Abstract
EF4, though similar structurally to the translocase EF-G, promotes back translocation of tRNAs on the ribosome, and is important for bacterial growth under certain conditions. Here, using a coordinated set of in vitro kinetic measures, including changes in the puromycin reactivity of peptidyl tRNA and in the fluorescence of labeled tRNAs and mRNA, we elucidate the kinetic mechanism of EF4-catalyzed back translocation and determine the effects of the translocation inhibitors spectinomycin and viomycin on the process. EF4-dependent back translocation proceeds from post-translocation complex (POST) to pre-translocation complex (PRE) via a four-step kinetic scheme, i.e., POST → I1 → I2 → I3 → PRE, that is not the simple reverse of translocation. During back translocation, movements of the tRNA core regions and of mRNA are closely coupled to one another, but are sometimes decoupled from movement of the 3′-end of peptidyl–tRNA. EF4 may be thought of as performing an interrupted catalysis of back translocation, stopping at the formation of I3 rather than catalyzing the complete process of back translocation culminating in PRE complex formation. The delay in polypeptide elongation resulting from transient accumulation of I3 is likely to be important for optimizing functional protein biosynthesis.
Keywords: EF4(LepA), back translocation, kinetic mechanism, fluorescence, puromycin
INTRODUCTION
Polypeptide elongation, the central phase of ribosome activity, is driven by two elongation factors, EF-Tu and EF-G. EF-Tu catalyzes aminoacyl-tRNA binding to the ribosome in response to a cognate codon in the A site. Following peptide bond formation, EF-G catalyzes translocation of the tRNAs in a pre-translocation (PRE) complex from the A and P to the P and E sites, respectively, forming a post-translocation (POST) complex. Both of these factors are GTPases and are present in all cells and in organelles such as mitochondria and chloroplasts. In addition to these universal elongation factors some non-canonical factors are known, including EF4 (LepA), a highly-conserved GTPase1, which is present in bacteria, mitochondria and chloroplasts but not in archaea or in the cytoplasm of eukaryotes2.
There has been a recent surge of interest in EF4, stemming largely from the recent demonstration of its ability to catalyze back translocation2, a process which occurs spontaneously, albeit quite slowly, following EF-G dissociation from the ribosome3,4. EF4 has been shown to have a conspicuous similarity to the translocation factor EF-G, since four out of the five EF-G domains - I, II, III, and V but not IV nor the G’ subdomain of I - are also found in EF4. EF4 also has a unique C-terminal domain with a new fold5. Domain IV of EF-G interacts with the A-site decoding center and is thought to prevent back translocation6. The structure of the PRE complex resulting from prolonged incubation of POST complex with EF4, has also been determined7. In this complex, denoted PRE(L), the 3′-end of peptidyl-tRNA occupies a position intermediate between those found for the classic A and A/T sites. Elucidation of the in vivo role of EF4 is also being pursued actively. Although ΔEF4 cells grown in rich medium have long been known to have no phenotype8, Nierhaus and co-workers (K. Nierhaus, private communication) have recently demonstrated that certain stress conditions, including high salt, cause a ΔEF4 strain to be overgrown by wild-type bacterial cells. In addition, Shoji et al.9 have shown a ΔEF4 E. coli strain to be hypersensitive to potassium tellurite and to penicillin.
Despite these recent studies, little is known about the detailed mechanism by which EF4 catalyzes back translocation. Here we report a coordinated set of in vitro kinetic measures showing that back translocation in the presence of EF4 proceeds from POST complex to PRE complex via at least three intermediates: i.e., POST → I1 → I2 → I3 → PRE(L). We show that I3 accumulates transiently in the presence of EF4, and that EF4 displays an “interrupted catalysis” in not catalyzing I3 to PRE(L) conversion. Our results further indicate that in various steps of the back translocation process, movement of the 3′-end of peptidyl–tRNA is decoupled from the movements of the rest of the core regions of the ribosome-bound tRNAs, as well as of mRNA, and that the 3′-end of peptidyl-tRNA has a position within the I3 complex that differs from its position within the PRE(L) complex. These results reinforce the notion that EF4 can transiently interrupt normal polypeptide elongation.
RESULTS
EF4 catalysis of back translocation measured by change in reactivity of peptidyl-tRNA toward puromycin
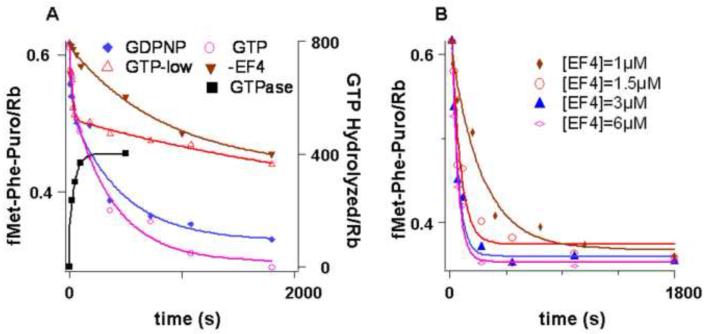
fMetPhe-tRNAPhe reacts with puromycin to form fMetPhe-puromycin at a rate which is 103- 104-fold higher when it is bound in the P-site following translocation in the POST complex than when it is bound in the A-site prior to translocation in the PRE complex10 - 13, permitting fMetPhe-tRNAPhe reactivity toward puromycin to be used as a measure of back translocation2. For the experiments shown in Figure 1, a POST complex was isolated by ultracentrifugation through a sucrose cushion, a procedure that removes all of EF-G, as shown by SDS-PAGE analysis, and ~60% of E-site bound tRNAfMet, while retaining P-site bound fMetPhe-tRNAPhe. The isolated POST complex was mixed with tRNAfMet, in the presence or absence of EF4 and either GTP or GDPNP and, at various times after mixing, was reacted with 1 mM puromycin for 15 s. Under these conditions, fMetPhe-tRNAPhe in the P-site is fully reacted (t1/2, 0.5 s), while fMetPhe-tRNAPhe in the A-site hardly reacts at all (t1/2, > 2000 s). The results presented in Figure 1A clearly establish that both EF4•GTP and EF4•GDPNP strongly catalyze a back translocation reaction that results in a decreased reactivity of fMetPhe-tRNAPhe toward puromycin. Catalysis thus depends on EF4•GTP binding, rather than on GTP hydrolysis, in accord with previous results2. Importantly, the amount of fMetPhe-puromycin formed as the EF4-catalyzed reaction reaches completion does not fall to zero, as would be expected for complete movement of fMetPhe-tRNAPhe from the P- to A-site, but rather to approximately half of the amount formed prior to EF4 addition, a point we return to below. A lesser drop in fMetPhe-puromycin formation is seen when the reaction is carried out in 0.5 mM rather than 2.0 mM GTP. This is directly attributable to the high rate of GTP turnover catalyzed by EF4 bound to the ribosome (Figure 1A)2, resulting in a depletion of GTP prior to completion of the EF4•GTP catalyzed process.
