Abstract
The N-myc downstream-regulated gene (NDRG) family consists of four related proteins, NDRG1-NDRG4, in mammals. We previously generated NDRG1-deficient mice that were unable to maintain myelin sheaths in peripheral nerves. This condition was consistent with human hereditary motor and sensory neuropathy, Charcot-Marie-Tooth disease type 4D, caused by a nonsense mutation of NDRG1. In contrast, the effects of genetic defects of the other NDRG members remain unknown. In this study, we focused on NDRG4, which is specifically expressed in the brain and heart. In situ mRNA hybridization on the brain revealed that NDRG4 was expressed in neurons of various areas. We generated NDRG4-deficient mice that were born normally with the expected Mendelian frequency. Immunochemical analysis demonstrated that the cortex of the NDRG4-deficient mice contained decreased levels of brain-derived neurotrophic factor (BDNF) and normal levels of glial cell line-derived neurotrophic factor, NGF, neurotrophin-3, and TGF-β1. Consistent with BDNF reduction, NDRG4-deficient mice had impaired spatial learning and memory but normal motor function in the Morris water maze test. When temporary focal ischemia of the brain was induced, the sizes of the infarct lesions were larger, and the neurological deficits were more severe in NDRG4-deficient mice compared with the control mice. These findings indicate that NDRG4 contributes to the maintenance of intracerebral BDNF levels within the normal range, which is necessary for the preservation of spatial learning and the resistance to neuronal cell death caused by ischemic stress.
Keywords: Brain, Gene Knockout, Mouse, Neurological Diseases, Neurotrophic Factor, BDNF, Cerebral Infarction, NDRG4, Neurotrophin, Spatial Learning
Introduction
N-myc downstream-regulated gene (NDRG)3 family members NDRG1-NDRG4 are intracellular proteins, consist of 340–394 amino acid residues, and share 53–65% sequence identity with each other. Furthermore, accumulating evidence implicates their roles in development, cancer metastasis, and the immune system (1–5).
We originally identified RTP (NDRG1) as a homocysteine-responsive gene in human umbilical vein endothelial cells (6), which is also called DRG1, Cap43, Rit42, Ndr1, and PROXY-1. NDRG1 expression is induced by a number of conditions, such as DNA damage, hypoxia, and intracellular calcium ion elevation (4). Overexpression of NDRG1 suppresses the metastatic potency of some types of cancer cells (4) and enhances the degranulation of mast cells in response to various stimuli (7). A nonsense mutation of NDRG1 causes hereditary motor and sensory neuropathy, Charcot-Marie-Tooth disease type 4D, which presents as distal muscle wasting and atrophy, foot and hand deformities, tendon areflexia, sensory loss, and deafness in afflicted individuals (8). We previously generated NDRG1-deficient mice and revealed the essential role of NDRG1 in the cytoplasm of Schwann cells for the maintenance of myelin sheaths in peripheral nerves (9). A frame shift deletion of Ndrg1 in Greyhounds also causes polyneuropathy (10).
Similar to NDRG1, the expression of NDRG2 is induced by stress conditions such as hypoxia (1). NDRG2 expression is up-regulated in cortical pyramidal neurons, senile plaques, and the cellular process of dystrophic neurons in the Alzheimer's brain, whereas expression is decreased in the rat frontal cortex after antidepressant treatment and electroconvulsive therapy (11). NDRG2 also plays a role in aldosterone-mediated epithelial sodium channel function (12), dendritic cell differentiation (13), and insulin action (14). NDRG3, on the other hand, may play a role in spermatogenesis because it is found in the outer layers of the seminiferous epithelium (3). Overexpression of NDRG3 contributes to the angiogenesis of tumors via up-regulation of chemokines (3).
In contrast to other NDRG members, NDRG4 expression is detected specifically in the brain and heart (15). In the embryonic mouse heart, NDRG4 expression is down-regulated under severe ventricular hypoplasia caused by Tbx2 misexpression, implying that NDRG4 is involved in cell growth and proliferation (16). However, information on the physiological function of NDRG4 is lacking. In the mouse brain, NDRG4 is identified in the neuronal cytoplasm of the cerebrum and cerebellum (17). Down-regulation of NDRG4 in PC12 cells results in extending shorter neurites in response to NGF (18). Considering that NDRG4 expression is induced by treatment with homocysteine in rat aortic smooth muscle cells (19), we speculated that NDRG4 is stress-related and has a cell-protective role in neurological disorders and cerebrovascular disease. This was supported by our finding that NDRG4 mRNA expression is decreased in the brain of patients with Alzheimer's disease (15).
Neurotrophins such as brain-derived neurotrophic factor (BDNF), glial cell line-derived neurotrophic factor (GDNF), NGF, neurotrophin-3 (NT-3), and TGF-β1 are essential for the survival and homeostatic maintenance of central neurons (20). BDNF, especially, is a potent modulator of synaptic connectivity in the central nervous system, influencing synaptic structure and function. The reduced levels of BDNF in the entorhinal cortex or forebrain are associated with poor memory (21, 22). BDNF also has neuroprotective action in models of ischemia. Increased BDNF levels in the brain for an appropriate period prior to the ischemic insult increases the resistance of the brain against lethal stresses caused by severe ischemia (23, 24). In contrast, a deficiency in endogenous BDNF renders the brain more susceptible to ischemic injury (25) and more suppressive to infarct tolerance by the preconditioning of spreading depression (26).
In this study, we generated NDRG4-deficient mice to reveal the roles of NDRG4 in the brain. As a result, we found that under the condition of NDRG4 deficiency, mice showed impaired phenotypes in spatial learning and neuroprotection with decreased levels of BDNF.
