Abstract
In the initial step of integration, retroviral integrase (IN) introduces precise nicks in the degenerate, short inverted repeats at the ends of linear viral DNA. The scissile phosphodiester bond is located immediately 3′ of a highly conserved CA/GT dinucleotide, usually 2 bp from the ends. These nicks create new recessed 3′-OH viral DNA ends that are required for joining to host cell DNA. Previous studies have indicated that unpairing, “fraying,” of the viral DNA ends by IN contributes to end recognition or catalysis. Here, we report that end fraying can be detected independently of catalysis with both avian sarcoma virus (ASV) and human immunodeficiency virus type 1 (HIV-1) IN proteins by use of fluorescence resonance energy transfer (FRET). The results were indicative of an IN-induced intramolecular conformational change in the viral DNA ends (cis FRET). Fraying activity is tightly coupled to the DNA binding capabilities of these enzymes, as follows: an inhibitor effective against both IN proteins was shown to block ASV IN DNA binding and end fraying, with similar dose responses; ASV IN substitutions that reduced DNA binding also reduced end fraying activity; and HIV-1 IN DNA binding and end fraying were both undetectable in the absence of a metal cofactor. Consistent with our previous results, end fraying is sequence-independent, suggesting that the DNA terminus per se is a major structural determinant for recognition. We conclude that frayed ends represent a functional intermediate in which DNA termini can be sampled for suitability for endonucleolytic processing.
Keywords: DNA Structure, Enzyme Inhibitors, Fluorescence Resonance Energy Transfer (FRET), HIV, Integrase, Avian Sarcoma Virus (ASV), DNA Binding, DNA Distortion, Enzymology, Structure Modeling
Introduction
Within hours after infection of a host cell, a colinear DNA copy of the retroviral RNA genome becomes integrated into the host chromosome, through a highly coordinated series of enzymatic events. The steps are carried out by the virus-encoded reverse transcriptase, which makes a double-stranded copy of the viral RNA genome, and the viral integrase (IN),2 which catalyzes the integration of this newly synthesized DNA into the DNA of the host cell. Following integration, an essential step for virus replication, the viral DNA is permanently associated with the genome of the host cell and all daughters and serves as a template for viral gene transcription and the production of progeny virus.
The underlying chemical steps in retroviral DNA integration are generally understood (1). The reaction can be divided into two steps, which can be reproduced in vitro using purified IN and model DNA substrates. (i) The nuclease activity of IN introduces nicks, 3′ of the highly conserved CA dinucleotide, usually 2 bp from the DNA terminus, in a “processing” step that releases a dinucleotide and generates new, recessed 3′-OH ends. (ii) In the second step, these newly exposed 3′-oxygen atoms are joined to staggered backbone phosphates on opposite strands of the host DNA, via concerted cleavage and ligation. Because the target phosphates are staggered, this “joining” step leaves gaps in the opposite strands of host DNA, and host mechanisms are required to repair the gaps and complete the covalent joining (for review, see Ref. 2). These combined viral and host-catalyzed enzymatic steps produce the characteristic sequence signatures of retroviral integration: loss of the terminal 2 bp from the linear viral DNA ends and the duplication, depending on the virus, of 4–6 bp of host DNA flanking the integration site.
Substrate Recognition by IN Does Not Depend on Sequence Alone
Sequences at the tips of the blunt-ended retroviral DNA (10–20 bp) are sufficient for IN recognition, processing, and joining. However, the mechanism by which such substrates are recognized is not completely understood, and it has been difficult to detect sequence-specific binding to viral DNA in vitro (3). Sequence identity of the generally imperfect terminal inverted repeats that are embedded in the U3 and U5 regions of the long terminal repeat (LTR) at the ends of each viral DNA can range from 16/18 for murine leukemia virus, to 12/18 for avian sarcoma virus (ASV), 11/18 for human immunodeficiency virus-1 (HIV-1), and only 5/18 for HTLV-1. A minimum length for the viral DNA substrate and roles for individual base pairs can be determined using in vitro integration assays. The conserved CA/GT sequence located 2 bp from the DNA end (immediately 5′ of the scissile phosphodiester bond) comprises an invariant feature of the terminal inverted repeats. Surprisingly, some studies have shown that even these conserved dinucleotide pairs are not absolutely essential for IN-mediated DNA processing in vitro (4) or integration in vivo (5, 6). Others have demonstrated cross-recognition of heterologous LTRs by retroviral IN proteins (7), which is typically accounted for by limited sequence identity between the cognate and heterologous DNA tips. Thus, several features of IN-substrate DNA interactions, including cross-recognition, the divergent sequences at the two viral tips, and the retention of viral DNA substrate function after significant base pair substitutions, all imply that DNA sequence per se plays only a partial role in recognition and processing of retroviral DNA ends by IN.
Role for DNA Structure in the Recognition of the Viral Substrate by IN
Results from several studies have indicated that DNA structure also contributes to substrate recognition by IN (8–13). Features that have been implicated include intrinsic properties of the highly conserved, subterminal CA/GT dinucleotide, which produces characteristically flexible base-paired structures (8–10). This dinucleotide sequence is conserved in the ends of all retroviral DNAs, LTR retrotransposons, and certain DNA transposons, suggesting that a sequence-driven DNA structure may provide shared cues for recognition or catalysis of the adjacent scissile bond. A role for the DNA terminus has also been implicated in the IN processing reaction. The first clue that properties of DNA termini may contribute to IN recognition or catalysis came from the observation that extension of the viral DNA duplex beyond the terminus reduced the efficiency of IN-mediated processing (11). The fact that such an embedded DNA tip sequence is resistant to this activity demonstrated that the structural context of the terminal sequence CA/GT is important for processing.
A seminal study by Scottoline et al. (12) evaluated the contributions of HIV-1 DNA end structure and sequence to HIV-1 IN processing of the viral DNA terminus. Using short model DNA substrates, these authors demonstrated that mismatching of terminal base pairs resulted in an increase in the efficiency of accurate processing by HIV-1 IN, whereas extending the duplex beyond the processing site inhibited this activity. They also found that mismatches at the junction between the viral DNA and the duplex extension could rescue the defect. These results implied that unpairing of DNA termini, denoted “fraying,” is an intermediate step in the processing reaction, and these authors suggested that pre-fraying of ends by mismatching enhanced processing by reducing an energetic barrier to end binding.
