Abstract
Endorepellin, the C-terminal module of perlecan, negatively regulates angiogenesis counter to its proangiogenic parental molecule. Endorepellin (the C-terminal domain V of perlecan) binds the α2β1 integrin on endothelial cells and triggers a signaling cascade that leads to disruption of the actin cytoskeleton. Here, we show that both perlecan and endorepellin bind directly and with high affinity to both VEGF receptors 1 and 2, in a region that differs from VEGFA-binding site. In both human and porcine endothelial cells, this interaction evokes a physical down-regulation of both the α2β1 integrin and VEGFR2, with concurrent activation of the tyrosine phosphatase SHP-1 and downstream attenuation of VEGFA transcription. We demonstrate that endorepellin requires both the α2β1 integrin and VEGFR2 for its angiostatic activity. Endothelial cells that express α2β1 integrin but lack VEGFR2, do not respond to endorepellin treatment. Thus, we provide a new paradigm for the activity of an antiangiogenic protein and mechanistically explain the specificity of endorepellin for endothelial cells, the only cells that simultaneously express both receptors. We hypothesize that a mechanism such as dual receptor antagonism could operate for other angiostatic fragments.
Keywords: Basement Membrane, Cell Surface Receptor, Heparan Sulfate, Protein-Protein Interactions, Proteoglycan Structure, α2β1 Integrin, VEGFR2, Endorepellin, Heparan Sulfate Proteoglycan, Perlecan
Introduction
Angiogenesis is regulated by opposing biological activities of stimulation and inhibition initiated by vascular endothelial growth factor (VEGF)4 (1, 2) and processed forms of matrix proteins such as thrombospondin, endostatin, and endorepellin (3). Most of these activities are mediated by VEGF receptor 2 (VEGFR2), which is located at strategic domains of the endothelial plasma membrane. This specialized topology facilitates rapid and efficient signal transduction through receptor homodimerization and association with co-receptors leading to recruitment of downstream signaling molecules (4). A central mechanism for modulating cellular responses to VEGFA occurs via activation of various integrin receptors (5), which, by functionally coupling with VEGFR2, regulate developmental and pathological angiogenesis (6). For example, endothelial cell binding to vitronectin through αvβ3 integrin positively regulates VEGFR2 (7, 8), whereas expression of active αvβ3 induces VEGF secretion thereby stimulating tumor growth and angiogenesis (9). Moreover, activated αvβ3 co-localizes with VEGFR2 on tumor endothelial cells (10) and angiogenesis in transgenic mice expressing a phosphorylation-defective β3 integrin is impaired due to an inability of β3 integrin to form a functional bipartite complex with VEGFR2 (11). These results demonstrate a functional interplay between αvβ3 integrin and VEGFR2 that leads to enhancement of ligand-induced activity of this receptor tyrosine kinase upon integrin engagement (12).
Several members of the β1 integrin family have also been involved in regulating angiogenesis and VEGFR2 activity (13–16). For instance, α9β1 integrin binds VEGFA and cooperates with VEGFR2 in promoting angiogenesis (17, 18). In contrast, α1β1 integrin induces T-cell protein tyrosine phosphatase that dephosphorylates and silences VEGFR2 (19) as well as caveolin-1 (20). A functional interaction between β1 integrins and VEGFR2 plays a role in the pathogenesis of infantile hemangiomas (21) and is required for matrix-bound VEGFA signaling in endothelial cells (22). Notably, VEGFR2 associates with two thrombospondin receptors, namely CD36 and β1 integrin, and activation of VEGFR2 by VEGFA is suppressed by antiangiogenic domains of thrombospondin (23).
Genetic ablation studies have revealed specialized functions for several cell-associated heparan sulfate proteoglycans (HSPGs) that depend on their ability to coordinate functional interactions between VEGFs and their cognate receptors (24). The current view is that cell surface HSPGs potentiate the duration and magnitude of VEGFA signaling through VEGFR2, the main receptor of vascular endothelial cells, and these activities have been corroborated by studies focused on vasculogenesis and tumor angiogenesis (25, 26). Less is known about the secreted HSPGs such as perlecan, which was originally shown to bind endothelial cell surface via β1 and β3 integrins (27). In addition to being a key component of basement membranes (28) and cell surfaces (29, 30), perlecan modulates several biological processes by regulating the activity of growth factors and receptors through high affinity interactions via its N-terminal HS or its modular protein core (31–33). Perlecan (34), acts as an early-response gene that is transcriptionally induced by TGFβ1 (35, 36) and cyclic mechanical strain (37) but repressed by interferon-γ (38). Deregulated perlecan expression plays a role in cancer progression (39–42), lipid uptake (43), and vascular injury and thrombosis (44–47).
Under homeostasis, perlecan binds VEGFA via its HS chains (48) where it is released by heparanase (49) and presented to VEGFR2 for signaling. Indeed, endothelial HS regulates VEGF-induced vascular permeability (50). Notably, VEGFA induces perlecan synthesis via activation of VEGFR2 in microvascular endothelial cells (51), indicating a positive feedback loop regulating VEGFA and perlecan biosynthesis. In contrast, sustained VEGFA blockade causes a marked up-regulation of perlecan in hepatoblastoma xenografts and sustained VEGFR2 activation (52). In agreement with these findings, perlecan knockdown attenuates the proliferative response to both VEGFA and FGF2 (53–55). However, in fibrosarcoma xenografts, suppression of perlecan favors tumor growth and invasion (56), suggesting that these biological responses are cell context-specific.
We have recently discovered that endorepellin, a C-terminal angiostatic fragment of perlecan, binds α2β1 integrin (57–60) and induces the tyrosine phosphatase SHP-1, which inactivates several receptor tyrosine kinases, including VEGFR2 (61). We further discovered that perlecan and α2β1 integrin are directly involved in vertebrate embryonic angiogenesis (62, 63), and attenuation of perlecan expression leads to an abnormal accumulation and localization of VEGFA (64). These data suggest that perlecan might be directly involved in modulating the VEGFA/VEGFR2 signaling axis. In this work, we discovered that perlecan binds with high affinity to both VEGFR1 and VEGFR2 via the C-terminal domain V/endorepellin. The concerted interaction of endorepellin, α2β1 integrin, and VEGFR2 leads to a transcriptional repression of VEGFA production, thereby contributing to the antiangiogenic activity of endorepellin. Our work provides a new paradigm for antiangiogenic fragments derived from large precursors, that is “a dual receptor” antagonism. We predict that a similar bioactivity could be operational for other processed forms of angiostatic matrix molecules.
EXPERIMENTAL PROCEDURES
Antibodies, Cells, and Other Reagents
The following antibodies were used in this study: anti-GAPDH from Advanced Immunochemical (Long Beach, CA), anti-integrin α2 I-domain blocking monoclonal antibody (1998Z) from Millipore (Billerica, MA), monoclonal rabbit anti-human VEGFR2 and anti-phospho-VEGFR2 at Tyr-1175 from Cell Signaling Technology (Beverly, MA), polyclonal rabbit antibodies against VEGFR2, SHP-1, and α2 integrin from Santa Cruz Biotechnology (Santa Cruz, CA), monoclonal mouse HRP-anti-phosphotyrosine (pTyr-20) from BD Biosciences (Franklin Lakes, NJ), and mouse anti-β-actin from Sigma-Aldrich. The rabbit anti-endorepellin antibody was described previously (58). Affinity purified goat anti-endorepellin antibody (AF2364) was purchased from R&D Systems (Minneapolis, MN). Secondary antibodies were as follows: HRP-conjugated goat anti-rabbit from Jackson ImmunoResearch Laboratories (West Grove, PA), and IR680 goat anti-mouse and IR800CW goat anti-rabbit IgG from LI-COR Biosciences (Lincoln, NE). Heparin from porcine intestinal mucosa, DAPI, SIGMAFASTTM O-phenylenediamine dihydrochloride and Na3VO4 were purchased from Sigma-Aldrich, NSC87877 from Calbiochem (Gibbstown, NJ), and rat tail collagen I from BD Biosciences. Human umbilical vein endothelial cells (HUVECs) were purchased from Lifeline Cell Technology (Walkersville, MD) and used only within the first five passages. Porcine aortic endothelial cells (PAE) and their transgenic cells expressing either VEGFR1 or VEGFR2 were described previously (65, 66). Low growth factor MatrigelTM was from BD Biosciences. Recombinant human endorepellin was produced as described previously (57). Recombinant human VEGFA (VEGF165) and recombinant placenta growth factor were purchased from R&D Systems. VEGFA was also obtained from the NIH repository (also made by R&D Systems). Recombinant bacterial VEGFE derived from the Orf parapoxvirus was purchased from Angio-Proteomie (Boston, MA). The following vascular endothelial growth factor receptors were used: VEGFR1-Fc (321-FL) and VEGFR2-Fc (357-KD) (R&D Systems), VEGFR1 containing Ig-like repeats 2–7 (Ig2–7, PF082) and VEGFR2 containing Ig-like repeats 1–7 (Ig1–7, 676490, Calbiochem). We utilized two sources of human perlecan. The first was human recombinant perlecan containing a His6 tag at its C terminus (like endorepellin), which was expressed in the FreeStyleTM 293 expression system from Invitrogen as described previously (67). The second source was human perlecan immunopurified from media conditioned by coronary artery endothelial cells (68). Perlecan was purified using an affinity column containing a monoclonal antibody against domain III (69) and using a protocol as described previously (70). Both preparations behaved identically in our binding assays.
Solid-phase Binding Assays, Slot Blot, and Overlay Assays
After each treatment, cells were washed three times for 5 min with PBS and then lysed in radioimmune precipitation assay buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mm EDTA, 1 mm EGTA, 1 mm Na3VO4, 10 mm β-glycerophosphate, and protease inhibitors (1 mm phenylmethanesulfonylfluoride, 10 μg/ml leupeptin, 10 μg/ml Tosyl phenylalanyl chloromethyl ketone, and 10 μg/ml aprotinin)). Lysates were then spun at 15,000 rpm for 5 min in a 4 °C temperature-controlled centrifuge, and the protein in the supernatant was collected. Proteins were separated by SDS-PAGE and electroblotted onto nitrocellulose membranes from Bio-Rad. The blots were developed with IR-labeled secondary antibodies and detected using the Odyssey (version 2.1, LI-COR). The conditioned medium was also collected for slot blotting. The proteins on slot blot membranes were detected using HRP-conjugated secondary antibodies and chemiluminescence. ELISAs were performed following a standard protocol. The substrates, either VEGFR1 (100 ng/well) or VEGFR2 (100 ng/well), were allowed to adhere overnight at 4 °C in the presence of carbonate buffer, pH 9.6. Plates were washed with PBS, blocked with 1% BSA, and incubated for 2 h with serial dilutions of endorepellin or VEGFA. In the quantitative competition experiments, endorepellin was kept at constant concentration (20 nm) and incubated with increasing concentrations of VEGFA. In the qualitative competition experiments, endorepellin and VEGFA concentrations were varied (20 or 200 nm) to create a 10-fold excess of each ligand over the other ligand. After ligand incubation, plates were extensively washed with PBS, blocked with 1% BSA, and incubated with primary and HRP-conjugated secondary antibodies. The immune complexes were revealed using SIGMAFASTTM O-phenylenediamine dihydrochloride. Absorbance at 490 nm was measured in a Victor3TM (PerkinElmer Life Sciences). For overlay assays, VEGFR1-Fc (100 ng), VEGFR2-Fc (100 ng) and VEGFA (100 ng) were either adsorbed directly onto the nitrocellulose membrane (for slot blot) or separated on SDS-PAGE (for overlay analysis). The membranes were then overlaid with endorepellin (1 μg) for 2–3 h at 25 °C. Endorepellin bound to the VEGF receptor chimera and/or to VEGFA was then detected by probing with an antibody against endorepellin. Endorepellin bound to progranulin (50 ng) was used as a positive control (71). Binding ability of adsorbed VEGFR chimera was assessed by overlaying the membranes (with adsorbed receptor chimera) with VEGFA followed by the detection of receptor-bound VEGFA.