Figure 1. Reactivity of peptidyl-tRNA toward puromycin and EF4-dependent GTPase.
A. Isolated POST complex (0.1 μM) containing fMetPhe-tRNAPhe in the P-site and tRNAfMet in the E–site was incubated for the times indicated with a solution containing tRNAfMet (0.15 μM) and either GTP or GDPNP in the presence or absence of EF4 (3 μM when present) at 25 °C. For fMetPhe-puromycin formation, the incubation period was followed by reaction for 15 s with 1 mM puromycin. ( ) -GTP, -EF4; (
) -GTP, -EF4; ( ) GTP, 2 mM, + EF4; (
) GTP, 2 mM, + EF4; ( ) GTP, 0.5 mM, + EF4; (
) GTP, 0.5 mM, + EF4; ( ) GDPNP, 0.5 mM + EF4. For GTP hydrolysis (■), the incubations were carried out with EF4 and γ-32P GTP (50 μM) followed directly by quenching. Concentrations are after mixing. B. fMetPhe-puromycin formation was carried out as in A. with varying concentrations of EF4, except that 2 mM puromycin was employed.
) GDPNP, 0.5 mM + EF4. For GTP hydrolysis (■), the incubations were carried out with EF4 and γ-32P GTP (50 μM) followed directly by quenching. Concentrations are after mixing. B. fMetPhe-puromycin formation was carried out as in A. with varying concentrations of EF4, except that 2 mM puromycin was employed.
The remainder of the experiments reported herein employ EF4•GDPNP as the catalyst, in order to avoid problems associated with GTP depletion. The experiments shown in Figure 1B demonstrate that the apparent rate of the EF4•GDPNP catalyzed reaction reaches saturation by 3 μM EF4, which is the concentration used for most of the experiments described below.
EF4 catalysis of back translocation measured by change in fluorescence of fMetPhe-tRNAPhe(prf) or tRNAfMet(prf)
Pan et al.13 showed that EF-G•GTP dependent translocation could be monitored by changes in the fluorescence of either fMetPhe-tRNAPhe(prf) or tRNAfMet(prf), in which each tRNA is labeled with proflavin within the dihydro(U) loop of the core region, and used such measurements in investigating the effects of the antibiotics viomycin (Vio) and spectinomycin (Spc) on translocation. We reasoned that reverse spectral changes might be found during EF4-catalyzed back translocation. Such is indeed the case.
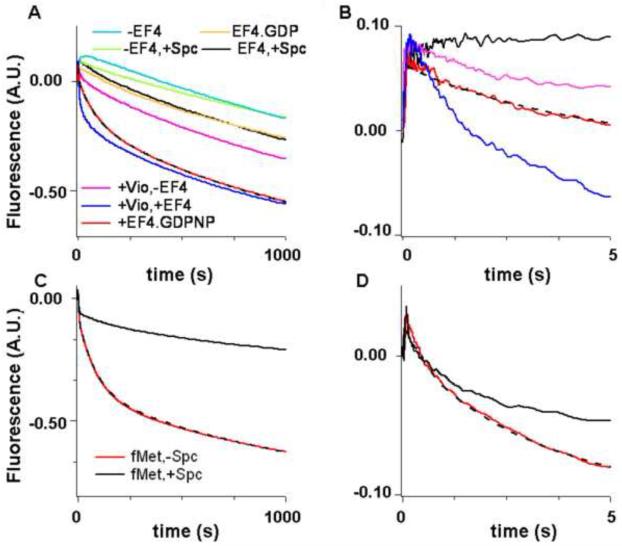
Rapid mixing of the isolated POST complex containing fMetPhe-tRNAPhe(prf) with tRNAfMet in the absence and presence of EF4•GDPNP in a stopped flow spectrophotometer gave the results shown in Figure 2A,B. In the absence of EF4•GDPNP the fluorescence change proceeds in two phases. A small increase in fluorescence intensity in the first phase (kapp1, 0.007 s−1) is followed by a large decrease in fluorescence intensity in the second. The rate constant for this second phase (kapp2, 0.00057s−1) is consistent with rate constants for uncatalyzed back translocation reported earlier3,4.
Figure 2. EF4-dependent back translocation measured by change in the fluorescence of fMetPhe-tRNAPhe(prf) and tRNAfMet(prf).
Fluorescence changes measured in a stopped-flow spectrofluorimeter over different time scales. The dashed lines through the red traces are the results of fits to Scheme 1 (Figure 5). A and B. fMetPhe-tRNAPhe(prf) (P-site) fluorescence change over different time scales. POST complex (0.1 μM) containing fMetPhe-tRNAPhe(Prf) in the P-site and tRNAfMet in the E–site was rapidly mixed with 0.15 μM tRNAfMet and either 3 μM EF4•GDPNP (red trace), or 3 μM EF4•GDP (orange trace); or 3 μM EF4•GDPNP and either 1 mM Spc (black trace) or 1 mM Vio (dark blue trace); or just 1 mM Spc alone (light green trace), or 1 mM Vio alone (pink trace), or GDPNP alone (teal trace). C and D. tRNAfMet(prf) (E-site) fluorescence change over different time scales. POST complexes (0.1 μM) containing fMetPhe-tRNAPhe in the P-site and tRNAfMet(prf) in the E–site were rapidly mixed with 0.15 μM tRNAfMet(prf) and 3 μM EF4•GDPNP in the absence (red) or presence (black) of 1 mM Spc. The small but rapid rise in fluorescence occurring immediately after mixing that is evident in parts B and D is likely due to EF4 binding (data not shown). It is complete within 0.15 s and may be considered as occurring instantaneously with respect to the subsequent reaction phases described in the text.
Fluorescence change in the presence of 3 μM EF4•GDPNP proceeds in three phases: an initial small decrease in fluorescence intensity is followed by two major decreases. The apparent rate constants for these three phases, kapp1, kapp2, and kapp3 are 0.60 s−1, 0.014 s−1, and 0.00082 s−1, respectively. Similar values were obtained at 1 μM EF4 (0.51 s−1, 0.010 s−1, 0.00072 s−1, respectively), consistent with a Kd for EF4•GDPNP binding to the ribosome below 1 μM, as earlier reported7. Interestingly, the last phase proceeds with an apparent rate constant that is only slightly higher than that found for the second phase of reaction in the absence of EF4. Replacement of EF4•GDPNP with EF4•GDP gives results similar to those obtained in the absence of added EF4 (Figure 2A,B, Table I), consistent with the interruption of EF4 catalysis that results from GTP depletion (Figure 1A).