Other independent findings have indicated that DNA end distortion takes place during IN catalysis. In the processing reaction, a water oxygen normally serves as the attacking nucleophile at the processing site. However, analysis of the products formed in the processing reaction in vitro indicate that an alternate nucleophile, the 3′-hydroxyl oxygen of the strand to be cleaved, can attack the internal phosphodiester bond to generate a cyclic dinucleotide product (13). Such a reaction would clearly require both fraying and distortion of the viral DNA end. Studies with HIV-1 IN and viral duplex substrates in which the ends of the DNA strands have been chemically linked showed that the processing reaction is reduced substantially, although not totally obliterated, by such a structural restriction (14).
Detection of the Physical Interaction of IN with DNA Ends
We previously applied a chemical probing approach with KMnO4 to measure directly the structural changes at viral DNA termini that were induced by interaction with ASV IN (4). We found that thymines at positions −1, −2, and −3 from the ends of both viral DNA strands (…CATT/…GTAA) became sensitive to modification by KMnO4 upon IN binding, consistent with both unpairing and unstacking of the terminal base pairs. We also demonstrated that mismatching of terminal bases increased the efficiency of the processing reaction, as had been observed with HIV-1 IN. Remarkably, we observed that the processing site at the −2 position could be specified in the absence of the highly conserved CA/GT, albeit at reduced efficiency. However, whereas DNA end fraying by HIV-1 IN was strongly predicted, we could not detect such activity by this protein via chemical probing. In this report we describe an approach based on fluorescence resonance energy transfer (FRET) for analyses of DNA end fraying by both HIV-1 and ASV IN proteins. We provide evidence that both proteins are indeed capable of promoting or stabilizing frayed DNA ends and uncover additional details concerning this important activity.
EXPERIMENTAL PROCEDURES
DNA Substrates
For FRET assays with ASV IN, a 22-bp duplex corresponding to the U3 end of the viral DNA was used (see Fig. 1), and with HIV IN, a 22-bp duplex corresponding to the U5 end of the viral DNA was used (cleaved strand, 5′-AGTGTGGAAAATCTCTAGCAGT; complement strand, 5′-ACTGCTAGAGATTTTCCACACT). Fluorophores were attached covalently to the 5′ ends, unless otherwise indicated. For DNA binding studies, ASV or HIV-1-derived viral DNA duplexes containing 6-carboxyfluorescein attached to the 5′ end of the cleaved strand were used. All fluorescently labeled oligodeoxynucleotides in our studies were HPLC-purified by the supplier, Integrated DNA Technologies (Coralville, IA). Unlabeled oligodeoxynucleotides were obtained from the Fox Chase Cancer Center Biotechnology Facility.
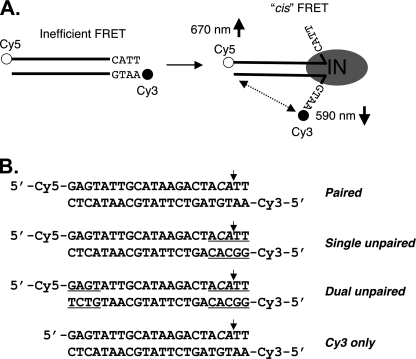
FIGURE 1.
DNA end fraying and experimental design. A, illustration of ASV viral DNA end fraying of terminal ∼4 bp by ASV IN as determined previously (4) and the predicted change in intramolecular (cis) FRET between donor (cyanine 3 (Cy3), filled circles) and acceptor (cyanine 5 (Cy5), open circles) fluorophores attached to the 5′ ends of a 22-bp oligodesoxyribonucleotide substrate. IN multimers, minimally dimers, are represented by a gray oval. B, sequences of the ASV-viral substrate duplexes used in these studies. Mismatched nucleotides at the termini are underlined. The small vertical arrows mark the normal sites of 3′ end processing by IN, in what is denoted the cleaved strand.
Protein Preparation
Methods for the expression and purification of ASV IN have been described (15). The HIV-1 IN used in the experiments was His-tagged, and methods for its expression and purification have also been reported previously (16).
DNA Binding Assays
DNA binding was measured by fluorescence polarization using an assay described previously (17).
FRET Detection
Interactions between IN proteins and substrates were monitored after mixing on ice, to prevent catalysis. Unless otherwise noted, 50 nm fluorescently labeled DNA was mixed with 400 or 800 nm IN in 150-μl reactions containing 150 mm NaCl, 10 mm Tris (pH 8.0). After 5 min on ice, readings were taken with a TECAN Genios Pro multimode microplate reader (Tecan Austira GmbH, Salzburg, Austria), set to Fluorescence Intensity mode, dual wavelength, with excitation at 535 nm. The Cy3 donor fluorophore has an excitation peak 535 nm and emission maximum (FD) at 590 nm. Fluorescence emission for the acceptor fluorophore Cy5 (FA), was measured at its peak of 670 nm, and for the donor fluorophore Cy3 at 590 nm upon excitation at 535 nm, a method that is optimal in terms of signal/noise (18). Three to five readings were normally taken at 2.5-min intervals. S.D. values for these readings were ≤5%. The values were averaged and FRET efficiencies determined according to the Förster equation, as described under “Results.”
Fluorescence emission spectra were recorded on a PTI scanning fluorometer (Photon Technology International, Lawrenceville, NJ), using an excitation wavelength of 535 nm and excitation and emission bandwidths of 2 and 4 nm, respectively.
