Abstract
Microarray analysis has provided a new understanding of pineal function by identifying genes that are highly expressed in this tissue relative to other tissues and also by identifying over 600 genes that are expressed on a 24-hour schedule. This effort has highlighted surprising similarity to the retina and has provided reason to explore new avenues of study including intracellular signaling, signal transduction, transcriptional cascades, thyroid/retinoic acid hormone signaling, metal biology, RNA splicing, and the role the pineal gland plays in the immune/inflammation response. The new foundation that microarray analysis has provided will broadly support future research on pineal function.
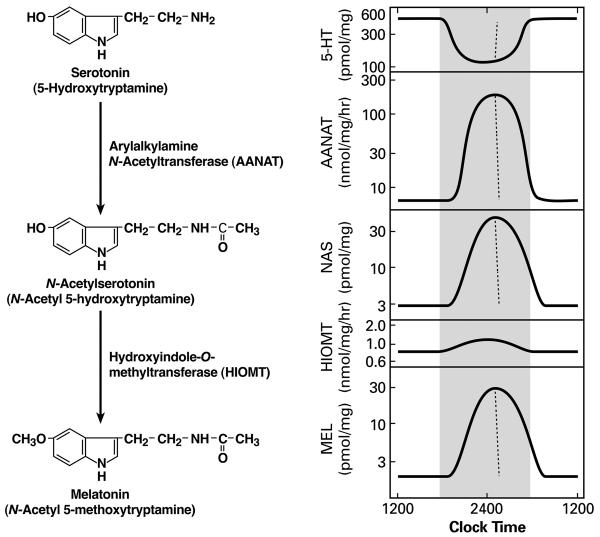
Decades of research on pineal function that focused on the daily rhythm in melatonin production generated a thorough understanding of the system that regulates the conversion of tryptophan to melatonin (Figure 1)(Klein, 1978, Klein, 1985, Maronde and Stehle, 2007). This included the delineation of the neural system which controls the pineal gland (Figure 2) and the transsynaptic/intracellular mechanisms which regulate this pathway (Figure 3)(Klein et al., 1997). A central and conserved feature of this process is that the daily rhythm in circulating melatonin in all vertebrates reflects a rhythm in melatonin production, and that this rhythm is due to daily changes in the activity of the next to last enzyme in this pathway, arylalkylamine N-acetyltransferase (Aanat)(Figure 1)(Klein, 2007).
Figure 1.
Daily rhythm in the serotonin to melatonin pathway. The chemical pathway is shown on the left and the dynamic changes in each element are shown on the right. This general pattern applies to all vertebrates; however the pattern seen in each species differs in amplitude and the shapes of the curves. The broken line represents the changes that are seen following exposure to light in the middle of the dark period. Modified after (Klein, 1974)
Figure 2.
The mammalian melatonin rhythm generating system. The circadian clock which controls the daily rhythm in melatonin production in mammals is in the suprachiasmatic nucleus of the hypothalamus (SCN), located immediately above the optic chiasm (OC). SCN neurons project to the paraventricular nucleus (PVN), making synaptic contact with neurons of the PVN that project caudally close to the midline via the mesencephalic periaqueductal gray (PAG) to make synaptic contact with the intermediolateral nuclei (IML) in the upper thoracic segments of the spinal cord. There, preganglionic neurons innervate a small subpopulation of cells in the superior cervical ganglia (SCG) that send projections to the pineal gland (P) via the internal carotid nerve (ICN) and the conarian nerves (CN). At night, stimulatory signals from the SCN cause release of NE from the postganglionic nerves structures terminating in the pineal gland. In darkness at night, signals from the SCN flow to the pineal gland. Stimulation of the pineal gland by the SCN gradually decreases during the course of the night. In constant darkness these rhythms persist, but will have periods that are greater or less than 24 h. Light controls the period of the rhythm by acting through the retina and a retinohypothalamic projection (RHP), a subpopulation of axons in the optic nerves, to entrain the circadian clock in the SCN to the environmental lighting cycle. Light also controls output from the SCN to the pineal gland so that exposure to light at night terminates SCN stimulation of the pineal gland, resulting in the changes depicted by broken lines in Figure 1. Constant lighting also prevents the changes shown here (Klein, 1985).
Figure 3.
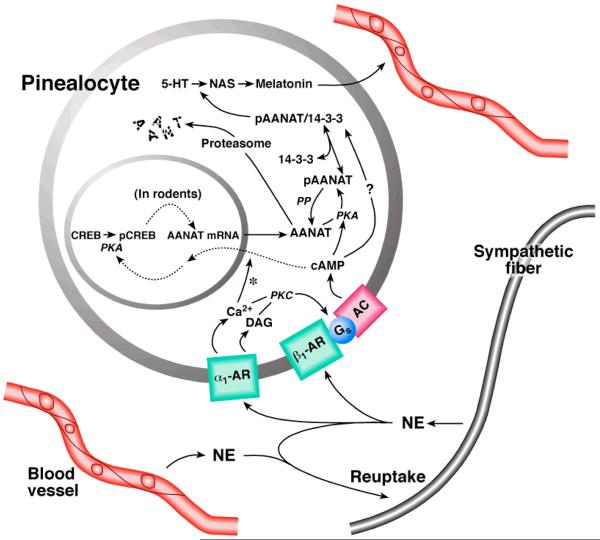
Control of Aanat in the rodent pineal gland. At night, NE is released from sympathetic nerves in the perivascular space in the pineal gland. NE interacts with adrenergic receptors on the pinealocyte membrane to increase intracellular levels of cyclic AMP. This results in activation of cyclic AMP dependent protein kinase (PKC) and phosphorylation of cyclic AMP response element binding protein (CREB), thereby promoting transcription of Aanat. PKA also phosphorylates the Aanat protein, which increases the affinity of the protein for 14-3-3 proteins. The phosphorylated Aaant (pAanat)/ 14-3-3 regulatory complex, protects the pAanat against dephosphorylation by protein phosphatase (PP) and destruction by proteosomal proteolysis; in addition, pAanat has higher affinity for serotonin ( 5-hydroxytryptamine, 5-HT) when complexed to 14-3-3. The complex exists in a dynamic equilibrium with pAANAT and 14-3-3; complex in which formation is favored by phosphorylation. Complex formation increases N-acetylation of 5-HT resulting in an increase in N-acetylserotonin (NAS), which in turn enhances melatonin (MEL) production by a mass action effect, as shown in Figure 1. The levels of hydroxyindole-O-methyltransferase (Asmt) HIOMT do not change significantly on a daily basis. Melatonin is not stored and rapidly passes through the membrane into the circulation. During the day, melatonin production is limited by low levels of Aanat activity. During the night, melatonin synthesis is limited by a availability of cofactors, the activity of other enzymes required for melatonin production, and availability of tryptophan. Termination of the release of NE rapidly reverses the activated system because NE dissociates from receptors and any NE in the perivascular space is rapidly taken back up into the nerve terminals. The drop in perivascular NE causes an immediate reduction to basal levels of cyclic AMP and PKA, leading to disassembly of the Aanat regulatory complex, dephosphorylation of pAANAT and proteosomal proteolysis of Aanat, thereby rapidly decreasing melatonin production and release. The decrease in melatonin release causes a decrease in circulating melatonin because melatonin in the circulation is rapidly destroyed in the liver. The reuptake function of the nerves explains why nonspecific stress or injection of NE have little influence on melatonin production (Klein, 2007).
The daily rhythm in Aanat reflects post translational control mechanisms and, of special interest in the context of this report, in some cases transcriptional control mechanisms are critical. This is seen in the rat, mouse, chicken and some fish (Klein, 2007). The night/day difference in the abundance of Aanat transcripts in the rat is large ( >100-fold), providing investigators with an attractive experimental model system to use to explore how neural signals control transcription. For more on AANAT the reader is referred to a recent review (Klein, 2007).
With a detailed understanding of Aanat regulation as a background, the question arose among investigators as to the scope of the influence of this regulatory system on gene expression in the rodent pineal gland: how many other genes are regulated in a similar manner. Studies on selected genes suggested that this regulatory mechanism was not, in fact, limited to Aanat. A more comprehensive account of the neural control of the rodent pineal transcriptome was pursued, starting in 2002 and has continued using a variety of microarray platforms (Humphries et al., 2002, Kim et al., 2005, Fukuhara and Tosini, 2008, Bailey et al., 2009).
The impact of this effort as of 2009 is the focus of this review. The reader is encouraged to refer to the original reports and related papers to obtain a detailed description of what has been discovered and how these advances have been interpreted. The current article should be considered a first step on a path leading towards a thorough understanding of how neural signals regulate gene expression in the rodent pineal gland.
Background - Control of Melatonin Production in Mammals
The body of research describing the control of the 24-hour rhythm in melatonin production and in Aanat that will be highlighted here will be limited to the mammalian pineal gland. It is of interest to note that regulation of melatonin production and Aanat in submammals differs from that in mammals, as described briefly below.
The neural circuit
Rhythmic production of melatonin is driven by a circadian clock, which functions autonomously in the absence of light/dark cues on a ~24 hour schedule. The location of the clock in mammals is the suprachiasmatic nucleus (SCN), The Mind's Clock(Figure 2)(Moore and Klein, 1974, Klein, 1978, Klein, 1985, Klein and Moore, 1979, Klein et al., 1983a).(Reppert et al., 1981b, Reppert et al., 1981a, Perlow et al., 1981, Perlow et al., 1980) This clock serves as a master oscillator integrating and maintaining circadian rhythms in sleep/wake activity, temperature, cardiovascular function and other physiological functions including the rhythm in melatonin production.
Although the clock in the SCN operates autonomously, in that it maintains a ~24 hour cycle, the clock is reset on a daily basis by light. Photic signals act through a circadian visual system in the retina and a retinohypothalamic projection to entrain SCN to environmental lighting(Klein, 1985, Klein and Moore, 1979). This insures that daily rhythms are optimally coordinated with the night/day cycle, including the melatonin rhythm. The system determines that the pineal gland and melatonin production are only stimulated at night, thereby earning melatonin the moniker `hormone of the night'. Light also acts via the retinohypothalamic projection to control output from the SCN to the pineal gland in a gating function. Through this mechanism, light exposure at night blocks transmission of SCN signals to the pineal gland and rapidly fine tunes melatonin production to reflect subtle changes in the environmental lighting cycle. These two effects of light – resetting and gating - are important elements in determining the integrity of melatonin rhythm as an reliable indicator of night and darkness, providing vertebrates with a circulating signal of time.
The SCN is linked to the pineal gland by a neural pathway which passes through central and peripheral neural structures, including the superior cervical ganglia (SCG), which innervate the glands(Klein et al., 1971, Klein, 1985). At night, SCN stimulation of this pathway causes the release of norepinephrine (NE) from the sympathetic nerves in the gland into pineal perivascular space (Figure 3).
As seen in mammals, in submammals an endogenous clock and light control melatonin production(Bernard et al., 1997, Wolfe et al., 1995). However, the organization of this system differs from that in mammals. In submammals, the clock is located within the same cell in which melatonin is synthesized. Similarly, these cells have two phototransduction systems, one of which resets the clock and another which gates clock-stimulation of Aanat activity(Zatz and Mullen, 1988b). In addition, in the bird, neural input to the pineal gland during the day suppresses pineal activity(Zatz and Mullen, 1988a, Cassone et al., 1986).
The mammalian retina is also capable of synthesizing melatonin, which serves a local messenger function. Melatonin production in the retina follows a circadian pattern that is driven by a clock in the retina, in a manner similar to that in submammalian pineal glands(Tosini et al., 2006, Iuvone et al., 2005).
In mammals, it has been claimed that a clock within the pineal gland controls a rhythmic in sensitivity to adrenergic stimulation(Chansard et al., 2006, Fukuhara et al., 2005, Vaughan and Reiter, 1987, Vaughan et al., 1987). It is not yet clear, however if this is a species specific innovation or a highly conserved among mammals. This is in sharp contrast to the abundant evidence from studies in many mammals that Aanat activity and melatonin production are regulated by the neural pathway described above and that the transsynaptic adrenergic “AND” gate signaling, described below plays a dominant regulatory role.
