Abstract
How the co-ordinated events of gene activation and silencing during cellular differentiation are influenced by spatial organization of the cell nucleus is still poorly understood. Little is known about the molecular mechanisms controlling subnuclear distribution of transcription factors, and their interplay with nuclear proteins that shape chromatin structure. Here we show that C/EBPβ not only associates with pericentromeric heterochromatin but also interacts with the nucleoskeleton upon induction of adipocyte differentiation of 3T3-L1 cells. Different C/EBPβ dimers localize in different nuclear domains. Using BiFC in living cells, we show that LAP (Liver Activating Protein) homodimers localize in euchromatin and heterochromatin. In contrast, LIP (Liver Inhibitory Protein) homodimers localize exclusively in heterochromatin. Importantly, their differential subnuclear distribution mirrors the site for interaction with HP1α. HP1α inhibits LAP transcriptional capacity and occupies the promoter of the C/EBPβ-dependent gene c/ebpα in 3T3-L1 preadipocytes. When adipogenesis is induced, HP1α binding decreases from c/ebpα promoter, allowing transcription. Thus, the equilibrium among different pools of C/EBPβ associated with chromatin or nucleoskeleton, as well as dynamic changes in their interaction with HP1α, play key roles in the regulation of C/EBP target genes during adipogenesis.
Keywords: cellular differentiation, subnuclear distribution, transcriptional regulation, C/EBPβ, HP1α
Introduction
Cellular differentiation is achieved by keeping a predetermined subset of genes in a state in which they can be expressed whereas the rest is silenced. While the coordinated sequence of transcription factors that contribute to changes in gene expression during adipocyte differentiation is well-established [1], the integration of gene activation and silencing events into the architectural framework of the cell nucleus is poorly understood. It is necessary, therefore, to understand the molecular mechanisms controlling the subnuclear distribution of proteins that regulate transcription together with their interplay with proteins that modulate chromatin structure. For insight into the complex relationships between transcription factors and nuclear architecture, this study examines subnuclear localization of the transcription factor CCAAT/Enhancer Binding Protein (C/EBP) β, a key factor in the adipogenic cascade, and its interaction with the Heterochromatin Protein (HP)1α.
C/EBPβ is involved in a wide variety of physiological events suggesting high versatility in mediating distinct biological outcomes. However, it is not known how such plasticity is achieved. It mediates gene expression in diverse processes of cell differentiation such as adipogenesis [2, 3], liver development and regeneration [4, 5], hematopoiesis [6, 7] and embryogenesis [8]. C/EBPβ also regulates gene expression in response to hormonal stimuli such as the activation of c-fos in response to growth hormone (GH) [9-11]. When adipogenesis is triggered, the expression of C/EBPβ and C/EBPδ is induced, leading to an increased expression of C/EBPα and PPARγ that in turn activate transcription of genes responsible for the acquisition of the adipocyte phenotype. Disruption of c/ebpβ gene in mice causes impaired development of adipose tissue [12]. Mouse embryonic fibroblasts (MEFs) derived from mice genetically lacking C/EBPβ are unable to undergo adipogenesis [13]. Recently, it has been shown that C/EBPβ is required for the cell fate switch from myoblastic precursors to brown fat cells [14]. C/EBPβ has alternative translation products (LAP and LIP) able to form homo- and heterodimers [15]. LAP harbors a highly conserved C-terminal region composed of a basic DNA binding domain and an amphipathic leucine zipper domain for dimerization (bZIP), and an N-terminal transactivation domain [15]. LIP possesses a truncated N-terminal transactivation domain. This truncation variant functions as a “dominant negative” form that inhibits transcription [15]. Regulation of C/EBPβ function has also been reported to be related to nuclear architecture. The regulation of gene expression by C/EBPβ correlates with its rapid up-regulation and relocalization to areas of heterochromatin upon GH, insulin and PDGF treatment of 3T3-F442A preadipocytes [16]. C/EBPβ also concentrates in heterochromatin when 3T3-L1 fibroblasts are induced to differentiate in adipocytes [17]. It is intriguing that C/EBPβ, a transcriptional activator, concentrates in heterochromatin, where there are few active genes. This raises the possibility that the functional activity of C/EBPβ may be controlled by its subnuclear distribution through spatial segregation mediated by its interacting regulator proteins.
The nucleus is considered to be structurally and functionally compartmentalized: DNA is packaged into chromatin that can be organized by interaction with the nuclear matrix, a fibrogranular ribonucleoprotein network [18, 19]. Chromatin has been classified in two structural and functional subsets [20, 21]. Euchromatin is partially decondensed in interphase and contains the majority of transcribed genes, while heterochromatin remains highly condensed throughout the cell cycle, and includes the centromeric and telomeric regions of chromosomes [20, 21]. Pericentromeric heterochromatin mainly consists of repetitive DNA sequences, replicates late in S-phase and contains few transcriptionally active genes [22]. Heterochomatin has a characteristic histone-modification profile, distinguished by histone hypoacetylation and H3K9 methylation which serves as a molecular anchor for the recruitment of structural proteins that modify chromatin and stabilize its compact structure. It is well established that trimethylated K9 of histone H3 (3MeK9H3) provides a binding site for Heterochromatin Protein (HP)1 which recruits modifier factors to heterochromatic loci and stabilizes its structure [23-26]. HP1 proteins are conserved throughout evolution from yeast to human and form part of the chromodomain superfamily [27]. In mammals three genes encode the proteins HP1α, β and γ. HP1 proteins have an N-terminal globular chromodomain (CD) and a C-terminal globular chromoshadow domain (CSD) linked by a less conserved and flexible hinge region (HR) [27]. HP1 proteins were initially implicated in gene silencing [28, 29], and are required for normal development of eukaryotic organisms [30]. HP1 has since been implicated not only in heterochromatin assembly and stability but also in centromere function, nuclear organization and gene regulation [27].
In the present study we mapped the nuclear distribution C/EBPβ homo- and heterodimers together with their interplay with HP1α and the nuclear matrix. The different dimers of C/EBPβ localize in different nuclear domains. Further, their interaction with HP1α also occurs in different nuclear domains. This interaction suppresses the ability of C/EBPβ to activate transcription. The rapid, dynamic changes in the distribution of C/EBPβ with respect to pericentromeric heterochromatin, HP1α and the nuclear matrix when adipogenesis is induced suggests that the spatio-temporal regulation of C/EBPβ may have a functional role during adipocyte differentiation. Taken together, these data highlight the importance of considering nuclear compartmentalization and protein-protein interactions to gain insight into the molecular mechanisms that control cellular differentiation.
Materials and Methods
Cell culture
Murine 3T3-L1 preadipocytes and human embryonic kidney 293T cells (from ATCC) were grown in Dulbecco's Modified Eagle's Medium containing 4.5 g/liter glucose and 10% v/v calf serum in an atmosphere of 10% CO2, 90% air at 37°C. 3T3-L1 cells were cultured until confluence, maintained for 48h (day 0), and differentiation was induced as previously described [31]. For TSA treatment, confluent 3T3-L1 preadipocytes were grown in the presence of either 87 nM TSA or vehicle (DMSO) for 72 h, and then induced to differentiate for 2 days. During the 5 days of TSA treatment, cell culture medium was replaced daily.
ImmunoFISH
Indirect Immunofluorescence
IIF was performed as previously described [16]. Briefly, 3T3-L1 cells were grown on coverslips and treated as indicated in figure legends. Cells were simultaneously fixed and permeabilized by immersion in cold methanol (-20°C) for 2 hours. Coverslips were washed three times with PBS, and inverted onto 50μl drop of PBS 1% BSA with anti-C/EBPβ dilution1/100 (SC-150, Santa Cruz), anti-P-C/EBPβ dilution 1/100 (Cell Signaling), anti-HP1α dilution 1/50 (05-689, UPSTATE), anti-NuMA dilution 1/50 (610562, BD Transduction Laboratories), anti-Flag dilution 1/ 100 (F3165, SIGMA) or anti-HA dilution 1/50 (MMS-101R, Covance) as indicated in figure legends. All IIF conditions were tested to avoid any staining artifacts. Nuclei were stained with DAPI or TO-PRO, and mounted in Vecta shield. Laser-scanning confocal microscopy was performed with LSM5 Pascal or a Meta microscope (Carl Zeiss) and images were taken in the middle section of the cell nucleus.
RNA interference
Knock down of C/EBPβ was performed as previously described [11]. Plasmids for siC/EBPβ or mU6pro (5μg each) were expressed in 3T3-L1 fibroblasts using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Forty-eight hours after transfection cells were induced to differentiate and twenty-four hours later cells were fixed and subjected to IIF as already described.
In situ Cell extraction
The extractions were performed directly on cells grown on coverslips (in situ extraction) using a protocol adapted from Fey et al. [34]. Briefly, 3T3-L1 cells grown on coverslips were washed with PBS followed by a quick wash with cytoskeleton (CSK) buffer [1mM Hepes, pH 6.8, 25 mM KCl, 5 mM MgCl2, 1 mM EGTA, 250 mM sucrose]. Then the cells were incubated with CSK buffer supplemented with 0.5% Triton X-100 for 5 min at room temperature for protein extraction. In the case of nuclease digestion, 2U/ μl DNase I (SIGMA) was added to PBS supplemented with 50 mM MgCl2 or 100μg/ml RNase A (SIGMA), and incubated 5 minutes at 20°C. After washes in PBS, cells were fixed in 4% paraformaldehyde, 4% sucrose in PBS during 15 min. at room temperature, washed three times with PBS, incubated with PBS 0.1% Triton X-100 for permeabilization prior incubation with the antibodies indicated in each condition.
Chromatin Immunoprecipitation (ChIP)Assay
ChIP was performed as previously described [11]. Briefly, 3T3-L1 cells were induced to differentiate in adipocytes for the times indicated; cells were washed with PBS and cross-linked with 1% formaldehyde. Cross-linking was stopped by addition of glycine to a final concentration of 125 mM. Cells were collected, nuclei were isolated and lysed by incubation in nuclear lysis buffer [50mM Tris-HCl, pH 8.1, 10mM EDTA, 1% SDS, and protease inhibitors]. Chromatin was fragmented in 500-800 bp fragments by sonication. The chromatin fractions were immunoprecipitated with the indicated antibodies or non-immune IgG at 4°C overnight. Following washes and elution, precipitates were heated overnight at 65°C to reverse cross-linking. DNA fragments were purified by QIAquick kit according to the manufacturer's instructions. A total of 4μl of purified DNA was subjected to PCR amplification using the primers for the C/EBP binding site in γ-satellite DNA [5′GGACCTGGAATATGGCGAGAAAACTGAA3′; 5′GGACGTGGAATATGGCAAGAAAACTGAA 3′] [16, 35], c/ebpα promoter [5′TGACTTAGAGGCTTAAAGGA 3′; 5′CGGGGACCGCTTTTATAGAG 3′] or c-fos promoter [5′GGCTGCAGCCGGCGAGCTG 3′; 5′AGAAGCGCTGTGAATGGATG 3′] [11]. PCR products were separated in 2% agarose gel and stained with ethidium bromide. Images were visualized using a Gel Doc XR (Bio-Rad, USA).
Immunoprecipitation and GST pulldown assays
293T cells were transiently transfected with the indicated cDNA by calcium phosphate co-precipitation assay as previously described [9, 10]. Two days after transfection cells were lysed, 5μl of anti-C/EBPβ (SC-150; Santa Cruz) or a non immune rabbit IgG was added, and samples were incubated for 2hs on ice. Then, 50 μl of Protein A-agarose was added and extracts were incubated with rotation at 4°C during 2h. The beads were washed four times with lysis buffer, 50 μl Laemmli buffer was added and samples were boiled for 5 min. SDS- PAGE and western blot were performed as previously described [9, 36].
