Abstract
Glioblastoma is the most common and malignant form of primary astrocytoma. Upon investigation of the insulin-like growth factor (IGF) pathway, we found the IGF2BP3/IMP3 transcript and protein to be up-regulated in GBMs but not in lower grade astrocytomas (p < 0.0001). IMP3 is an RNA binding protein known to bind to the 5′-untranslated region of IGF-2 mRNA, thereby activating its translation. Overexpression- and knockdown-based studies establish a role for IMP3 in promoting proliferation, anchorage-independent growth, invasion, and chemoresistance. IMP3 overexpressing B16F10 cells also showed increased tumor growth, angiogenesis, and metastasis, resulting in poor survival in a mouse model. Additionally, the infiltrating front, perivascular, and subpial regions in a majority of the GBMs stained positive for IMP3. Furthermore, two different murine glioma models were used to substantiate the above findings. In agreement with the translation activation functions of IMP3, we also found increased IGF-2 protein in the GBM tumor samples without a corresponding increase in its transcript levels. Also, in vitro IMP3 overexpression/knockdown modulated the IGF-2 protein levels without altering its transcript levels. Additionally, IGF-2 neutralization and supplementation studies established that the proproliferative effects of IMP3 were indeed mediated through IGF-2. Concordantly, PI3K and MAPK, the downstream effectors of IGF-2, are activated by IMP3 and are found to be essential for IMP3-induced cell proliferation. Thus, we have identified IMP3 as a GBM-specific proproliferative and proinvasive marker acting through IGF-2 resulting in the activation of oncogenic PI3K and MAPK pathways.
Keywords: Growth Factors, Insulin-like Growth Factor (IGF), MAP Kinases (MAPKs), Neurological Diseases, PI3K, RNA Binding Protein, Translation Control, Tumor Marker, Astrocytoma, Glioblastoma
Introduction
Gliomas, as the name suggests, are tumors of glial origin and are classified histologically depending on their cell type of origin as astrocytomas, oligodendrogliomas, or ependymomas (1). Based on the World Health Organization system of classification, gliomas are classified according to the hypothesized line of differentiation, i.e. whether they display features of astrocytic, oligodendroglial, or ependymal cells. They are further graded on a scale of I to IV according to their degree of malignancy as judged by the various histological features. Astrocytomas, accounting for >60% of the primary brain tumors diagnosed each year, are further classified into four grades based on their histology and immunohistochemical parameters as per the World Health Organization guidelines: grade I (pilocytic astrocytoma), grade II (diffuse astrocytoma; DA),3 grade III (anaplastic astrocytoma; AA), and grade IV (glioblastoma; GBM). Grade III and grade IV tumors are considered as malignant astrocytomas (2). The grade IV tumors present with significant intratumoral heterogeneity and remain to this day one of the most daunting therapeutic challenges due to their refractory responses to conventional therapeutics such as surgery, radiotherapy, and chemotherapy. Despite intensive therapeutic strategies and technical advances, the median survival of GBM patients has remained at a meager 15 months for the past decade (1, 3, 4).
The insulin-like growth factor (IGF) pathway arose early in evolution and has evolved to have key roles in regulating multiple aspects of organismal physiology ranging from energy metabolism, body size, longevity, cellular proliferation, and apoptosis, which has led to great interest in the relevance of this regulatory system to neoplasia (5). The IGF family includes the polypeptide ligands IGF-1 and IGF-2, two types of the cell membrane receptors (IGF-1R and IGF-2R), and an increasing number of binding proteins, IGFBPs. There are also a large group of IGFBP proteases, which hydrolyze the IGFBPs and modulate IGF bioavailability. Work done over the past decade or so has also identified novel RNA binding proteins (IGF-2BPs (insulin-like growth factor-2 binding proteins) or IMPs). This family of RNA binding proteins was identified based on their ability to bind to the IGF-2 mRNA and thereby regulate its stability and translation. Thus, the available information about the IGF pathway suggests it to be a complex signaling cascade with multiple layers of regulation. Interestingly, a large number of epidemiological studies have shown high levels of the IGF signaling to be associated with an increased risk for several common cancers, including those of the breast, prostate, lung, and colorectum (5–9). IGF-1 and IGF-2 have also been found to be strong mitogens for a wide variety of cancer cell lines (10, 11).
Upon investigation of the IGF family in glioma development, we identified IGF2BP3/IMP3, a translational activator of the IGF-2 mRNA, as a gene selectively overexpressed in GBMs. In addition to demonstrating a diagnostic and prognostic utility, we also establish a causal role for IMP3 in promoting proliferation, anchorage-independent growth, angiogenesis, and invasion. We also show that these pro-oncogenic roles of IMP3 are mediated through the translational activation of IGF-2, which further leads to the activation of its immediate downstream effectors, the PI3K and the MAPK pathways.
EXPERIMENTAL PROCEDURES
Reagents, Cell Lines, and Plasmids
Temozolomide, adriamycin, cisplatin, taxol, etoposide, and MTT were obtained from Sigma. LY294002 and U0126, the pharmacological inhibitors for the PI3K and the MAPK pathways were obtained from Alomone Biosciences. Purified IGF-2 protein was obtained from Abcam (Ab9575). A neutralizing antibody against human IGF-2 was obtained from Sigma-Aldrich (I-7276). U373, U138, H1299, HaCaT, LN18, U343, LN229, K562, U87-MG, MDA-MB231, Rat C6, B16F10 murine melanoma cells, and SVG cells were cultured in DMEM and minimum essential medium, respectively with 10% fetal bovine serum, penicillin, and streptomycin at 37 °C in a humidified atmosphere with 5% CO2. The cDNA for IGF2BP3/IMP3 (Open Biosystems) was subcloned into the mammalian expression vector pCEP4 with a hygromycin resistance gene.
Additional Methods can be found in the supplemental material.
Tumor Samples
Tumor samples were collected from patients who were operated on at the Sri Sathya Sai Institute of Higher Medical Sciences and National Institute of Mental Health and Neurosciences (Bangalore, India). Normal brain tissue samples (anterior temporal lobe) obtained during surgery for intractable epilepsy were used as control samples. The study has been scrutinized and approved by the ethics committee of the two clinical centers, and patient consent was obtained prior to initiation of the study as per the Institutional Ethics Committee guidelines and approval. Tissues were bisected, and one-half was snap-frozen in liquid nitrogen and stored at −80 °C until RNA isolation. The other half was fixed in formalin and processed for paraffin sections. These were used for histopathological grading of the astrocytoma and immunohistochemical studies. A total of 227 patient samples were used as follows: 121 glioblastoma (GBMs), 31 anaplastic astrocytoma, grade III (AAs); 26 anaplastic oligodendroglioma, grade III (AOs); 22 diffuse astrocytoma, grade II (DAs); seven diffuse oligodendroglioma, grade II (DOs); and 20 normal brain samples have been used in this study, of which a subset was used for the transcript analysis and the immunohistochemical and prognostic studies. The prognostic studies were limited to a subset of the GBM samples (n = 83).