Transsynaptic adrenergic “AND” gate signaling
NE released from the sympathetic nerves in the pineal gland moves through the perivascular space to the surface of pinealocytes. NE is a mixed α-adrenergic and β-adrenergic agonist and acts by binding to two GPCRs, Adra1b and Adrb1. Binding to Adr1b receptors partially activates adenyl cyclcase, resulting in the elevation of cyclic AMP(Auerbach et al., 1981a, Auerbach et al., 1981b, Vanecek et al., 1985, Sugden and Klein, 1984, Sugden et al., 1984, Klein et al., 1983b). Adra1b activation by NE elevates intracellular Ca++ and activates PLA2 and PLC(Ho et al., 1988a, Ho and Klein, 1987a). The effects on Ca++ PLC result in translocates PKC to the membrane where it acts to potentiates the effect of Adrb1 activation by NE(Sugden et al., 1985, Vanecek et al., 1985, Ho et al., 1988b) This appears to be through an action on adenylate cyclase, perhaps phoshorylation.
This action of NE is described as “AND” gate activation because activation of both Adra1b and Adrb1 are required for maximal stimulation. Interestingly, activation of VIP receptors can substitute for Adra1b activation in this system(Ho et al., 1987). Moreover, the role of Adrb1 activation by NE or VIP can be played by cholera toxin or forsolin(Chik et al., 1988b, Chik et al., 1988a). The former activates adenylate cyclase through an action of G-proteins and the latter via a direct action on adenylate cyclase. Likewise, the role of activation of Adra1b can be played by compounds that activate PKA (eg phorbol esters)(Sugden and Klein, 1988, Sugden et al., 1985).
The central role of cyclic AMP as a second messenger
Cyclic AMP mediates effects of NE on melatonin production through transcriptional and post translational mechanisms, as indicated above(Figure 3). Of special interest in the context of this review is that cyclic AMP induces expression of Aanat. Cyclic AMP acts by activating PKA, which in turn phosphorylates CREB. pCREB bound to CREs in the AANAT promoter initiates transcription(Klein, 1985, Klein, 2006a, Roseboom et al., 1996, Roseboom and Klein, 1995, Humphries et al., 2007, Baler et al., 1997, Baler et al., 1999).
The posttranslational effects of cyclic AMP on AANAT activity are highly conserved among vertebrate and are required for melatonin production to increase at night. The mechanism involves phosphorylation of AANAT at 3' and 5' flanking sites, which promotes binding to 14-3-3 proteins(Aitken, 2006), thereby preventing proteosomal proteolysis and increasing the affinity for serotonin (Klein, 2007, Ganguly et al., 2005, Gardino et al., 2006, Gastel et al., 1998, Obsil et al., 2001, Ganguly et al., 2002, Ganguly et al., 2001).
The Impact of Microarray
The finding that Aanat expression is induced by an NE/cAMP mechanism raised the question of whether expression of other genes is also induced by a similar mechanism. Several reports had suggested that this might be the case with a few genes(Tanaka et al., 1987, Murakami et al., 1989, Baler et al., 1996, Baler and Klein, 1995, Borjigin et al., 1999a, Borjigin et al., 1999b, Borjigin et al., 2003). To obtain a comprehensive answer to this question, microarray was used to profile night/day differences in gene expression in the pineal gland and also to determine whether gene expression was altered due to NE or cyclic AMP treatment(Bailey et al., 2009, Fukuhara et al., 2003, Fukuhara and Tosini, 2008, Humphries et al., 2002, Kim et al., 2005, Kim et al., 2009b)
Microarray was also used to identify genes that are highly expressed in the pineal gland relative to other tissues by calculating the relative expression in the pineal gland compared to the median level of expression in other tissues(Bailey et al., 2003). The resulting value, the rEx (relative Expression), provides an indication of the degree to which expression of a specific genes is enhanced in one tissue relative to median expression among other tissue and provides a indication of the functional potential of a tissue, independent of whether the gene is dynamically regulated. In the case of the pineal gland, examination of rEx values, for example reveal the high levels of expression of the genes which encode proteins dedicated to melatonin production, most of which doe not exhibit day/night changes in gene expression. This approach also disregards genes that are commonly expressed in all tissues at high levels, thereby selecting only those that are highly expressed in a specific tissue.
Global daily changes in gene expression
Physiological studies on the daily changes in gene expression in the rat pineal gland have indicated that there is a 2 to >100-fold change in the expression of over 600 genes, on a night/ day basis (Bailey et al., 2009). Of these, approximately most exhibit increases at night. The most highly rhythmic genes are listed in Table 1, which includes genes that are rhythmically expressed with an amplitude of 4-fold or greater. A more complete summary of these results including genes with a 2-fold rhythm, is available at http://sne.nichd.nih.gov/data/table_s3.xls.
Table 1. Genes that are rhythmically expressed in the pineal gland.
The genes listed are rhythmically expressed 4-fold or greater during the night or day (Bailey et al., 2009). A listing of all genes that are expressed rhythmically >2-fold is available at http://sne.nichd.nih.gov/data/table_s3.xls, which includes gene descriptions, probe set numbers and Entrez Gene numbers.
| Night 8-fold increase | Aanat, Arpp21, Atp7b, Cd24, Cebpb, Cited4, Crem, Cyp1a1, Dclk3*, Dio2, Drd4, Dusp1,Errfi1, Etnk1*, Fcer1a, Fgfr1, Fst, Gdf15, Hspa1a, Irak2, Irs2, Man2a1, Mfrp*, Mt1a, Nap1l5, nod3l, Nr4a1, Nr4a3, Pde10a, Per2, Prlr Ribc2, Slc15a1, Slc6a17, Snf1lk, Sostdc1, St8sia5, Syt4, Wnt10a*, Zrsr1 |
| Night 4 to 8-fold increase | Abca1, Ace, Adcy8, Ankrd52*, Anp32a, Atp2b3, Bex1, Btg2, Cacna1g, Camk1g, Ccrl2*, Cd8a, Chst2*, Cip98, Col5a3, Coq10b, Cry2, Dclk1, Ddit3, Dnm2, Dos*, Dsc2, Dscr1, Egf, Emd, Eomes, Fdx1, Folr1, Fosl2, Gadd45b, Galntl1, Gclm, Gem*,Gls, Gls2, Glt8d3, Grm1, Hcrtr1, Hhip, Hspa1b, Hsph1, Igf1r, Igfbp2, Kcnab2, Kctd3, Lamb3, Lcn7, Lphn2 Lrrn3, Lxn, Mat2a, Mcam, Mtac2d1, Nphp4, Padi4, Parvb*, Pcdh1*, Pde4b, Pde4d, Pde8b, Plagl1, Ptch1, PVR, Qscn6, Rab3ip, Reln, Rhobtb3*, Rock2, Scrt1*, Slc12a2, Slc17a6, Slc30a1, Slco1a5, Tbc1d1*, Tjp2, Top1, Tyro3, Xpot*, |
| Day 8-fold increase | Ccl9, Hs3st2, Ptpn21 |
| Day 4- to 8-fold increase | Cfl2*, Galnt14, Mina, Muc4, Sgk, Sorl1, St8sia3, Tfpi2, Tmed1, Vof16, Wdr89 |
Predicted gene.
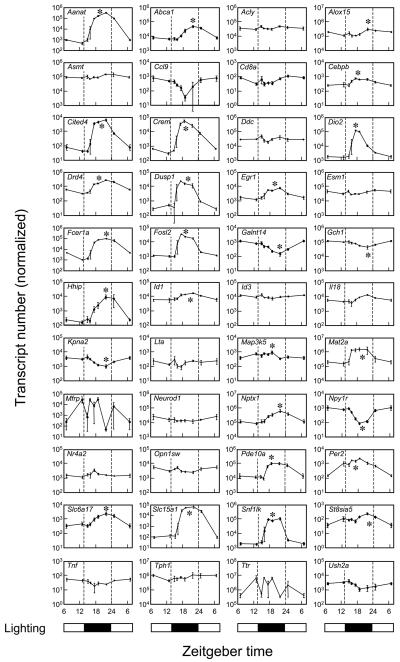
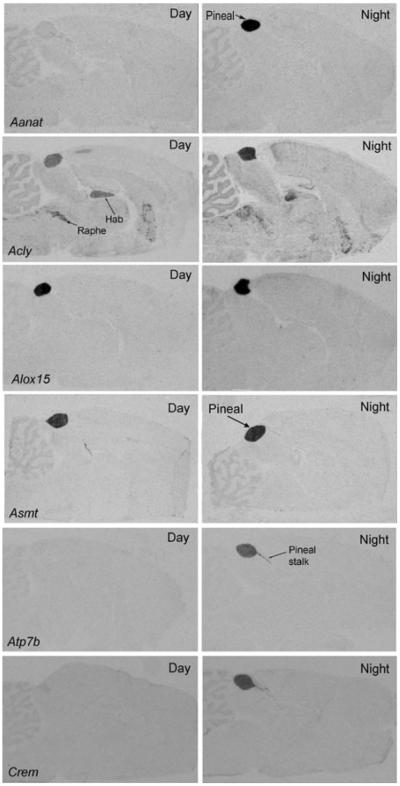
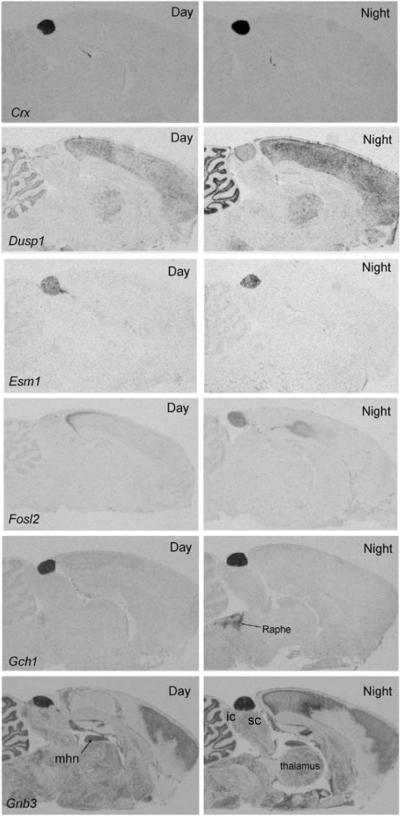
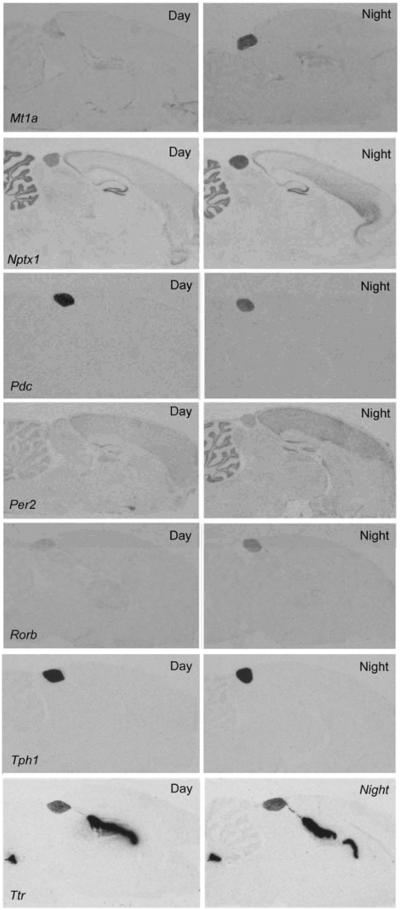
The magnitude of these changes is a conservative approximation; greater changes are often revealed by qRT-PCR analysis and with more frequent time sampling, which better describes the dynamics of gene expression (Figure 4). It can be seen that peak expression does not occur precisely at noon or midnight, and might occur towards dusk and in other towards dawn. Analysis of the night/day differences in expression by radiochemical in situ hybridization histology (Figure 5) reveals that the rhythmic pattern of expression is obvious in the pineal gland, as compared to the surrounding brain tissue.
