Summary
Ligand-dependent transcription by the nuclear receptor glucocorticoid receptor (GR) is mediated by interactions with co-regulators. The role of these interactions in determining selective binding of GR to regulatory elements remains unclear. Recent findings indicate a large fraction of genomic GR binding coincides with chromatin that is accessible prior to hormone treatment, suggesting that receptor binding is dictated by proteins that maintain chromatin in an open state. Combining DNaseI accessibility and chromatin immunoprecipitation with high-throughput sequencing, we identify the activator protein 1 (AP1) as a major partner for productive GR-chromatin interactions. AP1 is critical for GR-regulated transcription and recruitment to co-occupied regulatory elements, illustrating an extensive AP1-GR interaction network. Importantly, the maintenance of baseline chromatin accessibility facilitates GR recruitment and is dependent on AP1 binding. We propose a model where the basal occupancy of transcription factors act to prime chromatin and direct inducible transcription factors to select regions in the genome.
Introduction
Nuclear receptors, a class of ligand-inducible transcription factors, respond to environmental stimuli and mediate expression of genes involved in metabolic, developmental and inflammatory pathways. In the genomic context of eukaryotic cells, nuclear receptors and other DNA-binding factors interact with chromatin, the nucleoprotein complex of DNA and histones. The glucocorticoid receptor (GR), in conjunction with chromatin remodeling proteins, has been shown to be capable of dynamically regulating chromatin structure by ligand-induced remodeling of histone-DNA interactions which in turn leads to the recruitment of previously occluded secondary factors (Zaret and Yamamoto, 1984; Rigaud et al., 1991; Richard-Foy and Hager, 1987). Surprisingly, the global profiling of GR binding (So et al., 2007; Reddy et al., 2009) and accessible chromatin has shown (John et al., 2008; John et al., 2011) that GR binding events are almost always associated with open chromatin. The vast majority of GR binding events (up to 95%) are localized to regions of accessible chromatin that are open prior to hormone treatment (constitutive sites) while only a minority of GR peaks occupy inaccessible chromatin and trigger chromatin remodeling in a hormone-dependent fashion (inducible sites) (John et al., 2011). This suggests that other DNA-binding proteins prime the chromatin landscape in order to maintain an accessible chromatin environment and facilitate the recruitment of inducible transcription factors, such as GR (Herrera et al., 1989).
Co-operative transcription factor interactions have long been implicated in gene regulatory programs (Lin et al., 1990; Sogaard-Andersen and Valentin-Hansen, 1993). Nuclear receptors, including the glucocorticoid, androgen and estrogen receptors interact with a large repertoire of transcription factors (Schule et al., 1988; Norris et al., 2009; Hua et al., 2009; Lupien et al., 2008). The consequences of these interactions have largely been interpreted in the context of changes in interactions with the general transcription apparatus (De Bosscher et al., 2000; Luecke and Yamamoto, 2005).
In the global chromatin context, the contribution of specific transcription factor interactions in regulating GR binding to genomic elements remains a generally unanswered question. Here, we provide evidence that AP1 can facilitate the selective access of GR to specific sites in the genome by maintaining the local chromatin environment in an ‘open’ or accessible configuration. We find that AP1 and GR co-localize to the same regulatory elements in a large fraction of GR binding sites (51%). Using a dominant negative form of the AP1 protein (A-fos), we show that inhibition of the DNA binding activity of AP1 impedes the formation of accessible chromatin and reduces GR binding. These findings reveal a prerequisite for chromatin priming by one transcription factor for the secondary recruitment of other regulatory factors, suggesting that the overall complement of DNA-binding factors (AP1 amongst other proteins) in a given cell type can determine the baseline accessible chromatin landscape. This in turn orchestrates the binding profile and complex response program mediated by signal-dependent factors such as GR.
Results
Genomic binding of AP1 to regulatory elements
Upon hormone addition, 8,236 GR bound elements were identified by chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-Seq)(John et al., 2011) in the murine mammary epithelial cell line, 3134. To elucidate potential interacting partners at regulatory elements bound by GR, we performed an unsupervised de novo sequence motif analysis at GR binding sites and recovered, as expected, the glucocorticoid response element (GRE) at a large fraction of GR bound sites (>75%, at MAST P < 10−3). We also observed enrichment of the consensus AP1 binding site (John et al., 2011; Bailey et al., 2009). Indeed, GR and AP1 are capable of modulating each other’s activities through protein-protein interactions (Yang-Yen et al., 1990; Jonat et al., 1990; Diamond et al., 1990). Using c-Jun as a surrogate mark for bound AP1 (which is typically a complex of Jun-Jun or Jun-Fos oncoproteins), we examined the potential genome-wide interactions between glucocorticoid signaling and AP1 binding. We performed ChIP-seq analysis for AP1 occupancy in the presence and absence of hormone and observed that the preponderance of AP1 binding occurred within intergenic and intronic regions distal to promoters (Figure 1A and 1E).
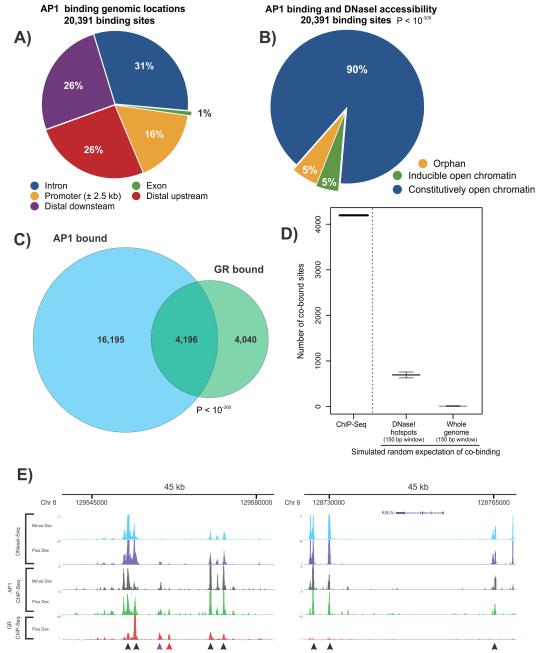
Figure 1. AP1 occupancy at genomic regulatory elements.
