Abstract
Plexin receptors regulate cell adhesion, migration, and guidance. The Rho GTPase binding domain (RBD) of plexin-A1 and -B1 can bind GTPases, including Rnd1. By contrast, plexin-C1 and -D1 reportedly bind Rnd2 but associate with Rnd1 only weakly. The structural basis of this differential Rnd1 GTPase binding to plexin RBDs remains unclear. Here, we solved the structure of the plexin-A2 RBD in complex with Rnd1 and the structures of the plexin-C1 and plexin-D1 RBDs alone, also compared with the previously determined plexin-B1 RBD.Rnd1 complex structure. The plexin-A2 RBD·Rnd1 complex is a heterodimer, whereas plexin-B1 and -A2 RBDs homodimerize at high concentration in solution, consistent with a proposed model for plexin activation. Plexin-C1 and -D1 RBDs are monomeric, consistent with major residue changes in the homodimerization loop. In plexin-A2 and -B1, the RBD β3-β4 loop adjusts its conformation to allow Rnd1 binding, whereas minimal structural changes occur in Rnd1. The plexin-C1 and -D1 RBDs lack several key non-polar residues at the corresponding GTPase binding surface and do not significantly interact with Rnd1. Isothermal titration calorimetry measurements on plexin-C1 and -D1 mutants reveal that the introduction of non-polar residues in this loop generates affinity for Rnd1. Structure and sequence comparisons suggest a similar mode of Rnd1 binding to the RBDs, whereas mutagenesis suggests that the interface with the highly homologous Rnd2 GTPase is different in detail. Our results confirm, from a structural perspective, that Rnd1 does not play a role in the activation of plexin-C1 and -D1. Plexin functions appear to be regulated by subfamily-specific mechanisms, some of which involve different Rho family GTPases.
Keywords: Biophysics, Cell Migration, Crystallography, Neurodevelopment, Protein-Protein Interactions, Signal Transduction, Thermodynamics, Axon Guidance, Rho GTPase Rnd1
Introduction
Members of the plexin family of transmembrane receptors bind to semaphorin guidance cues and perform critical functions in axonal growth cone migration during neural development (1–3), regulate blood vessel patterning in cardiovascular development (4, 5), are responsible for invasive growth of metastatic cancer cells (6, 7), and play a role in the immune response (8, 9). Vertebrate plexins can be classified into four subfamilies (A–D). Type A plexins transduce class 3 semaphorin signaling via the neuropilin co-receptor, whereas plexin type B receptors mediate class 4 semaphorin-induced growth cone collapse in hippocampal neurons and inhibition of cellular migration in various other cell types (10, 11). Different from type A and B plexins, plexin-C1 is a receptor for semaphorins that stimulate cytokine expression, inducing a proinflammatory response in monocytes (12). In dendritic cells, plexin-C1 stimulation by the Vaccinia virus semaphorin mimic, A39R, suppresses membrane extensions that are required for phagocytosis; this impairs dendritic cell migration, ultimately limiting the ability of dendritic cells to function as efficient antigen-presenting cells (13). In contrast to subfamily A and B plexins, which are predominantly expressed in neuronal systems, plexin-D1 is detected in the vascular endothelium, as well as in neuronal systems, and plays an essential role in the formation of the vasculature during early embryogenesis (14). In endothelial cells, plexin-D1 associates with neuropilin to transduce multiple semaphorin signals (5, 15). Furthermore, plexin-D1 is expressed at high levels in a wide range of human tumor tissues (16), whereas other plexins, such as plexin-B1, function as tumor suppressors in some settings. Because the signaling pathways are diverse and additional mechanisms of plexin action probably remain to be discovered, structural characterization of plexins will elucidate the structure-function relationships and the potential of these receptors as therapeutic targets.
The four plexin subfamilies share a similar domain structure (1). The intracellular region of the plexins exhibits sequence similarity to the GTPase-activating protein (GAP)5 domain of p120 RasGAP (17, 18). The uniqueness of the GAP domain lies in that this part of plexins is split into two segments by a region of ∼200 residues. This intervening domain is overall well conserved and has been identified as the binding location of several Rho GTPases, including Rnd1 (19–21), and hereafter is referred to as the Rho GTPase binding domain (RBD).
Rho family GTPases play important roles in the regulation of the cell's actin cytoskeleton and mediate the repulsive and attractive effects of guidance molecules (22). The Rnd proteins are a distinct subgroup of Rho GTPases. Rnd1 and Rnd3 lack intrinsic GTPase activity due to amino acid substitutions at critical positions, whereas Rnd2 may have partial GTPase activity (23). It has been established that three Rho GTPases (i.e. Rac1, Rnd1, and RhoD), when in the active GTP bound form, directly associate with the RBDs of A and B family plexins (19, 21). Moreover, it has been reported recently that Rnd2 is involved in the Sema3E/plexin-D1-induced inhibition of axon outgrowth in cortical neurons (24). However, the mechanisms by which Rho GTPases regulate plexin function remain to be fully elucidated, particularly in regard to the role of the Rnd1 GTPase in plexin signaling.
Recent studies have revealed the atomic details of the entire intracellular domain of plexin-B1 and -A3 (25, 26). In both structures, the two RasGAP homologous segments are intertwined and form a compact fold, similar to that seen in other RasGAPs, whereas the RBD domain forms an independent folding unit and is linked to the core GAP domain via a so-called “coupling loop” (25). Rnd1 binding has been shown to be essential for the GAP activity of plexin-A1 and -B1 (15, 17, 19), suggesting the importance of the RBD domain and its interaction with Rho family GTPases.
Here we report the structures of the RBDs of plexin-C1 and plexin-D1 as well as that of plexin-A2 in complex with Rnd1. The analysis of the binding interfaces of the plexin-A2 and also of the previously published plexin-B1 RBD·Rnd1 complex and the structure-based alignment of other plexin RBDs reveal the primary factors in the protein-protein interaction between plexin RBDs and Rnd1. These results provide insight into the specificity of Rnd1 binding and suggest a differential use of this GTPase in the activation mechanism of plexins.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The genes encoding the RBDs of human plexin-A2 (residues 1490–1600), plexin-B1 (residues 1746–1852), plexin-C1 (residues 1198–1305), plexin-D1 (residues 1553–1678), and Rnd1 GTPase (residues 5–200) and Rnd2 (residues 1–227) were amplified by PCR and subcloned into the expression vector pET28-MHL, which encodes an N-terminal His6 tag followed by the tobacco etch virus protease cleavage site. All recombinant proteins were overexpressed in Escherichia coli strain BL21(DE3) (Stratagene) upon overnight induction with 1.0 mm isopropyl 1-thio-β-d-galactopyranoside at 16 °C. Proteins were purified using nickel-NTA affinity chromatography and gel filtration chromatography using Superdex 75 (GE Healthcare). Rnd1 and Rnd2 GTPases were loaded with non-hydrolyzable nucleotide by overnight incubation with 5 mm GMPPNP. Protein samples were stored at 4 °C in gel filtration buffer.
Crystallization
Proteins were crystallized using the sitting drop vapor diffusion method. Equal volumes (0.5 μl) of protein solution and reservoir solution were mixed for crystallization. Plexin-A2 RBD and Rnd1 were mixed in a 1:1 ratio in the buffer used for protein purification. The crystal of the complex grew in 25.5% polyethylene glycol 3350 with 0.2 m MgCl2 and 0.1 m HEPES at pH 7.5. Plexin-C1 RBD was crystallized using an in situ proteolysis method (27). A 0.5-μl protein sample containing chymotrypsin (1:100, w/w) was mixed with 0.5 μl of well solution containing 0.1 m HEPES (pH 7.5), 2.0 m (NH4)2SO4, and 0.2 m NaCl. The plexin-D1 RBD was overexpressed in E. coli using a prepacked M9 selenomethionine growth medium kit (Medicilon) and the N-terminal His6 tag was removed by tobacco etch virus protease cleavage before crystallization. The crystal of the plexin-D1 RBD was grown in 0.1 m sodium cacodylate buffer (pH 5.2) containing 1.39 m sodium citrate and 15 mm tris(2-carboxyethyl)phosphine, using the in situ proteolysis method with chymotrypsin (1:100, w/w). All crystals grew to mountable size in 3–5 days.
Data Collection and Structure Determination
Diffraction data were collected at beam line 19-ID at the Advanced Photon Source (Argonne, IL) with a Quantum-315 CCD detector (ADSC) and reduced with the HKL3000 suite (28). The structure of the plexin-A2 RBD·Rnd1 complex was solved by molecular replacement with MOLREP (29) using models of Rnd1 (PDB code 2REX) (25) and murine plexin-A3 RBD (PDB code 3IG3) (26) as starting models. Initial phases for the plexin-D1 structure were obtained by single-wavelength anomalous diffraction, using the programs SHELX (30), MLPHARE (31), and DM (32) through the HKL3000 interface. The structure of plexin-C1 was solved by molecular replacement with the program PHASER (33) and a composite search model of plexin-D1 (described here) and the mouse plexin-A3 RBD coordinates (26) (PDB code 3IG3). ARP/wARP was used for automated model building, PHENIX (34) and REFMAC (35) for coordinate/B-factor refinement, COOT (36) for manual model adjustments, and MOLPROBITY (37) for validation of geometry.