Clinical Cohort for Survival Analysis
Newly diagnosed glioblastoma patients (n = 83) who underwent surgery in the two clinical centers (National Institute of Mental Health and Neurosciences/Sri Sathya Sai Institute of Higher Medical Sciences) were included prospectively. The study was approved by the ethics committee, and patient consent was obtained prior to initiation of the study. Adult patients (ages between 18 to 65 years) with a supratentorial lobar tumor who underwent maximal safe resection of the tumor with minimal residue noted on post-operative MRI scans and those with a post-operative Karnofsky's performance score of ≥70 were included in the study. All patients were treated uniformly with adjuvant radiotherapy and chemotherapy. Radiotherapy was administered with a total dose of 59.4 gray, given in 33 fractions along with concomitant chemotherapy with temozolomide, administered at the dose of 100 mg/day, which was continued daily for 45 days. Subsequently, five cycles of cyclical chemotherapy with temozolomide (150 mg/m2 body surface area for 5 days every 28 days) was administered. The patients were followed up clinically and with MRIs. Additional surgery was offered for those who developed symptomatic recurrences. The maximum follow-up period was 34 months.
RESULTS
IMP3 Is Up-regulated in GBM
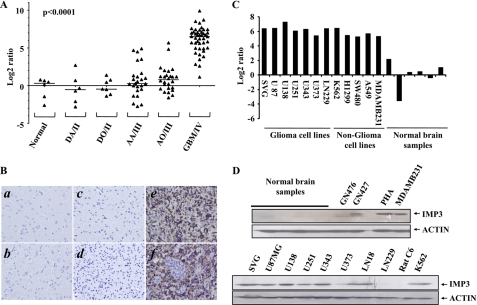
To dissect the role of the IGF pathway during glioma development, we analyzed the transcript levels of all the members (ligands, receptors, ligand binding proteins, etc.) of the IGF pathway across the different grades of glioma (supplemental Fig. S1, A–L). Further analysis identified IGF2BP3/IMP3 (insulin-like growth factor-2 binding protein-3/IGF-2 mRNA binding protein-3) as an IGF family member whose transcript levels were dramatically up-regulated only in the GBMs (supplemental Table S1). We further investigated the IMP3 transcript levels in an independent cohort of patient samples and found the transcript levels of IMP3 to be up-regulated to very high levels specifically in the GBMs (median log2 ratio = 6.49) as compared with DA, AA, DO, AO, and normal brain samples (median log2 ratios = −0.52, 0.28, −0.46, 0.78, and 0.30, respectively) (Fig. 1A). Furthermore, cytoplasmic expression of IMP3 protein was present in 87.60% (85/97) of GBMs (Fig. 1B, e and f) compared with AA (14.2%; 3/21), DA (5%; 1/20), and normal brain samples (0.0%; 0/20) with p < 0.0001 (Fig. 1B, a–d). The GBM-specific up-regulation of IMP3 protein is also confirmed by immunoblotting (data not shown). We also found IMP3 to be up-regulated in most glioma and non-glioma-derived cell lines (transcript and protein) (Fig. 1, C and D, and data not shown).
FIGURE 1.
IMP3 transcript and protein expression analysis. A, log2-transformed gene expression ratios obtained from RT-qPCR analysis of indicated samples are plotted for IMP3. B, sections from normal brain. Panels a (gray matter), b (white matter (negative)), c (DA (negative)), d (AA (negative)), e (secondary GBM (positive)), and f (primary GBM (positive)) were stained for IMP3 protein by immunohistochemistry. C, log2-transformed gene expression ratios obtained from RT-qPCR analysis of indicated cell lines or normal brain samples are plotted for IMP3. D, equal amounts of total protein lysates from indicated cell lines, patient samples, or normal brain samples were subjected to Western blotting to detect levels of IMP3 (anti-IMP3, Santa Cruz Biotechnology) and actin (anti-actin-HRP, Sigma) proteins.
IMP3 Induces Cell Proliferation, Anchorage-independent Growth, Invasion, and Chemoresistance
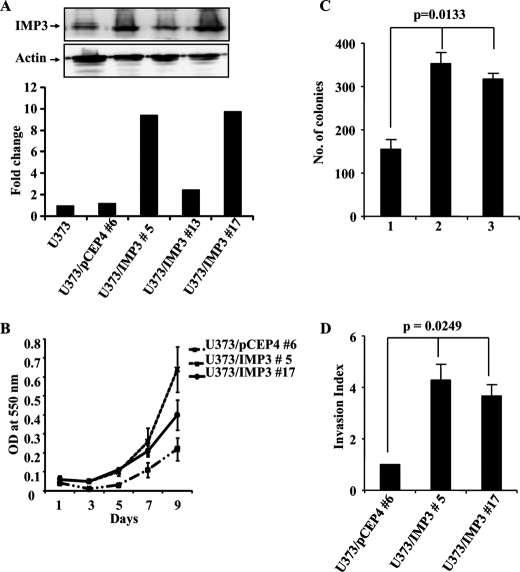
To determine the importance of IMP3 overexpression in GBMs, we developed IMP3 overexpressing U373 stable clones because the parental cell line had relatively low levels of the IMP3 protein. IMP3 stable clones (U373/IMP3#5 and U373/IMP3#17) showed increased IMP3 transcript and protein levels compared with the vector stable (U373/pCEP4#6) clone (Fig. 2A). Also, U373/IMP3#5 and U373/IMP3#17 cells grow significantly faster than the U373/pCEP4#6 clone (Fig. 2B). In addition, IMP3 stable clones formed more colonies in soft agar than the vector stable clone (Fig. 2C, compare lane 1 with lanes 2 and 3). Furthermore, IMP3 stable clones invaded 3.5 to 4.5 times more through a Matrigel matrix than the corresponding vector stable cells (Fig. 2D). IMP3 overexpressing stable cells of H1299, a lung carcinoma cell line with comparatively low levels of IMP3, also showed similar phenotypes (supplemental Fig. S2, A–E). Moreover, adenovirus-mediated IMP3 overexpression induced proliferation in immortalized human keratinocytes (HaCaT), immortalized human astrocytes (SVG) and lung carcinoma cells (H1299) (supplemental Fig. S3, A–C, respectively). Thus, IMP3 appears to promote cell proliferation, anchorage-independent growth and tumor cell invasion in multiple cell types.