A. Genomic location of AP1 binding sites identified by ChIP-Seq. Binding profile of AP1 was determined by c-Jun ChIP-Seq after dexamethasone induction (1hr). Promoter regions are defined as +/− 2.5 kb from an annotated transcription start site.
B. AP1 binding is associated with regions of baseline accessible chromatin. Most AP1 occupancy (90%, blue) occurs within pre-hormone (constitutive) accessible chromatin. A small fraction of AP1 peaks (~5%) are associated with either inducible chromatin or closed chromatin (orphans). Overlapping sites are defined by an intersection of ≥10bp. Statistical significance was determined by binomial distribution.
C. AP1 and GR binding exhibit a high degree of overlap. Venn diagram summarizing global overlap between AP1 and GR binding. Overlapping peaks are defined by an intersection of ≥10bp. A major fraction of GR peaks (51%) overlap sites of AP1 binding. Statistical significance was determined by binomial distribution.
D. Global overlap of AP1 and GR binding is statistically significant. Random sampling simulations in silico were employed to compute the chance overlap between GR and AP1 binding and compared with in vivo experimental data (GR and AP1 ChIP-Seq). Genomic regions for simulated binding were DNaseI hotspots windowed into 150 bp or the entire mouse genome windowed into 150 bp. The overlap determined by ChIP-Seq is significantly enriched over random chance. Figure shows the median and error bars show the minimum and maximum values. See Figure S1H for methodology.
E. Genomic regions of GR and AP1 show significant overlap. Examples [UCSC browser shots; (Kent et al., 2002)] of AP1 ChIP-Seq in the absence and presence of hormone and DNaseI-Seq and GR ChIP-Seq in the absence and presence of A-fos. The DNaseI and GR ChIP experiments were performed after 1hr hormone treatment. Left panel (45 kb region) shows numerous sites of GR and AP1 overlap, all coincident with open chromatin. Right panel (45 kb region) contains AP1 peaks, independent of GR occupancy but coincident with open chromatin. Black arrows denote GR peaks (left panel) or AP1 peaks (right panel) in constitutively accessible chromatin. Purple arrow denotes GR binding at inducible sites. Red arrow denotes orphan GR binding. Inducible DHS sites are markedly weaker than constitutive DHS sites (John et al., 2011).
To understand the role of chromatin structure in regulating AP1 occupancy, we profiled sites of localized chromatin remodeling marked by DNaseI hypersensitive sites (DHSs). DHSs can be robustly identified by sequencing small DNA fragments released from nuclease-treated chromatin (Hesselberth et al., 2009; John et al., 2011). The vast majority of AP1 occupancy (90%) was independent of hormone and occurred predominantly at constitutively open chromatin (Figure 1B, P < 10−300). This latter observation is consistent with a recent report that GR binding itself invariably coincides with open chromatin domains, with the majority of GR binding coincident with pre-existing sites of accessible chromatin (John et al., 2011). These findings highlight a critical role for baseline chromatin accessibility in regulating interactions of sequence-specific transcription factors with their cognate sequences.
We next determined the extent of GR and AP1 co-localization and found a high degree of co-occupancy between the two transcription factors (51% of GR peaks were co-occupied with AP1, Figure 1C, Figure 1E). Both GR and AP1 peaks were significantly enriched over input (Figure S1B and Supplementary Methods). To test the possibility that GR and AP1 can simultaneously co-occupy chromatin, we performed a sequential ChIP for GR followed by AP1, showing at the selected loci, that both factors co-occupy cis-regulatory elements (Figure S1C-G).
Analysis of DNaseI cleavage patterns in deeply (>100 million uniquely mapped tags) sequenced samples enables visualization of regulatory factor footprints within DHSs (Hesselberth et al., 2009). At GR bound elements harboring the AP1 motif, we readily visualized footprinting of AP1 prior to and following hormone treatment (Figure S1A). The protected domain centered on the AP1 motif is approximately 22bp, in agreement with traditional DNaseI footprints of AP1 occupied elements (Lowrey et al., 1992). Collectively, the motif analysis, ChIP-Seq and footprinting results strongly suggest a role for AP1 in the cis-regulatory control of GR binding and function. Specifically, our data suggests that the binding of AP1 prior to hormone might act to preset the chromatin landscape and facilitate GR binding to accessible regulatory elements. Additionally, we profiled GR binding in a murine hepatocyte cell line (Hepa1c1c7). De novo motif analysis for cis-elements within hepatocyte GR peaks were enriched for the AP1, SP1 and forkhead motifs (Figure S1I). This observation suggests that AP1 and other regulatory factors likely contribute to cell-type specific regulation of GR function.
AP1 occupancy regulates global GR binding and function
GR is known to rapidly regulate hundreds of genes, both positively and negatively, in a hormone-dependent manner (John et al., 2009). To explore the role of AP1 interactions on GR mediated gene expression, we exploited the use of a dominant-negative Fos mutant, Acidic-fos (A-fos). A-fos is a truncated Fos protein consisting of the leucine zipper domain and a substitution of the basic region with an acidic extension that maintains dimerization with Jun but inhibits its ability to bind DNA (Olive et al., 1997). We generated a congenic cell line expressing the dominant-negative Fos (A-fos) under conditional control (tet-off expression)(Walker et al., 1999). This enabled the near complete inhibition of endogenous AP1 binding only in the absence of tetracycline (select examples in Figure 3A, Figure S2).