Analytical Gel Filtration Assay
Analytical gel filtration was performed at 4 °C using a Superdex75 10/300 GL column (GE Healthcare). The column was run in 20 mm Tris/HCl, pH 7.5, 0.5 m NaCl, 5 mm MgCl2, and 5% (v/v) glycerol at a flow rate of 0.5 ml/min. GMPPNP (5 mm) was added to all of the protein samples for internal reference and binding factor. Each injection contained a 200-μl protein sample with a concentration of ∼5 mg/ml, and the protein absorbance was monitored at 280 nm. For gel filtration to study the dimerization behavior of the plexin-A2 RBD, we used 500 μl of protein at concentrations up to 3 mm. The protein absorbance of the concentrated samples was followed at 254 nm.
Isothermal Titration Calorimetry
Binding interactions of plexin RBDs and Rnd1 or Rnd2 were performed at 25 °C using a MicroCal iTC200 microcalorimeter. The experiments were carried out in 50 mm phosphate buffer, pH 7.0, 50 mm NaCl, 4 mm MgCl2, 0.5 mm tris(2-carboxyethyl)phosphine. The Plexin RBDs were kept in the syringe (volume 40 μl, concentration 400 μm), whereas the Rnd1 or Rnd2 was placed in the cell (volume 250 μl, concentration 40 μm). The interval between injections was 120 s. The data were analyzed in Origin 7.0, and the binary equilibrium association constant (Ka = 1/Kd) was fitted (where Kd is equal to [GTPase][plexin]/[GTPase-plexin complex]).
In order to confirm the differences in binding affinity for key interactions, we used a second technique, plasmon surface resonance. The method and data are shown in the supplemental material.
RESULTS
Structure of the Plexin-B1 RBD in Complex with Rnd1
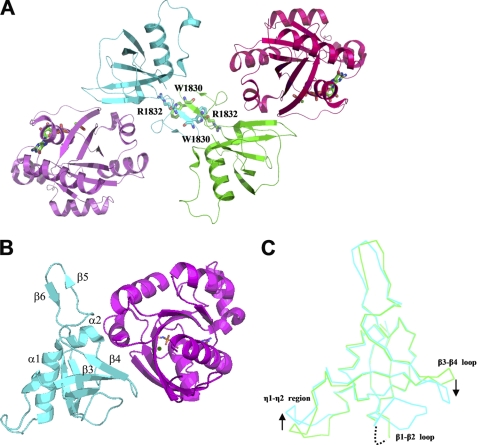
We previously solved and reported the structure of the plexin-B1 RBD in complex with Rnd1 at a resolution of 2.3 Å (PDB code 2REX) (25). However, the interface had not been analyzed. The plexin-B1 RBD in the complex forms a homodimer by the C2 symmetric antiparallel β-sheet (β5-β6), with a π-cation interaction between Trp1830 and Arg1832, in an asymmetric unit (Fig. 1A). A similar homodimerization interface has been observed in the structure of the Rnd1-free form of plexin-B1 RBD (21).
FIGURE 1.
Structures of the complex of plexin-B1 RBD and Rnd1. A, ribbon diagram of the plexin-B1 RBD·Rnd1 complex homodimer/heterotetramer. The asymmetric unit consists of plexin-B1 RBD chains A (cyan) and C (green) (residues 1746–1852), forming a homodimer, and Rnd1 chains B (violet) (residues 13–188) and D (hot pink) (residues 7–188), bound on opposite sides to the RBD dimer. The homodimer interface is stabilized by a π-cation interaction between Trp1830 and Arg1832 (21). B, one unit of the plexin-B1 RBD (cyan) and Rnd1 (violet) complex. Several of the β strands (β1–β7) and two α-helices (α1 and α2) are labeled in plexin-B1 RBD. C, superimposition of the RBDs of the plexin-B1 RBD·Rnd1 complex (cyan) with the free plexin-B1 RBD (green) illustrates conformational changes that take place in the β1-β2 loop, the β3-β4 loop, and the η1-η2 region upon complex formation with Rnd1.
Rnd1 binding to the plexin-B1 RBD appears to promote ordering of a flexible region, the loop β3-β4, in the plexin-B1 RBD-Rnd1 interface in the crystal (Fig. 1, B and C). In the free RBD, the B-factors in this loop are ∼20% above those seen for Cα atoms in the regular secondary structure. In the bound form, they are similar to those of the α-helices and β-strand regions, whereas a rotational movement of the β3-β4 loop appears to create a surface for optimal Rnd1 binding. The RBD β1-β2 loop, which is not directly involved in the binding of Rnd1, was also ordered in the crystal. By contrast, in the free form of the plexin-B1 RBD, the corresponding loop is disordered (Fig. 1C). However, NMR data and molecular dynamics calculations on RBD-GTPase complexes suggest that this loop remains highly flexible in solution (38).6 The conformation of a region on the opposite side of the RBD that contains the 310-helices η1 and η2 was pushed away from the main structure, although, by comparison with other RBD structures, this movement may be independent of the ordering of the two loop regions mentioned above.
In addition, Rnd1 binding caused an appreciable twisting of the conformation of the β-hairpin (β5 and β6 and the intervening loop), weakening the dimerization interaction, which is part of the mechanism that maintains monomer-dimer equilibrium in plexin-B1 signaling (25). In the complex, two Rnd1 molecules bind to opposite sides of the plexin-B1 RBD homodimer interface (Fig. 1A). An equimolar ratio of the two proteins is seen in solution, based on gel filtration data, which is consistent with previous observations that Rnd1 binding weakens the RBD dimerization (21, 39).
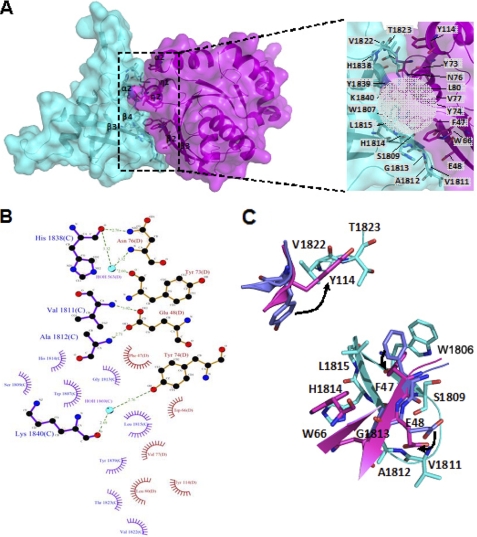
Rnd1 binding to the plexin-B1 RBD buries a solvent-accessible surface area of ∼1900 Å2 as calculated using NACCESS (by S. J. Hubbard and J. M. Thornton available on the World Wide Web). Binding involves twelve residues of plexin-B1 located in strands β3 and β4, a short α2-helix, and a loop toward the dimerization β-hairpin as well as nine residues of Rnd1 located in the β2 and β3 strands, the 310-helices η1 and η2, and one α-helix of the GTPase (Fig. 2, A and B). The composition of the heterodimer interface residues indicates that the core interface is hydrophobic. Mainly, Trp1807 and Leu1815 of the plexin-B1 RBD interact with Phe47 and Val77 of Rnd1; the interacting residues are surrounded by Ser1809, Gly1813, Tyr1839, and Lys1840 of the plexin-B1 RBD and Trp66, Tyr73, Tyr74, Asn76, and Leu80 of Rnd1 (Fig. 2A). Fold-X calculations (41) substituting residues at the interface for Ala (or Ser in case the residue is Ala already) predict seven hot spots (with substitution energy greater than 1 kcal/mol) on the side of the plexin-B1 RBD (Ser1809, Gly1810, Gly1813, His1814, Leu1815, Val1822, and Thr1823) and six hot spots on the side of Rnd1 (Phe47, Trp66, Tyr73, Tyr74, Val77, and Leu80).
FIGURE 2.
Interface of the plexin-B1 RBD·Rnd1 complex. A, transparent surface representation of the plexin-B1 RBD·Rnd1 complex. The plexin-B1 RBD and Rnd1 are shown in cyan and violet, respectively. The β3 and β4 strands and helix α2 of the plexin-B1 RBD are located in the interface, as are the β2 and β3 strands, the 310-helices η1 and η2, and one α-helix of Rnd1. The residues in the interface are represented by a stick model. The close-up view of the interface is shown on the right, where the hydrophobic core is highlighted by a gray transparent circle. B, amino acid interactions at the interface are shown in a LIGPLOT diagram (53). Hydrogen bonds are presented as dashed lines; interatomic distances are in angstroms. “Radiating” curves indicate hydrophobic contacts between the corresponding atoms or residues and the surrounding residues. C, two areas of the interface contact are shown, illustrating that the three residues of Rnd1, Phe47, Glu48, and Tyr114, are flipped toward plexin-B1 (shown in cyan), as seen in the superimposition of the bound (magenta for complex with plexin-B1) and the free (blue) structures of Rnd1.
Several hydrogen bonds were also observed in the interface (Fig. 2B). The side-chain amide nitrogen atom of Asn76 of Rnd1 forms a hydrogen bond to the main-chain carboxyl oxygen atom of His1838 of the plexin-B1 RBD (2.8 Å), and their side chains stack on each other. The side chain of Glu48 of Rnd1 forms hydrogen bonds to the main-chain amides of Val1811 (3.0 Å) and Ala1812 (2.7 Å) of the plexin-B1 RBD, maintaining the rotated conformation of the β3-β4 loop. The side chain of Trp66 of Rnd1 forms a T-shaped interaction with one face of His1814 in the plexin-B1 RBD, whereas the main chain of the ordered β1-β2 loop interacts with the other face of His1814 in plexin-B1 RBD. His1814 is also part of a network of hydrogen bonds that includes Glu1806 and Arg1808.
