Abstract
The neural cell adhesion molecule (NCAM) was recently shown to be involved in the progression of various tumors with diverse effects. We previously demonstrated that NCAM potentiates the cellular invasion and metastasis of melanoma. Here we further report that the growth of melanoma is obviously retarded when the expression of NCAM is silenced. We found that the proliferation of murine B16F0 melanoma cells, their colony formation on soft agar, and growth of transplanted melanoma in vivo are clearly inhibited by the introduction of NCAM siRNA. Interestingly, change of NCAM expression level is shown to regulate the activity of Wnt signaling molecule, β-catenin, markedly. This novel machinery requires the function of FGF receptor and glycogen synthase kinase-3β but is independent of the Wnt receptors, MAPK-Erk and PI3K/Akt pathways. In addition, NCAM is found to form a functional complex with β-catenin, FGF receptor, and glycogen synthase kinase-3β. Moreover, up-regulation of NCAM140 and NCAM180 appears more potent than NCAM120 in activation of β-catenin, suggesting that the intracellular domain of NCAM is required for facilitating the β-catenin signaling. Furthermore, the melanoma cells also exhibit distinct differentiation phenotypes with the NCAM silencing. Our findings reveal a novel regulatory role of NCAM in the progression of melanoma that might serve as a new therapeutic target for the treatment of melanoma.
Keywords: β-Catenin, Cell Adhesion, Cell Differentiation, Tumor, Wnt Pathway, Cell Proliferation, Melanoma, NCAM
Introduction
Melanoma arises from skin neural crest-derived pigmented melanocytes and accounts for around 80% of mortality of skin cancer (1) with less than 5% of a 5-year survival rate (2). The rapid growth of melanoma cells by overriding senescence and activation of pro-proliferating signal transduction were shown to play a key role in the development of melanoma into metastatic stage (3). Even though many cell growth promoting factors have been identified to be involved in the progression of melanoma and targeted for therapeutic intervention, the treatment efficacy of advanced melanoma with vertical growth and metastasis has not been significantly improved over the past few decades (1). Hence, further study to elucidate the molecular mechanisms underlying the proliferation of melanoma cells is required for the finding of novel potential intervention targets to improve the treatment of melanoma.
It has been demonstrated that the adhesion molecules that mediate intercellular and cell-matrix interactions extensively participate in the progression of melanoma (4, 5) by altering the adhesion status and signal transduction of the cells. Neural cell adhesion molecule (NCAM),2 a member of the immunoglobulin superfamily, has been well characterized in cell proliferation, differentiation, migration, neurite outgrowth, and synaptic plasticity in the nervous system (6–9). However, recent studies have also revealed that the expression of NCAM drastically fluctuates in many tumors and affects tumor progression and prognosis (10). In small cell lung cancer (11) and the majority of multiple myelomas (12), the up-regulation of NCAM was shown to associate with a more aggressive tumor phenotype and poor prognosis. In contrast, the down-regulation of NCAM was observed in the development of colon cancer and some of myeloma (12, 13) and all neuroblastoma (14), suggesting that the role of NCAM on tumor progression is largely dependent on tumor types. In melanoma, the earlier studies demonstrated that melanoma from central nervous system (CNS) metastases and non-CNS-derived metastases exhibit distinct levels of NCAM (15) and high expression of NCAM was detected in aggressive metastasizing uveal melanoma (16). In addition, we also found that NCAM facilitates the cellular invasion and metastasis in a recent study (17). However, whether NCAM also promote melanoma growth is not known.
Besides mechanically mediating cell adhesion, NCAM also induces diverse cell signaling cascades (8). It has been well demonstrated that NCAM is able to co-activate FGFR-associated signaling pathways (18, 19) such as mitogen-activated protein kinases (MAPKs) (20, 21) and PKC (22). NCAM was also shown to trigger the activation of cAMP-dependent protein kinase (PKA) (23), phosphatidylinositide 3-kinase (PI3K)/Akt (24), Src-family of nonreceptor-tyrosine kinases Fyn, and focal adhesion kinase (25, 26). Interestingly, both MAPK and PI3K/Akt pathways are shown to participate in the initiation and development of melanoma (27–30). However, up to date, which pathways might be employed by NCAM to affect the progression of melanoma is completely unknown.
The canonical Wnt pathway plays an important role in lineage determination and proliferation of melanocytes (31, 32). Through inactivation of GSK-3β, Wnt pathway can further activate the β-catenin and downstream gene expression (33). β-Catenin has been found remarkably accumulated in a large number of human melanoma specimens (34). In cultured melanoma cells, activation of Wnt/β-catenin signaling was shown to promote cell proliferation (35–37), suggesting the important role of Wnt/β-catenin pathway in tumorigenesis of melanoma. Besides the Wnt pathway, the activity of β-catenin can also be regulated by other molecules such as MAPK/ERK and PI3K/Akt pathways (38, 39). In addition to the role of activation of transcription, β-catenin is also involved in cell adhesion by binding to cadherin family members (40, 41). Given that NCAM plays important roles in cell adhesion and tumor development, it would be very interesting to determine if there is any level of cross-talk between NCAM and the Wnt/β-catenin pathways and the contribution of this interaction in the progression of melanoma.
In the present study we demonstrated that the reduction of NCAM expression can retard the proliferation of murine B16F0 melanoma cells in vitro and in vivo that is attributed to the impaired β-catenin pathway. Moreover, β-catenin was shown to bind with NCAM in the complex containing FGFR and GSK-3β, suggesting a novel mechanism by which NCAM participates in the progression of melanoma.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
Murine B16F0 melanoma cells were obtained from American Type Culture Collection (Manassas, VA) and routinely maintained in high glucose Dulbecco's modified Eagle's medium (DMEM) with 4500 mg/liter d-glucose, l-glutamine, and 110 mg/liter sodium pyruvate (Invitrogen) supplemented with 10% fetal calf serum (FCS) (Invitrogen), 20 mm sodium bicarbonate (Sigma), 5 mm HEPES (Sigma), and 1% penicillin/streptomycin mix (Invitrogen). Cells were kept at 37 °C in a 5% CO2 incubator. Specific inhibitors for AKT (AKTI) and FGFR (SU5402) were purchased from Calbiochem. Lipofectamine 2000, G418, and puromycin were from Invitrogen.
Plasmids and DNA Constructs
Two sequences specific to mouse ncam were selected to generate siRNA fragments: target 1 (5′-CGA CTT CTT TGG CCA CTA TAC-3′) and target 2 (5′-GGA CAT ACT CTA CCA GTG CAA-3′). All targets and scrambled control oligonucleotides duplexes were cloned into pSilencer3.1-U6 and pSilencer4.1-CMV vectors, respectively, according to the manufacturer's instructions (Ambion). The cDNAs for full-length mouse NCAM120, NCAM140, and NCAM180 and the intracellular domain of NCAM140 and NCAM180 were cloned into pcDNA4/myc-His A vector (Invitrogen). The primers used for amplification of NCAM inserts include the following: NCAM extracellular forward (5′-CCC AAG CTT GCC ACC ATG CTG CGA ACT AAG GAT CTC-3′), NCAM extracellular reverse (5′-GCT CTA GAC GAG AAA GCA GCC TTG CC-3′), NCAM intracellular forward (5′-CCCAAG CTT GCC ACC ATG GCA GAG TAT GAA GTC TAT GTG-3′), and NCAM intracellular reverse (5′-GCT CTA GAT GCT TTG CTC TCA TTC TCT TTT G-3′). Plasmids of wild type and dominant negative β-catenin have been described previously (42). The dominant negative TCF4 (DN-TCF4) was generously provided by Dr. Alman, University of Toronto. The dominant negative Akt (43) was subcloned into pIRES2-EGFP and tagged with c-Myc epitope (Clontech). Transfection was conducted using Lipofectamine 2000 following the manufacturer's instructions. Stable cell lines expressing plasmids of NCAM siRNA were obtained by selecting the transformed cells with 1 μg/ml puromycin and 800 μg/ml G418. For transient transfection of other plasmids, cells were fed with fresh medium containing 10% FCS 6 h post transfection and further incubated for 24 h before further experiments. β-Catenin (sc-29210) and the control (sc-37007) siRNAs were purchased from Santa Cruz Biotechnology and transfected according to the manufacturer's instructions.