Immunoblotting and Immunoprecipitation
Following specified treatment, endothelial cells were rinsed twice in ice-cold phosphate-buffered saline and lysed in radioimmune precipitation assay buffer for 20 min on ice. Insoluble material was removed by centrifugation at 14,000 × g for 10 min at 4 °C. Equivalent levels of protein, determined using the DC protein assay reagent (Bio-Rad), were used. For immunoprecipitation, protein A-Sepharose magnetic beads from GE Healthcare were absorbed with antibodies for 4 h at 4 °C, and precleared cell lysates were added to the beads for 18 h at 4 °C. After extensive washing, the beads were boiled in reducing buffer, and supernatants were separated by SDS-PAGE. Proteins were then transferred to nylon membranes (Bio-Rad), probed with indicated antibodies, and developed with either enhanced chemiluminescence technique from Thermo Fisher Scientific (Waltham, MA) or IR-labeled secondary antibodies and detected with the use of either image Quant LAS 4000 (GE Healthcare) or Odyssey (version 2.1, LI-COR).
Preparation of IR800-labeled Endorepellin, Pulldown, and In-cell Binding Assays
Purified endorepellin was labeled with the IR800 dye using IRDye® 800CW labeling kit (LI-COR) according to the manufacturer's instructions. Briefly, endorepellin was mixed with dye in a molar ratio of 1:1 and kept for 2 h at 20 °C while protecting the vial from light. IRDye 800CW dye bears an N-hydroxysuccinimide ester-reactive group that couples to aliphatic amines, especially lysine residues, thereby forming stable conjugates. Free dye was removed from the labeled endorepellin by using 0.5-ml Pierce Zeba Desalting spin columns. A 10% SDS-PAGE was run to check the labeled endorepellin. For pulldown assays, protein A-Sepharose magnetic beads (GE Healthcare) were washed with TBS and absorbed with 2 μg of VEGFR1-Fc or VEGFR2-Fc in 0.1% BSA-TBS (with protease inhibitors) for 18 h at 4 °C. The beads were blocked with 1% BSA-TBS for 1 h at 25 °C, washed, and incubated for 3 h with 1 μg of IR800-labeled endorepellin at 25 °C. Following extensive washing, the beads were boiled in reducing buffer, and supernatants were separated by SDS-PAGE and scanned using Odyssey (version 2.1, LI-COR). For in-cell binding assays, confluent wild-type and VEGFR1- or VEGFR2-expressing PAE cells were serum-starved for 3 h and incubated with IR800-endorepellin (50 nm) in 0.1% BSA/DMEM for 1 h on ice and in the dark. The cells were extensively washed, fixed in 10% formaldehyde, and scanned with the Odyssey Image system at 800 nm. The cells were then washed again and incubated with the far-red fluorescent DNA dye DRAQ5TM (1:10,000) from Biostatus Limited (Leicestershire, UK) in 0.1% BSA/PBS. After washing a further three times with PBS, the cells were scanned at 700 nm, and these values were used to normalize the data.
In-cell Western and Quantitative Real-time PCR (qPCR)
For in-cell Western assays, HUVECs grown on collagen were treated with 1 μm NSC87877 (SHP-1 inhibitor) for various time intervals. After treatment, the cells were washed with ice-cold PBS and fixed in 100% methanol at −20 °C for 15 min. The cells were washed a further three times with PBS and blocked at 4 °C for 2 h with 5% BSA/PBS. Once blocked, primary antibody (rabbit anti-VEGFA) was added for 2 h at a dilution of 1:200 in 1% BSA/PBS. The cells were then washed three times with PBS. Anti-rabbit IR800-labeled secondary antibody was added for 1 h and used to visualize VEGFA levels. The values were normalized using the far-red fluorescent DNA dye DRAQ5TM (1:10,000) in 0.1% BSA/PBS. After three washes with PBS, the cells were viewed using the LI-COR Odyssey. Gene expression analysis by qPCR was carried out as described previously (72). Briefly, subconfluent (∼3 × 105 cells) six-well plates of HUVECs or PAE-VEGFR2 cells were treated with PBS (mock) or with 200 nm endorepellin for 2 or 4 h. After incubation, cells were lysed directly in 500 μl of TRIzol reagent (Invitrogen). Total RNA (1 μg) was annealed with oligo(dT)18–21 primers, and cDNA was synthesized utilizing the SuperScript Reverse Transcriptase II (Invitrogen) according to the manufacturer's instructions. Gene-specific primers for VEGFA, FGF2, and firefly luciferase were verified before use. (See supplemental Table 1 for primer sequences.) The target genes and endogenous housekeeping gene, ACTB, were amplified via independent reactions using the Brilliant SYBR Green Master Mix II (Agilent Technologies, Cedar Creek, TX). All samples were run in quadruplicate on the Mx3005P real-time PCR platform (Agilent) and cycle number (Ct) was obtained for each independent amplicon reaction. Fold change determinations were made utilizing the comparative Ct method for gene expression analysis. Briefly, Delta Ct (ΔCt) values are representative of the normalized gene expression levels (VEGFA, FGF2, and luciferase) with respect to ACTB (β-actin, endogenous housekeeping control). ΔΔCt values represent the experimental cDNA (samples treated with 200 nm endorepellin at the indicated time points) minus the corresponding gene levels of the calibrator sample (samples treated with PBS). Finally, the reported fold change represents an average of the fold changes as calculated using the double ΔCt method (2−ΔΔCT).
Immunofluorescence and Confocal Microscopy
Immunofluorescence microscopy was performed as described previously (61, 73). Approximately 5 × 104 of HUVECs or PAE cells were plated on four-well chamber slides (BD Biosciences), that were coated with gelatin from Genlantis (San Diego, CA), and grown to full confluence in 10% FBS at 37 °C. Cells were switched to serum-free medium 2 h prior to each treatment. Slides were rinsed twice with Dulbecco's phosphate-buffered saline and fixed/permeabilized with ice-cold methanol for 10 min. Subsequently, slides were subjected to standard immunofluorescence protocols and mounted with VECTASHIELD from Vector Laboratories (Burlingame, CA). PAE cells were subjected to various assays, including capillary morphogenesis in Matrigel and actin disassembly assays using rhodamine-labeled phalloidin (Sigma-Aldrich) as described previously (58, 74). Images were acquired using a LEICA DM5500B microscope with Leica Application Suite, Advanced Fluorescence software (version 1.8, Leica Microsystems, Inc., Wetzlar, Germany). For confocal microscopy, cells were washed with PBS, fixed with 4% paraformaldehyde for 15 min at room temperature, permeabilized with 0.1% Triton X-100 for 30 s, and blocked with 5% BSA. Following incubation with various primary antibodies, detection was determined using goat anti-mouse IgG Alexa Fluor® 488 and goat anti-rabbit IgG Alexa Fluor® 568 (Invitrogen), and, when appropriate, incubated with DAPI to visualize the nuclei. Slides were examined using a confocal laser scanning microscope (LSM 510 META; Carl Zeiss MicroImaging, Inc., Oberkochen, Germany) with a Neofluar 40×/1.3 oil immersion objective. Merged images represent single optical sections (< 0.8 μm), collected with the pinhole set to 1 Airy Unit for the red channel, and adjusted to give the same optical slice thickness in the green and blue channels. Images were acquired in single confocal planes to more precisely determine co-localization. Image processing and analysis were done using Zeiss LSM 510 software (version 3.2). Z-Stacks were acquired using a 63× oil objective of an Olympus IX70 microscope. Filters were set to 488 and 568 nm for dual channel imaging, and Z stacks were acquired at 0.36-μm intervals. All the images were analyzed using Laser Sharp 2000 in conjugation with ImageJ software (NIH, Bethesda, MD) and Adobe Photoshop CS3 (Adobe Systems, San Jose, CA).
VEGFA Promoter Luciferase Assays
PAE-VEGFR2 cells were stably transfected with a reporter plasmid containing a 2.6 Kb genomic fragment encompassing the human VEGFA promoter, from −2361 to +298 relative to the transcriptional start site cloned upstream of the firefly luciferase (75). About 8 × 106 cells were co-transfected with pcDNA3.1/hygromycin at a ratio of 20:1 for antibiotic selection. We isolated several positive clones as well as mass culture of PAE-VEGFR2VEGF-Luc cells using 500 μg/ml of hygromycin B. Stable cells were serum-starved for 2 h and then treated individually or in combination with 100 nm endorepellin or 50 ng/ml of VEGFA for various time points. Luciferase activity was assayed utilizing the Dual-Luciferase Reporter Assay System from Promega (Madison, WI) and normalized to cell number.
Quantification and Statistical Analysis
Immunoblots were quantified by scanning densitometry using ImageJ software or using Odyssey software for the infrared-labeled secondary antibodies. Significance of differences was determined by unpaired Student's t test using SigmaPlot (version 11.0) and SigmaStat for Windows (version 3.10; Systat Software, Inc., Port Richmond, CA). Mean differences were considered statistically significant at p < 0.05.
RESULTS
Endorepellin Inhibits VEGFA Production in Endothelial Cells
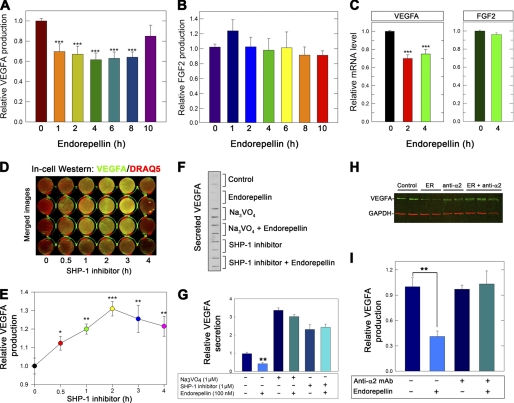
Based on the profound antiangiogenic activity of endorepellin, we hypothesized that endorepellin could modulate endogenous VEGFA, a major proangiogenic factor. Thus, we measured intracellular VEGFA protein levels after exposing HUVECs to endorepellin (100 nm) for various time points. We found a time-dependent and highly significant inhibition of endogenous VEGFA production in endothelial cells (p < 0.001, Fig. 1A), which lasted for up to 8 h before returning to the base line. Notably, under the same experimental conditions, we found no change in endogenous FGF2 (Fig. 1B), indicating that endorepellin was specifically affecting VEGFA levels. In agreement with these findings, qPCR utilizing cDNAs synthesized from the reported time points revealed a significant down-regulation in the expression of VEGFA (p < 0.001, Fig. 1C, left panel) at both 2 and 4 h of endorepellin treatment relative to vehicle-treated (PBS) cells. In contrast, FGF2 transcripts did not appreciably change at 2 h (data not shown) and 4 h (Fig. 1C, right panel). Collectively, these data demonstrate a role for endorepellin as a negative regulator of the VEGFA locus, further substantiating its role as a potent antiangiogenic molecule and linking the C terminus of perlecan protein core to the VEGFA/VEGF receptor axis.