In vitro translation of CMV-LAP or CMV-LIP was performed with 35S-methionine (Perkin Elmer, Boston, MA) and a TNT-coupled transcription translation system, according to the manufacturer's instructions (Promega, Madison, WI). GST- HP-1α, GST-1-116-HP-1α, GST-116-191-HP-1α (kind gift of A. Dejean (Institut Pasteur, Paris, France)) or GST alone were expressed in E. coli, and purified on glutathione-sepharose beads (SIGMA). For in vitro binding GST fusions or GST alone were incubated with the labeled protein in 1 ml binding buffer containing 300 mM NaCl, 0.5% Triton-X-100, 50 mM Tris pH 8, and 2 mM EDTA, and protease inhibitors for 1 h at room temperature. Beads were washed five times with the same buffer. Proteins were separated by SDS-PAGE electrophoresis, electrotransfered and visualized by autoradiography.
Analysis of Bimolecular Fluorescence Complementation (BiFC)
The sequences encoding amino acids residues 1-155 or 156-264 of enhanced yellow fluorescent protein, designated -YN and -YC respectively, cloned into pCMV-HA or pCMV-FLAG2 [37, 38] were fused to the 3′ end of the coding regions for full length HP1α (kindly provided by T. Misteli, Nat. Cancer Inst., NIH, Bethesda, USA), HP1 α-1-112 (HP1α –CD/HR), HP1α-1-77 (HP1α-CD), HP1α- 82-112 (HP1α-HR), HP1α- 113-191 (HP1α-CSD), LAP, LIP, and the basic region and leucine zipper domains (amino acids 194 to 276 common to LAP and LIP), referred as bZIP. Constructs designated LAP ΔZIP and LIPΔZIP in which the leucine zipper (amino acids 228 to 276) was deleted were also subcloned in the YN/HA- or YC/Flag- tag vectors, respectively. To construct LIPΔbZIP both the basic and leucine zipper domains (amino acids 194 to 276) were deleted and a oligonucleotide encoding the SV40 nuclear localization signal [PKKKRKV] [39] was inserted in frame at the C-terminal of LIP and subcloned in the YN/HA or YC/Flag vector. The sequence of all constructs was confirmed by sequencing.
3T3-L1 preadipocytes were grown in six well plates to 50% confluence and transfected with 0.15- 0.5 μg of the plasmids encoding the fusion proteins as YN- or YC- indicated in figure legends using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. BiFC analysis was performed as previously described [37, 38]. The fluorescence complementation was observed by using a LSM5 Pascal or a Meta confocal microscope (Carl Zeiss) and images were taken in the middle section of the cell nucleus. YFP fluorescence emission was measured at 530 ± 20 nm during excitation at 490 ± 5nm. Image analyses for fluorescence complementation localization were performed using the 3D-surface plot software application plug in included in the Image-J program (v.1.42) available from the NIH, as previously described [40].
Gene expression assays
293T cells were transiently transfected by calcium phosphate co-precipitation assay as described [9] with C/EBP-Luc or c-fos-Luc (0.5 μg), and RSV-β-galactosidase (0.1 μg) plasmids, in the absence or presence of CMV-LAP (0.1 μg or 1 ng), p300 (0.5 μg) and increasing amounts of HP1α, as indicated in figure legend. Twenty-four hours after transfection, cell lysates were prepared, and luciferase or β-galactosidase activity was measured using a Veritas™ microplate luminometer (Turner Biosystems, USA). Luciferase values were normalized to β-galactosidase activity. Each condition was tested in duplicate in each experiment. A two-sample t-test (SigmaStat) was used to judge statistical significance where a value of p < 0.05 was considered statistically significant.
Results
C/EBPβ interacts with C/EBP sites in pericentromeric major satellite DNA
To assess the nuclear distribution of C/EBPβ during adipogenesis, 3T3-L1 cells were induced to differentiate and IIF was performed using specific antibodies raised against an epitope in the carboxy terminal domain common to both forms of C/EBPβ, LAP and LIP. C/EBPβ is detected in the nucleus within 2 h of induction of adipocyte differentiation. C/EBPβ is diffusely distributed throughout the nucleus and in speckles. Areas of intense C/EBPβ staining coincide with areas intensely stained by DAPI, which in murine cells correspond to chromocenters [41]. The same distribution was observed at all time points post-induction of adipogenesis analyzed. C/EBPβ staining was specific since no signal was observed in cells where C/EBPβ was knocked-down by siRNA (Fig. 1A, C/EBPβ siRNA). To determine whether C/EBPβ specifically localizes in pericentromeric heterochromatin, we performed immuno-FISH that allow detection of DNA and protein together in structurally preserved nuclei [32]. C/EBPβ was almost undetectable in preadipocytes (Fig. 1B) and none appeared to be associated with pericentromeric heterochromatin, identified by rhodamine labeled γ-satellite probe (Fig. 1C, merged image panel D). Twenty-four hours post- induction of adipogenesis, C/EBPβ concentration was increased and areas of intense signal as well as areas of a more diffuse distribution could be seen (Fig. 1E). The areas of intense C/EBPβ signal (Fig 1E) consistently colocalized with γ-satellite DNA (Fig. 1F), seen as yellow when both images were merged (Fig. 1G). Thus, these results demonstrate that C/EBPβ localizes to clustered pericentromeric heterochromatin upon induction of adipogenesis.
Figure 1. C/EBPβ localizes in pericentromeric heterochromatin upon binding to C/EBPsites present in satellite DNA when adipogenesis is induced.
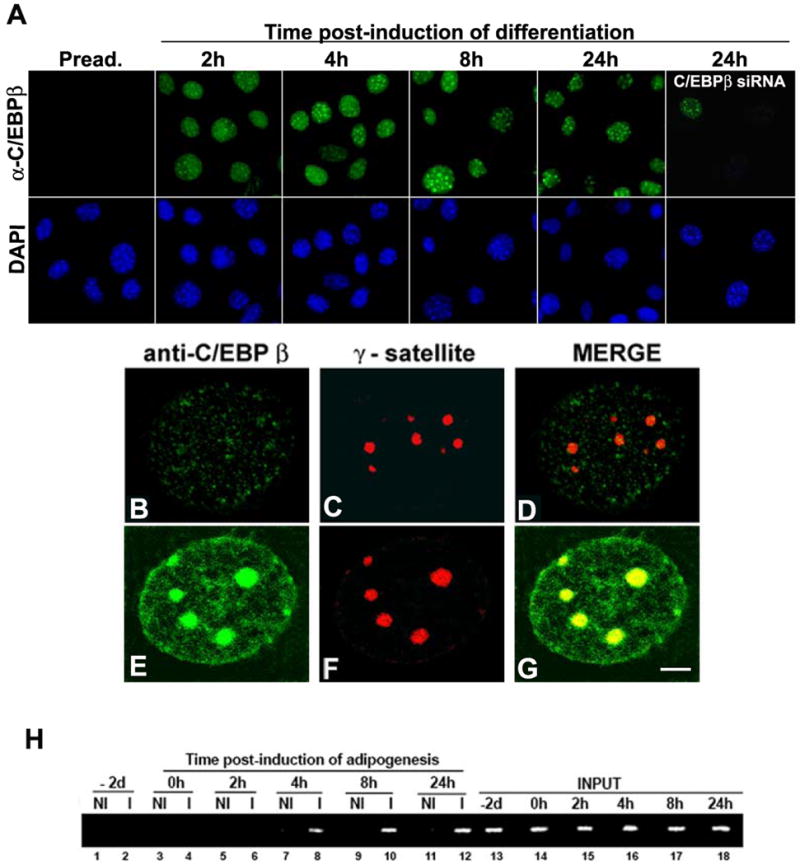
A- 3T3-L1 preadipocytes were grown on coverslips and adipocyte differentiation was induced for the indicated time, cells were fixed, IIF was performed using anti-C/EBPβ and nuclei were counterstained with DAPI. B-G Immuno-FISH was performed as described in Materials and Methods using anti-C/EBPβ and a probe for major satellite DNA labeled with Rhodamine. Scale bar, 2 μm. H- 3T3-L1 cells were induced to differentiate to adipocytes for the indicated periods of time. ChIP was performed using anti-C/EBPβ (I) or non-immune IgG (NI). 1% Input is shown (lanes 13 to 18). Similar results were obtained in two independent experiments.
The nucleotide sequences of the major γ-satellite DNA contain eight repeats of a consensus C/EBP binding site (TT/GXXGXAAT/G) to which C/EBPβ can bind as LAP or LIP homo- or heterodimers [16]. To examine in vivo whether endogenous C/EBPβ concentrates in pericentric heterochromatin upon its interaction to the C/EBP consensus sites present in major satellite DNA, ChIP assays were performed. Chromatin–bound C/EBPβ was immunoprecipitated from nuclei of 3T3-L1 cells at different time points after induction of adipogenesis, and immunoprecipitated DNA fragments were amplified using primers specific for major satellite DNA. No binding of C/EBPβ to major satellite DNA was detected in preadipocytes (Fig. 1H, lanes 2 and 4) or 2 hours after induction of adipogenesis (Fig. 1G, lane 6). However, C/EBPβ bound to satellite DNA within 4 hours of induction of adipogenesis (Fig. 1G, lane 8), and it was detected after 8 and 24 hours of induction of adipocyte differentiation (lanes 10 and 12). Taken together these results demonstrate that C/EBPβ associates with pericentromeric heterochromatin when 3T3-L1 cells undergo adipogenesis.
C/EBPβ subnuclear distribution depends on chromatin structure
In heterochromatin, histones are hypoacetylated and highly methylated [42, 43]. Long-term treatment of cells with the histone deacetylase inhibitor Trichostatin A (TSA) reversibly disrupts the compaction of pericentromeric heterochromatin (44). In order to determine whether the structural integrity of chromatin is required for its association with C/EBPβ, 3T3-L1 preadipocytes were grown for three days in the presence of TSA or vehicle (DMSO), induced to differentiate for 48 hours with MDI in the presence or absence of TSA, and the subnuclear distribution of C/EBPβ was assessed by IIF. In the absence of TSA, C/EBPβ was detected as intense foci as well as diffusely distributed in the nucleus (Fig. 2A) as expected [16, 17]. C/EBPβ signal coincided with areas intensely stained with DAPI (Fig. 2C), corresponding to pericentromeric heterochromatin (merged image, Fig. 2D). C/EBPβ co-localized with HP1α (Fig. 2B, merged image Fig. 2H). In marked contrast, when 3T3-L1 cells were grown for three days and induced to differentiate for 48h in the presence of TSA (total of five days with TSA) C/EBPβ appeared to be distributed throughout the nucleus as small speckles (Fig. 2E). Delocalization of C/EBPβ with TSA treatment was accompanied by some disruption of chromocenters revealed by a less focused HP1α distribution (Fig. 2F) and by a more diffuse DAPI staining (Fig. 2G), as previously reported [44, 45]. C/EBPβ was substantially excluded from the remaining areas of intense HP1α and DAPI staining (Fig. 2 inset, arrowheads). Prolonged TSA treatment did not affect C/EBPβ or HP1α protein level as demonstrated by WB (Fig. 2J). Further, TSA treatment increased the level of acetyl histone H4 and decreased the level of 3MeK9H3 (Fig. 2J) that favor chromatin decondensation, as previously shown [45]. To address whether the subnuclear delocalization of C/EBPβ is a consequence of failure of its binding to C/EBP consensus sites in satellite DNA, we performed ChIP assays. Prolonged TSA treatment abrogated the binding of C/EBPβ to γ satellite DNA (Fig. 2K, lane 2 +TSA vs. –TSA). Taken together, these results imply that C/EBPβ requires the integrity of chromatin structure for proper subnuclear distribution when 3T3-L1 preadipocytes are induced to differentiate. TSA treatment is known to block the differentiation of 3T3-L1 cells into adipocytes [46]. Our results suggest that alteration of the higher-order organization of pericentromeric foci, accompanied by nuclear redistribution of C/EBPβ, may contribute to this blockade in adipogenesis.