Molecular Modeling of an ASV Intasome Complex
The sequence of ASV IN was aligned with the PFV IN intasome template (Protein Data Bank ID code 3L2Q) using the program MolIDE (19), and side chain conformations were predicted with the program SCWRL 4 (20). The inner dimer of PFV was chosen for modeling a dimer of ASV bound to viral DNA ends. The DNA base orientations were maintained while converting the viral DNA to ASV U3-specific bases with the SWISS-PROT DeepView software. To compare the position of the inhibitor Y-3, the ASV IN catalytic core structure with bound Y-3 (Protein Data Bank ID code 1A5V) was superimposed on the intasome model to observe changes in loop orientation and clashes between Y-3 and viral DNA strands. The ASV IN-Y3 structure was also used to predict the position of Y-3 in a model of HIV-1 IN (21). Structure figures were produced with Chimera (22).
RESULTS
Experimental Strategy and Validation
FRET is a distance-dependent interaction in which excitation is transferred from a donor to an acceptor fluorophore. The efficiency of such transfer depends on the inverse sixth power of the distance between them and can be calculated using the Förster equation,
where EFRET is the FRET efficiency, R is the distance between the fluorophores, and R0 is the characteristic Förster distance for the particular donor-acceptor pair at which EFRET equals 0.5. For the Cy3-Cy5 pair used in our studies, the Förster distance is 53 Å. The terminal donor and acceptor in our standard 22-bp DNA duplexes are approximately 81 Å apart, a distance that generates a weak FRET signal for these fluorophores, allowing for optimal detection of FRET increases (Fig. 1A). FRET efficiency, and changes thereof, can be monitored experimentally by determining the fluorescence of the donor in the absence (FD) and presence (FDA) of the acceptor using the formula EFRET = 1 − FDA/FD.
To validate our assay system, we measured the emission ratios for fluorophores in DNA duplexes (Fig. 1B) modeled after the sequence at the U3 end of unintegrated DNA of the ASV. The viral sequences were either fully base-paired or contained mismatches of 4–5 bp at one or both ends; a control duplex contained only one fluorophore. The results (Fig. 2A) showed the same background signal for the control, an unlabeled duplex, and a single-stranded duplex with the same donor, neither of which contained an acceptor fluorophore. Increased FRET efficiency was detected with the fully base-paired duplex containing both donor and acceptor fluorophores. This value includes the signal from the expected, normal breathing that takes place at the ends of DNA duplexes (23). Further increases in FRET efficiencies were observed with the duplexes that contained either one or two unpaired ends. Because Cy3 and Cy5 dyes have been shown to stack at duplex DNA termini (24, 25), such increases could be due to changes in the relative orientation of the dyes, as well as their distance from one another. We interpret the results with mispaired substrates to indicate that release of terminal stacking of the dyes promotes orientational flexibility, allowing an ensemble of dye-dye orientations with higher transfer efficiency. In addition or, alternatively, the average dye-dye distance may decrease due to strand peel-back. Regardless of the interpretation, these experiments demonstrate that end fraying can be monitored using this FRET-based methodology.
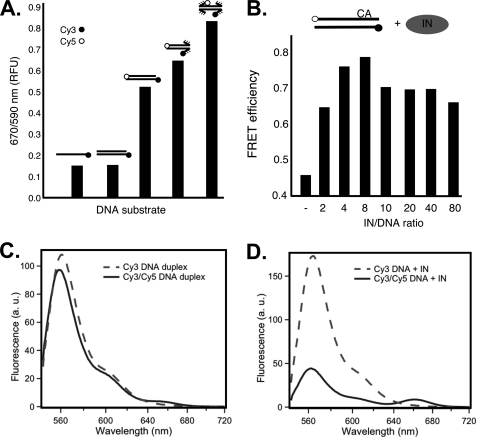
FIGURE 2.
FRET changes depend on disruption of base pairing in DNA termini and the ratio of ASV IN to substrate DNA. A, FRET measured in the absence of IN. The bars show relative FRET values obtained with single- or double-stranded ASV-DNA end duplex substrates that contain only a donor fluorophore and duplex oligonucleotides that contain both donor and acceptor fluorophores with paired, or singly or doubly unpaired ends. B, FRET increase of the paired ASV-DNA end duplex substrate, optimal in IN excess, peaking as the ratio approaches 4. Concentration of the paired ASV-DNA end oligonucleotide was kept constant at 50 nm. Sequence is shown in Fig. 1. C, fluorescence emission spectra of matching (50 nm) samples of ASV-DNA duplex labeled with donor only (Cy3, dashed line) or donor and acceptor (Cy3/Cy5, solid line). D, emission spectra of the same DNA samples as in C, but in the presence of 400 nm ASV IN. Instrumental parameters, including excitation wavelength (535 nm) and bandwidth (2 nm for excitation and 4 nm for emission), were identical in both C and D. Labeling conventions in A and B are as in Fig. 1, with CA above the top line in B representing the conserved dinucleotide in the cleaved strand.
To determine how the presence of IN affects DNA end fraying, the fully base-paired ASV duplex substrate was mixed with an increasing molar amount of the cognate ASV IN, on ice and in the absence of metal cofactor to prevent catalysis. The results show a dramatic increase in FRET efficiency, reaching close to maximum at an IN:DNA ratio of 4:1, consistent with evidence that the functional form of IN is a tetramer (26). Increasing the ratio beyond 8:1 led to a slight decrease, possibly due to quenching by the excess protein. We also note that the IN-induced increase in FRET efficiency is significantly greater than the increase triggered by mispairing of the substrate ends (Fig. 2A). This difference is consistent with the predicted severe IN-mediated distortion of DNA ends by ASV IN (4) or with an IN-stabilized, favorable dye orientation. However, the increased FRET is inconsistent with DNA curvature changes induced by IN (27), and below we address the possible contribution of intermolecular interactions.