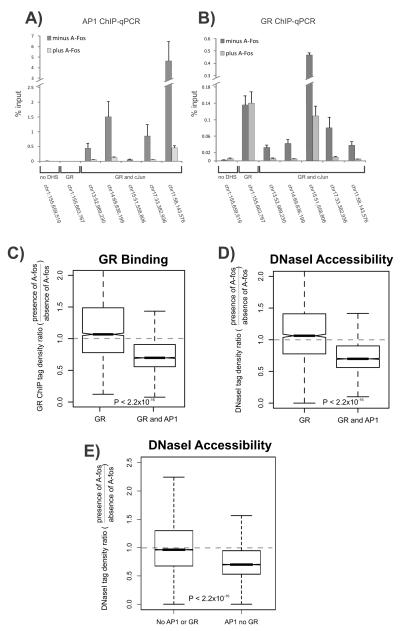
Figure 3. Abrogating AP1 binding attenuates genome-wide GR binding and chromatin accessibility.
A-B. A-fos expression reduces both GR and AP1 binding at co-bound sites. Shown are AP1 (A) and GR ChIP (B) q-PCRs performed in the absence or presence of A-fos and treated with hormone for 1hr at select sites. The following regions were analyzed: a control site which is a non-DNaseI hypersensitive site (no DHS) with no GR or AP1 binding (Chr1:155,659,519); a GR only bound site (Chr1:155,663,767) and five sites bound by AP1 and GR. Data represent the average of three biological replicates. Error bars represent standard error of the mean.
C-D. A-fos expression reduces global GR binding (C) and chromatin accessibility (D) at GR and AP1 co-bound sites. Shown are box plots of the effect of A-fos on GR binding and chromatin accessibility at sites bound by GR alone or sites bound by both GR and AP1. A-fos selectively affects sites bound by AP1 and compromises GR binding and chromatin accessibility at co-bound regions.
E. A-fos expression reduces global chromatin accessibility at AP1 bound sites, independent of GR binding. Shown are box plots of the effect of A-fos on chromatin accessibility at sites not bound by either AP1 or GR or sites bound by AP1 alone. A-fos selectively compromises endogenous AP1 binding and affects chromatin accessibility at sites bound by AP1 (see Figure S3B). For boxplots (C-E), the effect of A-fos on GR binding and chromatin accessibility is expressed as a ratio of tag densities: minus Tet (A-fos expressing) over plus Tet (no A-fos expression). Dotted line is indicative of no A-fos effect. Statistical significance was determined using a KS-test. Boxplots show the median and upper and lower quartiles. Whiskers show the minimum and maximum values. Notches denote the 95% confidence interval of the median.
To address the role of AP1 in GR function, we employed microarray analysis of global gene expression. We identified 651 genes regulated by GR upon hormone activation. Inhibiting AP1 binding with A-fos, affected transcription for 48% of all GR regulated genes (Figure 2A, P < 10−150, Figure S2F). Specifically, 46% of GR induced genes and 51% of GR repressed genes (Figure S2G-H) were compromised in expression upon the inhibition of AP1 binding. In contrast, only 8% of all expressed genes were affected by A-fos (Figure S2E). Analysis of nascent RNA levels (q-PCR) using primers designed to intron-exon boundaries corroborated the microarray data (Figure S2I-J). These results demonstrate that ablation of AP1 binding results in a substantial attenuation of the GR response and provide strong functional evidence of a role for AP1 in transcriptional pathways moderated by GR.
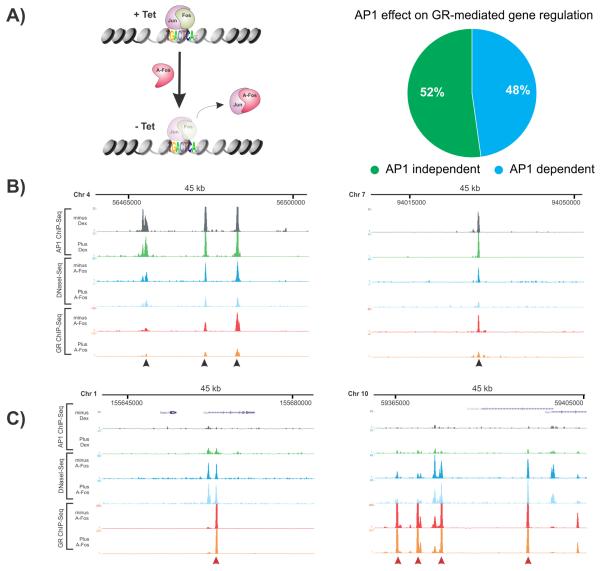
Figure 2. Abrogating AP1 binding attenuates GR activity and chromatin accessibility.
A. A-fos inhibits expression of GR regulated genes. Schematic of a cell line generated to express A-fos (dominant negative Fos) using a tetracycline (tet-off) conditional expression system (left panel). Expression of A-fos results in the strong inhibition of endogenous AP1 through the formation of DNA-binding incompetent heterodimers. Genome-wide effect of the dominant negative activity of A-fos on gene expression was examined by microarray. A-fos mediated changes in gene expression were calculated at a log2 difference of ≥1. 48% of all 651 GR-regulated genes (induced and repressed) were compromised in the hormone response.
B. A-fos expression attenuates GR binding at sites co-bound by GR and AP1. Black arrows denote GR peaks.
C. A-fos expression does not affect GR binding sites that do not co-localize with AP1. Red arrows denote GR peaks. B and C show examples [UCSC browser shots (Kent et al., 2002)] of AP1 ChIP-Seq in the absence and presence of hormone and DNaseI-Seq and GR ChIP-Seq in the absence and presence of A-fos. The DNaseI and GR ChIP experiments were performed after 1hr hormone treatment.