Cell Proliferation Study
Cells were plated into 96-well plates at 2000 cells per well and grew in complete media for 96 h. Relative proliferation rate was determined with Cell Proliferation Kit I (Roche Applied Science). To minimize the interference with absorbance reading by secreted melanin, cells were incubated with 3-{4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) in fresh medium for 4 h followed by further incubation in solubilization solution overnight. Absorbance was measured at 570 nm with the reference wavelength of 660 nm with Tecan GENios microplate reader (Tecan Group Ltd). All results were from three independent experiments.
Cell Cycle Analysis
For quantification of DNA content, the isolated cells were permeabilized with 70% cold ethanol at 4 °C for 1 h followed by incubation with 50 μg/ml RNase A at 37 °C for 30 min. The cells were then labeled with 50 μg/ml propidium iodide (Sigma). DNA content was analyzed in a FACSCalibur flow cytometer (BD Biosciences) with the use of ModFIT software (Vantage Software, Topsham, ME). All results were from three independent experiments.
Cell Growth in Soft Agar
2 × 104 cells were seeded in 0.3% agar, 10% FCS in high glucose DMEM. Dishes were precoated with 0.35% agar in DMEM to prevent cells from attaching the plastic bottom. Cell colonies were stained with 0.5% crystal violet for count at day 14, and the results were obtained from three independent experiments.
Tumor Growth in Vivo
A total of 12 female C57BL/6 mice 6–8 weeks old (15–20 g) were used in this study. Dark/light cycles of 12-h duration were maintained with food and water available ad libitum. Animals were randomly selected for experiment groups. The mice were injected at the back subcutaneously with 75 × 104 B16F0 cells carrying control or NCAM siRNA plasmids. Mice were sacrificed for tumor measurement after 3 weeks.
Immunoblotting
Cells were lysed for 1 h on ice with lysis buffer (50 mm Tris, 100 mm NaCl, 5 mm EDTA, 1 mm EGTA, 5 mm MgCl2, 1% Triton X-100, 1% glycerol, and 1× complete protease inhibitor (Roche Applied Science)). Lysates were cleared by centrifugation (15,000 × g 15 min, 4 °C), and protein concentration was quantified by the Bradford assay (Bio-Rad). 20–40 μg of lysate was resolved by SDS-PAGE and transferred to a polyvinylidene fluoride membrane (Millipore). After blocking with 5% skimmed milk for 1 h, membranes were incubated with specific primary antibodies in either 5% skimmed milk or 5% bovine serum albumin (BSA) followed by incubation with corresponding HRP-conjugated secondary antibodies. Detection was carried out with ImmobilonTM Western Chemiluminescent HRP substrate system (Millipore). The primary antibodies were purchased from Millipore (NCAM 5032), Cell Signaling Technology (E-cadherin 4065, β-catenin 9562, phospho-β-catenin (Ser-33/37) 2009, GSK-3β 9315, phospho-GSK-3β (Ser-9) 9323, Akt 9272, phospho-Akt (Ser-473) 4051, CREB 9197, phospho-CREB (Ser-133) 9191, phospho-p44/42 MAPK (Thr-202/Tyr-204) 9101, phospho-LRP5/6 (Ser-1490) 2568, cyclin D1 2926, Santa Cruz Biotechnology (ERK1 sc-93, LRP5/6 sc-57354, Lamin B sc-6216), Upstate (myc tag, 06-549), and Sigma (β-acin A5441, GAPDH G8795). All results were from three independent experiments.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde for 15 min and then permeabilized with 0.2% Triton X-100/PBS for 30 min. After blocking with 4% BSA, PBS for 1 h, cells were incubated with primary antibodies against NCAM, GSK-3β, β-catenin, or c-Myc tag in 1% BSA, Triton X-100, PBS overnight at 4 °C. After washing, cells were incubated for 3 h at room temperature with secondary antibodies including Alexa Fluor 568 goat anti-rabbit IgG and Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen). The cell nuclei were then stained with 4′6-diamidino-2-phenlylindole (0.1 μg/ml) (Invitrogen) in 4% BSA, PBS for 10 min followed and mounted with FluorSaveTM reagent (Calbiochem). Next the slides were scrutinized field by field with a confocal microscope (LSM710, Carl Zeiss).
Quantitative PCR
The reactions of real-time PCR were performed with the KAPA SYBR® qPCR kit (KAPA Biosystems) according to the manufacturer's instructions with an initial denaturation step at 95 °C for 20 s followed by 40 cycles with denaturation at 95 °C for 3 s and annealing and elongation at 60 °C for 1 min. At the end of each cycle the fluorescence emitted by SYBR Green was measured. The primers used were as follows: E-cadherin (sense, 5′-CAG GTC TCC TCA TGG CTT TGC-3′; antisense, 5′-CTT CCG AAA AGA AGG CTG TCC-3′); tyrosinase (sense, 5′-CTC TGG GCT TAG CAG TAG GC-3′; antisense, 5′-GCA AGC TGT GGT AGT CGT CT-3′); Wnt3A (sense, 5′-CTC CTC TCG GAT ACC TCT TAG TG-3′; antisense, 5′-GCA TGA TCT CCA CGT AGT TCC TG-3′); β-catenin (sense, 5′-ATG GAG CCG GAC AGA AAA GC-3′; antisense, 5′-CTT GCC ACT CAG GGA AGG A-3′); LRP5 (sense, 5′-AAG GGT GCT GTG TAC TGG AC-3′; antisense, 5′-AGA AGA GAA CCT TAC GGG ACG-3′); LRP6 (sense, 5′-TTG TTG CTT TAT GCA AAC AGA CG-3′; antisense, 5′-GTT CGT TTA ATG GCT TCT TCG C-3′); Frizzled7 (sense, 5′-CGG GGC CTC AAG GAG AGA A-3′; antisense, 5′-GTC CCC TAA ACC GAG CCA G-3′), Frizzled8 (sense, 5′-ATG GAG TGG GGT TAC CTG TTG-3′; antisense, 5′-CAC CGT GAT CTC TTG GCA C-3′), microphthalmia-associated transcription factor (MITF) (sense, 5′-ACT TTC CCT TAT CCC ATC CAC C-3′; antisense, 5′-TGA GAT CCA GAG TTG TCG TAC A-3′), Axin2 (sense, 5′-CAA TGA CAC CAC TCC AGA TGA G-3′; antisense, 5′-GGC CAA AGA AGT CGT TGC G-3′); β-actin (sense, 5′-GAG ACC TTC AAC ACC CCA GCC-3′; antisense, 5′-AAT GTC ACG CAC GAT TTC CC-3′). Samples were obtained from three independent experiments, and each sample was used for three reactions of real-time PCR.
Co-immunoprecipitation
250 μg of cell lysate was incubated with 3 μg of rabbit anti NCAM antibody (Millipore) on a roller mixer overnight at 4 °C. After that, 20 μl of protein A+G-agarose beads (Calbiochem) were added and incubated for 4 h at 4 °C. Beads were pelleted by centrifugation at 12,000 × g for 30 s and boiled in 40 μl of 2× sample buffer for 5 min. Proteins were collected by centrifugation at 12,000 × g for 1min for Western blotting analysis. All results were from three independent experiments.
Statistical Analyses
Results are reported as the mean ± S.D. Statistical comparisons were performed using Student's t test or one-way analysis of variance. Significance was set at p < 0.05.