Figure 4.
qRT-PCR analysis of transcripts that are night/day differentially expressed or have high rEx values or both. The lighting cycle is represented at the bottom of each column. Transcripts are identified by gene symbol. Each value is the mean ± S.E. of three determinations. Values were normalized to Actb, Gapdh, Hrpt1, and Rnr1. A single asterisk identifies statistically significant rhythmic patterns of gene expression (p< 0.01) based on log transformed raw values analyzed by one-way analysis of variance. From (Bailey et al., 2009) , which contains technical details.
Figure 5. Radiochemical in situ hybridization histology images.
Each panel contains autoradiographs prepared from sections of rat brains through the pineal gland. The sections on the left are from animals killed during the day and those on the right are from animals killed during the night. The sections were incubated with antisense probes identified in the bottom left-hand corner of the Day image. c Hab, habenula; ic, inferior colliculus; mhn, medial habenular nucleus; Raphe, dorsal raphe nucleus; sc, superior colliculus. These figures are available in high resolution at http://science.nichd.nih.gov/confluence/display/sne/Daily+Changes+Gallery Taken from (Bailey et al., 2009), which contains technical details..
Control of global changes in gene expression by adrenergic cyclic AMP signalling
Organ culture studies have demonstrated that >95% of the genes that exhibit night/day differences are also induced by treatment with norepinephrine in organ culture (Bailey et al., 2009). Similar studies done with dibutyryl cyclic AMP have indicated that >95% of the changes causes by NE treatment are also seen following treatment with dibutyryl cyclic AMP . These studies are available at http://sne.nichd.nih.gov/data/table_s3.xls,. Forskolin has also been used as a cyclic AMP elevating agent to demonstrate that genes stimulated by NE are stimulated by elevation of cyclic AMP (Bailey et al., 2009).
The mechanism through which cyclic AMP controls transcription of the hundreds of genes may involve PKA dependent phosphorylation of CREB and cyclic AMP response elements in each gene. However, recent studies have demonstrated that some effects of cyclic AMP may an epigenetic mechanism in which the acetylation and phosphorylation state of histones is altered. This can alters chromatin organization and unmask buried regulatory elements, allowing transcription factors access to bind and promote transcription(Kanyo et al., 2009, Ho et al., 2007).
Genes expressed at unchanging selectively high levels in the pineal gland
As discussed above, the purpose of identifying genes that are highly expressed in the pineal gland relative to other tissues, those with high rEx values, is to obtain a more complete impression of the functional potential of this tissue as compare to others and to eliminate from consideration those genes essential for cell functioning in general. The most highly expressed genes (rEx >8-fold) appear in Table 2; a more complete list of genes appears at http://sne.nichd.nih.gov/data/table_s4.xls. The high expression of genes with high rEx values is apparent from the results of radiochemical in situ hybridization histology (Figure 5).
Table 2. Genes highly expressed in the pineal gland relative to other tissues.
Genes were selected by rEx values (Bailey et al., 2009).. A complete listing of genes with pineal rEx values >4 is available at http://sne.nichd.nih.gov/data/table_s4.xls, which includes gene descriptions, probe set numbers and Entrez Gene numbers.
| Pineal rEx | Gene symbol |
|---|---|
| >16 | A2m, Aanat, Abca1, Abhd14b, Adra1b, Adrb1, Aipl1, Alox15, Arhgap24, Arr3, Asl, Asmt, Atp7b, Ca3, Cabp1, Cacna1f, Camk1g, Ccl9, Cd1d1,Cd24, Cdh22, Chga, Chma3, Chrnb4, Cnga1, Cngb1, Cntrob,a Col8a1*, Cplx3, Cpt1b, Crem, Crocc*, Crtac1, Crx, Ctsc, Cyp1b1, Dclk3,a Ddc,Defb24, Drd4, Dusp1, Efemp1, Egflam, Esm1, Eya2, Fcer1a, Fdx1, Fkbp4, Fkbp5, Frmpd1*, Fst, Fzd4, Gch, Gdf15, Gem*, Gnat2*, Gnb3, Grk1, Guca1*, Hs3st2, Hspa1a, Hspa1b, Hspb1, Igfbp6, Impg1, Impg2, Irak2, Irs1, Isl2, Ka15, Kcne2, Kcnh6, Kcnj14, Krt1–19, Lamp3, Lgals1, Lgals3, Lhx4,a Lix1*, Lpl, Lrrc21, M6prbp1, Map4k1*, Mat2a, Mcam, Me2*, Miox, Mitf, Morn1, Mpp3, Mpp4, Mtac2d1, Mx2, Ncaph,Neurod1, Nphp4, Nphs1, Nptx1, Opn1sw, Osap, Otx2, Padi4, Pax4, Pax6, Pcbd1, Pcdh21, Pdc, Pde4b, Pla2g5, Plscr1, Rbp3, Rds, Ribc2, Rom1, Rorb*, Rxrg, Sag, Scn7a, Serping1, Slc12a5, Slc15a1, Slc17a6, Slc24a1, Slc30a1, Slc39a4*, Slc6a6, Snap25, Snf1lk, Sorl1, Spink4,Stk22s1, Sv2b, Tm7sf2, Tph1, Ttr, Tulp1*, Unc119, Vof16 |
| 8–16 | Accn4, Acsl1, Acvr1, Adam2, Ak3l1, Als2cr4*, Ampd2, Anp32e, Anpep, Atp1b2, Atp6v1c2, Baiap2l1, Bmp6, Bzrp, Cacna1h, Ccdc125, Ccl2, Ccl6, Ccnd2, Cd63, Cd74, Cd8a, Cebpb, Cfd, Cflar, Chst2*, Cip98, Col15a1, Col1a1, Cr16, Crcp, Cyp1a1, Dcn, Depdc7, Dhrs8, Dnajc12, Dnm2, Dnm3, Dpt*, Dsc2, Dscr1, Epb4.1, Errfi1, Etnk1*, Exoc5, F5, Farp2*, Fosl2, Foxd1, Frmd4b, G0s2, Gabrr1, Gale, Galnt4, Galntl1, Gla, Gls, Gmds, Gnas, Grm1, Hcn1, Hk2, Hsd3b7, Hspb6, Id1, Ifitm3, Igfbp2, Igsf1, Igsf4a, Il13ra2, Il17re, Irf7, Itgb2, Kctd14*, Kctd3, Kit, Klhl4,Lad1*, Lama2*, Lamb1*, Lmbr1l, Lmod1*, Lnx1*, Lox, Loxl1, Lrrc8e, Lum, Lxn, Mad2l2, Mak, Mak10, Man2a1, Mapk6, Msrb2, Msx1,Mt1a, Muc4, Mylk*, Myo5b, Nacad, Nr4a1, Nradd, Nrap*, Nup107, Oasl1, Orai1, Pcbp3, Pde10a, Pde6b*, Pdp2, Pgam2, Pgm1, Pid1, Pik3r3, Pla2g1b, Pla2r1*, Plcd1, Plcd4, Postn*, Pqlc1, Prkar2b, Prkca, Prtg, Psph, Ptgis, Ptms, Ptprn, PVR, Qscn6, Rab3c, Rarres1, Rasgrf2, Rax, Rere, Resp18, Rnase1, Rreb1*, RT1-Aw2, Rtbnd*, Sall1*, Sema3a, Slc12a2, Slc19a2, Slc1a5, Slc25a10, Slc47a1, Slc4a2, Slc7a6*, Slco4a1, Sod3, Spint2, Svop, Tagln2, Tcn2, Tex14,a Timp1, Tmepai,a Tnfrsf9, Ugdh, Wnt10a,a Zmat2, Zrsr1 |
Functional implications
Analysis of the combined lists of highly expressed genes and those with high rEx values provides interesting insights into pineal function, including highly special capabilities and also more general processes(Table 4). It is not clear yet whether the gene expression patterns obtained from analysis of the rat pineal gland will be valid when considering other mammals and submammalian vertebrates. It remains to be determined whether all pineal glands have the same genetic potential, or if some of the observations made with the rat are rat specific evolutionary innovations. Similarly, it is not clear whether the genes which confer specific functionalities in the rat do this in other species, or if their role is played by other genes. These uncertainties require further investigation. Below are presented several functional associations of the genes of interest – those with high amplitude dynamic changes and those with high rEx values, which appear in Table 4. The identification of novel genes of interest which have not appeared in the pineal literature has resulted in extended analysis, and where this has occurred, this is reviewed.
Table 4. Functional grouping of genes that are night/day differentially expressed (N/D > 2 or < ½) or at high levels (rEx > 4) in the pineal gland.
Modified from (Bailey et al., 2009), which contains details regarding the creation of this table.