Table I.
Fluorescence changes [POST ➜ PRE or PRE(L)]a
| Expt | EF4 | GXP | Fluorophore | Anti- biotic |
kapp1, s−1 | Relative ΔF1b |
kapp2, s−1 | Relative ΔF2b |
kapp3, s−1 | Relative ΔF3b |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | + | GDPNP | tRNAPhe(prf) | − | 0.60 ± 0.04 |
−0.036 ±0.006 |
0.014 ± 0.002 |
−0.26 ±0.04 |
8.2 ± 0.3 × 10−4 |
−(1.00) |
| 2c | − | GDPNP | tRNAPhe(prf) | − | 0.007 ± 0.001 |
+0.035± 0.008 |
5.7 ± 0.3 × 10− |
−0.77 ± 0.08 | ||
| 3c | + | GDP | tRNAPhe(prf) | − | 0.009 ± 0.001 |
−0.029 ±0.005 |
8.4 ± 0.6 × 10−4 |
−0.69 ± 0.08 | ||
| 4c | + | GDPNP | tRNAPhe(prf) | Spc, 1 mM |
0.003± 0.0003 |
−0.046 ±0.007 |
6.3 ± 0.4 × 10−4 |
−0.73 ± 0.07 | ||
| 5c | − | GDPNP | tRNAPhe(prf) | Spc, 1 mM |
0.003± 0.0003 |
−0.059 ±0.012 |
5.2 ± 0.6 × 10−4 |
−0.91 ± 0.12 | ||
| 6 | + | GDPNP | tRNAPhe(prf) | Vio, 1mM |
1.22 ± 0.08 |
−0.13± 0.03 |
0.034 ± 0.001 |
−0.17 ±0.06 |
7.5 ± 0.3 × 10−4 |
−0.98 ± 0.10 |
| 7 | − | GDPNP | tRNAPhe(prf) | Vio, 1mM |
1.37 ± 0.11 |
−0.034 ± 0.003 |
0.012 ± 0.001 |
−0.10 ±0.02 |
8.0 ± 0.2 × 10−4 |
−0.87 ± 0.08 |
| 8 | + | GDPNP | tRNAfMet(prf) | − | 0.54 ± 0.01 |
−0.077 ±0.011 |
0.0136 ± 0.003 |
−0.35 ±0.05 |
9.7 ± 0.7 × 10−4 |
−(1.00) |
| 9 | + | GDPNP | tRNAfMet(prf) | Spc, 1 mM |
0.65 ± 0.05 |
−0.08 ±0.02 |
0.005 ± 0.001 |
−0.07 ±0.02 |
5.7 ± 0.8 × 10−4 |
−0.71 ± 0.08 |
| 10 | + | GDPNP | Flu-mRNA | − | 0.062 ± 0.008 |
−0.014 ±0.002 |
0.018 ±0.004 |
−0.29 ±0.03 |
9.8 ± 0.8 × 10−4 |
−(1.00) |
| 11c | − | GDPNP | Flu-mRNA | − | 0.002 ± 0.0004 |
−0.023 ±0.004 |
8.2 ± 0.5 × 10−4 |
−0.59± 0.07 | ||
| 12c | + | GDPNP | Flu-mRNA | Spc, 5 mM |
0.0052 ± 0.0005 |
−0.058 ±0.010 |
9.1± 0.5 × 10−4 |
−0.62± 0.07 | ||
| 13 | + | GDPNP | Flu-mRNA | Vio, 1mM |
0.95 ± 0.08 |
−0.028 ±0.008 |
0.040± 0.010 |
−0.36 ±0.05 |
9.5 ± 0.7 × 10−4 |
−0.97± 0.06 |
Figures 2A,B, 4A-C; kapp1, kapp2, and kapp3 are apparent rate constants and ΔF1, ΔF2, and ΔF3 are relative fluorescence changes for phases one, two and three of back translocation.
ΔF values for experiments 1-7, 8-9, and 10-13 are normalized to ΔF3 for experiments 1, 8, and 10, respectively.
Phases one and two not resolved in these experiments. Parameter values listed as kapp1, kapp3, ΔF1, ΔF3.
Phases 1 and 2 could not be resolved in the presence of EF4•GDPNP and Spc, although it is clear that this antibiotic virtually abolishes EF4-dependent back translocation (experiments #4 and #5, Table I, Figure 2A,B) by strongly inhibiting the initial fluorescence decrease, while having a lesser effect on the last phase of fluorescence decrease (Table I). All three phases are resolvable in the presence of Vio, which increases both kapp1 and kapp2, in each case by about 2-fold, while little affecting kapp3 (Table I, experiment #6). Three phases of fluorescence change are also seen in the presence of Vio when EF4 is omitted (Table I, experiment #7), but the magnitude of fluorescent change in the first two phases is strongly reduced, as is the value of kapp2.
Back translocation could also be monitored by the change in fluorescence observed on rapid mixing with EF4•GDPNP of the isolated POST complex containing tRNAfMet(prf) in the E-site and fMetPhe-tRNAPhe in the P-site (Figure 2C,D). Here again there is a three-phase change, in which each phase is characterized by a decrease in fluorescence intensity, with kapp1, kapp2, and kapp3 values (0.54 s−1, 0.014 s−1, 0.00097 s−1) that are virtually identical to those found when translocation is monitored by changes in fMetPhe-tRNAPhe(prf) fluorescence (see above). This result provides strong evidence that motion of the core regions of the two tRNAs in EF4-catalyzed back translocation are strongly coupled, paralleling results obtained for EF-G-catalyzed translocation13. Using tRNAfMet(prf) fluorescence as the measure of back translocation, it is possible to demonstrate that while Spc has little effect on either the apparent rate constant or the magnitude of fluorescence change in the first phase of reaction, it strongly decreases the values of both parameters in the second phase, while having a lesser effect on the third phase (Table 1).