RESULTS
NCAM Silencing Decreases the Proliferation and Promotes Differentiation of B16F0 Melanoma Cells
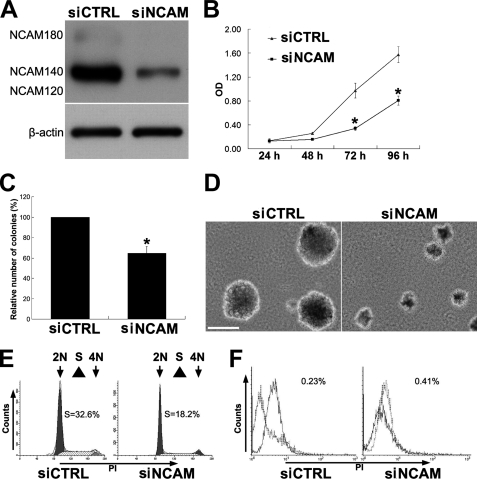
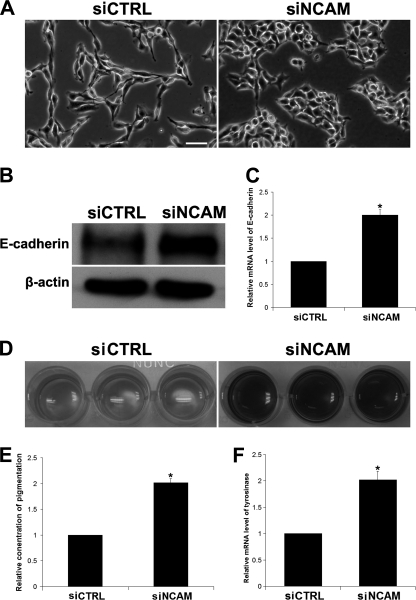
To determine the role of NCAM on the growth of melanoma, we tried to silence the expression of NCAM in murine melanoma B16F0 cells with two siRNA-incorporated plasmids targeting different sites of mouse ncam mRNA. As shown in Fig. 1A, the expression levels of NCAM in cells stably transfected with siRNA plasmids (siNCAM) were drastically knocked down compared with those in the cells transfected with scrambled siRNA (siCTRL). We then compared the growth capacity of these cells and found that NCAM silencing significantly reduced the growth of melanoma cells (Fig. 1B). Consistently, results from experiments of cell colony formation on soft agar also showed that the siNCAM cells produced much fewer and smaller cell colonies than the siCTRL cells (Fig. 1, C and D). In addition, cell cycle analysis also revealed that NCAM silencing resulted in a decrease of the S-phase fraction in siNCAM cells (Fig. 1E). To exclude the possibility of NCAM silencing rendered cell vulnerability (44) thus affecting cell growth, we measured the cell apoptosis by flow cytometry but did not detect obvious differences of cell death between siCTRL and siNCAM cells (Fig. 1F). These findings demonstrated that NCAM knockdown can retard the growth of B16F0 melanoma cells. Similar effects were also confirmed in NCAM-deficient mouse bone marrow stromal cells (data not shown). Furthermore, we also observed that NCAM siRNA caused a more flatten morphology of knock out cells compared with the spindle shaped wild type cells (Fig. 2A). With growth, the siNCAM cells exhibited as clusters or colonization, in contrast to the randomly distribution of the siCTRL cells (Fig. 2A). Such morphological differences may indicate the mesenchymal-epithelial transition (45) of melanoma cells by NCAM silencing. Because the up-regulation of E-cadherin has been considered as one of the markers for mesenchymal-endothelial transition (45), we then compared the expression levels of E-cadherin between these cells by real-time PCR and Western blot. As expected, the expression of E-cadherin was found to be significantly up-regulated after NCAM knockdown (Fig. 2, B and C).
FIGURE 1.
NCAM silencing decreases the proliferation of B16F0 melanoma cells. A, pSilence3.1-U6 and pSilence4.1-CMV vectors carrying siNCAM or siCTRL were co-transfected into B16F0 melanoma cells and generated stable cell lines by selection. NCAM expression was determined by immunoblotting. β-Actin was used as loading control. B, both siNCAM- and siCTRL-transfected cells were plated in 96-well plates. An MTT assay was performed to determine the proliferation of cells. *, p < 0.05; n = 6. C and D, siNCAM and siCTRL cells were seeded in agar dishes and incubated for 14 days. The colonies were stained with crystal violet and counted in nine randomly chosen areas under the microscopy. *, p < 0.05; n = 9. Scale bar, 100 μm. E, cell cycle analysis is shown. F, flow cytometry was used to measure apoptosis in siCTRL and siNCAM cells. Dotted lines indicate background staining.
FIGURE 2.
NCAM silencing promotes the differentiation of B16F0 cells. A, morphology of siCTRL and siNCAM cells is shown. Scale bar, 25 μm. Expression of E-cadherin was determined by immunoblotting (B) and by real-time PCR (C). *, p < 0.05. D, pigmentation was seen in cells cultured for 3 days. E, quantitative analysis of pigment in medium is shown. *, p < 0.05; n = 3. F, mRNA level of tyrosinase by real-time PCR is shown. *, p < 0.05.
Melanocytes are shown to be responsible for the production of the dark pigment melanin of skin. The capacity of melanin production was suggested to correlate with the differentiation and maturation status of melanocytes (46). Thus, we examined the levels of melanin in both cell types and found that NCAM silencing increased the pigmentation of siNCAM cells, and the production of melanin was also consistently increased in medium (Fig. 2, D and E). To further confirm the change of melanin synthesis, we applied real-time PCR to investigate mRNA level of tyrosinase, a key enzyme to regulate the synthesis of melanin (47). As expected, the mRNA level of tyrosinase was also increased significantly after NCAM silencing (Fig. 2F).
The Growth of Melanoma in Mice Is Retarded by NCAM Silencing
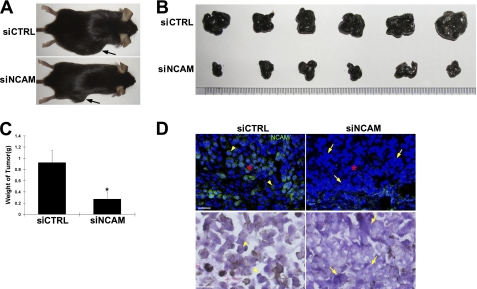
To further investigate the role of NCAM on the growth of melanoma cells in vivo, siCTRL and siNCAM cells were subcutaneously transplanted into mice respectively. After 1 week, some small tumor nodes were detected in the mice transplanted with siCTRL cells but not in the ones with siNCAM cells. At week 3, the size of melanoma in siNCAM cell-grafted mice was increased but still much smaller than those in siCTRL cell-grafted mice (Fig. 3, A and B). The differences of tumor mass were further confirmed by measuring the weight of melanoma. The tumors from siNCAM cells were much lighter than those from siCTRL cells (Fig. 3C), suggesting that NCAM silencing indeed retarded the growth of melanoma cells not only in vitro but also in vivo. More convincingly, results from immunohistochemical experiments confirmed that the melanoma tumors from siNCAM cells expressed much less NCAM than those from siCTRL cells (Fig. 3D, upper panels, red asterisks). Interestingly, the growth pattern of siNCAM cells in tumor tissues also exhibited more compacted clusters (Fig. 3D, lower panels, arrows) than the siCTRL cells as shown by hematoxylin and eosin staining (Fig. 3D, lower panels, arrowheads).
FIGURE 3.