Minor conformational changes are observed in Rnd1 upon binding to plexin-B1 RBD (Fig. 2C) in comparison with the structure of free homodimeric Rnd1 (PDB code 2CLS). In the complex the side chain of Phe47 of Rnd1 is flipped toward Trp1807 of the plexin-B1 RBD, whereas the rotation of the β3-β4 loop of plexin-B1 RBD (residues Ser1809–Gly1813) exposes the same side chain of Trp1807 so that it can form a π-π interaction with the side chain of Phe47 of Rnd1. The side-chain rotation of Tyr114 is obvious because it forms hydrophobic contacts with Val1822 and with the side-chain methyl group of Thr1823 of plexin-B1. The movement of the side chain of Rnd1 Glu48 is also observed because it forms hydrogen bonds to the β3-β4 loop of plexin-B1 RBD.
Structure of the Plexin-A2 RBD in Complex with Rnd1
The structure of the plexin-A2 RBD was solved in complex with Rnd1 at a resolution of 2.0 Å (PDB code 3Q3J) (Fig. 3A and Table 1). In the crystal structure, the plexin-A2 RBD does not form a homodimer seen in the plexin-B1 RBD (PDB code 2REX) (25). In fact, the equivalent region of the plexin-B1 homodimerization loop is disordered in the plexin-A2·Rnd1 complex (the electron density of residues 1575–1578, which include the equivalent Trp and Arg residues, is very poor). The electron density surrounding these residues arises from regions of symmetry-related molecules in the crystal lattice that are remote from their own putative dimerization segment. Thus, this region is not involved in a dimer formation in the crystal; however, free plexin-A2 RBD homodimerizes at high protein concentration in solution (see Fig. 6B). These findings are again consistent with a weakened dimerization of the RBD once a Rho family GTPase is bound.
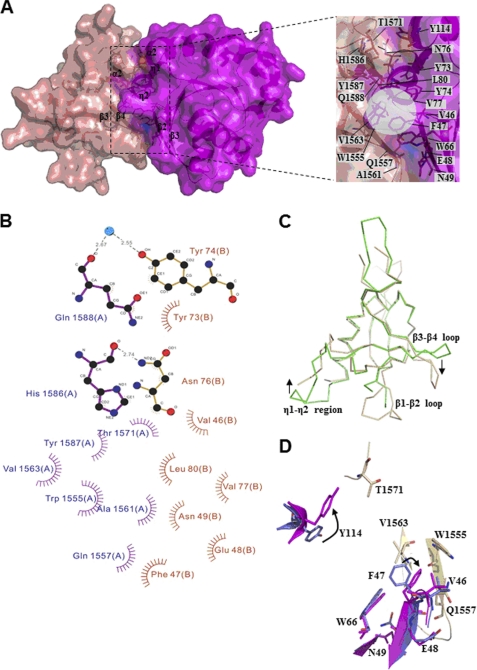
FIGURE 3.
Interface of the plexin-A2 RBD·Rnd1 complex. In part similar to Fig. 2A, a transparent surface representation of plexin-A2 RBD·Rnd1 complex is shown, with the interface displayed in an expanded view. B, LIGPLOT diagram of contacts at the interface. Rnd1 residues Trp66 and Tyr114 are omitted for clarity (but are shown in D). C, superimposition of the RBDs of the plexin-A2 RBD·Rnd1 complex (gold) and free plexin-B1 (green) illustrates conformational changes that take place. D, superposition of two regions in contact showing perturbations similar to those of the Rnd1 side-chain structure upon association with plexin-A2 RBD as seen in the case of plexin-B1 RBD binding. Gold, RBD residues; blue, free Rnd1; magenta, bound Rnd1 residues.
TABLE 1.
Statistics for x-ray diffraction data and structure refinement of the plexin-C1 and plexin-D1 RBDs and the plexin-A2·Rnd1 complex
| Plexin-C1 | Plexin-D1 | Plexin-A2·Rnd1 | |
|---|---|---|---|
| PDB code | 3KUZ | 3H6N | 3Q3J |
| Diffraction data | |||
| Unit cell parameters (Å) | a = 72.25 | a = 81.78 | a = 59.02 |
| b = 72.25 | b = 27.05 | b = 67.13 | |
| c = 116.24 | c = 52.65 | c = 145.29 | |
| Unit cell parameters (degrees) | β = 114.06.0 | ||
| Space group | P43212 | C2 | I222 |
| No. of copies/asymmetric unit | 2 | 1 | 2 |
| Resolution range (Å)a | 20–2.30 (2.34–2.30) | 30–2.00 (2.03–2.00) | 40–1.97 (2.00–19.7) |
| Unique reflections | 14,277 (697) | 7,245 (381) | 20,734 (1,018) |
| Data completeness (%) | 99.6 (98.3) | 99.5 (99.7) | 99.9 (100.0) |
| Rsym (%)b | 18.7 (96.3) | 8.6 (49.9) | 7.0 (96.4) |
| Redundancy | 9.0 (6.2) | 4.4 (3.6) | 7.2 (7.2) |
| Average I/σ(I) | 13.2 (1.5) | 28.1 (2.6) | 36.3 (2.4) |
| Refinement statistics | |||
| Rwork (%)c/Rfree (%) | 25.0/28.0 | 24.3/27.6 | 21.1/24.8 |
| No. of atoms | |||
| Protein | 1,608 | 775 | 2073 |
| Ligand/ion | 0 | 2 | 33 |
| Water | 44 | 11 | 38 |
| r.m.s. deviation from ideal | |||
| Bond length (Å) | 0.015 | 0.012 | 0.013 |
| Bond angles (degrees) | 1.5 | 1.1 | 1.2 |
| Average B-factors (Å2) | |||
| Protein | 24.6 | 38.7 | 39.0 |
| Ligand/ion | N/A | 52.5 | 26.9 |
| Water | 21.1 | 40.3 | 34.7 |
| Ramachandran plot (%) (55) | |||
| Most favored | 94.8 | 96.8 | 91.9 |
| Additional allowed | 5.2 | 3.2 | 8.1 |
| Generously allowed | None | None | None |
| Disallowed | None | None | None |
a Numbers in parentheses are for the outer shell.
b Rsym = Σ|I − 〈I〉|/ΣI.
c Rwork = Σ‖Fo| − |Fc‖/Σ|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively. Rfree was calculated as Rwork by using 3.8% of the data selected in thin resolution shells with SFTOO.
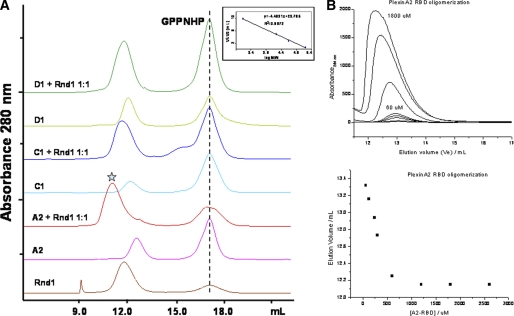
FIGURE 6.
Analytical gel filtration assay for binding of Rnd1 to GTPases (A) and plexin-A2 RBD dimerization (B). A, the blue star indicates the peak corresponding to the plexin-A2 RBD·Rnd1 complex. The data show that there is no binding between plexin-C1 or plexin-D1 and Rnd1. The inset shows the elution volumes of standard proteins (158, 44, 17, and 1.4 kDa) on a Superdex 75 10/300 GL column (GE Healthcare), plotted against log molecular masses, which were used to estimate the molecular masses of samples. B, plexin-A2 RBD dimerization measured under similar conditions as above. The data show that the RBD dimerizes, albeit at a slightly higher concentration than observed for plexin-B1 RBD (40).
The RBD has the same ubiquitin fold as other family members and the structure has an r.m.s. deviation of 1.00 and 0.93 Å, as calculated using SSM (29), to the Rnd1 bound plexin-B1 and to the unbound plexin-A3 RBD in the intracellular regions of plexin-A3 (26), respectively. In the plexin-A2 complex, the RBD-Rnd1 interface closely resembles that of the plexin-B1 complex. Similarly to the observation made above for the RBD-B1·Rnd1 complex, loop β3-β4 is also bent away from Rnd1 in the complex with RBD-A2 (Fig. 3C) and has relatively low B-factors. In another part of the protein, and different from the plexin-B1 RBD·Rnd1 complex, the RBD-A2 β1-β2 loop is three residues shorter than in the RBD of plexin-B1 but is not moved toward Rnd1 and has high B-factors. Another loop region, following the RBD α1 helix and opposite to the binding interface, is disordered in the RBD-A2·Rnd1 complex.
