE2 | Asp491 OD* | 2.7 | 3.3 | 5.5 (<1) | 4.8 (<1) | <1 | 4.1 (43) | 5.1 (<1) | 4.8 (1) | 15 |
| Trp7 NE1 | Met/Thr457 CO | 2.6 | 2.6 | 2.9 (100) | 2.9 (100) | 100 | 3.1 (84) | 2.9 (100) | 3.0 (97) | 94 |
| Ala9 N | Asp491 OD* | 3.2 | 3.0 | 2.9 (100) | 3.0 (99) | 100 | 2.9 (98) | 3.2 (83) | 2.9 (99) | 93 |
| His10 N | Asp491 OD* | 2.8 | 2.6 | 3.0 (97) | 2.9 (99) | 98 | 3.0 (88) | 3.7 (46) | 3.1 (89) | 78 |
| His10ND1 | Asp491 OD* | 5.6 | 5.3 | 3.6 (61) | 3.7 (60) | 61 | 5.7 (<1) | 6.0 (1) | 3.0 (92) | 31 |
| Intramolecular Atom Pairs | ||||||||||
| Val3 N | His10 CO | 3.0 | 3.0 | 2.8 (100) | 2.9 (99) | 100 | 5.0 (11) | 3.0 (97) | 4.5 (20) | 43 |
| Val3 CO | Cys12 N | 3.4 | 3.0 | 3.0 (95) | 3.0 (98) | 97 | 5.0 (3) | 2.9 (96) | 3.0 (91) | 63 |
| Val3 CO | Gln5 N | 2.9 | 3.0 | 3.0 (98) | 3.0 (98) | 98 | 3.4 (65) | 3.2 (93) | 3.7 (31) | 63 |
| Gln5 NE2 | Trp7 CO | 3.3 | 4.5 | 3.4 (58) | 4.0 (0.3) | 32 | 3.0 (93) | 3.4 (60) | 3.5 (53) | 69 |
All distances are in Å; hydrogen-bond occupancies (%) are in parentheses. All values have been computed by analysis of 700 snapshots (per run), extracted from the 7-ns simulations at 10-ps intervals. A hydrogen bond was present if the donor (D)–acceptor (A) distance was less than 3.5 Å and the corresponding angle (D–H···A) was larger than 90°. Columns X1 and X2 report the corresponding distances, respectively, in the first and second complex of the asymmetric crystallographic unit.3
| (1) |
where PL, P, and L denote, respectively, the complex, the protein, and the ligand. The individual free energies were estimated in the Molecular Mechanics/Generalized Born Surface Area (MM-GB/SA) approximation,47 by removing all water molecules and ions from the simulation trajectories and applying the relation
| (2) |
where X corresponds to PL, P, or L. The first four terms on the right-hand side of Eq. (2) are, respectively, the bonded, Coulomb, generalized-Born and van der Waals energy, and Sx is the solvent-accessible surface area of state X. The GBSW generalized-Born model was used.48,49 The coefficient σ was set to 0.005 Kcal mol−1 E−1,2 for consistency with the GBSW parameterization. Contributions due to the protein and ligand entropy changes on association were not included in the association free-energy calculations. These terms are associated with large errors (Ref. 50 and references, therein, for a detailed discussion) and partially cancel when comparing the two complexes. Furthermore, it is experimentally established that W4A9 is active against human C3c and inactive against rat C3c. Thus, our intended calculation of the compstatin association-free energies for human and rat C3c is qualitative; indeed, we show that the W4A9 affinity for rat C3c is predicted to be weaker, in agreement with the loss of compstatin–rat C3c interactions (Results) and with the experimental lack of activity against rat C3c.6,26 The interaction energies between two groups of atoms (R and R′; Fig. 3) were computed by the relation
Figure 3.
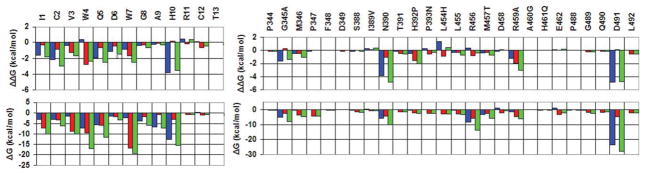
Residue intermolecular interaction energies for compstatin (left panel) and C3c (right panel). For each complex, the energies are computed by Eq. (3) and are averaged over all runs. Lower row: Human C3c complex; Upper row: Energy differences (human—rat). Blue, red and green bars (color online) correspond to polar, non-polar and total free-energy components, respectively. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
| (3) |
The generalized-Born energies and the atomic accessible-surface areas depend on the environment surrounding the groups R and R′. To apply Eq. (3), we considered that all protein and ligand atoms were present and set the charges of atoms outside the two groups R and R′ as zero. In our calculations, R corresponded to a compstatin residue and R′ to the entire C3c model; alternatively, R was a C3c residue, and R′ was the entire ligand.
RESULTS
We conducted seven independent 7-ns runs of the human and rat C3c:W4A9 complexes and the corresponding free human and rat C3c proteins. The simulations are summarized in Table I; details of the simulation protocols are presented in the Methods section.
Table I.
Summary of Simulations Conducted in the Present Work
| Runs | Systems | Starting conformationa | Duration (ns)b |
|---|---|---|---|
| H1 | Human complex | First molecule | 7 |
| H2 | Human complex | Second molecule | 7 |
| R1 | Rat complex | First molecule | 7 |
| R2 | Rat complex | Second molecule | 7 |
| R3 | Rat complex | Second moleculec | 7 |
| FH1 | Free human C3c protein | First molecule | 7 |
| FR1 | Free rat C3c protein | First molecule | 7 |
The starting conformations corresponded to the first or second molecule in the asymmetric unit of the human C3c:W4A9 crystallographic structure (PDB entry 2QKI3); details are presented in the Methods section.