FIGURE 1.
Endorepellin evokes VEGFA down-regulation mediated by α2β1 integrin and SHP-1. A and B, quantification of intracellular VEGFA and FGF2 levels from total HUVEC lysates stimulated with 100 nm endorepellin for 0–10 h as indicated. The values were obtained by immunoblotting and quantified using the Odyssey (LI-COR). C, qPCR of VEGFA and FGF2 transcript levels in response to endorepellin for the specified periods of treatment. Data represent the average fold changes normalized to ACTB (β-actin) ± S.E. (n = 10), ***, p < 0.001. D, in-cell Western assay of HUVECs grown on collagen and treated with the SHP-1 inhibitor NSC87877 (1 μm) for 0–4 h before being fixed. VEGFA levels were determined by immunoblotting with anti-VEGFA followed by IR800-labeled secondary antibody (green) and were normalized to DNA labeled with the far-red fluorescent DNA dye DRAQ5TM (red). Note that merged images produce a yellow color when VEGFA increases with time upon blocking SHP-1 with NSC87877. E, quantification of VEGFA levels as described in D using the Odyssey Imaging system (LI-COR). F, slot blot of secreted VEGFA from HUVECs treated with endorepellin for 6 h. Where indicated, cells were pretreated for 1 h with Na3VO4 (1 μm) or SHP-1 inhibitor (1 μm). G, quantification of secreted VEGFA as described in F normalized on total cell number. H, immunoblot of HUVEC lysates treated with endorepellin (100 nm) and α2β1 blocking antibody (10 μg/ml, mAb 1998Z, Millipore) either alone or in combination. In the latter case, HUVECs were preincubated for 1 h with the blocking antibody before the addition of endorepellin. The lysates were then probed with an anti-VEGFA antibody. I, quantification of VEGFA levels as described in panel H. The values for A and B and D–I represent the mean ± S.E. from four independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Endorepellin-evoked Down-regulation of VEGFA Is Associated with SHP-1 and Requires α2β1 Integrin
We have shown recently that endorepellin evokes an integrin α2β1-mediated activation of the tyrosine phosphatase SHP-1, found to be physically associated via the cytoplasmic domain of the α2 integrin subunit. This interaction leads to a dynamic activation of SHP-1 and dephosphorylation of various receptor tyrosine kinases, including VEGFR2 (61). Thus, we tested the effects of a SHP-1 inhibitor (NSC87877, 1 μm) (76) on VEGFA production using in-cell Western assay and quantification of the infrared signal normalized on DNA content, as determined by DRAQ5TM staining of nuclear DNA. Theoretically, a phosphatase inhibitor should prolong VEGFA levels and proangiogenic signaling by indirectly increasing VEGFA production due to a positive feedback loop on receptor tyrosine kinases. We discovered a significant enhancement of endogenous VEGFA levels evoked by the SHP-1 inhibitor, and these changes lasted for 4 h with a maximum at 2 h (Fig. 1, D and E). We note that NSC87877 is a dual inhibitor of SHP-1 and SHP-2 activity (76). However, we have shown that siRNA-mediated knockdown of SHP-2 in endothelial cells did not dampen the response to endorepellin in contrast to siRNA-mediated SHP-1 knockdown (61).
If endorepellin was acting through activation of SHP-1 or another Tyr phosphatase to down-regulate the production and secretion of VEGFA, then both global and SHP-1-specific inhibitors should recover VEGFA production, even in the presence of endorepellin. Thus, we determined the levels of secreted VEGFA using slot blot assays of media conditioned by endothelial cells treated with endorepellin in the presence or absence of Na3VO4, a general Tyr phosphatase inhibitor, or the SHP-1 inhibitor NSC87877. We found a marked suppression of VEGFA secretion by endorepellin and block of endorepellin-mediated effects by both Na3VO4 and the SHP-1 inhibitor (Fig. 1F). A similar, albeit less dramatic recovery was obtained with intracellular levels of VEGFA (data not shown). Quantification of four independent experiments showed not only the two Tyr phosphatase inhibitors effectively blocked endorepellin-mediated down-regulation of VEGFA but also the overall secretion was increased 2–3-fold (Fig. 1G).
To confirm that the effects on VEGFA were mediated by the known receptor for endorepellin (73), we used a function blocking monoclonal antibody against the α2β1 integrin. The results showed a marked attenuation of endorepellin activity on endogenous VEGFA levels in endothelial cells (Fig. 1, H and I), thus confirming the requirement for the α2β1 integrin in mediating the biological activity of endorepellin.
Endorepellin Evokes a Concurrent Internalization and Down-regulation of α2β1 Integrin and VEGFR2
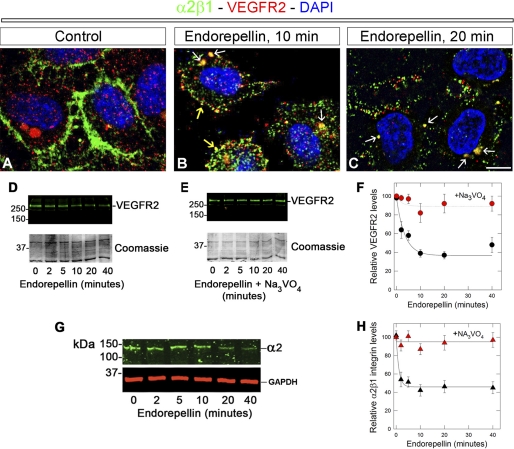
The results presented above suggest that both the α2β1 integrin and VEGFR2 could be affected by endorepellin. Therefore, we performed confocal laser microscopy on endothelial cells challenged with endorepellin. We discovered that both α2β1 integrin (green) and VEGFR2 (red) were internalized within 10 min following endorepellin treatment and co-localized in small vesicles near the inner leaflets of the plasma membrane (Fig. 2B, yellow arrows). In addition, already at 10 min, we observed larger vesicles in perinuclear regions, which became more prominent at 20 min (Fig. 2, B and C, white arrows). In contrast, untreated endothelial cells showed no basal co-localization of the two receptors (Fig. 2A). High magnification confocal microscopy using Z-stacks showed progressive co-localization of both receptors evoked by endorepellin (supplemental Fig. S1A, white arrows). In contrast to endorepellin, VEGFA evoked internalization of VEGFR2 but did not affect the α2β1 integrin (supplemental Fig. S1B). Immunoblotting analysis showed that the levels of VEGFR2 were rapidly and progressively down-regulated by endorepellin (Fig. 2D), and these effects were completely abolished by Na3VO4 (Fig. 2E). Quantification of three independent experiments showed that the levels of VEGFR2 declined rapidly (t½ ∼ 5 min, Fig. 2F). Concurrent down-regulation of the α2β1 integrin was also established (Fig. 2G), and this decline was slightly faster than that of VEGFR2 (t½ ∼ 3.5 min, Fig. 2H). Thus, endorepellin could interact with both α2β1 integrin and VEGFR2, causing rapid internalization and degradation of both receptors. These findings further strengthen the concept of dual receptor antagonism.
FIGURE 2.
Endorepellin evokes a concurrent internalization and down-regulation of α2β1 integrin and VEGFR2. A–C, representative confocal images of human endothelial cells before or after endorepellin treatment for 10 and 20 min as indicated. The cells were permeabilized with 0.1% Triton X-100 for 10 s and immunostained with an antibody against the α2 integrin subunit (green), VEGFR2 (red), or DAPI (blue). Note the progressive co-localization of α2β1 integrin and VEGFR2 within subplasmalemmal (B, yellow arrows) and perinuclear (B and C, white arrows) vesicles. All images were captured with the same exposure and gain. Bar ∼ 5 μm. D and E, immunoblotting with anti-VEGFR2 of total HUVEC lysates following endorepellin treatment in the absence (D) or presence (E) of Na3VO4. The bottom parts of the gels were stained with Coomassie Blue and serve as loading control. F, quantification of VEGFR2 levels following endorepellin treatment for the indicated time intervals in the absence (black circles) or presence (red circles) of Na3VO4. Data represent the mean ± S.E. from three experiments. G, immunoblotting with an antibody against the α2 integrin subunit or against GAPDH as indicated following treatment with endorepellin for various time intervals. H, quantification of α2β1 integrin levels following endorepellin treatment for the indicated time intervals in the absence (black triangles) or presence (red triangles) of Na3VO4. Data represent the mean ± S.E. from three experiments.
Perlecan Interacts Directly with VEGFR1 and VEGFR2 via Its C-terminal Domain V Endorepellin
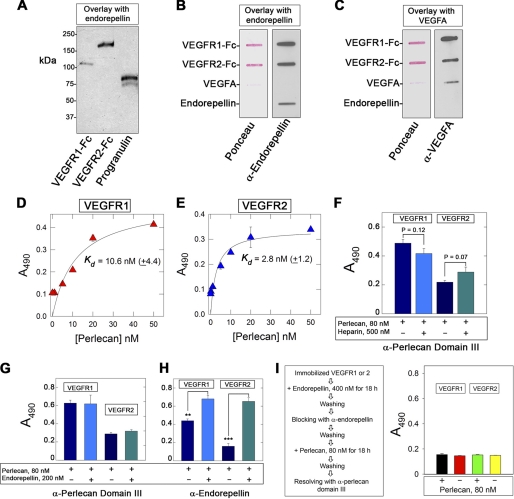
In light of the fact that VEGFR2 and α2β1 were co-down-regulated by endorepellin and due to the association of VEGFR2 with β1-containing integrins (21–23), we hypothesized that endorepellin could promote the formation of a multireceptor signaling complex by acting as a molecular “bridge” between the α2β1 integrin and VEGFR2. First, we separated by gel electrophoresis VEGFR1-Fc and VEGFR2-Fc, which contain the extracellular ligand-binding domain of either receptor fused to the Fc portion of IgG. Second, we transferred the gels to nitrocellulose membranes, and lastly, we overlaid the membranes with soluble endorepellin. Endorepellin bound both receptors (Fig. 3A) as well as progranulin, a known avid partner of endorepellin, which served as internal positive control (71).
FIGURE 3.
Perlecan interacts directly with VEGFR1 and VEGFR2 via endorepellin. A, representative SDS-PAGE overlay assay demonstrating the binding of endorepellin to VEGFR1-Fc and VEGFR2-Fc. Note that endorepellin binds also to progranulin, a known avid partner of endorepellin. Following SDS-PAGE, the gels were electroblotted into a nitrocellulose membrane, blocked with 5% BSA, overlaid with endorepellin (∼12 nm), and incubated with an antiendorepellin antibody followed by an HRP-conjugated secondary antibody, and detected by chemiluminescence. B and C, representative slot blot overlay assays demonstrating that endorepellin binds to both VEGFR1-Fc and VEGFR2-Fc but not to VEGFA. The slot blots were incubated with either antiendorepellin or anti-VEGFA antibodies as indicated, followed by HRP-conjugated secondary antibody and detection by chemiluminescence. D and E, ligand-binding assays using human recombinant perlecan as soluble ligand and the ectodomains of VEGFR1 or VEGFR2 as immobilized substrates. F, ligand-binding assays using human perlecan as soluble ligand and the ectodomains of VEGFR1 or VEGFR2 as immobilized substrates. Note that perlecan cannot be displaced by excess molar amounts of heparin. G and H, ligand-binding assays using human perlecan or endorepellin as soluble ligands and the ectodomains of VEGFR1 or VEGFR2 as immobilized substrates. The reactions were visualized with the antibodies listed at the bottom. I, schematic diagram showing the outline of the experimental protocol (left panel) and data from ligand binding assays (right panel). Note that binding of perlecan to either receptor is abrogated by blocking endorepellin with antiendorepellin rabbit polyclonal antiserum. In D–I, the values represent the mean ± S.E. from three to four experiments. **, p < 0.01; ***, p < 0.001.