The occupancy of AP1 in chromatin prior to hormone treatment might act as a regulating interface for GR recruitment to response elements upon hormone activation. To examine this, we performed an unbiased genome-wide analysis of GR binding by ChIP-Seq in the absence and in the presence of A-fos. At loci co-occupied by GR and AP1, the expression of A-fos resulted in a marked attenuation of GR recruitment (Figure 2B, Figure 3B and Figure S3A). We analyzed the genome-wide effect of ablating AP1 binding by measuring GR binding at co-bound elements. We detected a significant reduction in GR occupancy (Figure 3C, P < 2.2×10−16) suggesting that AP1 binding is an important regulator for GR recruitment. Sites that are bound only by GR and not AP1 would be predicted to be unaffected by A-fos. Indeed, we found that GR binding at these sites remained unchanged in the presence of A-fos (dotted line, Figure 3C, Figure 2C).
To explore the possibility that AP1 helps maintain chromatin accessibility at GR bound elements, we assayed DNaseI sensitivity genome-wide using DNaseI-Seq in the context of A-fos expression. At individual sites displaying attenuated GR binding in the presence of A-fos, we observed a concomitant strong reduction in chromatin accessibility (Figure 2B). A global analysis for chromatin accessibility by DNaseI-Seq confirmed this reduction in accessible chromatin upon A-fos expression at loci co-bound by AP1 and GR (Figure 3D, P < 2.2×10−16). We also used siRNAs to deplete endogenous c-Jun and confirm effects on GR and chromatin accessibility thereby validating c-Jun contributions by an independent strategy (Supplementary Figure S3C). We also found that AP1 binding that is not coincident with GR binding also shows a net global loss in chromatin accessibility upon A-fos expression (Figure 3E, P < 2.2×10−16, see Figure S3B for examples). These observations demonstrate a global requirement for AP1 binding in the maintenance of chromatin accessibility.
Importantly, at response elements bound by GR alone (but not AP1) we observed no change in either DNaseI accessibility or GR ChIP signal upon A-fos expression (Figure 2C, 3C and 3D). To determine whether all A-fos induced losses in GR binding correlate with losses in chromatin accessibility, we fit a linear regression model for DNaseI and GR binding and indeed found a significant correlation (R=0.31, P < 2.2×10−16) (Figure S3D). This suggests that while a proportion of GR binding events attenuated by A-fos show a loss of chromatin accessibility, a number of affected GR binding sites are independent of changes in chromatin accessibility (Figure S3A, S3D-F), suggesting that accessory factors might contribute to the overall maintenance of open chromatin (see Discussion).
DNA sequence and chromatin structure specify classes of co-bound elements
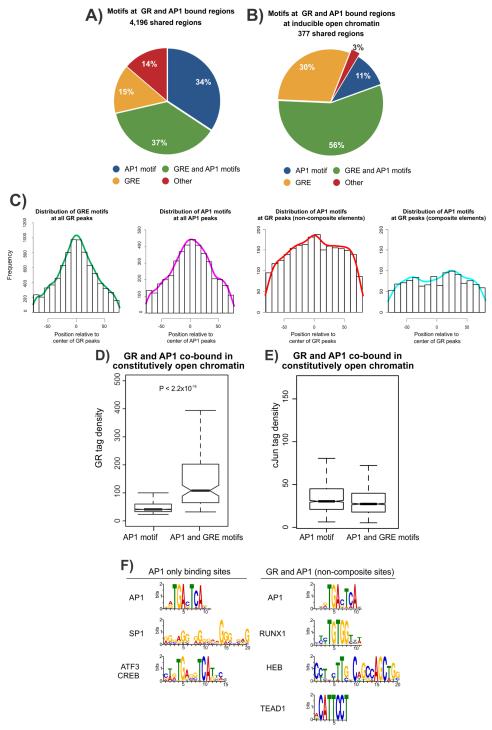
The mode of interaction between GR and AP1 is believed to depend on the sequence context of their cognate recognition motifs. GR can interact with AP1 in a DNA-dependent manner, binding to composite regulatory elements harboring both the glucocorticoid response element (GRE) and AP1 motif (Yang-Yen et al., 1990; Diamond et al., 1990). Alternatively, GR can modulate AP1 activity indirectly through protein-protein interactions (tethering) at non-composite elements (co-bound sites containing an AP1 motif but not a GRE) (Schule et al., 1990). To differentiate between these alternate mechanisms of transcription factor interactions, we analyzed the GRE and AP1 motif instances for all 4,196 elements bound by GR and AP1. We find an equal proportion of co-bound peaks that only have the AP1 motif (ie. no GRE, 34%) or both AP1 and GRE motifs (37%) (Figure 4A, MAST P < 10−3). However, co-bound elements that are associated with hormone-dependent chromatin remodeling contain a higher proportion of matches to the canonical GRE (86% in inducible vs 48% in constitutive) and harbor fewer instances of non-composite elements when compared to constitutive sites (AP1 motif only, 11% in inducible vs 37% in constitutive, Figure 4B vs S4B). The ‘pioneering’ potential of GR manifests at a substantial fraction of GR occupancy sites in the absence of AP1 (47% compared to 6% at co-bound sites, Figure S4D), at which 96% contain the GRE. These results suggest that induced remodeling of chromatin by GR might require a GRE with GR acting as a classic ‘pioneer’ factor that facilitates chromatin remodeling and the binding of secondary factors. However, this modality of GR activity represents a minority of all GR interactions with chromatin (~15%) (John et al., 2011). The mechanisms that direct GR to remodel chromatin at this small subset of GREs are unclear, but likely involve additional epigenetic contributions such as DNA methylation or histone modifications. Conversely, at constitutive sites (> 70% of all GR sites), the dominant mechanism of GR-chromatin interactions at co-bound sites involves the utilization of AP1 to mediate interactions with GR at either composite or non-composite elements.
Figure 4. Motif composition at GR and AP1 co-bound sites.