| Functional group | Gene Symbol |
|---|---|
| Specialized processes | |
| Immune response/inflammation | Abhd2*, And, Ahcy, Alms1*, ArHgef9, Bbs7, Bcar1, Btg2, C3, Ccl2, Ccl6, Ccl7, Ccl9, Ccrl2*, Cd1d1, Cd47, Cd74, Cd8a, Crcp, Ctsc, Ctss, Defb24, Dscr1, Fcer1a, Fras1*, Gdf15, Gem* Hivep1, Hivep2, Icsbp1, Ifi35, IfiTm1*, Ifnar1*, Igsf4a, Igsf9*, Igha, II13ra2, II17re, II18, IIrI1I, IIk, Impdh2, Inhbb, Irak2, Irf7, Ler3, Litaf, Lrrc8, Lta4h, Mal2, Mdk, Mina, Mmd2, Mox2, Mx2, Oit1*, Optn, Pcna, Plscr1, Pvr, Pvrl2, RT1-A1, RT1-A2, RT1-A3, RT1-Aw2, RT1-Bb, RT1-Da, Sct2, Sema3a, Serping1, Slfn3, Stch, Stip1, Tfp12, Tpm4, Ush2a, Vof16 |
| Melatonin Synthesis | Aanat, Acly, Asmt, Ddc, Gch1, Gchfr, Mat2a, Pcbd1, Tph1 |
| Photo-detection | Genes linked to photodetection in the pineal gland are considered to be highly expressed in both the pineal gland and retina; they are listed in Table 4. |
| T3/RA Signaling | Dio2, Hr, Rbp3, Rdh12*, Rorb*, Rxrg, Thrb, Ttr |
| Non-specialized processes | |
| Adhesion | Cdh22, Celsr32, Cml5, Cntn4, Dsc2, Eva*, Gja12*, Glycam1, Grn, Hnt, Mcam, Mfap4, Mpp4, Muc4, Nell2, Parvb, Pcdh21, Prph2, Pvr, Scarb2, Sdc4, Spon1, Ssx21p |
| Cell cycle/cell death | Acom1, Acvr1, Aprin, Bag1*, Giklk, Casp7, Ccnd2, Cdc25a, Cdc5l, Cdk5, Cdkn1b, Cdkn1c, Cflar, Ches1*, Commd5, Csnk2a2, Ddit3, Dnm1, Dnm2, Elmo3, Faim, Gos2, Gadd45a, Gadd45b, Igf1r, Igfbpl1*, Jag1, Junb, Mad2l2, Mak10, Ntf3, Pafah1b1, Pard3, Pdia3, Plagl1, Ptgs2, Qscn6, Rarres1, Rgc32, Fhob, Slc31a1, Strn3, Tacc3, Vegfc |
| Cytoskeleton | Ap1g1, Baiapw, Bbs4*, Catna1, Clasp2, Clta, Col14a1*, Col3a1, Col4a3, Col8a1*, Cope*, Cpg2, Dnch1, Dncl2b*, Dncl2b*, Emilin1*, Emls, Fgd2*, lnb*, Flnc*, Fmod, Fni, Fscn2*, Hdac11*, Ka15, Kif1b, Kif22, Kif2c, Krt1-18, Krt1-19, Krt25, Lad1*, Lama2*, Lamb1-1*, Lap1b, Lcp1, Lix1, Lmod1, Lumk, Mapt, Marcks, Mfap5*, Mgp, Mrgl19, Mtap2, Mylip*, Nrap*, Pgea1, Rpl3, Sas, Selpl*, Sdo3, Spna2, Tctex1, Thbs4, Tmem16a*, Tmem22, Tpm4, Tuba4, Tubb5, Unc119, Vil2, Vim |
| DNA modification | Adprt, Blm*, Bnc2*, Cntn1, Commd1*,Ctps*, Herc3*, Hmgb2, Kpna2, Mcm4, Pcna, Prc1*, Prim1, Ptms, Rere, Thap4, Tlk1*, Top1, Tspyl4, Zdhhc22, Zfp143, Zfp162, Zfp238, Zfp36l1, Zhx1, Znf444, Zswim5* |
| Endothelium | Esm1, Vegfb, Vegfc, Vwf |
| Growth | Efemt1, Egf, Egfr, Egfr1, Fgf1, Fgfr1, Gadd45g, Gdf15, Gfer, Grb2, Igf1r, Igfbp2, Igfbp6, Pdgfrl, Pgf, Tgfb1, Tgfbi, Vegfb, Vegfc |
| Signaling | Calcium: Atp2b3, Cabp1, Cacna1f, Cacna1g, Calm1, Camk1g, Camk2b, Cip98, Dcamkl1, Dcamkl3 |
| Cyclic nucleotide: Adcy8, Akap11, Cnga1, Cngb1, Creb3, Guca1a, Gucy1a3, Hcn1, Pde4b, Pde4d, Pde6b, Pde8b, Pde10a, Prkar2b, Prkca | |
| G-protein: Arf3, Arr3, Arl2bp, Arl6ip5, Gem, Gna12, Gnaq, Gnas, Gnat2, Gnaz, Gnb1, Gnb3, Gng11, Grk1, Pdc1, Rgs2, Rgs4, Rgs7, Rgs9, Rgs17, Sag1, Tyro3 | |
| Membrane receptors/ligands: Acvr1, Adra1b, Adrb1, Agtrap, Bmp6, Chrna3, Chrnb1, Chrnb4, Crcp, Drd1a, Drd4, Ece1, Ednrb, Egf, Egfr, Fgf, Fgfr1, Fst, Fzd4, Grip2, Grm1, Grm2, Hcrtr1, Htr2c, Igf1r ,Igfbp2, Igfbp3, Igfbp5, Igfbp6, Lepr, Nog, Opn1sw, Prlr, Sort1,Vipr2 | |
| Lipid/ Phospholipid/cholesterol: Abca1,Alox15, Cyp27a1, Ephx1, Inpp5e, Itpr1, Lta4h, Ltb4dh, Pa2g1b, Pik3r3, Pla2g5, Plcb1, Plcd4, Ptgds, Ptgis | |
| MAP kinase: Dusp1, Errfi1, Map3k5, Map3k6, Map4k1, Mapk14, Mapk6 | |
| Protein phosphorylation, serine/threonine: Calm1, Camk1g, Camk2b, Cdk5, Cdkn1b, Crkas, Dcamkl1, Enh, Fez1, Gsk3b, Nell2, Pak2, Prkar2b, Prkca, Prkcdbp, Prkce, Prkcl1, Rock2, Sik2, Snrk, Stk2, Stk39 | |
| Protein phosphorylation, tyrosine: Crkas, Efna5, Jak1, Kit, Ntrk2, Ntrk3, Ptp2E, Ptp4a1, Ptpn16, Ptprj, Ptprr, Ptp-Td14, Tyro3 | |
| RNA modification | Ankrd24*, Bfsp1, Bop1, Bzw2, Eif2ak4*, Eif2c2, Eif3s9, Eif4g2, Ell2, Hdac5, Mbnl2, Polr2d*, Qtrt1, Rnase1, Rnase2*, Rpat1, Sfpq, Xpot* |
| Small molecule biology | Metal homeostasis, Atp7b, Chordc1, Mt1a, Mt2, Slc30a1 Slc39a4 |
| Ion homeostasis: Atp1a1, Atp1b1, Atp1b2, Atp2a2, Atp2b1, Cacna1h, Cacnb2, Clcn3, Cnga1, Cngb1, Hcn1, Kcnab2, Kcne2, Kcnh6, Kcnj14, Kctd3, Scn7a, Slc12a2, Slc12a5, Slc17a6, Slc24a1 | |
| Solute transport: Slc2a1, Slc2a4, Slc3a1, Slc4a2, Slc4a4, Slc6a6, Slc7a1, Slc7a7, Slc12a2, Slc12a5, Slc14a1, Slc15a1, Slc16a1, Slc16a6, Slc21a1, Slc21a7, Slc22a1, Slc25a10, Slc29a1, Slc30a1, Slc34a1 | |
| Transcription factors | Arntl, Bhlhb3, Cebpb, Crem, Cbx5, Cry2, Crx, Datf1, Eya2, Fosl2, Foxd1, Hdac5, Homer1, Homer2, Hr, Isl2, Jun, Junb, Mitf, Msx1, Neurod1, Nr1d2, Nr1h4, Nr2f6, Nr4a1, Nr4a3, Otx2, Pax4, Pax6, Per2, Ptch1, Rax, Rorb, Rxrg, Thrb, |
| Vesicle biology | Cadps, Chga, Chgb, Clta, Cltb, Dnm1, Dnm2, Dnm3, Lphn2, Ptprn, Scg2, Scg3, Snap23, Snap25, Sny2, Stx3, Sv2b, Syt4 |
Melatonin production
Among the genes that are rhythmically expressed or expressed with high rEx values are those that encode proteins dedicated to melatonin synthesis, including Tph1, Ddc,Aanat, and Asmt (also known as Hiomt) (Bailey et al., 2009) In addition, genes encoding enzymes which generate cofactors for enzymes in the melatonin pathway are also among these genes of interest, including three associated with synthesis of biopterin (Gch, Cchfr, and Pcbd1), which is required for Tph1; one that is dedicated to AcCoa(Acl)) synthesis, required for Aanat; and, another that encodes an enzyme that synthesizes S-adenosylmethionine (Mat2A) which is required by Asmt.
Of these, Mat2A has been studied in detail (Kim et al., 2005). It exhibits a daily rhythm with high levels at night, at a time when S-adenosylmethionine is in greatest demand for melatonin synthesis. The nocturnal increase in Mat2A mRNA is associated with an increase in protein and enzyme activity; both of which appear to be due to neural stimulation. Moreover, these increases are dependent on NE acting through a cAMP mechanism involving PKA.
Adrenergic cyclic AMP-Signal transduction
Consistent with previous work on the pineal gland, microarray revealed that the tissue expresses high levels of the two G-protein linked adrenergic receptors which mediate the effects of NE, Adrb1b and Adra1b AR; in both cases, the rEx values for these receptors are >16 (Bailey et al., 2009). GTP binding proteins were also found to be expressed at relatively high levels, as well as several phosphodiesterase genes, which are likely to influence effect of NE on cyclic AMP.
Analysis of one phosphodiesterase, PDE4B2(Kim et al., 2007), has indicated that the gene is expressed in pinealocytes and that the daily rhythm in expression of this gene is circadian and under control of the SCN/SCG neural system which regulates melatonin production. In addition, the increase in mRNA is associated with an increase in both protein and PDE activity. The increase in PDE activity peaks at the end of the night period, perhaps as part of a programmed system which turns off NE/cAMP stimulation of the pineal gland.
Several signal transduction related genes (Sag, Pdc, Grk1) are known to be expressed in the pineal gland. Microarray expanded this list (Table 3) by determining that they are expressed at similar levels in retina and has expanded this list of genes. These genes may represent evolutionary extra baggage. However, it is more likely that some if not all play common roles in the pineal gland and the retinal photoreceptor. This seems reasonable based on their common origin (Klein, 2004) and the likelihood that genes functioned in the primitive ancestral photodetector and that their functional role was retained in both tissues during their independent evolution.
Table 3. Genes highly expressed only in the pineal gland and retina.
The relative expression (rEx) of these genes in both the pineal gland and the retina is >8-fold relative to the median expression level in 7 tissues (rEx > 8)(Bailey et al., 2009). A listing of these genes and genes with rEx values >8 in either the pineal gland or the retina, is available at http://sne.nichd.nih.gov/data/table_s5.xls. which includes gene descriptions, probe set numbers and Entrez Gene numbers.
| Aipl1, Arr3, Cacna1f, Cnga1, Cngb1, Cplx3, Crocc*, Crx, Drd4, Egflam, Frmpd1*, Gabrr1, Gnat2*, Gnb3, Grk1, Guca1a*, Hspa1b, Impg1,sImpg2, Kcnh6, Kcnj14, Lamp3, Lrrc21, Mak, Mpp4, Neurod1, Opn1sw, Osap, Otx2, Pax4, Pcbp3, Pcdh21, Pdc, Pde6b*, Pla2r1*, Rax, Rbp3, Rds, Rom1, Rtbnd*, Sag, Slc24a1, Slc6a6, Slco4a1, Stk22s1, Tulp1*, Unc119 |
The genes encoding downstream proteins which mediate effects of cyclic AMP were also found to be highly expressed, including PKA. Of special interest was the discovery that pineal gland expresses high levels of SNF1-LK (SIK), a kinase which is known to indirectly modulate pCREB dependent transcription. SNF1-LK has a marked daily rhythm in the pineal gland, which is under adrenergic/ cyclic AMP control (Table 1, Figure 4)(Bailey et al., 2009) . The substrates of this kinase are TORCs which bind to and inhibit CREB activity. Phosphorylation of TORCs promotes their binding to 14–3–3 proteins, which results in their transport out of the nucleus. The increase in SNF1-LK expression is likely to result in increased phosphorylation and decreased TORC binding to CREB, thereby enhancing transcription. This is consistent with the results of studies in which SNF1-LK has been overexpressed and knocked down in pinealocytes, and appears to control the transcription of Aanat and two other genes known to be under adrenergic cyclic AMP control, Dio2 and Mkp1. (Kanyo et al., 2009) Accordingly, SNF1-LK/TORC input appears to play a broad role in the daily changes in gene expression in the pineal gland.
Phototransduction related genes
It is well established that the melatonin producing cells in the submammalian pineal gland – pineal photoreceptors - detect light(Klein, 2006b, Klein, 2004). This is based on electrophysiological and biochemical studies. In addition, anatomical studies have established that pineal photoreceptors have anatomical features similar to those of retinal photoreceptors. Prior genetic studies have also indicated that the mammalian pineal gland expresses genes that are closely associated with phototransduction in the retina, including Sag, Pdc and Grk1.(Ho et al., 1986, Somers and Klein, 1984, Babila et al., 1992, Schaad et al., 1991, Reig et al., 1990)
Consistent with this background, microarray analysis significantly expanded the list of genes that are highly expressed in both the pineal gland and retina (Table 3; a more complete listing appears at http://sne.nichd.nih.gov/data/table_s5.xls.)(Bailey et al., 2009) This revealed that the mammalian pineal gland and retina express a cluster of genes and that these genes are not expressed in other tissues to a significant degree. Hence, they can be considered to be pineal/retina “specifi”. Some of these are linked to photoreception directly and others are recognized as transcription factors with known or likely roles in controlling cell fate and preservation of phenotype.
Of special note is that the analysis that identified this set of pineal/retinal expressed genes includes some with little or no mention in the pineal and retinal liteterature; and, that there are many unannotated sequences that require identification. Based on their pattern of expression, it is reasonable to predict that these genes play a conserved role in phototransduction.