FIGURE 2.
Stable overexpression of IMP3 in U373 glioma cells leads to increased proliferation, invasion, and anchorage-independent growth. A, fold change in the IMP3 transcript levels obtained from RT-qPCR analysis of the parent cell line (U373), the vector stable clone (#6), and the IMP3 stable clones (#5, 13 and 17) are plotted. The inset shows the Western blotting analysis for the vector and IMP3 stable clones with the indicated antibodies. B, viability as an indicator of cell proliferation was measured by MTT assay at indicated time points for vector and IMP3 overexpressing U373 stable clones and plotted. C, U373 vector (#6) and IMP3 stable clones (# 5, 17) were subjected to soft agar colony formation assay, and the number of colonies in the soft agar assay are counted and shown. ANOVA for difference in the colony formation abilities of the vector and IMP3 stable clones is significant (p = 0.0133). D, the invasion index of the vector and IMP3 stable U373 cells was determined based on the BDTM Matrigel assay as per the manufacturer's instructions 24 h after seeding. ANOVA for difference in the invasion index of the vector and IMP3 stable clones is significant (p = 0.0249).
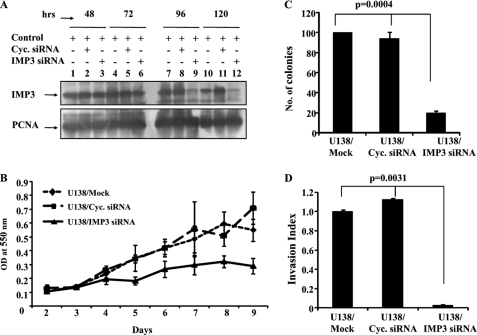
To confirm these findings, we tested whether the above-mentioned properties are reversed upon IMP3 silencing. Transfection with an IMP3-specific siRNA but not a cyclophilin siRNA resulted in an efficient reduction in the IMP3 transcript and protein levels in U138 cells, a glioma cell line with relatively higher IMP3 levels (supplemental Fig. S4, A and B, and Fig. 3A). IMP3 knockdown in U138 cells reduced cell proliferation and viability (Fig. 3B and supplemental Fig. S4C), soft agar colony formation (Fig. 3C), and invasion (Fig. 3D). Furthermore, IMP3 knockdown also made U138 cells more sensitive to taxol, temozolomide, and adriamycin (supplemental Fig. S4, D–F). Thus, these results establish a role for IMP3 in promoting cell proliferation, anchorage-independent growth, invasion, and chemoresistance.
FIGURE 3.
siRNA-mediated IMP3 knockdown in U138 glioma cells reduces proliferation, anchorage-independent growth, and invasion. A, equal amounts of total protein lysates from mock, cyclophilin, and IMP3 siRNA-treated U138 cells at the indicated time points were subjected to Western blotting to detect levels of IMP3 and proliferating cell nuclear antigen proteins. B, viability was measured by MTT assay as an indicator of cell proliferation at the indicated time points for either mock, cyclophilin, or IMP3 siRNA transfected U138 cells. C, U138 cells were either mock, cyclophilin, or IMP3 siRNA-transfected and subjected to soft agar colony formation assay. The number of colonies in the soft agar assay are counted and shown. ANOVA for difference in their colony formation abilities is significant (p = 0.0004). D, the invasion index of either mock, cyclophilin (Cyc.), or IMP3 siRNA-transfected U138 cells was determined based on the BDTM Matrigel assay as per the manufacturer's instructions 24 h after seeding. ANOVA for difference in their invasion index is significant (p = 0.0031).
IMP3 Promotes Tumor Growth, Angiogenesis, and Metastasis, Resulting in Poor Survival
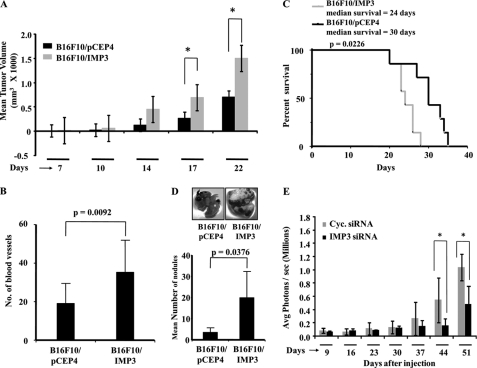
To confirm these IMP3 functions in vivo, we used B16F10 murine melanoma cells. The IMP3 stable transfectants of B16F10 cells (B16F10/IMP3) expressed very high levels of the IMP3 transcript and protein (supplemental Fig. S5A). B16F10/IMP3 cells also showed increased proliferation and anchorage-independent growth as compared with the vector stable B16F10 (B16F10/PCEP4) cells (supplemental Fig. S5, B–D). In a subcutaneous tumor model, B16F10/IMP3 tumors grew faster and had more blood vessels than B16F10/pCEP4 tumors (Fig. 4, A and B). Increased expression of the IMP3 protein in B16F10/IMP3-derived tumors was confirmed by immunohistochemistry (supplemental Fig. S5E, compare c and d with a and b). In the same experiment, when the survival of mice was followed, we found that mice bearing IMP3-overexpressing tumors showed shorter survival than the vector stable tumor-bearing mice (Fig. 4C). In a lung metastasis model, tail vein injection of B16F10/IMP3 cells resulted in significantly more tumor nodules compared with the B16F10/pCEP4 cells (Fig. 4D). We have further extended these studies and monitored the effect of IMP3 down-regulation on intracranial glioma tumor growth. For this experiment, U87-MG-Luc cells were transfected with the IMP3 or cyclophilin siRNA (efficacy of knockdown was validated; supplemental Fig. S5F) and injected into the cortex of NIH nude/nude mice, and growth of the implanted tumor cells was subsequently followed through bioluminescence imaging. Similar to our earlier observations, we found that the tumors with IMP3 knockdown show retarded growth compared with the control tumors (Fig. 4E, compare black bars versus gray bars). These set of data thus validate our in vitro observations and suggest that IMP3 contributes to increased tumor growth, angiogenesis, and metastasis resulting in the poor survival of mice carrying IMP3-overexpressing tumors.
FIGURE 4.