All runs had an additional equilibration phase of 280 ps.
Run R3 was started after extending for 50 ps the equilibration phase of run R2.
Human C3c:W4A9 complex
Crystallographic structure
Native compstatin is highly flexible in solution and possesses a thermodynamically dominant conformer, with residues 5–8 in a Type I β-turn (probability 42–63%) 7 and the terminal residues 1–4 and 12–13 in a hydrophobic cluster.10 NMR and computational studies suggest that analog W4A9 has a similar conformation with native compstatin in solution.16,18 The same analog W4A9 binds to human C3c in a different conformation.3 Region 5–8 is extended, enabling the ligand to contact four protein sectors [Fig. 2(a)]. The N-terminal residues 1–5 are parallel to C3c main-chain segments 345–349 and 388–393; Trp7 is intercalated between main-chain segments 455–458 and 488–491, and residues 9–10 interact with residues 488–492; 3 and the W4A9 main-chain forms a new β-turn in the region 8–11, enabling the segments 10–12 and 1–4 to interact via two hydrogen bonds (Val3 NH-His10 CO and Val3 CO-Cys12 NH).
Simulation conformations
We conducted two simulations of the complex (H1 and H2 in Table I) and one of the free protein (FH1). The simulations of the complex preserve faithfully the crystallographic structure and interactions (PDB Code 2QKI).3 RMSDs between the simulation and crystallographic conformations are reported in Table II and a trajectory corresponding to run H1 is shown in Supporting Information Video 1. The RMSD values in the last 1 ns are 0.71–0.94 Å for the C3c main-chain heavy atoms and less than 1.15 Å for the four proximal to W4A9 sectors (Table II). In the free C3c simulation, sectors 344–349 and 454–462 have somewhat larger RMSD values from the experimental conformation of the complex (1.33 and 1.47 Å, respectively). However, when compared with the experimental structure of free C3c (PDB code: 2A74),35 the corresponding RMSD values become somewhat smaller (1.16 and 1.06 Å). Visual inspection of the free C3c trajectories verified that parts of these sectors (344–347 and 457–462, respectively) tend to approach the free C3c experimental conformation during the simulation. This suggests that the ligand interactions probably assist these two sectors to retain their observed position in the C3c:W4A9 complex.
Table II.
Root Mean Square Difference (RMSD) Between the Simulation Coordinates of Main-Chain Heavy Atoms (N, Cα, and C) and their Corresponding Coordinates in the Crystallographic Structure3
| Runsa | C3cb | 344–349c Moiety | 388–393c Moiety | 454–462c Moiety | 488–492c Moiety | Compstatin | Compstatind |
|---|---|---|---|---|---|---|---|
| H1 | 0.90 (0.90) | 0.93 (0.95) | 1.04 (0.90) | 1.15 (1.09) | 1.09 (1.23) | 1.73 (1.57) | 1.01 (0.85) |
| H2 | 0.71 (0.70) | 0.86 (0.77) | 0.99 (0.97) | 0.80 (0.87) | 0.64 (0.72) | 1.80 (1.60) | 1.20 (0.90) |
| R1 | 0.93 (0.85) | 0.82 (0.75) | 1.51 (1.42) | 1.14 (1.00) | 2.03 (1.59) | 2.15 (2.12) | 1.63 (1.46) |
| R2 | 0.89 (0.88) | 1.28 (1.21) | 2.11 (1.81) | 0.84 (0.90) | 0.73 (0.73) | 2.66 (2.33) | 0.84 (0.80) |
| R3 | 1.12 (1.04) | 1.00 (0.91) | 2.13 (1.88) | 0.94 (1.08) | 1.72 (1.58) | 2.98 (2.23) | 1.20 (1.19) |
| FH1 | 0.94 (0.84) | 1.33 (0.97) | 1.03 (0.88) | 1.47 (1.17) | 1.02 (0.98) | — | — |
| FH1e | 1.04 (0.99) | 1.16 (1.07) | 0.99 (0.77) | 1.06 (0.93) | 1.06 (0.99) | — | — |
| FR1 | 0.89 (0.86) | 1.12 (1.14) | 1.21 (1.50) | 1.42 (1.14) | 1.43 (1.05) | — | — |
All values (except the last column) are computed without rotation/translation and are averaged over the last 1 ns of the runs. In parentheses are the corresponding average RMSD values over the entire length of each run (7 ns). All values are reported in Å.
H1–H2 and R1–R3 denote, respectively, the simulations of the human and rat C3c:W4A9 complexes; FH1 and FR1 denote the simulations of the free human and free rat C3c proteins. Details of the runs are in Table I.
All C3c main-chain heavy atoms, excluding the external, harmonically restrained protein shell (Methods).
Residues in these sectors contain atoms within 7 Å from compstatin.
RMSD values after alignment of the compstatin main-chain atoms N, Cα, and C with respect to the experimental conformation of bound compstatin.
RMSD values with respect to the experimental coordinates of the free C3c protein (PDB code 2A74).
The RMSD values for the ligand backbone are 1.73–1.80 Å, demonstrating that W4A9 stays very near the experimental docking conformation throughout the runs. After alignment with the crystallographic conformation of the ligand backbone, the corresponding RMSD values are 1.0–1.2 Å. Thus, the bound conformation is very well preserved; the Turn 8–11 and the intramolecular β-bridge Val3-Arg11 are always present, and an additional 2–5 β-turn is occasionally formed.