Dopamine signal transduction
Micorarray analysis has revealed the pineal gland expresses the gene encoding the dopamine type 4 receptor (Drd4) and that expression follows a 24-hour pattern(Bailey et al., 2009, Humphries et al., 2002, Fukuhara and Tosini, 2008). An in depth study of Drd4 expression found that expression of the genes is highest in the pineal gland relative to other tissues, except the retina(Kim et al., 2009a). In addition, it was found that expression of Drd4 is >100-fold higher than that of other dopamine receptors, making it the dominant receptor for this transmitter in the pineal gland. In vivo and in vitror studies revealed that Drd4 expression is controlled by an adrenergic/cyclic AMP mechanism that also requires thyroid hormone via a cyclic AMP/T3 “AND” gate which required both messengers for maximal effectiveness. This advance establishes a new paradigm for control of the Drd4 expression.
The functional question of whether this receptor plays a role in pineal function has not been answered. It is striking that the Drd4 gene is also expressed in the retina, because this suggests that the association of this receptor with the pineal gland and retina is rooted in their common ancestral photodetector cell(Jackson et al., 2009, Nir et al., 2002). In addition, it is reasonable to predict that dopamine will be coreleased with NE from nerves in the pineal gland, because DA is the precursor of NE and occurs with NE in pineal nerve terminals.(Pellegrino de Iraldi and Zieher, 1966, Hyyppa, 1971, Kvetnansky et al., 1979)
Zn++ and Cu++ biology
A number of genes encoding proteins dedicated to metal homeostatis have high rEx values or are rhythmically expressed. These include metalothionine, which binds and buffers intracellular Zn++ and Cu++, and the transporters for these metals, the zinc transporter Slc39a13 and ATP7B and the copper transporting ATPase. Zn++ is of special importance in melatonin synthesis because it is required by Gch, (Auerbach et al., 2000). an enzyme dedicated to the generation of biopterin, the cofactor of the first enzyme in melatonin synthesis Tph1.
Thyroid/retinoic acid signaling
Prior to the use of microarray, a sizable literature existed on the expression in the pineal gland of the enzyme which converts T4 into T3, Dio2. It was known that expression of Dio2 exhibits a daily rhythm that is rapidly translated into changes in enzyme activity(Tanaka et al., 1986, Tanaka et al., 1987, Murakami et al., 1988). It was also known that Dio2 expression is controlled by NE/cyclic AMP signaling. However, there was little indication of the function of T3 on the pineal, other than effects of high concentrations on melatonin production(Nir and Hirschmann, 1978).
The findings of microarray confirmed the rhythmic expression of Dio2 and also revealed that the pineal gland expresses relatively high levels of other genes which encode proteins dedicated to thyroid hormone signaling. These include the genes that encode thyroid binding protein Ttr, the thyroid hormone receptor Thrb and genes that encode retinoic acid receptors that form functional heterodimers with thyroid hormone receptors. In addition, the pineal gland expressed genes that encode proteins dedicated to retinoic acid biology, including the binding protein Rbp2 and the retinol dehydrogenase Rdh12.
The evidence of a potential for thyroid hormone signaling directed investigation of the possibility that T3 was involve din in the adrenergic control of expression of the gene that encodes Drd4. As detailed above, this led to the description of a dual regulatory mechanism thorugh which cyclic AMP and T3 control Drd4 expression.
Inflammation immune response
One of the most rhythmically expressed genes and one with a high rEx value identified by microarray analysis was Fcer1a, the alpha subunit of the high affinity immunoglobulin E receptor (FcεRIa)(Bailey et al., 2009). In subsequent work it was found that the encoded protein also exhibits a large daily rhythm, both of which are control of the same neural NE/cyclic AMP system which regulates AANAT activity(Ganguly et al., 2007). This discovery led to the finding the gamma subunit of the IgE receptor is also expressed. This and the finding that from microarray studies of the large number of other gene expressed in the pineal gland that are known to be involved in the inflammation/immune response (Table 4) points to the possibility that the pineal gland supports a previously unrecognized link between the neuroendocrine and immune systems, through which IgE receptors detect increased levels of circulating allergens, which initiates events that result in T cells being attracted to the pineal gland, where they would be exposed to high levels of chemicals synthesized in gland, thereby activating the T cells.
Transcription factors
Microarray analysis has revealed high levels of a number of transcription factors in the pineal gland, many of which are also expressed in photoreceptors, consistent with the common origin of vertebrated pinealocytes and photoreceptors (Table 4). The transcription factors expressed at similar levels in both tissues includes NeuroD1, Crx, Otx, rax, Pax6, and Pax 4. In addition, it was found the high levels of transcripts of a homolog of the drosophila transcript factor Eya2 is occur in the pineal gland, as is Mitf.
These findings have stimulated significant investigations of the expression of most of these genes(Rath et al., 2009a, Rath et al., 2009b). It is known the Otx and Pax6 are essential for the pineal to develop. These genes are also expressed in the adult pineal gland, suggesting a role in maintenance of phenotype. Future work will elucidate the role that these and other transcription factors play in pineal cell fate determination of maintenance of phenotype. Moreover, the results of microarray have determined that Six6 , which encodes a transcription factor, is present in the retina but not the pineal gland. With this information, it may be possible to manipulate `missing' transcription factors and coax adult pinealocytes to detect light.
Lipid signaling
Evidence of potential for lipid signaling via several pathways was provided by microarray analays, which indicated relatively high levels of expression of genes which encode proteins dedicated to signaling via inositols (Plcb1, Plcd4, Inpp5e, Itpr1,Pik3r3 ), and derivatives of arachidonic acid (Pla2g1b,Pla2g5) including prostaglandins (Ptgds, Ptgis), leukotrienes, (Lta4h, Ltb4dh, and hepoxilins (Alox15, Ephx1) . The role of inositol signaling in the pineal gland appears to be the mediation of Adra1b control of Ca++ and PKC(Ho et al., 1988b, Ho et al., 1988a, Ho and Klein, 1987a, Ho and Klein, 1987b). The regulatory role of the arachadonic acid derivatives requires further study. The question of role of Alox15 is especially compelling because it is unusually highly expressed in the pineal gland, relative to other tissue (Yoshimoto et al., 1984, Hada et al., 1994). Alox15 products in the pineal gland have been studied, providing a foundation for future investigations of the functional role they play in this tissue.
RNA modification
Microarray studies revealed that the pineal gland expresses high levels of a cluster of genes dedicated to RNA modification, including Mbnl2, a gene first identified in Drosophila, where it is necessary for normal photoreceptor development (Kim et al., 2009b). This is a zinc finger protein that is thought to be dedicated to RNA splicing and to act by binding to specific repeat sequences in RNA. Mbnl2 transcripts and protein exhibit large daily rhythms; the abundance of both are regulated by NE/cyclic AMP signaling. The identification of specific transcripts that are spliced under control of Mbnl2 presents the next challenge in this line of investigation.
Circadian Clock Genes
Two clock genes that are highly expressed and rhythmic in the pineal gland are Per2 and Rorb. Per2 might impact gene expression in the pineal gland via interactions with E-boxes; the regulation of Per2 expression is not established and does not appear to involve NE/cyclic AMP signaling (Bailey et al., 2009, Fukuhara et al., 2002). Reports in the literature have established that clock genes Arntl, Clock, Per1, Per2, Per3, Cry1, and Cry2t are expressed in the pineal gland, some following a 24-h pattern(Namihira et al., 1999, Fukada and Okano, 2002, Karolczak et al., 2004, Simonneaux et al., 2004) . Microarray found they are expressed at levels that are less than 4-fold the median tissue level of expression. Although the circadian clock in the pineal gland does not play a dominant role in controlling melatonin production, it is possible that it functions in conjunction with clock input from the SCN to modulate NE stimulation of the gland.
MAP kinase signaling
A sizable literature exists on the role of MAP kinase signaling in the pineal gland (Price et al., 2007, Ho et al., 2005, Price et al., 2004b, Price et al., 2004a, Chik et al., 2004, Ho et al., 2003). Micorarray analysis extended this by indicating that there was a large daily rhythm in the expression of Dusp1, also known as Mkp1 (MAP kinase phosphatase 1)(Bailey et al., 2009). This enzyme inactivates MAP kinase by the concomitant dephosphorylation of both its phosphothreonine and phosphotyrosine residues, thereby terminating MAP kinase signaling. The finding of a rhythm in Dusp1 expression led to the discovery that Dusp1 expression is controlled by NE acting through cyclic AMP(Price et al., 2004b, Price et al., 2004a). This is of special interest because it establishes a new link between these two signaling pathways in the pineal gland.
Oliogopeptide transporter Slc15a1
Microarray analysis revealed that one of the most highly rhythmic genes expressed the pineal gland is Slc15a1 with a >100-fold amplitude(Bailey et al., 2009, Gaildrat et al., 2005) The gene, also known as Pept1, encodes a twelve membrane spanning oligopeptide transporter that is highly expressed in the intestine. Expression in the pineal gland is equally high, but the transcript expressed in the pineal gland encodes a truncated three membrane spanning protein. Expression of the gene in the pineal gland is controlled by an intronic promoter which has characteristics of the promoter which regulates expression of Aanat. Slc15a1 protein levels follow the dynamics of the transcript. The function of this protein has not been established. However the observation that it is targeted for the cell membrane and that it can interact with another transporter (Gaildrat et al., 2005) indicates it is reasonable to pursue the possibility that it modulates the function of other membrane proteins, perhaps transporters, receptors or channels, through a direct interaction.
The Future
The identification of a large number of selectively expressed(higher Ex) and rhythmically expressed genes has initiated a new phase of pineal research, one in which investigators are faced with hundreds of interesting genes and many questions. There are numberous new puzzles to solve and the number of puzzle pieces is nearly overwhelming. Determining what lines of research to pursue will be made easier, perhaps, as the transcriptomes of other species become available and fully annotated. The beginnings of the description of the chicken and zebrafish pineal transcriptomes have appeared in the literature.(Alon et al., 2009, Bailey et al., 2004, Toyama et al., 2009) and the human, mouse and rhesus pineal transcriptomes are currently under investigation by the Section on Neuroendocrinology. The results of these efforts will allow investigators to identify highly conserved genes associated with pineal function, allowing them to select for study an important evolutionarily conserved gene, rather than an evolutionary innovation found in only one or a few species.
It is anticipated that an unrealized function of the pineal gland may be deduced from this work, perhaps one involving the immune/inflammation response. It is also possible that the understanding of subtle differences in gene expression patterns in the pineal gland and retina may impact efforts to develop strategies to create photoreceptors to use in implantation efforts to restore sight in subjects whose photoreceptors have degenerated. Progress towards this goal will be hastened as we obtain a better understanding of the regulatory cascades the control gene expression in these tissues.
Microarray will soon be displaced as the discovery tool of choice by advanced sequencing methodologies, which will expand our knowledge of the genes expressed in tissues, and also identify non-coding mRNAs expressed in the pineal gland. These advances will form the foundation of a new genetic understanding of pineal function and will propel efforts aimed at understanding the role that the pineal gland plays in biology and human health.
Acknowledgments
This work was supported by funds from the Intramural Research Programs of the National Institute of Child Health and Human Development, (D.C.K., J.L.W., S.L.C.) and the Center for Information Technology (Z. G. R. and P. J. M.), National Institutes of Health; The Wellcome Trust (D.C.); Canadian Institutes of Health Research (A.K.Hand C.C.). The Lundbeck Foundation, Danish Medical Research Council Grants 271-07-0412 and 271-06-0754, The Novo Nordisk Foundation, The Carlsberg Foundation, Fonden til Lægevidenskabens Fremme, and Simon Fougner Hartmanns Familiefond. (M.F.R. and M.M. ); Research Equipment Initiative Grant BB/D52503X/1 from the Biotechnology and Biological Sciences Research Council (D.S.).