EXPERIMENTAL PROCEDURES
Antibodies
Anti-NDRG1 rabbit antiserum was raised against recombinant glutathione S-transferase-fusion protein of human NDRG1 (27). Anti-NDRG2, anti-NDRG3, and anti-NDRG4 rabbit antisera were raised against the synthetic peptides Q351SSESGTLPSGPPGH365 for mouse NDRG2 (17), F343SRSVTSNQSDGTQE357 for mouse NDRG3 (17), and C-N214RPGTVPNAKTLR226-CONH2 for mouse NDRG4, respectively, which were conjugated with keyhole limpet hemocyanin. Polyclonal antibodies in the antisera were purified by antigen-immobilized affinity column chromatography. Anti-NeuN and anti-glial fibrillary acidic protein (GFAP) were purchased from Millipore and Dako, respectively.
Construction of the Targeting Vector
We previously isolated and characterized genomic clones carrying NDRG4 (15). The NDRG4-B and NDRG4-Bvar isoforms are the alternative splicing products, whereas the NDRG4-H isoform is produced by the alternative promoter usage. The initiating Met codons for NDRG4-B/Bvar and NDRG4-H exist in exons 5 and 3, respectively. Exon 6 is common to all isoforms. The loxP-flanked pSTneoB cassette (28) was inserted within exon 6. The ∼11-kb sequence was inserted into the diphtheria toxin A fragment cassette vector (29), and the DNA was linearized by SalI digestion for electroporation.
Generation of NDRG4-deficient Mice
R1 mouse embryonic stem cells (30) were electroporated with the targeting vector and selected in medium containing G418. Targeted clones were identified by Southern blotting using the Gene Images Random-prime system (GE Healthcare) with 5′- and 3′-external probes. These cells were injected into blastocysts to obtain chimeras, which were crossed with wild-type C57BL/6 mice (Japan SLC) for germ line transmission of the disrupted Ndrg4 allele. The genotypes of the offspring were examined by PCR analysis of DNA isolated from ear biopsy using three primers; P1 (CATCTCTCCAAGAGCCAGAGTGT), P2 (AAGATGCAGCCACACTTACGATT), and P3 (AACAGTAACAGCTTCCCACATC). Heterozygous mice with the disrupted Ndrg4 allele were backcrossed with wild-type C57BL/6 mice. The mouse experiments were approved by the Animal Care and Use Committee of the National Cerebral and Cardiovascular Center in Japan, and were performed in accordance with the institutional and national guidelines and regulations.
Western Blotting Analysis
Protein expression was analyzed by Western blotting as described previously (31). Briefly, organs perfused with PBS (10 mm sodium phosphate, 150 mm NaCl (pH 7.4)) were homogenized in SDS sample buffer (10 mm Tris-HCI, 2% SDS, 50 mm DTT, 2 mm EDTA, 0.02% bromphenol blue, 6% glycerol (pH 6.8)), boiled for 7 min, and subjected to SDS-PAGE. Proteins in the gels were transferred to an immunoblot PVDF membrane (Bio-Rad). Following a blocking step with 5% skim milk, the membrane was incubated with anti-NDRG4, anti-NDRG1, anti-NDRG2, or anti-NDRG3 and probed with HRP-labeled goat anti-rabbit IgG (Kirkegaard and Perry Laboratories). The membrane was developed using Immobilon Western chemiluminescent HRP substrate (Millipore), and chemiluminescence was detected by a LAS-3000 image analyzer (GE Healthcare).
In Situ mRNA Hybridization
Digoxigenin-labeled riboprobes were prepared for nucleotide positions 1269–1777 (NDRG4-a) and 1811–2343 (NDRG4-b) of mouse NDRG4 (NM_145602). The paraffin-embedded brain sections (6 μm thick) were dewaxed, rehydrated, and treated with proteinase K (8 μg/ml) for 30 min at 37 °C. The sections were acetylated by 0.25% acetic anhydride and hybridized with the riboprobes (300 ng/ml) for 16 h at 60 °C. Following treatment with RNase A (50 μg/ml) for 30 min at 37 °C and 0.5% blocking reagent (Roche), the sections were incubated with anti-DIG alkaline phosphate conjugate (Roche). Colorimetric reactions were performed with nitro blue tetrazolium chloride/5-bromo-4-chloro-3-indolyl phosphate solution (Sigma), and then the sections were counterstained with Kernechtrot stain solution (Mutoh). Because NDRG4-a and NDRG4-b riboprobes exhibited quite similar performance, only the data from NDRG4-a riboprobes are shown.
Immunohistochemistry
Serial sections for in situ mRNA hybridization were deparaffinized, rehydrated, and boiled by microwave irradiation in 10 mm citrate buffer (pH 6.0). After incubation with 0.3% hydrogen peroxide, the sections were blocked, incubated with anti-NeuN, and stained using Histofine MOUSESTAIN Kit (Nichirei) and diaminobenzidine according to the manufacturer's instructions. Alternate sections were blocked with Protein Block Serum-Free (Dako) and the Avidin/Biotin blocking kit (Vector Laboratories), and then incubated with anti-GFAP. Following incubation with biotin-conjugated goat anti-rabbit Ig (Dako), the sections were treated with HRP-conjugated streptavidin (Nichirei) and stained with diaminobenzidine. The sections were counterstained with Mayer's hematoxylin (Mutoh).
Measurement of Neurotrophin Levels
Protein levels of BDNF, GDNF, NGF, NT-3, and TGF-β1 were measured as described (32). The PBS-perfused cerebral cortex described above was excised from each mouse (7–18 weeks old) and homogenized. The protein levels were measured using a two-site sandwich ELISA, Emax Immunoassay System (Promega). The protein concentration in each sample was measured using a BCA protein assay kit (Thermo Scientific).