C3c—W4A9 interactions
Mutational studies have shown that residues Val3, Gln5, and Trp7 are critical for compstatin activity.10,12–14,16 The substitutions Val4Trp and His9Ala increase the W4A9 activity by 45 times, compared with native compstatin.16 Our simulations show that these residues make several strong polar and nonpolar contacts with the protein. Figure 3 plots intermolecular interaction energies for W4A9 and C3c residues, computed with Eq. (3) of the Methods section. The individual values for the compstatin and C3c residues of Figure 3 are reported in the Supporting Information Table ST1. Table III lists the statistics of intermolecular and intramolecular (W4A9) hydrogen-bonding pairs. Supporting Information Figure SF1 contains contact maps for selected protein-ligand side-chain pairs.
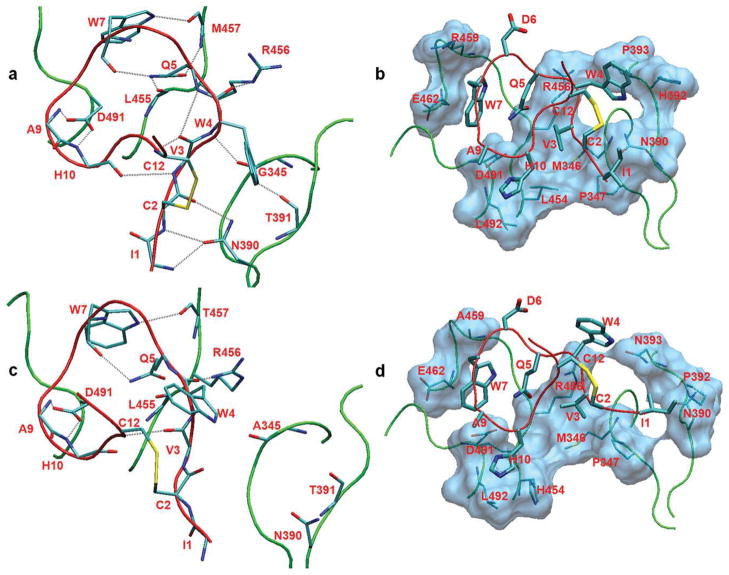
Inspection of the lower panel of Figure 3 shows that W4A9 residues 1–5 and 7–10 interact strongly with residues in the proximal sectors 345–349, 388–393, 454–462, and 488–492. Figure 4(a) shows a representative simulation conformation of the binding site, illustrating the important hydrogen-bonding interactions. The N-terminal residues Ile1 and Cys2 form hydrogen bonds with the side chain of Asn390. The Trp4 main chain makes a very stable hydrogen bond with Gly345 CO and a somewhat weaker interaction with the side chain of Arg456. The Gln5 side chain makes two intermolecular hydrogen bonds with main chain groups of Leu455 and Met457 and an additional intramolecular hydrogen bond with the Trp7 main chain. These bonds participate in a cluster of interactions, involving the groups Trp7 NE1, Met457 CO, Met457 NH, Gln5 OE1, Gln5 NE2, and Trp7 CO [Fig. 4(a)]. The Trp7 side chain intercalates between sectors 455–458 and 488–491, making a stable hydrogen bond with Met457 CO and a more distant interaction with Gln5 OE1. The main-chain NH groups of Ala9 and His10 form very stable hydrogen bonds with the Asp491 side chain. The side chain of His10 makes an additional, less stable interaction with the Asp491 side chain.
Figure 4.
Upper panel: Typical simulation structure of the human C3c:W4A9 complex, showing important (a) hydrogen bonds and (b) nonpolar contacts. Lower panel: Conformation at the end of run R3 of the rat C3c:W4A9 complex, showing the retained (c) hydrogen-bonds, and (d) nonpolar contacts. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Figure 4(b) illustrates important nonpolar contacts, contributing to the affinity of the complex. The Val3 side-chain forms a stable hydrophobic cluster with Met346, Pro347, and Leu454. The Trp4 side chain packs between the Cys2–Cys12 disulfide-bridge and Pro393. The Trp7 side chain makes contacts with the nonpolar parts of Gln5, Met457, Arg459, and Glu462. The His10 side chain is also near a hydrophobic nucleus formed by Leu454 and Leu492. Possibly, the His9 side chain interferes with the interactions of His10 in the nativecompstatin:C3 complex. The remaining residues (Asp6, Arg11, and Thr13) are exposed to the solvent and contribute little to the stability of the complex.
Rat C3c:W4A9 complex
We performed three simulations for the complex (R1–R3 in Table I) and one simulation for the free protein (FR1). As explained in the Methods, to avoid building into the rat model any a priori structural differences from the human complex, we initiated the rat simulations from the experimental conformation of the human complex. With this choice, our simulations investigated whether the rat C3c complex was able to maintain the conformation of the human C3c:W4A9 complex or had the propensity to undergo conformational changes associated with a decrease in compstatin affinity.