Abbreviations used
The gene symbols used have been taken from Entrez Gene
- CRE
cyclic AMP response element
- CREB
Cyclic AMP response element binding protein
- pCREB
phosphorylated CREB
- rEx
relative expression value in one tissue compared to median expression in other tissues
- Cyclic AMP
3' 5' cyclic adenosine monophosphate
- GPCR
G protein-coupled receptors
- NE
norepinephrine
- PKA
cyclic AMP dependent protein kinase
- PKC
Ca++, phospholipid dependent protein kinase
- PLA
phospholipase A
- PLC
phospholipase C
- SCN
suprachiasmatic nucleus
- SCG
superior cervical ganglia
REFERENCES
- AITKEN A. 14-3-3 proteins: a historic overview. Semin Cancer Biol. 2006;16:162–72. doi: 10.1016/j.semcancer.2006.03.005. [DOI] [PubMed] [Google Scholar]
- ALON S, EISENBERG E, JACOB-HIRSCH J, RECHAVI G, VATINE G, TOYAMA R, COON SL, KLEIN DC, GOTHILF Y. A new cis-acting regulatory element driving gene expression in the zebrafish pineal gland. Bioinformatics. 2009;25:559–62. doi: 10.1093/bioinformatics/btp031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AUERBACH DA, KLEIN DC, KIRK KL, CANTACUZENE D, CREVELING CR. Effects on fluorine analogs of norepinephrine on stimulation of cyclic adenosine 3',5'-monophosphate and binding to beta-adrenergic receptors in intact pinealocytes. Biochem Pharmacol. 1981a;30:1085–9. doi: 10.1016/0006-2952(81)90446-9. [DOI] [PubMed] [Google Scholar]
- AUERBACH DA, KLEIN DC, WOODARD C, AURBACH GD. Neonatal rat pinealocytes: typical and atypical characteristics of [125I]iodohydroxybenzylpindolol binding and adenosine 3',5'-monophosphate accumulation. Endocrinology. 1981b;108:559–67. doi: 10.1210/endo-108-2-559. [DOI] [PubMed] [Google Scholar]
- AUERBACH G, HERRMANN A, BRACHER A, BADER G, GUTLICH M, FISCHER M, NEUKAMM M, GARRIDO-FRANCO M, RICHARDSON J, NAR H, HUBER R, BACHER A. Zinc plays a key role in human and bacterial GTP cyclohydrolase I. Proc Natl Acad Sci U S A. 2000;97:13567–72. doi: 10.1073/pnas.240463497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BABILA T, SCHAAD NC, SIMONDS WF, SHINOHARA T, KLEIN DC. Development of MEKA (phosducin), G beta, G gamma and S-antigen in the rat pineal gland and retina. Brain Res. 1992;585:141–8. doi: 10.1016/0006-8993(92)91199-o. [DOI] [PubMed] [Google Scholar]
- BAILEY MJ, BEREMAND PD, HAMMER R, BELL-PEDERSEN D, THOMAS TL, CASSONE VM. Transcriptional profiling of the chick pineal gland, a photoreceptive circadian oscillator and pacemaker. Mol Endocrinol. 2003;17:2084–95. doi: 10.1210/me.2003-0121. [DOI] [PubMed] [Google Scholar]
- BAILEY MJ, BEREMAND PD, HAMMER R, REIDEL E, THOMAS TL, CASSONE VM. Transcriptional profiling of circadian patterns of mRNA expression in the chick retina. J Biol Chem. 2004;279:52247–54. doi: 10.1074/jbc.M405679200. [DOI] [PubMed] [Google Scholar]
- BAILEY MJ, COON SL, CARTER DA, HUMPHRIES A, KIM JS, SHI Q, GAILDRAT P, MORIN F, GANGULY S, HOGENESCH JB, WELLER JL, RATH MF, MOLLER M, BALER R, SUGDEN D, RANGEL ZG, MUNSON PJ, KLEIN DC. Night/day changes in pineal expression of >600 genes: central role of adrenergic/cAMP signaling. J Biol Chem. 2009;284:7606–22. doi: 10.1074/jbc.M808394200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BALER R, COON S, KLEIN DC. Orphan nuclear receptor RZRbeta: cyclic AMP regulates expression in the pineal gland. Biochem Biophys Res Commun. 1996;220:975–8. doi: 10.1006/bbrc.1996.0517. [DOI] [PubMed] [Google Scholar]
- BALER R, COVINGTON S, KLEIN DC. The rat arylalkylamine N-acetyltransferase gene promoter. cAMP activation via a cAMP-responsive element-CCAAT complex. J Biol Chem. 1997;272:6979–85. doi: 10.1074/jbc.272.11.6979. [DOI] [PubMed] [Google Scholar]
- BALER R, COVINGTON S, KLEIN DC. Rat arylalkylamine N-acetyltransferase gene: upstream and intronic components of a bipartite promoter. Biol Cell. 1999;91:699–705. [PubMed] [Google Scholar]
- BALER R, KLEIN DC. Circadian expression of transcription factor Fra-2 in the rat pineal gland. J Biol Chem. 1995;270:27319–25. doi: 10.1074/jbc.270.45.27319. [DOI] [PubMed] [Google Scholar]
- BERNARD M, KLEIN DC, ZATZ M. Chick pineal clock regulates serotonin N-acetyltransferase mRNA rhythm in culture. Proc Natl Acad Sci U S A. 1997;94:304–9. doi: 10.1073/pnas.94.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BORJIGIN J, DENG J, SUN X, DE JESUS M, LIU T, WANG MM. Diurnal pineal 3-O-sulphotransferase 2 expression controlled by beta-adrenergic repression. J Biol Chem. 2003;278:16315–9. doi: 10.1074/jbc.M300828200. [DOI] [PubMed] [Google Scholar]
- BORJIGIN J, DENG J, WANG MM, LI X, BLACKSHAW S, SNYDER SH. Circadian rhythm of patched1 transcription in the pineal regulated by adrenergic stimulation and cAMP. J Biol Chem. 1999a;274:35012–5. doi: 10.1074/jbc.274.49.35012. [DOI] [PubMed] [Google Scholar]
- BORJIGIN J, PAYNE AS, DENG J, LI X, WANG MM, OVODENKO B, GITLIN JD, SNYDER SH. A novel pineal night-specific ATPase encoded by the Wilson disease gene. J Neurosci. 1999b;19:1018–26. doi: 10.1523/JNEUROSCI.19-03-01018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CASSONE VM, TAKAHASHI JS, BLAHA CD, LANE RF, MENAKER M. Dynamics of noradrenergic circadian input to the chicken pineal gland. Brain Res. 1986;384:334–41. doi: 10.1016/0006-8993(86)91169-8. [DOI] [PubMed] [Google Scholar]
- CHANSARD M, LIANG J, IWAHANA E, BAKER T, WHITTAKER J, FUKUHARA C. Role of calcium in the gating of isoproterenol-induced arylalkylamine N-acetyltransferase gene expression in the mouse pineal gland. J Pineal Res. 2006;41:85–94. doi: 10.1111/j.1600-079X.2006.00341.x. [DOI] [PubMed] [Google Scholar]
- CHIK CL, HO AK, KLEIN DC. Alpha 1-adrenergic potentiation of vasoactive intestinal peptide stimulation of rat pinealocyte adenosine 3',5'- monophosphate and guanosine 3',5'-monophosphate: evidence for a role of calcium and protein kinase-C. Endocrinology. 1988a;122:702–8. doi: 10.1210/endo-122-2-702. [DOI] [PubMed] [Google Scholar]
- CHIK CL, HO AK, KLEIN DC. Dual receptor regulation of cyclic nucleotides: alpha 1-adrenergic potentiation of vasoactive intestinal peptide stimulation of pinealocyte adenosine 3',5'-monophosphate. Endocrinology. 1988b;122:1646–51. doi: 10.1210/endo-122-4-1646. [DOI] [PubMed] [Google Scholar]
- CHIK CL, MACKOVA M, PRICE D, HO AK. Adrenergic regulation and diurnal rhythm of p38 mitogen-activated protein kinase phosphorylation in the rat pineal gland. Endocrinology. 2004;145:5194–201. doi: 10.1210/en.2004-0864. [DOI] [PubMed] [Google Scholar]
- FUKADA Y, OKANO T. Circadian clock system in the pineal gland. Mol Neurobiol. 2002;25:19–30. doi: 10.1385/MN:25:1:019. [DOI] [PubMed] [Google Scholar]
- FUKUHARA C, DIRDEN JC, TOSINI G. Regulation of period 1 expression in cultured rat pineal. Neurosignals. 2002;11:103–14. doi: 10.1159/000058547. [DOI] [PubMed] [Google Scholar]
- FUKUHARA C, DIRDEN JC, TOSINI G. Analysis of gene expression following norepinephrine stimulation in the rat pineal gland using DNA microarray technique. J Pineal Res. 2003;35:196–203. doi: 10.1034/j.1600-079x.2003.00078.x. [DOI] [PubMed] [Google Scholar]
- FUKUHARA C, TOSINI G. Analysis of daily and circadian gene expression in the rat pineal gland. Neurosci Res. 2008;60:192–8. doi: 10.1016/j.neures.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FUKUHARA C, YAMAZAKI S, LIANG J. Pineal circadian clocks gate arylalkylamine N-acetyltransferase gene expression in the mouse pineal gland. J Neurochem. 2005;93:156–62. doi: 10.1111/j.1471-4159.2004.03008.x. [DOI] [PubMed] [Google Scholar]
- GAILDRAT P, MOLLER M, MUKDA S, HUMPHRIES A, CARTER DA, GANAPATHY V, KLEIN DC. A novel pineal-specific product of the oligopeptide transporter PepT1 gene: circadian expression mediated by cAMP activation of an intronic promoter. J Biol Chem. 2005;280:16851–60. doi: 10.1074/jbc.M414587200. [DOI] [PubMed] [Google Scholar]
- GANGULY S, COON SL, KLEIN DC. Control of melatonin synthesis in the mammalian pineal gland: the critical role of serotonin acetylation. Cell Tissue Res. 2002;309:127–37. doi: 10.1007/s00441-002-0579-y. [DOI] [PubMed] [Google Scholar]
- GANGULY S, GASTEL JA, WELLER JL, SCHWARTZ C, JAFFE H, NAMBOODIRI MA, COON SL, HICKMAN AB, ROLLAG M, OBSIL T, BEAUVERGER P, FERRY G, BOUTIN JA, KLEIN DC. Role of a pineal cAMP-operated arylalkylamine N-acetyltransferase/14–3–3-binding switch in melatonin synthesis. Proc Natl Acad Sci U S A. 2001;98:8083–8. doi: 10.1073/pnas.141118798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GANGULY S, GRODZKI C, SUGDEN D, MOLLER M, ODOM S, GAILDRAT P, GERY I, SIRAGANIAN RP, RIVERA J, KLEIN DC. Neural adrenergic/cyclic AMP regulation of the immunoglobulin E receptor alpha-subunit expression in the mammalian pinealocyte: a neuroendocrine/immune response link? J Biol Chem. 2007;282:32758–64. doi: 10.1074/jbc.M705950200. [DOI] [PubMed] [Google Scholar]
- GANGULY S, WELLER JL, HO A, CHEMINEAU P, MALPAUX B, KLEIN DC. Melatonin synthesis: 14–3–3-dependent activation and inhibition of arylalkylamine N-acetyltransferase mediated by phosphoserine-205. Proc Natl Acad Sci U S A. 2005;102:1222–7. doi: 10.1073/pnas.0406871102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GARDINO AK, SMERDON SJ, YAFFE MB. Structural determinants of 14–3–3 binding specificities and regulation of subcellular localization of 14–3–3-ligand complexes: a comparison of the X-ray crystal structures of all human 14–3–3 isoforms. Semin Cancer Biol. 2006;16:173–82. doi: 10.1016/j.semcancer.2006.03.007. [DOI] [PubMed] [Google Scholar]