To verify the novel interaction between endorepellin and VEGFR1/2, we utilized slot-blot assays in which the receptors are not denatured as in the overlay assay shown above. We also utilized VEGFA and endorepellin as bound substrates. In these assays, endorepellin specifically bound to both VEGFR1-Fc and VEGFR2-Fc (Fig. 3B), and VEGFA bound to both receptors as expected (Fig. 3C). We further found that there was no binding of endorepellin to VEGFA (Fig. 3, B and C) in agreement with our previous results (64).
To establish whether the parent HSPG perlecan could also bind VEGFR1 and VEGFR2, we utilized human perlecan in solid-phase binding assays. To avoid the possibility of nonspecific binding of perlecan to the Fc fragment of the chimeric receptors, we used, as immobilized substrates, recombinant ectodomains of VEGFR1 (Ig2–7) and VEGFR2 (Ig1–7). These soluble ectodomains contain the ligand-binding sites for VEGFA, that is, Ig2–3. We discovered that perlecan bound to both receptors with relatively high affinity (Kd = 2.8 to 10.6 nm, Fig. 3, D and E). Notably, the perlecan bound to either receptor ectodomain could not be displaced by a 6-fold molar excess of heparin (Fig. 3F).
Next, we determined whether endorepellin could compete out perlecan binding. To distinguish between the C-terminal domain V/endorepellin and whole perlecan, we employed a mouse monoclonal antibody specific for domain III of perlecan protein core (69) or a rabbit anti-perlecan domain V (58), which recognizes both perlecan and endorepellin. Notably, similar amounts of perlecan bound to either receptor in the presence or absence of a molar excess of endorepellin when detected with the anti-domain III antibody (Fig. 3G). However, an additive effect was noted when the same experiments were repeated using the anti-domain V/endorepellin (Fig. 3H). These findings raised the possibility that other regions of perlecan protein core could also mediate a specific binding to VEGFR1 or VEGFR2. An alternate possibility is that perlecan could interact with endorepellin bound to the ectodomains of the receptors, a notion supported by the fact that perlecan has a tendency to self-assemble into dimeric and oligomeric forms via their C termini (77). To test this hypothesis, we incubated for 18 h the receptors with molar excess of endorepellin (400 nm), blocked the bound endorepellin with the rabbit anti-endorepellin polyclonal antibody, and then incubated the complexes with perlecan. Following an additional 18-h incubation, the plates were reacted with the monoclonal anti-domain III antibody (Fig. 3I, left panel). The results showed abrogation of perlecan binding (Fig. 3I, right panel). An unrelated mouse monoclonal Ig was incapable of blocking perlecan binding (data not shown). We conclude that perlecan utilizes its C-terminal domain V/endorepellin as the primary binding module for VEGFR1 and VEGFR2.
Detailed Analysis of Endorepellin Binding to VEGFR1 and VEGFR2
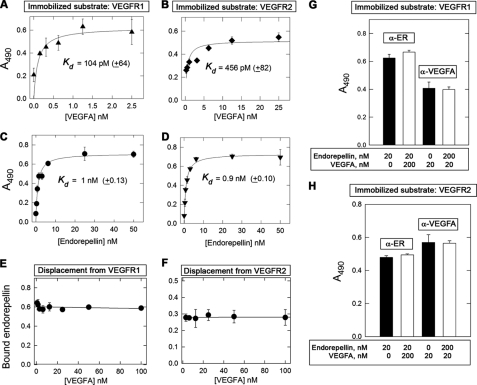
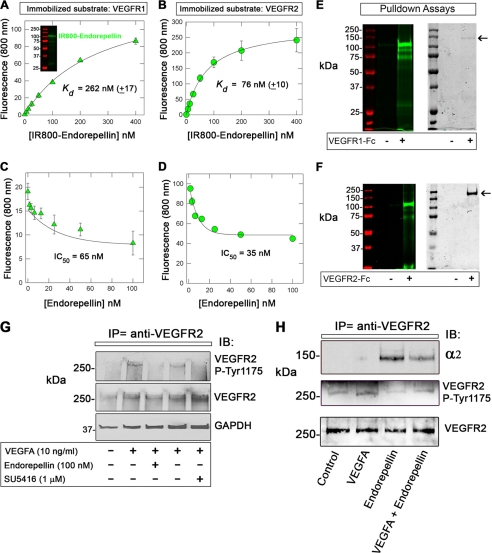
To determine in detail the binding affinity and specificity of endorepellin for VEGFR1 and VEGFR2, we performed solid-phase binding assays using immobilized ectodomains of either receptor. To validate the findings presented above, we measured the binding affinity of dimeric and fully functional recombinant VEGFA to VEGFR1/2 and found that VEGFA bound in a saturable and high affinity manner to the ectodomains of both receptors (Fig. 4, A and B). In agreement with previous studies using iodinated VEGFA and PAE or HUVECs (65), VEGFA binding to either receptor was in the picomolar range, with ∼5-fold stronger affinity for VEGFR1 as compared with VEGFR2, Kd ∼ 100 and ∼ 460 pm, respectively (Fig. 4, A and B). Using this experimental strategy, we found that endorepellin bound to both VEGFR1 and VEGFR2 in a saturable manner with a similar affinity (Kd ∼ 1 nm, Fig. 4, C and D), even higher than the parent HSPG perlecan.
FIGURE 4.
Affinity interactions between endorepellin and the ectodomains of VEGFR1/2. A–D, ligand-binding assays using endorepellin or VEGFA as soluble ligands and the ectodomains of VEGFR1 or VEGFR2 as immobilized substrates. E and F, competition experiments using constant molar amounts of endorepellin (10 nm) and increasing molar amounts of VEGFA as indicated. G and H, competition experiments using a 10-fold molar excess of endorepellin or VEGFA, as indicated. Soluble ligands were recognized with either antiendorepellin (α-ER) or anti-VEGFA (α-VEGFA). Values represent the mean ± S.E. of three experiments run in triplicate.
Specificity of binding to VEGFR2 was further proven by utilizing recombinant LG3, the terminal globular domain of endorepellin (74). Under identical experimental conditions and using the same antibody, LG3 did not bind to VEGFR1 (supplemental Fig. S2A) or VEGFR2 (data not shown). These negative controls are important because they rule out the involvement of the His6 tag in binding to the VEGFR1 and VEGFR2 and suggest that more proximal portions of endorepellin are responsible for VEGFR binding.
To eliminate the possibility that the endorepellin-VEGFR interaction might be due to a charge effect, we performed competition experiments in which a constant amount of endorepellin (100 nm) was bound to immobilized receptors and then incubated with increasing molar amounts of heparin. We found that even high concentrations (∼20-fold molar excess) of heparin did not appreciably displace endorepellin from either VEGFR1 or VEGFR2 (supplemental Fig. S2, B and C). These results indicate that the binding of endorepellin to the ectodomains of VEGFR1/2 is specific and not due to electrostatic interactions.
Next, we performed similar competition binding experiments using a molar excess of recombinant VEGFA. In three independent experiments, we found no appreciable displacement of endorepellin by VEGFA from either receptor (Fig. 4, E and F). To further corroborate these findings, we utilized recombinant VEGFE, an Orf virus VEGFA-like product that specifically binds VEGFR2 but not VEGFR1 (78). Under the same experimental conditions, a molar excess of VEGFE could not displace bound endorepellin from either VEGFR1 or VEGFR2 (supplemental Fig. S3, A and B). Analogous to the noncanonical binding to the α2β1 integrin (73), the binding of endorepellin to VEGFR1 or VEGFR2 was independent of the cations Ca2+ or Mg2+ (supplemental Fig. S3, C and D).
Finally, we utilized human recombinant placenta growth factor, a member of the VEGF family, which specifically binds to the second Ig-like repeat of VEGFR1 but not to VEGFR2 (79, 80). Again, under the same conditions, a 10-fold molar excess of recombinant placenta growth factor could not displace the VEGFR1-bound endorepellin (data not shown). The lack of endorepellin displacement by VEGFA, VEGFE, and recombinant placenta growth factor indicates that endorepellin binds to a region within the ectodomain of both receptors that does not overlap with that of natural VEGFR1/2 ligands.
Endorepellin and VEGFA Bind Concurrently on Ectodomain of Either VEGFR1 or VEGFR2
The previous results raised the possibility that endorepellin and VEGFA could simultaneously bind on the ectodomains of VEGFR1 and VEGFR2 in a noncompetitive fashion. To test this hypothesis, we performed binding experiments using a low concentration (20 nm) of either endorepellin or VEGFA and incubated together with a 10-fold molar excess (200 nm) of either ligand. At the end of the co-incubation, the wells were exposed to either antiendorepellin or anti-VEGFA antibodies. Notably, neither ligand was capable of competing with each other in receptor binding, and both bound to VEGFR1 and VEGFR2 in a similar fashion (Fig. 4, G and H). We conclude that endorepellin and VEGFA bind to non-overlapping regions on the VEGFR1 and VEGFR2 ectodomains.
IR800-labeled Endorepellin Binds to VEGFR1 and VEGFR2
To further investigate the novel binding of endorepellin to VEGFR1 or VEGFR2 and to avoid the possibility that a nonspecific antibody/antigen reactions could induce false positive results, we established a novel in vitro binding assay devoid of antibodies. First, we labeled endorepellin with the infrared dye IRDye® 800CW and monitored IR800-endorepellin by gel electrophoresis. IR800-endorepellin was properly labeled and migrated at ∼100 kDa (Fig. 5A, inset), as expected for a protein of ∼85 kDa conjugated with several dye molecules. Notably, IR800-endorepellin bound in a saturable manner to both VEGFR1 (Fig. 5A) and VEGFR2 (Fig. 5B) with Kd of 262 nm and 76 nm, respectively. In contrast, IR800-endorepellin bound in a nonsaturable, linear fashion to immobilized BSA (supplemental Fig. S4A). Decreased affinity for either receptor is likely derived from the modifications caused by the covalent linkage of the infrared dye. That is, the binding of N-hydroxysuccinimide residues to primary aliphatic amines such as lysine side chains might have interfered with binding. However, the bound IR800-endorepellin could be efficiently displaced by unlabeled endorepellin (Fig. 5, C and D), indicating specificity of binding. Similar to the unlabeled endorepellin, VEGFA (data not shown) and heparin could not appreciably displace IR800-endorepellin from VEGFR2 (supplemental Fig. S4B). Even a 22-fold molar excess of heparin did not appreciably displace IR800-labeled endorepellin from the immobilized VEGFR2. This is important because it excludes ionic interactions between the positively charged IR800 and negatively charged amino acid residues within the ectodomain of VEGFR2.
FIGURE 5.