We also measured fluorescence emission spectra for two of the duplex substrates, using a scanning fluorometer. Fig. 2C compares the spectra (upon excitation of the donor at 535 nm) for matched samples of the ASV-paired duplex labeled with the Cy3 donor only or both donor and acceptor (Cy3/Cy5) (Fig. 1B). The observed decrease in donor emission (main band at ∼560 nm) is consistent with a relatively low FRET efficiency (EFRET = 0.098). Assuming a Förster distance of 53 Å, this corresponds to an average donor-acceptor distance of 77 Å, close to that expected for a duplex of 22 bp. A weak band at 660 nm, near the emission maximum of the Cy5 acceptor, further supports this interpretation. The spectra in Fig. 2D were obtained under identical conditions, but in the presence of 8-fold molar excess of ASV IN. The low level of donor emission observed for the double-labeled duplex substrate compared with that containing only the Cy3 donor, confirms that IN binding results in more efficient energy transfer (EFRET = 0.75), consistent with a substantial decrease in apparent donor-acceptor distance (from ∼77 Å to 44 Å). As expected, in the presence of IN the double-labeled duplex also shows an enhanced acceptor emission band at 660 nm, compared with same duplex shown in C in the absence of IN (note that emission intensities above 600 nm are attenuated due to decreasing detector efficiency and other instrument factors).
ASV IN-mediated FRET Is Primarily an Intramolecular Interaction
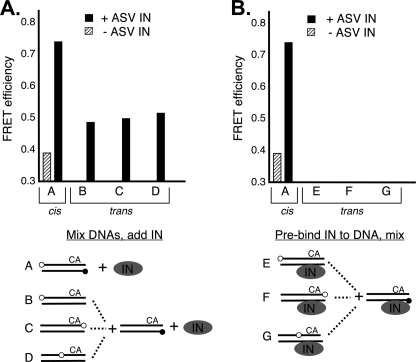
ASV IN is capable of catalyzing the concerted integration of two short duplex viral substrates into a target DNA, although with very low efficiency (28). However, assembly of a complex in which the two viral ends are held in position for insertion would not result in trans FRET because the Cy3 FRET donors would be brought in juxtaposition (head-to-head). Nevertheless, as part of IN-mediated sampling of assembly complexes, it is possible that head-to-tail complexes might form, which would juxtapose the Cy3 and Cy5 dyes in separate duplexes. As ensemble FRET provides an average signal, a small fraction of head-to-tail molecules in which Cy3 and Cy5 would be in close proximity could contribute to the overall signal. To distinguish between FRET signals derived from cis or trans interactions between IN-bound DNA duplexes, we devised the mixing experiments described in Fig. 3 in which the optimal 4:1 ratio of IN to DNA was used. To measure trans FRET between head-to-head ends we used substrates in which the Cy3 FRET donor was placed at the 5′ end of the noncleaved strand and a set of duplexes in which the Cy5 acceptor was placed at three alternative positions. The FRET efficiency obtained with the duplex that contained both fluorophores (in cis) was consistent with the value of 0.75 obtained in Fig. 2D. We found that in the presence of IN, the FRET efficiency for any of the combinations of Cy5-labeled duplexes with the fixed Cy3-labeled duplex produced a much reduced, but measurable signal, indicating that there is some contribution from intermolecular interactions. Surprisingly, this trans FRET signal was independent of the position of the FRET acceptor (Fig. 3A). We conclude that trans FRET is produced by randomly positioned DNA molecules as might occur in a complex in which one DNA molecule is bound at the position of a viral DNA end and the second at the target DNA binding site. This would be consistent with the fact that in vitro reactions that contain only short viral DNA duplexes produce products corresponding to one duplex integrating into another. To test this interpretation, we pre-formed complexes containing IN and Cy5-labled duplexes or Cy3-labeled duplexes and then mixed them prior to measuring FRET. In this case, the FRET efficiency was at background levels (<0.3). These results can be explained by saturation of sites in which duplexes are held as both viral ends and target DNA. We conclude that the major FRET signal produced in the dual Cy3-Cy5 substrate is due primarily to intramolecular repositioning of the dyes.
FIGURE 3.
Distinguishing cis and trans FRET. A, duplex substrates and conditions were designed to distinguish cis versus trans IN-induced FRET. One fixed duplex contained only the FRET donor at the 5′ end of the noncleaved strand, as in the standard duplex substrate. A second set of duplexes contained only the acceptor fluorophore at three different positions on the cleaved strand (substrates B, C, and D). These substrates were designed to detect orientation-specific trans FRET. Equal amounts of separately labeled duplexes were first mixed, and then IN was added and the mixture left on ice for 5 min before FRET readings were taken. B, measurement of trans FRET efficiencies between duplexes containing only the acceptor or donor fluorophore and incubated separately with IN on ice for 5 min before mixing. FRET readings were taken immediately thereafter. The total substrate DNA concentration was 50 nm, and the ASV IN to DNA ratio was 4 in all experiments. The experimental schemes are illustrated below each graph with labeling conventions as in Figs. 1 and 2.
FRET Depends on Interaction with Residues in Both the C-terminal Domain (CTD) and the Catalytic Core Domain (CCD) of IN
To determine which domains of ASV IN are critical for the increase in cis FRET efficiency, we tested the activities of IN fragments that lacked either the N-terminal domain (NTD; IN(49–286)), the CTD (IN(1–207)), or both (IN(52–207)). The results showed a substantial increase in FRET efficiency with increasing amounts of the fragment that contained both the core and CTD, but no significant activity with the fragment that contained the NTD and core, or with the core domain alone (Fig. 4A). We conclude from these results that FRET depends on simultaneous interaction of the DNA with both of these domains. We note that our previous results showed that the ASV IN core domain alone was sufficient to promote end fraying as detected by KMnO4 modification (4). We speculate that stacking of the fluorophores at the termini in our FRET substrates (24, 25) may partially stabilize these DNA duplexes, leading to a requirement for interaction with additional DNA-binding determinants to promote their fraying.