- GASTEL JA, ROSEBOOM PH, RINALDI PA, WELLER JL, KLEIN DC. Melatonin production: proteasomal proteolysis in serotonin N-acetyltransferase regulation. Science. 1998;279:1358–60. doi: 10.1126/science.279.5355.1358. [DOI] [PubMed] [Google Scholar]
- HADA T, HAGIYA H, SUZUKI H, ARAKAWA T, NAKAMURA M, MATSUDA S, YOSHIMOTO T, YAMAMOTO S, AZEKAWA T, MORITA Y, et al. Arachidonate 12-lipoxygenase of rat pineal glands: catalytic properties and primary structure deduced from its cDNA. Biochim Biophys Acta. 1994;1211:221–8. doi: 10.1016/0005-2760(94)90272-0. [DOI] [PubMed] [Google Scholar]
- HO AK, CHIK CL, KLEIN DC. Transmembrane receptor cross-talk: concurrent VIP and alpha 1-adrenergic activation rapidly elevates pinealocyte cGMP greater than 100-fold. Biochem Biophys Res Commun. 1987;146:1478–84. doi: 10.1016/0006-291x(87)90816-3. [DOI] [PubMed] [Google Scholar]
- HO AK, CHIK CL, KLEIN DC. Permissive role of calcium in alpha 1- adrenergic stimulation of pineal phosphatidylinositol phosphodiesterase (phospholipase C) activity. J Pineal Res. 1988a;5:553–64. doi: 10.1111/j.1600-079x.1988.tb00798.x. [DOI] [PubMed] [Google Scholar]
- HO AK, KLEIN DC. Activation of alpha 1-adrenoceptors, protein kinase C, or treatment with intracellular free Ca2+ elevating agents increases pineal phospholipase A2 activity. Evidence that protein kinase C may participate in Ca2+-dependent alpha 1-adrenergic stimulation of pineal phospholipase A2 activity. J Biol Chem. 1987a;262:11764–70. [PubMed] [Google Scholar]
- HO AK, KLEIN DC. Phosphatidylinositol phosphodiesterase (phospholipase C) activity in the pineal gland: characterization and photoneural regulation. J Neurochem. 1987b;48:1033–8. doi: 10.1111/j.1471-4159.1987.tb05622.x. [DOI] [PubMed] [Google Scholar]
- HO AK, MACKOVA M, CHO C, CHIK CL. Regulation of 90-kilodalton ribosomal S6 kinase phosphorylation in the rat pineal gland. Endocrinology. 2003;144:3344–50. doi: 10.1210/en.2003-0215. [DOI] [PubMed] [Google Scholar]
- HO AK, MCNEIL L, TERRIFF D, PRICE DM, CHIK CL. Role of protein turnover in the activation of p38 mitogen-activated protein kinase in rat pinealocytes. Biochem Pharmacol. 2005;70:1840–50. doi: 10.1016/j.bcp.2005.09.013. [DOI] [PubMed] [Google Scholar]
- HO AK, PRICE DM, DUKEWICH WG, STEINBERG N, ARNASON TG, CHIK CL. Acetylation of histone H3 and adrenergic-regulated gene transcription in rat pinealocytes. Endocrinology. 2007;148:4592–600. doi: 10.1210/en.2007-0578. [DOI] [PubMed] [Google Scholar]
- HO AK, SOMERS RL, KLEIN DC. Development and regulation of rhodopsin kinase in rat pineal and retina. J Neurochem. 1986;46:1176–9. doi: 10.1111/j.1471-4159.1986.tb00634.x. [DOI] [PubMed] [Google Scholar]
- HO AK, THOMAS TP, CHIK CL, ANDERSON WB, KLEIN DC. Protein kinase C: subcellular redistribution by increased Ca2+ influx. Evidence that Ca2+-dependent subcellular redistribution of protein kinase C is involved in potentiation of beta-adrenergic stimulation of pineal cAMP and cGMP by K+ and A23187. J Biol Chem. 1988b;263:9292–7. [PubMed] [Google Scholar]
- HUMPHRIES A, KLEIN D, BALER R, CARTER DA. cDNA array analysis of pineal gene expression reveals circadian rhythmicity of the dominant negative helix-loop-helix protein-encoding gene, Id-1. J Neuroendocrinol. 2002;14:101–8. doi: 10.1046/j.0007-1331.2001.00738.x. [DOI] [PubMed] [Google Scholar]
- HUMPHRIES A, WELLS T, BALER R, KLEIN DC, CARTER DA. Rodent Aanat: intronic E-box sequences control tissue specificity but not rhythmic expression in the pineal gland. Mol Cell Endocrinol. 2007;270:43–9. doi: 10.1016/j.mce.2007.02.003. [DOI] [PubMed] [Google Scholar]
- HYYPPA M. Hypothalamic monoamines and pineal dopamine during the sexual differentiation of the rat brain. Experientia. 1971;27:336–7. doi: 10.1007/BF02138182. [DOI] [PubMed] [Google Scholar]
- IUVONE PM, TOSINI G, POZDEYEV N, HAQUE R, KLEIN DC, CHAURASIA SS. Circadian clocks, clock networks, arylalkylamine N-acetyltransferase, and melatonin in the retina. Prog Retin Eye Res. 2005;24:433–56. doi: 10.1016/j.preteyeres.2005.01.003. [DOI] [PubMed] [Google Scholar]
- JACKSON CR, CHAURASIA SS, ZHOU H, HAQUE R, STORM DR, IUVONE PM. Essential roles of dopamine D4 receptors and the type 1 adenylyl cyclase in photic control of cyclic AMP in photoreceptor cells. J Neurochem. 2009;109:148–57. doi: 10.1111/j.1471-4159.2009.05920.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KANYO R, PRICE DM, CHIK CL, HO AK. Salt-inducible kinase 1 in the rat pinealocyte: adrenergic regulation and role in arylalklyamine N-acetyltransferase gene transcription. Endocrinology. 2009 doi: 10.1210/en.2009-0275. [DOI] [PubMed] [Google Scholar]
- KAROLCZAK M, BURBACH GJ, STIES G, KORF HW, STEHLE JH. Clock gene mRNA and protein rhythms in the pineal gland of mice. Eur J Neurosci. 2004;19:3382–8. doi: 10.1111/j.0953-816X.2004.03444.x. [DOI] [PubMed] [Google Scholar]
- KIM JS, BAILEY MJ, HO AK, MOLLER M, GAILDRAT P, KLEIN DC. Daily Rhythm in Pineal Phosphodiesterase Activity Reflects Adrenergic/cAMP Induction of the PDE4B2 Variant. Endocrinology. 2007 doi: 10.1210/en.2006-1420. [DOI] [PubMed] [Google Scholar]
- KIM JS, BAILEY MJ, WELLER JL, SUGDEN D, RATH MF, MOLLER M, KLEIN DC. Thyroid hormone and adrenergic signaling interact to control pineal expression of the dopamine receptor D4 gene (Drd4) Mol Cell Endocrinol. 2009a doi: 10.1016/j.mce.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM JS, COON SL, BLACKSHAW S, CEPKO CL, MOLLER M, MUKDA S, ZHAO WQ, CHARLTON CG, KLEIN DC. Methionine adenosyltransferase:adrenergic-cAMP mechanism regulates a daily rhythm in pineal expression. J Biol Chem. 2005;280:677–84. doi: 10.1074/jbc.M408438200. [DOI] [PubMed] [Google Scholar]
- KIM JS, COON SL, WELLER JL, BLACKSHAW S, RATH MF, MOLLER M, KLEIN DC. Muscleblind-like 2: circadian expression in the mammalian pineal gland is controlled by an adrenergic-cAMP mechanism. J Neurochem. 2009b doi: 10.1111/j.1471-4159.2009.06184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLEIN DC. Circadian rhythms in indole metabolism in the rat pineal gland. In: Schmitt FO, Worden FG, editors. The Neurosciences; Third Study Program. MIT Press; Cambridge, Massachusetts: 1974. pp. 509–515. [Google Scholar]
- KLEIN DC. The pineal gland: a model of neuroendocrine regulation. Res Publ Assoc Res Nerv Ment Dis. 1978;56:303–27. [PubMed] [Google Scholar]
- KLEIN DC. Photoneural regulation of the mammalian pineal gland. Ciba Found Symp. 1985;117:38–56. doi: 10.1002/9780470720981.ch4. [DOI] [PubMed] [Google Scholar]
- KLEIN DC. The 2004 Aschoff/Pittendrigh lecture: Theory of the origin of the pineal gland--a tale of conflict and resolution. J Biol Rhythms. 2004;19:264–79. doi: 10.1177/0748730404267340. [DOI] [PubMed] [Google Scholar]
- KLEIN DC. Arylalkylamine N-acetyltransferase: “The timezyme”. J Biol Chem. 2006a doi: 10.1074/jbc.R600036200. [DOI] [PubMed] [Google Scholar]
- KLEIN DC. Evolution of the vertebrate pineal gland: the AANAT hypothesis. Chronobiol Int. 2006b;23:5–20. doi: 10.1080/07420520500545839. [DOI] [PubMed] [Google Scholar]
- KLEIN DC. Arylalkylamine N-acetyltransferase: “the Timezyme”. J Biol Chem. 2007;282:4233–7. doi: 10.1074/jbc.R600036200. [DOI] [PubMed] [Google Scholar]
- KLEIN DC, COON SL, ROSEBOOM PH, WELLER JL, BERNARD M, GASTEL JA, ZATZ M, IUVONE PM, RODRIGUEZ IR, BEGAY V, FALCON J, CAHILL GM, CASSONE VM, BALER R. The melatonin rhythm-generating enzyme: molecular regulation of serotonin N-acetyltransferase in the pineal gland. Recent Prog Horm Res. 1997;52:307–57. discussion 357–8. [PubMed] [Google Scholar]
- KLEIN DC, MOORE RY. Pineal N-acetyltransferase and hydroxyindole-O-methyltransferase: control by the retinohypothalamic tract and the suprachiasmatic nucleus. Brain Res. 1979;174:245–62. doi: 10.1016/0006-8993(79)90848-5. [DOI] [PubMed] [Google Scholar]
- KLEIN DC, SMOOT R, WELLER JL, HIGA S, MARKEY SP, CREED GJ, JACOBOWITZ DM. Lesions of the paraventricular nucleus area of the hypothalamus disrupt the suprachiasmatic leads to spinal cord circuit in the melatonin rhythm generating system. Brain Res Bull. 1983a;10:647–52. doi: 10.1016/0361-9230(83)90033-3. [DOI] [PubMed] [Google Scholar]
- KLEIN DC, SUGDEN D, WELLER JL. Postsynaptic alpha-adrenergic receptors potentiate the beta-adrenergic stimulation of pineal serotonin N-acetyltransferase. Proc Natl Acad Sci U S A. 1983b;80:599–603. doi: 10.1073/pnas.80.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLEIN DC, WELLER JL, MOORE RY. Melatonin metabolism: neural regulation of pineal serotonin: acetyl coenzyme A N-acetyltransferase activity. Proc Natl Acad Sci U S A. 1971;68:3107–10. doi: 10.1073/pnas.68.12.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KVETNANSKY R, KOPIN IJ, KLEIN DC. Stress increases pineal epinephrine. Commun Psychopharmacol. 1979;3:69–72. [PubMed] [Google Scholar]
- MARONDE E, STEHLE JH. The mammalian pineal gland: known facts, unknown facets. Trends Endocrinol Metab. 2007;18:142–9. doi: 10.1016/j.tem.2007.03.001. [DOI] [PubMed] [Google Scholar]
- MOORE RY, KLEIN DC. Visual pathways and the central neural control of a circadian rhythm in pineal serotonin N-acetyltransferase activity. Brain Res. 1974;71:17–33. doi: 10.1016/0006-8993(74)90188-7. [DOI] [PubMed] [Google Scholar]
- MURAKAMI M, GREER MA, HJULSTAD S, GREER SE, TANAKA K. The role of the superior cervical ganglia in the nocturnal rise of pineal type-II thyroxine 5'-deiodinase activity. Brain Res. 1988;438:366–8. doi: 10.1016/0006-8993(88)91365-0. [DOI] [PubMed] [Google Scholar]