Affinity interaction between IR800-endorepellin and the VEGFR1/2 and attenuation of VEGFA-evoked activation of VEGFR2 in HUVECs. A and B, ligand-binding assays using IR800-endorepellin as a soluble ligand and VEGFR1 or 2 as immobilized substrates. C and D, displacement assay using constant molar amounts of IR800-endorepellin (10 nm) and increasing molar amounts of unlabeled endorepellin as indicated. Values represent the mean ± S.E. of three independent experiments run in triplicates. E, pulldown of IR800-endorepellin utilizing VEGFR1-Fc immobilized to protein A-Sepharose magnetic beads. Note that IR800-endorepellin is specifically bound to the magnetic beads linked to the VEGFR1-Fc (+). The left panel is visualized with the Odyssey infrared image analysis system. Note that the molecular mass markers emit at ∼680 nm and are visualized in red. The right panel shows the same gel after staining with Coomassie Blue to detect the VEGFR1-Fc (arrow). F, similar experiment as described in E. Note that also in this case, IR800-endorepellin binds specifically to VEGFR2-Fc (+) (left panel) and co-localizes with the presence of VEGFR2-Fc (arrow in right panel). Notice also that VEGFR2-Fc migrates slower than VEGFR1-Fc due to its larger size (i.e. an extra Ig repeat). G, representative immunoblot of total proteins from HUVECs treated with VEGFA, endorepellin, or the VEGFR2 kinase inhibitor SU5416 as indicated. Notice that endorepellin treatment inhibits the phosphorylation of VEGFR2 at Tyr-1175 analogous to SU5416. Total VEGFR2 and GAPDH levels provide loading controls. H, representative co-immunoprecipitation studies using anti-VEGFR2 antibodies. Note that the α2 integrin subunit can be co-immunoprecipitated only in the presence of endorepellin. Induction of Tyr-1175 phosphorylation by VEGFA and blockage by endorepellin indicate that both ligands were biologically active. The experiments were repeated three times with comparable results.
To further validate the interaction of IR800-labeled endorepellin with VEGFR1 and VEGFR2, we performed pulldown assays. First, we linked the VEGFR1-Fc and VEGFR2-Fc to protein A-Sepharose magnetic beads; then, after blocking with 1% BSA, the complexes were incubated with IR800-endorepellin. The majority (>95%) of IR800-endorepellin bound to both receptors when analyzed by Odyssey (Fig. 5, E and F, left panels). The gels were then stained with Coomassie Blue and revealed that both VEGFR1-Fc and VEGFR2-Fc were efficiently pulled down by the protein A-coated magnetic beads (Fig. 5, E and F, arrows in right panels) and corresponded to the lanes where IR800-endorepellin was detected by Odyssey. We believe this is robust evidence for the specific interaction of endorepellin with VEGF receptors insofar as both receptors are vectorially bound to the protein A-beads via their C-terminal Fc portions, thereby allowing their N-terminal ligand-binding domains to be exposed to the solvent where soluble endorepellin can interact.
Endorepellin Attenuates VEGFA-evoked Activation of VEGFR2 in Human Endothelial Cells
To establish whether endorepellin would interfere with VEGFA signaling through VEGFR2, we performed immunoprecipitation studies using an anti-VEGFR2 antibody in endothelial cells treated for 10 min with VEGFA and/or endorepellin. We found that endorepellin attenuated VEGFA-evoked phosphorylation of VEGFR2 at Tyr-1175 (p < 0.05, in three independent experiments) analogous to the efficient block by the specific VEGFR2 kinase inhibitor SU5416 (Fig. 5G) (81). It is important to note that Tyr-1175 within the intracellular VEGFR2 domain is a key phosphorylation site that binds phospholipase Cγ and various adaptor proteins such as Sck and Shb, which lead to sustained downstream signaling including stimulation of cell proliferation, migration, survival, and vascular permeability (1).
In further experiments, we were able to co-immunoprecipitate the α2 integrin subunit in the presence of endorepellin but not in the presence of VEGFA (Fig. 5H). As an internal control, VEGFA was capable of inducing phosphorylation at Tyr-1175, and endorepellin counteracted this activity (Fig. 5H).
Moreover, we were able to perform the reverse co-immunoprecipitation using the anti-α2 integrin subunit. In the reciprocal experiment, using antibodies directed against the α2 integrin subunit for the immunoprecipitation step, VEGFR2 co-precipitated with the α2 integrin subunit only when endorepellin was present but not when VEGFA alone was used (supplemental Fig. S5). Thus, we conclude that endorepellin induces the recruitment of α2β1 integrin to the VEGFR2 coincident with antagonism of VEGFA activity.
Effects of Endorepellin on PAE Cells and Their Transgenic Counterparts Expressing Either VEGFR1 or VEGFR2
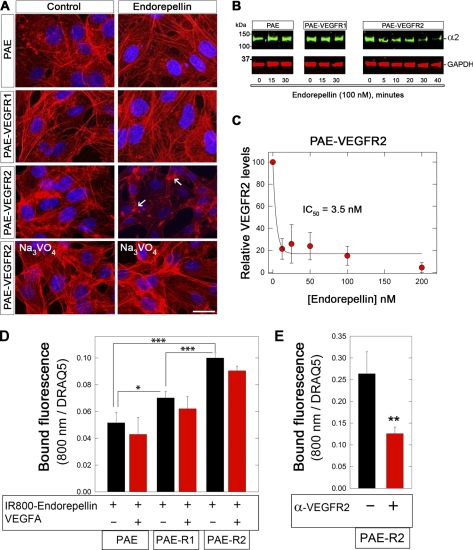
Having established a physical link for endorepellin to both the integrin α2β1 and VEGFR2 in human endothelial cells, we decided to utilize a simpler cell system in which both VEGFR1 and VEGFR2 are barely detectable, that is, the PAE and the two transgenic cells expressing either VEGFR1 or VEGFR2 (supplemental Fig. S6A). These cells have been extensively utilized for various investigations regarding the biology of VEGF/VEGFRs and co-receptors such as neuropilin-2 (65, 66, 82, 83). Initially, we determined the ability of endorepellin to evoke actin destabilization (58, 63, 73) and to block capillary-like morphogenesis on Matrigel (57). Surprisingly, endorepellin caused actin disassembly only in the PAE-VEGFR2 cells but not in the wild-type PAE or PAE-VEGFR1 cells, and these effects were blocked by Na3VO4 (Fig. 6A). Similarly, capillary-like morphogenesis was blocked by endorepellin only in the PAE-VEGFR2 cells (supplemental Fig. S6B).
FIGURE 6.
Effects of endorepellin on PAE cells and their transgenic counterparts expressing either VEGFR1 or VEGFR2. A, fluorescent images of PAE cells following staining with rhodamine phalloidin to visualize the actin cytoskeleton (red) and DAPI to visualize the nuclei (blue). Note that only PAE-VEGFR2 cells respond to endorepellin (100 nm, 30 min as seen through actin cytoskeleton disassembly (white arrows)) and this effect could be blocked by 1 μm Na3VO4. The experiments were repeated three times with similar results. Bar, 10 μm. B, representative immunoblots with antibodies against the α2 integrin subunit of total lysates from various PAE cells following treatment with endorepellin (100 nm) at various time points. Note that the α2 integrin subunit is down-regulated only in the PAE-VEGFR2 cells at 30 min with further decreases at 40 min. C, endorepellin induces down-regulation of VEGFR2 in a dose-dependent manner. PAE-VEGFR2 cells were treated with endorepellin (0–200 nm) for 10 min. D, in-cell binding assays using IR800-labeled endorepellin (50 nm) in the presence or absence of VEGFA (500 nm) on PAE, PAE-VEGFR1 (PAE-R1), or PAE-VEGFR2 (PAE-R2). The bound endorepellin fluorescence (800 nm) was normalized on far-red dye DRAQ5 (700 nm), which binds DNA. E, in-cell binding using a 10-min preincubation with a neutralizing mouse monoclonal antibody directed toward the ectodomain of VEGFR2 (10 μg/ml) followed by a 1-h incubation with IR800-endorepellin. In C–E, values represent the mean ± S.E. of three experiments run in triplicate. *, p < 0.05; **, p < 0.01.
When the three cell types were exposed to endorepellin (100 nm) for various periods of time, only the PAE-VEGFR2 showed a progressive down-regulation of the integrin α2 subunit, and thus the integrin α2β1 (Fig. 6B). Concurrently, VEGFR2 was significantly down-regulated by endorepellin (IC50 ∼ 3.5 nm, Fig. 6C) with similar kinetics (data not shown).
Next, we performed in-cell binding assays using IR800-labeled endorepellin in the presence or absence of a molar excess of VEGFA. The results showed that endorepellin could not be displaced by VEGFA (Fig. 6D) in agreement with the cell-free binding experiments shown above. Moreover, PAE cells expressing VEGFR1 and VEGFR2 bound significantly more IR800-endorepellin than PAE cells (Fig. 6D). Most importantly, preincubation of the PAE-VEGFR2 with a monoclonal antibody directed against the ectodomain of VEGFR2 significantly inhibited IR800-endorepellin binding (p < 0.01, Fig. 6E).
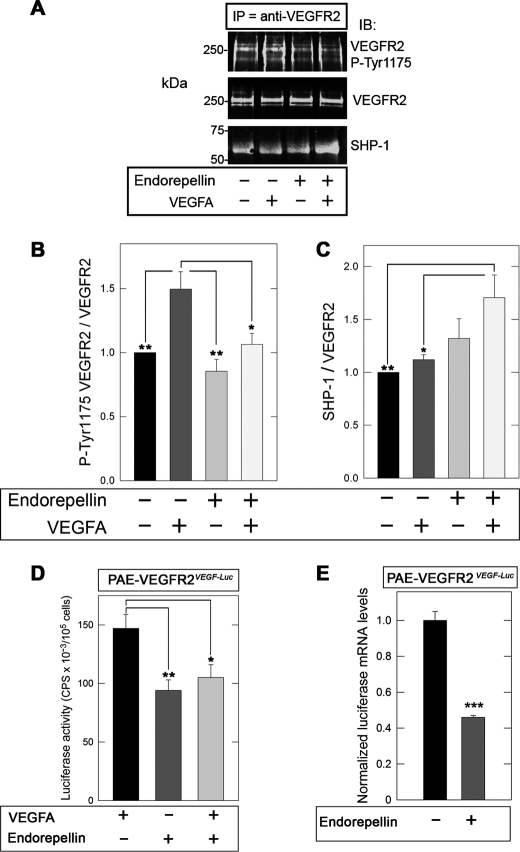
Finally, PAE-VEGFR2 responded to endorepellin in a similar fashion as HUVECs. Specifically, VEGFA-evoked phosphorylation of VEGFR2 at Tyr-1175 was markedly suppressed by co-incubation with endorepellin (Fig. 7, A and B), and this correlated with an increased recruitment of SHP-1 as detected by co-immunoprecipitation and immunoblotting (Fig. 7, A and C). Our results suggest that endorepellin requires both the integrin α2β1 and VEGFR2 to exert its angiostatic activity. The similarity of the overall response between HUVECs and PAE-VEGFR2 further strengthens this concept.
FIGURE 7.