Figure 2. TSA disrupts subnuclear distribution of C/EBPβ.
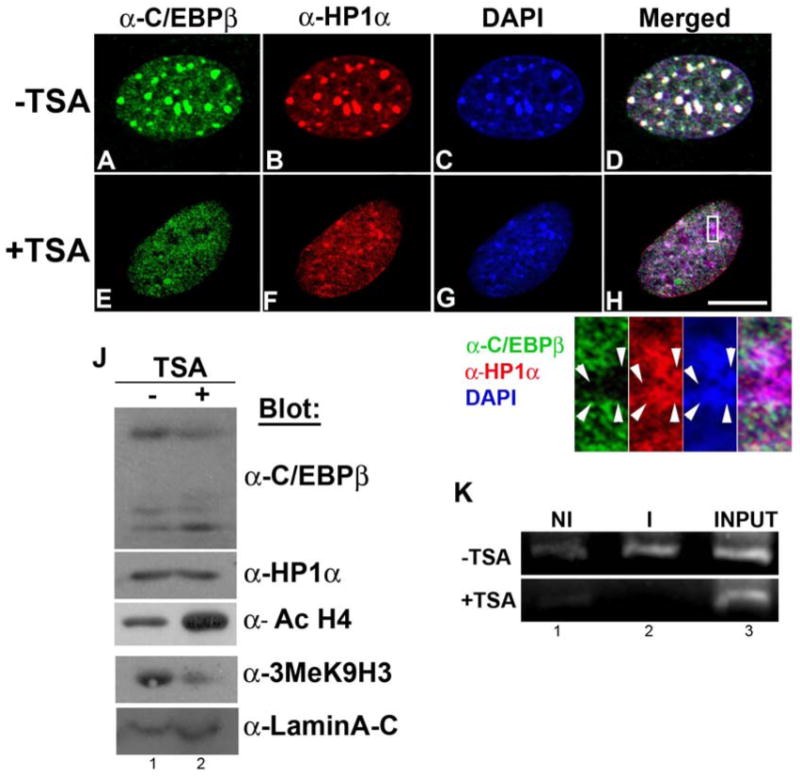
3T3-L1 preadipocytes were grown on coverslips for three days and induced to differentiate by treatment with MDI (48 h) in the presence of vehicle (-TSA) or 87 nM Trichostatin A (+TSA). Under these experimental conditions: A-H Cells were fixed and permeabilized in cold methanol, subjected to IIF with the indicated antibodies, and nuclei were counterstained with DAPI. Scale bar, 5 μm. J- Cell lysates were subjected to WB analysis with the indicated antibodies. K- ChIP was performed using anti-C/EBPβ (I) or non-immune IgG (NI) to evaluate C/EBPβ binding to consensus sites present in γ-satellite DNA. 1% Input is shown. Similar results were obtained in three independent experiments.
C/EBPβ interacts not only with chromatin but also with the nuclear matrix
In the nucleus, chromatin can be organized by its interaction with the nuclear matrix, a fibrogranular ribonucleoprotein network to which many nuclear factors also interact [19]. In order to investigate whether C/EBPβ is retained in the nucleus by its interaction with chromatin and/or the nuclear matrix, 3T3-L1 cells were subjected to in situ extraction with Triton X-100. This treatment extracts proteins from the nucleoplasm together with proteins weakly bound to chromatin and/or the nuclear matrix [34, 44, 47]. When 3T3-L1 cells were induced to differentiate and then subjected to in situ extraction with Triton X-100, C/EBPβ was retained in the nucleus (Fig. 3A) and remained associated with HP1α-rich pericentromeric heterochromatic foci (Fig 3B, and merged Fig. 3D). This implies that C/EBPβ is normally tightly associated with chromatin and/or the nuclear matrix and not a protein easily extractable from the nucleus. HP1α was also resistant to Triton X-100 extraction (Fig. 3B). To analyze whether proper C/EBPβ subnuclear distribution depends on its interaction with ribonucleoproteins, in situ extraction was followed by RNase A treatment. We found that C/EBPβ retained its subnuclear distribution after such treatment (Fig. 3E- Fig. 3H), suggesting that C/EBPβ localization is independent of its interaction with ribonucleoprotein(s) or RNA. In contrast, HP1α pericentromeric localization was diminished after RNase A treatment (Fig. 3F), as previously reported [48]. Next, to analyze whether C/EBPβ interacts not only with chromatin but also with the nuclear matrix, cells were incubated with DNase I after Triton X-100 treatment [47]. No DAPI staining was observed after DNase I treatment (Fig. 3K) indicating that chromatin was completely removed. The nuclear matrix, however, was not removed by DNase I digestion, shown by the presence of distinct NuMA foci (Fig. 3M), here used as a control [19]. Interestingly, some C/EBPβ (Fig. 3I), and HP1α (Fig. 3J) could still be detected in the nucleus after chromatin digestion by DNase I, implying that a fraction of C/EBPβ and HP1α could be retained through their interaction with the nuclear matrix. Further, C/EBPβ and HP1α associated to the nuclear matrix did not co-localize (Fig. 3L). In addition, when cells were grown for three days and induced to differentiate for 48 h in the presence of TSA, C/EBPβ was no longer retained in the nucleus after in situ (data not shown), suggesting that TSA treatment weakened the interaction of C/EBPβ with chromatin as well as the nuclear matrix, resulting in more soluble C/EBPβ in the nucleoplasm. Thus, these results indicate that C/EBPβ subnuclear distribution depends both on its interaction with chromatin and the nuclear matrix, and the existence of different subsets of C/EBPβ and HP1α that may be functionally distinct.
Figure 3. C/EBPβ interacts both with chromatin and the nuclear matrix.
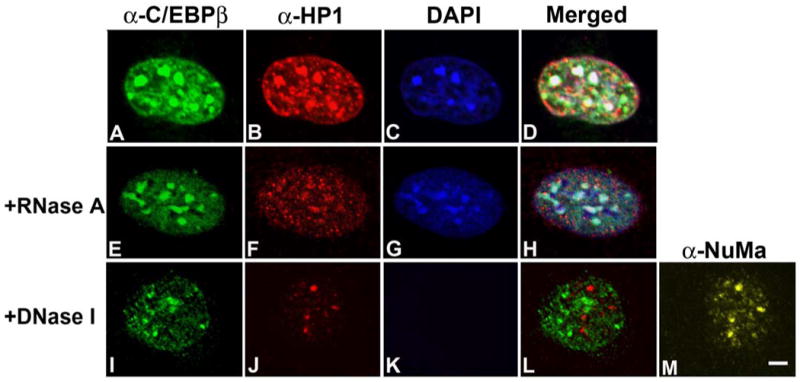
3T3-L1 cells grown on coverslips were induced to differentiate with MDI for 48 h and then subjected to in situ extraction alone (panels A to D), or followed by RNase (panels E to H) or DNase treatment (panels I to M). Then cells were fixed, and subjected to IIF with the indicated antibodies. Results are representative of three independent experiments. Scale bar, 2 μm.
LAP and LIP homo- and heterodimers localize in different nuclear domains
We applied Bimolecular Fluorescence Complementation (BiFC) in living cells to visualize the precise subnuclear localization of LAP or LIP homo- and heterodimers, circumventing limitations of using antibodies directed against an epitope common to both C/EBPβ forms. BiFC visualizes interactions between proteins in their native location through complementation between two non-fluorescent fragments of the yellow fluorescent protein (YFP) that takes place when they are brought together by interactions between proteins to which the YFP fragments are fused [37, 49]. Thus, complementary fragments of YFP (-YN and -YC) were fused to the carboxy-terminal end of LAP, and LIP Different combinations of plasmids encoding the fusion proteins were transfected in 3T3-L1 cells, and fluorescence complementation was monitored by confocal microscopy in living cells. When LAP-YN and LAP-YC were co-expressed, fluorescence complementation was observed in foci and throughout the nucleus (Fig. 4A-I). The BiFC signal was analyzed by 3D-surface plot analysis, confirming the presence of fluorescence both in foci and throughout the nucleus (Fig. 4A-II). The punctate pattern of fluorescence exhibited by LAP homodimers coincided with heterochromatic areas intensely stained by TO-PRO-3 when BiFC was analyzed in fixed cells (Supplementary Fig. 1). In contrast, when LIP-YN and LIP-YC were co-expressed the fluorescent signal was exclusively restricted to foci (Fig. 4B -III) that coincided with areas of intense TO-PRO-3 that stain pericentromeric heterocromatin in fixed cells (Supplementary Fig.1). Image analysis using Image-J software confirms the presence of BiFC fluorescent signal only in foci (Fig.4B- IV). Further, when LAP-YC and LIP-YN were co-expressed fluorescence was observed in foci and diffusely throughout the nucleus (Fig. 4C-V) exhibiting a similar pattern of nuclear distribution to LAP homodimers and confirmed by image analysis using Image-J program (Fig. 4C- VI). No detectable fluorescence was observed in cells expressing LIP-YC with LIPΔZIP-YN, in which the leucine zipper required for dimerization was deleted (Fig. 4D) The fusion proteins were present in overlapping nuclear domains as shown by IIF since LIPΔZIP-YN has an HA tag and LIP-YC has a FLAG tag. Similar results were obtained when LAP-YC and LAPΔZIP were co-expressed (data not shown). The expression level of the different fusion proteins was comparable (Supplementary Fig. 1) as assessed by WB. In addition, no fluorescence was detected when LAP or LIP as -YN or -YC fusion proteins were expressed alone. Thus, formation of the bimolecular fluorescent complex required specific dimerization between LAP or LIP proteins reflecting dimer formation. Taken together, we demonstrate for the first time that LAP and LIP homo- and heterodimers are distributed in different nuclear domains. LAP homodimers localized in heterochromatin and in euchromatic nuclear domains where LAP can activate the expression of C/EBPβ target genes. In contrast, LIP homodimers concentrate exclusively in pericentromeric heterochromatin, possibly as a mechanism that maintains an inhibitor of transcription distant from target genes.
Figure 4. Visualization of C/EBPβ homo- and heterodimers formation in living cells.
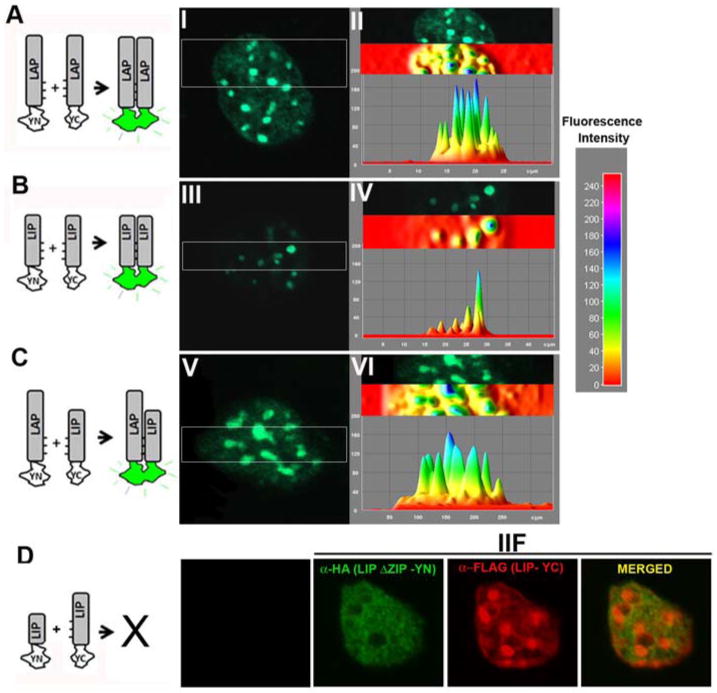
LAP and LIP as -YC or -YN fusion proteins were expressed in 3T3-L1 cells, and the fluorescence emission of the cells was imaged 24h after transfection by confocal microscopy. A-C Images of representative nucleus that correspond to more than 98% of the fluorescent cells in each population are shown. Fluorescence distribution was evaluated using 3D-surface plot analysis. The color code for fluorescence intensity is depicted on the right. The diagrams to the left of the images represent the experimental strategies used. D- IIF shows the expression of LIP-ΔZIP-YN and LIP-YC that do not show recovery of fluorescence emission because they lack the capacity to form dimers.