Morris Water Maze (MWM) Test
We conducted the MWM test (33) using modifications as described previously (32). In a 64 × 91 cm-sized pool of opaque water (from a non-toxic agent), a 10 × 10 cm-square-shaped platform was hidden at a fixed position 2 cm under the surface of the water. The temperature of the water was kept at 24–25 °C during the procedure. Each mouse (6–8 weeks old) performed four trials per day, over five consecutive days, without any prior or subsequent training. We defined a successful escape, i.e. standing on the platform, as a stop for more than 1 s with all limbs on the platform. The cut-off time in a trial was set at 300 s. Mice that failed to reach the platform in 300 s were removed from the water, and the time needed to escape to the platform (escape latency) became 300 s. In each trial, the escape latency, the total path length needed to navigate to the platform, and the maximum swimming speed were analyzed using a video-tracking system, Smart (Panlab).
Middle Cerebral Artery Occlusion (MCAO) Model
Temporary focal ischemia was induced using the three-vessel occlusion technique as described previously (34). Briefly, under halothane-inhalation anesthesia, the left middle cerebral artery of each mouse (8–19 weeks old) was cauterized at the lateral border of the olfactory tract, and bilateral common carotid arteries were clip-occluded for 15 min followed by reperfusion. After opening the skull and subsequent cauterization of the MCA, the wound for the surgical MCA obstruction was closed within 3 min to avoid hypothermic neuroprotection against reperfusion injury (35). The rectal temperature was regulated so that it stayed within the physiological range (36.5–37.5 °C) using a temperature controller (NS-TC10, Neuroscience) during the operation. The heart rate and mean blood pressure were monitored via the tail artery using indirect blood pressure meter BP-98AW (Softron).
Regional Cerebral Blood Flow (rCBF) in the Penumbra-like Peripheral Area
The rCBF was monitored using the laser-Doppler blood flowmetry meter TBF-LN1 (Unique Medical) (34). The measurement area was set in the penumbra-like peripheral area of the ischemic region at 2 mm caudal and 1 mm dorsal to the crossover point of the left middle cerebral artery and the lateral surface of the olfactory tract. The rCBF was measured just before (control), during, and after MCAO.
Cerebral Function
Twenty-four hours or 7 days after MCAO, neurological deficits were examined according to a published scoring scale, with some modifications (35). Balance in the body trunk while being lifted by the tail was graded according to the following criteria: 0, no deficit (no twisting of the body); 1, mild deficit (asymmetric twisting tendency of the body); and 2, severe deficit (repeated asymmetric twisting of the body). Motor function of the extremities while being lifted by the tail was graded as follows: 0, no deficit (symmetrical movement of the forelimbs); 1, mild deficit (intermittent asymmetrical flexion of the forelimbs); and 2, severe deficit (continuous asymmetrical flexion of the forelimbs). The neurological deficit score (from 0 to 4) comprises the sum of the grades of the balance in body trunk and motor function of extremities.
Measurement of Infarcted Volume
Mice were perfused transcardially with heparinized PBS at 24 h or 7 days after MCAO to wash out any blood components from the brain tissue, which visualizes intraluminal blood coagulation or thrombosis formation, if any. The brain was removed and cut from the frontal tip into 1-mm-thick slices. Viable tissue was stained red with 2% 2,3,5-triphenyltetrazolium chloride followed by fixation with 4% paraformaldehyde in PBS. The infarct and total hemispheric areas of each slice were measured by tracing the borders in a computer-assisted image-analysis system, WinROOF (Mitani). To assess the total infarct volume after MCAO, an edema index was calculated by dividing the total volume of the hemisphere ipsilateral to the MCAO by the volume of the contralateral hemisphere. The infarcted volume was adjusted by dividing the volume by the edema index. The value of edema index at 7 days after MCAO could be considered 1.00, as found in our previous study (35).
Statistical Analysis
Data are presented as the means ± S.D. We used unpaired Student's t tests for comparisons within each parameter. Probability values of < 0.05 were considered statistically significant.
RESULTS
Localization of NDRG4 in Mouse Brain
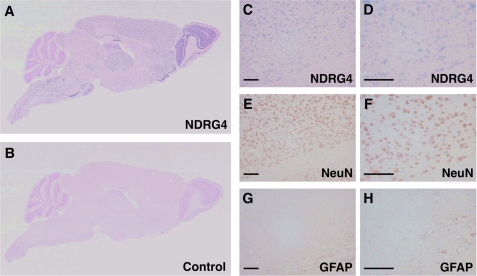
We performed in situ mRNA hybridization to investigate the cellular localization of NDRG4 in the adult mouse brain. NDRG4 mRNA was widely distributed in various parts of brain (Fig. 1, A and B), including the olfactory bulb, olfactory tuberculum, cerebral cortex, striatum, hippocampus, dentate gyrus, thalamus, hypothalamus, mesencephalon, cerebellum, pons, and medulla oblongata (supplemental Fig. S1). To identify the cell types that were positive for the NDRG4 riboprobe, we analyzed serial sections by in situ mRNA hybridization for NDRG4 (Fig. 1, C and D) in combination with immunostaining for NeuN, a marker of neurons (E and F) or GFAP, a marker of astrocytes (G and H). NDRG4 expression was mainly observed in NeuN-positive cells but not in GFAP-positive cells, indicating that NDRG4 was specifically expressed in neurons. This was consistent with our previous finding that NDRG4 protein was expressed in neurons of the cerebral cortex and Purkinje cells of the cerebellum (17).
FIGURE 1.