A. Motifs within GR peaks at co-bound sites contain both composite and non-composite elements. The GRE and AP1 motif position-weight matrices derived from de novo motif discovery were used to scan the genome at a p-value of 10−3. The presence of a motif within the peak was defined as an overlap between peak and motif by ≥1bp.
B. GR peaks at inducible chromatin sites are highly enriched for the GRE motif. Motif composition of GR and AP1 co-bound sites associated with hormone-dependent inducible DHS. The GRE motif is represented in 86% of all GR peaks in inducible sites, in contrast to co-bound regions in constitutively open chromatin (see Figure S4B).
C. Composite and non-composite elements show distinct motif distribution profiles at GR and AP1 co-bound sites. Distribution of the GRE and AP1 motif sequence motifs were determined relative to the center of peaks. Motifs were determined from de novo motif discovery at a p-value of 10−3 where at least half the motif overlapped the peak. Shown are the distribution profiles for the GRE motif at all GR peaks, AP1 motif at all AP1 peaks, AP1 motif uniquely present at non-composite elements and the AP1 motif uniquely present at composite elements at co-bound sites.
D. GR binding is more robust at composite than non-composite elements. Tag density (a measure of DNA binding strength) at GR peaks was determined for GR binding in constitutively accessible chromatin at all co-bound sites containing either the AP1 motif alone (non-composite) or both AP1 and GRE motifs (composite). Statistical significance was determined using a KS-test.
E. AP1 binding strength is independent of motif composition. Tag density at AP1 peaks was determined for AP1 binding in constitutive sites at all co-bound sites containing either the AP1 motif alone (non-composite) or both AP1 and GRE motifs (composite). In D-E, boxplots show the median and upper and lower quartiles. Whiskers show the minimum and maximum values. Notches denote the 95% confidence interval of the median.
F. DNA motifs at AP1 only versus non-composite sites. De novo motif discovery was performed for AP1 binding sites lacking GR binding and AP1 sites that are co-bound with GR. Shown are results that match known sequence motifs at a significance of P < 10−4. Non-composite GR and AP1 co-bound sites are enriched by different DNA sequence motifs compared to AP1 only occupied sites. The AP1 motif was highly enriched in both cases (E < 1.6×10−353).
To extract positional information of motifs within GR or AP1 peaks (Figure S4E), we analyzed the genome-wide distribution of motifs within GR and AP1 peaks and observed that the GRE and AP1 motifs show a trend towards the center of their respective peaks, as expected (Figure 4C). At GR peaks lacking a GRE (non-composite element), the AP1 motif shows a trend towards the center of the GR peak, while composite element containing peaks have the AP1 motif in a broad distribution flanking the center of GR peaks (Figure 4C). In addition, we find that GR binding at elements containing the AP1 motif alone have lower tag densities than elements composed of both AP1 and GRE motifs (P < 2.2×10−16, Figure 4D). These results suggest that GR interactions at non-composite elements might indeed involve indirect interactions with AP1 (Figure 4D). In contrast, the AP1 tag density at AP1 motif only peaks are unchanged when compared with AP1 peaks containing AP1 and GRE motifs, suggesting that, in general, GR is not a major effector of AP1 binding (Figure 4E). We next analyzed the effect of inhibiting AP1 binding on GR recruitment at composite and non-composite elements and observed that both classes of interactions show attenuated GR binding upon A-fos expression (Figure S4F). We additionally analyzed chromatin accessibility at these elements and similarly observed that both classes show an equivalent reduction in DNaseI sensitivity (Figure S4G). The interaction of AP1 with chromatin is, therefore, required for the maintenance of chromatin accessibility and GR binding at shared binding sites and can operate through either composite or non-composite binding mechanisms. As GR specifically interacts with a subset of non-composite AP1 binding sites, we compared motifs enriched within AP1 and GR co-bound non-composite sites versus AP1 only sites (no GR binding). As expected, we enriched for the AP1 motif in both groups, however, non-composite co-bound sites were enriched with distinct motifs compared to AP1 only sites (Figure 4F), suggesting that additional co-regulators contribute to the interaction between GR and AP1 at these elements.
Discussion
Gene regulatory pathways, such as those activated by nuclear receptors, involve the co-ordinated recruitment of transcription factors and co-factors to regulatory elements in chromatin (Rigaud et al., 1991; Ohlsson and Edlund, 1986). Nuclear receptors are known to interact with a diverse group of transcriptional regulators (Schule et al., 1988; Norris et al., 2009; Carroll et al., 2005). In this study, we detail an extensive interaction network between GR and AP1 on a genome-wide scale. These interactions occur predominantly at regulatory elements that are in accessible chromatin. The binding of factors to a pre-determined open chromatin landscape suggests that the maintenance of chromatin accessibility is a critical checkpoint for the recruitment of sequence-specific factors to DNA (John et al., 2008; John et al., 2011).
Utilizing a dominant-negative approach to interfere with the binding activity of the transcription factor AP1, we have explored the interaction between AP1 and GR for receptor recruitment and chromatin accessibility in an unbiased manner on a genome-wide scale. Genomic binding analysis reveals a widespread attenuation of GR loading at AP1 and GR co-occupied elements upon inhibiting AP1 binding. The loss of GR binding is specific to elements bound by AP1, thereby providing evidence for a direct effect of AP1 on GR binding at shared sites.