C/EBPβ interacts with HP1α
C/EBPβ co-localizes with HP1α mainly in pericentromeric heterochromatin and in some euchromatic areas (Figs. 2 and 3). Since HP1α has been implicated in the regulation of gene transcription, we asked whether C/EBPβ and HP1α interact. To test this possibility, LAP or LIP was expressed in 293T cells in the absence or presence of GFP-HP1α. As shown in figure 5A, HP1α co-immunoprecipitated with both LAP (lane 2) and LIP (lane 4). The co-immunoprecipitation of HP1α was specific since no band was detected when LAP or LIP were immunoprecipitated from cells that did not co-express GFP-HP1α (Fig. 5A, lanes 1 and 3) or when GFP-HP1α was expressed alone (Fig. 5A, lane 5). To determine whether interaction between C/EBPβ and HP1α is direct, we performed GST-pull down binding assays with bacterially expressed GST-HP1α fusion protein and in vitro translated LAP or LIP (Fig. 5B). GST-HP1α interacted with [35S] methionine-labeled LAP (Fig. 5B, lane 2), as well as LIP (lane5). No interaction was detected when [35S] methionine-labeled LAP or LIP were incubated with GST alone (Fig. 5B, lanes 3 and 6, respectively). Taken together these results show that both of the major translational forms of C/EBPβ, LAP and LIP, interact with HP1α.
Figure 5. C/EBPβ interacts with HP-1α.
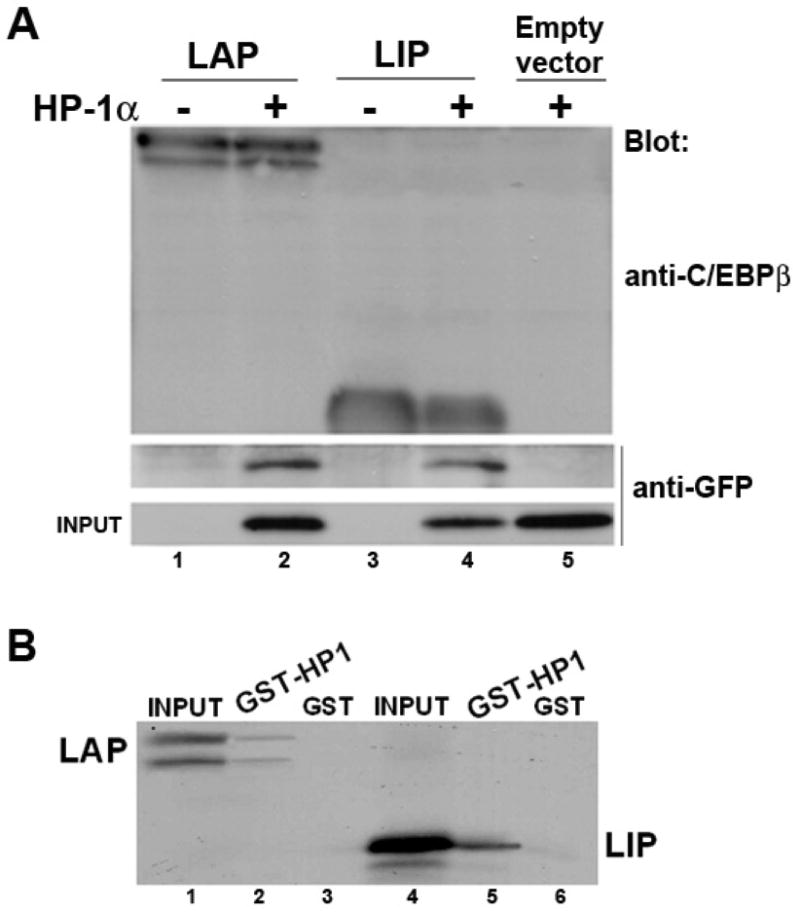
A- LAP or LIP was over-expressed in the absence or presence of GFP-HP1α in 293T cells. 48h after transfection cells were lysed, LAP and LIP were immunoprecipitated with anti-C/EBPβ. Immunoprecipitated proteins were separated by SDS-PAGE and then immunobloted with anti-C/EBPβ or anti-GFP. B- GST-HP1α or GST alone was expressed in E. coli, conjugated to glutathione-agarose beads, and incubated with [35S] methionine-labeled LAP or LIP, as described in Materials and Methods. Associated proteins were resolved by SDS-PAGE and analyzed by autoradiography. As a control, 10% of the [35S] methionine-labeled LAP or LIP was applied to the gel (lanes 1 and 4, respectively).
Visualization of C/EBPβ and HP1α interaction in the nucleus of living cells
Next we investigated the interaction between C/EBPβ and HP1α in living cells using BiFC assay to determine the precise nuclear domain where their interaction occurs. Different combinations of plasmids encoding LAP, LIP and HP1α as YN- or YC- fusion proteins were transfected in 3T3-L1 cells, and fluorescence complementation was monitored in living cells by confocal microscopy. When LAP-YN and HP1-YC were co-expressed, fluorescence complementation was observed in foci and throughout the nucleus (Fig. 6A). In contrast, when LIP-YN and HP-1α-YC were co-expressed the fluorescent signal was exclusively restricted to foci (Fig. 6C). The analysis of BiFC signal using 3D-surface imaging software, confirmed the distribution of fluorescence signal detected due to LAP-HP1α (Fig. 6B) or LIP-HP1α (Fig. 6C) interaction. The same results were obtained by digital analysis of 50 cells co-expressing LAP- or LIP-HP1α, respectively. No fluorescence was observed when LAP, LIP or HP1α fusion proteins were expressed alone (data not shown). As a control, HP1α-YN and HP-1α-YC were co-expressed, and dimerization of HP-1α rendered a fluorescent signal both in heterochromatic and euchromatic nuclear domains (data not shown), in agreement with subnuclear distribution of endogenous HP1α (Fig. 2F). Thus, formation of the bimolecular fluorescent complex was the result of specific interactions between LAP or LIP and HP1α, interaction that takes place in discrete and different subnuclear domains.
Figure 6. Visualization of the interaction between C/EBPβ and HP1α in living cells.
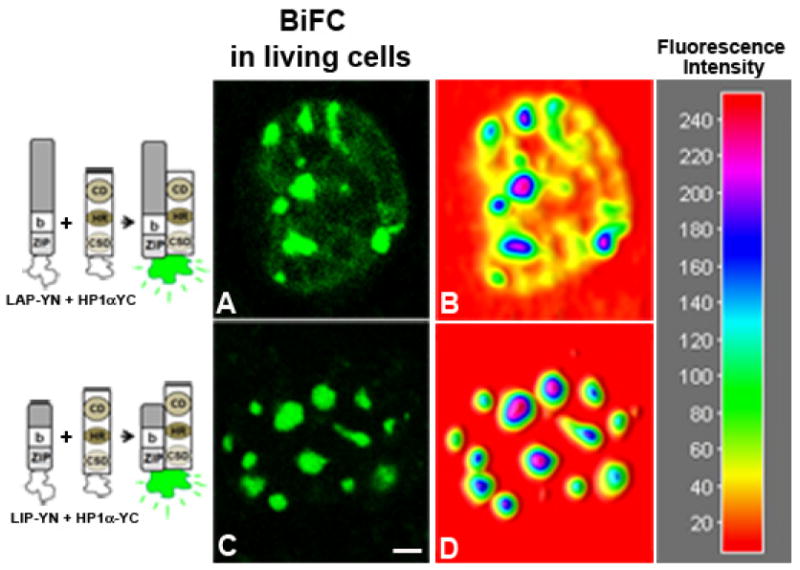
HP1α-YC was co-expressed with LAP-YN (panel A) or LIP-YN (panel C) in 3T3-L1 cells, and the fluorescence emission in living cells was imaged 24h after transfection. Images of representative nucleus that correspond to more than 95% of the fluorescent cells are shown. Fluorescence distribution was evaluated using Image-J program (panels B and D, respectively), and color code for fluorescence intensity is depicted on the right. Scale bar represents 2 μm.
The chromodomain and hinge region of HP-1α mediate the interaction with C/EBPβ
HP1α possesses three domains, the chromodomain (CD), hinge region (HR) and chromoshadow domain (CSD) (schematic representation in Fig. 7A). To analyze the domain of HP-1α required for its interaction with C/EBPβ, GST-pull down assays were performed with bacterially expressed full length GST-HP-1α, GST-HP1α-1-116 (corresponding to the CD and HR) or GST- HP1α-116-191 (corresponding to the CSD), and 35S-labeled LAP or LIP. Both, LAP (Fig. 7B, lane 2), and LIP (shown in Fig 5B, lane 5) interacted directly with HP1α full length, but not with GST alone (Fig. 5B, lane 1). LAP interacted with HP1α-1-116 (Fig. 7B, lane 3) but not with HP1α-116-191 (Fig. 7B, lane 4), suggesting that the chromodomain and/or hinge region are the domains of HP1α required for this interaction. The same results were obtained when 35S-labeled LIP was assayed (data not shown). To elucidate the precise domain of HP1α necessary for its interaction with C/EBPβ we performed BiFC assays using constructs that correspond to the different domains of HP1α as YC fusion proteins. As expected based on the results obtained in the GST-pull down experiments, LIP-YN interacted with HP1-CD/HR-YC, (Fig. 7C) and their interaction was detected in heterochromatic areas of intense TO-PRO staining (yellow signal in panel E that corresponds to the overlay of panels C and D). Similar results were obtained when LAP-YN was co-expressed with HP1-CD/HR-YC (data not shown). In addition, and as expected based on the results of the GST-pull down assay (Fig 7B, line 4), when LIP-YN (Fig. 7F) or LAP-YN (data not shown) was co-expressed with HP1-CSD-YC, no recovery of fluoresce was observed. LIP-YN, and HP1-CSD-YC were properly expressed as assessed by IIF (Fig. 7G and H respectively), and the fusion proteins were present in overlapping nuclear domains; however since they do not interact no fluorescence complementation was detected. Taken together these results indicate that the CD and/or the HR of HP1α are the domains that mediate the interaction of HP1α with the different C/EBPβ proteins. Deletion of the CSD, domain responsible for HP1 dimerization, does not abrogate HP1-C/EBPβ interaction indicating that dimerization of HP1α is not required for its interaction with LAP and LIP. Our results also provide evidence that BiFC can be used as a useful tool to determine the protein domains involved in protein-protein interactions in the cell milieu.
Figure 7. HP1α interacts through the chromodomain and hinge region with C/EBPβ.