The total RMSD of the protein main-chain heavy atoms from the starting (crystallographic) structure were 0.89–1.12 Å in the various runs (Table II), comparable with the corresponding values for the human C3c simulations. However, specific sectors of the protein tend to be more flexible and deviate more from the conformation of the human complex. This is illustrated in Figure 2(b), which displays the crystal structure colored according to the average residue-RMSD values of the final conformations in runs R1–R3 (blue regions indicate low and white-to-red high RMSD values). The conformations at the end of the three runs R1–R3 are also included in the figure. Interestingly, in all three rat C3c complex runs, the protein main-chain moiety 388–393 has a noticeably larger RMSD (1.51–2.13 Å), compared with the human complex [0.99–1.04 Å; Fig. 2(b) and Table II]. Sector 388–393 contains four mutations, including the displacement of a proline residue from position 393 in human C3c to position 392 in rat (alignment in Fig. 1). In the human complex, it packs against the C3c main-chain segment 345–349 and the W4A9 N-terminal moieties 1–4 on one side but is relatively exposed to solvent on the other side. In the rat simulations, this sector is displaced in the same direction, away from the ligand [Fig. 2(b) and Supporting Information Video 2, which displays a trajectory corresponding to run R3). This behavior is also observed in independent simulations of seven other complexes of rat C3c with a series of compstatin analogs (Bellows et al., in preparation) and in the free rat C3c run of the this study. Sector 488–492 has also a larger deviation from its initial conformation in two of the rat runs. The RMSD values of the other two sectors proximal to the ligand 344–349, 454–462 are closer to the corresponding values in the human simulations.
The RMSD values of the W4A9 main-chain heavy atoms are 2.15–2.98 Å, suggesting that the ligand is less tightly bound to rat complex. This will be also confirmed below, with an analysis of the interactions and binding energies. In runs R1 and R3, the 8–11 turn is maintained, but the ligand conformation tends to become somewhat more open and the intramolecular hydrogen bonds Val3 NH-His10 CO and Val3 CO-Cys12 NH disappear.
Rat C3c—W4A9 interactions
The intermolecular difference (human–rat) interaction energies, decomposed in protein and ligand residue contributions, are displayed in the upper panel of Fig. 3. The values of these residue interaction energy differences are also listed in the Supporting Information Table ST2. Errors in these values are also included in Table ST2; they are estimated as the standard deviation of average values, computed over four successive trajectory segments of equal length (1.75 ns).
The interaction energy differences are associated with large fluctuations (Table ST2) but are consistently negative, reflecting the weaker affinity of W4A9 for the rat C3c protein. In accordance with this, the average W4A9–rat C3c hydrogen-bonding occupancies (column 11 of Table III) are consistently smaller than the corresponding average occupancies in the human runs (column 7 of Table III) and most protein-ligand side-chain contacts are reduced (Supporting Information Fig. SF1). Figures 4(c,d) show the remaining intermolecular hydrogen-bonding and nonpolar interactions at the end of run R3. A similar behavior is observed in the other runs.
The displacement of protein segment 388–393 disrupts the hydrogen-bonding interactions of Ile1 and Cys2 main-chain moieties with the Asn390 side-chain and of the Trp4 side-chain with Thr391 [Table III and Fig. 4(c)].
The mutation Leu454His disrupts the hydrophobic cluster of residues Val3, Met346, Pro347, and Leu454 of the human complex [Figs. 4(b,d)] and weakens the non-polar interactions of the Val3 side-chain with the protein (Fig. 3). In R2 and R3, the contact between Val3 and Met346 is lost. As a result, the Val3 side chain is not maintained in a stable orientation. This is shown in Supporting Information Figure SF2, which contains dial plots with the time evolution of selected ligand and protein torsional angles. In R1 and R2, the side chain of His454 is distanced from Val3 and explores several conformations, occasionally interacting with His10.
The substitutions His392Pro and Pro393Asn disrupt the nonpolar contacts of the Trp4 side chain with His392 and Pro393 in the human complex [compare Figs. 4(b,d)]. As the sector 388–393 moves away, the Trp4 side chain explores alternative conformations (Supporting Information Fig. SF2).
The Gln5 side chain makes weaker polar interactions with rat C3c, due to the partial loss of hydrogen bonds with residues Leu455, Thr457, Asp491, and suboptimal interactions with the side chain of His454. Trp7 makes weaker polar and nonpolar interactions with the rat protein (Fig. 3). Its side chain remains intercalated between the sectors 455–458 and 488–491 but loses its contact with the side chain of residue Arg459, which is converted to Ala in the rat protein. His10 has the largest reduction in interaction energy, mainly due to the partial loss of its hydrogen bonds with the Asp491 side chain. The nonpolar contacts of the His10 side chain with Leu492 (invariant) and Leu454 (converted to histidine) are also weaker in the rat runs.
Comparison of the stability of the human and rat complexes
The loss of several hydrogen bonding and nonpolar interactions in the rat simulations suggests that compstatin has a smaller affinity for rat C3c, in accordance with its lack of inhibitory function against rat C3c. The association free energies of the human and rat complexes, computed by Eqs. (1) and (2) of the Methods section by the end-point MM-GB/SA approximation,47 are reported in Table IV.
Table IV.