EF4 catalysis of back translocation measured by change in the fluorescence of Flu-mRNA014
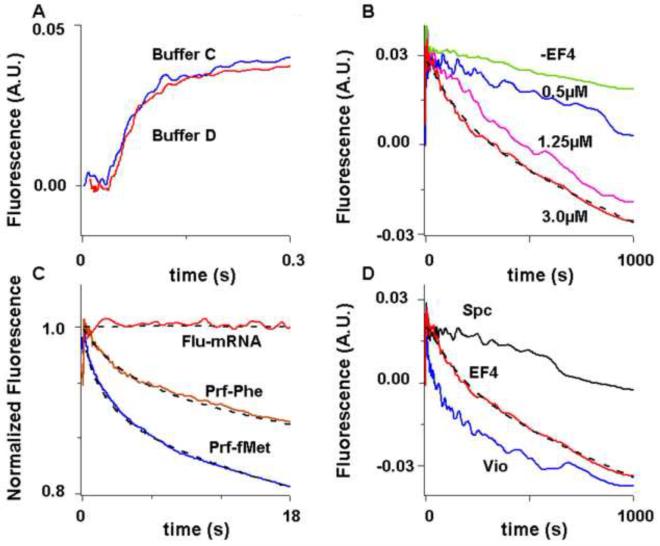
Changes in the fluorescence of 3′-labeled mRNA has been used to monitor the movement of mRNA during translocation14, 15. We find (Figure 3A) that, following a brief lag period, the fluorescence of Flu-mRNA014 in a PRE complex increases during translocation induced by EF-G•GTP, in good agreement with previous results15. The kcat and Km,EF-G values for the first and second phases of reaction (Supplementary Table I) correspond very well to our previous measurements based on tRNA fluorescence changes13.
Figure 3. EF-G-dependent translocation and EF4-dependent back translocation measured by change in the fluorescence of Flu-mRNA014.
Fluorescence changes measured in a stopped-flow spectrofluorometer. Ribosomes were programmed with Flu-mRNA14 except as otherwise indicated. The dashed lines through traces are the results of fits to Scheme 1 (Figure 5). A. Translocation. PRE complexes (0.1 μM) were rapidly mixed with 3 μM EFG•GTP in either Buffer C (blue trace) or D (50 mM Tris-HCl [pH 7.5], 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2, and 1 mM DTT) (red trace). Buffer D was used in Pan et al (2007). B - D. Back translocation. POST complexes (0.1 μM) containing 0.15 μM added tRNAfMet were rapidly mixed with EF4•GDPNP. B. Using varying EF4•GDPNP concentrations. C. The initial phase of back translocation, as monitored by the fluorescence change of Flu-mRNA14 (red trace), fMetPhe-tRNAPhe(prf) (brown trace), or tRNAfMet(prf) (blue trace). In the latter two cases, ribosomes were programmed with mRNA MFK. For ease of comparison, fluorescence changes are normalized to the total change for full back translocation. POST complex concentration was increased to 0.3 μM for Flu-mRNA14 to increase signal-to-noise ratio. Very similar results were obtained at 0.1 μM. D. The effects of added Spc (5 mM) (black trace) and Vio (1 mM) (blue trace) on EF4-dependent back translocation (no added antibiotic) (red trace).
Back translocation, as monitored using Flu-mRNA014, is accelerated in the presence of EF4•GDPNP, with a saturated rate being reached by 3 μM, in accord with results presented above (Figure 3B). The reaction proceeds in three phases, with a brief initial lag phase followed by two phases of clear fluorescence decrease (Table 1). While the apparent second and third phase rate constants (0.018 s−1 and 0.001 s−1) are quite similar to the corresponding constants determined for back translocation monitored with the prf-labeled tRNAs, the apparent rate constant for the first phase, 0.062 s−1, is ~10-fold slower (Table 1, Figure 3C). Added Spc prolongs the lag phase while having very little effect on the rate of the last phase. By contrast, added Vio almost abolishes the lag phase, increasing kapp1 by 15-fold to a value, 0.95 s−1, that is similar to the value of kapp1 measured in the presence of Vio as monitored using tRNAPhe(prf), while increasing kapp2 by 2.0-fold and having little effect on kapp3 (Figure 3D, Table I).
Reactivity toward puromycin of intermediates seen by fluorescence changes
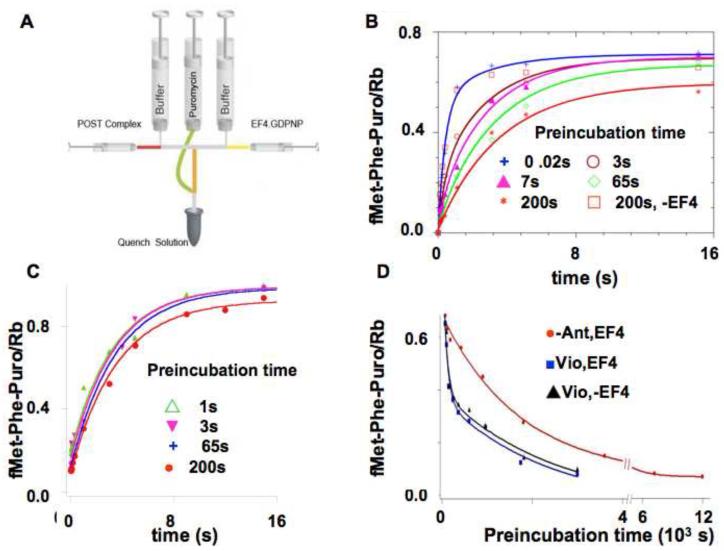
The fluorescence results shown in Figures 2 and 3 provide evidence for the formation of intermediate species on POST complex to PRE(L) complex conversion. The reactivity of these intermediates toward puromycin was determined by an experimental protocol involving two rapid mixing steps and a quenching step (Figure 4A). In the first mixing step, isolated POST complex was mixed with a solution of EF4•GDPNP and tRNAfMet. This was followed by pre-incubation for several different fixed amounts of time, chosen to permit characterization of intermediate reactivity toward puromycin, after which puromycin (5 mM) was added with mixing. The puromycin reaction was allowed to continue for various times prior to quenching.
Figure 4. EF4-dependent back translocation measured by the change in puromycin reactivity of fMetPhe-tRNAPhe.