- IMP3 overexpression in B16F10 murine melanoma cells increases its tumorigenic and invasive potential. A, B16F10 vector and IMP3 stable clones were injected subcutaneously into the flank of NIH nu/nu mice, and the tumor sizes were measured at indicated times points and plotted. An unpaired t test for difference in their average tumor value on days 17 and 22 is significant (p = 0.0312 and 0.0209, respectively). p ≤ 0.05 is represented with an asterisk. B, the H&E-stained section of tumors derived from B16F10 vector and IMP3 stable clones (from A) were used for counting the number of blood vessels microscopically in three representative fields. The mean number of blood vessels for four of the subcutaneous tumors with the S.D. is plotted. An unpaired t test for difference in their number of blood vessels is significant (p = 0.0092). C, the animals as described in A were monitored for their survival. A Kaplan-Meier curve was generated based on the survival of the mice in the two groups (the vector and IMP3-stable clones) and was found to be significant (Gehan-Breslow, p = 0.0226). D, the vector or IMP3-stable B16F10 cells were injected into the lateral tail vein of C57BL/6 mice. The total number of macro metastatic nodules formed in each group after 30 days of injection was counted and plotted. An unpaired t test for difference in their number of nodules is significant (p = 0.0376). The inset shows representative pictures of the lungs from the two groups. E, U87-MG-Luc cells were either transfected with cyclophilin (Cyc.) or IMP3 siRNA and implanted into the cortex of NIH nude/nude mice (n = 5) (0.5 million cells per animal). The subsequent growth of the implanted tumor cells was monitored through bioluminescent imaging based on luciferase activity, and the average photon flux for each group of animals is plotted in the figure along with the S.D. in each case. An unpaired t test for difference in their average photon flux on days 44 and 51 is significant (p = 0.0436 and 0.0327, respectively). p ≤ 0.05 is represented with an asterisk.
IMP3 Expression and Its Localization, Implications for Invasion and Prognosis
Neoplastic glial cells have been proposed to exhibit perineuronal, perivascular, subpial, and intrafascicular spread as unique mechanisms of invasion adapted for the structure and composition of the CNS (12). Interestingly, among the 83 GBM samples, in addition to the tumor core, a perivascular accumulation of IMP3 stained cells was seen in the infiltrating front of the tumor in 68 cases (81.93%) (supplemental Fig. S6A, c and d). Furthermore, clusters of positively stained cells were noted invading the gray matter and the subpial regions in 83.7% cases (supplemental Fig. S6A, a and b). Also, survival analysis revealed that the median survival of patients positive for IMP3 expression (black line) was lesser than that of the group negative for IMP3 expression (red line) on univariate analysis (supplemental Fig. S6B). However, on multivariate analysis considering the age of patients, IMP3 expression did not sustain its significance.
When tested in a mouse model of glioma (13), we found that the majority of mouse glioma samples showed high IMP3 transcript and protein levels (supplemental Fig. S7, A and B, respectively). In addition to the strong positivity in the tumor core, IMP3 positive cells were seen in the subpial regions (supplemental Fig. S7C, boxes A and B, respectively). Thus, these results establish an association of IMP3 expression with tumor invasion and patient survival.
Involvement of IGF-2 and PI3K/MAPK Pathways in IMP3-induced Cell Proliferation
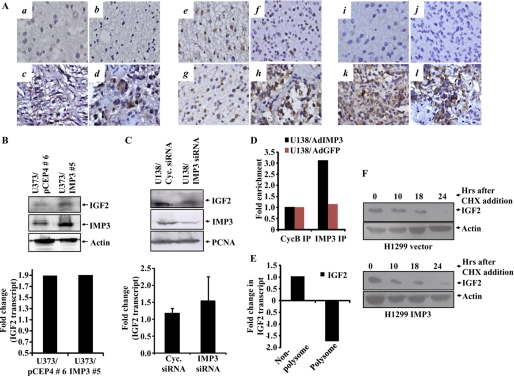
Because IMP3 has been shown to be a translational activator of IGF-2 mRNA (14), we hypothesized that IGF-2 protein levels may increase in GBMs without a corresponding increase in the IGF-2 mRNA levels as a result of the abundance of IMP3. Indeed, we found that although the IGF-2 transcript levels do not change between normal brain samples and the different grades of astrocytoma (supplemental Fig. S8), there is, however, a substantial significant increase in the IGF-2 protein levels in the GBMs (Fig. 5A, compare panels c and d with a and b) as analyzed by immunohistochemistry. Binding of IGF-1/2 to IGF-1R further activates the oncogenic PI3K and MAPK signaling cascades (5). In good agreement, the increased IGF-2 protein levels in the GBMs also correlated with positive staining for phospho-Akt (active form of Akt) (Fig. 5A, compare panels k and l with i and j), whereas total Akt (Akt2) levels remain unchanged across all grades of astrocytoma (Fig. 5A, e–h). This observed increase in the IGF-2 protein levels in GBMs without a corresponding increase in its transcript levels strongly implicates IMP3 as a possible candidate involved in augmenting IGF-2 signaling in the GBMs.
FIGURE 5.
IMP3 exerts its proproliferative effects through the translational activation of IGF-2 mRNA. A, immunohistochemical of staining for IGF-2 for section from DA (a), 3/9 (33.3%) positive for staining; AA, 2/10 (20%) positive for staining (b); secondary GBM (c) and primary GBM (d), positive for staining (29/50; 58.00%). Including four normal brain samples (all four negative), ANOVA for difference in their IGF-2 positivity was carried out (p = 0.0239). Immunohistochemical staining for AKT expression for a section from DA (e), all cases positive for staining (9/9); AA, all cases positive for staining (10/10) (f); secondary GBM (g), and primary GBM (h), (30/34; 88.20%) positive for staining. Including four normal brain samples (all cases positive) ANOVA for difference in their Akt positivity was carried out (p = 0.4139). Immunohistochemical staining for phospho Akt for section from DA, 1/9 (11.1%) positive for staining (i); AA, 2/10 (20%) cases positive for staining (j); secondary GBM (k) and primary GBM (l), 23/34 (67.60%) positive for staining. Including four normal brain samples (all four negative), ANOVA for difference in their pAkt positivity was carried out. p = 0.0008). B, equal amounts of lysates for the vector and IMP3 overexpressing stable clones of U373 were subjected to Western blotting to detect the levels of IMP3 (anti-IMP3, Sigma), IGF-2 (anti-IGF-2, Santa Cruz Biotechnology), and actin (anti-actin HRP, Sigma). Fold change in the IGF-2 transcript levels obtained from RT-qPCR analysis in the IMP3-overexpressing and vector-stable clones relative to the control cells is depicted below. There is a significant increase in the IGF-2 protein levels upon IMP3 overexpression without a change in the IGF-2 transcript levels (compare lane 2 with 1). C, equal amounts of total protein lysates from cyclophilin (Cyc.) and IMP3 siRNA-transfected U138 cells at 120 h after transfection were subjected to Western blotting to detect levels of IGF-2 (anti-IGF-2, Abcam), IMP3 (anti-IMP3, Sigma), and proliferating cell nuclear antigen (anti-proliferating cell nuclear antigen, oncogene) proteins. The bottom panel shows the fold change in the IGF-2 transcript levels obtained from RT-qPCR analysis of the siRNA transfected U138 cells at 120 h. There is a significant decrease in the IGF-2 protein levels upon IMP3 knockdown without a change in the IGF-2 transcript levels (compare lane 2 with lane 1). D, fold change in the real-time RT-qPCR-based quantitation of the IGF-2 transcript levels in the IMP3 and control antibody immunoprecipitates is plotted. LN229 cells were infected with a control adenovirus and an Ad-IMP3 adenovirus and IMP3 and the control antibody immunoprecipitates were used to analyze for the IGF-2 transcript levels. E, U138 cells were transfected with IMP3 or cyclophilin siRNA, and 4 days after siRNA transfection, the cell lysates were made and subjected to polysomal fractionation. Relative fold change in the IGF-2 transcript levels between cyclophilin and IMP3 siRNA-transfected cells within polysome or non-polysome fractions are shown. F, H1299 vector and IMP3-stable cells were treated with 50 μg/ml cycloheximide (CHX) for 10, 18, and 24 h. Equal amounts of lysates were subjected to Western blotting to detect the levels of IGF-2 and actin.