The growth of melanoma in mice is retarded by NCAM silencing. siCTRL and siNCAM cells were transplanted subcutaneously into the back of C57BL/6J mice respectively. A, representative mice were injected with siCTRL and siNCAM cells 3 weeks post-transplantation. Arrows indicate the subcutaneous melanoma. B, specimen of melanoma tumors is shown. C, quantitative analysis of the weight of melanoma tumors is shown. *, p < 0.05; n = 6. D, expression of NCAM in tumors determined by immunohistochemistry is shown. Scale bars: 25 μm; Hematoxylin and eosin staining scale bars, 50 μm.
β-Catenin Is Involved in NCAM-facilitated Melanoma Growth Yet Is Independent of Wnt and PI3K-Akt Pathways
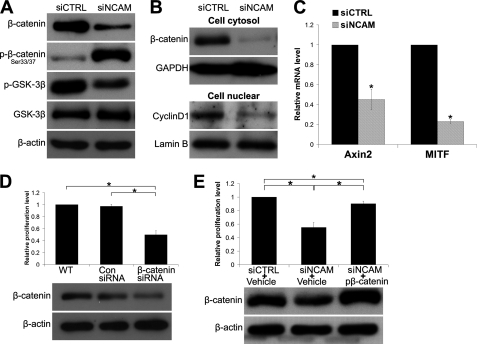
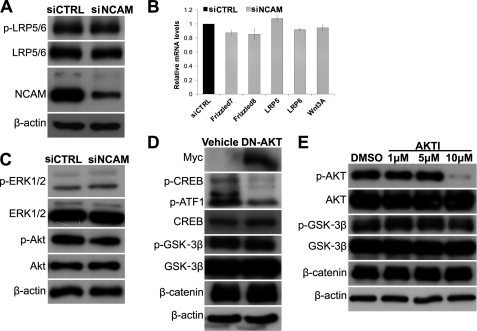
As revealed by previous studies, the β-catenin pathway participates in the progression of melanoma and cell adhesion by associating with cadherin (48), and we therefore set to determine the contribution of β-catenin in this scenario. As shown in Fig. 4A, the expression level of β-catenin was found to be drastically decreased in siNCAM cells. Because β-catenin had been shown to be phosphorylated by GSK-3β at serines 33 and 37 in the absence of Wnt ligands, thus leading to its ubiquitin-dependent degradation (49), we then determined the level of phosphorylated GSK-3β and β-catenin in siNCAM and siCTRL cells and found that the level of the inactive form of phosphorylated GSK-3β is much lower in siNCAM cells compared with siCTRL cells. Consistently, the level of phosphorylated β-catenin (Ser-33/37) is much higher in siNCAM cells than siCTRL cells (Fig. 4A), implying that NCAM facilitates the β-catenin pathway via inactivation of GSK-3β. β-Catenin is also found to bind tightly to the cytoplasmic domain of cadherins and plays an essential role in the cell adhesion (48). Only the free form of β-catenin co-activates gene transcription stimulated by Wnt pathway (50, 51). We thus further examined the status of cytosol β-catenin (signaling β-catenin) by cell fractionation experiment and found that cytosol β-catenin was significantly reduced in siNCAM cells (Fig. 4B). Additionally, cyclin D1, a key downstream effector in the Wnt/β-catenin signaling pathway, was also found decreased (Fig. 4B). Moreover, results of real-time PCR demonstrated that Axin2 and MITF, direct targets of signaling β-catenin (37, 52), were also down-regulated by NCAM silencing (Fig. 4C). These findings strongly suggested that β-catenin-mediated signaling pathway is retarded by NCAM knockdown in B16F0 cells. This function was also observed in B16F10 cells (data not shown).
FIGURE 4.
β-Catenin is involved in the reduced proliferation of B16F0 cells. A, immunoblotting shows the reduced expression level of β-catenin and phosphorylation of GSK-3β at Ser-9 as well as the elevated phosphorylation of β-catenin at Ser 33/37 in cells transfected with NCAM siRNA. β-Actin was used as loading control. B, shown is expression of cytosol β-catenin and nuclear Cyclin D1 by immunoblotting. GAPDH and Lamin B were used as loading control respectively. C, the expression levels of Axin2 and MITF by real-time PCR are shown. *, p < 0.05. D, expression of β-catenin in cells transiently transfected with β-catenin siRNA and their proliferation are shown. *, p < 0.05. E, cell proliferation was examined in siNCAM cells overexpressing wild type of β-catenin or the empty vehicle. *, p < 0.05. Representative pictures of three independent experiments are shown.
To verify the contribution of β-catenin to the growth of B16F0 cells, we transfected the cells with β-catenin siRNA duplex. As expected, β-catenin silencing significantly reduced the proliferation of B16F0 cells (Fig. 4D). In addition, overexpression of wild type β-catenin was also transfected transiently into the siNCAM cells and effectively rescued the decreased cell proliferation by NCAM silencing (Fig. 4E). To clarify the mechanism underlying the NCAM facilitated β-catenin pathway, we examined the activity of Wnt receptors and found no obvious difference of the phosphorylation of low density protein 5/6 (p-LRP5/6) between these two cell lines (Fig. 5A). In addition, results of real-time PCR did not reveal any significant changes of mRNA levels of Wnt3A and its receptors Frizzle7, Frizzle8, LRP5, and LRP6 (Fig. 5B), suggesting that the NCAM does not affect the activity of Wnt receptors.
FIGURE 5.
NCAM-silencing reduced β-catenin signaling is independent of canonical Wnt and PI3K-Akt pathways. A, immunoblotting showed the phosphorylation of LRP5/6 in cells. β-Actin was used as loading control. B, mRNA levels of Wnt3A and its receptors Frizzle7, Frizzle8, LRP5, and LRP6 were determined by real-time PCR. C, immunoblotting showed the levels of p-Akt and p-ERK in cells. D, the Myc-tagged DN-Akt and vehicle were transiently transfected into B16F0 cells followed by detection of Myc, p-CREB, p-GSK-3β, and β-catenin by immunoblotting. E, B16F0 cells were treated with AKTI (AKT inhibitor) at different concentrations for 12 h. The levels of p-AKT, p-GSK-3β, and β-catenin were determined by immunoblotting. Representative pictures of three independent experiments are shown.
Previous studies had revealed that both PI3K-Akt and MAPK-Erk pathways can be activated by NCAM (20, 24), and interestingly, the PI3K-Akt pathway was shown to regulate the activity of GSK-3β (53). In the present study we found that the phosphorylation of Akt (p-Akt) was decreased in siNCAM cells, whereas phosphorylation of Erk (p-Erk) remained unchanged (Fig. 5C). Thus, we set out to examine whether the down-regulated PI3K-Akt pathway contributes to the reduction of p-GSK-3β and β-catenin by NCAM knockdown. We found that transfection of the DN-Akt plasmid drastically reduced the phosphorylation of its target gene, CREB (Fig. 5D). However, both p-GSK-3β and β-catenin remained unchanged in the cells transfected with DN-AKT. Similar results were also obtained from the cells treated with the AKT-specific inhibitor, AKTI, as shown in Fig. 5E, suggesting that both MAPK and PI3K-Akt pathways are not responsible for the decreased β-catenin by NCAM silencing.