Endorepellin attenuates VEGFR2 and VEGFA transcription in PAE-VEGFR2 cells. A, representative co-immunoprecipitation (IP) studies of VEGFR2 from PAE-VEGFR2 immunoblotted (IB) with antibodies against VEGFR2 p-Tyr-1175, total VEGFR2, or SHP-1. Note that endorepellin attenuates the ligand-mediated activation of VEGFR2 but also enhances the recruitment of SHP-1 to VEGFR2. B and C, quantification of co-immunoprecipitation studies similar to those shown in A. The values represent the mean ± S.E. of four independent experiments. *, p < 0.05; **, p < 0.01. D, transcriptional inhibition of VEGFA by endorepellin. Mass cultures of stably transfected PAE-VEGFR2VEGF-Luc cells were exposed for 4 h to VEGFA, endorepellin, or in combination. Note that endorepellin suppresses VEGFA-driven luciferase activity and counteracts the stimulatory effects of VEGFA. The values were normalized on PBS-treated controls and represent the means + S.E. from four experiments run in triplicate. *, p < 0.05; **, p < 0.01. E, qPCR trials evaluating firefly luciferase expression in PAE-VEGFR2VEGF-Luc after treatment with 100 nm endorepellin for 4 h. The values were normalized on ACTB (β-actin) mRNA levels and are representative of two trials run in quadruplicate. ***, p < 0.001.
Endorepellin Inhibits VEGFA-mediated Stimulation of VEGFA Transcription
To investigate in detail the function of endorepellin following its high affinity binding to VEGFR2, we analyzed VEGFA transcriptional regulation as an output of luciferase activity. To this end, we made stable transfectants of PAE-VEGFR2 cells using a reporter plasmid containing a 2.6-Kb genomic fragment encompassing the human VEGFA promoter, from −2361 to +298 relative to the transcriptional start site, cloned upstream of firefly luciferase (75). Mass cultures of stably transfected PAE-VEGFR2VEGF-Luc cells were exposed for 4 h to VEGFA, endorepellin, or a combination of both in serum-free conditions. Endorepellin suppressed VEGFA promoter activity and could counteract the stimulatory effects of VEGFA (Fig. 7D). In agreement with the experiments reported above for both HUVECs and PAE-VEGFR2 cells, the effect of VEGFA and endorepellin, either alone or in combination, was totally abolished by co-incubation with 1 μm Na3VO4 (data not shown). Next, we measured luciferase mRNA levels by qPCR and found that they were also significantly down-regulated following a 4-h treatment with endorepellin (p < 0.001, Fig. 7E). Finally, we measured porcine VEGFA mRNA levels in PAE- VEGFR2VEGF-Luc following a 4-h treatment with endorepellin and obtained a significant down-regulation (data not shown) similar to the transcriptional inhibition of the reporter gene described above. These combined functional and biochemical data corroborate the results presented above and strongly support the notion that endorepellin interacts with VEGFR2 to partially block VEGFA-mediated activation of the receptor and downstream induction of VEGFA transcription.
DISCUSSION
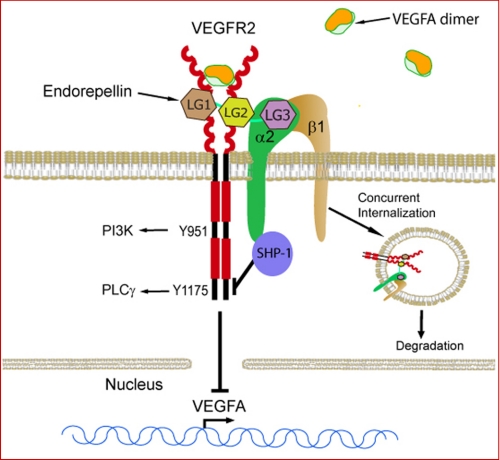
About 24 years ago, Yurchenco and co-workers (77) described the macromolecular architecture of perlecan protein core as a “flexible tandem array of globular domains” and speculated that, in analogy to other modular proteins such as fibronectin, each perlecan domain would prove in time to have its own function. The multiple functions of one of the perlecan protein core domains certainly confirm this prediction. The fact that perlecan domain V/endorepellin binds with high affinity not only cell surface receptors but also proteins involved in cancer and angiogenesis such as FGF7 (84), FGF-binding protein (85), endostatin (57), extracellular matrix protein 1 (86), and progranulin (71), point to an important biological role for this multimodular proteoglycan (87, 88). In the present study, we discovered that perlecan binds to VEGFR2 via its C-terminal domain V/endorepellin and can counteract the bioactivity of VEGFA through a portion of its protein core opposite to where the proangiogenic HS chains are located. Thus, we provide evidence for a bivalent activity of a basement membrane and pericellular HSPG that could act in a concerted manner to affect the α2β1 integrin receptor and VEGFR2, which are both key players in angiogenesis. A current working model (Fig. 8) summarizes our present findings. Endorepellin could act as an allosteric inhibitor of VEGFR2 by binding to a region different than VEGFA, which is known to bind Ig2–3. This binding likely occurs via the two proximal LG1 and LG2 domains, whereas LG3 wound bind to the α2β1 integrin. This dual receptor binding leads to rapid internalization of both receptors and degradation. On the other hand, endorepellin activates the phosphatase SHP-1, which in turn would dephosphorylate key tyrosine residues in the VEGFR2, thereby blocking prosurvival and proangiogenic downstream signaling pathways. Moreover, this dual antagonistic activity of endorepellin in attenuating both α2β1 integrin and VEGFR2 causes a transcriptional repression of VEGF gene transcription leading to reduced VEGFA mRNA and protein production and reduced VEGFA secretion by endothelial cells. The activity of endorepellin is analogous to that of TIMP-2, which also involves a SHP-1-dependent mechanism of heterologous receptor inactivation. Specifically, TIMP-2 binds to the α3β1 integrin and induces SHP-1, which in turn dephosphorylates several receptor tyrosine kinases, including VEGFR2 and FGFR1, via a mechanism that is independent of its antiprotease activity (89–91).
FIGURE 8.
Dual receptor antagonism for endorepellin. The schematic depicts our current working model. Endorepellin might act as an allosteric inhibitor of VEGFR2 by binding to a region different than VEGFA, which is known to bind Ig2–3. This binding likely occurs via the two proximal LG1 and LG2 domains, whereas LG3 wound bind to the α2β1 integrin. This dual receptor binding leads to rapid internalization of both receptors and degradation. On the other hand, endorepellin activates the phosphatase SHP-1, which in turn would dephosphorylate key tyrosine residues in the VEGFR2, thereby blocking prosurvival and proangiogenic downstream signaling pathways. Attenuation of VEGFR2 signaling also leads to a repression of VEGF gene transcription, which causes reduced VEGFA protein production and secretion by endothelial cells.
Abrogation of the Hspg2 gene in both mice (92) and zebrafish (62) causes abnormal vessel formation, particularly evident in the latter animal model where the intersegmental vessels, produced by angiogenic sprouting from the dorsal aorta, are blunted and nonfunctional. The similar phenotypes between the perlecan morphants and those with knockdown of Vegfa (93) and the α2β1 integrin (63) as well as the overlapping vascular pathology observed between the perlecan and the Plcγ1 morphants (94), a major downstream target of VEGFR2, indicate that perlecan is a key upstream component of the VEGFA/VEGFR2 signaling pathway. We discovered previously that perlecan knockdown causes a paradoxical increase in total VEGFA protein with abnormal deposits of VEGFA often away from normal vascular patterning and that the perlecan morphants could be partially rescued by microinjections of human recombinant VEGFA (64). Consistent with this hypothesis, combined administration of perlecan and VEGFA to HUVECs enhances VEGFR2 phosphorylation at Tyr-951 (64). Similarly, a soluble form of perlecan domain I harboring HS chains enhances VEGFA activity on VEGFR2 phosphorylation at Tyr-951 and downstream Akt activation (95). Collectively, these findings support a dual role for perlecan as a regulator of “proper” VEGFA localization and as a mediator of signaling through VEGFR2. Our current results support the novel hypothesis that perlecan and endorepellin evoke the recruitment of a larger signaling complex encompassing at least two transmembrane receptors and several intracellular binding partners, including, among others, SHP-1. This key tyrosine phosphatase is constitutively associated with the α2 integrin subunit in HUVECs (61) and with VEGFR2 in HUVECs and PAE-VEGFR2 cells (82, 96). Thus, SHP-1 could function as the intracellular proximal bridge between the two receptors. It is known that SHP-1 needs steric activation to remove the autoinhibiting SH-2 domain from its active site as well as the activating phosphorylation site (97). Endorepellin binding to the integrin-VEGFR2 complex could evoke this steric change through conformational changes in the integrin, whereas VEGFR2 could promote Tyr phosphorylation through its intracellular kinase domain.
In the present study, we provide detailed analysis of endorepellin binding to VEGFR1 and VEGFR2. Molar excess of VEGFA or heparin do not appreciably displace endorepellin from either receptor, indicating that binding of endorepellin to VEGFR1/2 is located in a region of the receptor ectodomain that does not overlap with VEGFA binding and that the endorepellin-VEGFR1/2 interaction is not due to electrostatic interactions. These results are important because it has been shown previously that the soluble ectodomain of VEGFR2, similar in structure to that used here, binds heparin (98) via a short stretch of basic amino acid residues located between Ig6 and Ig7 (99). Moreover, a highly basic decapeptide derived from this stretch inhibits VEGFA binding to VEGFR2 in the presence of heparin, suggesting a specific interaction rather than competition for VEGFR2 (99). As VEGFA binds to Ig2–3 of VEGFR1 (100), then endorepellin might bind to Ig4–7. However, the lack of displacement by heparin suggests that endorepellin might bind between Ig4 and Ig5. Notably, structural studies have confirmed that VEGFA by binding to Ig2–3 of VEGFR2 causes receptor dimerization and promotes further interaction via the membrane-proximal Ig7 (101), which has been recently to be critical for receptor dimerization/activation and signaling (102). Thus, endorepellin could potentially inhibit VEGFR2 dimerization as recently shown for antibodies targeting Ig4–7 (103, 104).
Another important finding of this report is that the terminal globular domain of endorepellin, LG3, previously shown to directly interact with the α2 I domain of the α2β1 integrin (58, 73), does not bind to either VEGFR1 or -2. This result suggests that endorepellin could simultaneously bind to α2β1 integrin via LG3 and VEGFR2 via LG1–LG2 moieties (Fig. 8). Thus, our results provide a plausible explanation for the selective bioactivity of endorepellin for endothelial cells as only these cells express both the α2β1 integrin receptor and VEGFR2. This is a reasonable explanation for our previous in vitro and in vivo results, which have shown that cells expressing α2β1 even at high levels, such as fibroblasts, HT1080 fibrosarcoma, or A431 squamous carcinoma cells, do not respond to endorepellin both in migration assays and in disruption of the actin cytoskeleton (57, 58, 105).
It is notable that, as for the case of other angiogenic integrins, genetic ablation of α2 integrin subunit leads to an enhanced angiogenesis in both tumor xenograft development (73, 106) and wound healing (107, 108). This anomalous phenotype could be explained in part by our previous and current findings. The lack of α2β1 integrin in mice is associated with lower levels and activity of SHP-1, which requires the intracellular domain of the α2 integrin subunit to be properly anchored in endothelial cells and liver (61). This would lead to a protracted hyperactivation state for various receptor tyrosine kinases involved in prosurvival and proangiogenic activities. This is true for melanoma tumor xenografts grown in mice lacking the α2β1 integrin. Consistent with this hypothesis, mice lacking the α2β1 integrin and harboring syngeneic melanoma tumor xenografts exhibit an increase in tumor angiogenesis and a concurrent up-regulation of VEGFR1 in the tumor endothelium (106). Moreover, lack of the α2β1 integrin could be also important because the endogenous circulating endorepellin would not be able to interact with its primary functional receptor. Indeed, endorepellin and its C-terminal LG3 fragments have been found in many body fluids, including blood, urine, and amniotic fluid (88). Endorepellin is secreted by preapoptotic endothelial cells via cathepsin L (109), whereas LG3 is found in the secretome of various tumor cells likely generated by BMP-1 (74, 110). Finally, low circulating levels of LG3 in breast cancer patients have been correlated with poor prognosis (111).