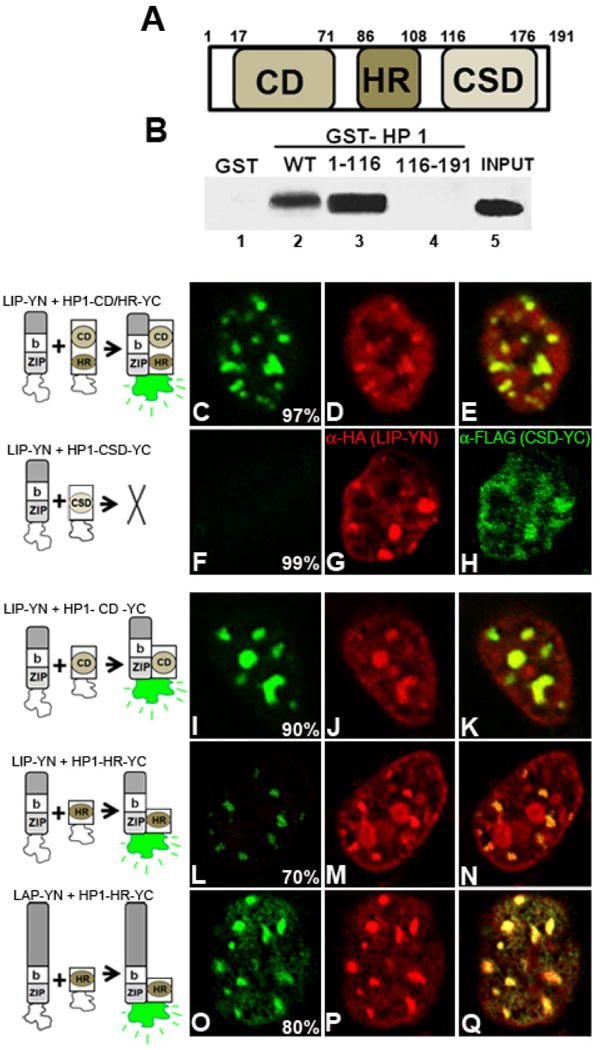
A- Schematic representation of HP1α: CD: chromodomain, HR: Hinge region, CSD: chromoshadow domain. B- GST alone, GST-HP1α full length (WT), GST- HP1α 1-116, or GST- HP1α 116-191 was expressed in E. coli, conjugated to glutathione-agarose beads, and incubated with [35S] methionine-labeled LAP, as described in Materials and Methods. Associated proteins were resolved by SDS-PAGE and analyzed by autoradiography. As control, 10% of the [35S] methionine-labeled LAP was applied to the gel (INPUT). Panels C to Q - Proteins indicated in front of each panel were co-expressed in 3T3-L1 cells, and fluorescence emission was imaged 24h after transfection in fixed cells. Nuclei were counterstained with TO-PRO. Percentage of cells with the pattern of BiFC is indicated in each panel. The images are representative of four independent experiments.
To investigate which domain of HP1α is mediating its interaction with C/EBPβ, LIP-YN or LAP-YN was expressed in combination with HP1-CD-YC. LIP-YN interacted with HP1-CD-YC in foci (Fig. 7I) that correspond to heterochromatin intensely stained with TO-PRO (Fig. 7J, and yellow signal in panel K). LAP-YN also interacted with HP1-CD-YC in heterochromatic areas (data not shown). LIP-YN and LAP-YN interacted with HP1-HR–YC, indicating that HP1α may interact through its CD or alternatively the HR with LAP and LIP. Interestingly, LIP-YN and HP1-HR–YC interacted exclusively in heterochromatic domains (Fig. 7L-M) in 70± 6 % of the cells in which fluorescent complementation was observed. In contrast, when LAP-YN was co-expressed with HP1-HR–YC the fluorescent signal was observed both in euchromatic as well as in heterochromatic nuclear domains (Fig 7O-Q) in 80 ± 4 % of the cells. Thus, these results suggest that HP1α may have the possibility to interact with C/EBPβ alternatively through its chromodomain and hinge domain. It is tempting to speculate that this may enable to HP1α to interact selectively with different subsets of LAP located in heterochromatic or euchromatic nuclear domains.
The basic region of C/EBPβ is required for its interaction with HP-1α
To determine the domain of C/EBPβ necessary for the interaction with HP1α, we engineered a series of constructs. First we tested bZIP-YN that corresponded only to the basic DNA binding domain and the leucine zipper (bZIP), domains common to LAP and LIP, fused to YN fragment of YFP. In 3T3-L1 cells co-expressing bZIP-YN and HP1α-YC (Fig. 8A) fluorescent signal was exclusively detected in foci that corresponded to pericentromeric heterochromatin intensely stained with DAPI (Fig. 8B, and overlay of images A-B in panel C). Next, to determine whether the leucine zipper (ZIP) was required for their interaction with HP1α, we used LAPΔZIP and LIPΔZIP that lack the leucine zipper. In cells expressing LAPΔZIP-YC and HP1α-YN (Fig. 8D), recovery of fluorescence was detected in foci that coincided with heterochromatin intensely stained with DAPI (panels E and G), as well as diffusely distributed throughout the nucleus suggesting that the leucine zipper does not mediate the interaction with HP1α and that dimerization of LAP is not required for its interaction with HP1α. Similar results were obtained when LIPΔZIP-YC was co-expressed with HP-1α-YN. Both LAP- and LIPΔZIP do not dimerize and consequently cannot bind to C/EBP consensus sites, thus we analyzed by IIF whether LAP- and LIPΔZIP exhibited the same subnuclear distribution as the corresponding full-length form. In cells co-expressing LAP- or LIPΔZIP and HP1α, both LAPΔZIP and LIPΔZIP were detected in euchromatic well as in heterochromatic domains co-localizing with HP1α (data not shown). The subnuclear distribution of LAP- and LIPΔZIP was the same as the full-length forms (shown in Fig. 5D and J). In contrast, when LAP- or LIPΔZIP was expressed alone, IIF analysis revealed a diffuse subnuclear distribution mostly excluded from pericentromeric heterochromatin (data not shown), suggesting that HP1α may recruit LAP or LIP monomers to heterochromatin. This recruitment is independent of the complementation of the amino- and carboxy-terminal fractions of YFP since co-expression of LAP- or LIPΔZIP-YN with GFP-HP1α showed the same results (data not shown). In order to test whether the basic region for DNA binding is responsible for C/EBPβ interaction with HP1α, we generated LIPΔbZIP-YN by deleting both the basic and the leucine zipper domains of LIP. Since the basic domain of C/EBPβ possesses the NLS (Williams, 1997 #2947), we included the SV40 NLS sequence for LIPΔbZIP-YN to ensure localization to the nucleus. When LIPΔbZIP-YN and HP1α-YC were co-expressed, no fluorescence complementation was observed (Fig. 8G) indicating that the basic region of C/EBPβ is the domain responsible for its interaction with HP1α. Indirect immunofluorescence showed that both LIPΔbZIP-YN (Fig. 8J) and HP1α-YC (Fig. 8K) were properly expressed. HP1α-YC exhibited the same subnuclear distribution as endogenous HP1α (Fig. 8K). In contrast, LIPΔbZIP-YN exhibited a diffuse subnuclear distribution, and was excluded from pericentromeric heterochromatin (Fig. 8J and L). Taken together these results demonstrate that LAP and LIP, isoforms of C/EBPβ, interact with HP1α through their basic region. Moreover, HP1α may interact with monomers or dimers of C/EBPβ since deletion of the leucine zipper that abrogates LAP and LIP dimerization does not prevent their interaction with HP1α.
Figure 8. The basic domain of C/EBPβ is required for the interaction with HP1α.
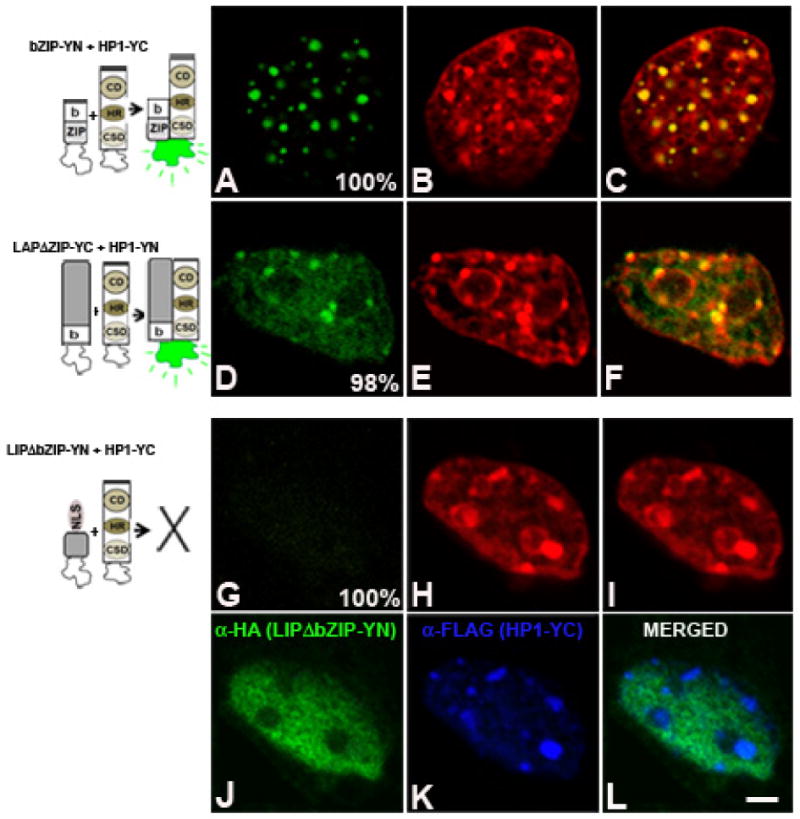
bZIP-YN (panels A to C), LAPΔZIP-YC (panels D to F) or LIPΔbZIP-YN (panels G to L) were co-expressed with HP1α-YN or -YC in 3T3-L1 cells, and fluorescence emission was imaged 24h after transfection in fixed cells stained with TO-PRO. The nuclear distribution of LIPΔbZIP-YN and HP1α-YC is shown by IIF (panels J and K, respectively). Percentage of cells with the pattern of BiFC is indicated in each panel. Images are representative of four independent experiments. Scale bar, 2 μm.
The nuclear pattern of phosphorylated C/EBPβ changes as adipogenesis progresses
Murine C/EBPβ is phosphorylated on Thr188 upon GH-, and insulin-dependent MAPK activation as well as upon induction of adipogenesis [10, 50]. Phosphorylation of Thr 188 of C/EBPβ is required for transcriptional activation [10, 51]. Thus, we examined the subnuclear distribution of C/EBPβ phosphorylated on Thr188 using IIF. In preadipocytes, a negligible signal with anti-phospho-C/EBPβ was observed (Fig. 9A). HP1α was mainly concentrated in heterochromatin (Fig. 9B). Western blot analysis revealed no band for total and phosphorylated LAP and LIP (Fig. 9M, lane 1). However, C/EBPβ phosphorylated on Thr 188 (P-C/EBPβ) is detected mainly in foci of intense staining as well as in minute speckles through the nucleus (Fig. 9D) within 4 hours after induction of adipocyte differentiation of 3T3-L1 cells. The intense P-C/EBPβ foci co-localized with HP1α (Fig. 9E and merged Fig. 9F). The signal detected with anti-phospho-C/EBPβ and antibodies that recognize total C/EBPβ render the same pattern (data not shown). Results obtained by IIF and confocal microscopy correlated with those obtained by Western blot analysis that showed increase in the level of expression as well as phosphorylation of LAP on Thr188 and LIP on Thr37 within 4 hours of induction of adipogenesis (Fig 9M, lane 2 vs. 1). By 24 hours after induction of adipogenesis, phosphorylated C/EBPβ is mainly distributed in minute speckles throughout the nucleus (Fig. 9G and Fig. 9J). The amount of P-C/EBPβ is decreased in pericentromeric heterochromatin and co-localization with HP-1α is reduced (Fig. 9G-I). However, C/EBPβ is still detected in pericentromeric heterochromatin as reveal by antibodies that recognize phosphorylated and unphosphorylated C/EBPβ (Fig. 9K). Thus, when adipocyte differentiation of 3T3-L1 cells is induced, the level of C/EBPβ phosphorylated on Thr188 increases, and transcriptionally active C/EBPβ co-localizes with HP1α. Since we found that HP1α interacts with C/EBPβ (Figs. 4 - 7), this observation implies that HP1α may modulate C/EBPβ transcriptional capacity, and in this way participates in the regulation of C/EBP target genes during adipogenesis.