Average Association Free Energies of the Human and Rat C3c:Compstatin Complexes, Computed From the MD Simulations (All Values in kcal mol−1)
| Runs | Total ΔGa | Intermolecular contributions to ΔGb |
|||
|---|---|---|---|---|---|
| Total | Nonpolar components | Polar components | Polar interactionsc | ||
| H1 | −43 (6) | −55 (1) | −62 (1) | 7 (2) | −48 (5) |
| H2 | −53 (6) | −54 (2) | −59 (1) | 5 (0) | −47 (1) |
| Average | −48 (6) | −55 (1) | −61 (1) | 6 (2) | −47 (4) |
| R1 | −25 (5) | −47 (1) | −51 (2) | 4 (2) | −38 (4) |
| R2 | −36 (5) | −45 (1) | −55 (1) | 10 (1) | −35 (2) |
| R3 | −24 (5) | −45 (7) | −49 (9) | 4 (2) | −36 (4) |
| Average | −29 (5) | −46 (4) | −52 (5) | 6 (1) | −36 (4) |
| ΔΔG | −19 (8) | −9 (4) | −9 (5) | 0 (2) | −11 (5) |
| X1 | −54 | −64 | 10 | −42 | |
| X2 | −57 | −59 | 2 | −46 | |
The association-free energies were computed by application of Eqs. (1) and (2) for the complex, free protein, and free ligand (Methods). Values were averaged over 700 snapshots, spanning the 7-ns runs. Errors (in parentheses) correspond to the standard deviation of mean values, evaluated over four successive trajectory segments of equal length (1.75 ns). X1 and X2 are the two independent complexes of the asymmetric crystallographic unit.3
Intermolecular components were computed by application of Eqs. (1) and (2), assuming that the free protein and free ligand had the same conformation as in the complex. Polar and nonpolar components are defined in Eq. (2).
The polar interaction components measure the strength of intermolecular polar interactions (Coulomb + GB) in the complex. They are evaluated by Eq. (3) (Methods).
End-point methods such as MM-GB/SA and the related Molecular Mechanics/Poisson Boltzmann Surface Area (MM-PB/SA) approximation have been used extensively to compute affinities for protein-ligand complexes (Ref. 51 and references therein) and their connection with statistical thermodynamics has been outlined in Ref. 52. Their use should be done with caution, as they are based on several assumptions; they combine a molecular mechanics energy function with an implicit treatment of solvation effects and include solute conformational entropy effects in an approximate manner (Ref. 50 for a recent criticism and references therein). In this study, we employ the MM-GB/SA method to verify that W4A9 has a weaker affinity for rat C3c, relative to human C3c. This qualitative estimate is sufficient, as it has been established experimentally that W4A9 is active against human C3c and inactive against rat C3c.6,26
The formation of the human complex is associated with an estimated free-energy decrease of −48 kcal mol−1; −55 kcal mol−1 correspond to the formation of intermolecular interactions; and +7 kcal mol−1 (the difference) to conformational changes in the protein and ligand on association. In the rat complex, the estimated association-free energy is −29 kcal mol−1, with −46 kcal mol−1 contributed by intermolecular interactions and +17 kcal mol−1 by conformational changes in protein and ligand. Additional contributions due to changes in the protein and ligand conformational entropies are neglected in our calculations. Overall, the human complex is estimated to be more stable, both due to stronger intermolecular interactions and due to less strain of the protein and ligand on association. Further decomposition shows that the more negative intermolecular contribution in the human complex is due to improved nonpolar interactions. This is clearly seen in Supporting Information Figure SF3, which plots the nonpolar interaction energy of W4A9 with the four C3c proximal sectors 344–349, 388–393, 454–462, and 488–492. The polar intermolecular component opposes the formation of both complexes to the same extent (+6 kcal mol−1). Note that the polar interactions between C3c and W4A9 [evaluated with Eq. (3) and listed in the last column of Table IV] are stronger by 11 kcal mol−1 in the human complex, in accordance with the higher hydrogen-bond occupancies reported above; however, the burial of polar groups on association raises the free energy more in the human complex, yielding a similar total polar-associated component in both complexes.
DISCUSSION AND CONCLUSION
Studies based on NMR experiments, rational design, experimental combinatorial design, and computational combinatorial design have examined systematically the properties of compstatin and several mutant derivatives in solution and have produced a number of compstatin analogs of variable activity.4,7–17,19,20,23 Despite the insights from these studies and the crystallographic structure of the human C3c:W4A9 complex, several questions still remain. In particular, compstatin fails to inhibit C3 activation in nonprimate mammals.26
The present simulations of the human complex are in agreement with the recently determined crystallographic structure and interactions.3 Residues Trp7, Trp4, His10, Gln5, Val3, Ile1, Cys2, and Ala9 contribute significantly (in decreasing order) to the stability of the complex (Fig. 3). In agreement with this, a large body of mutational studies have shown that residues Trp7, Gln5, and Val3 are critical for compstatin activity10,12–14,16 and that the double substitution of Val4Trp and His9Ala increases activity by 45 times, compared with native compstatin.16 Computational design studies suggest that the placement of an aromatic residue at position 4 increases the ligand activity.13,23 Furthermore, a recent pharmacophore model has suggested that the aromatic ring on the fourth residue and the hydrophobic character of the Cys2-Cys12 disulfide bond are important properties of active inhibitors.25 Analysis of the simulation trajectories provides an interpretation of these results. Trp7 is inserted between sectors 455–459 and 488–491 and makes a stable hydrogen bond with Met457 CO and nonpolar contacts with Met457, Arg459, and Glu462 [Fig. 4(a,b)]. Trp4 packs against the Cys2-Cys12 disulfide-bridge on one side and makes nonpolar contacts with Pro393 and the Cα atom of Gly345 on the other side. It also makes a very stable hydrogen bond with Gly345 CO. His10 forms stable hydrogen bonds with the Asp491 and contacts Leu454 and Leu492. Gln5 forms hydrogen bonds with Leu455 and Met457. Ile1 and Cys2 make main-chain hydrogen bonds with Asn390 and the Val3 side-chain participates in a hydrophobic cluster with Met346, Pro347, and Leu454.