FGFR Facilitates the Activation of β-Catenin Induced by NCAM
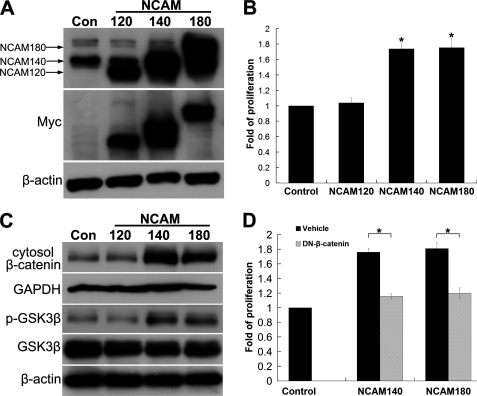
To further clarify the mechanisms underlying NCAM induced activation of β-catenin, we first determined the effects of different members of NCAM on the cell growth by transfecting the cells with three major NCAM isoforms, NCAM120, NCAM140, and NCAM180 (Fig. 6A). MTT proliferation assay revealed that overexpression of NCAM140 and NCAM180, but not NCAM120, promoted cell proliferation (Fig. 6B). As expected, only NCAM140 and NCAM180 significantly increase the amount of p-GSK-3β and cytosol β-catenin (Fig. 6C). Notably, overexpression of various isoforms of NCAM does not obviously increase the level of p-Akt (data not shown), further demonstrating that NCAM does not rely on the Akt pathway in activating β-catenin pathway. To further determine the contribution of β-catenin in this scenario, the DN-β-catenin was co-transfected into the cells with NCAM140 or NCAM180. The function of DN-β-catenin was confirmed by the down-regulation of β-catenin-targeted genes, Axin2 and MITF (data not shown). The increased cell proliferation by NCAM140/180 was repressed in DN-β-catenin-transfected cells (Fig. 6D). Similar results were also obtained from the experiments of transfection with dominant negative TCF4, the β-catenin co-transcription factor (data not shown), suggesting that β-catenin pathway participates in the NCAM-induced proliferation of B16F0 cells.
FIGURE 6.
NCAM140/180 facilitates the growth of B16F0 melanoma cells via increasing β-catenin levels. A, pcDNA4-NCAM/myc-His expression vector encoding the full-length NCAM120, NCAM140, or NCAM180 was transiently transfected into B16F0 cells. The cells transfected with an empty vector were used as control. Myc tag and NCAM expression were examined by immunoblotting. B, cell proliferation was measured by MTT assay. C, p-GSK-3β and cytosol β-catenin were examined in B16F0 cells with overexpressing of NCAM120, NCAM140, or NCAM180. D, pcDNA4-NCAM140 or NCAM180 and DN-β-catenin were transiently co-transfected into the B16F0 cells. The cells transfected with empty vectors were used as the control. An MTT assay was used to measure cell proliferation. *, p < 0.05.
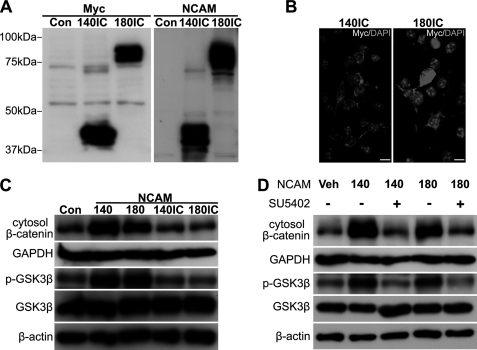
Compared with NCAM120, both NCAM140 and NCAM180 have intracellular domains that are likely to be responsible for triggering the β-catenin pathway. In an attempt to verify this hypothesis, we cloned and transfected the cells with the intracellular domains of NCAM140 (140IC) and NCAM180 (180IC) containing the transmembrane domain. As demonstrated by Western blot and immunocytochemistry results, the intracellular domains of NCAM were successfully expressed and localized on the cellular membrane (Fig. 7, A and B). Unexpectedly, we did not observe any stimulation of β-catenin and p-GSK-3β by the intracellular domains compared with the full-length NCAM140/180 (Fig. 7C), suggesting that the coordination between the intra- and extracellular domains of NCAM is required for the activity of β-catenin pathway.
FIGURE 7.
FGFR contributes to the NCAM promoted the β-catenin signaling. A and B, pcDNA4/myc-His vector containing 140IC or 180IC was transiently transfected into B16F0 cells. Western blot showed the expression of Myc tag and NCAM (A). Localization of expressed proteins was determined by immunocytochemistry (B). Scale bar: 10 μm. C, p-GSK-3β and signaling β-catenin were examined by immunoblotting in cells transfected with NCAM140, NCAM180, 140IC, or 180IC. D, B16F0 cells were pretreated with FGFR inhibitor, SU5402, at 10 μm for 4 h before transfection of NCAM140 or -180. Western blot results showed the levels of β-catenin and p-GSK-3β upon NCAM140/180 up-regulation. Veh, vehicle.
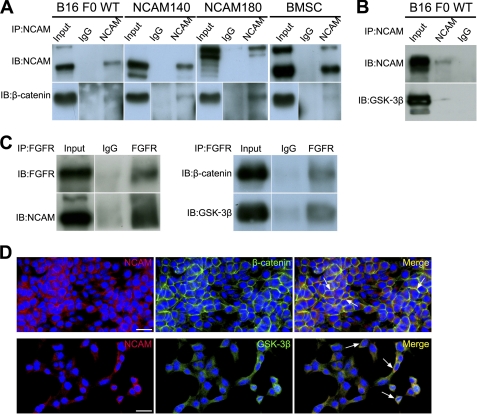
NCAM has been well documented to interact with FGFR extracellularly to elicit downstream signaling (18, 19). To examine whether FGFR is involved in the NCAM-induced β-catenin signaling, we pretreated cells with the FGFR-specific inhibitor SU5402 for 4 h followed by transfection of NCAM140/180 constructs. Intriguingly, as shown in Fig. 7D, SU5402 completely abrogated the increased β-catenin and p-GSK-3β by NCAM140/180 up-regulation, indicating that the NCAM-induced β-catenin signaling is FGFR-dependent. Because NCAM was demonstrated to regulate β-catenin activity in our study, we wondered whether NCAM could bind with β-catenin directly. As expected, co-immunoprecipitation experiments show that NCAM is capable of interacting with β-catenin in B16F0 cells (Fig. 8A). To verify this result, we also performed co-immunoprecipitation experiments in NCAM140- and NCAM180-overexpressing B16F0 cells and bone marrow stromal cells. Notably, the co-immunoprecipitation data further confirmed that NCAM is able to associate with β-catenin (Fig. 8A). Given the previous finding that the activity of GSK-3β can be regulated by NCAM, we thus speculated whether NCAM also binds with GSK-3β (Fig. 8B). As expected, such interaction was confirmed by their co-localization by immunohistochemistry staining (Fig. 8D). Similarly, our co- immunoprecipitation experiments also revealed the association of FGFR with the complex of NCAM, β-catenin, and GSK-3β in B16 F0 cells (Fig. 8C). These findings clearly demonstrated that NCAM could cooperate with FGFR for activation of β-catenin in promoting the growth of melanoma cells.
FIGURE 8.
NCAM is able to interact with FGFR, GSK-3β, and β-catenin. A, cell lysates were immunoprecipitated (IP) with anti-NCAM antibody or unrelated isotype IgG and analyzed for the presence of NCAM and β-catenin by Western blot (IB). B, cell lysates were immunoprecipitated with anti-NCAM antibody or IgG and analyzed for NCAM and GSK-3β. C, cell lysates were immunoprecipitated with anti-FGFR antibody or IgG and analyzed for the presence of FGFR, NCAM, β-catenin and GSK-3β. D, immunocytochemistry results indicated the co-localization (yellow, arrows) of NCAM (red) with β-catenin (upper panel, green) and GSK-3β (lower panel, green). Scale bars, 20 μm. Representative pictures of three independent experiments are shown.
DISCUSSION
NCAM has been well known for regulating cell adhesion, proliferation, differentiation, migration, neurite outgrowth, and synaptic plasticity in nervous system. Recent evidence shows that NCAM also participates in the development of various cancers with different effects. In this report we demonstrated that NCAM promotes the proliferation of B16F0 melanoma cells by activation of β-catenin signaling through FGFR rather than the canonical Wnt pathway. Furthermore, NCAM could interact with FGFR, β-catenin, and GSK-3β. These findings may enrich our understanding of the role of NCAM-mediated pathways in the progression of melanoma and benefit the improvement of melanoma treatment.