In vivo, transcription factors bind to only a small complement of potential recognition sequences within the genome (Hesselberth et al., 2009; Carroll et al., 2005). This selectivity is largely due to the requirement for co-operativity with other factors, resulting in the generation of accessible chromatin. This in turn may potentiate binding of additional factors, as has been observed for GR (John et al., 2008, John et al., 2011). Here, we demonstrate that ablating AP1 binding attenuates chromatin accessibility, suggesting a fundamental role for AP1 in the maintenance of chromatin accessibility and, by extension, in the recruitment of GR to chromatin. However, not all regions of accessible chromatin are dependent on AP1 occupancy (Figure 2C and S3A), implicating the activity of other transcription factors in the maintenance of chromatin accessibility. Indeed, de novo motif discovery at such sites show enrichment for Ets and other transcription factor motifs (data not shown). Other DNA-binding factors might, therefore, regulate open chromatin either independently or in concert with AP1. However, the recruitment of GR at these sites might still require AP1 binding by a mechanism potentially mediated by direct interactions that are distinct from the modulation of chromatin accessibility. Furthermore, the regulatory factors that contribute to GR recruitment are likely to be cell-specific. We have identified the AP1 motif at GR binding sites in murine hepatocytes, and additionally show enrichment for other elements such as the SP1 and forkhead motifs. The forkhead motif has also been observed at GR binding sites in pituitary cells. Taken together, these results suggest a role for multiple factors in directing GR binding to chromatin in a cell and region-specific manner (John et al., 2011).
The mode of interaction between nuclear receptors and other transcription factors has been classically categorized into DNA-binding dependent and DNA-binding independent (Yang-Yen et al., 1990; Jonat et al., 1990; Diamond et al., 1990). The DNA-independent activity of GR has been described as a ‘tethering’ mechanism through indirect interactions with AP1 (or other proteins) in the absence of a GRE (Yang-Yen et al., 1990; Reichardt et al., 2001). Such a mechanism of transcriptional modulation is not unique to GR and has been observed for other nuclear receptors (Norris et al., 2009; Saatcioglu et al., 1997; Schule et al., 1991; Adler et al., 1988). We have classified the global mechanisms of GR and AP1 interactions and find that interactions at composite or non-composite elements operate at comparable frequencies. The prerequisite for AP1 occupancy in recruiting GR binding is independent of the motif composition, as we find a dependence of GR binding on AP1 at both types of elements. Intriguingly, non-composite sites are enriched for DNA sequence motifs distinct from AP1 sites lacking GR binding (Figure 4F). GR might therefore be influenced by other co-regulators (eg. RUNX1) on chromatin to interact with AP1 in a DNA-independent manner. Recently, RUNX1 (AML) has been shown to be enriched at non-composite ER sites and is thought to interact with ER through ‘tethering’ mechanisms (Stender et al., 2010).
The classic model of nuclear receptor interactions with other transcription factors such as AP1 and NF-κB involves modulated recruitment and interactions of co-factors with the transcriptional machinery (De Bosscher et al., 2000; Kassel et al., 2004; Rogatsky et al., 2002; Luecke and Yamamoto, 2005; Nissen and Yamamoto, 2000). We propose an alternate model whereby the binding of a transcription factor prior to hormone acts to direct the binding of nuclear receptors. Using unbiased approaches to study the interaction between the ligand inducible transcription factor GR and the constitutively expressed transcription factor AP1, we have revealed a prerequisite for AP1 binding in the recruitment of GR to regulatory elements genome-wide. These interactions occur in the context of chromatin, where the accessibility of the response element to factor binding requires remodeled chromatin. The vast majority of GR occupancy occurs at chromatin regions accessible prior to hormone treatment, suggesting that AP1 occupancy attracts GR to specific functional regulatory elements. The necessity for pre-bound factors and open chromatin in GR function is likely to extend to other nuclear receptors and inducible transcription factors. In breast cancer cells, FOXA1 has been linked to ER binding (Hurtado et al., 2010). During differentiation, C/EBP-α is associated with PPARγ recruitment and PU.1 in liver X receptor binding (Siersbaek et al., 2011; Lefterova et al., 2008; Heinz et al., 2010). The occupancy of these distinct regulatory factors might maintain chromatin accessibility and contribute to the recruitment of nuclear receptors to genomic elements. We provide a conceptual framework for chromatin priming by transcription factors which act to direct and localize the recruitment of inducible transcription factors to genomic elements in order to mediate the regulation of transcriptional programs in response to external stimuli.
Experimental Procedures
Cell growth conditions and generation of congenic lines
The 3134 murine mammary epithelial cell line was grown in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen) and was supplemented with 10% fetal bovine serum (Invitrogen), sodium pyruvate, non-essential amino acids and 2 mM glutamine maintained in a humidifier at 37°C and 5% CO2. A-fos was placed under tetracycline (tet) regulation (tet-off), stably integrated into the 3134 cell line containing the tet regulator (tTa). Cells were plated for experiments in DMEM growth medium supplemented with 10% charcoal-dextran treated serum without tetracycline for 48 hours to induce expression of A-fos, prior to vehicle or hormone treatment (see Supplementary Methods for details).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as per standard protocols (Upstate) with minor modifications using the following antibodies: GR cocktail (PA-510A and PA-511A, Affinity BioReagents; sc-1004 (M-20), Santa Cruz Biotechnology) and c-Jun (sc-1694 (H-79), Santa Cruz Biotechnology). Cells were treated with vehicle or induced with 100nM dexamethasone (Sigma) for 1 hr. Five samples of parallel ChIP experiments from two biological replicates were pooled as a single replicate before generating sequencing libraries. GR and c-Jun ChIPs were validated by quantitative PCR (q-PCR) using SyBr green on a real-time detection system following manufacturer instructions (iCycler IQ; Bio-Rad Laboratories). siRNA (for ChIP and FAIRE) and sequential ChIP (re-ChIP) experiments were performed according to standard protocols (see Supplementary Methods for details).