A. Design of the preincubation-incubation experiment (adapted from a figure presented in reference 26). POST complex was rapidly mixed with EF4•GDPNP and E. coli tRNAfMet and preincubated prior to rapid mixing with puromycin, further incubation and quenching. B. POST complex was rapidly mixed with EF4•GDPNP and E. coli tRNAfMet and preincubated for the indicated times [0.02 s ( ); 3 s (
); 3 s ( ); 7 s (
); 7 s ( ); 65 s (
); 65 s ( ); 200 s (
); 200 s ( )] prior to rapid mixing and incubation with 5 mM puromycin for up to 50 s, and quenching. The preincubation mixtures contained 0.1 μM POST complex, 3 μM EF4•GDPNP and 0.15 μM E.coli tRNAfMet. Lines through the data are fits to Scheme 1 (Figure 5). A control sample in which preincubation was carried out for 200 s in the absence of EF4 showed no loss in puromycin reactivity (
)] prior to rapid mixing and incubation with 5 mM puromycin for up to 50 s, and quenching. The preincubation mixtures contained 0.1 μM POST complex, 3 μM EF4•GDPNP and 0.15 μM E.coli tRNAfMet. Lines through the data are fits to Scheme 1 (Figure 5). A control sample in which preincubation was carried out for 200 s in the absence of EF4 showed no loss in puromycin reactivity ( ).C. As described in B, but in the presence of 5 mM spectinomycin. Preincubation times were: 1 s (
).C. As described in B, but in the presence of 5 mM spectinomycin. Preincubation times were: 1 s ( ); 3 s (
); 3 s ( ); 65 s (
); 65 s ( ); 200 s (
); 200 s ( ). All traces are fit by single exponential, with an apparent rate constant of 0.30 ± 0.03 s−1. D. The effect of long pre-incubation times. 0.1 μM of POST complex was preincubated for the times indicated with 0.15 μM E.coli tRNAfMet and various other combinations [3 μM EF4•GDPNP (
). All traces are fit by single exponential, with an apparent rate constant of 0.30 ± 0.03 s−1. D. The effect of long pre-incubation times. 0.1 μM of POST complex was preincubated for the times indicated with 0.15 μM E.coli tRNAfMet and various other combinations [3 μM EF4•GDPNP ( ); 3 μM EF4•GDPNP plus 1 mM Vio (
); 3 μM EF4•GDPNP plus 1 mM Vio ( ); 1 mM Vio in the absence of EF4 (▲)], prior to incubation with puromycin (5 mM) for 20 s prior to quenching. The EF4•GDPNP trace is fit to a single exponential, giving an apparent rate constant of 7.9± 0.5 × 10−4 s−1. The EF4•GDPNP plus 1 mM Vio and 1 mM Vio alone traces are fit by double exponentials, giving identical values of 0.011 ± 0.005 s−1 and 8.8 ± 0.9 × 10−4 s−1 for each trace.
); 1 mM Vio in the absence of EF4 (▲)], prior to incubation with puromycin (5 mM) for 20 s prior to quenching. The EF4•GDPNP trace is fit to a single exponential, giving an apparent rate constant of 7.9± 0.5 × 10−4 s−1. The EF4•GDPNP plus 1 mM Vio and 1 mM Vio alone traces are fit by double exponentials, giving identical values of 0.011 ± 0.005 s−1 and 8.8 ± 0.9 × 10−4 s−1 for each trace.
The results of these experiments (Figure 4B) demonstrate that although the apparent rate constant for reaction with puromycin decreases even after rather short times of pre-incubation (e.g., from 3.29 ± 0.07 s−1 for POST complex down to 0.99 ± 0.03 s−1 after a pre-incubation of 7 s – see also Supplementary Table II), the decrease in overall reaction stoichiometry is only evident at the longest time of pre-incubation (200 s) prior to puromycin addition. A control experiment lacking EF4 showed no change in puromycin reactivity even after 200 s of preincubation. Added spectinomycin retards the decrease in the rate and stoichiometry of fMetPhe-puromycin formation seen over the preincubation period 3 s – 200 s (compare Figures 4B and 4C).
The decrease in overall reaction stoichiometry in the presence of EF4•GDPNP was further examined by measuring the amount of fMetPhe-puromycin formed when extended preincubation periods (up to 200 min) were followed by reaction with puromycin (Figure 4D) for a time period allowing full reaction of the POST complex and the intermediate species, but only minor reaction of the PRE(L) complex. This procedure results in a monophasic decrease in the amount of fMetPhe-puromycin formed, allowing calculation of an apparent rate constant for PRE(L) complex formation, 0.00079 s−1, which is quite similar to the values obtained for the last phases of fluorescence change in both the presence and absence of EF4 (Table I, experiments # 1, #2, #8, #10, #11). Added Vio, whether in the presence or absence of EF4•GDPNP, induces a change to a biphasic reaction, in which the second phase proceeds with a rate constant (kapp2, 0.00088 s−1) quite similar to that observed in the presence of EF4•GDPNP alone, while the initial phase proceeds some 12 times faster (kapp1, 0.011 s−1). Very similar rate constants are seen for phases 2 and 3 of Vio-induced fluorescence changes of fMetPhe-tRNAPhe(prf) in the absence of EF4•GDPNP (Table 1, #7).
A quantitative model for back translocation in the presence of EF4
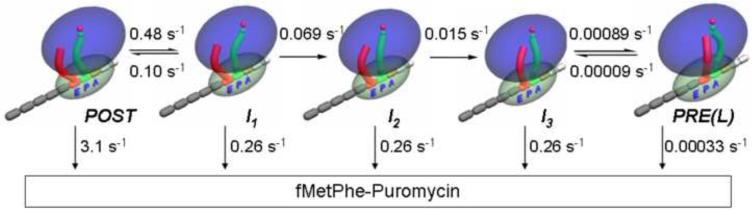
All of the results measuring EF4-dependent back translocation in Figures 2 – 4 can be fit quantitatively to the minimal four step kinetic model shown in Figure 5 (Scheme 1). Here I1, I2, and I3, formed via steps 1 - 3, respectively, are intermediates in the overall conversion of POST complex to PRE(L) complex, which is formed via step 4.
Figure 5. A quantitative kinetic scheme for interrupted EF4 catalysis of back translocation.
EF4 catalyzes back translocation via steps 1 - 3, but has little effect on step 4. The rate constants shown are the results of global fitting to the results, measured at 3 μM EF4•GDPNP, in the absence of antibiotic, presented in Figures 2, 3B,C and 4B,D. Apparent rate constants for fMetPhe-puromycin formation were measured at 5 mM puromycin.
Back translocation, as monitored by each of the three fluorescent probes used in this study (fMetPhe-tRNAPhe(prf), tRNAfMet(prf), Flu-mRNA14), proceeds in a triphasic manner (Table 1). The values of kapp2 and kapp3 are very similar for each of the three probes [the change in puromycin reactivity (Figure 4D) provides a fourth independent estimate of kapp3], an indication that the rates of these two phases are unaffected by the introduction of fluorescent labels into tRNA and mRNA. However, the values of kapp1 as determined using fMetPhe-tRNAPhe(prf) or tRNAfMet(prf), while similar to each other (~0.6 s−1), are each 10 times faster than the kapp1 value determined with Flu-mRNA14 (0.06 s−1). This decrease in kapp1 is unlikely to be due to perturbations arising from the presence of the fluorescein group in Flu-mRNA14 for the following reasons: i) the position of fluorescein group attachment, at the 3′-end of the mRNA, is well-removed from the A, P, and E sites of the ribosome; ii) addition of Vio, which binds to a region of the ribosome quite distant from the 3′-end of Flu-mRNA14 (whether in the PRE or POST complex, as reviewed in reference 16), increases kapp1 as measured with Flu-mRNA14 to a value similar to that seen using unlabeled mRNA (Table 1, experiments #7, #13); iii) translocation using Flu-mRNA14 (Figure 3A) proceeds at a rate very similar to that seen with unlabeled mRNA. A more likely interpretation, which is incorporated into Scheme 1, is that the difference in kapp1 values reflects the formation of two intermediates, I1 and I2, which are likely to have similar structures, since they have the same puromycin reactivity and show only relatively minor differences in their tRNA and mRNA fluorescence intensities (Supplementary Table III).