It has been well established that abnormal proliferation plays a key role in melanocyte transformation (1), and the growth capacity determines the progression and prognosis of melanoma in clinic (54). Previous studies revealed the important role of cell surface molecules on the progression of melanoma (55). NCAM was shown to participate in the progression of various tumors with different effects (10). We pioneered the study previously that NCAM potentiates the cellular invasion and metastasis of melanoma (17). In the current work we further reported that NCAM silencing reduced the progression of murine melanoma including decreased cell proliferation, clonogenicity, and tumor growth in vivo. Not only NCAM, but its family members, the adhesion molecule L1 (CD171) (56) and cell adhesion molecule (MCAM) (57), have been demonstrated to promote the progression of melanoma, which strongly supports the notion of the adhesion molecules as the progression marker of melanoma (58). In addition, we also found that silencing of NCAM was associated with the differentiation and mesenchymal-epithelial transition of melanoma cells. Because proliferation and differentiation are mutually exclusive events and mesenchymal-epithelial transition correlated with the reduction of tumor invasiveness, these findings suggest that silencing of NCAM would drastically decrease the progression of melanoma by changing various cell behaviors.
Through homo- or heterophilic binding, NCAM is well demonstrated to trigger multiple signaling cascades in neuronal cells, of which aberrant activation of MAPK and PI3K/Akt has been shown to associate with the development of melanoma (27–30). However, in the present study both PI3K-Akt and MAPK/Erk pathways are not shown to be significantly implicated in the NCAM-mediated proliferation of B16F0 melanoma cells. Instead, we found that β-catenin played a crucial role down-stream NCAM in melanoma cell proliferation. The β-catenin pathway has been demonstrated to either promote (37, 59–61) or decrease the progression of melanoma (62–65). Chien et al. (66) recently found that the Wnt3 could increase the level of β-catenin and reduce the proliferation of melanoma cells. However, the role of β-catenin in this study seems not conclusive because the application of β-catenin siRNA could not rescue the Wnt3 decreased cell proliferation. This study also indicated that the roles of Wnt and β-catenin pathways may not consist in the context of melanoma. In present study we applied gain or loss of function of signaling β-catenin and indicated that β-catenin is indeed able to promote the proliferation of B16 F0 melanoma cells. These controversial reports indicated that the role of β-catenin on the development of melanoma is affected by various regulatory factors. As demonstrated by previous study, the activity of β-catenin was sequestered by binding with E-cadherin (39). Here, we demonstrated that NCAM could activate the β-catenin pathway to promote the progression of melanoma. β-Catenin is known as an essential component in canonical Wnt signaling and is phosphorylated by the serine/threonine kinases casein kinase I and GSK-3β in a scaffolding destruction complex in the absence of Wnt (67). Once Wnt binds to its receptors, Frizzled and LRP5/6, the inhibitory GSK-3β is removed, and β-catenin translocates into the nucleus for activation of transcription factors (33). Notably, the inactivation of GSK3β in the canonical Wnt pathway may be regulated by its interaction with other proteins involving FRAT (68, 69) and LRP6 (70) rather than by its phosphorylation at Ser-9 (71). However, in the present study we found that phosphorylation of GSK3β at Ser-9 is in parallel with the change of β-catenin upon NCAM knockdown or overexpression, indicating a novel Wnt-independent regulatory pathway.
NCAM is well established to cooperate with FGFR to evoke various downstream signaling cascades, such as PLC-γ, MAPK, and PI3K/Akt pathways (20, 72–74). FGFR has recently been extensively documented to be involved in the activation of β-catenin during tumor development (75–77). It has been found that a variety of Erk-mediated signaling components such as MAP kinase-activated protein kinase-1, p70 S6 kinases, and p90RSK-1 (39, 78, 79) were shown to inactivate GSK-3β. Akt was also shown to be able to phosphorylate GSK-3β and facilitate the β-catenin signaling (38), suggesting that NCAM might prime the FGFR-induced activation of β-catenin. Moreover, FGF signaling activation also leads to β-catenin released from binding with E-cadherin, thus, increasing the pool of free β-catenin (76). Interestingly, our experiments further revealed that FGFR signaling is required for the NCAM-mediated activation of β-catenin in melanoma cells. The cooperative effects of FGFR and NCAM on the activation of β-catenin could also be supported by the finding that bFGF-induced inactivation of GSK-3β was compromised by NCAM knocked down (data not shown). However, among the signaling molecules downstream of FGFR, we only detected that PI3K/Akt pathway could be affected by NCAM in these cells, which is not shown to contribute to the regulation of β-catenin pathway. Obviously, some other unidentified mechanisms might be involved in the NCAM-FGFR-induced activity of β-catenin.
Although β-catenin is mainly located in cytosol, it is also found in association with endothelial-specific cell adhesion molecule (80) and N-cadherin (40). Because NCAM was shown to bind with N-cadherin, this motivates us to investigate whether NCAM is also associated with β-catenin. As expected, we found that NCAM is able to interact with β-catenin and also GSK-3β. Not surprisingly, FGFR is also found in this complex with GSK-3β and β-catenin, as FGFR is an important NCAM interaction partner and can directly bind to the extracellular domain of NCAM (19). Thus, we proposed a novel model that NCAM could regulate the β-catenin pathway via FGFR-dependent machinery in the progression of melanoma. Among the three major members of NCAM, NCAM140 and 180 but not NCAM120 were found to promote the β-catenin signaling, suggesting the important role of the intracellular domain of NCAM in the activation of β-catenin. However, our results further showed that the intracellular domains of NCAM140 and NCAM180 alone are not sufficient to promote the activation of β-catenin, indicating that the extracellular domain of NCAM is also indispensible for this machinery. Given that overexpression of NCAM120 could activate the FGFR in previous studies (18), we proposed that the extracellular domain of NCAM mainly activates FGFR, whereas the intracellular domain could facilitate the activation of β-catenin by FGFR. A similar scenario was also shown by the finding that the interaction between NCAM and FGFR extracellularly leads to the recruitment of intracellular PKC to NCAM-spectrin complex (81), demonstrating the importance of the extracellular domain of NCAM in the activation of its intracellular downstream factors (82). However, this notion is different from the model of endothelial-specific cell adhesion molecule in which the intracellular domain of endothelial-specific cell adhesion molecule is oncogenic and is able to trigger β-catenin-mediated signaling (80). Taken together, our study suggests a novel regulatory effect of NCAM, largely via FGFR, on activation of β-catenin to promote the melanoma progression. Because the NCAM- specific antibodies have been used in tumor therapy (83, 84) and the antagonist peptides are also developed (85), our findings regarding the role of NCAM on melanoma progression would help for novel treatment development.
Footnotes
- NCAM
- neural cell adhesion molecule
- siNCAM
- NCAM siRNA
- siCTRL
- scrambled siRNA
- IC
- intracellular domain
- FGFR
- FGF receptor
- GSK-3β
- glycogen synthase kinase-3β
- MTT
- 3-{4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide
- MITF
- microphthalmia-associated transcription factor
- CREB
- cAMP-response element-binding protein
- DN
- dominant negative.