The simulations of the rat C3c complex elucidate the lack of compstatin activity against nonprimate C3.6,26 The rat C3 protein undergoes local conformational changes, which disrupt polar and nonpolar interactions with compstatin and reduce the stability of the complex. These changes involve mainly the proximal to compstatin sectors 388–393 and 488–492. The conformational changes are reproducible, as they are observed in all simulations conducted here [Fig. 2(b)], and in independent simulations of seven additional complexes of rat C3c with various compstatin analogs (Bellows, et al., in preparation).
In the rat complex, all intermolecular hydrogen bonds have consistently smaller occupancies (Table III), most protein-ligand side-chain contacts are reduced (Supporting Information Fig. SF1), and the intermolecular interactions of all W4A9 residues (with the exception of Arg11) are consistently weaker, compared with the human complex (Fig. 3). The strongest residue interaction energy differences correspond to residues His10, Cys2, Trp4, Gln5, Trp7, Ile1, and Val3, in decreasing order. As discussed above, these differences are partly due to protein and ligand structural rearrangements, and partly due to differences in the human and rat C3 primary sequences. The displacement of sector 488–492 causes residue His10 to lose a hydrogen bond with the Asp491 side chain and a nonpolar contact with Leu492. The displacement of sector 388–393 and the residue substitutions His392Pro and Pro393Ala disrupt hydrogen bonding and nonpolar interactions with Cys2, Trp4, and Ile1 (Figs. 4c-d). Trp7 loses contacts with the side chain of residue 459, which is converted from Arg to Ala.
The rat C3c mutations His392Pro and Pro393Asn are also observed in many nonprimate mammals. Key residue Asp491 is mutated to alanine and valine in Guinea pig and Sus scrofa, respectively; Arg459 is converted to proline in most nonprimate mammals (Fig. 1). Presumably, these mutations also contribute to the loss of compstatin affinity for C3c in these nonprimate mammals.
The analysis (Fig. 3) showed that residues Asp6, Gly8, Ala9, Arg11, and Thr13 make weak interactions with the protein. Among these, residues Asp6 and Gly8 have been deemed indispensable for activity by earlier alanine-scanning studies; the alanine-substitution effect of Gly8 was detrimental on activity whereas that of Asp6 significantly reduced activity.7 Substitutions of the weakly interacting residues Ala9, Arg11 and Thr13, His10 (whose interactions with C3 are affected by the amino-acid type at position 9) and the N-terminal residue Ile1 may improve inhibition. In a recent de novo computational design study, we examined systematically the affinity for human C3 of a large number of compstatin variants with substitutions at these positions.23 Positions 4 and 13 were found to favor Trp, whereas positions 1, 9, and 10 were dominated by Asn and position 11 by Gln. Experimental binding studies with three of the designed sequences showed improved C3 binding, compared with native compstatin.
Our simulations also provide insights on the design of active inhibitors against rat (or other nonprimate) C3 proteins. Introduction of a charged or polar side chain in position 1 may compensate for the loss of Ile1-Asn390 and Cys2-Asn390 hydrogen-bonding interactions, with proximal residues, such as Asp349, Ser387, and Ser436. Similarly, introduction of a positively charged residue to position 9 or 10 (e.g., a move of the Arg side chain from position 11) may restore some of the lost interactions between Ala9/His10 and the Asp491 side chain. Position 13 can also be improved; the present residue (Thr) does not interact strongly with the protein in the rat or human complex; finally, some of the native compstatin residues of segment 5–8, which were considered critical for activity against the human complex, may be tolerant for mutations in the nonprimate mammal proteins. We are currently investigating new compstatin analogs by computational and experimental methods.
Supplementary Material
Acknowledgments
Grant sponsor: National Science Foundation; Grant number: CTS-0426691; Grant sponsor: National Institutes of Health; Grant number: 5R01GM052032-10.
The simulations were performed on Linux clusters of the Biophysics group at the University of Cyprus. Some simulations were performed at an IBM cluster at the Cyprus Institute. GA and PhT would like to thank Prof. Ro-land Dunbrack for the SCWRL4 program, and Prof. Vassilis Promponas for useful discussions and assistance with the Modeller program.