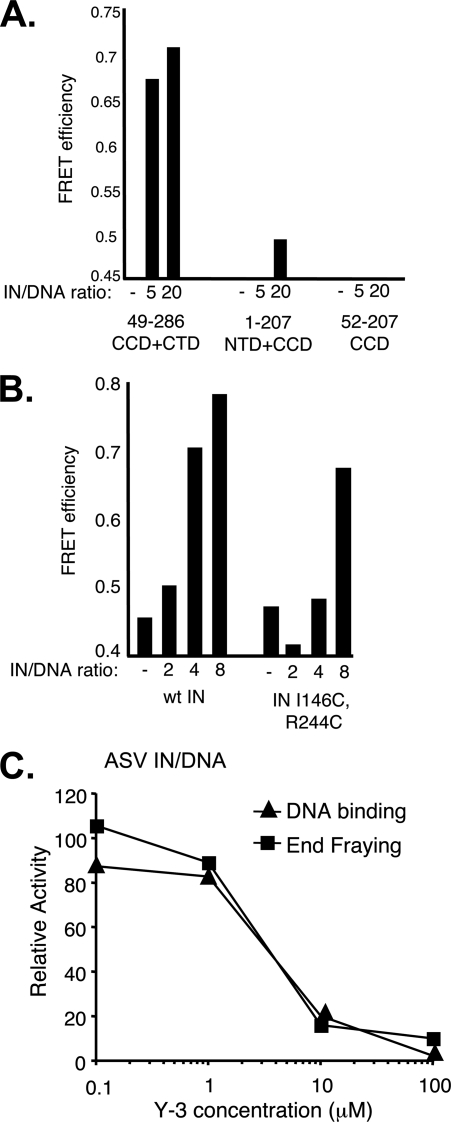
FIGURE 4.
ASV IN CCD + CTD two-domain fragment retains fraying activity, and substitutions that reduce DNA binding also reduce end fraying. A, FRET efficiency as a function of the ratio of ASV IN-derived fragments to the viral DNA duplex. B, comparison of FRET efficiencies in the presence of increasing ratios of wild-type (wt) ASV IN and a full-length derivative that contains cysteine substitutions in the CCD and CTD, which reduce DNA binding. C, inhibition of ASV IN fraying and DNA binding by the Y-3. DNA binding was measured using a fluorescence polarization assay (17) in reactions that contained 50 nm of a 28- + 30-nucleotide substrate representing the recessed, processed U3 end of ASV DNA labeled with 6-carboxyfluorescein at the 5′ end of the cleaved strand, and 200 nm ASV IN. Y-3 was obtained from the NCI Chemical Repository through the Drug Synthesis and Chemistry Branch. The FRET was measured at the same IN:DNA ratio of 4. Data are plotted relative to control reactions that contained no Y-3. The same FRET substrate, at a constant 50 nm, was used in all three panels.
Previous studies have implicated specific HIV-1 IN residues, Gln148 in the flexible loop in the core domain and Glu246 in the CTD of HIV-1 IN, in binding to the cognate viral DNA substrate (29, 30). The corresponding PFV IN residues, Thr210 in the flexible loop and Asn348 in the CTD, are in close association with the noncleaved strand of viral DNA in the crystal structure of the intasome complex (31). We reasoned that if end fraying is related to substrate binding, then substitution of residues in analogous positions of ASV IN might lead to a reduction in FRET efficiency. In separate IN-DNA cross-linking studies, for which we constructed a number of cysteine-substituted derivatives (data not shown), we confirmed that the analogous ASV IN residues, Ile146 and Arg244 are in close proximity to the viral DNA component in a joined viral-target DNA structure that represents an integration intermediate (26). As shown in Fig. 4B, the FRET efficiency obtained with an ASV IN derivative that contains the double substitution, IN I146C/R244C, is close to background at an IN:DNA ratio of 4. FRET efficiency with this derivative does not exceed the background level until a higher ratio of IN to DNA is included in the reaction. Results from fluorescence anisotropy measurements (17) indicate that DNA binding efficiency of the substituted derivative is also reduced compared with wild type (data not shown). These findings are consistent with the notion that duplex binding to the viral substrate site on ASV IN is associated with end fraying.
We previously identified a small molecule, the bisulfonated naphthalene compound called Y-3, which inhibits processing and joining by ASV IN and HIV-1 IN with similar effectiveness (32). The binding site for Y-3 on ASV IN was determined by crystallography with the catalytic core to include the flexible loop above the metal cofactor binding residues in the catalytic center, and similar binding could be modeled for HIV-1 IN; however, the mechanism of Y-3 inhibition remained unclear. In experiments described in Fig. 4C, we preincubated ASV IN with increasing concentrations of Y-3 and measured the effects on both DNA binding and FRET with the cognate, fully paired ASV-DNA duplex. As almost identical curves for inhibition were obtained, we conclude that the binding of Y-3 interferes with DNA-IN interactions that are equally critical for both activities.
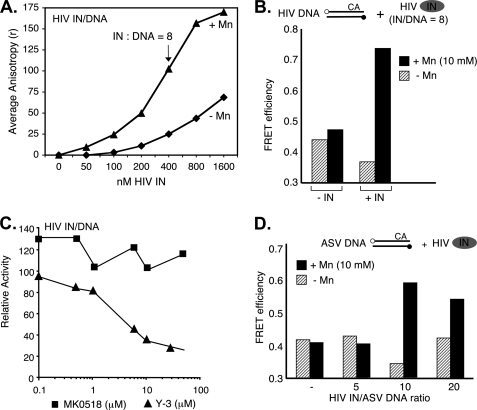
HIV-1 IN Induces DNA End Fraying, but Only in the Presence of a Metal Cofactor
We have reported previously that in the case of HIV-1 IN, unlike ASV IN, the presence of a metal cofactor (Mg2+ or Mn2+) induces a conformational change that is required for its activity (33). Results from a fluorescence polarization assay in which HIV-1 IN was mixed with its cognate DNA duplex on ice to prevent catalysis show that DNA binding is dependent on both IN concentration and the presence of a metal cofactor (Fig. 5A). The effect of the metal on the FRET efficiency of an HIV-1-DNA duplex in the presence of HIV-1 IN is shown in Fig. 5B. In the absence of metal, FRET is essentially at background level, whereas a strong signal is detected in the presence of metal. Results in Fig. 5C shows that Y-3 inhibits end fraying by HIV-1 IN with a dose response similar to that observed with ASV IN (Fig. 4C). In contrast, under the same conditions, there is no reduction in end fraying by HIV-1 IN with the strand transfer inhibitor, raltegravir (MK0518). The latter result is consistent with the known binding properties of this class of inhibitors (34). Taken together, the results in Fig. 5, A–C, indicate that, as with ASV IN, end fraying by HIV-1 IN is associated with DNA binding. However, this reaction is clearly not sequence-specific because in the presence of the metal cofactor, HIV-1 IN can also promote FRET with the ASV-DNA duplex (Fig. 5D). Because HIV-1 IN is unable to process ASV sequences efficiently (35), this result indicates that DNA fraying/binding activity is necessary but not sufficient for catalysis.