Mapping chromatin accessibility by DNaseI-Seq
DNaseI digestion was performed as previously described, with minor modifications (John et al., 2011; Hesselberth et al., 2009). Briefly, cells were vehicle treated or hormone induced with 100nM dexamethasone for 1 hr. Cells expressing A-fos were induced for 48 hrs prior to hormone treatment. Nuclei were isolated and digested with DNaseI (Roche) 60 units (U) /ml for 3 mins at 37°C. Cleaved chromatin was incubated with 10 ug/ml RNase A (Roche) and proteinase K (Ambion) overnight at 55°C. DNA was purified by phenol-chloroform extraction and size fractionated by sucrose gradient centrifugation to isolate DNaseI cleavage fragments (100-500 bp) (Sabo et al., 2006) suitable for direct assembly into sequencing libraries.
Library construction and sequencing
DNA from ChIP and DNaseI digestions (for ChIP-Seq and DNaseI-Seq analysis respectively), were assembled into libraries for sequencing according to the Illumina genomic prep kit protocols with minor modifications. Libraries were sequenced on the Illumina genome analyzer platform to generate 27 or 36 bp reads by standard manufacturer’s sequencing-by-synthesis protocols (see Supplementary Methods for details of sequencing data analysis).
Supplementary Material
Acknowledgments
We thank Stephanie Morris, Lars Grontved and Raffaella Nativio for critical reading of the manuscript. This research was supported, in part, by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research and NIH Grant #1RC2HG005654 to J.A.S. S.C.B was supported, in part, by The Needham Cooper Postgraduate Medicine Scholarship. The authors thank Michael Radonovich for technical assistance and Richard Sandstrom for help with data submission. The authors do not have any financial conflict of interest that might be construed to influence the results or interpretation of their manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Codes
All DNaseI and ChIP-Seq data are available through the UCSC genome browser (http://genome.ucsc.edu/) and through NCBI Sequence Read Archive (SRA) under study number SRP007111 and the following accession codes: SRS211887, SRS211888, SRS211889, SRS211890, SRS211891, SRS211892, SRS211894, SRS211895, SRS211896, SRS211897, SRS211898 and SRS211899. All expression array data are available from the Gene Expression Omnibus (GEO) database under study number GSE29983 and the following accession codes: GSM742082, GSM742083, GSM742084, GSM742085, GSM742086, GSM742087, GSM742088 and GSM742089. The deep sequenced DHS data sets are available through ENCODE DCC (UCSC genome browser) under UW mouse DGF (Digital Genomic Footprinting).
Supplemental Information
Supplemental information includes four figures and one table and can be found with this article at doi:
References
- Adler S, Waterman ML, He X, Rosenfeld MG. Steroid receptor-mediated inhibition of rat prolactin gene expression does not require the receptor DNA-binding domain. Cell. 1988;52:685–695. doi: 10.1016/0092-8674(88)90406-0. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- De Bosscher K, Vanden Berghe W, Vermeulen L, Plaisance S, Boone E, Haegeman G. Glucocorticoids repress NF-kappaB-driven genes by disturbing the interaction of p65 with the basal transcription machinery, irrespective of coactivator levels in the cell. Proc. Natl. Acad. Sci. USA. 2000;97:3919–3924. doi: 10.1073/pnas.97.8.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MI, Miner JN, Yoshinaga SK, Yamamoto KR. Transcription factor interactions: selectors of positive or negative regulation from a single DNA element. Science. 1990;249:1266–1272. doi: 10.1126/science.2119054. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera RE, Shaw PE, Nordheim A. Occupation of the c-fos serum response element in vivo by a multi-protein complex is unaltered by growth factor induction. Nature. 1989;340:68–70. doi: 10.1038/340068a0. [DOI] [PubMed] [Google Scholar]
- Hesselberth JR, Zhang Z, Sabo PJ, Chen X, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat. Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua S, Kittler R, White KP. Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell. 2009;137:1259–1271. doi: 10.1016/j.cell.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet. 2010;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Johnson TA, Sung MH, Biddie SC, Trump S, Koch-Paiz CA, Davis SR, Walker R, Meltzer P, Hager GL. Kinetic complexity of the global response to glucocorticoid receptor action. Endocrinology. 2009;150:1766–1774. doi: 10.1210/en.2008-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Johnson TA, Sung MH, Biddie SC, Lightman SL, Voss TC, Davis SR, Meltzer PS, Stamatoyannopoulos JA, Hager GL. Interaction of the glucocorticoid receptor with the global chromatin landscape. Mol. Cell. 2008;29:611–624. doi: 10.1016/j.molcel.2008.02.010. [DOI] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonat C, Rahmsdorf HJ, Park KK, Cato ACB, Gebel S, Ponta H, Herrlich P. Antitumor promotion and antiinflammation: Down-modulation of AP-1 (Fos/Jun) activity by glucocorticoid hormone. Cell. 1990;62:1189–1204. doi: 10.1016/0092-8674(90)90395-u. [DOI] [PubMed] [Google Scholar]
- Kassel O, Schneider S, Heilbock C, Litfin M, Göttlicher M, Herrlich P. A nuclear isoform of the focal adhesion LIM-domain protein Trip6 integrates activating and repressing signals at AP-1 and NF-kappaB-regulated promoters. Genes Dev. 2004;18:2518–2528. doi: 10.1101/gad.322404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ, Jr., Liu XS, Lazar MA. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YS, Carey M, Ptashne M, Green MR. How different eukaryotic transcriptional activators can cooperate promiscuously. Nature. 1990;345:359–361. doi: 10.1038/345359a0. [DOI] [PubMed] [Google Scholar]