To further validate our hypothesis, we examined the levels of the IGF-2 mRNA and protein in the IMP3 over expressing stable clones of U373 and found increased IMP3 expression to correlate with increased IGF-2 protein levels without an equivalent change in the IGF-2 transcript levels (Fig. 5B). Additionally, we knocked down IMP3 levels in U138 cells and monitored the effect on IGF-2 levels in the cells. Upon analysis, we found a significant reduction in the IGF-2 protein levels upon siRNA-mediated IMP3 knockdown without a corresponding change in the IGF-2 transcript levels (Fig. 5C). We also carried out RNA immunoprecipitation studies upon adenovirus-mediated overexpression of IMP3 and assayed for enrichment of the IGF-2 mRNA. Real-time RT-qPCR based quantitation of the immunoprecipitates indicates a 3.1-fold enrichment of the IGF-2 mRNA in the IMP3-overexpressing sample upon immunoprecipitation with anti-IMP3 antibody, thereby establishing a direct association between the IMP3 protein and the IGF-2 mRNA (Fig. 5D). To further substantiate the translational activation functions of IMP3 in regulating IGF-2 levels, we have monitored the polysomal and nonpolysomal association of the IGF-2 mRNA under conditions of IMP3 knockdown (supplemental Fig. S9). As shown, IMP3 silencing resulted in a 1.8-fold reduction in polysome-associated IGF-2 mRNA with a concomitant 1.0-fold increase in nonpolysome-associated IGF-2 mRNA, thereby suggesting reduced translation of the IGF-2 transcript under IMP3 knockdown conditions (Fig. 5E). The IGF-2 protein essentially followed a similar stability pattern upon cycloheximide treatment in both vector and IMP3 stable cells (Fig. 5F), which ruled out the possibility that increased stability is a cause of increased IGF-2 protein in IMP3-stable clones. These data further corroborate well with our observations from the patient samples and establish IMP3 as a translational activator of the IGF-2 mRNA in our model system as well.
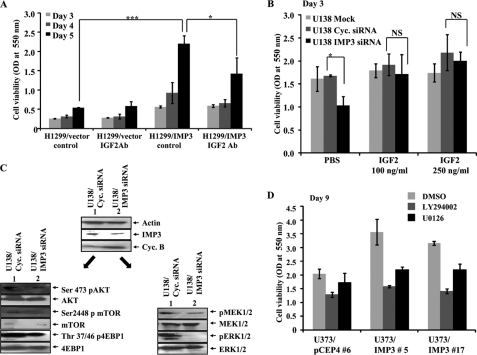
Based on this evidence, we next speculated that the proproliferative effects of IMP3 could be mediated through the actions of IGF-2 and its downstream effectors. To validate this, we monitored the effect of antibody mediated IGF-2 neutralization on the growth of IMP3 overexpressing clones (H1299). In good agreement, we found that an IGF-2 antibody significantly abrogated the growth of IMP3-overexpressing stable clones and not the vector clones, thereby suggesting that the proproliferative effects of IMP3 are indeed mediated through IGF-2 (Fig. 6A). In a converse experiment, we find that exogenously supplied IGF-2 is able to rescue the growth inhibition caused by IMP3 knockdown in U138 glioma cells. We found that although IMP3 knockdown compromises the growth of cells as compared with the mock or cyclophilin siRNA transfection, this inhibition was relieved by the exogenous addition of IGF-2 (Fig. 6B). These studies implicate IGF-2 as one of the primary effectors of IMP3 function.
FIGURE 6.
IMP3 exerts its proproliferative effects through the IGF-2-PI3K and IGF-2-MAPK cascades. A, viability was measured by MTT assay at indicated time points for H1299 vector and IMP3 stable clones treated with a neutralizing antibody for IGF-2 as indicated. The assays were carried out in triplicate, and the mean value for each cell type at each time point was used to generate the graph. A one-way analysis of variance was carried out to test the significance of the observed differences between the groups and a p ≤ 0.05 is represented with an asterisk, p ≤ 0.01 is represented as double asterisks and p ≤ 0.0001 is represented as triple asterisks. B, viability was measured by MTT assay as an indicator of cell proliferation at the indicated time points for either mock, cyclophilin (cyclo.), or IMP3 siRNA-transfected U138 cells. IGF-2 was exogenously supplemented as indicated to monitor its ability to rescue the effects of IMP3 knockdown. A one-way analysis of variance was carried out to test the significance of the observed differences between the groups. A p value ≤ 0.05 is represented as an asterisk, which is significant, and NS refers to nonsignificant difference. C, equal amounts of total protein lysates from cyclophilin and IMP3 siRNA-treated U138 cells were subjected to Western blotting with the indicated antibodies. Note that the intensity of phospho-mTOR, phospho-Akt, phospho-4EBP1, phospho-MEK1/2, and phospho-ERK1/2 bands reduces in lane 2 compared with lane 1, whereas that of total the corresponding total proteins remain unchanged. D, viability was measured by MTT assay on day 9 after plating for the U373 vector clone (#6) and IMP3 stable clones (#5 and 17) treated with dimethyl sulfoxide (vehicle), LY294002 (20 μm), or U0126 (10 μm). Please note that dimethyl sulfoxide (DMSO)-treated IMP3 stable clone (#5 and #17) grow faster than vector stable clone (compare light gray bars). Both LY294002 and U0126 treated IMP3 stable clone (#5 and #17) failed to show faster growth than vector stable clone (compare dark gray and black bars with light gray bars).