Localization of NDRG4 in the mouse brain. Sagittal sections were prepared from 8-week-old wild-type mice and subjected to in situ mRNA hybridization of NDRG4 (A and B). Digoxigenin-labeled antisense riboprobes for NDRG4 (A) but not sense riboprobes (B, negative control) produced positive signals (blue). The nuclei were counterstained in red. C–H, serial coronal sections of wild-type mouse brain were subjected to in situ mRNA hybridization of NDRG4 (C and D, blue signals) and immunostaining of NeuN (E and F, brown signals) or GFAP (G and H, brown signals). The nuclei were counterstained in red (C and D) and blue (E–H). D, F, and H are higher-magnification images of C, E, and G, respectively. Scale bars = 100 μm.
Generation of NDRG4-deficient Mice
To elucidate the effects of NDRG4 deficiency in neuronal cells, we generated NDRG4 knockout mice using gene targeting strategies (Fig. 2A). The Ndrg4 gene covers all NDRG4 protein isoforms, NDRG4-B, NDRG4-Bvar, and NDRG4-H (15). The genomic DNA fragment encompassing exon 6, which is the most upstream common coding region of NDRG4 isoforms, was used to construct the targeting vector. The genotype was confirmed by genomic PCR analysis (Fig. 2B). The F1 mice with one Ndrg4-disrupted allele (Ndrg4+/−) were backcrossed with wild-type C57BL/6 mice (Ndrg4+/+). Ndrg4+/− mice were then crossed to generate the NDRG4-deficient mice (Ndrg4−/−). Ndrg4−/− mice were born normally with the expected Mendelian distribution. The numbers of Ndrg4+/+, Ndrg4+/−, and Ndrg4−/− live births were 56, 115, and 51, respectively (p = 0.77, chi-square test). Both male and female Ndrg4−/− mice were fertile.
FIGURE 2.
Generation of NDRG4-deficient mice and expression pattern of NDRG family proteins in mouse organs. A, targeting strategy for the Ndrg4 gene knockout. Solid boxes represent open reading frames of Ndrg4. The loxP-flanked (open triangles) pSTneoB cassette with a polyadenylation signal (STPro Neo pA) was inserted into exon 6, and the diphtheria toxin A fragment cassette (DT-A) was included at the 3′ end of the vector. The 5′- and 3′-external probes used for Southern blotting selection of ES clones are shown by bars. The PCR primers (P1, P2, and P3) for genotyping are shown by arrows. S, SalI; H, HindIII; E, EcoRI; V, EcoRV. B, genotyping of wild-type (Ndrg4+/+), heterozygous NDRG4-deficient (Ndrg4+/−), and homozygous NDRG4-deficient (Ndrg4−/−) mice. PCR amplification of the Ndrg4+/+ and Ndrg4−/− alleles resulted in products of 171 and 246 bp, respectively. C, expression patterns of NDRG family proteins in mouse organs. Equal protein amount of organ homogenates from 17-week-old Ndrg4−/− and Ndrg4+/+ mice were subjected to Western blotting analysis using each antibody. Anti-NDRG4 detected NDRG4-B (38 kDa), NDRG4-Bvar (39 kDa), and NDRG4-H (41 kDa) in the Ndrg4+/+ brain but not in the Ndrg4−/− brain. The expression of NDRG1, NDRG2, and NDRG3 was not affected by the lack of NDRG4 in the brain.
NDRGs Expression in Mouse Organs
To examine the expression patterns of NDRG family proteins in Ndrg4+/+ and Ndrg4−/− mice, we performed a Western blotting analysis of their organs as adults. NDRG4 was specifically expressed in the brain, and little or no signal was detected in other tissues (Fig. 2C). The three isoforms, NDRG4-B (38 kDa), NDRG4-Bvar (39 kDa), and NDRG4-H (41 kDa), were detectable in the brain of Ndrg4+/+ mice, whereas they were absent in the Ndrg4−/− brain. As described previously (15), NDRG1 was ubiquitously expressed in all tested organs. NDRG2 was mainly expressed in the brain, heart, and liver, with weaker expression in the kidney and colon. NDRG3 was observed in the brain, kidney, and small intestine. The expression levels of NDRG1, NDRG2, and NDRG3 in the brain were not affected by a lack of NDRG4, suggesting that there were no compensatory up-regulation mechanisms of gene expression.
Neurotrophin Levels
To investigate whether NDRG4 deficiency impacts brain function, we measured the protein levels of major neurotrophins in the brain (Fig. 3). The quantification of BDNF in the cortex homogenates revealed a significant decrease of BDNF in Ndrg4−/− (49.4 ± 18.8 pg/mg protein, n = 9) compared with Ndrg4+/+ (71.1 ± 19.5 pg/mg protein, n = 9) mice. In contrast, the levels of GDNF, NGF, NT-3, and TGF-β1 in the cortex were not significantly different between the Ndrg4+/+ and Ndrg4−/− mice (GDNF, 28.0 ± 3.0 versus 26.0 ± 3.0 pg/mg protein; NGF, 10.9 ± 1.4 versus 10.6 ± 1.7 pg/mg protein; NT-3, 7.6 ± 2.3 versus 7.7 ± 2.0 pg/mg protein; TGF-β1, 3.8 ± 1.8 versus 4.5 ± 1.7 pg/mg protein; n = 9). Therefore, we expected that abnormal regulation of BDNF protein levels may be involved in the development of the phenotypes of Ndrg4−/− mice.
FIGURE 3.
Neurotrophins in the mouse cortex. The protein levels of BDNF, GDNF, NGF, NT-3, and TGF-β1 in the cortex isolated from the Ndrg4+/+ and Ndrg4−/− mice were measured by ELISA. Data are expressed as the mean ± S.D. (n = 9). NS, p > 0.05.