Rnd1 binding to the plexin-A2 RBD buries a surface area of ∼1300 Å2 according to NACCESS, which is considerably smaller than for the plexin-B1 RBD·Rnd1 complex, in part due to missing side chains in the A2 complex (e.g. in the β3-β4 loop). According to the crystal structure, binding involves eight residues on the side of the plexin-A2 RBD in direct contacts with Rnd1. On the side of Rnd1, eleven residues are involved (Fig. 3, B and D). These Rnd1 residues are located in the same regions as in the plexin-B1 RBD-Rnd1 interaction, and many make the same contacts with the A2 RBD. Accordingly, hydrophobic side chains of Trp1555 and Val1563 of plexin-A2 interact with Phe47 and Val77 of Rnd1, and this non-polar patch is surrounded by Gln1557, Ala1561, Tyr1587, and Gln1588 on the side of plexin and by Trp66, Tyr73, Tyr74, Ans76, and Leu80 of Rnd1 (Fig. 3A).
Fold-X calculations (41) utilizing an Ala (or in the case of a preexisting Ala, a Ser) scan predict six hot spot residues on the side of the RBD (Trp1555, Arg1559, Ala1561, Val1563, Ile1570, and Thr1571) and four hot spot residues on the side of Rnd1 (Phe47, Tyr74, Val77, and Leu80) as having substitution energy differences greater than 1 kcal/mol. It should be noted that although all four of the Rnd1 hot spot residues are involved in both complexes, only four of the seven hot spot RBD-A2 residues are hot spots in the case of the plexin-B1 RBD·Rnd1 complex, suggesting that despite a similarity of the overall features, some of the detailed energetic contributions of residues to the complex differ considerably between the two RBD proteins. In addition, the relative contribution of RBD and Rnd1 residues to the interface appears to be different in the two complexes. Specifically, two more Rnd1 residues, Trp66 and Tyr73, as well as one less RBD residue are involved in the case of the complex plexin-B1 RBD·Rnd1 compared with the complex with plexin-A2.
A hydrogen bond connects the Rnd1 Asn76 side-chain amino group and the plexin-A2 RBD main-chain carbonyl oxygen of His1586, as it does in the plexin-B1 RBD·Rnd1 structure. There is less hydrogen bonding across the interface, however, compared with the plexin-B1 complex (Fig. 3B, compare with Fig. 2B). The Rnd1 Glu48 side chain is farther away from the β3-β4 turn of the plexin-A2 RBD and does not form hydrogen bonds with the main-chain segment. In fact, this region may be more flexible in the plexin-A2 complex because side chains of Arg1559 and Ile1560 at the equivalent position (Val1811 and Ala1812 in plexin-B1) do not have electron density beyond Cβ. On the other hand, Fold-X calculations (where the full side chains are built in) suggest that Arg1559 and Ala1561 have a considerable effect on the stability of the complex and, thus, may be responsible for the rotation of the loop region away from the interaction with Rnd1. His1814 in the RBD-B1 has been replaced by Arg1562. The hydrocarbon portion of its side chain weakly interacts with Trp66 of Rnd1, whereas the guanidinium moiety of Arg1562 participates in a salt bridge network with Glu1554 and Arg1556, a +/−/+ charge motif that appears conserved across all plexin-A and -B family members, with the exception of plexin-B2.
As in the case of plexin-B1 RBD binding, several minor conformational changes occur in Rnd1 (Fig. 3D). The same observations are made for the plexin-A2 RBD·Rnd1 complex as were made for the plexin-B1 RBD upon binding to Rnd1 regarding a side-chain flip of Rnd1 Phe47 and Tyr114. In the case of Tyr114, one of the interaction partners is changed from a Val1822 in plexin-B1 to Ile1570 in plexin-A2. The most significant change is that the side chain of Glu48 is not hydrogen-bonded to the β3-β4 loop in the plexin-A2 RBD·Rnd1 complex.
Structure of the Plexin-C1 RBD
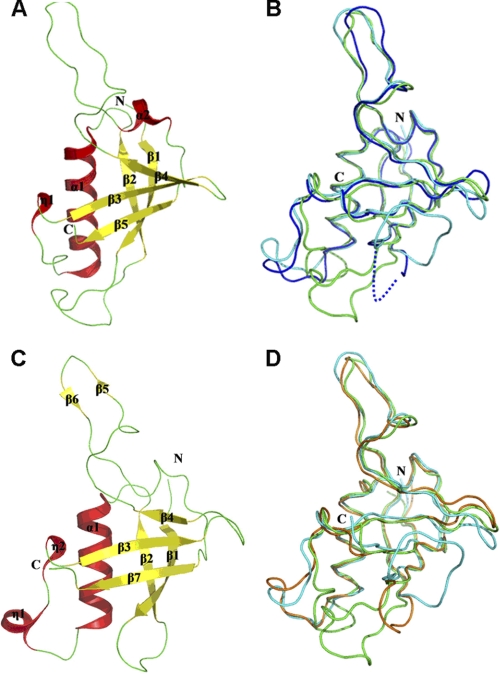
We solved the structure of plexin-C1 RBD at 2.3Å (residues 1198–1305, PDB code 3KUZ) (Table 1). Superimposition of the plexin-C1 RBD with the free plexin-B1 RBD structure (PDB code 2R2O) gives a root mean square deviation of 1.5 Å over 94 aligned residues (29). The plexin-C1 RBD consists of a central α-helix, two short 310-helices, and a five-stranded β-sheet, presenting a ubiquitin-like fold (Fig. 4A). In the core region, a five-stranded β-sheet has a left-handed twist, forming a groove where the central α-helix (α1) is bound. The central α-helix of the plexin-C1 RBD structure, whose equivalent in the plexin-B1 intracellular region is involved in RBD binding to the RasGAP domain, is longer by four residues than that of the plexin-B1 RBD.
FIGURE 4.
Crystal structure of plexin-C1 and -D1 RBD. A, ribbon diagram of plexin-C1 RBD. B, superimposition of the plexin-C1 RBD (green) and the free (blue) and bound (cyan) forms of the plexin-B1 RBD. C, plexin-D1 RBD. D, superimposition of the plexin-D1 RBD (gold) and the free (blue) and bound (cyan) forms of the plexin-B1 RBD in a ribbon diagram.
Additional differences between this and the plexin-B1 RBD are observed in two other regions (Fig. 4B). First, the β1-β2 loop (Glu1208–Arg1218) is ordered in the crystal and is bound to the elongated helix α1 as well as to its C-terminal loop region through hydrogen bonds. In contrast, the corresponding β1-β2 loop of the plexin-B1 RBD is invisible in the free form and becomes ordered in the complex with Rnd1, as noted above. Second, the region of plexin-C1 RBD (Leu1270–Ile1289), which corresponds to the homodimerization β-hairpin of the plexin-B1 RBD, is not engaged in dimerization. Several residues involved in homodimerization of the plexin-B1 RBD are different in the plexin-C1 RBD. In particular, Ile1283 of plexin-C1 RBD replaces Trp1830 of plexin-B1 RBD, eliminating an important π-stacking interaction and, thereby, reducing the possibility of homodimerization through this region. This is consistent with the monomeric state of plexin-C1 in solution, as shown by gel filtration data and also by NMR (data not shown). In the asymmetric unit of the crystal, we observe that two molecules of the plexin-C1 RBD are linked by an intermolecular disulfide bond through Cys1227. Disulfide bonds are not generally found in cytosolic proteins due to the reducing environment. Therefore, the dimerization of plexin-C1 RBD via the intermolecular disulfide bond appears to be a crystallization artifact.
Structure-based sequence analysis shows that plexin-C1 has unique residues at the putative Rnd1 binding site. All side chains are different from plexin-B1 with the exception of two residues (His1291 and Tyr1292) in helix α2 (Fig. 5A). The difference in sequence results in a series of repulsive effects if plexin-C1 were to interact with Rnd1 with the same interface as plexin-B1. Briefly, Trp1807 of plexin-B1 is replaced by Leu1260 in plexin-C1; the presence of leucine at this site precludes the π-π interaction with Phe47 of Rnd1 and results in the additional loss of several hydrophobic contacts. In addition to the changes in the hydrophobic interface, the shorter turn between the β3 and β4 strands of plexin-C1 may result in significant steric hindrance involving Glu48 of Rnd1. The long side chains of Met1262 and Arg1265 of plexin-C1 in this location, compared with those of Ser1809 and Gly1813 in plexin-B1, also could prevent binding to Rnd1. These changes are likely to be responsible for the absence of binding of Rnd1 to the plexin-C1 RBD, as demonstrated by the gel filtration chromatography and isothermal titration calorimetry (see below).
FIGURE 5.
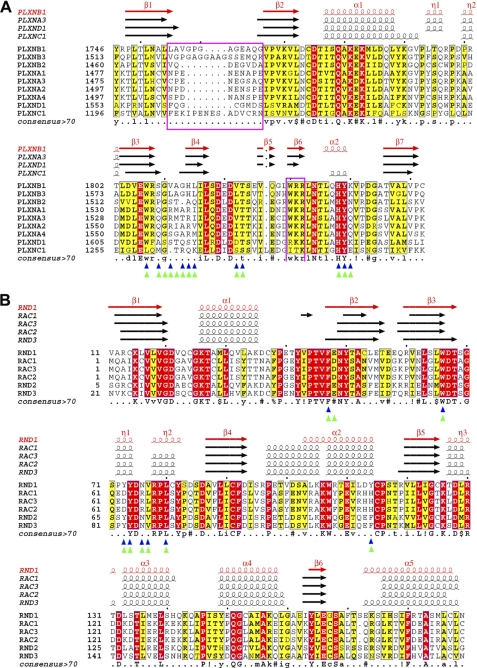
Structure-based sequence alignments. Structure-based sequence alignment of human plexin RBDs (A) and Rnd1 to -3 and Rac1 to -3 Rho GTPase (B). Alignments were generated using ESPript 2.1 (54). Sequence similarities are highlighted in yellow, and sequence identities are shown as white letters on a red background. Secondary structural elements (arrows for β-strands and coils for α-helices) are indicated at the top. Blue and green triangles indicate residues at the plexin-A2 and -B1 RBD·Rnd1 complex interface, respectively, as discussed under “Results.”