REFERENCES
- 1. Gray-Schopfer V., Wellbrock C., Marais R. (2007) Nature 445, 851–857 [DOI] [PubMed] [Google Scholar]
- 2. Cummins D. L., Cummins J. M., Pantle H., Silverman M. A., Leonard A. L., Chanmugam A. (2006) Mayo Clin. Proc. 81, 500–507 [DOI] [PubMed] [Google Scholar]
- 3. Miller A. J., Mihm M. C., Jr. (2006) N. Engl. J. Med. 355, 51–65 [DOI] [PubMed] [Google Scholar]
- 4. Li G., Satyamoorthy K., Herlyn M. (2001) Cancer Res. 61, 3819–3825 [PubMed] [Google Scholar]
- 5. van Kilsdonk J. W., Wilting R. H., Bergers M., van Muijen G. N., Schalkwijk J., van Kempen L. C., Swart G. W. (2008) Cancer Res. 68, 3671–3679 [DOI] [PubMed] [Google Scholar]
- 6. Amoureux M. C., Cunningham B. A., Edelman G. M., Crossin K. L. (2000) J. Neurosci. 20, 3631–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Crossin K. L., Krushel L. A. (2000) Dev. Dyn. 218, 260–279 [DOI] [PubMed] [Google Scholar]
- 8. Maness P. F., Schachner M. (2007) Nat Neurosci 10, 19–26 [DOI] [PubMed] [Google Scholar]
- 9. Rønn L. C., Berezin V., Bock E. (2000) Int. J. Dev. Neurosci. 18, 193–199 [DOI] [PubMed] [Google Scholar]
- 10. Zecchini S., Cavallaro U. (2010) Adv. Exp. Med. Biol. 663, 319–333 [DOI] [PubMed] [Google Scholar]
- 11. Miyahara R., Tanaka F., Nakagawa T., Matsuoka K., Isii K., Wada H. (2001) J. Surg. Oncol. 77, 49–54 [DOI] [PubMed] [Google Scholar]
- 12. Ely S. A., Knowles D. M. (2002) Am. J. Pathol. 160, 1293–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huerta S., Srivatsan E. S., Venkatesan N., Peters J., Moatamed F., Renner S., Livingston E. H. (2001) Surgery 130, 834–843 [DOI] [PubMed] [Google Scholar]
- 14. Blaheta R. A., Hundemer M., Mayer G., Vogel J. U., Kornhuber B., Cinatl J., Markus B. H., Driever P. H., Cinatl J., Jr. (2002) Cell Commun. Adhes. 9, 131–147 [DOI] [PubMed] [Google Scholar]
- 15. Geertsen R., Zenklusen R., Kamarashev J., Burg G., Dummer R. (1999) Int. J. Cancer 83, 135–140 [DOI] [PubMed] [Google Scholar]
- 16. Mooy C. M., Luyten G. P., de Jong P. T., Jensen O. A., Luider T. M., van der Ham F., Bosman F. T. (1995) Hum. Pathol. 26, 1185–1190 [DOI] [PubMed] [Google Scholar]
- 17. Shi Y., Liu R., Zhang S., Xia Y. Y., Yang H. J., Guo K., Zeng Q., Feng Z. W. (2011) Int. J. Biochem. Cell Biol. 43, 682–690 [DOI] [PubMed] [Google Scholar]
- 18. Cavallaro U., Niedermeyer J., Fuxa M., Christofori G. (2001) Nat. Cell Biol. 3, 650–657 [DOI] [PubMed] [Google Scholar]
- 19. Kiselyov V. V., Skladchikova G., Hinsby A. M., Jensen P. H., Kulahin N., Soroka V., Pedersen N., Tsetlin V., Poulsen F. M., Berezin V., Bock E. (2003) Structure 11, 691–701 [DOI] [PubMed] [Google Scholar]
- 20. Kolkova K., Novitskaya V., Pedersen N., Berezin V., Bock E. (2000) J. Neurosci. 20, 2238–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schmid R. S., Graff R. D., Schaller M. D., Chen S., Schachner M., Hemperly J. J., Maness P. F. (1999) J. Neurobiol. 38, 542–558 [PubMed] [Google Scholar]
- 22. Kolkova K., Stensman H., Berezin V., Bock E., Larsson C. (2005) J. Neurochem. 92, 886–894 [DOI] [PubMed] [Google Scholar]
- 23. Jessen U., Novitskaya V., Pedersen N., Serup P., Berezin V., Bock E. (2001) J. Neurochem. 79, 1149–1160 [DOI] [PubMed] [Google Scholar]
- 24. Ditlevsen D. K., Køhler L. B., Pedersen M. V., Risell M., Kolkova K., Meyer M., Berezin V., Bock E. (2003) J. Neurochem. 84, 546–556 [DOI] [PubMed] [Google Scholar]
- 25. Beggs H. E., Baragona S. C., Hemperly J. J., Maness P. F. (1997) J. Biol. Chem. 272, 8310–8319 [DOI] [PubMed] [Google Scholar]
- 26. Beggs H. E., Soriano P., Maness P. F. (1994) J. Cell Biol. 127, 825–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ackermann J., Frutschi M., Kaloulis K., McKee T., Trumpp A., Beermann F. (2005) Cancer Res. 65, 4005–4011 [DOI] [PubMed] [Google Scholar]
- 28. Cohen C., Zavala-Pompa A., Sequeira J. H., Shoji M., Sexton D. G., Cotsonis G., Cerimele F., Govindarajan B., Macaron N., Arbiser J. L. (2002) Clin. Cancer Res. 8, 3728–3733 [PubMed] [Google Scholar]
- 29. Satyamoorthy K., Li G., Gerrero M. R., Brose M. S., Volpe P., Weber B. L., Van Belle P., Elder D. E., Herlyn M. (2003) Cancer Res. 63, 756–759 [PubMed] [Google Scholar]
- 30. Stahl J. M., Sharma A., Cheung M., Zimmerman M., Cheng J. Q., Bosenberg M. W., Kester M., Sandirasegarane L., Robertson G. P. (2004) Cancer Res. 64, 7002–7010 [DOI] [PubMed] [Google Scholar]
- 31. Dunn K. J., Williams B. O., Li Y., Pavan W. J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10050–10055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Larue L., Kumasaka M., Goding C. R. (2003) Pigment Cell Res. 16, 312–317 [DOI] [PubMed] [Google Scholar]
- 33. Cadigan K. M., Nusse R. (1997) Genes Dev. 11, 3286–3305 [DOI] [PubMed] [Google Scholar]
- 34. Rimm D. L., Caca K., Hu G., Harrison F. B., Fearon E. R. (1999) Am. J. Pathol. 154, 325–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Delmas V., Beermann F., Martinozzi S., Carreira S., Ackermann J., Kumasaka M., Denat L., Goodall J., Luciani F., Viros A., Demirkan N., Bastian B. C., Goding C. R., Larue L. (2007) Genes Dev. 21, 2923–2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moon R. T., Kohn A. D., De Ferrari G. V., Kaykas A. (2004) Nat. Rev. Genet. 5, 691–701 [DOI] [PubMed] [Google Scholar]
- 37. Widlund H. R., Horstmann M. A., Price E. R., Cui J., Lessnick S. L., Wu M., He X., Fisher D. E. (2002) J. Cell Biol. 158, 1079–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Desbois-Mouthon C., Cadoret A., Blivet-Van Eggelpoël M. J., Bertrand F., Cherqui G., Perret C., Capeau J. (2001) Oncogene 20, 252–259 [DOI] [PubMed] [Google Scholar]
- 39. Ding Q., Xia W., Liu J. C., Yang J. Y., Lee D. F., Xia J., Bartholomeusz G., Li Y., Pan Y., Li Z., Bargou R. C., Qin J., Lai C. C., Tsai F. J., Tsai C. H., Hung M. C. (2005) Mol. Cell 19, 159–170 [DOI] [PubMed] [Google Scholar]
- 40. Heasman J., Crawford A., Goldstone K., Garner-Hamrick P., Gumbiner B., McCrea P., Kintner C., Noro C. Y., Wylie C. (1994) Cell 79, 791–803 [DOI] [PubMed] [Google Scholar]
- 41. Sanson B., White P., Vincent J. P. (1996) Nature 383, 627–630 [DOI] [PubMed] [Google Scholar]