To further validate the involvement of IGF-2, we have analyzed the role of the PI3K and MAPK pathways, which are the known immediate downstream effectors of IGF-2. Treatment with IMP3 siRNA and not the cyclophilin siRNA, in U138 glioma cells, resulted in a substantial reduction in the activation status of both the PI3K and MAPK arms of IGF signaling. Levels of Ser473 phospho-Akt, Ser2448 phospho-mTOR, Thr37/46 phospho-4EBP1, phospho-MEK1/2, and phospho-ERK1/2 were reduced upon treatment with the IMP3 siRNA without a corresponding change in the levels of the total proteins, respectively (Fig. 6C, compare lane 2 with 1). To further investigate the importance of these pathways in IMP3-induced cell proliferation, we monitored the growth of IMP3 overexpressing cells under conditions wherein either of these pathways was pharmacologically inhibited. As shown above, U373/IMP3 clones (#5 and #17) showed increased proliferation when compared with the U373 vector (#6) clone (Fig. 6D, compare light gray bars). However, when the cells were treated with the PI3K inhibitor (LY294002) or MEK1/2 inhibitor (UO126), the proliferation of IMP3 overexpressing cells was severely abrogated unlike the vector stable cells, which were only modestly affected (Fig. 6D, compare dark gray bars and black bars with corresponding light gray bars). Thus, these results together establish IMP3 as a causal factor involved in the activation of these two major pro-oncogenic pathways through the translational activation of IGF-2 in GBMs, thereby augmenting glioma cell proliferation.
DISCUSSION
In this study, we found IMP3, a novel RNA binding protein, to be specifically up-regulated in GBMs. IMP3, also known as L523S or KOC (K homologous domain-containing protein overexpressed in cancer), is a member of the IMP family that also consists of IMP1 and -2. Originally identified from a pancreatic tumor cDNA screen, IMP3 has since been found to be expressed in a number of solid tumors and in many fetal tissues wherein the IMP family members have been reported to play a pivotal role in RNA trafficking and stabilization, cell growth, and migration during embryogenesis (15–17). Recent studies have also identified IMP3 to be overexpressed in other human malignancies of thyroid, bladder, pancreas, esophagus, endometrial and cervical carcinoma, melanoma, non-small cell lung carcinoma, osteosarcoma, and etc. (18–26). IMP3/IGF2BP3 has also been found to be associated with aggressive and advanced carcinomas in colon, kidney, neuroendocrine skin, liver, bladder, breast, ovary, and other soft tissue sarcomas (25, 27–39). However, there are no reports of IMP3 involvement in any type of brain cancer, including glioma. In our studies, we find IMP3 to be overexpressed to high levels specifically in the GBMs, potentiating its use as a diagnostic marker, which could be an invaluable aid in the accurate classification of the tumor and subsequently in the treatment protocol to be followed. We could also demonstrate a possible prognostic utility for IMP3 as patients overexpressing the protein have a poor median survival. This GBM-specific expression of IMP3 also suggests that IMP3 may not play a role in the early events of transformation and may instead be essential for the features seen in the more advanced grades of cancer such as neoangiogenesis, invasion, migration, chemoresistance, stress tolerance, etc.
Unlike the previous gene-phenotype association studies (14, 18, 22–40), we have also established a causal role for IMP3 in cancer by overexpression and knockdown based approaches in glioma and non-glioma-derived cell lines. Through in vitro studies, we show that IMP3 leads to increased proliferation, anchorage-independent growth, and invasion. Concordantly, in vivo, we find that overexpression of IMP3 leads to faster tumor growth with greater angiogenesis and a consequent poor prognosis for the mice in an intracranial and a subcutaneous tumor model. IMP3 overexpression also resulted in greater invasive potential in vivo as evidenced by the increased formation of nodules in a lung metastasis model.
Glioma cells have been proposed to have unique mechanisms of invasion as they migrate through the normal parenchyma, collect just below the pial margin (subpial spread), surround neurons and vessels (perineuronal and perivascular satellitosis), and migrate through the white matter tracts (intrafascicular spread) (12, 41, 42). The tumor-infiltrating front represents the invading glioma cells and the genes expressed in this region are likely to provide an advantage to the invading tumor cells (43). Indeed, we found IMP3-positive cells in the perivascular areas, the infiltrating front of the tumor, and in the subpial regions. We have further extended our studies to a mouse model of glioma where, similar to the human GBMs, the majority of the tumor samples showed high levels of the IMP3. Also, similar to the patient samples, many tumors derived from this mouse model of glioma showed IMP3 positivity in the subpial regions. Thus, these results collectively establish IMP3 as a protumorigenic gene overexpressed in the more malignant grades of glioma with a role in promoting proliferation, invasion, migration, angiogenesis, and chemoresistance.
The embryonic expression of IMP3 and its reappearance in malignant cancers may correspond to the reacquisition of the neoplastic cell of primitive migratory behavior, which is seen during development of the central nervous system, leading to the diffuse spread of individual tumor cells over long distances, resulting in the extreme mortality associated with the disease (17, 34, 42–44).
Structurally, studies reveal IMP3 to be an RNA binding protein with two RNA recognition motifs and four KH domains, which bind to the IGF-2 mRNA, thereby activating its translation (14). We therefore hypothesized that the increased levels of IMP3 in the GBMs could lead to greater levels of the IGF-2 protein without an increase in its transcript levels. We also found higher levels of IGF-2 protein in GBMs as compared with the lower grades without a corresponding increase in the IGF-2 transcript levels. Furthermore, through knockdown and overexpression studies, we found IMP3 to play a causal role in increasing the levels of IGF-2 protein in the cells without a corresponding increase in the IGF-2 transcript levels. Through IGF-2 neutralization or exogenous supplementation in the background of IMP3 overexpression or knockdown, we also establish that the proproliferative effects of IMP3 are indeed mediated through the actions of IGF-2. The mitogenic effects of the IGF pathway are known to be mediated through the PI3K and the MAPK pathways. We have further established a causal role for IMP3 in the activation of these pathways as the siRNA-mediated knockdown of IMP3 adversely affected these signaling cascades. We also establish their role in mediating the proproliferative effects of IMP3 by the use of pharmacological inhibitors of these pathways, which abrogated IMP3-induced proliferation specifically. This becomes increasingly relevant as activation of the Akt pathway has been identified as a key event responsible for the conversion of the grade III AA to the grade IV GBMs (45). Our studies establish IMP3 as a critical oncogenic factor expressed solely in the GBMs with the potential to activate the PI3K/Akt pathway through the translational activation of IGF-2 levels where it could play a vital role in the progressive conversion from lower grade astrocytomas to GBMs.