FIGURE 5.
Detection of end fraying by HIV-1 IN. A, DNA binding measured using a fluorescence polarization assay (17) in reactions that contained 50 nm of a 20- + 22-nucleotide substrate representing the processed U5 end of HIV DNA, labeled with 6-carboxyfluorescein at the 5′ end of the cleaved strand as described in Fig. 4. Triangles, data for reactions that contained 10 mm MnCl2. Diamonds, data for reactions that contained no metal cofactor. B, relative FRET activity with a fully paired 22-bp HIV-1-DNA substrate duplex in the absence or presence a metal cofactor and IN:DNA ratio of 8. C, relative FRET activity of HIV-1 IN on an HIV-DNA substrate in the presence of Y-3 or MK0518 (raltegravir). D, FRET efficiencies with a fully paired 22-bp ASV-substrate duplex and HIV-1 IN at increasing IN:DNA ratios, in the absence or presence of a metal cofactor. Experimental designs are illustrated above the graphs, with labeling conventions as in Figs. 1 and 2. In both cases DNA duplex concentrations were at 50 nm.
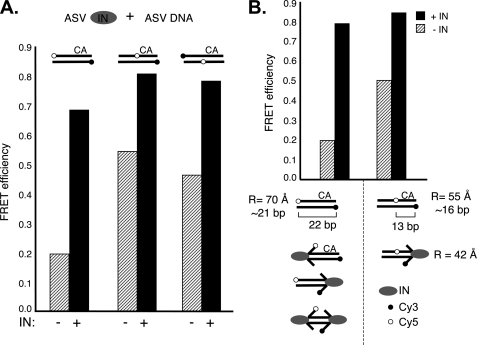
Estimate of the Degree of Fraying at a Single End of a Viral DNA Duplex by ASV IN
As noted above, relative distances between donor and acceptor fluorophore pairs can be calculated from determinations of FRET efficiencies. However, interpretation of such calculations could be complicated by breathing or IN-stabilized fraying at either or both ends of our test duplex, as well as previously documented fluorophore orientation effects (24, 36). To minimize the impact of these potential problems, we designed ASV-derived DNA duplexes in which the acceptor fluorophore was placed at an internal position, thereby allowing an estimate of IN-mediated fraying independently at both ends of the viral duplex substrate (Fig. 6A). As expected from previous studies (4) and results in Fig. 5D, fraying by ASV IN could be detected at both DNA ends. Therefore, to measure changes in dye distances, upon IN binding, we compared substrates in which the Cy5 dye was at an end or internal (Fig. 6B). Based on our FRET efficiency data, the distances between the dyes in the internally labeled duplex and the duplex with two terminal dyes were estimated at 55 Å and an expected 70 Å, respectively. Given the potential complications cited above, these values are reasonably close to the distance derived in Fig. 2D for the duplex with terminal dyes, and the values of 45 Å and 81 Å, respectively, that can be calculated from the known lengths of the duplexes and the predicted positions of the fluorophores in a B-DNA structure. FRET efficiency with the duplex in which the acceptor fluorophore was placed at an internal position was shown to increase from 0.5 in the absence of IN to 0.95 in the presence of IN, equivalent to an apparent distance change of 55 Å to 42 Å.
FIGURE 6.
ASV IN-mediated FRET changes are consistent with viral DNA end fraying. A, effects of dye positions on FRET efficiencies. Positioning of the Cy5 dye internally, 9 bp from either end of the 22-bp ASV substrate DNA, resulted in the expected increased FRET in the absence of IN compared with the substrate with terminal dyes. In the presence of ASV IN, there was a further increase in FRET with all substrates. The substrates with the internal dye allowed assessment of FRET changes at both ends of the viral substrate. Notably, an increase in FRET was detected on the non-CA end. B, FRET efficiencies and distance calculations compared between the ASV-DNA end duplex containing terminal or internal dyes in the absence or presence of ASV IN. Diagrams below highlight interactions that could contribute to increased FRET in the presence of IN. Use of the substrate with the internal Cy5 dye allows measurements uniquely at the CA end of the substrate, and this substrate was used to measure distance changes in the presence of IN. Apparent distances between the fluorophores (R) were calculated from the FRET efficiencies, in the presence or absence of IN, and are indicated below. Labeling conventions are as in Figs. 1 and 2.
DISCUSSION
We have developed a FRET-based assay to detect viral DNA end fraying by retroviral IN proteins. Unlike our previously described chemical probing method (4), this assay can detect end fraying by HIV-1 IN which requires the presence of a metal cofactor to bind viral DNA efficiently and effectively. By use of the FRET assay we not only confirmed the previously detected end fraying activity of ASV IN (4) (Fig. 2), but also demonstrated that combined determinants in both the catalytic core and CTDs are essential to detect activity in this assay. We also showed that an inhibitor or specific amino acid substitutions that reduce such DNA end fraying also reduce DNA binding (Fig. 4). DNA end fraying and DNA binding by HIV-1 IN were also coupled (Fig. 5, A and B).
Experiments in which donor and acceptor fluorophores were attached to either the same or separate molecules showed that under optimal conditions of IN excess, the IN-induced FRET increases are due primarily to intramolecular (cis) FRET. In the presence of short viral DNA duplexes, such as used in our experiments, both ASV IN and HIV-1 IN catalyze mainly single-end processing and joining reactions (37, 38). Consequently, we do not expect two viral ends to be held in trans under these conditions, a configuration that could produce intermolecular FRET. We conclude, therefore, that in the process of binding each viral DNA end, retroviral IN proteins promote disruption of terminal base pairs in preparation for catalysis. It is noteworthy, however, that although HIV-1 IN can mediate fraying of an ASV-viral DNA duplex substrate (Fig. 5D), such a duplex is not an efficient substrate for HIV-1 IN processing (35). Furthermore, we have demonstrated that ASV IN can fray either end of a viral DNA duplex with equal efficiency (Fig. 6A). These observations establish a clear distinction between the functions of DNA end fraying and actual catalysis and imply that distinct recognition criteria must be satisfied for each.