Structure of the Plexin-D1 RBD
The structure of the plexin-D1 RBD (residues 1553–1678) was solved at a resolution of 2.0 Å (Table 1). The final model of the plexin-D1 RBD contains residues 1553–1656 with 22 residues invisible at the C terminus of the recombinant protein (PDB code 3H6N). The plexin-D1 RBD also adopts a ubiquitin-like fold (Fig. 4C) with the r.m.s. deviation to the plexin-B1 RBD of 1.6 Å over 97 aligned residues (29).
The β5-β6 segment of the plexin-D1 RBD has a conformation similar to the homo-dimerization β-hairpin loop of plexin-B1 RBD as a result of interactions between the antiparallel β5 and β6 strands and a tight turn conformation connecting these two strands. Unlike plexin-B1, however, the plexin-D1 RBD appears to be monomeric by gel filtration and NMR (data not shown). In the plexin-B1 RBD homodimer, the Trp1830 residue of one molecule interacts with an arginine residue of the second molecule through a π-cation interaction, which is critical for homodimerization (23). In contrast, Trp1830 is replaced by arginine in the plexin-D1 RBD, abolishing the critical π-cation interaction, thus preventing homodimerization (Fig. 5A).
Sequence alignments show that five of the plexin-B1 RBD residues that interact with Rnd1 are conserved in the plexin-D1 RBD (e.g. tryptophan in the hydrophobic interface and the tripeptide motif HYK in helix α2) (Fig. 5A). In contrast, the more hydrophilic nature of the β3-β4 loop of plexin-D1 may interfere with Rnd1 binding. Val1811 and Ala1812 of plexin-B1 are replaced with Ser1614 and Ser1615 in plexin-D1. In a structural alignment with Rnd1 in the same position as it is found in the plexin-B1 RBD·Rnd1 complex, Ser1614 would have a steric clash with Rnd1 Glu48, whereas the replacement of Ala1812 of plexin-B1 with Thr1615 in plexin-D1 results in steric hindrance of the interaction with Rnd1 Arg49. The replacement of Leu1815 of plexin-B1 by Tyr1618 in plexin-D1 could promote a π-π interaction of Tyr1618 with Phe47 of Rnd1, but the bulky side chain of the tyrosine can also provide steric hindrance. Consistent with this idea, an oncogenic form of plexin-B1 that contains a mutation of Leu1815 to a bulky phenylalanine residue causes a substantial reduction in affinity for Rnd1 (42). Furthermore, the side-chain hydroxyl group of plexin-D1 Tyr1618 is likely to interfere with the hydrophobic nature of this interface region. Both steric hindrance and weakened hydrophobicity would negatively affect the formation of the plexin-D1 RBD·Rnd1 complex.
Analysis of Binding of Rnd1 in Vitro by Size Exclusion Chromatography and Isothermal Titration Calorimetry
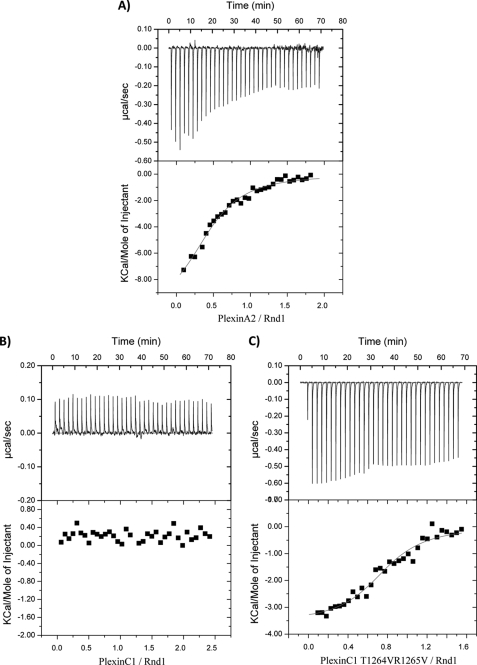
We examined the binding of plexin RBDs (plexin-A2, plexin-B1, plexin-C1, and plexin-D1) to Rnd1 by gel filtration chromatography. The data show that, in the presence of the non-hydrolyzable GTP analog GMPPNP, plexin-A2 and plexin-B1 RBDs bind to Rnd1, whereas plexin-C1 and plexin-D1 RBDs do not interact with Rnd1 (Fig. 6A). The isothermal titration calorimetry measurements also show that plexin-C1 and plexin-D1 do not interact with Rnd1, whereas plexin-A2 interacts with Rnd1 with a very similar binding affinity (Kd = 7.0 μm) as compared with plexin-B1 (Kd = 5.5 μm) (Fig. 7 and Table 2, top). Within the confines of the quantitative differences typically observed in such measurements, we confirmed the relative differences in binding behavior of the RBDs and key mutants (see below) with a second technique, surface plasmon resonance spectroscopy (see supplemental material).
FIGURE 7.
Representative ITC data showing binding of Rnd1 to plexin RBDs. A, ITC raw data (base line-corrected) and fitted data for the interaction of Rnd1 with the RBD of plexin-A2. No interaction is detected between RBD of plexin-C1 and Rnd1 (B), whereas a modest binding is generated by the T1264V/R1265V mutation in RBD of plexin-C1 toward Rnd1 (C).
TABLE 2.
Thermodynamic parameters for the association of Rnd1/2 with RBDs derived from ITC measurements
Uncertainty in measurement is given in parentheses. NI, no interaction.
| Ka | Kd | ΔH | ΔG | TΔS | |
|---|---|---|---|---|---|
| 105m−1 | μm | kcal/mol | kcal/mol | kcal/mol | |
| Rnd1-GTP | |||||
| Plexin-A2 WT | 1.4 (0.1) | 7.0 (0.7) | −8.0 (0.7) | −7.0 (0.1) | −1.0 (0.1) |
| Plexin-B1 WT | 1.8 (0.1) | 5.5 (0.6) | −6.7 (0.3) | −7.1 (0.1) | +0.5 (0.1) |
| L1815K | NI | NI | NI | NI | NI |
| L1815Y | 0.7 (0.5) | 14.1 (8) | −1.4 (0.2) | −6.6 (0.3) | +5.2 (0.4) |
| Plexin-C1 WT | NI | NI | NI | NI | NI |
| K1267L | 1.0 (0.8) | 10.0 (5.5) | −1.9 (0.7) | −6.8 (0.4) | +4.9 (0.4) |
| T1264V/R1265V | 3.6 (0.8) | 2.6 (0.4) | −3.4 (0.1) | −7.6 (0.2) | +4.2 (0.5) |
| Plexin-D1 WT | NI | NI | NI | NI | NI |
| Y1618L | 1.9 (0.6) | 5.2 (1.8) | −3.1 (0.1) | −7.2 (0.1) | +4.1 (0.2) |
| S1614V/T1615A | 0.5 (0.35) | 20 (14) | +1.3 (0.2) | −6.4 (0.3) | +7.7 (0.3) |
| S1613G/Q1616A | NI | NI | NI | NI | NI |
| Rnd2-GMPPNP | |||||
| Plexin-A2 WT | 0.9 (0.4) | 10.7 (4.1) | −1.2 (0.2) | −6.7 (0.3) | +5.5 (0.3) |
| Plexin-B1 WT | 0.4 (0.4) | >20 | −0.2 (0.2) | −6.3 (0.4) | +6.1 (0.4) |
| L1815K | 0.8 (0.2) | 20.0 (3.6) | −3.2 (1.2) | −6.5 (0.2) | +3.5 (0.3) |
| L1815Y | 0.9 (0.4) | 9.8 (3.9) | −1.6 (0.5) | −6.7 (0.4) | +5.0 (0.4) |
| Plexin-C1 WT | NI | NI | NI | NI | NI |
| K1267L | 0.9 (0.7) | 11.1 (5.1) | −0.6 (0.4) | −6.7 (0.3) | +6.1 (0.4) |
| T1264V/R1265V | 1.2 (0.4) | 8.3 (2.1) | −1.5 (0.4) | −6.9 (0.3) | +5.4 (0.3) |
| Plexin-D1 WT | 0.6 (0.4) | 16.6 (12) | −0.4 (0.2) | −6.5 (0.4) | +6.1 (0.5) |
| Y1618L | 0.7 (0.2) | 14.2 (3.2) | −1.6 (0.4) | −6.6 (0.2) | +5.0 (0.3) |
| S1614V/T1615A | 1.3 (0.4) | 7.7 (2.5) | −1.4 (0.2) | −6.9 (0.2) | +5.5 (0.2) |
| Rnd1-GTP Y114F | |||||
| Plexin-A2 WT | 1.0 (0.2) | 10.0 (2.4) | −4.1 (0.1) | −6.7 (0.3) | +2.7 (0.2) |
| Plexin-B1 WT | 1.8 (0.5) | 5.5 (1.5) | −2.5 (0.3) | −6.7 (0.1) | +4.2 (0.1) |
| Plexin-C1 WT | NI | NI | NI | NI | NI |
| Plexin-D1 WT | NI | NI | NI | NI | NI |
Experimental Verification of the Structure-based Predictions
The mutation of residue Trp1807, Asp1810, Ala1812, Gly1813, His1814, Leu1815, or Val1822 in plexin-B1 RBD to the corresponding residue in plexin-C1 or plexin-D1 RBD highly destabilizes the plexin-B1 RBD·Rnd1 complex, as shown by computational modeling with FoldX, suggesting the importance of these side chains (results not shown). In order to test these and the other structure-based predictions experimentally, we mutated several residues in plexin-B1, such as Leu1815 to Lys or Tyr (the equivalent residues in the plexin-C1 and -D1 RBDs), in order to demonstrate that these changes abolish or significantly weaken the plexin-B1 RBD-Rnd1 interaction (Table 2, top). Indeed, binding to the L1815K is no longer detectable by ITC, and binding of L1815Y is almost 3-fold weaker compared with wild type.