Figure 9. C/EBPβ phosphorylated on Thr188 co-localizes with HP1α in pericentromeric heterochromatin upon induction of adipogenesis.
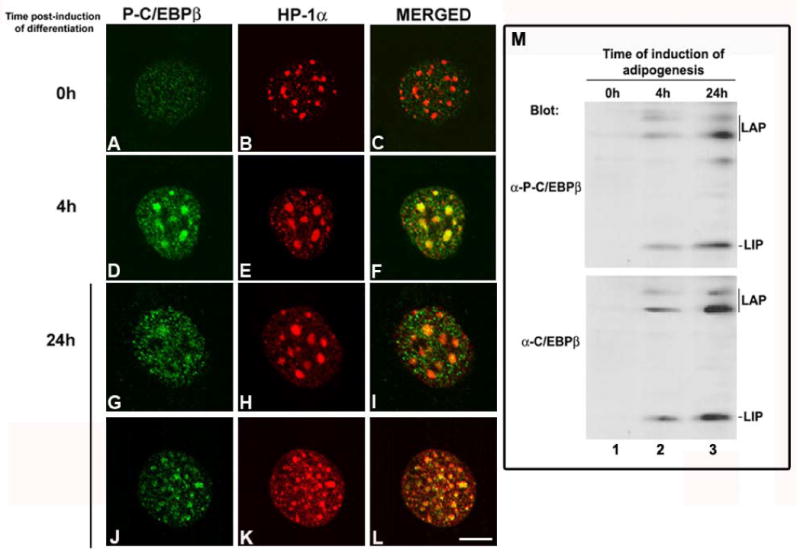
3T3-L1 cells were grown on coverslips, induced to differentiate in adipocytes for the indicated periods of time, and subjected to indirect immunofluorescence using anti-phospho-C/EBPβ and anti-HP1α. Merged of C/EBPβ and HP1α immunostaining is shown in panels C, F and I. Images are representative of three independent experiments. Scale bar, 5μm. J- 3T3-L1 preadipocytes were lysed at the indicated period of time after induction of adipocyte differentiation with MDI. The total lysates were subjected to SDS-PAGE and then immunoblotted with anti-phospho-C/EBPβ (1/500) or anti-C/EBPβ (1/1000). Results are representative of three independent experiments.
HP1α restrains LAP transcriptional activity
To investigate the functional importance of LAP and HP1α interaction, LAP was co-expressed with increasing amounts of HP1α in the presence of a luciferase reporter gene driven by four C/EBP consensus sites (C/EBP-Luc) (Fig 10A) or by the c-fos promoter (c-fos-Luc) (Fig. 10B) in 293T cells. Over-expression of LAP activated the C/EBP-Luc and c-fos-Luc promoter activity 7 and 16 fold over the basal levels, respectively, (Fig. 10 A and B). In contrast, the co-expression of increasing amounts of HP1α restrained LAP transcriptional capacity to activate the C/EBP- and c-fos-Luc promoter activities in a dose-dependent manner (Fig. 10A and B, respectively). The expression levels of LAP and HP1α were monitored by WB, and the level of LAP expression remained constant in all conditions, as shown in panel A. It has been demonstrated that p300 interacts and co-activates LAP-mediated c-fos promoter activation [11]. Here, we show that p300 increases LAP-mediated c-fos-Luc promoter activation, and p300 effect was progressively reduced in the presence of increasing amounts of HP1α (Fig. 10C). Taken together, these results suggest that HP1α interacts with LAP and this interaction inhibits LAP transcriptional capacity. Next, ChIP assays were performed to examine in vivo whether changes in C/EBPβ occupancy of target genes are accompanied by changes in HP1α. In preadipocytes, binding of C/EBPβ was detected in the c/ebpα promoter (Fig. 10D, day 0 - c/ebpα); however, no detectable level of C/EBPα was detected in preadipocytes, as assessed by WB and as previously shown [31]. Further, HP1α was detected bound to the c/ebpα promoter (Fig 10D, day 0 - c/ebpα) possibly inhibiting C/EBPβ capacity to induce C/EBPα expression. Importantly, when adipogenesis was induced the increase of C/EBPβ binding to the c/ebpα promoter was accompanied by a decreased binding of HP1α and a decreased 3MeK9H3 (Fig. 10D, day 2 - c/ebpα). In addition, the amount of active RNA-polymerase II phosphorylated in Ser2 associated with c/ebpα promoter increased (Fig. 10D, day 2 vs. 1 – c/ebpα). These changes in the c/ebpα promoter occupancy were accompanied by increased of C/EBPα protein level without changes in HP1α, as shown by WB. ChIP analysis of c-fos promoter also showed the presence of C/EBPβ, HP1α and 3MeK9H3 in 3T3-L1 preadipocytes (Fig 10D, day 0 – c-fos). Interestingly, two days after induction of adipocyte differentiation, a slight increase in C/EBPβ and active RNA-polymerase II binding was observed, but without changes in HP1α and 3MeK9H3 (Fig 10D, day 2 - c-fos). This was accompanied by no change in c-fos expression (data not shown). Taken together, these results show for the first time that HP1α occupies the promoters of C/EBPβ target genes to inhibit their transcription. When adipogenesis is induced, HP1α is released from C/EBPβ target genes such as c/ebpα, consistent with its transcription required for cell differentiation.
Figure 10. Expression of HP1α restrains LAP transcripcional activity.
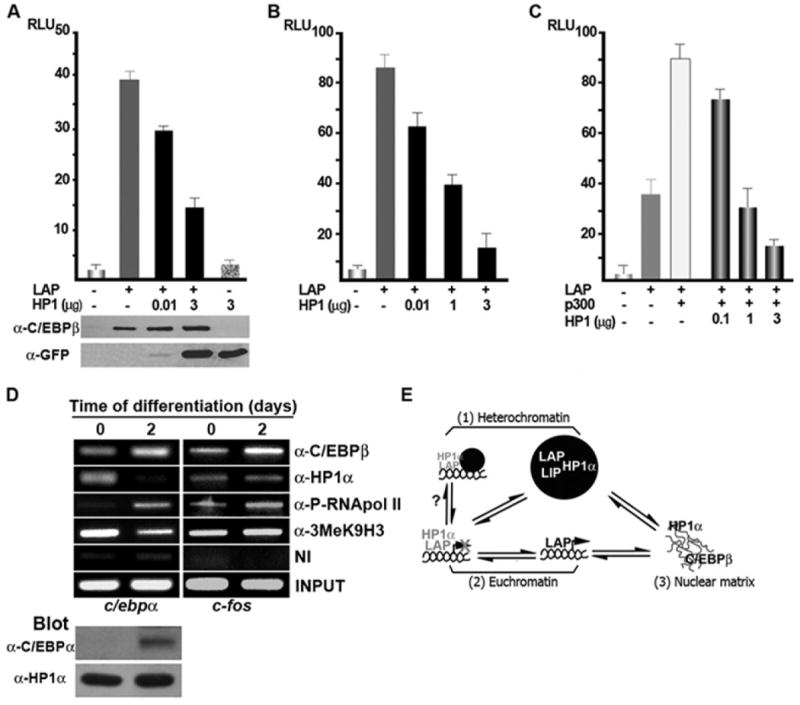
A- C 293T cells were transiently transfected with LAP (0.1μg, panels A and B, or 1ng panel C), p300 (0.5 μg), and the indicated amounts of GFP-HP-1α, along with C/EBP-Luc (panel A), c/fos-Luc (panels B and C), and RSV-β-galactosidase plasmids. After 48 h of transfection luciferase activity was measured and normalized to β-galactosidase activity. Each bar represents the mean ± S.E. for five independent experiments. The level of LAP and GFP-HP-1α expression was analyzed by Western blot. D- 3T3-L1 cells before or after 2 days of induction of differentiation were subjected to ChIP assays with α-C/EBPβ, α-HP1α, α- P-Ser2-RNApol II, α-3MeK9H3 antibodies. The precipitated DNA fragments were subjected to PCR analysis to test the presence of sequences corresponding to the c/ebpα or c-fos promoter respectively. Input material (1%) is shown for comparison. A representative of three independent experiments is shown. Protein expression level was assessed by WB with the indicated antibodies. E- Model for C/EBPβ- HP1a and their subnuclear compartmentalization. C/EBPβ is distributed in three different nuclear compartments: 1-Pericentromeric heterochromatin (●) where LAP and LIP homo- and heterodimers may interact with HP1α; 2- Euchromatin where LAP may interact with HP1α and this interaction restrains LAP transcriptional capacity or alternatively the C/EBPβ target gene be repositioned from a permissive environment to a repressive context to be silenced; and 3- Nuclear matrix where C/EBPβ does not colocalize with HP1α. The interplay of the equilibrium between different pools of C/EBPβ bound to chromatin or to the nuclear matrix, as well as its interaction with HP1α in different subnuclear domains, may play a key role in the regulation of C/EBP target genes when adipogenesis is induced as shown in this study for c/ebpα and c-fos genes.
Discussion
The structural and functional complexity of the eukaryotic cell nucleus is one of its striking features. During cellular differentiation, certain subsets of genes are activated while others are silenced generating a pattern of gene expression as cell fate decision is taken. These gene expression patterns are not only regulated at the level of expression, and post-translational modification of transcriptional regulators but also at the epigenetic level by changes in chromatin structure [43] and by dynamic changes in nuclear compartmentalization favoring either transcriptional activation or silencing [32, 33, 52]. The murine cell line 3T3-L1 can be induced to undergo adipogenesis in culture [53] and provides an ideal model system in which to study the dynamic changes in nuclear organization relative to the expression patterns of well-characterized genes. C/EBPβ is a transcription factor essential for the progression of adipogenesis that is rapidly up-regulated when 3T3-L1 preadipocytes are induced to differentiate [1, 54]. Here, we show that C/EBPβ associates with clusters of pericentromeric heterochromatin within 4 hours of induced adipogenesis in 3T3-L1 cells, through binding to consensus sites (Fig. 1). Binding coincides with the MAPK-dependent phosphorylation of C/EBPβ on Thr 188, a modification enabling C/EBPβ to properly bind to chromatin and activate transcription of target genes (Fig. 9) [10, 17, 50]. Since clustered pericentromeric heterochromatin is thought to form domains of transcriptional inactivation in mammalian nuclei [32], it was an unexpected finding that a transcriptional activator should adopt such a nuclear location. The structure and functions of centromeric and pericentromeric heterochromatin are very sensitive to long term exposure to histone deacetylase inhibitors, both in yeast and mammalian cells. Long term TSA treatment promotes hyperacetylation of histones and delocalization of HP1 proteins (Fig 2) [44]. Treatment with TSA blocks differentiation of myoblasts and 3T3-L1 preadipocytes [46, 55]. Here, we show that TSA treatment of 3T3-L1 cells induced to differentiate leads to disruption of chromatin structure revealed by decreased size of areas intensely stained with DAPI (Fig. 2) which may also contribute to the blockade of differentiation. Importantly, when 3T3-L1 cells are induced to differentiate in the presence of TSA both C/EBPβ and HP1α lose their proper subnuclear distribution. These results suggest that C/EBPβ may require intact chromatin structure to acquire its proper subnuclear distribution that will ultimately contribute to C/EBPβ function during adipocyte differentiation. The fact that C/EBPβ is not simply redistributed in the nucleus along with the disorganized heterochromatin as consequence of TSA treatment was revealed by its lost of binding to consensus sites present in satellite DNA. TSA has wider effects altering the network of interactions that C/EBPβ possesses with chromatin, the nucleoskeleton and nuclear factors that leads to C/EBPβ increased extractability by in situ extraction
Interestingly, we show for the first time that a fraction of C/EBPβ interacts with the nuclear matrix which is revealed by its nuclear retention when chromatin is digested by DNase I treatment. Different nuclear factors have been shown to be associated to the nuclear matrix, e.g. histone deacetylases, steroid hormone receptors, and oncogene proteins like c-myb [19]. It has been proposed that actively transcribing RNA polymerases are located on the nuclear matrix [56] near actively transcribed genes together with bound transcription factors, facilitating accessibility for binding to the promoter and regulating the expression of their target genes [57]. We show that a fraction of C/EBPβ associates with the nuclear matrix (Fig. 3), raising the possibility that it constitutes a pool in close contact with the transcriptional machinery. We also demonstrate for the first time that a fraction of HP1α is associated to the nuclear matrix and, interestingly, C/EBPβ and HP1α do not co-localize in this nuclear compartment (Fig. 3). The matrix-associated sub-fraction of C/EBPβ that did not co-localize with the remaining matrix-associated HP1α, may represent a potentially active sub-fraction that up-regulates tissue-specific genes that are activated early in adipogenesis such c/ebpα gene, a possibility that is under investigation. Further, the fraction of C/EBPβ associated to the nuclear matrix is also sensitive to TSA treatment suggesting that acetylation of C/EBPβ and possibly other nuclear factors may participate in the regulation of the equilibrium of C/EBPβ bound to chromatin and the nuclear matrix compartments.