Spatial Learning Ability
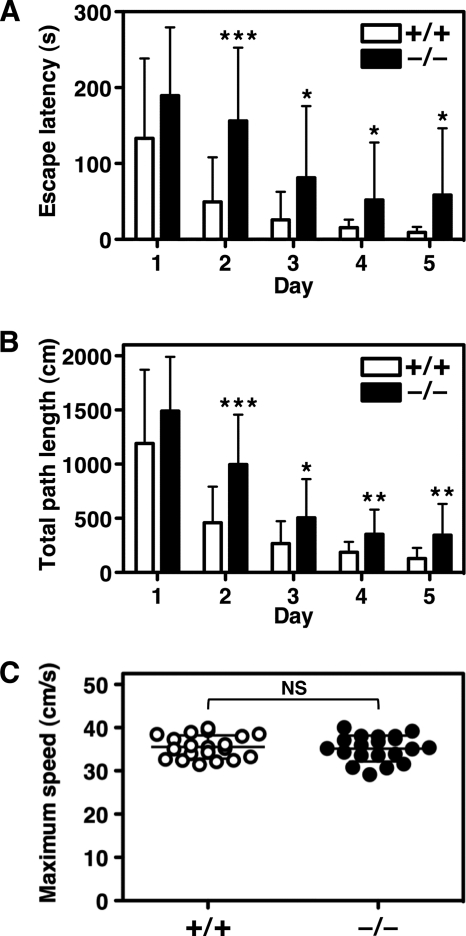
To confirm whether the lack of NDRG4 affects the ability of spatial learning and memory, we analyzed the performance of the mice in the MWM task. We found that escape latency to the hidden platform was significantly longer after the first trial for Ndrg4−/− mice compared with Ndrg4+/+ mice (Fig. 4A). The total path length needed to navigate to the platform was also significantly longer in Ndrg4−/− than in Ndrg4+/+ mice after the first trial (Fig. 4B). In contrast, there were no significant differences in the maximum swimming speed between Ndrg4+/+ (35.5 ± 2.7 cm/s) and Ndrg4−/− (35.2 ± 3.0 cm/s) mice, indicating that Ndrg4−/− mice have normal sensorimotor function (Fig. 4C). These results indicated that poor performance of Ndrg4−/− mice in the MWM test was caused by the attenuation of spatial learning ability accompanied with BDNF reduction.
FIGURE 4.
MWM test. Spatial learning and memory function of the Ndrg4+/+ and Ndrg4−/− mice were tested in the MWM task. The escape latency (A), total path length (B), and maximum swimming speed (C) to the hidden platform on trials over five consecutive days are shown. The Ndrg4−/− mice exhibited inferior performance in escape latency and total path length in the MWM task as compared with Ndrg4+/+ mice. However, the maximum swimming speed was equivalent between groups. Data are mean with error bars of S.D. (n = 20 in each experimental group). *, p < 0.05; **, p < 0.01; ***, p < 0.001; NS, p > 0.05.
Neuronal Damage after Focal Ischemia
To elucidate whether NDRG4 is involved in the neuroprotective actions of BDNF, we explored the effect of NDRG4 deficiency on the development of neuronal damage after MCAO. We first performed transcardiac perfusion of PBS 24 h or 7 days after ischemia and confirmed that there was no thrombus formation except for the coagulated point in the proximal part of the middle cerebral artery in both Ndrg4+/+ and Ndrg4−/− mice by visual inspection. A 2,3,5-triphenyltetrazolium chloride staining assay for viable cells at 24 h after a 15 min of MCAO demonstrated larger infarct lesion sizes in Ndrg4−/− mice compared with in Ndrg4+/+ mice (Fig. 5, A and B). There were no differences in the edema index between the groups (1.07 ± 0.04 in Ndrg4+/+ and 1.06 ± 0.03 in Ndrg4−/−, n = 10). Corroborating the histological results, MCAO-treated Ndrg4−/− mice showed more severe neurological deficits compared with Ndrg4+/+ mice in the cerebral function scoring test (Fig. 5C). At 7 days after the 15-min MCAO, the infarction volumes were significantly larger in Ndrg4−/− mice compared with Ndrg4+/+ mice, as seen in the acute phase study (Fig. 5B). However, differences in neurological deficit scores between Ndrg4+/+ and Ndrg4−/− mice decreased at the end of the observation period (Fig. 5C).
FIGURE 5.
Induction of temporary focal ischemia. A, representative images of six corresponding coronal sections from Ndrg4+/+ and Ndrg4−/− mouse brains at 24 h after MCAO. The 1-mm-thick slices were stained with 2,3,5-triphenyltetrazolium chloride. The sizes of infarcted region (white area) were larger in the brain slices of Ndrg4−/− mice compared with Ndrg4+/+ mice. B, quantification of infarct volumes (mm3) at 24 h and 7 days after MCAO. Ndrg4−/− mice (●) had larger infarct volumes than Ndrg4+/+ mice (○). Data are mean ± S.D. (n = 10). C, neurological deficit scored at 24 h and 7 days after MCAO. Ndrg4−/− mice had a severe neurological deficit score compared with Ndrg4+/+ mice. Data are mean ± S.D. (n = 10). D, rCBF in the penumbra-like peripheral area of the ischemic legion. The rCBF was measured by laser-Doppler blood flowmetry system. The rCBFs were expressed as percentages of their preischemic normal values. During MCAO, rCBF was reduced to an equivalent level, and reperfusion was achieved in both groups, although the rCBF values of Ndrg4−/− mice were lower compared with Ndrg4+/+ mice. Data are mean with error bars of S.D. in Ndrg4+/+ (○, n = 7) and Ndrg4−/− mice (●, n = 5). The differences in rCBF between Ndrg4+/+ and Ndrg4−/− mice at each point were not significant.