Furthermore, we mutated residues in plexin-C1 and plexin-D1 to the corresponding plexin-B1 residues and showed a substantial gain in binding affinity for Rnd1. Specifically, we generated RBD-D1 Y1618L, S1614V/T1615A, and S1613G/Q1616A mutants and RBD-C1 K1267L and T1264V/R1265V mutants, reverting key residues to their equivalents in plexin-B1, and measured possible binding interactions with Rnd1 (Table 2, top). As noted above, neither wild type RBDs of plexin-C1 nor -D1 bind to Rnd1 with affinities detectable by ITC. Remarkably, the single and double mutations, Y1618L and S1614V/T1615A, are sufficient to generate a plexin-D1 RBD-Rnd1 interaction with a modest binding affinity, detectable by ITC (Kd = 5–20 μm, compared with 5.5 μm for the plexin-B1 RBD-Rnd1 interaction). A K1267L rescue mutant generates weak binding affinity for Rnd1 in plexin-C1, and a similar double mutant, T1264V/R1265V, yields a protein with strong affinity for Rnd1 (Kd 2.6 μm), despite the fact that Lys1267 at the interface would still be weakening an interaction. The result suggests that not all interactions that are seen in the plexin-B1 and -A2 RBD complexes with Rnd1 are equally important and that hydrophobic contacts between the RBD β3-β4 loop and Rnd1 play a major role in complex formation.
Binding of the RBDs to Rnd2, a close homologue of Rnd1 (with 62% sequence identity and 83% similarity over residues 5–180) was also examined by ITC (Table 2, middle). Several mutations were studied in order to explore differences in the interaction between RBDs and Rnd2, compared with Rnd1. First, we tested the affinity for the four wild type RBD proteins, and ITC showed weaker binding of Rnd2 to plexin-A2 and -B1 compared with Rnd1. Similarly to Rnd1, no binding was evident by ITC to the plexin-C1 wild type RBD. However, the plexin-D1 wild type RBD showed a weak affinity for Rnd2 (whereas no affinity to Rnd1 was detected). Surprisingly, L1815K and L1815Y mutations in plexin-B1 did not weaken the interaction with Rnd2 as strongly as for Rnd1. Similarly, Y1618L barely increased binding of the plexin-D1 RBD protein to Rnd2, whereas this rescue mutation had a strong effect for Rnd1 binding. By contrast, the K1267L mutation rescued Rnd2 binding in plexin-C1 similarly as with Rnd1, and the plexin-C1 and -D1 RBD β3-β4 loop mutants also showed similar increases in affinity for Rnd2. In conclusion then, compared with Rnd1 binding, the central β-strand 4 location (Leu1815 in plexin-B1) does not play as critical a role for Rnd2 binding to the plexin-D1 RBD, suggesting that the mode of Rnd2 binding is somewhat different from that for the plexin-A1 and -B1 RBD·Rnd1 complexes whose structures we obtained so far.
Finally, as mentioned above, a singular difference in terms of the interacting residues is the mutation of Tyr114 to Phe between Rnd1 and Rnd2. We therefore wondered whether mutation of this residue in Rnd1 would be sufficient to mimic the binding of Rnd2 to plexin-D1 for instance. Table 2 (bottom) shows that this is not the case; in fact, the binding affinity of this mutant is unchanged also for the plexin-A2 and -B1 wild-type RBDs.
DISCUSSION
Properties of the Rho GTPase Binding Site of Plexin-A2 and Plexin-B1
The RBD residues at the Rnd1 binding interface in the complexes are mostly non-polar and constitute a hydrophobic patch, with a few polar residues that form hydrogen bonds. In addition, it appears that charges on histidines play a role in Rnd1 binding. A thermodynamic analysis of the plexin-B1 RBD-Rnd1 interaction has shown that the affinity of the interaction is pH-dependent. The tendency of the plexin-B1 RBD to form complexes increases at low pH (e.g. the Kd is ∼3.1 μm at pH 6.4 compared with ∼5.5 μm at pH 6.8), whereas no appreciable binding is detected at pH 8.0 (39). His1814 and His1838 of the plexin-B1 RBD form π-π stacking and T-shaped interactions with Rnd1 residues as well as with residues within plexin-B1 RBD. Because it is known that protonation of histidine enhances the strength of both its stacking and T-shaped interactions with DNA bases (43); for example, we hypothesize that protonation of these two interface histidines at low pH increases the affinity of binding to Rnd1, whereas their deprotonation at high pH has the opposite effect. As noted, the histidine just following the β3-β4 turn (His1814 in plexin-B1) is conserved in all B family plexins except for plexin-B2. The second His in the second short α-helix (His1838 in plexin-B1) is conserved in all plexins, and its main chain carbonyl is hydrogen-bonded across the interface to Rnd1 in both complexes.
The Plexin-B1 RBD·Rnd1 Complex as a Prediction Target for Computational Docking Assessment (CAPRI)
The plexin-B1 RBD·Rnd1 complex was entered as target 30 into the “blind” computational docking exercise (CAPRI) at the end of 2008 (44). A total of 35 groups, including several automated docking servers, participated using the unbound x-ray structures of the plexin-B1 RBD dimer and Rnd1 as starting points. Each predictor submitted 10 models. Of these, only two structure predictions for the complex were scored as acceptable (the lowest of three scoring classes requiring either >30% of the contact residues to be correct or a ligand r.m.s. deviation of <10 Å or an interface r.m.s. deviation of <4.0 Å) (45). A remarkable observation in both Rnd1-bound plexin-A2 and -B1 RBDs is the conformational change in the β3-β4 loop. At its farthest point, the loop has been pushed 3.2 and 5.8 Å away from the Rnd1 binding interface (measured between the bound and unbound positions of the main-chain nitrogen of Ile1560/Met1539 (human A2/murine A3) and Ala1812/Ala1812 (human B1/human B1), respectively). The extent of the conformational change made this target very difficult for traditional docking methods. The two groups that produced the adequate models indeed employed algorithms that incorporate molecular dynamics or normal mode calculations (46, 47), allowing the β3-β4 loop region to partially adjust (the fraction of correct interface contacts was 43 and 39%, the interface r.m.s. deviation was 2.5 and 4.3 Å, and the ligand r.m.s. deviation was 7.8 and 8.4 Å for the predictions by the Zacharias and Bates groups, respectively).
Predictions for Rnd1 Binding to Other Plexin-A and -B Family Members
Non-polar residues in the β3-β4 loop region at one side of the interaction interface are important for Rnd1 binding, and the polar residues seen in plexin-C1 and -D1 significantly diminish binding affinity, as shown by the generation of binding upon substitution of these residues to those found in the RBD of plexin-B1 (Val1822 and Ala1823). This region is part of the segments that were previously defined as plexin-specific Rho GTPase association motifs (PRAMs). The first of these motifs, originally defined as aEWXXGXX(G/A)+aaa (21), needs to be corrected in that our data show that glycine residues are not as important as the non-polar (aliphatic) nature of the mid-loop residues. Also, the Trp residue in β3 is not strictly required. Thus, the first PRAM should be redefined as aEaXXX(a/R)aXX(a/R/K)XL, at least for Rnd protein binding, where a indicates aliphatic residues, X represents an unknown preference/any residue, + is a positively charged residue, and (a/b/c) indicates a preference for amino acid a, b, or c.
In light of this change, the following predictions can be made about the Rnd1 binding affinity of the plexin-A and -B families. As mentioned above, the plexin-A3 structure and sequence matches the plexin-A2 RBD closely (74% BLAST sequence identity over the length of the model; Fig. 5A). Several key residues of the revised first PRAM are structurally conserved in the plexin-A family members. Specifically, the turn β3-β4 sequence RIARV in the plexin-A2 RBD is replaced by RMTRI in plexin-A3, whereas it is similar in the plexin-A1 RBD with a sequence of RMARI. In all cases, the two Arg residues are expected to point away from the interface because their side chains have partial hydrophobic character, and the second Arg forms a charged triad with the highly conserved Glu and Arg that surround the conserved Trp on β-strand 3 of the RBD structure. As stipulated above, the second and third positions also have aliphatic character. The exception to this is the sequence of the plexin-A4 RBD, which reads SGARM in this region. No information exists yet on Rnd1 binding to plexin-A4, and our analysis predicts that binding is possibly weakened.