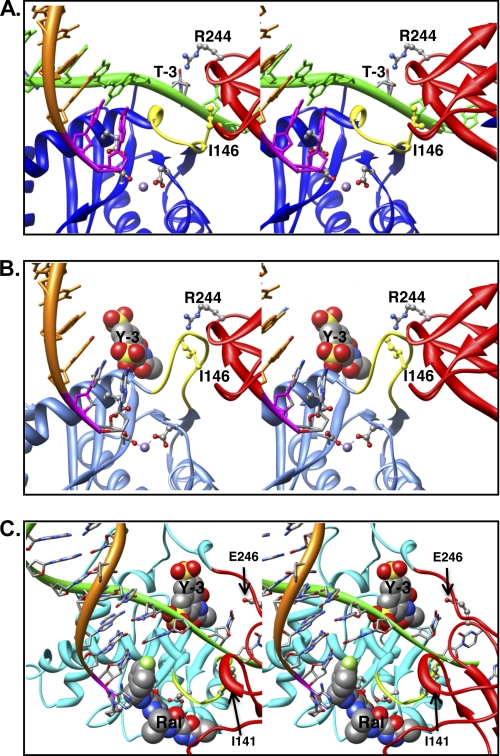
Although FRET provides a powerful method to analyze native or induced nucleic acid structures, it is sometimes difficult to determine precise distances, due primarily to uncertainty regarding dye orientation. Based on our previous studies (4), we expected that the unpairing and severe DNA distortion induced by IN would be detectable by FRET. With respect to DNA end unpairing or partial melting, unpaired terminal regions would have significant structural freedom because single-strand DNA is less constrained than duplex DNA. Furthermore, we showed that mispaired ends could be further distorted by ASV IN (4). However, both Cy3 and Cy5 participate in base pair-like stacking at DNA termini, and unpairing could lead to an increase in FRET signal due to favorable, parallel dipole interactions caused by loss of this constraint. Recently, a crystal structure of PFV IN complexed with viral DNA ends was reported (31). The viral DNA substrates used for crystallography had recessed 3′ ends and lacked the dinucleotide normally removed during IN-mediated processing. Consequently, the terminal nucleotides on the noncleaved strand are already unpaired. However, in the structure, the viral DNA end is further unpaired extending to position 3 within the conserved CA/GT. As we previously showed that ASV IN could promote unpairing through position 3 (4) we believe that the PFV structure and our biochemical analyses have revealed a similar activity.
In the PFV-DNA crystal structure, the terminal bases of the noncleaved strand are flipped out of what would be the helical frame in the duplex. However, the structure of the unpaired strand does not show severe distortion whereby the strand trajectory is reversed or severely “peeled back” (see model in Fig. 7A). The trajectory of the DNA in the PFV structure would not explain the increase in FRET observed in our experiments, as the dye-dye distance would not be expected to decrease. If we assume that the distortion that we have detected biochemically is a general feature of IN-DNA end interactions, the static view in the PFV crystal structure may not capture more severe distortions that occur in solution.
FIGURE 7.
Model of IN bound to a viral DNA end in the absence or presence of the inhibitors. A, the model of ASV IN was prepared using crystal structure of PFV IN bound to viral DNA as a template (Protein Data Bank ID code 3L2Q) (31, 40) and is portrayed as a stereo pair. The cleaved strand is shown as an orange ribbon, with the conserved CA residues in magenta. The noncleaved strand is in green and is held by cooperation of the CTD (red ribbon) and the flexible loop (yellow ribbon) adjacent to the active site in the CCD (blue ribbon). For clarity, the linkers between the NTD, core, and CTD domains are not shown. Active site residues that bind the divalent metal cofactors (purple sphere) as well as the two residues proposed to participate in positioning the noncleaved strand are portrayed in ball and stick: Ile146 (yellow) in the flexible loop and Arg244 in the CTD. The PFV residue Asn348, which is analogous to ASV Arg244, has been shown to make base-specific contacts; the Arg244 rotamer shown is in a position to make similar contact with the thymine at the −3 position in the noncleaved viral DNA strand. B, model of ASV IN bound to inhibitor Y-3. Color code is the same as in A except that the core domain ribbon is now shown in light blue. The Y-3 inhibitor is shown in space-filling representation in its previously established binding site (32), which induces a change in the position of the flexible loop and Ile146 (yellow) that preclude proper binding of the noncleaved DNA strand. C, model of HIV-1 IN bound to both Y-3 and raltegravir. Colors are as above, except that the HIV-1 CCD is in cyan. To position Y-3, the HIV-1 intasome model with bound raltegravir (21) was superimposed onto the ASV IN catalytic core bound to Y-3. As with ASV IN, Y-3 is predicted to interfere sterically with binding of the noncleaved strand of HIV viral DNA. In contrast, the presence of viral DNA in the complex enhances raltegravir binding to HIV-1 IN, but such binding is predicted to interfere with target DNA binding. Importantly, Y-3 and raltegravir are predicted to bind on opposite sides of the flexible loop.
To determine how specific amino acid side chains of ASV IN can contribute to DNA binding and end fraying, we modeled this protein bound to its viral substrate DNA using the PFV crystal structure as a template and methods described previously (39). As shown in Fig. 7, the ASV model provides insight into the roles of residues in the catalytic core and CTDs with respect to DNA binding and end fraying, as well as the basis for Y-3 inhibition. Residues Ile146 and Arg244, from the CCD and CTDs, are in positions close to the end of the noncleaved strand (Fig. 7A), consistent with the effects observed on end fraying when these residues were substituted with cysteines (Fig. 4B). It is also clear from the model how the presence of Y-3 in its binding site (Fig. 7B), and the concomitant change in conformation of the flexible loop above the active site that includes Ile146, preclude access and proper gripping of the noncleaved strand. Y-3 also inhibits end fraying by HIV-1 IN (Fig. 5C). However, raltegravir, which binds at the active site in a position that blocks binding of target DNA but not viral DNA, has no effect on end fraying (Fig. 7C).