Physiological measures of Ndrg4+/+ and Ndrg4−/− mice, heart rate (24 h, 477 ± 100 versus 464 ± 95 beat/min, n = 10; 7 days, 497 ± 57 versus 483 ± 131 beat/min, n = 7 and 5, respectively) and mean blood pressure (24 h, 46 ± 8 versus 55 ± 13 mm Hg, n = 10; 7 days, 68 ± 10 versus 63 ± 25 mm Hg, n = 7 and 5, respectively) were not significantly different during ischemic treatment. These results indicate that NDRG4 is essential for the acquisition of normal resistance to the acute and chronic phase of cerebral ischemia through the retention of BDNF levels.
rCBF
We monitored the rCBF in the penumbra-like peripheral area of the ischemic legion to exclude the possibility that the larger infarct in the Ndrg4−/− brain was due to a decrease of rCBF during MCAO. Using laser-Doppler blood flowmetry, we found that rCBF values during the 15-min MCAO were equivalently reduced in Ndrg4+/+ (8.4 ± 3.2%) and Ndrg4−/− (7.1 ± 2.0%) mice (Fig. 5D). After MCAO, perfusion was observed in both groups as expected, but the rCBF values were relatively lower in Ndrg4−/− than in Ndrg4+/+ mice, which indicates that the sensitivity to ischemic stress is increased under a deficiency of NDRG4.
DISCUSSION
In this study, we revealed that NDRG4 was involved in the retaining of BDNF levels in the cortex. We also revealed that NDRG4-deficient mice showed cognitive deficits and impaired cerebral infarction tolerance. Although these phenomena in the brain seem to be physiologically distinct from each other, the abilities in learning/memory and neuroprotection are both appropriate indicators of biological activities involving BDNF (20).
BDNF participates in synaptic plasticity and memory processing in the adult brain (22). Indeed, mice that lack BDNF in their forebrain fail to learn the MWM task (21), whereas an increase in BDNF levels in the brain improves spatial learning and memory (22, 26). These observations are consistent with our findings that Ndrg4−/− mice have lower amounts of cortex BDNF than Ndrg4+/+ and impaired spatial learning and memory function. Because BDNF also increases the survivability of neurons against ischemia, decreased levels of BDNF in the Ndrg4−/− cortex can explain the enlarged lesion sizes that appeared after the stress induced by temporary focal ischemia (23–26). BDNF-mediated production of prostacyclin (36) may be associated with the neuronal vulnerabilities of the Ndrg4−/− mice because prostacyclin has a potent neuroprotective effect against focal cerebral ischemia (37). Despite the decreased levels of BDNF in the cortex of Ndrg4−/− mice, the neurological deficits were recovered at 7 days after ischemia. It needs further investigations to clarify the mechanisms of neurologic recovery in Ndrg4−/− mice. Some signaling pathways mediated by BDNF receptors such as tropomyosin-related kinase B might be up-regulated by a sustained decrease of BDNF in Ndrg4−/− mice.
The expression of NDRG4 is decreased in the brains of patients with Alzheimer's disease (15), and BDNF expression is also decreased in the cortex of Alzheimer's patients (22), observations consistent with our current finding of decreased levels of BDNF in the Ndrg4−/− mouse brain. Therefore, it is likely that NDRG4 exists upstream of the BDNF production. A decrease of NDRG4 may cause neuronal vulnerability via an associated reduction of BDNF levels and thus may be a potential contributor or a risk factor in the pathogenesis of Alzheimer's disease.
Although the molecular mechanisms by which NDRG4 influences cerebral BDNF levels are unknown, NDRG4-mediated signaling pathways may play an essential role in BDNF synthesis and secretion. BDNF secretion is dependent on the activation of voltage-gated Na+ channels and the subsequent of Ca2+ influx through voltage-gated N-type Ca2+ channels (38). In addition, BDNF release is involved in caffeine/ryanodine-sensitive Ca2+ release from intracellular stores. These findings support the idea that NDRG4 might regulate BDNF secretion via Ca2+ mobilization.
In contrast to the dysfunctional effects of NDRG4 deficiency on the central nervous system, NDRG1 deficiency results in peripheral nervous system defects. Although a brain magnetic resonance imaging study demonstrated subcortical white matter abnormalities in sibling patients with Charcot-Marie-Tooth disease type 4D (39), the lack of NDRG1 exhibited no adverse effects on higher brain functions (9) and on brain anatomy (17), suggesting that other NDRG members may compensate for the NDRG1 deficiency in the central nervous system. Similarly, the mild phenotypes of the Ndrg4−/− mice may be due to the compensatory action of the other NDRG members. Further analysis using double-knockout mice such as Ndrg1−/−Ndrg4−/− may reveal the overlapping roles of the NDRG members.
Although the NDRG4 mRNA is abundantly expressed in the human brain and heart (15, 16), Western blotting analysis in the present study could only detect the NDRG4 protein isoforms in the brain but not in the heart of the wild-type mice. This was probably due to the extremely low levels of NDRG4 protein in the heart. This unexpected finding may be caused by the low translational efficiency or the instability of NDRG4 mRNA in the heart. However, recent reports implicate biological roles of NDRG4 in the heart. The knockdown of NDRG4 during embryonic development in zebrafish results in phenotypes such as a hypoplastic heart with pericardial edema, a dilated atrium, looping defects, reduced circulation, and a slower heart rate with weaker contraction (40). Severe ventricular hypoplasia down-regulates NDRG4 expression in the mouse embryonic heart (16). These reports indicate that NDRG4 is necessary for the normal regulation of myocardial proliferation and cardiac growth during early cardiogenesis. In addition, human chromosome 16q21 near NDRG4 was identified as the locus that influences QT interval duration (41, 42). Although we currently do not find any histological and functional abnormalities for the Ndrg4−/− heart, more detailed studies may reveal the roles of NDRG4 on cardiac function.