All three intermediates in Scheme 1 have ~12-fold less reactivity toward puromycin than the POST complex. I3 is converted to PRE(L) complex via step 4, which is clearly rate-determining for the overall process. The value of k4, 9 × 10−4 s−1, is only slightly higher than the overall rate constant for PRE formation measured in the absence of EF4, 5.7 × 10−4 s−1. Thus, EF4 may be thought of as performing an interrupted catalysis, stopping at the formation of I3 rather than catalyzing the complete process of back translocation culminating in PRE complex formation.
Scheme 1 rationalizes the results presented in Figure 1, since incubation of POST complex with EF4•GDPNP for ~ 300 s should lead to a ribosome population containing mostly I3 complex, ~ 50% of which should react with 1 mM puromycin in 15 s (the kapp for reaction of I2 or I1 with 1 mM puromycin is ~0.06 s−1). Scheme 1 also provides a framework for rationalizing antibiotic effects in the presence of EF4. Thus, spectinomycin appears to stabilize I1 and/or I2, since it allows normal I1/I2 formation while retarding further I1/I2 conversion (Table 1, experiments #8, #9 and #10, #12, Figure 4C). In contrast, Vio accelerates I1, I2, and I3 formation while little affecting the rate of PRE(L) complex formation (Table 1, experiments #1, #6 and #10, #13, Figure 4D), consistent with recent results13, 16, 17 indicating that Vio stabilizes hybrid structures that are possible intermediates in both translocation and back translocation.
DISCUSSION
Here we present the first detailed study of the kinetic mechanism of EF4-dependent back translocation, allowing formulation of a quantitative kinetic scheme (Figure 5, Scheme 1) for the process of POST complex conversion to PRE(L) complex, as measured by changes in fluorescence intensities of fluorescent derivatives of mRNA, tRNAfMet and fMetPhe-tRNAPhe, and by the puromycin reactivity of the latter. This process proceeds via four steps leading from POST complex to I1 (step 1), I2 (step 2), I3 (step 3) and PRE(L) complex (step 4) formation. In common with EF-G dependent forward translocation13, EF4-dependent back translocation requires the conformation of EF4 bound to GTP, or to its structural analogue GDPNP, with EF4•GDP giving little or no apparent catalysis (Figure 2A,B).
Our results indicate that during steps 1 and 3, movement of the flexible 3′-end of peptidyl–tRNA, as monitored by puromycin reactivity, is largely decoupled from movements of the core region and of mRNA, as monitored by fluorescence intensity changes. Thus, step 1 is accompanied by a large change in puromycin reactivity with only minor changes in both fluorescence intensities, while step 3 involves considerable changes in these intensities with no change in puromycin reactivity. In contrast, step 4 is accompanied by large changes in all three experimental parameters, implying substantial concerted movement of the entire tRNA molecule together with mRNA.
The effects of added Vio, which is known to induce intersubunit movement within the ribosome16, provide another example of the decoupling of the movements of the 3′-end and core regions of peptidyl–tRNA. When added in the absence of EF4, Vio has only a modest effect on tRNA fluorescence intensity over the same time scale (~100 s) that it has a major impact on puromycin reactivity (Figures 2A,B and 4D). The structural basis for this selectivity is, however, not obvious, given the evidence that Vio binds to the ribosome near the intersubunit bridge B2a that is formed between 16S rRNA helix 44 and 23S rRNA helix 6916, a region of the ribosome that is quite removed from the peptidyl transferase center. Perhaps Vio binds to more than one site on the ribosome, as has been shown for other antibiotics18 - 20.
Pan et al.13 have shown that EF-G-catalyzed translocation proceeds via a three step reaction, PRE → (P/E) → INT → POST. In monitoring the reaction via changes in the fluorescence of fMetPhe-tRNAPhe(prf), they showed that the second step was accompanied by an increase in fluorescence, which was followed by a decrease in fluorescence corresponding to the third step. They further showed that the Vio and Spc inhibited translocation by specifically stabilizing the intermediates P/E and INT, respectively (P/E does not accumulate in the absence of Vio). It is tempting to speculate that the microscopic reverse of this process might be similar to the EF4-dependent back translocation process described in this paper, POST → I1 → I2 → I3 → PRE, if INT and P/E resembled I1/I2 and I3, respectively. Several observations are consistent with this notion: i) Spc addition results in the accumulation of both I1/I2 in back translocation and of INT in translocation13, which is likely a result of Spc interference with 30S head swiveling, a ribosomal motion that appears to be required in both directions of translocation21, ii) the reactivities toward puromycin of I1/I2 [8% as compared to POST complex, Figure 5] and INT (5%)13 are similar, and iii) Vio addition both accelerates EF4-dependent POST → I3 conversion and stabilizes the P/E complex13.
However, other results, summarized in Table II, provide strong evidence against the validity of this speculation. For example, INT → POST conversion proceeds with a large increase in Flu-mRNA014 fluorescence (Figure 3A), so that POST → INT conversion must proceed with a large decrease. In contrast, POST → I1/I2 conversion proceeds with little change in Flu-mRNA014 fluorescence (Supplementary Table 3). Similarly, I1/I2 → I3 conversion does not alter the puromycin reactivity of fMetPhe-tRNAPhe, whereas INT → P/E conversion would result in a drastic loss of such activity. Also, I3 → PRE conversion, a step not catalyzed by EF4, results in a large decrease in fMetPhe-tRNAPhe(prf) fluorescence but P/E → PRE conversion would result in essentially no change. A movie illustrating the decoupled movement of the 3′-end of peptidyl-tRNA from the rest of the tRNA molecule during back translocation, and contrasting back translocation with translocation, can be found at http://media.sas.upenn.edu/chemistry/rwertz/EF4.avi.
Table II.