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- 1.Muller-Eberhard HJ. Complement-Chemistry and pathways. In: Gallin JI, Goldstein IM, Synderman R, editors. Inflammation: basic principles and clinical correlates. 2. New York: Raven Press; 2002. pp. 21–54. [Google Scholar]
- 2.Sahu A, Lambris JD. Complement inhibitors: a resurgent concept in anti-inflammatory therapeutics. Immunopharmacology. 2000;49:133–148. doi: 10.1016/s0162-3109(00)80299-4. [DOI] [PubMed] [Google Scholar]
- 3.Janssen BJC, Halff EF, Lambris JD, Gros P. Structure of Compstatin in Complex with Complement Component C3c Reveals a New Mechanism of Complement Inhibition. J Biochem. 2007;282:29241–29247. doi: 10.1074/jbc.M704587200. [DOI] [PubMed] [Google Scholar]
- 4.Morikis D, Lambris JD. Structure, dynamics, activity and function of compstatin and design of more potent analogs. In: Morikis D, Lambris JD, editors. Structural biology of the complement system. Boca Raton, FL: CRC Press/Taylor & Francis Group; 2005. [Google Scholar]
- 5.Soulika AM, Holland MC, Sfyroera G, Sahu A, Lambris JD. Compstatin inhibits complement activation by binding to the β-chain of complement factor 3. Mol Immunol. 2006;43:2023–2029. doi: 10.1016/j.molimm.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Sahu A, Ray BK, Lambris JD. Inhibition of human complement by a C3-binding peptide isolated from a phage-displayed random peptide library. J Immunol. 1996;157:884–891. [PubMed] [Google Scholar]
- 7.Morikis D, Assa-Munt N, Sahu A, Lambris JD. Solution structure of compstatin, a potent complement inhibitor. Protein Sci. 1998;7:619–627. doi: 10.1002/pro.5560070311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klepeis JL, Floudas CA, Morikis D, Lambris JD. Predicting peptide structures using NMR data and deterministic global optimization. J Comput Chem. 1999;20:1354–1370. [Google Scholar]
- 9.Sahu A, Soulika A, Morikis D, Spruce L, Moore WT, Lambris JD. Binding kinetics, structure-activity relationship, and biotransformation of the complement inhibitor compstatin. J Immunol. 2000;165:2491–2499. doi: 10.4049/jimmunol.165.5.2491. [DOI] [PubMed] [Google Scholar]
- 10.Morikis D, Roy M, Sahu A, Troganis A, Jennings PA, Tsokos GC, Lambris JD. The structural basis of compstatin activity examined by structure-function-based design of peptide analogs and NMR. J Biol Chem. 2002;277:14942–14953. doi: 10.1074/jbc.M200021200. [DOI] [PubMed] [Google Scholar]
- 11.Morikis D, Lambris JD. Structural aspects and design of low-molecular-mass complement inhibitors. Biochem Soc Trans. 2002;30:1026–1036. doi: 10.1042/bst0301026. [DOI] [PubMed] [Google Scholar]
- 12.Soulika AM, Morikis D, Sarrias MR, Roy M, Spruce LA, Sahu A, Lambris JD. Studies of structure-activity relations of complement inhibitor compstatin. J Immunol. 2003;170:1881–1890. doi: 10.4049/jimmunol.171.4.1881. [DOI] [PubMed] [Google Scholar]
- 13.Klepeis J, Floudas CA, Morikis D, Tsokos CG, Argyropoulos E, Spruce L, Lambris JD. Integrated computational and experimental approach for lead optimization and design of compstatin variants with improved activity. J Am Chem Soc. 2003;125:8422–8423. doi: 10.1021/ja034846p. [DOI] [PubMed] [Google Scholar]
- 14.Morikis D, Soulika AM, Mallik B, Klepeis JL, Floudas CA, Lambris JD. Improvement of the anti-C3 activity of compstatin using rational and combinatorial approaches. Biochem Soc Trans. 2004;32:28–32. doi: 10.1042/bst0320028. [DOI] [PubMed] [Google Scholar]
- 15.Klepeis JL, Floudas CA, Morikis D, Tsokos CG, Lambris JD. Design of peptide analogs with improved activity using a de novo protein design approach. Ind Eng Chem Res. 2004;43:3817–3826. [Google Scholar]
- 16.Mallik B, Katragadda M, Spruce LA, Carafides C, Tsokos CG, Morikis D, Lambris JD. Design and NMR characterization of active analogues of compstatin containing non-natural amino acids. J Med Chem. 2005;48:274–286. doi: 10.1021/jm0495531. [DOI] [PubMed] [Google Scholar]
- 17.Morikis D, Mallik B, Zhang L. Biophysical and bioengineering methods for the study of the complement system at atomic resolution. WSEAS Trans Biol Biomed. 2006;6:408–413. [Google Scholar]
- 18.Tamamis P, Skourtis S, Morikis D, Lambris JD, Archontis G. Conformational analysis of compstatin analogues with molecular dynamics simulations in explicit water. J Mol Graph Model. 2007;26:571–580. doi: 10.1016/j.jmgm.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 19.Mallik B, Lambris JD, Morikis D. Conformational interconversion in compstatin probed with molecular dynamics simulations. Proteins. 2003;52:130–141. doi: 10.1002/prot.10491. [DOI] [PubMed] [Google Scholar]
- 20.Magotti P, Ricklin D, Qu H, Wu Y-Q, Kaznessis YN, Lambris JD. Structure-kinetic relationship analysis of the therapeutic complement inhibitor compstatin. J Mol Recognit. 2009;22:495–505. doi: 10.1002/jmr.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holland MC, Morikis D, Lambris JD. Synthetic small molecule complement inhibitors. Curr Opin Investig Drugs. 2004;5:1164–1173. [PubMed] [Google Scholar]
- 22.Morikis D, Floudas CA, Lambris JD. Structure-based integrative computational and experimental approach for the optimization of drug design. In: Sunderam VS, van Albada GD, Sloot PMA, Dongarra JJ, editors. ICCS 2005, lecture notes in computer science: computational science. Berlin: Springer-Verlag; 2005. pp. 680–688. [Google Scholar]
- 23.Bellows ML, Fung HK, Taylor MS, Floudas CA, López De Victoria A, Morikis D. New compstatin variants through two de novo protein design frameworks. Biophys J. 2010;98:2337–2346. doi: 10.1016/j.bpj.2010.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mallik B, Morikis D. Development of a quasi-dynamic pharmacophore model for anti-complement peptide analogues. J Am Chem Soc. 2005;127:10967–10976. doi: 10.1021/ja051004c. [DOI] [PubMed] [Google Scholar]