In conclusion, we provide a new paradigm for the activity of an antiangiogenic protein by demonstrating that endorepellin requires two receptors for its angiostatic activity, namely the α2β1 integrin and VEGFR2. As perlecan and endorepellin have been present for >500 million years of evolution, we speculate that a mechanism such as “dual receptor antagonism,” could be operational for other members of the extracellular matrix or blood derivatives that are specifically acting on endothelial cell homeostasis. Moreover, our findings support endorepellin as a novel antiangiogenic modality that could supplement established therapies in clinical use or in preclinical stages.
Supplementary Material
Acknowledgments
We thank A. McQuillan for excellent technical assistance, G. Purkins for help with the confocal imaging, L. Claesson-Welsh (Uppsala University) for generously providing the PAE cells, A. Nyström and Z. P. Shaik for help in the initial stages of this project, C. Y. Chuang and C. Shimono for help in purifying perlecan, D. Mukhopadhyay (Mayo Clinic) for generously providing the VEGF promoter-luciferase construct, and D. Braddock (Yale University) for valuable advice.
This work was supported in part by National Institutes of Health Grants RO1 CA39481, RO1 CA47282, and RO1 CA120975. This work was also supported by a grant from the Mizutani Foundation for Glycoscience (to R. V. I.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S6.
- VEGF
- vascular endothelial growth factor
- LG
- laminin-like globular domain of endorepellin
- VEGFR
- VEGF receptor
- HSPG
- heparan sulfate proteoglycan
- HUVEC
- human umbilical vein endothelial cell
- PAE
- porcine aortic endothelial
- SHP-1
- src homology-2 domain containing protein phosphatase 1
- qPCR
- quantitative real time PCR.
REFERENCES
- 1. Olsson A. K., Dimberg A., Kreuger J., Claesson-Welsh L. (2006) Nat. Rev. Mol. Cell Biol. 7, 359–371 [DOI] [PubMed] [Google Scholar]
- 2. Senger D. R. (2010) Mol. Biol. Cell 21, 377–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Iozzo R. V. (2005) Nat. Rev. Mol. Cell Biol. 6, 646–656 [DOI] [PubMed] [Google Scholar]
- 4. Ferrara N., Gerber H. P., LeCouter J. (2003) Nat. Med. 9, 669–676 [DOI] [PubMed] [Google Scholar]
- 5. Byzova T. V., Goldman C. K., Pampori N., Thomas K. A., Bett A., Shattil S. J., Plow E. F. (2000) Mol. Cell 6, 851–860 [PubMed] [Google Scholar]
- 6. Humphries M. J., Travis M. A., Clark K., Mould A. P. (2004) Biochem. Soc. Trans. 32, 822–825 [DOI] [PubMed] [Google Scholar]
- 7. Soldi R., Mitola S., Strasly M., Defilippi P., Tarone G., Bussolino F. (1999) EMBO J. 18, 882–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Borges E., Jan Y., Ruoslahti E. (2000) J. Biol. Chem. 275, 39867–39873 [DOI] [PubMed] [Google Scholar]
- 9. De S., Razorenova O., McCabe N. P., O'Toole T., Qin J., Byzova T. V. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 7589–7594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mahabeleshwar G. H., Chen J., Feng W., Somanath P. R., Razorenova O. V., Byzova T. V. (2008) Cell Cycle 7, 335–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mahabeleshwar G. H., Feng W., Phillips D. R., Byzova T. V. (2006) J. Exp. Med. 203, 2495–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Somanath P. R., Malinin N. L., Byzova T. V. (2009) Angiogenesis 12, 177–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Senger D. R., Claffey K. P., Benes J. E., Perruzzi C. A., Sergiou A. P., Detmar M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 13612–13617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Senger D. R., Perruzzi C. A., Streit M., Koteliansky V. E., de Fougerolles A. R., Detmar M. (2002) Am. J. Pathol. 160, 195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sweeney S. M., DiLullo G., Slater S. J., Martinez J., Iozzo R. V., Lauer-Fields J. L., Fields G. B., San Antonio J. D. (2003) J. Biol. Chem. 278, 30516–30524 [DOI] [PubMed] [Google Scholar]
- 16. Davis G. E., Senger D. R. (2008) Curr. Opin. Hematol. 15, 197–203 [DOI] [PubMed] [Google Scholar]
- 17. Vlahakis N. E., Young B. A., Atakilit A., Hawkridge A. E., Issaka R. B., Boudreau N., Sheppard D. (2007) J. Biol. Chem. 282, 15187–15196 [DOI] [PubMed] [Google Scholar]
- 18. Oommen S., Gupta S. K., Vlahakis N. E. (2011) J. Biol. Chem. 286, 1083–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mattila E., Auvinen K., Salmi M., Ivaska J. (2008) J. Cell Sci. 121, 3570–3580 [DOI] [PubMed] [Google Scholar]
- 20. Borza C. M., Chen X., Mathew S., Mont S., Sanders C. R., Zent R., Pozzi A. (2010) J. Biol. Chem. 285, 40114–40124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jinnin M., Medici D., Park L., Limaye N., Liu Y., Boscolo E., Bischoff J., Vikkula M., Boye E., Olsen B. R. (2008) Nature Med. 14, 1236–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen T. T., Luque A., Lee S., Anderson S. M., Segura T., Iruela-Arispe M. L. (2010) J. Cell Biol. 188, 595–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang X., Kazerounian S., Duquette M., Perruzzi C., Nagy J. A., Dvorak H. F., Parangi S., Lawler J. (2009) FASEB J. 23, 3368–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bishop J. R., Schuksz M., Esko J. D. (2007) Nature 446, 1030–1037 [DOI] [PubMed] [Google Scholar]
- 25. Fuster M. M., Wang L., Castagnola J., Sikora L., Reddi K., Lee P. H., Radek K. A., Schuksz M., Bishop J. R., Gallo R. L., Sriramarao P., Esko J. D. (2007) J. Cell Biol. 177, 539–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jakobsson L., Kreuger J., Holmborn K., Lundin L., Eriksson I., Kjellén L., Claesson-Welsh L. (2006) Dev. Cell 10, 625–634 [DOI] [PubMed] [Google Scholar]
- 27. Hayashi K., Madri J. A., Yurchenco P. D. (1992) J. Cell Biol. 119, 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yurchenco P. D., Amenta P. S., Patton B. L. (2004) Matrix Biol. 22, 521–538 [DOI] [PubMed] [Google Scholar]
- 29. Iozzo R. V. (1984) J. Cell Biol. 99, 403–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Iozzo R. V., Cohen I. R., Grässel S., Murdoch A. D. (1994) Biochem. J. 302, 625–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Iozzo R. V., San Antonio J. D. (2001) J. Clin. Invest. 108, 349–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Whitelock J. M., Melrose J., Iozzo R. V. (2008) Biochemistry 47, 11174–11183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Iozzo R. V., Sanderson R. D. (2011) J. Cell Mol. Med. 15, 1013–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cohen I. R., Grässel S., Murdoch A. D., Iozzo R. V. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 10404–10408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dodge G. R., Kovalszky I., Hassell J. R., Iozzo R. V. (1990) J. Biol. Chem. 265, 18023–18029 [PubMed] [Google Scholar]
- 36. Iozzo R. V., Pillarisetti J., Sharma B., Murdoch A. D., Danielson K. G., Uitto J., Mauviel A. (1997) J. Biol. Chem. 272, 5219–5228 [DOI] [PubMed] [Google Scholar]
- 37. Baker A. B., Ettenson D. S., Jonas M., Nugent M. A., Iozzo R. V., Edelman E. R. (2008) Circ. Res. 103, 289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sharma B., Iozzo R. V. (1998) J. Biol. Chem. 273, 4642–4646 [DOI] [PubMed] [Google Scholar]
- 39. Cohen I. R., Murdoch A. D., Naso M. F., Marchetti D., Berd D., Iozzo R. V. (1994) Cancer Res. 54, 5771–5774 [PubMed] [Google Scholar]
- 40. Iozzo R. V., Cohen I. (1993) Cell Mol. Life Sci. 49, 447–455 [Google Scholar]
- 41. Zhou Z., Wang J., Cao R., Morita H., Soininen R., Chan K. M., Liu B., Cao Y., Tryggvason K. (2004) Cancer Res. 64, 4699–4702 [DOI] [PubMed] [Google Scholar]
- 42. Iozzo R. V. (1998) Annu. Rev. Biochem. 67, 609–652 [DOI] [PubMed] [Google Scholar]
- 43. Fuki I. V., Iozzo R. V., Williams K. J. (2000) J. Biol. Chem. 275, 25742–25750 [DOI] [PubMed] [Google Scholar]
- 44. Nugent M. A., Nugent H. M., Iozzo R. V., Sanchack K., Edelman E. R. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6722–6727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kinsella M. G., Tran P. K., Weiser-Evans M. C., Reidy M., Majack R. A., Wight T. N. (2003) Arterioscler. Thromb. Vasc. Biol. 23, 608–614 [DOI] [PubMed] [Google Scholar]
- 46. Tran P. K., Tran-Lundmark K., Soininen R., Tryggvason K., Thyberg J., Hedin U. (2004) Circ. Res. 94, 550–558 [DOI] [PubMed] [Google Scholar]
- 47. Tran-Lundmark K., Tran P. K., Paulsson-Berne G., Fridén V., Soininen R., Tryggvason K., Wight T. N., Kinsella M. G., Borén J., Hedin U. (2008) Circ. Res. 103, 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Whitelock J. M., Iozzo R. V. (2005) Chem. Rev. 105, 2745–2764 [DOI] [PubMed] [Google Scholar]
- 49. Whitelock J. M., Murdoch A. D., Iozzo R. V., Underwood P. A. (1996) J. Biol. Chem. 271, 10079–10086 [DOI] [PubMed] [Google Scholar]
- 50. Xu D., Fuster M. M., Lawrence R., Esko J. D. (2011) J. Biol. Chem. 286, 737–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kaji T., Yamamoto C., Oh-i M., Fujiwara Y., Yamazaki Y., Morita T., Plaas A. H., Wight T. N. (2006) Biochim. Biophys. Acta 1760, 1465–1474 [DOI] [PubMed] [Google Scholar]
- 52. Kadenhe-Chiweshe A., Papa J., McCrudden K. W., Frischer J., Bae J. O., Huang J., Fisher J., Lefkowitch J. H., Feirt N., Rudge J., Holash J., Yancopoulos G. D., Kandel J. J., Yamashiro D. J. (2008) Mol. Cancer Res. 6, 1–9 [DOI] [PubMed] [Google Scholar]