In conclusion, we found that NDRG4 has an essential role in retaining normal spatial learning and memory, in protecting cerebral neurons against severe ischemic stress, and in maintaining BDNF levels in the brain within the normal range. Although the mechanisms by which NDRG4 influences intracerebral BDNF levels are yet unidentified, the decreased level of cortical BDNF may induce impairments in the central nervous system of Ndrg4−/− mice. Further investigation of Ndrg4−/− mice, including brain vasculature characterization and neurogenesis, may provide insight into effective therapies for some central nervous system diseases, including Alzheimer's disease and ischemic stroke.
Supplementary Material
Acknowledgment
We thank Dr. Yuka Eura for experimental materials.
This work was supported by grants-in-aid from the Ministry of Health, Labor, and Welfare of Japan; from the Ministry of Education, Culture, Sports, Science and Technology of Japan; and from the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NIBIO) of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- NDRG
- N-myc downstream-regulated gene
- BDNF
- brain-derived neurotrophic factor
- GDNF
- glial cell line-derived neurotrophic factor
- NT-3
- neurotrophin-3
- GFAP
- glial fibrillary acidic protein
- MCAO
- middle cerebral artery occlusion
- rCBF
- regional cerebral blood flow.
REFERENCES
- 1. Yao L., Zhang J., Liu X. (2008) Acta Biochim. Biophys. Sin. 40, 625–635 [DOI] [PubMed] [Google Scholar]
- 2. Schilling S. H., Hjelmeland A. B., Radiloff D. R., Liu I. M., Wakeman T. P., Fielhauer J. R., Foster E. H., Lathia J. D., Rich J. N., Wang X. F., Datto M. B. (2009) J. Biol. Chem. 284, 25160–25169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang W., Li Y., Li Y., Hong A., Wang J., Lin B., Li R. (2009) Int. J. Cancer 124, 521–530 [DOI] [PubMed] [Google Scholar]
- 4. Kitowska A., Pawelczyk T. (2010) Acta Biochim. Pol. 57, 15–21 [PubMed] [Google Scholar]
- 5. Melotte V., Qu X., Ongenaert M., van Criekinge W., de Bruïne A. P., Baldwin H. S., van Engeland M. (2010) FASEB J. 24, 4153–4166 [DOI] [PubMed] [Google Scholar]
- 6. Kokame K., Kato H., Miyata T. (1996) J. Biol. Chem. 271, 29659–29665 [DOI] [PubMed] [Google Scholar]
- 7. Taketomi Y., Sunaga K., Tanaka S., Nakamura M., Arata S., Okuda T., Moon T. C., Chang H. W., Sugimoto Y., Kokame K., Miyata T., Murakami M., Kudo I. (2007) J. Immunol. 178, 7042–7053 [DOI] [PubMed] [Google Scholar]
- 8. Kalaydjieva L., Gresham D., Gooding R., Heather L., Baas F., de Jonge R., Blechschmidt K., Angelicheva D., Chandler D., Worsley P., Rosenthal A., King R. H., Thomas P. K. (2000) Am. J. Hum. Genet. 67, 47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Okuda T., Higashi Y., Kokame K., Tanaka C., Kondoh H., Miyata T. (2004) Mol. Cell. Biol. 24, 3949–3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Drögemüller C., Becker D., Kessler B., Kemter E., Tetens J., Jurina K., Jäderlund K. H., Flagstad A., Perloski M., Lindblad-Toh K., Matiasek K. (2010) PLoS ONE 5, e11258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Takahashi K., Yamada M., Ohata H., Momose K., Higuchi T., Honda K., Yamada M. (2005) Int. J. Neuropsychopharmacol. 8, 381–389 [DOI] [PubMed] [Google Scholar]
- 12. Boulkroun S., Fay M., Zennaro M. C., Escoubet B., Jaisser F., Blot-Chabaud M., Farman N., Courtois-Coutry N. (2002) J. Biol. Chem. 277, 31506–31515 [DOI] [PubMed] [Google Scholar]
- 13. Choi S. C., Kim K. D., Kim J. T., Kim J. W., Yoon D. Y., Choe Y. K., Chang Y. S., Paik S. G., Lim J. S. (2003) FEBS Lett. 553, 413–418 [DOI] [PubMed] [Google Scholar]
- 14. Burchfield J. G., Lennard A. J., Narasimhan S., Hughes W. E., Wasinger V. C., Corthals G. L., Okuda T., Kondoh H., Biden T. J., Schmitz-Peiffer C. (2004) J. Biol. Chem. 279, 18623–18632 [DOI] [PubMed] [Google Scholar]
- 15. Zhou R. H., Kokame K., Tsukamoto Y., Yutani C., Kato H., Miyata T. (2001) Genomics 73, 86–97 [DOI] [PubMed] [Google Scholar]
- 16. Dupays L., Kotecha S., Angst B., Mohun T. J. (2009) Dev. Biol. 333, 121–131 [DOI] [PubMed] [Google Scholar]
- 17. Okuda T., Kokame K., Miyata T. (2008) J. Histochem. Cytochem. 56, 175–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ohki T., Hongo S., Nakada N., Maeda A., Takeda M. (2002) Brain Res. Dev. Brain Res. 135, 55–63 [DOI] [PubMed] [Google Scholar]