In the plexin-B1 RBD·Rnd1 complex, the Val1811 and His1814 side chains of the plexin-B1 VAGHL sequence also point away from the Rnd1 binding interface. In plexin-B3, the corresponding sequence reads LAGHL, again highly conducive for binding, whereas in plexin-B2, there is a point deletion at the first position, and the remainder reads STAQ. This is a significant departure from the motif, probably leading to reduced Rnd1 binding, although residues in other regions, such as Leu1519, may compensate (see discussion on Rdn1/Rnd2 binding to plexin-C1 and -D1 RBDs below).
In summary, plexins have evolved a region at the edge of the Rnd1 binding interface that shows significant sequence variations in some family members and between families. The structural and mutagenesis data show that the character of the β3-β4 loop appears to have a profound effect on Rnd binding affinity.
Absence of Rnd1 Binding and Weak Binding of Rnd2 by the RBDs of Plexin-C1 and -D1
The lack of non-polar residues in key positions of the β3-β4 loop of the plexin-C1 and -D1 RBDs explains why Rnd1 neither binds to these two plexins nor plays a regulatory role in their activities. On the other hand, it has been reported that Rnd2 binding to plexin-D1 is required for Sema3E-induced inhibition of axonal outgrowth in cortical neurons (24). Table 2 (middle) shows that GMPPNP-loaded Rnd2 indeed binds the plexin-D1 RBD weakly (Kd = 16.6 μm). It should be noted that the structure of Rnd2 has not yet been determined. Based on the analysis of interface residues in the plexin-B1 and -A2 RBD·Rnd1 complexes, only one residue differs between Rnd1 and Rnd2 in the region of contact with the RBDs; Tyr114 of Rnd1 is replaced by Phe120 in Rnd2 (Fig. 5B). We made this mutation in Rnd1 in order to mimic Rnd2, but the modified GTPase showed no ITC-detectable interaction with either the RBD of plexin-C1 or -D1, whereas overall binding affinity with plexin-B1 was unperturbed, as shown in Table 2 (bottom). (Interestingly, the changes in enthalpy and entropy contributions for this interaction suggest an increase in burial of hydrophobic area relative to wild type, whereas enthalpy is decreased, possibly due to lack of a hydrogen bond to Tyr114).
Thus, the Tyr-to-Phe change fails to explain how Rnd2 can bind to the plexin-D1 RBD because, based upon the model derived from the plexin-B1 RBD·Rnd1 complex, the RBD region that would contact the Rnd2 GTPase in the region of Phe114 is actually largely hydrophilic (plexin-C1 and -D1 present a Ser and Asp, respectively; by contrast, these residues are Val1822 in the plexin-B1 and Ile1570 in the plexin-A2 RBD). Two neighboring residues, Glu1820 and Glu1825, in plexin-B1 have been replaced by Leu and Val, respectively, in plexin-C1 and by Ile and Val in plexin-D1. (Glu1820 is also replaced by a Leu in plexin-B2.)
Another example is mutation L1815K at the center of strand β4 of the plexin-B1 RBD, made to mimic plexin-C1. This abrogated binding to Rnd1, but the mutant still weakly binds to Rnd2. Similarly, the mutation Y1618L (the same β4 site), which generated binding in the plexin-D1 RBD for Rnd1, has little effect on the affinity of the same protein for Rnd2. This finding suggests that the site is not as important for Rnd2 binding and, again, implies that the mode of Rnd2 binding to plexin RBDs is likely to be different at least in some detail from that of Rnd1; not all interactions at the interface will be the same. At the same time, essentially identical interface contacts on the side of Rnd1 are observed when this GTPase interacts with the two different RBDs of plexin-A2 and -B1, strongly suggesting that these independent crystal structures are not an artifact.
Recently, it has been found that the modes of interaction even involving homologous proteins can be considerably different (42).7 In fact, there are several residues in the switch 1 and 2 regions, outside the putative binding region, that differ between Rnd1 and Rnd2. These could bind directly in a slightly different binding mode, as postulated above, or may communicate a conformational change to the residues that are directly bound. There is considerable evidence for longer range determinants for GTPase functional specificity, based upon mutagenesis studies (48), that would support the latter hypothesis.
The current model of plexin-B1 activation involves a shift in the equilibrium between a state of the intracellular protein region that is dimerized through the RBD and one that is not. The latter state is stabilized by Rnd1 binding to the plexin-B1 RBD (25). However, because a dimeric form of the plexin-D1 RBD is not observed, the RBD dimer-monomer equilibrium model may not apply to plexin-D1 activation by Rnd2. Instead, Rnd2 binding to the plexin-D1 RBD could induce a conformational change in a different region that affects the interactions between the RBD and the GAP region, helping to activate the receptor.
The binding affinity of either Rnd1 or Rnd2 to the wild type plexin-C1 RBD is very weak (Kd > 50 μm as estimated from ITC and also confirmed by surface plasmon resonance), consistent with cell-based studies that suggest that this interaction may not be required for the activation of GAP function in plexin-C1 (24). In addition, plexin-C1 occurs as a monomer in solution, suggesting that a mechanism that involves destabilization of RBD dimers, as seen for plexin-B1, is not applicable. This implies a different activation mechanism for plexin-C1 (e.g. one that requires the binding of yet to be identified agonist proteins).
Functional Implications for Rnd1 GTPase Binding to Plexins
At present, it is known that Rnd1 binding is required for certain functions of the plexin-A1 and -B1 receptors. The structures reported here, together with the analysis of the interface, predict that also plexin-B3 and plexin-A3 will bind to Rnd1. Whether (or how well) the RBDs of plexin-B2 and -A4 bind this GTPase is presently unknown. It is possible that other GTPases, including Rnd2, substitute for Rnd1 in order to stimulate plexin function. Such specificity is not unexpected; For instance, good binding of RhoD and weak binding of Rac1 were seen for plexin-A1 (19), whereas in plexin-B1, these affinities are reversed (21). It should be noted that the plexin RBD-Rnd GTPase interactions are of lower affinity compared with other effector-small Rho GTPase interactions. However, modest affinities have been observed in other systems, such as for the WASP-Cdc42 interaction with a Kd of 3–7 μm (49). In the case of Rnd1 (50) and of the plexin-B1 receptor, both partners are largely membrane-associated, and it is known that proteins can be concentrated by their membrane localization up to several thousand-fold over their density in the cytoplasm (51, 52), making these binding affinities physiologically relevant. In addition, protein preorientation, reduced entropy, and allosteric effects can enhance complex formation at membranes. Further work is needed to expand the present study into how other Rho and Ras GTPases interact in detail with plexins. This knowledge, together with measurements of active GTPase levels and their co-localization with the receptor in the cell, will ultimately lead to a detailed understanding of the plexin-mediated signaling mechanisms in different developmental and also pathogenic settings.
In conclusion, all RBDs structurally characterized thus far exhibit a typical ubiquitin-like fold that, except for loop regions, is highly conserved structurally. The comparative analysis of the plexin-A2 and -B1 RBD·Rnd1 complexes shows that the binding interface is formed mainly by the RBD β4 strand and a second short α-helix, with a conformational adjustment of the β3-β4 loop. Our mutagenesis studies show that the character of residues in this latter, less well conserved loop and of a residue at the center of strand β4 is critical for Rnd1 binding but less so for Rnd2 binding. In fact, it was possible to recover Rnd1 binding in the plexin-C1 and -D1 RBD by making this loop more non-polar. We also show that the plexin-A2 RBD tends to dimerize, whereas dimerization is absent in plexin-C1 and -D1 RBDs. This is consistent with the previous finding that Rnd1 destabilizes RBD dimers but does not play a role in the activation of plexin-C1 and -D1. Overall, then, evidence is mounting that plexin functions are regulated by subfamily-specific mechanisms, some of which involve different Rho family GTPases.
Supplementary Material
Acknowledgments
We thank Shufen Cao for assistance with Rnd1 and Rnd2 protein preparation and Drs. Christina Kiel and Luis Serrano (EMBL-CRG, Barcelona) for introducing us to Fold-X. Crystallographic data shown in this report were derived from work performed at Argonne National Laboratory, Structural Biology Center, beam line 19ID, at the Advanced Photon Source. Data collection at The National Institute of General Medical Sciences and National Cancer Institute Collaborative Access Team, beam line 23ID-B, was supported by NCI Grant Y1-CO-1020 and NIGM Grant Y1-GM-1104 from the National Institutes of Health. Use of the Advanced Photon Source was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02–06CH11357.
This work was supported, in whole or in part, by National Institutes of Health Grants R01GM73071, K02HL084384, and R01GM092851 (to M. B.). This work was supported in part by the Structural Genomics Consortium, a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S1.
The atomic coordinates and structure factors (codes 3Q3J, 3KUZ, and 3H6N) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
P. K. Hota, L. Zhang, and M. Buck, unpublished data.
P. K. Hota and M. Buck, manuscript in preparation.
- GAP
- GTPase-activating protein
- RBD
- Rho GTPase binding domain
- GMPPNP
- 5′-guanylyl imidodiphosphate
- ITC
- isothermal titration calorimetry
- PRAM
- plexin-specific Rho GTPase association motif
- PDB
- Protein Data Bank.