- 53. Aviezer D., Iozzo R. V., Noonan D. M., Yayon A. (1997) Mol. Cell Biol. 17, 1938–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sharma B., Handler M., Eichstetter I., Whitelock J. M., Nugent M. A., Iozzo R. V. (1998) J. Clin. Invest. 102, 1599–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Savorè C., Zhang C., Muir C., Liu R., Wyrwa J., Shu J., Zhau H. E., Chung L. W., Carson D. D., Farach-Carson M. C. (2005) Clin. Exp. Metastasis 22, 377–390 [DOI] [PubMed] [Google Scholar]
- 56. Mathiak M., Yenisey C., Grant D. S., Sharma B., Iozzo R. V. (1997) Cancer Res. 57, 2130–2136 [PubMed] [Google Scholar]
- 57. Mongiat M., Sweeney S. M., San Antonio J. D., Fu J., Iozzo R. V. (2003) J. Biol. Chem. 278, 4238–4249 [DOI] [PubMed] [Google Scholar]
- 58. Bix G., Fu J., Gonzalez E. M., Macro L., Barker A., Campbell S., Zutter M. M., Santoro S. A., Kim J. K., Höök M., Reed C. C., Iozzo R. V. (2004) J. Cell Biol. 166, 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bix G., Castello R., Burrows M., Zoeller J. J., Weech M., Iozzo R. A., Cardi C., Thakur M. L., Barker C. A., Camphausen K., Iozzo R. V. (2006) J. Natl. Cancer Inst. 98, 1634–1646 [DOI] [PubMed] [Google Scholar]
- 60. Bix G., Iozzo R. A., Woodall B., Burrows M., McQuillan A., Campbell S., Fields G. B., Iozzo R. V. (2007) Blood 109, 3745–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nyström A., Shaik Z. P., Gullberg D., Krieg T., Eckes B., Zent R., Pozzi A., Iozzo R. V. (2009) Blood 114, 4897–4906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zoeller J. J., McQuillan A., Whitelock J., Ho S. Y., Iozzo R. V. (2008) J. Cell Biol. 181, 381–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. San Antonio J. D., Zoeller J. J., Habursky K., Turner K., Pimtong W., Burrows M., Choi S., Basra S., Bennett J. S., DeGrado W. F., Iozzo R. V. (2009) Am. J. Pathol. 175, 1338–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zoeller J. J., Whitelock J. M., Iozzo R. V. (2009) Matrix Biol. 28, 284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Waltenberger J., Claesson-Welsh L., Siegbahn A., Shibuya M., Heldin C. H. (1994) J. Biol. Chem. 269, 26988–26995 [PubMed] [Google Scholar]
- 66. Ito N., Claesson-Welsh L. (1999) Angiogenesis 3, 159–166 [DOI] [PubMed] [Google Scholar]
- 67. Li S., Shimono C., Norioka N., Nakano I., Okubo T., Yagi Y., Hayashi M., Sato Y., Fujisaki H., Hattori S., Sugiura N., Kimata K., Sekiguchi K. (2010) J. Biol. Chem. 285, 36645–36655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chuang C. Y., Lord M. S., Melrose J., Rees M. D., Knox S. M., Freeman C., Iozzo R. V., Whitelock J. M. (2010) Biochemistry 49, 5524–5532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Murdoch A. D., Liu B., Schwarting R., Tuan R. S., Iozzo R. V. (1994) J. Histochem. Cytochem. 42, 239–249 [DOI] [PubMed] [Google Scholar]
- 70. Whitelock J. M., Graham L. D., Melrose J., Murdoch A. D., Iozzo R. V., Underwood P. A. (1999) Matrix Biol. 18, 163–178 [DOI] [PubMed] [Google Scholar]
- 71. Gonzalez E. M., Mongiat M., Slater S. J., Baffa R., Iozzo R. V. (2003) J. Biol. Chem. 278, 38113–38116 [DOI] [PubMed] [Google Scholar]
- 72. Buraschi S., Pal N., Tyler-Rubinstein N., Owens R. T., Neill T., Iozzo R. V. (2010) J. Biol. Chem. 285, 42075–42085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Woodall B. P., Nyström A., Iozzo R. A., Eble J. A., Niland S., Krieg T., Eckes B., Pozzi A., Iozzo R. V. (2008) J. Biol. Chem. 283, 2335–2343 [DOI] [PubMed] [Google Scholar]
- 74. Gonzalez E. M., Reed C. C., Bix G., Fu J., Zhang Y., Gopalakrishnan B., Greenspan D. S., Iozzo R. V. (2005) J. Biol. Chem. 280, 7080–7087 [DOI] [PubMed] [Google Scholar]
- 75. Mukhopadhyay D., Knebelmann B., Cohen H. T., Ananth S., Sukhatme V. P. (1997) Mol. Cell Biol. 17, 5629–5639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chen L., Sung S. S., Yip M. L., Lawrence H. R., Ren Y., Guida W. C., Sebti S. M., Lawrence N. J., Wu J. (2006) Mol. Pharmacol. 70, 562–570 [DOI] [PubMed] [Google Scholar]
- 77. Yurchenco P. D., Cheng Y. S., Ruben G. C. (1987) J. Biol. Chem. 262, 17668–17676 [PubMed] [Google Scholar]
- 78. Ogawa S., Oku A., Sawano A., Yamaguchi S., Yazaki Y., Shibuya M. (1998) J. Biol. Chem. 273, 31273–32282 [DOI] [PubMed] [Google Scholar]
- 79. Park J. E., Chen H. H., Winer J., Houck K. A., Ferrara N. (1994) J. Biol. Chem. 269, 25646–25654 [PubMed] [Google Scholar]
- 80. Christinger H. W., Fuh G., de Vos A. M., Wiesmann C. (2004) J. Biol. Chem. 279, 10382–10388 [DOI] [PubMed] [Google Scholar]
- 81. Fong T. A., Shawver L. K., Sun L., Tang C., App H., Powell T. J., Kim Y. H., Schreck R., Wang X., Risau W., Ullrich A., Hirth K. P., McMahon G. (1999) Cancer Res. 59, 99–106 [PubMed] [Google Scholar]
- 82. Kroll J., Waltenberger J. (1997) J. Biol. Chem. 272, 32521–32527 [DOI] [PubMed] [Google Scholar]
- 83. Gluzman-Poltorak Z., Cohen T., Shibuya M., Neufeld G. (2001) J. Biol. Chem. 276, 18688–18694 [DOI] [PubMed] [Google Scholar]
- 84. Mongiat M., Taylor K., Otto J., Aho S., Uitto J., Whitelock J. M., Iozzo R. V. (2000) J. Biol. Chem. 275, 7095–7100 [DOI] [PubMed] [Google Scholar]
- 85. Mongiat M., Otto J., Oldershaw R., Ferrer F., Sato J. D., Iozzo R. V. (2001) J. Biol. Chem. 276, 10263–10271 [DOI] [PubMed] [Google Scholar]
- 86. Mongiat M., Fu J., Oldershaw R., Greenhalgh R., Gown A. M., Iozzo R. V. (2003) J. Biol. Chem. 278, 17491–17499 [DOI] [PubMed] [Google Scholar]
- 87. Iozzo R. V. (1994) Matrix Biol. 14, 203–208 [DOI] [PubMed] [Google Scholar]
- 88. Iozzo R. V., Zoeller J. J., Nyström A. (2009) Mol. Cells 27, 503–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Seo D. W., Li H., Guedez L., Wingfield P. T., Diaz T., Salloum R., Wei B. Y., Stetler-Stevenson W. G. (2003) Cell 114, 171–180 [DOI] [PubMed] [Google Scholar]
- 90. Seo D. W., Li H., Qu C. K., Oh J., Kim Y. S., Diaz T., Wei B., Han J. W., Stetler-Stevenson W. G. (2006) J. Biol. Chem. 281, 3711–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lee S. J., Tsang P. S., Diaz T. M., Wei B. Y., Stetler-Stevenson W. G. (2010) Lab. Invest. 90, 374–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Costell M., Carmona R., Gustafsson E., González-Iriarte M., Fässler R., Muñoz-Chápuli R. (2002) Circ. Res. 91, 158–164 [DOI] [PubMed] [Google Scholar]
- 93. Nasevicius A., Larson J., Ekker S. C. (2000) Yeast 17, 294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lawson N. D., Mugford J. W., Diamond B. A., Weinstein B. M. (2003) Genes Dev. 17, 1346–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Muthusamy A., Cooper C. R., Gomes R. R., Jr. (2010) BMC Biochemistry 11, 43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Bhattacharya R., Kwon J., Wang E., Mukherjee P., Mukhopadhyay D. (2008) J. Mol. Signal. 3, 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Poole A. W., Jones M. L. (2005) Cell Signalling 17, 1323–1332 [DOI] [PubMed] [Google Scholar]
- 98. Chiang M. K., Flanagan J. G. (1995) Growth Factors 12, 1–10 [DOI] [PubMed] [Google Scholar]
- 99. Dougher A. M., Wasserstrom H., Torley L., Shridaran L., Westdock P., Hileman R. E., Fromm J. R., Anderberg R., Lyman S., Linhardt R. J., Kaplan J., Terman B. I. (1997) Growth Factors 14, 257–268 [DOI] [PubMed] [Google Scholar]
- 100. Wiesmann C., Fuh G., Christinger H. W., Eigenbrot C., Wells J. A., de Vos A. M. (1997) Cell 91, 695–704 [DOI] [PubMed] [Google Scholar]
- 101. Ruch C., Skiniotis G., Steinmetz M. O., Walz T., Ballmer-Hofer K. (2007) Nat. Struct. Mol. Biol. 14, 249–250 [DOI] [PubMed] [Google Scholar]
- 102. Yang Y., Xie P., Opatowsky Y., Schlessinger J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 1906–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Tvorogov D., Anisimov A., Zheng W., Leppänen V. M., Tammela T., Laurinavicius S., Holnthoner W., Heloterä H., Holopainen T., Jeltsch M., Kalkkinen N., Lankinen H., Ojala P. M., Alitalo K. (2010) Cancer Cell 18, 630–640 [DOI] [PubMed] [Google Scholar]
- 104. Kendrew J., Eberlein C., Hedberg B., McDaid K., Smith N. R., Weir H. M., Wedge S. R., Blakely D. C., Foltz I., Zhou J., Kang J. S., Barry S. T. (2011) Mol. Cancer Ther. 10, 770–784 [DOI] [PubMed] [Google Scholar]
- 105. Bix G., Iozzo R. V. (2005) Trends Cell Biol. 15, 52–60 [DOI] [PubMed] [Google Scholar]
- 106. Zhang Z., Ramirez N. E., Yankeelov T. E., Li Z., Ford L. E., Qi Y., Pozzi A., Zutter M. M. (2008) Blood 111, 1980–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Grenache D. G., Zhang Z., Wells L. E., Santoro S. A., Davidson J. M., Zutter M. M. (2007) J. Invest. Dermatol. 127, 455–466 [DOI] [PubMed] [Google Scholar]
- 108. Zweers M. C., Davidson J. M., Pozzi A., Hallinger R., Janz K., Quondamatteo F., Leutgeb B., Krieg T., Eckes B. (2007) J. Invest. Dermatol. 127, 467–478 [DOI] [PubMed] [Google Scholar]
- 109. Cailhier J. F., Sirois I., Laplante P., Lepage S., Raymond M. A., Brassard N., Prat A., Iozzo R. V., Pshezhetsky A. V., Hébert M. J. (2008) J. Biol. Chem. 283, 27220–27229 [DOI] [PubMed] [Google Scholar]
- 110. Grønborg M., Kristiansen T. Z., Iwahori A., Chang R., Reddy R., Sato N., Molina H., Jensen O. N., Hruban R. H., Goggins M. G., Maitra A., Pandey A. (2006) Mol. Cell Proteom. 5, 157–171 [DOI] [PubMed] [Google Scholar]
- 111. Chang J. W., Kang U. B., Kim D. H., Yi J. K., Lee J. W., Noh D. Y., Lee C., Yu M. H. (2008) Proteomics Clin. Appl. 2, 23–32 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.