- MURAKAMI M, GREER SE, MCADAMS S, GREER MA. Comparison of isoproterenol and dibutyryl adenosine cyclic 3',5'-monophosphate stimulation of thyroxine 5'-deiodinase activity in cultured pineal glands from euthyroid and hypothyroid rats. Life Sci. 1989;44:425–9. doi: 10.1016/0024-3205(89)90267-1. [DOI] [PubMed] [Google Scholar]
- NAMIHIRA M, HONMA S, ABE H, TANAHASHI Y, IKEDA M, HONMA K. Daily variation and light responsiveness of mammalian clock gene, Clock and BMAL1, transcripts in the pineal body and different areas of brain in rats. Neurosci Lett. 1999;267:69–72. doi: 10.1016/s0304-3940(99)00324-9. [DOI] [PubMed] [Google Scholar]
- NIR I, HARRISON JM, HAQUE R, LOW MJ, GRANDY DK, RUBINSTEIN M, IUVONE PM. Dysfunctional light-evoked regulation of cAMP in photoreceptors and abnormal retinal adaptation in mice lacking dopamine D4 receptors. J Neurosci. 2002;22:2063–73. doi: 10.1523/JNEUROSCI.22-06-02063.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIR I, HIRSCHMANN N. The effect of thyroid hormones on rat pineal indoleamine metabolism in vitro. J Neural Transm. 1978;42:117–26. doi: 10.1007/BF01675350. [DOI] [PubMed] [Google Scholar]
- OBSIL T, GHIRLANDO R, KLEIN DC, GANGULY S, DYDA F. Crystal structure of the 14–3–3zeta:serotonin N-acetyltransferase complex. a role for scaffolding in enzyme regulation. Cell. 2001;105:257–67. doi: 10.1016/s0092-8674(01)00316-6. [DOI] [PubMed] [Google Scholar]
- PELLEGRINO DE IRALDI A, ZIEHER LM. Noradrenaline and dopamine content of normal, decentralized and denervated pineal gland of the rat. Life Sci. 1966;5:149–54. doi: 10.1016/0024-3205(66)90127-5. [DOI] [PubMed] [Google Scholar]
- PERLOW MJ, REPPERT SM, BOYAR RM, KLEIN DC. Daily rhythms in cortisol and melatonin in primate cerebrospinal fluid. Effects of constant light and dark. Neuroendocrinology. 1981;32:193–6. doi: 10.1159/000123157. [DOI] [PubMed] [Google Scholar]
- PERLOW MJ, REPPERT SM, TAMARKIN L, WYATT RJ, KLEIN DC. Photic regulation of the melatonin rhythm: monkey and man are not the same. Brain Res. 1980;182:211–6. doi: 10.1016/0006-8993(80)90848-3. [DOI] [PubMed] [Google Scholar]
- PRICE DM, CHIK CL, HO AK. Norepinephrine induction of mitogen-activated protein kinase phosphatase-1 expression in rat pinealocytes: distinct roles of alpha- and beta-adrenergic receptors. Endocrinology. 2004a;145:5723–33. doi: 10.1210/en.2004-0880. [DOI] [PubMed] [Google Scholar]
- PRICE DM, CHIK CL, TERRIFF D, WELLER J, HUMPHRIES A, CARTER DA, KLEIN DC, HO AK. Mitogen-activated protein kinase phosphatase-1 (MKP-1): >100-fold nocturnal and norepinephrine-induced changes in the rat pineal gland. FEBS Lett. 2004b;577:220–6. doi: 10.1016/j.febslet.2004.09.083. [DOI] [PubMed] [Google Scholar]
- PRICE DM, WLOKA MT, CHIK CL, HO AK. Mitogen-activated protein kinase phosphatase-1 (MKP-1) preferentially dephosphorylates p42/44MAPK but not p38MAPK in rat pinealocytes. J Neurochem. 2007;101:1685–93. doi: 10.1111/j.1471-4159.2007.04557.x. [DOI] [PubMed] [Google Scholar]
- RATH MF, BAILEY MJ, KIM JS, COON SL, KLEIN DC, MOLLER M. Developmental and daily expression of the Pax4 and Pax6 homeobox genes in the rat retina: localization of Pax4 in photoreceptor cells. J Neurochem. 2009a;108:285–94. doi: 10.1111/j.1471-4159.2008.05765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RATH MF, BAILEY MJ, KIM JS, HO AK, GAILDRAT P, COON SL, MOLLER M, KLEIN DC. Developmental and diurnal dynamics of Pax4 expression in the mammalian pineal gland: nocturnal down-regulation is mediated by adrenergic-cyclic adenosine 3',5'-monophosphate signaling. Endocrinology. 2009b;150:803–11. doi: 10.1210/en.2008-0882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REIG JA, YU L, KLEIN DC. Pineal transduction. Adrenergic----cyclic AMP-dependent phosphorylation of cytoplasmic 33-kDa protein (MEKA) which binds beta gamma-complex of transducin. J Biol Chem. 1990;265:5816–24. [PubMed] [Google Scholar]
- REPPERT SM, PERLOW MJ, TAMARKIN L, ORLOFF D, KLEIN DC. The effects of environmental lighting on the daily melatonin rhythm in primate cerebrospinal fluid. Brain Res. 1981a;223:313–23. doi: 10.1016/0006-8993(81)91144-6. [DOI] [PubMed] [Google Scholar]
- REPPERT SM, PERLOW MJ, UNGERLEIDER LG, MISHKIN M, TAMARKIN L, ORLOFF DG, HOFFMAN HJ, KLEIN DC. Effects of damage to the suprachiasmatic area of the anterior hypothalamus on the daily melatonin and cortisol rhythms in the rhesus monkey. J Neurosci. 1981b;1:1414–25. doi: 10.1523/JNEUROSCI.01-12-01414.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROSEBOOM PH, COON SL, BALER R, MCCUNE SK, WELLER JL, KLEIN DC. Melatonin synthesis: analysis of the more than 150-fold nocturnal increase in serotonin N-acetyltransferase messenger ribonucleic acid in the rat pineal gland. Endocrinology. 1996;137:3033–45. doi: 10.1210/endo.137.7.8770929. [DOI] [PubMed] [Google Scholar]
- ROSEBOOM PH, KLEIN DC. Norepinephrine stimulation of pineal cyclic AMP response element-binding protein phosphorylation: primary role of a beta-adrenergic receptor/cyclic AMP mechanism. Mol Pharmacol. 1995;47:439–49. [PubMed] [Google Scholar]
- SCHAAD NC, SHINOHARA T, ABE T, KLEIN DC. Photoneural control of the synthesis and phosphorylation of pineal MEKA (phosducin) Endocrinology. 1991;129:3289–98. doi: 10.1210/endo-129-6-3289. [DOI] [PubMed] [Google Scholar]
- SIMONNEAUX V, POIREL VJ, GARIDOU ML, NGUYEN D, DIAZ-RODRIGUEZ E, PEVET P. Daily rhythm and regulation of clock gene expression in the rat pineal gland. Brain Res Mol Brain Res. 2004;120:164–72. doi: 10.1016/j.molbrainres.2003.10.019. [DOI] [PubMed] [Google Scholar]
- SOMERS RL, KLEIN DC. Rhodopsin kinase activity in the mammalian pineal gland and other tissues. Science. 1984;226:182–4. doi: 10.1126/science.6091271. [DOI] [PubMed] [Google Scholar]
- SUGDEN D, KLEIN DC. Rat pineal alpha 1-adrenoceptors: identification and characterization using [125I]iodo-2-[beta-(4-hydroxyphenyl)-ethylaminomethyl]tetralone. Endocrinology. 1984;114:435–40. doi: 10.1210/endo-114-2-435. [DOI] [PubMed] [Google Scholar]
- SUGDEN D, KLEIN DC. Activators of protein kinase C act at a postreceptor site to amplify cyclic AMP production in rat pinealocytes. J Neurochem. 1988;50:149–55. doi: 10.1111/j.1471-4159.1988.tb13242.x. [DOI] [PubMed] [Google Scholar]
- SUGDEN D, VANECEK J, KLEIN DC, THOMAS TP, ANDERSON WB. Activation of protein kinase C potentiates isoprenaline-induced cyclic AMP accumulation in rat pinealocytes. Nature. 1985;314:359–61. doi: 10.1038/314359a0. [DOI] [PubMed] [Google Scholar]
- SUGDEN D, WELLER JL, KLEIN DC, KIRK KL, CREVELING CR. Alpha-adrenergic potentiation of beta-adrenergic stimulation of rat pineal N-acetyltransferase. Studies using cirazoline and fluorine analogs of norepinephrine. Biochem Pharmacol. 1984;33:3947–50. doi: 10.1016/0006-2952(84)90006-6. [DOI] [PubMed] [Google Scholar]
- TANAKA K, MURAKAMI M, GREER MA. Type-II thyroxine 5'- deiodinase is present in the rat pineal gland. Biochem Biophys Res Commun. 1986;137:863–8. doi: 10.1016/0006-291x(86)91159-9. [DOI] [PubMed] [Google Scholar]
- TANAKA K, MURAKAMI M, GREER MA. Rhythmicity of triiodothyronine generation by type II thyroxine 5'-deiodinase in rat pineal is mediated by a beta-adrenergic mechanism. Endocrinology. 1987;121:74–7. doi: 10.1210/endo-121-1-74. [DOI] [PubMed] [Google Scholar]
- TOSINI G, CHAURASIA SS, MICHAEL IUVONE P. Regulation of arylalkylamine N-acetyltransferase (AANAT) in the retina. Chronobiol Int. 2006;23:381–91. doi: 10.1080/07420520500482066. [DOI] [PubMed] [Google Scholar]
- TOYAMA R, CHEN X, JHAWAR N, AAMAR E, EPSTEIN J, REANY N, ALON S, GOTHILF Y, KLEIN DC, DAWID IB. Transcriptome analysis of the zebrafish pineal gland. Dev Dyn. 2009;238:1813–26. doi: 10.1002/dvdy.21988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VANECEK J, SUGDEN D, WELLER J, KLEIN DC. Atypical synergistic alpha 1- and beta-adrenergic regulation of adenosine 3',5'-monophosphate and guanosine 3',5'-monophosphate in rat pinealocytes. Endocrinology. 1985;116:2167–73. doi: 10.1210/endo-116-6-2167. [DOI] [PubMed] [Google Scholar]
- VAUGHAN GM, PRUITT BA, JR., MASON AD., JR. Nyctohemeral rhythm in melatonin response to isoproterenol in vitro: comparison of rats and Syrian hamsters. Comp Biochem Physiol C. 1987;87:71–4. doi: 10.1016/0742-8413(87)90183-6. [DOI] [PubMed] [Google Scholar]
- VAUGHAN GM, REITER RJ. The Syrian hamster pineal gland responds to isoproterenol in vivo at night. Endocrinology. 1987;120:1682–4. doi: 10.1210/endo-120-4-1682. [DOI] [PubMed] [Google Scholar]
- WOLFE MS, LEE NR, ZATZ M. Properties of clock-controlled and constitutive N-acetyltransferases from chick pineal cells. Brain Res. 1995;669:100–6. doi: 10.1016/0006-8993(94)01267-l. [DOI] [PubMed] [Google Scholar]
- YOSHIMOTO T, KUSAKA M, SHINJO F, YAMAMOTO S, DRAY F. 12- and 15-lipoxygenases in rat pineal gland. Prostaglandins. 1984;28:279–85. doi: 10.1016/0090-6980(84)90063-7. [DOI] [PubMed] [Google Scholar]
- ZATZ M, MULLEN DA. Norepinephrine, acting via adenylate cyclase, inhibits melatonin output but does not phase-shift the pacemaker in cultured chick pineal cells. Brain Res. 1988a;450:137–43. doi: 10.1016/0006-8993(88)91553-3. [DOI] [PubMed] [Google Scholar]
- ZATZ M, MULLEN DA. Two mechanisms of photoendocrine transduction in cultured chick pineal cells: pertussis toxin blocks the acute but not the phase-shifting effects of light on the melatonin rhythm. Brain Res. 1988b;453:63–71. doi: 10.1016/0006-8993(88)90143-6. [DOI] [PubMed] [Google Scholar]