- 25.Chiu TL, Mulakala C, Lambris JD, Kaznessis YN. Development of a new pharmacophore model that discriminates active compstatin analogs. Chem Biol Drug Des. 2008;72:249–256. doi: 10.1111/j.1747-0285.2008.00709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sahu A, Morikis D, Lambris JD. Compstatin, a peptide inhibitor of complement, exhibits species-specific binding to complement component C3. Mol Immunol. 2003;39:557–566. doi: 10.1016/s0161-5890(02)00212-2. [DOI] [PubMed] [Google Scholar]
- 27.Polydoridis S, Oikonomakos NG, Leonidas DD, Archontis G. Recognition of ribonuclease A by 3′-5′-pyrophosphate-linked dinucleotide inhibitors: a molecular dynamics/continuum electrostatics analysis. Biophys J. 2007;92:1659–1672. doi: 10.1529/biophysj.106.093419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Archontis G, Watson KA, Xie Q, Andreou G, Chrysina ED, Zographos SE, Oikonomakos NG, Karplus M. Glycogen phosphorylase inhibitors: a free energy perturbation analysis of glucopyranose spirohydantoin analogues. Proteins. 2005;61:984–998. doi: 10.1002/prot.20641. [DOI] [PubMed] [Google Scholar]
- 29.Brooks BR, Brooks CL, III, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis ME, Madura JD, Luty BA, McCammon JA. Electrostatics and diffusion of molecules in solution: simulations with the University of Houston Brownian Dynamics Program. Comput Phys Commun. 1991;62:187–197. [Google Scholar]
- 31.Thompson D, Lazennec C, Plateau P, Simonson T. Probing electrostatic interactions and ligand binding in aspartyl-tRNA synthetase through site-directed mutagenesis and computer simulations. Proteins. 2008;71:1450–1460. doi: 10.1002/prot.21834. [DOI] [PubMed] [Google Scholar]
- 32.Thompson D, Plateau P, Simonson T. Free-energy simulations and experiments reveal long-range electrostatic interactions and substrate-assisted specificity in an aminoacyl-tRNA synthetase. Chem-biochem. 2006;7:337–344. doi: 10.1002/cbic.200500364. [DOI] [PubMed] [Google Scholar]
- 33.Eswar N, Marti-Renom MA, Webb B, Madhusudhan MS, Eramian D, Shen M, Pieper U, Sali A. Current protocols in bioinformatics. Suppl 15. New York: John Wiley; 2006. Comparative protein structure modeling with MODELLER; pp. 5.6.1–5.6.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krivov GG, Shapovalov MV, Dunbrack RL., Jr Improved prediction of protein side-chain conformations with SCWRL4. Proteins. 2009;77:778–795. doi: 10.1002/prot.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janssen BJC, Huizinga EG, Raaijmakers HC, Roos A, Daha MR, Nilsson-Ekdahl K, Nilsson B, Gros P. Structures of complement component C3 provide insights into the function and evolution of immunity. Nature. 2005;437:505–511. doi: 10.1038/nature04005. [DOI] [PubMed] [Google Scholar]
- 36.Mackerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FKT, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkewicz-Kuczera J, Yin D, Karplus M. An all-atom empirical potential for molecular modelling and dynamics study of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 37.Mackerell AD, Jr, Feig M, Brooks CL., III Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 38.Macias AT, Mackerell AD., Jr CH/pi interactions involving aromatic amino acids: refinement of the CHARMM tryptophan force field. J Comput Chem. 2005;26:1452–1463. doi: 10.1002/jcc.20281. [DOI] [PubMed] [Google Scholar]
- 39.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 40.Neria E, Fischer S, Karplus M. Simulation of activation free energies in molecular systems. J Chem Phys. 1996;105:1902–1921. [Google Scholar]
- 41.Darden T, York D, Pedersen L. Particle Mesh Ewald: an N log (N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 42.Nose S. A unified formulation of the constant temperature molecular dynamics method. J Chem Phys. 1984;81:511–519. [Google Scholar]
- 43.Hoover W. Canonical dynamics: equilibrium phase-space distributions. Phys Rev A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 44.Feller S, Zhang Y, Pastor RW, Brooks B. Constant-pressure molecular-dynamics simulation: the Langevin piston method. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- 45.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 46.Seeber M, Cecchini M, Rao F, Settanni G, Caflisch A. Wordom: a program for efficient analysis of molecular dynamics simulations. Bioinformatics. 2007;23:2625–2627. doi: 10.1093/bioinformatics/btm378. [DOI] [PubMed] [Google Scholar]
- 47.Massova I, Kollman PA. Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspect Drug Discov Des. 2000;18:113–135. [Google Scholar]
- 48.Im W, Lee MS, Brooks CL., III Generalized Born model with a simple smoothing function. J Comput Chem. 2003;24:1691–1702. doi: 10.1002/jcc.10321. [DOI] [PubMed] [Google Scholar]
- 49.Chen J, Im W, Brooks CL., III Balancing solvation and intramolecular interactions: toward a consistent generalized Born force field. J Am Chem Soc. 2006;128:3728–3736. doi: 10.1021/ja057216r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh N, Warshel A. Absolute binding free energy calculations: on the accuracy of computational scoring of protein-ligand interactions. Proteins. 2010;78:1705–1723. doi: 10.1002/prot.22687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gilson MK, Zhou HX. Calculation of protein-ligand binding affinities. Annu Rev Biophys Biomol Struct. 2007;36:21–42. doi: 10.1146/annurev.biophys.36.040306.132550. [DOI] [PubMed] [Google Scholar]
- 52.Swanson JMJ, Henchmann R, McCammon JA. Revisiting free energy calculations: a theoretical connection to MM/PBSA and direct calculation of the association free energy. Biophys J. 2004;86:67–74. doi: 10.1016/S0006-3495(04)74084-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 54.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamic. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.