- Lowrey CH, Bodine DM, Nienhuis AW. Mechanism of DNase I hypersensitive site formation within the human globin locus control region. Proc. Natl. Acad. Sci. USA. 1992;89:1143–1147. doi: 10.1073/pnas.89.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecke HF, Yamamoto KR. The glucocorticoid receptor blocks P-TEFb recruitment by NFkappaB to effect promoter-specific transcriptional repression. Genes Dev. 2005;19:1116–1127. doi: 10.1101/gad.1297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen RM, Yamamoto KR. The glucocorticoid receptor inhibits NFkappaB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 2000;14:2314–2329. doi: 10.1101/gad.827900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris JD, Chang CY, Wittmann BM, Kunder RS, Cui H, Fan D, Joseph JD, McDonnell DP. The homeodomain protein HOXB13 regulates the cellular response to androgens. Mol. Cell. 2009;36:405–416. doi: 10.1016/j.molcel.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson H, Edlund T. Sequence-specific interactions of nuclear factors with the insulin gene enhancer. Cell. 1986;45:35–44. doi: 10.1016/0092-8674(86)90535-0. [DOI] [PubMed] [Google Scholar]
- Olive M, Krylov D, Echlin DR, Gardner K, Taparowsky E, Vinson C. A dominant negative to activation protein-1 (AP1) that abolishes DNA binding and inhibits oncogenesis. J. Biol Chem. 1997;272:18586–18594. doi: 10.1074/jbc.272.30.18586. [DOI] [PubMed] [Google Scholar]
- Reddy TE, Pauli F, Sprouse RO, Neff NF, Newberry KM, Garabedian MJ, Myers RM. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19:2163–2171. doi: 10.1101/gr.097022.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt HM, Tuckermann JP, Gottlicher M, Vujic M, Weih F, Angel P, Herrlich P, Schutz G. Repression of inflammatory responses in the absence of DNA binding by the glucocorticoid receptor. EMBO J. 2001;20:7168–7173. doi: 10.1093/emboj/20.24.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard-Foy H, Hager GL. Sequence specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 1987;6:2321–2328. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaud G, Roux J, Pictet R, Grange T. In vivo footprinting of rat TAT gene: dynamic interplay between the glucocorticoid receptor and a liver-specific factor. Cell. 1991;67:977–986. doi: 10.1016/0092-8674(91)90370-e. [DOI] [PubMed] [Google Scholar]
- Rogatsky I, Luecke HF, Leitman DC, Yamamoto KR. Alternate surfaces of transcriptional coregulator GRIP1 function in different glucocorticoid receptor activation and repression contexts. Proc. Natl. Acad. Sci. USA. 2002;99:16701–16706. doi: 10.1073/pnas.262671599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saatcioglu F, Lopez G, West BL, Zandi E, Feng W, Lu H, Esmaili A, Apriletti JW, Kushner PJ, Baxter JD, Karin M. Mutations in the conserved C-terminal sequence in thyroid hormone receptor dissociate hormone-dependent activation from interference with AP-1 activity. Mol. Cell Biol. 1997;17:4687–4695. doi: 10.1128/mcb.17.8.4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo PJ, Kuehn MS, Thurman R, Johnson BE, Johnson EM, Cao H, Yu M, Rosenzweig E, Goldy J, Haydock A, et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat. Methods. 2006;3:511–518. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- Schule R, Muller M, Kaltschmidt C, Renkawitz R. Many transcription factors interact synergistically with steroid receptors. Science. 1988;242:1418–1420. doi: 10.1126/science.3201230. [DOI] [PubMed] [Google Scholar]
- Schule R, Rangarajan P, Kliewer S, Ransone LJ, Bolado J, Yang N, Verma IM, Evans RM. Functional antagonism between oncoprotein c-Jun and the glucocorticoid receptor. Cell. 1990;62:1217–1226. doi: 10.1016/0092-8674(90)90397-w. [DOI] [PubMed] [Google Scholar]
- Schule R, Rangarajan P, Yang N, Kliewer S, Ransone LJ, Bolado J, Verma IM, Evans RM. Retinoic acid is a negative regulator of AP-1-responsive genes. Proc. Natl. Acad. Sci. USA. 1991;88:6092–6096. doi: 10.1073/pnas.88.14.6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbaek R, Nielsen R, John S, Sung MH, Baek S, Loft A, Hager GL, Mandrup S. Adipogenic development is associated with extensive early remodeling of the chromatin landscape and establishment of transcription factor ‘hotspots’. EMBO J. 2011;30:1–14. doi: 10.1038/emboj.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So AY, Chaivorapol C, Bolton EC, Li H, Yamamoto KR. Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS. Genet. 2007;3:e94. doi: 10.1371/journal.pgen.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogaard-Andersen L, Valentin-Hansen P. Protein-protein interactions in gene regulation: the cAMP-CRP complex sets the specificity of a second DNA-binding protein, the CytR repressor. Cell. 1993;75:557–566. doi: 10.1016/0092-8674(93)90389-8. [DOI] [PubMed] [Google Scholar]
- Stender JD, Kim K, Charn TH, Komm B, Chang KC, Kraus WL, Benner C, Glass CK, Katzenellenbogen BS. Genome-wide analysis of estrogen receptor alpha DNA binding and tethering mechanisms identifies Runx1 as a novel tethering factor in receptor-mediated transcriptional activation. Mol Cell Biol. 2010;30:3943–3955. doi: 10.1128/MCB.00118-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker D, Htun H, Hager GL. Using inducible vectors to study intracellular trafficking of GFP-tagged steroid/nuclear receptors in living cells. Methods (Companion to Methods in Enzymology) 1999;19:386–393. doi: 10.1006/meth.1999.0874. [DOI] [PubMed] [Google Scholar]
- Yang-Yen HF, Chambard JC, Sun YL, Smeal T, Schmidt TJ, Drouin J, Karin M. Transcriptional interference between c-Jun and the glucocorticoid receptor: mutual inhibition of DNA binding due to direct protein-protein interaction. Cell. 1990;62:1205–1215. doi: 10.1016/0092-8674(90)90396-v. [DOI] [PubMed] [Google Scholar]
- Zaret KS, Yamamoto KR. Reversible and persistent changes in chromatin structure accompany activation of a glucocorticoid-dependent enhancer element. Cell. 1984;38:29–38. doi: 10.1016/0092-8674(84)90523-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.