C/EBPβ has alternative translation products, LAP and LIP, able to form homo- and heterodimers [15]. The lack of antibodies that can individually recognize them and the inability to individually target their expression by knockdown strategies (LAP and LIP are translated from the same mRNA molecule) imposed technical limitations to study the nuclear distribution of these different dimers, as well as their interaction with factors that might regulate their function. We overcame part of these limitations by the use of BiFC, a powerful strategy for visualizing the interactions occurring within protein complexes in living cells, thus enabling the investigation of protein behavior in their normal milieu [49, 58]. We show for the first time that different C/EBPβ dimers exhibit a differential subnuclear distribution (Fig. 4). LAP homodimers are present in heterochromatin in euchromatic nuclear domains (Fig. 10E). Since LAP has the capacity to activate the expression of genes localized in euchromatic domains, it is possible that LAP homodimers are retained in the heterochromatic compartment as a reservoir of LAP with little capacity to activate the transcription of target genes by spatial restriction from such genes (which are expected to be located in euchromatic regions of the nucleus) (Fig. 10E). In contrast, LIP homodimers which lack the N-terminal transactivation domain and inhibit transcription, localize exclusively in pericentromeric heterochromatin, possibly as a mechanism that maintains an inhibitor of transcription away from target genes (Fig. 10E). The bZIP construct that has only the basic region and the leucine zipper domain, also concentrates in pericentromeric heterochromatin, suggesting that exclusive localization of LIP in heterochromatin is independent of its N-terminal domain. Further, LAP-LIP heterodimers distribute similarly to LAP homodimers, raising the possibility that LAP may be “recruiting” LIP to euchromatic domains. LAP-LIP heterodimers are proposed to be less transcriptionally active than LAP homodimers [15], thus the control of the subnuclear distribution of C/EBPβ heterodimers constitutes an important step in the regulation of C/EBPβ target genes. Subcellular localization and more precisely subnuclear distribution of regulatory factors have functional relevance in the control of gene expression [59]. Nuclear receptors (NR) constitute a very well studied example of how changes in subcellular distribution upon hormonal signaling are integrated to regulate the expression of target genes. In this regard, binding of different agonists to NR such as the glucocorticoid receptor or the mineralocorticoid receptor has shown to determine a differential recruitment of specific cytoplasmic and nuclear factors leading to different nuclear pattern of NR distribution as well as a different capacity for being retained in the nucleus, events that have functional consequences at transcriptional level thus in the biological responses to steroid hormones [60, 61]. The differential distribution of LAP and LIP homo- and heterodimers in the nucleus may be regulated by posttranslational modifications and by C/EBPβ protein- protein interactions, and consequently playing a role in the control of C/EBP target genes, possibilities that are under current investigation.
In addition to distinct epigenetic modifications present in the histone tails, euchromatin and heterochromatin contain different non- histone components such as HP1. HP1α binds to methylated histone H3K9 stabilizing the compact structure of heterochromatin. However, the distribution of HP1 is not limited to heterochromatic and telomeric domains, but it is also present in euchromatic sites [62, 63]. Based on these observations it is proposed that HP1 may play an important role in regulating the expression within many different euchromatic regions. Here we show that the different forms of C/EBPβ, LAP and LIP, interact with HP1α and interestingly their interaction occurs in different nuclear domains, as we demonstrated in living cells by BiFC (Fig. 6). LAP interacts with HP1α in pericentromeric heterochromatin and euchromatic domains. In contrast, LIP interacts with HP1α exclusively in pericentromeric heterochromatin. These results markedly contrast with those obtained by IIF that simply showed that C/EBPβ co-localized with HP1α both in euchromatic (Fig. 2) and heterochromatic subnuclear domains without being able to specify which form of C/EBPβ is present in each compartment. The nuclear domains where LAP and LIP interact with HP1α coincide with the domains where the different dimers localize, reinforcing the notion of the importance of the subnuclear localization of nuclear factors for proper interaction and therefore function; however, from our results, it cannot be established whether in the cell nucleus their interaction is direct or as part of a heterocomplex. Further, HP1α interacts with the basic DNA binding domain common to LAP and LIP (Fig. 7), which raises the possibility that HP1α may modulate the capacity of C/EBPβ to bind to its consensus sites in different chromatin contexts, possibility that is under current investigation. HP1α interacts though the CD with LAP and LIP exclusively in heterochromatin. In contrast, HP1α interacts through the HR with LAP in heterochromatin and euchromatin raising the possibility that HP1α may interact through the CD or the HR with different pools of LAP. It has been reported that some factors may required more than one HP1 domain for their interaction. One of such example is the origin recognition complexes (ORCs) that require the integrity of both the CD and the CSD for interacting with HP1 [64]. C/EBPβ is the first transcription factor to which HP1α has been shown to interact alternatively through different domains, and this possibility may give plasticity to HP1α to form complexes with C/EBPβ in different nuclear domains. Since HP1α inhibits LAP transcriptional capacity (Fig. 10) the regulation of LAP-HP1α interaction in different subnuclear domains may play an important role in the regulation of C/EBP target genes. ChIP assays showed that C/EBPβ and HP1α are bound to c/ebpα promoter in 3T3-L1 preadipocytes when this gene is not expressed, reinforcing the notion of HP1a as a repressor of C/EBP target genes. Importantly, when adipocyte differentiation is induced binding of C/EBPβ increases and of HP1α decreases, and this change in occupancy of c/ebpα promoter is required for transcription to proceed. On the other hand, HP1α is detected bound to the c-fos promoter in preadipocytes as well as after induction of adipogenesis, possibly repressing the expression of c-fos gene. It has been previously shown that 243 cells, generated from primary embryonic fibroblasts from c-fos null mouse embryos, spontaneously differentiate in adipocytes [65], supporting the notion that c-fos gene needs to be kept repressed for preadipocytes to undergo differentiation.
Conclusions
we have presented evidence indicating that C/EBPβ is present in the nucleus in three different compartments: heterochromatin, euchromatin, and the nuclear matrix (Fig. 10E). Changes in the equilibrium among the different pools of C/EBPβ may account for its versatility in regulating different biological outcomes. LAP and LIP form homo- and heterodimers that are differentially distributed in the nucleus. This differential subnuclear distribution it is likely to restrict their interaction with HP1α in precise nuclear domains which ultimately may have different functional consequences. C/EBPβ is detected bound to the promoter of target genes in the absence of active transcription as shown here and previously [11]. Since HP1α interacts with LAP in euchromatic domains and inhibits its transcriptional capacity, the presence of LAP-HP1α complex may keep a euchromatic gene repressed until the appropriate signal releases HP1α to allow transcription, as shown here for c/ebpα gene. It was previously demonstrated that during B cell development Ikaros protein localizes to centromeric heterochromatin and that the proximity of lymphoid-associated genes to Ikaros complexes inversely correlates with its transcriptional status [32]. Thus, it is also possible that HP1α may function as a“recruiter” of C/EBPβ target genes to a more repressive nuclear domain such as pericentromeric heterochromatin, as a mechanism to silence genes when adipogenesis is induced. These scenarios are not mutually exclusive, and are under current investigation. Intriguingly, a fraction of both C/EBPβ and HP1α is located in the nuclear matrix; however, they do not interact in this nuclear compartment. It is possible that the fraction of C/EBPβ that does not interact with HP1α and is present in the nuclear matrix may be in contact with the transcriptional machinery; thereby forming a pool of C/EBPβ engaged in active transcription. C/EBPβ and HP1α are nuclear factors that possess differential capacity to interact in different compartments of the cell nucleus and this delicate equilibrium ultimately plays a key role in the regulation of C/EBP target genes required for adipocyte differentiation.