- 42. Wang Z., Havasi A., Gall J. M., Mao H., Schwartz J. H., Borkan S. C. (2009) J. Am. Soc. Nephrol. 20, 1919–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Choudhury G. G. (2001) J. Biol. Chem. 276, 35636–35643 [DOI] [PubMed] [Google Scholar]
- 44. Zhang S., Xia Y. Y., Lim H. C., Tang F. R., Feng Z. W. (2010) Neurochem. Int. 56, 919–929 [DOI] [PubMed] [Google Scholar]
- 45. Thiery J. P., Sleeman J. P. (2006) Nat. Rev. Mol. Cell Biol. 7, 131–142 [DOI] [PubMed] [Google Scholar]
- 46. Chamberlain A. J., Fritschi L., Kelly J. W. (2003) J. Am. Acad. Dermatol. 48, 694–701 [DOI] [PubMed] [Google Scholar]
- 47. Van Woert M. H., Korb F., Prasad K. N. (1971) J. Invest. Dermatol. 56, 343–348 [DOI] [PubMed] [Google Scholar]
- 48. Jamora C., Fuchs E. (2002) Nat. Cell Biol. 4, E101–E108 [DOI] [PubMed] [Google Scholar]
- 49. Logan C. Y., Nusse R. (2004) Annu. Rev. Cell Dev. Biol. 20, 781–810 [DOI] [PubMed] [Google Scholar]
- 50. Fang D., Hawke D., Zheng Y., Xia Y., Meisenhelder J., Nika H., Mills G. B., Kobayashi R., Hunter T., Lu Z. (2007) J. Biol. Chem. 282, 11221–11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Young C. S., Kitamura M., Hardy S., Kitajewski J. (1998) Mol. Cell. Biol. 18, 2474–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jho E. H., Zhang T., Domon C., Joo C. K., Freund J. N., Costantini F. (2002) Mol. Cell. Biol. 22, 1172–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cross D. A., Alessi D. R., Cohen P., Andjelkovich M., Hemmings B. A. (1995) Nature 378, 785–789 [DOI] [PubMed] [Google Scholar]
- 54. Balch C. M., Buzaid A. C., Atkins M. B., Cascinelli N., Coit D. G., Fleming I. D., Houghton A., Jr., Kirkwood J. M., Mihm M. F., Morton D. L., Reintgen D., Ross M. I., Sober A., Soong S. J., Thompson J. A., Thompson J. F., Gershenwald J. E., McMasters K. M. (2000) Cancer 88, 1484–1491 [DOI] [PubMed] [Google Scholar]
- 55. McGary E. C., Lev D. C., Bar-Eli M. (2002) Cancer Biol. Ther. 1, 459–465 [DOI] [PubMed] [Google Scholar]
- 56. Meier F., Busch S., Gast D., Göppert A., Altevogt P., Maczey E., Riedle S., Garbe C., Schittek B. (2006) Int. J. Cancer 119, 549–555 [DOI] [PubMed] [Google Scholar]
- 57. Xie S., Luca M., Huang S., Gutman M., Reich R., Johnson J. P., Bar-Eli M. (1997) Cancer Res. 57, 2295–2303 [PubMed] [Google Scholar]
- 58. Denton K. J., Stretch J. R., Gatter K. C., Harris A. L. (1992) J. Pathol. 167, 187–191 [DOI] [PubMed] [Google Scholar]
- 59. Rubinfeld B., Robbins P., El-Gamil M., Albert I., Porfiri E., Polakis P. (1997) Science 275, 1790–1792 [DOI] [PubMed] [Google Scholar]
- 60. Takahashi Y., Nishikawa M., Suehara T., Takiguchi N., Takakura Y. (2008) Int. J. Cancer 123, 2315–2320 [DOI] [PubMed] [Google Scholar]
- 61. Takahashi Y., Nishikawa M., Takakura Y. (2006) J. Control Release 116, 90–95 [DOI] [PubMed] [Google Scholar]
- 62. Chien A. J., Conrad W. H., Moon R. T. (2009) J. Invest. Dermatol. 129, 1614–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Omholt K., Platz A., Ringborg U., Hansson J. (2001) Int. J. Cancer 92, 839–842 [DOI] [PubMed] [Google Scholar]
- 64. Pollock P. M., Hayward N. (2002) Melanoma Res. 12, 183–186 [DOI] [PubMed] [Google Scholar]
- 65. Reifenberger J., Knobbe C. B., Wolter M., Blaschke B., Schulte K. W., Pietsch T., Ruzicka T., Reifenberger G. (2002) Int. J. Cancer 100, 549–556 [DOI] [PubMed] [Google Scholar]
- 66. Chien A. J., Moore E. C., Lonsdorf A. S., Kulikauskas R. M., Rothberg B. G., Berger A. J., Major M. B., Hwang S. T., Rimm D. L., Moon R. T. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 1193–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kimelman D., Xu W. (2006) Oncogene 25, 7482–7491 [DOI] [PubMed] [Google Scholar]
- 68. Li L., Yuan H., Weaver C. D., Mao J., Farr G. H., 3rd, Sussman D. J., Jonkers J., Kimelman D., Wu D. (1999) EMBO J. 18, 4233–4240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Farr G. H., 3rd, Ferkey D. M., Yost C., Pierce S. B., Weaver C., Kimelman D. (2000) J. Cell Biol. 148, 691–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mi K., Dolan P. J., Johnson G. V. (2006) J. Biol. Chem. 281, 4787–4794 [DOI] [PubMed] [Google Scholar]
- 71. Ding V. W., Chen R. H., McCormick F. (2000) J. Biol. Chem. 275, 32475–32481 [DOI] [PubMed] [Google Scholar]
- 72. Ditlevsen D. K., Povlsen G. K., Berezin V., Bock E. (2008) J. Neurosci. Res. 86, 727–743 [DOI] [PubMed] [Google Scholar]
- 73. Niethammer P., Delling M., Sytnyk V., Dityatev A., Fukami K., Schachner M. (2002) J. Cell Biol. 157, 521–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ong S. H., Hadari Y. R., Gotoh N., Guy G. R., Schlessinger J., Lax I. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6074–6079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Holnthoner W., Pillinger M., Groger M., Wolff K., Ashton A. W., Albanese C., Neumeister P., Pestell R. G., Petzelbauer P. (2002) J. Biol. Chem. 277, 45847–45853 [DOI] [PubMed] [Google Scholar]
- 76. Katoh M., Katoh M. (2006) Cancer Biol. Ther. 5, 1059–1064 [DOI] [PubMed] [Google Scholar]
- 77. Pai R., Dunlap D., Qing J., Mohtashemi I., Hotzel K., French D. M. (2008) Cancer Res. 68, 5086–5095 [DOI] [PubMed] [Google Scholar]
- 78. Stambolic V., Woodgett J. R. (1994) Biochem. J. 303, 701–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sutherland C., Leighton I. A., Cohen P. (1993) Biochem. J. 296, 15–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Maetzel D., Denzel S., Mack B., Canis M., Went P., Benk M., Kieu C., Papior P., Baeuerle P. A., Munz M., Gires O. (2009) Nat. Cell Biol. 11, 162–171 [DOI] [PubMed] [Google Scholar]
- 81. Leshchyns'ka I., Sytnyk V., Morrow J. S., Schachner M. (2003) J. Cell Biol. 161, 625–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bodrikov V., Leshchyns'ka I., Sytnyk V., Overvoorde J., den Hertog J., Schachner M. (2005) J. Cell Biol. 168, 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Klehr M., Koehl U., Mühlenhoff M., Tawadros S., Fischer T., Schomäcker K., Heuckmann J. M., Bochennek K., Jensen M. (2009) J. Immunother. 32, 442–451 [DOI] [PubMed] [Google Scholar]
- 84. Lutz R. J., Whiteman K. R. (2009) MAbs 1, 548–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Berezin V., Bock E. (2010) Adv. Exp. Med. Biol. 663, 337–353 [DOI] [PubMed] [Google Scholar]