Our current understanding suggests a pro-oncogenic role for the IGF pathway in multiple cancers. The IGF pathway is known to be hyperactivated in a number of cancers through diverse mechanisms such as the increased expression of the ligands (IGF-1, IGF-2) and the receptor (IGF-1R); alterations in the IGFBPs; and loss of the negative regulator IGF-2R (10, 11, 46–50). In this light, our work reveals a novel mechanism for activation of the IGF pathway at the level of mRNA translation. We find IMP3, a novel RNA binding protein to be dramatically up-regulated in GBMs, wherein this protein exerts its pro-oncogenic functions through the translational activation of the IGF-2 mRNA. However, it is also conceivable that IMP3 has other IGF-2-independent functions in regulating cell physiology and tumorigenesis.
Supplementary Material
Acknowledgments
We thank professors Inder Verma, M. R. S. Rao, P. Kondaiah, and U. Varshney for help and valuable suggestions. We also thank A. A. Srinivas, Cini Samuel, K. Premkumar, and K. Chandrasekar. Infrastructural support by funding from Indian Council of Medical Research, Department of Biotechnology, Department of Science, and University Grants Commission to the Department of Microbiology and Cell Biology is acknowledged. The Central Animal Facility of the Indian Institute of Science is acknowledged for animal experiments.
This work was supported by a grant from Council of Scientific and Industrial Research, Government of India under the New Millennium Indian Technology Leadership Initiative program.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Table S1, Figs. S1–S9, and additional references.
- DA
- diffuse astrocytoma
- IMP3
- IGF-2 mRNA binding protein
- qPCR
- quantitative PCR
- IGF-2
- insulin-like growth factor-2
- GBM
- glioblastoma
- AA
- anaplastic astrocytoma
- AO
- anaplastic oligodendroglioma
- DO
- diffuse oligodendroglioma
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- ANOVA
- analysis of variance.
REFERENCES
- 1. Wen P. Y., Kesari S. (2008) N. Engl. J. Med. 359, 492–507 [DOI] [PubMed] [Google Scholar]
- 2. Furnari F. B., Fenton T., Bachoo R. M., Mukasa A., Stommel J. M., Stegh A., Hahn W. C., Ligon K. L., Louis D. N., Brennan C., Chin L., DePinho R. A., Cavenee W. K. (2007) Genes Dev. 21, 2683–2710 [DOI] [PubMed] [Google Scholar]
- 3. Meyer M. A. (2008) N. Engl. J. Med. 359, 1850; author reply 1850 [DOI] [PubMed] [Google Scholar]
- 4. Stupp R., Hegi M. E., Mason W. P., van den Bent M. J., Taphoorn M. J., Janzer R. C., Ludwin S. K., Allgeier A., Fisher B., Belanger K., Hau P., Brandes A. A., Gijtenbeek J., Marosi C., Vecht C. J., Mokhtari K., Wesseling P., Villa S., Eisenhauer E., Gorlia T., Weller M., Lacombe D., Cairncross J. G., Mirimanoff R. O. (2009) Lancet Oncol. 10, 459–466 [DOI] [PubMed] [Google Scholar]
- 5. Pollak M. (2008) Nat. Rev. Cancer 8, 915–928 [DOI] [PubMed] [Google Scholar]
- 6. Gennigens C., Menetrier-Caux C., Droz J. P. (2006) Crit. Rev. Oncol. Hematol. 58, 124–145 [DOI] [PubMed] [Google Scholar]
- 7. Lφnning P. E., Helle S. I. (2004) Novartis Found. Symp. 262, 205–212 [PubMed] [Google Scholar]
- 8. Manousos O., Souglakos J., Bosetti C., Tzonou A., Chatzidakis V., Trichopoulos D., Adami H. O., Mantzoros C. (1999) Int. J. Cancer 83, 15–17 [DOI] [PubMed] [Google Scholar]
- 9. Moschos S. J., Mantzoros C. S. (2002) Oncology 63, 317–332 [DOI] [PubMed] [Google Scholar]
- 10. Baserga R., Peruzzi F., Reiss K. (2003) Int. J. Cancer 107, 873–877 [DOI] [PubMed] [Google Scholar]
- 11. Toretsky J. A., Helman L. J. (1996) J. Endocrinol. 149, 367–372 [DOI] [PubMed] [Google Scholar]
- 12. Holland E. C. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6242–6244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marumoto T., Tashiro A., Friedmann-Morvinski D., Scadeng M., Soda Y., Gage F. H., Verma I. M. (2009) Nat. Med. 15, 110–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liao B., Hu Y., Herrick D. J., Brewer G. (2005) J. Biol. Chem. 280, 18517–18524 [DOI] [PubMed] [Google Scholar]
- 15. Nielsen F. C., Nielsen J., Christiansen J. (2001) Scand. J. Clin. Lab. Invest. Suppl. 234, 93–99 [PubMed] [Google Scholar]
- 16. Nielsen F. C., Nielsen J., Kristensen M. A., Koch G., Christiansen J. (2002) J. Cell Sci. 115, 2087–2097 [DOI] [PubMed] [Google Scholar]
- 17. Nielsen J., Christiansen J., Lykke-Andersen J., Johnsen A. H., Wewer U. M., Nielsen F. C. (1999) Mol. Cell. Biol. 19, 1262–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bellezza G., Cavaliere A., Sidoni A. (2009) Hum. Pathol. 40, 1205–1206 [DOI] [PubMed] [Google Scholar]
- 19. Do S. I., Kim Y. W., Park H. R., Park Y. K. (2008) Oncol. Res. 17, 269–272 [DOI] [PubMed] [Google Scholar]
- 20. Li C., Rock K. L., Woda B. A., Jiang Z., Fraire A. E., Dresser K. (2007) Mod. Pathol. 20, 242–247 [DOI] [PubMed] [Google Scholar]