Comparison of EF4-dependent back translocation and the microscopic reverse of EF-G dependent translocation
| Processa | ΔF, fMetPhe- tRNAPhe(prf) |
ΔF, Flu-mRNA014 | Relative puromycin Reactivityb |
|---|---|---|---|
|
Back translocation
(EF4) |
|||
| POST ➜ I1/I2 | small decrease | small decrease | 0.08 |
| I1/I2 ➜ I3 | moderate decrease | moderate decrease | 0.08 |
| I3 ➜ PRE | large decrease | large decrease | 0.0001 |
|
Reverse of
translocation (EF-G) a |
|||
| POST ➜ INT | small increase | large decrease | 0.05 |
| INT ➜ P/Ec | large decrease | no change | 0.0003 |
| P/E ➜ PRE | no change | no change | 0.0003 |
It thus appears, perhaps not unsurprisingly, that reaction pathways for back translocation and reverse translocation differ in detail, due likely to differences in the interactions of EF4 and EF-G with the POST and PRE complexes, as well as with intermediates involved in their interconversions. For example, EF4 contacts the 3′-end and D-stem of ribosome-bound fMetPhe-tRNAPhe via subdomain V’ and the CTD, respectively, both of which are lacking in EF-G5, 7. In addition, there is a steric interference between fMetPhe-tRNAPhe and domain IV of EF-G, that is lacking in EF4. It is tempting to speculate that the failure of EF4 to accelerate I3 → PRE(L) conversion might result from a strong EF4 interaction with fMetPhe-tRNAPhe in the I3 complex.
The position of peptidyl-tRNA in the I3 complex is likely to be different from the A/L position identified in the PRE(L) complex7. In this latter work a POST complex was preincubated with EF4•GDPNP for 15 min at 37 °C prior to rapid freezing for cryoelectronmicroscopy studies. This protocol should convert virtually all of the peptidyl-tRNA to a form having very low reactivity toward puromycin, making the reasonable assumption that k4 (Scheme 1, Figure 5) increases 2 – 3-fold between 25 °C and 37 °C. Very low reactivity toward puromycin is consistent with the positioning of A/L peptidyl-tRNA in between the classic A–site and the A/T site7, the latter being further displaced from the puromycin-reactive P-site than the A-site itself. By contrast, peptidyl-tRNA in the I3 complex, which is formed with a t½ of ~ 1 min at 25 °C as a transient in overall POST to PRE(L) conversion, has intermediate reactivity toward puromycin and so is likely to occupy a position falling in between the A- and P-sites. Similarly, our results suggest that tRNAfMet might occupy a position within the 30S subunit that is intermediate between the P- and E-sites, possibly similar to the position that fMet-tRNAfMet is thought to adopt during 70S initiation complex formation22.
What biological purpose might I3 formation serve? Qin et al.2 have shown that added EF4 significantly increases the fraction of active protein made in a cell-free coupled transcription-translation system. EF4 capture of the POST complex, by competing with aa-tRNA•EF-Tu•GTP ternary complex binding, will at least transiently interrupt normal polypeptide elongation and could, by facilitating co-translational protein folding23, increase the fraction of active protein produced, as suggested by Shoji et al.9 Alternatively, or in addition, I3 formation could allow for correction of a defective translocation, possibly arising from high-salt induced perturbation of ribosome structure, following a putative displacement of EF4•GDP by EF-G•GTP (EF-G is present in considerably higher concentration in bacterial cells than EF4), as suggested by Nierhaus and co-workers (K. Nierhaus, private communication, and references 2 and 6). Here it is worth noting that added EF-G.GTP readily converts I3 back to POST complex, as measured by a rapid increase in reactivity of peptidyl-tRNA toward puromycin (H. Liu and B. S. Cooperman, in preparation).
A possible concern regarding the putative biological role of I3 is the apparently large energy expenditure that would be required in vivo to form I3 from POST complex, given the high catalytic activity of ribosome-bound EF4 as a GTPase (Figure 1) and the rather slow overall rate constant for EF4-catalyzed POST conversion to I3 (0.012 s−1, Scheme 1) that we have measured. Further studies will be needed to determine whether I3 might be formed more rapidly if EF4 acted in concert with other factors, and/or whether back translocation of the first POST complex, having initiator tRNA in the E-site, might proceed significantly more slowly than back translocation from subsequent POST complexes.
MATERIALS AND METHODS
The following materials were prepared as described previously: EF42, tight-coupled ribosomes from E. coli MRE600 cells, cloned E. coli His-tagged proteins EF-G, EF-Tu, IF1, IF2, IF3, E. coli [35S]-fMet-tRNAfMet, E. coli [35S]-fMet-tRNAfMet(prf), E. coli [3H]-Phe-tRNAPhe, and E. coli [3H]-Phe-tRNAPhe(prf)13. Protein concentrations were determined by Bradford assay24. mRNA MFK and mRNA MFK014 (Flu) were purchased from Dharmacon (Lafayette, CO) with the following sequences: GGG AAG GAG GUA AAA AUG UUU AAA CGU AAA UCU ACU and GGG AAG GAG GUA AAA AUG UUU AAA CGU AA-Flu, respectively (initiator codon underlined).
Complex preparation
Complexes were made up in Buffer C (20 mM HEPES-KOH (pH 7.5 at 0 °C), 150 mM NH4Ac, 4.5 mM MgAc2, 4 mM 2-mercaptoethanol, 0.05 mM spermine, and 2 mM spermidine) at 37 °C. Initiation complex was formed by incubating ribosomes (2 μM) with mRNA MFK (8 μM) or Flu-mRNA14 (8 μM), IF1 (3 μM), IF2 (3 μM), IF3 (3 μM), GTP (2 mM) and [35S]-fMet-tRNAfMet (3 μM) for 25 min. Ternary complex was formed by incubating EF-Tu (6 μM) with [3H]-Phe-tRNAPhe(prf16/20) (6 μM), GTP (6 mM), phosphoenolpyruvate (Roche Diagnostics) (1.5 mM), pyruvate kinase (Roche Diagnostics) (0.015 mg/mL) for 15 min. Pre-translocation complex (PRE) was formed by mixing initiation complex (2 μM) with ternary complex (3 μM) and incubating for 45 sec. PRE complexes were translocated in the presence of EF-G (molar ratio to 70S 0.2:1) and GTP (1 mM) during an incubation at 37 °C for 10 min to form post-translocation complex (POST). POST complexes were purified by ultracentrifugation through a 1.1 M Sucrose cushion in Buffer C (110,000 rpm, 40 min centrifugation). SDS-PAGE analysis confirmed the total removal of EF-G. POST complex concentration was calculated from the amount of ribosome-bound fMet-[3H]-Phe-tRNAPhe(prf16/20) or fMet-[3H]-Phe-tRNAPhe.