REFERENCES
- 1. Tamagnone L., Artigiani S., Chen H., He Z., Ming G. I., Song H., Chedotal A., Winberg M. L., Goodman C. S., Poo M., Tessier-Lavigne M., Comoglio P. M. (1999) Cell 99, 71–80 [DOI] [PubMed] [Google Scholar]
- 2. Negishi M., Oinuma I., Katoh H. (2005) Cell Mol. Life Sci. 62, 1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Patel B. N., Van Vactor D. L. (2002) Curr. Opin. Cell Biol. 14, 221–229 [DOI] [PubMed] [Google Scholar]
- 4. Gitler A. D., Lu M. M., Epstein J. A. (2004) Dev. Cell 7, 107–116 [DOI] [PubMed] [Google Scholar]
- 5. Serini G., Valdembri D., Zanivan S., Morterra G., Burkhardt C., Caccavari F., Zammataro L., Primo L., Tamagnone L., Logan M., Tessier-Lavigne M., Taniguchi M., Püschel A. W., Bussolino F. (2003) Nature 424, 391–397 [DOI] [PubMed] [Google Scholar]
- 6. Giordano S., Corso S., Conrotto P., Artigiani S., Gilestro G., Barberis D., Tamagnone L., Comoglio P. M. (2002) Nat. Cell Biol. 4, 720–724 [DOI] [PubMed] [Google Scholar]
- 7. van Beijnum J. R., Petersen K., Griffioen A. W. (2009) Front. Biosci. 1, 216–225 [DOI] [PubMed] [Google Scholar]
- 8. O'Connor B. P., Ting J. P. (2008) Immunol. Res. 41, 217–222 [DOI] [PubMed] [Google Scholar]
- 9. Moretti S., Procopio A., Boemi M., Catalano A. (2006) Curr. Neurovasc. Res. 3, 295–305 [DOI] [PubMed] [Google Scholar]
- 10. Takahashi T., Fournier A., Nakamura F., Wang L. H., Murakami Y., Kalb R. G., Fujisawa H., Strittmatter S. M. (1999) Cell 99, 59–69 [DOI] [PubMed] [Google Scholar]
- 11. Okuno T., Nakatsuji Y., Moriya M., Takamatsu H., Nojima S., Takegahara N., Toyofuku T., Nakagawa Y., Kang S., Friedel R. H., Sakoda S., Kikutani H., Kumanogoh A. (2010) J. Immunol. 184, 1499–1506 [DOI] [PubMed] [Google Scholar]
- 12. Comeau M. R., Johnson R., DuBose R. F., Petersen M., Gearing P., VandenBos T., Park L., Farrah T., Buller R. M., Cohen J. I., Strockbine L. D., Rauch C., Spriggs M. K. (1998) Immunity 8, 473–482 [DOI] [PubMed] [Google Scholar]
- 13. Walzer T., Galibert L., De Smedt T. (2005) Eur. J. Immunol. 35, 391–398 [DOI] [PubMed] [Google Scholar]
- 14. Gu C., Yoshida Y., Livet J., Reimert D. V., Mann F., Merte J., Henderson C. E., Jessell T. M., Kolodkin A. L., Ginty D. D. (2005) Science 307, 265–268 [DOI] [PubMed] [Google Scholar]
- 15. Toyofuku T., Zhang H., Kumanogoh A., Takegahara N., Suto F., Kamei J., Aoki K., Yabuki M., Hori M., Fujisawa H., Kikutani H. (2004) Genes Dev. 18, 435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roodink I., Verrijp K., Raats J., Leenders W. P. (2009) BMC Cancer 9, 297–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oinuma I., Ishikawa Y., Katoh H., Negishi M. (2004) Science 305, 862–865 [DOI] [PubMed] [Google Scholar]
- 18. Kruger R. P., Aurandt J., Guan K. L. (2005) Nat. Rev. Mol. Cell Biol. 6, 789–800 [DOI] [PubMed] [Google Scholar]
- 19. Zanata S. M., Hovatta I., Rohm B., Püschel A. W. (2002) J. Neurosci. 22, 471–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Driessens M. H., Hu H., Nobes C. D., Self A., Jordens I., Goodman C. S., Hall A. (2001) Curr. Biol. 11, 339–344 [DOI] [PubMed] [Google Scholar]
- 21. Tong Y., Chugha P., Hota P. K., Alviani R. S., Li M., Tempel W., Shen L., Park H. W., Buck M. (2007) J. Biol. Chem. 282, 37215–37224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Puschel A. W. (2007) Adv. Exp. Med. Biol. 600, 12–23 [DOI] [PubMed] [Google Scholar]
- 23. Fujita H., Katoh H., Ishikawa Y., Mori K., Negishi M. (2002) J. Biol. Chem. 277, 45428–45434 [DOI] [PubMed] [Google Scholar]
- 24. Uesugi K., Oinuma I., Katoh H., Negishi M. (2009) J. Biol. Chem. 284, 6743–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tong Y., Hota P. K., Penachioni J. Y., Hamaneh M. B., Kim S., Alviani R. S., Shen L., He H., Tempel W., Tamagnone L., Park H. W., Buck M. (2009) J. Biol. Chem. 284, 35962–35972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. He H., Yang T., Terman J. R., Zhang X. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 15610–15615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dong A., Xu X., Edwards A. M., Chang C., Chruszcz M., Cuff M., Cymborowski M., Di Leo R., Egorova O., Evdokimova E., Filippova E., Gu J., Guthrie J., Ignatchenko A., Joachimiak A., Klostermann N., Kim Y., Korniyenko Y., Minor W., Que Q., Savchenko A., Skarina T., Tan K., Yakunin A., Yee A., Yim V., Zhang R., Zheng H., Akutsu M., Arrowsmith C., Avvakumov G. V., Bochkarev A., Dahlgren L. G., Dhe-Paganon S., Dimov S., Dombrovski L., Finerty P., Jr., Flodin S., Flores A., Gräslund S., Hammerström M., Herman M. D., Hong B. S., Hui R., Johansson I., Liu Y., Nilsson M., Nedyalkova L., Nordlund P., Nyman T., Min J., Ouyang H., Park H. W., Qi C., Rabeh W., Shen L., Shen Y., Sukumard D., Tempel W., Tong Y., Tresagues L., Vedadi M., Walker J. R., Weigelt J., Welin M., Wu H., Xiao T., Zeng H., Zhu H. (2007) Nat. Methods 4, 1019–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 [DOI] [PubMed] [Google Scholar]
- 29. Vagin A., Teplyakov A. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 30. Schneider T. R., Sheldrick G. M. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 31. Otwinowski Z. (1991) Proceedings of the CCP4 Study Weekend (Evans P. R., Leslie A. G. W. eds) pp. 80–86, Daresbury Laboratory, Warrington, U.K [Google Scholar]
- 32. Cowtan K. D., Main P. (1996) Acta Crystallogr. D Biol. Crystallogr. 52, 43–48 [DOI] [PubMed] [Google Scholar]
- 33. McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 34. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 35. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 36. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 37. Davis I. W., Murray L. W., Richardson J. S., Richardson D. C. (2004) Nucleic Acids Res. 32, W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bouguet-Bonnet S., Buck M. (2008) J. Mol. Biol. 377, 1474–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hota P. K., Buck M. (2009) Protein Sci. 18, 1060–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tong Y., Hughes D., Placanica L., Buck M. (2005) Structure 13, 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L. (2005) Nucleic Acids Res. 33, W382–W388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tong Y., Hota P. K., Hamaneh M. B., Buck M. (2008) Structure 16, 246–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Churchill C. D., Wetmore S. D. (2009) J. Phys. Chem. B 113, 16046–16058 [DOI] [PubMed] [Google Scholar]
- 44. Janin J. (2010) Proteins 78, 3067–3072 [DOI] [PubMed] [Google Scholar]
- 45. Lensink M. F., Wodak S. J. (2010) Proteins 78, 3085–3095 [DOI] [PubMed] [Google Scholar]
- 46. Fiorucci S., Zacharias M. (2010) Proteins 78, 3131–3139 [DOI] [PubMed] [Google Scholar]
- 47. Li X., Moal I. H., Bates P. A. (2010) Proteins 78, 3189–96 [DOI] [PubMed] [Google Scholar]
- 48. Heo W. D., Meyer T. (2003) Cell 113, 315–328 [DOI] [PubMed] [Google Scholar]
- 49. Buck M., Xu W., Rosen M. K. (2004) J. Mol. Biol. 338, 271–285 [DOI] [PubMed] [Google Scholar]
- 50. Nobes C. D., Lauritzen I., Mattei M. G., Paris S., Hall A., Chardin P. (1998) J. Cell Biol. 141, 187–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Giesen P. L., Willems G. M., Hemker H. C., Hermens W. T. (1991) J. Biol. Chem. 266, 18720–18725 [PubMed] [Google Scholar]
- 52. Goryachev A. B., Pokhilko (2006) PLoS Comput. Biol. 12, e172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wallace A. C., Laskowski R. A., Thornton J. M. (1995) Protein Eng. 8, 127–134 [DOI] [PubMed] [Google Scholar]
- 54. Gouet P., Robert X., Courcelle E. (2003) Nucleic Acids Res. 31, 3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Cryst. 26, 283–291 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.