- 21. Li C., Zota V., Woda B. A., Rock K. L., Fraire A. E., Jiang Z., Lu D., Xu B., Dresser K., Lutman C. V., Fischer A. H. (2007) Mod. Pathol. 20, 1263–1268 [DOI] [PubMed] [Google Scholar]
- 22. Li L., Xu H., Spaulding B. O., Cheng L., Simon R., Yao J. L., di Sant'Agnese P. A., Bourne P. A., Huang J. (2008) Hum. Pathol. 39, 1205–1211 [DOI] [PubMed] [Google Scholar]
- 23. Lu D., Vohra P., Chu P. G., Woda B., Rock K. L., Jiang Z. (2009) Am. J. Surg. Pathol. 33, 521–525 [DOI] [PubMed] [Google Scholar]
- 24. Slosar M., Vohra P., Prasad M., Fischer A., Quinlan R., Khan A. (2009) Endocr. Pathol. 20, 149–157 [DOI] [PubMed] [Google Scholar]
- 25. Xu H. (2008) Expert. Rev. Mol. Diagn. 8, 557–558 [DOI] [PubMed] [Google Scholar]
- 26. Zheng W., Yi X., Fadare O., Liang S. X., Martel M., Schwartz P. E., Jiang Z. (2008) Am. J. Surg. Pathol. 32, 304–315 [DOI] [PubMed] [Google Scholar]
- 27. Hoffmann N. E., Sheinin Y., Lohse C. M., Parker A. S., Leibovich B. C., Jiang Z., Kwon E. D. (2008) Cancer 112, 1471–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jiang Z., Chu P. G., Woda B. A., Liu Q., Balaji K. C., Rock K. L., Wu C. L. (2008) Clin. Cancer Res. 14, 5579–5584 [DOI] [PubMed] [Google Scholar]
- 29. Jiang Z., Chu P. G., Woda B. A., Rock K. L., Liu Q., Hsieh C. C., Li C., Chen W., Duan H. O., McDougal S., Wu C. L. (2006) Lancet Oncol. 7, 556–564 [DOI] [PubMed] [Google Scholar]
- 30. Jiang Z., Lohse C. M., Chu P. G., Wu C. L., Woda B. A., Rock K. L., Kwon E. D. (2008) Cancer 112, 2676–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kapoor S. (2008) Clin. Cancer Res. 14, 5640; author reply 5640–5641 [DOI] [PubMed] [Google Scholar]
- 32. Köbel M., Xu H., Bourne P. A., Spaulding B. O., Shih IeM., Mao T. L., Soslow R. A., Ewanowich C. A., Kalloger S. E., Mehl E., Lee C. H., Huntsman D., Gilks C. B. (2009) Mod. Pathol. 22, 469–475 [DOI] [PubMed] [Google Scholar]
- 33. Li D., Yan D., Tang H., Zhou C., Fan J., Li S., Wang X., Xia J., Huang F., Qiu G., Peng Z. (2009) Ann. Surg. Oncol. 16, 3499–3506 [DOI] [PubMed] [Google Scholar]
- 34. Mueller F., Bommer M., Lacher U., Ruhland C., Stagge V., Adler G., Gress T. M., Seufferlein T. (2003) Br. J. Cancer 88, 699–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Noske A., Faggad A., Wirtz R., Darb-Esfahani S., Sehouli J., Sinn B., Nielsen F. C., Weichert W., Buckendahl A. C., Röske A., Müller B., Dietel M., Denkert C. (2009) Int. J. Gynecol. Pathol. 28, 203–210 [DOI] [PubMed] [Google Scholar]
- 36. Riener M. O., Fritzsche F. R., Clavien P. A., Pestalozzi B. C., Probst-Hensch N., Jochum W., Kristiansen G. (2009) Hum. Pathol. 40, 1377–1383 [DOI] [PubMed] [Google Scholar]
- 37. Walter O., Prasad M., Lu S., Quinlan R. M., Edmiston K. L., Khan A. (2009) Hum. Pathol. 40, 1528–1533 [DOI] [PubMed] [Google Scholar]
- 38. Yantiss R. K., Woda B. A., Fanger G. R., Kalos M., Whalen G. F., Tada H., Andersen D. K., Rock K. L., Dresser K. (2005) Am. J. Surg. Pathol. 29, 188–195 [DOI] [PubMed] [Google Scholar]
- 39. Yuan R. H., Wang C. C., Chou C. C., Chang K. J., Lee P. H., Jeng Y. M. (2009) Ann. Surg. Oncol. 16, 1711–1719 [DOI] [PubMed] [Google Scholar]
- 40. Sitnikova L., Mendese G., Liu Q., Woda B. A., Lu D., Dresser K., Mohanty S., Rock K. L., Jiang Z. (2008) Clin. Cancer Res. 14, 1701–1706 [DOI] [PubMed] [Google Scholar]
- 41. Bellail A. C., Hunter S. B., Brat D. J., Tan C., Van Meir E. G. (2004) Int. J. Biochem. Cell Biol. 36, 1046–1069 [DOI] [PubMed] [Google Scholar]
- 42. Giese A., Bjerkvig R., Berens M. E., Westphal M. (2003) J Clin Oncol 21, 1624–1636 [DOI] [PubMed] [Google Scholar]
- 43. Giese A., Westphal M. (1996) Neurosurgery 39, 235–250; discussion 250–232 [DOI] [PubMed] [Google Scholar]
- 44. Mueller-Pillasch F., Pohl B., Wilda M., Lacher U., Beil M., Wallrapp C., Hameister H., Knöchel W., Adler G., Gress T. M. (1999) Mech. Dev. 88, 95–99 [DOI] [PubMed] [Google Scholar]
- 45. Sonoda Y., Ozawa T., Aldape K. D., Deen D. F., Berger M. S., Pieper R. O. (2001) Cancer Res. 61, 6674–6678 [PubMed] [Google Scholar]
- 46. Frasca F., Pandini G., Sciacca L., Pezzino V., Squatrito S., Belfiore A., Vigneri R. (2008) Arch. Physiol. Biochem. 114, 23–37 [DOI] [PubMed] [Google Scholar]
- 47. Grimberg A. (2003) Cancer Biol. Ther. 2, 630–635 [PMC free article] [PubMed] [Google Scholar]
- 48. Jerome L., Shiry L., Leyland-Jones B. (2003) Endocr. Relat. Cancer 10, 561–578 [DOI] [PubMed] [Google Scholar]
- 49. Monzavi R., Cohen P. (2002) Best. Pract. Res. Clin. Endocrinol. Metab. 16, 433–447 [DOI] [PubMed] [Google Scholar]
- 50. van Roozendaal C. E., Gillis A. J., Klijn J. G., van Ooijen B., Claassen C. J., Eggermont A. M., Henzen-Logmans S. C., Oosterhuis J. W., Foekens J. A., Looijenga L. H. (1998) FEBS Lett. 437, 107–111 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.