Supplementary Material
Acknowledgments
We thank Dr. P. Murphy (Univ. of Seattle, WA) for critical reading of the manuscript, and Drs. N. Dillon, A Dejean, J. Seeler, and T. Misteli for kindly providing us with plasmids. This work was supported by grants to G.P.P. from Agencia Nacional de Promoción Científica y Tecnológica (PICT 26495, PICT 02109, and PICT 00640), by a Fogarty International Research Collaboration Award R03TW008143-01A1 to J.S. and G.P.P., and NIH grant DK51563 to O.A.M. L.P.P., M.A.D. and N.C. were recipients of CONICET doctoral fellowships, and SS is a recipient of a CONICET postdoctoral fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7:885–96. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 2.MacDougald OA, Lane MD. Transcriptional regulation of gene expression during adipocyte differentiation. Ann Rev Biochem. 1995;64:345–373. doi: 10.1146/annurev.bi.64.070195.002021. [DOI] [PubMed] [Google Scholar]
- 3.Lane MD, Tang QQ, Jiang MS. Role of the CCAAT enhancer binding proteins (C/EBPs) in adipocyte differentiation. Biochem Biophys Res Comm. 1999;266:677–683. doi: 10.1006/bbrc.1999.1885. [DOI] [PubMed] [Google Scholar]
- 4.Diehl AM, Michaelson P, Yang SQ. Selective induction of CCAAT/enhancer binding protein isoforms occurs during rat liver development. Gastroenterology. 1994;106:1625–1637. doi: 10.1016/0016-5085(94)90420-0. [DOI] [PubMed] [Google Scholar]
- 5.Diehl AM. Roles of CCAAT/enhancer-binding proteins in regulation of liver regenerative growth. J Biol Chem. 1998;273:30843–30846. doi: 10.1074/jbc.273.47.30843. [DOI] [PubMed] [Google Scholar]
- 6.Scott LM, Civin CI, Rorth P, Friedman AD. A novel temporal expression pattern of three C/EBP family members in differentiating myelomonocytic cells. Blood. 1992;80:1725–1735. [PubMed] [Google Scholar]
- 7.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–76. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 8.Begay V, Smink J, Leutz A. Essential requirement of CCAAT/enhancer binding proteins in embryogenesis. Mol Cell Biol. 2004;24:9744–51. doi: 10.1128/MCB.24.22.9744-9751.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piwien-Pilipuk G, Van Mater D, Ross SE, MacDougald OA, Schwartz J. Growth hormone regulates phosphorylation and function of C/EBP beta by modulating Akt and glycogen synthase kinase-3. J Biol Chem. 2001;276:19664–19671. doi: 10.1074/jbc.M010193200. [DOI] [PubMed] [Google Scholar]
- 10.Piwien-Pilipuk G, MacDougald OA, Schwartz J. Dual regulation of phosphorylation and dephosphorylation of C/EBPb modulate its transcriptional activation and DNA binding in response to growth hormone. J Biol Chem. 2002;277:44557–44565. doi: 10.1074/jbc.M206886200. [DOI] [PubMed] [Google Scholar]
- 11.Cui TX, Piwien-Pilipuk G, Huo JS, Kaplani J, Kwok R, Schwartz J. Endogenous CCAAT/enhancer binding protein beta and p300 are both regulated by growth hormone to mediate transcriptional activation. Mol Endocrinol. 2005;19:2175–86. doi: 10.1210/me.2004-0502. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka T, Yoshida N, Kishimoto T, Akira S. Defective adipocyte differentiation in mice lacking the C/EBPb and/or C/EBPd gene. EMBO J. 1997;16:7432–7443. doi: 10.1093/emboj/16.24.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang QQ, Otto TC, Lane MD. CCAAT/enhancer-binding protein beta is required for mitotic clonal expansion during adipogenesis. Proc Nat Acad Sci USA. 2003;100:850–855. doi: 10.1073/pnas.0337434100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, Spiegelman BM. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–8. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Descombes P, Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991;67:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- 16.Piwien Pilipuk G, Galigniana MD, Schwartz J. Subnuclear localization of C/EBP beta is regulated by growth hormone and dependent on MAPK. J Biol Chem. 2003;278:35668–77. doi: 10.1074/jbc.M305182200. [DOI] [PubMed] [Google Scholar]
- 17.Tang QQ, Lane MD. Activation and centromeric localization of CCAAT/enhancer-binding proteins during the mitotic clonal expansion of adipocyte differentiation. Genes & Devel. 1999;13:2231–2241. doi: 10.1101/gad.13.17.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smetana K, Steele WJ, Busch H. A nuclear ribonucleoprotein network. Experimental Cell Research. 1963;31:198–201. [Google Scholar]
- 19.Nickerson J. Experimental observations of a nuclear matrix. J Cell Sci. 2001;114:463–74. doi: 10.1242/jcs.114.3.463. [DOI] [PubMed] [Google Scholar]
- 20.Heitz E. Das heterochromatin der Moose. Jahrb Wiss Botanik. 1928;69:762–818. [Google Scholar]
- 21.Dillon N, Festenstein R. Unravelling heterochromatin: competition between positive and negative factors regulates accessibility. Trends Genet. 2002;18:252–258. doi: 10.1016/s0168-9525(02)02648-3. [DOI] [PubMed] [Google Scholar]
- 22.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 23.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–4. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 24.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–20. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 25.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–3. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 26.Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299:721–5. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- 27.Hiragami K, Festenstein R. Heterochromatin protein 1: a pervasive controlling influence. Cell Mol Life Sci. 2005;62:2711–26. doi: 10.1007/s00018-005-5287-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eissenberg JC, James TC, Foster-Hartnett DM, Hartnett T, Ngan V, Elgin SC. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1990;87:9923–7. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Festenstein R, Sharghi-Namini S, Fox M, Roderick K, Tolaini M, Norton T, Saveliev A, Kioussis D, Singh P. Heterochromatin protein 1 modifies mammalian PEV in a dose- and chromosomal-context-dependent manner. Nat Genet. 1999;23:457–61. doi: 10.1038/70579. [DOI] [PubMed] [Google Scholar]
- 30.Eissenberg JC, Hartnett T. A heat shock-activated cDNA rescues the recessive lethality of mutations in the heterochromatin-associated protein HP1 of Drosophila melanogaster. Mol Gen Genet. 1993;240:333–8. doi: 10.1007/BF00280383. [DOI] [PubMed] [Google Scholar]
- 31.Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, MacDougald OA. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- 32.Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 33.Brown KE, Baxter J, Graf D, Merkenschlager M, Fisher AG. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol Cell. 1999;3:207–217. doi: 10.1016/s1097-2765(00)80311-1. [DOI] [PubMed] [Google Scholar]
- 34.Fey EG, Krochmalnic G, Penman S. The nonchromatin substructures of the nucleus: the ribonucleoprotein (RNP)-containing and RNP-depleted matrices analyzed by sequential fractionation and resinless section electron microscopy. J Cell Biol. 1986;102:1654–65. doi: 10.1083/jcb.102.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stephanova E, Russanova V, Chentsov Y, Pashev I. Mouse centromeric heterochromatin: isolation and some characteristics. Exp Cell Res. 1988;179:545–553. doi: 10.1016/0014-4827(88)90292-3. [DOI] [PubMed] [Google Scholar]
- 36.Liao J, Piwien-Pilipuk G, Ross SE, Hodge CL, Sealy L, MacDougald OA, Schwartz J. CCAAT/Enhancer-binding protein beta (C/EBP beta) and C/EBPd contribute to growth homone-regulated transcription of c-fos. J Biol Chem. 1999;274:31597–31604. doi: 10.1074/jbc.274.44.31597. [DOI] [PubMed] [Google Scholar]
- 37.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–98. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 38.Grinberg AV, Hu CD, Kerppola TK. Visualization of Myc/Max/Mad family dimers and the competition for dimerization in living cells. Mol Cell Biol. 2004;24:4294–308. doi: 10.1128/MCB.24.10.4294-4308.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aagaard L, Schmid M, Warburton P, Jenuwein T. Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres. J Cell Sci. 2000;113(Pt 5):817–29. doi: 10.1242/jcs.113.5.817. [DOI] [PubMed] [Google Scholar]
- 40.Quinta HR, Maschi D, Gomez-Sanchez C, Piwien-Pilipuk G, Galigniana MD. Subcellular rearrangement of hsp90-binding immunophilins accompanies neuronal differentiation and neurite outgrowth. J Neurochem. doi: 10.1111/j.1471-4159.2010.06970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guenatri M, Bailly D, Maison C, Almouzni G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J Cell Biol. 2004;166:493–505. doi: 10.1083/jcb.200403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henikoff S. Heterochromatin function in complex genomes. Biochem Biophys Acta. 2000;1470:1–8. doi: 10.1016/s0304-419x(99)00034-7. [DOI] [PubMed] [Google Scholar]
- 43.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 44.Taddei A, Maison C, Roche D, Almouzni G. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat Cell Biol. 2001;3:114–20. doi: 10.1038/35055010. [DOI] [PubMed] [Google Scholar]
- 45.Bartova E, Pachernik J, Harnicarova A, Kovarik A, Kovarikova M, Hofmanova J, Skalnikova M, Kozubek M, Kozubek S. Nuclear levels and patterns of histone H3 modification and HP1 proteins after inhibition of histone deacetylases. J Cell Sci. 2005;118:5035–46. doi: 10.1242/jcs.02621. [DOI] [PubMed] [Google Scholar]
- 46.Lagace DC, Nachtigal MW. Inhibition of histone deacetylase activity by valproic acid blocks adipogenesis. J Biol Chem. 2004;279:18851–60. doi: 10.1074/jbc.M312795200. [DOI] [PubMed] [Google Scholar]
- 47.Todorov IT, Attaran A, Kearsey SE. BM28, a human member of the MCM2-3-5 family, is displaced from chromatin during DNA replication. J Cell Biol. 1995;129:1433–45. doi: 10.1083/jcb.129.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muchardt C, Guilleme M, Seeler JS, Trouche D, Dejean A, Yaniv M. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO Rep. 2002;3:975–81. doi: 10.1093/embo-reports/kvf194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kerppola TK. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu Rev Biophys. 2008;37:465–87. doi: 10.1146/annurev.biophys.37.032807.125842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park BH, Qiang L, Farmer SR. Phosphorylation of C/EBPbeta at a consensus extracellular signal-regulated kinase/glycogen synthase kinase 3 site is required for the induction of adiponectin gene expression during the differentiation of mouse fibroblasts into adipocytes. Mol Cell Biol. 2004;24:8671–80. doi: 10.1128/MCB.24.19.8671-8680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakajima T, Kinoshita S, Sasagawa T, Sasaki K, Naruto M, Kishimoto T, Akira S. Phosphorylation at threonine-235 by a ras-dependent mitogen-activated protein kinase cascade is essential for transcription factor NF-IL6. Proc Natl Acad Sci USA. 1993;90:2207–2211. doi: 10.1073/pnas.90.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–7. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 53.Green H, Kehinde O. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell. 1975;5:19–27. doi: 10.1016/0092-8674(75)90087-2. [DOI] [PubMed] [Google Scholar]
- 54.Yeh WC, Cao Z, Classon M, McKnight SL. Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev. 1995;9:168–181. doi: 10.1101/gad.9.2.168. [DOI] [PubMed] [Google Scholar]
- 55.Iezzi S, Cossu G, Nervi C, Sartorelli V, Puri PL. Stage-specific modulation of skeletal myogenesis by inhibitors of nuclear deacetylases. Proc Natl Acad Sci U S A. 2002;99:7757–62. doi: 10.1073/pnas.112218599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cook PR. The organization of replication and transcription. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- 57.Sutherland H, Bickmore WA. Transcription factories: gene expression in unions? Nat Rev Genet. 2009;10:457–66. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- 58.Kerppola TK. Visualization of molecular interactions by fluorescence complementation. Nat Rev Mol Cell Biol. 2006;7:449–56. doi: 10.1038/nrm1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carmo-Fonseca M. The contribution of nuclear compartmentalization to gene regulation. Cell. 2002;108:513–521. doi: 10.1016/s0092-8674(02)00650-5. [DOI] [PubMed] [Google Scholar]
- 60.Vicent GP, Pecci A, Ghini A, Piwien-Pilipuk G, Galigniana MD. Differences in nuclear retention characteristics of agonist-activated glucocorticoid receptor may determine specific responses. Exp Cell Res. 2002;276:142–154. doi: 10.1006/excr.2002.5532. [DOI] [PubMed] [Google Scholar]
- 61.Gallo LI, Ghini AA, Piwien Pilipuk G, Galigniana MD. Differential recruitment of tetratricorpeptide repeat domain immunophilins to the mineralocorticoid receptor influences both heat-shock protein 90-dependent retrotransport and hormone-dependent transcriptional activity. Biochemistry. 2007;46:14044–57. doi: 10.1021/bi701372c. [DOI] [PubMed] [Google Scholar]
- 62.Piacentini L, Fanti L, Berloco M, Perrini B, Pimpinelli S. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J Cell Biol. 2003;161:707–14. doi: 10.1083/jcb.200303012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fanti L, Berloco M, Piacentini L, Pimpinelli S. Chromosomal distribution of heterochromatin protein 1 (HP1) in Drosophila: a cytological map of euchromatic HP1 binding sites. Genetica. 2003;117:135–47. doi: 10.1023/a:1022971407290. [DOI] [PubMed] [Google Scholar]
- 64.Pak DT, Pflumm M, Chesnokov I, Huang DW, Kellum R, Marr J, Romanowski P, Botchan MR. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell. 1997;91:311–23. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- 65.Bennett CN, Hodge CL, MacDougald OA, Schwartz J. Role of Wnt10b and C/EBP alpha in spontaneous adipogenesis of 243 cells. Biochem Biophys Res Commun. 2003;302:12–16. doi: 10.1016/s0006-291x(03)00092-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