Rate measurements
All rate measurements and associated incubation steps were carried out in Buffer C except as otherwise indicated at 25 °C. Stopped-flow fluorescence experiments were performed using an SX.18MV stopped-flow spectrofluorometer (Applied Photophysics). Proflavin and fluorescein were excited at 462 nm and 460 nm, respectively, and fluorescence was monitored using a 495-nm long-pass filter. Rapid quench experiments were performed out using a KinTek Chemical-Quench-Flow Model RQF-3 machine. Puromycin reactions were quenched with 0.3 M NaAc solution at pH 5.0. GTPase reactions were quenched with 1.8 mM KH2PO4 in 0.6 M HClO4 and 32Pi formation was determined as described25. Puromycin reactions employing two mixing steps (Figure 4). POST complex was rapidly mixed with EF4•GDPNP and E. coli tRNAfMet and preincubated for various times prior to rapid mixing with puromycin and further incubation for up to 50 s, after which reaction mixtures were quenched by expelling the final solution into a chilled test tube (0 °C) containing 0.3 M NaAc, pH 5.0 with a dead-time of 0.08 s.
Rate Constant Estimation
Global fitting of data presented in Figures 2 - 4 to Scheme 1 (Figure 5) was carried out using the program Scientist (MicroMath Research, LC). Fitting of the data presented in Figures 1 - 4 to single-, double- or triple-exponential equations using the program Igor-Pro (Wavemetrics) yielded the apparent rate constants presented in Table I and in Supplemental Tables I and II.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH grant GM071014 to B. S. C. We thank Knud Nierhaus for several interesting discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.March PE, Inouye M. GTP-binding membrane protein of Escherichia coli with sequence homology to initiation factor 2 and elongation factors Tu and G. Proc Natl Acad Sci U S A. 1985;82:7500–7504. doi: 10.1073/pnas.82.22.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin Y, Polacek N, Vesper O, Staub E, Einfeldt E, Wilson DN, Nierhaus KH. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell. 2006;127:721–733. doi: 10.1016/j.cell.2006.09.037. [DOI] [PubMed] [Google Scholar]
- 3.Shoji S, Walker SE, Fredrick K. Back translocation of tRNA in the ribosome. Mol Cell. 2006;24:931–942. doi: 10.1016/j.molcel.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Konevega AL, Fischer N, Semenkov YP, Stark H, Wintermeyer W, Rodnina MV. Spontaneous reverse movement of mRNA-bound tRNA through the ribosome. Nat Struct Mol Biol. 2007;14:318–324. doi: 10.1038/nsmb1221. [DOI] [PubMed] [Google Scholar]
- 5.Evans RN, Blaha G, Bailey S, Steitz TA. The structure of LepA, the ribosomal back translocase. Proc Natl Acad Sci U S A. 2008;105:4673–4678. doi: 10.1073/pnas.0801308105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connell SR, Takemoto C, Wilson DN, Wang H, Murayama K, Terada T, Shirouzu M, Rost M, Schuler M, Giesebrecht J, et al. Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol Cell. 2007;25:751–764. doi: 10.1016/j.molcel.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 7.Connell SR, Topf M, Qin Y, Wilson DN, Mielke T, Fucini P, Nierhaus KH, Spahn CM. A new tRNA intermediate revealed on the ribosome during EF4-mediated back-translocation. Nat Struct Mol Biol. 2008;15:910–915. doi: 10.1038/nsmb.1469. [DOI] [PubMed] [Google Scholar]
- 8.Dibb NJ, Wolfe PB. lep operon proximal gene is not required for growth or secretion by Escherichia coli. J Bacteriol. 1986;166:83–87. doi: 10.1128/jb.166.1.83-87.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shoji S, Janssen BD, Hayes CS, Fredrick K. Translation factor LepA contributes to tellurite resistance in Escherichia coli but plays no apparent role in the fidelity of protein synthesis. Biochimie. 2010 Nov 17; doi: 10.1016/j.biochi.2009.11.002. (2009) [Epub ahead of print]PMID: 19925844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Semenkov YP, Shapkina TG, Kirillov SV. Puromycin reaction of the A-site bound peptidyl-tRNA. Biochimie. 1992;74:411–417. doi: 10.1016/0300-9084(92)90080-x. [DOI] [PubMed] [Google Scholar]
- 11.Sharma D, Southworth DR, Green R. EF-G-independent reactivity of a pre-translocation-state ribosome complex with the aminoacyl tRNA substrate puromycin supports an intermediate (hybrid) state of tRNA binding. RNA. 2004;10:102–113. doi: 10.1261/rna.5148704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J Mol Biol. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 13.Pan D, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Studer SM, Feinberg JS, Joseph S. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J Mol Biol. 2003;327:369–381. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- 15.Savelsbergh A, Katunin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 16.Ermolenko DN, Spiegel PC, Majumdar ZK, Hickerson RP, Clegg RM, Noller HF. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat Struct Mol Biol. 2007;14:493–497. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- 17.Szaflarski W, Vesper O, Teraoka Y, Plitta B, Wilson DN, Nierhaus KH. New features of the ribosome and ribosomal inhibitors, non-enzymatic recycling, misreading and back-translocation. J Mol Biol. 2008;380:193–205. doi: 10.1016/j.jmb.2008.04.060. [DOI] [PubMed] [Google Scholar]
- 18.Brodersen DE, Clemons WM, Jr, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- 19.Pioletti M, Schlünzen F, Harms J, Zarivach R, Glühmann M, Avila H, Bashan A, Bartels H, Auerbach T, Jacobi C, Hartsch T, Yonath A, Franceschi F. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 2001;20:1829–1839. doi: 10.1093/emboj/20.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schluenzen F, Takemoto C, Wilson DN, Kaminishi T, Harms JM, Hanawa-Suetsugu K, Szaflarski W, Kawazoe M, Shirouzu M, Nierhaus KH, Yokoyama S, Fucini P. The antibiotic kasugamycin mimics mRNA nucleotides to destabilize tRNA binding and inhibit canonical translation initiation. Nat Struct Mol Biol. 2006:13871–13878. doi: 10.1038/nsmb1145. [DOI] [PubMed] [Google Scholar]
- 21.Borovinskaya MA, Shoji S, Holton JM, Fredrick K, Cate JH. A steric block in translation caused by the antibiotic spectinomycin. ACS Chem Biol. 2007;2:545–552. doi: 10.1021/cb700100n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell. 2005;121:703–712. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 23.Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol. 2009;16:274–280. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
- 24.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 25.Pan D, Kirillov S, Zhang CM, Hou YM, Cooperman BS. Rapid ribosomal translocation depends on the conserved 18-55 base pair in P-site transfer RNA. Nat Struct Mol Biol. 2006;13:354–359. doi: 10.1038/nsmb1074. [DOI] [PubMed] [Google Scholar]
- 26.Shcherbakova I, Mitra S, Beer RH, Brenowitz M. Following molecular transitions with single residue spatial and millisecond time resolution. Methods in Cell Biology. 2008;84:589–615. doi: 10.1016/S0091-679X(07)84019-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.