- 19. Nishimoto S., Tawara J., Toyoda H., Kitamura K., Komurasaki T. (2003) Eur. J. Biochem. 270, 2521–2531 [DOI] [PubMed] [Google Scholar]
- 20. Mattson M. P. (2008) Ann. N.Y. Acad. Sci. 1144, 97–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gorski J. A., Balogh S. A., Wehner J. M., Jones K. R. (2003) Neuroscience 121, 341–354 [DOI] [PubMed] [Google Scholar]
- 22. Nagahara A. H., Merrill D. A., Coppola G., Tsukada S., Schroeder B. E., Shaked G. M., Wang L., Blesch A., Kim A., Conner J. M., Rockenstein E., Chao M. V., Koo E. H., Geschwind D., Masliah E., Chiba A. A., Tuszynski M. H. (2009) Nat. Med. 15, 331–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schäbitz W. R., Schwab S., Spranger M., Hacke W. (1997) J. Cereb. Blood Flow Metab. 17, 500–506 [DOI] [PubMed] [Google Scholar]
- 24. Yanamoto H., Nagata I., Sakata M., Zhang Z., Tohnai N., Sakai H., Kikuchi H. (2000) Brain Res. 859, 240–248 [DOI] [PubMed] [Google Scholar]
- 25. Endres M., Fan G., Hirt L., Fujii M., Matsushita K., Liu X., Jaenisch R., Moskowitz M. A. (2000) J. Cereb. Blood Flow. Metab. 20, 139–144 [DOI] [PubMed] [Google Scholar]
- 26. Yanamoto H., Xue J. H., Miyamoto S., Nagata I., Nakano Y., Murao K., Kikuchi H. (2004) Brain Res. 1019, 178–188 [DOI] [PubMed] [Google Scholar]
- 27. Agarwala K. L., Kokame K., Kato H., Miyata T. (2000) Biochem. Biophys. Res. Commun. 272, 641–647 [DOI] [PubMed] [Google Scholar]
- 28. Takagi T., Moribe H., Kondoh H., Higashi Y. (1998) Development 125, 21–31 [DOI] [PubMed] [Google Scholar]
- 29. Yagi T., Nada S., Watanabe N., Tamemoto H., Kohmura N., Ikawa Y., Aizawa S. (1993) Anal. Biochem. 214, 77–86 [DOI] [PubMed] [Google Scholar]
- 30. Nagy A., Rossant J., Nagy R., Abramow-Newerly W., Roder J. C. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 8424–8428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kokame K., Agarwala K. L., Kato H., Miyata T. (2000) J. Biol. Chem. 275, 32846–32853 [DOI] [PubMed] [Google Scholar]
- 32. Nakajo Y., Miyamoto S., Nakano Y., Xue J. H., Hori T., Yanamoto H. (2008) Brain Res. 1241, 103–109 [DOI] [PubMed] [Google Scholar]
- 33. Morris R. (1984) J. Neurosci. Methods 11, 47–60 [DOI] [PubMed] [Google Scholar]
- 34. Yanamoto H., Nagata I., Niitsu Y., Xue J. H., Zhang Z., Kikuchi H. (2003) Exp. Neurol. 182, 261–274 [DOI] [PubMed] [Google Scholar]
- 35. Yanamoto H., Nagata I., Niitsu Y., Zhang Z., Xue J. H., Sakai N., Kikuchi H. (2001) Stroke 32, 232–239 [DOI] [PubMed] [Google Scholar]
- 36. Santhanam A. V., Smith L. A., Katusic Z. S. (2010) Stroke 41, 350–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lin H., Lin T. N., Cheung W. M., Nian G. M., Tseng P. H., Chen S. F., Chen J. J., Shyue S. K., Liou J. Y., Wu C. W., Wu K. K. (2002) Circulation 105, 1962–1969 [DOI] [PubMed] [Google Scholar]
- 38. Kolarow R., Brigadski T., Lessmann V. (2007) J. Neurosci. 27, 10350–10364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Echaniz-Laguna A., Degos B., Bonnet C., Latour P., Hamadouche T., Lévy N., Leheup B. (2007) Neuromuscul. Disord. 17, 163–168 [DOI] [PubMed] [Google Scholar]
- 40. Qu X., Jia H., Garrity D. M., Tompkins K., Batts L., Appel B., Zhong T. P., Baldwin H. S. (2008) Dev. Biol. 317, 486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Newton-Cheh C., Eijgelsheim M., Rice K. M., de Bakker P. I., Yin X., Estrada K., Bis J. C., Marciante K., Rivadeneira F., Noseworthy P. A., Sotoodehnia N., Smith N. L., Rotter J. I., Kors J. A., Witteman J. C., Hofman A., Heckbert S. R., O'Donnell C. J., Uitterlinden A. G., Psaty B. M., Lumley T., Larson M. G., Stricker B. H. (2009) Nat. Genet. 41, 399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pfeufer A., Sanna S., Arking D. E., Müller M., Gateva V., Fuchsberger C., Ehret G. B., Orrú M., Pattaro C., Köttgen A., Perz S., Usala G., Barbalic M., Li M., Pütz B., Scuteri A., Prineas R. J., Sinner M. F., Gieger C., Najjar S. S., Kao W. H., Mühleisen T. W., Dei M., Happle C., Möhlenkamp S., Crisponi L., Erbel R., Jöckel K. H., Naitza S., Steinbeck G., Marroni F., Hicks A. A., Lakatta E., Müller-Myhsok B., Pramstaller P. P., Wichmann H. E., Schlessinger D., Boerwinkle E., Meitinger T., Uda M., Coresh J., Kääb S., Abecasis G. R., Chakravarti A. (2009) Nat. Genet. 41, 407–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.