In summary, the FRET assay described herein provides physical evidence for viral substrate end fraying by both HIV-1 IN and ASV IN and has identified a new assayable activity for HIV-1 IN that can be used both in studies of enzyme mechanism and the design of novel inhibitors.
Acknowledgments
We thank Drs. Eileen Jaffe and RaviShankar Bojja for critical comments and helpful discussions and Marie Estes for help in preparing the manuscript. We thank Drs. Yves Pommier and Christophe Marchand for the raltegravir and Dr. Alan Engelman for providing coordinates for the HIV intasome model described by Krishnan et al. (21). We also acknowledge the use of the Fox Chase Cancer Center Biotechnology and Molecular Modeling Facilities in this research.
This work was supported, in whole or in part, by National Institutes of Health Grants AI40385, CA71515, and CA006927.
- IN
- integrase
- ASV
- avian sarcoma virus
- CCD
- catalytic core domain
- CTD
- C-terminal domain
- FA
- fluorescence of acceptor
- FD
- fluorescence of donor in absense of acceptor
- FDA
- fluorescence of donor in presence of acceptor
- NTD
- N-terminal domain
- PFV
- prototype foamy virus
- Y-3
- 4-acetylamino-5-hydroxynapthalene-2,7-disulfonic acid.
REFERENCES
- 1. Vandegraaff N., Engelman A. (2007) Expert Rev. Mol. Med. 9, 1–19 [DOI] [PubMed] [Google Scholar]
- 2. Skalka A. M., Katz R. A. (2005) Cell Death Differ. 12, 971–978 [DOI] [PubMed] [Google Scholar]
- 3. Yi J., Asante-Appiah E., Skalka A. M. (1999) Biochemistry 38, 8458–8468 [DOI] [PubMed] [Google Scholar]
- 4. Katz R. A., DiCandeloro P., Kukolj G., Skalka A. M. (2001) J. Biol. Chem. 276, 34213–34220 [DOI] [PubMed] [Google Scholar]
- 5. Oh J., Chang K. W., Hughes S. H. (2008) J. Virol. 82, 11480–11483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oh J., Chang K. W., Wierzchoslawski R., Alvord W. G., Hughes S. H. (2008) J. Virol. 82, 503–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Katzman M., Katz R. A. (1999) Adv. Virus Res. 52, 371–395 [DOI] [PubMed] [Google Scholar]
- 8. Lee I., Harshey R. M. (2003) J. Mol. Biol. 330, 261–275 [DOI] [PubMed] [Google Scholar]
- 9. Lee I., Harshey R. M. (2003) Nucleic Acids Res. 31, 4531–4540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Montaño S. P., Coté M. L., Roth M. J., Georgiadis M. M. (2006) Nucleic Acids Res. 34, 5353–5360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vink C., van Gent D. C., Elgersma Y., Plasterk R. H. (1991) J. Virol. 65, 4636–4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Scottoline B. P., Chow S., Ellison V., Brown P. O. (1997) Genes Dev. 11, 371–382 [DOI] [PubMed] [Google Scholar]
- 13. Engelman A., Mizuuchi K., Craigie R. (1991) Cell 67, 1211–1221 [DOI] [PubMed] [Google Scholar]
- 14. Agapkina J., Smolov M., Zubin E., Mouscadet J. F., Gottikh M. (2004) Eur. J. Biochem. 271, 205–211 [DOI] [PubMed] [Google Scholar]
- 15. Andrake M. D., Ramcharan J., Merkel G., Zhao X. Z., Burke T. R., Jr., Skalka A. M. (2009) AIDS Res. Ther. 6, 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Taganov K. D., Cuesta I., Daniel R., Cirillo L. A., Katz R. A., Zaret K. S., Skalka A. M. (2004) J. Virol. 78, 5848–5855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Merkel G., Andrake M. D., Ramcharan J., Skalka A. M. (2009) Methods 47, 243–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Woehler A., Wlodarczyk J., Neher E. (2010) Biophys. J. 99, 2344–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Canutescu A. A., Shelenkov A. A., Dunbrack R. L., Jr. (2003) Protein Sci. 12, 2001–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Q., Canutescu A. A., Dunbrack R. L., Jr. (2008) Nat. Protoc. 3, 1832–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krishnan L., Li X., Naraharisetty H. L., Hare S., Cherepanov P., Engelman A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 15910–15915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 23. Ramreddy T., Rao B. J., Krishnamoorthy G. (2007) J. Phys. Chem. B 111, 5757–5766 [DOI] [PubMed] [Google Scholar]
- 24. Levitus M., Ranjit S. (2011) Q. Rev. Biophys. 123–151 [DOI] [PubMed] [Google Scholar]
- 25. Iqbal A., Wang L., Thompson K. C., Lilley D. M., Norman D. G. (2008) Biochemistry 47, 7857–7862 [DOI] [PubMed] [Google Scholar]
- 26. Bao K. K., Wang H., Miller J. K., Erie D. A., Skalka A. M., Wong I. (2003) J. Biol. Chem. 278, 1323–1327 [DOI] [PubMed] [Google Scholar]
- 27. Spring B. Q., Clegg R. M. (2007) J. Phys. Chem. B 111, 10040–10052 [DOI] [PubMed] [Google Scholar]
- 28. Aiyar A., Hindmarsh P., Skalka A. M., Leis J. (1996) J. Virol. 70, 3571–3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gao K., Butler S. L., Bushman F. (2001) EMBO J. 20, 3565–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Johnson A. A., Santos W., Pais G. C., Marchand C., Amin R., Burke T. R., Jr., Verdine G., Pommier Y. (2006) J. Biol. Chem. 281, 461–467 [DOI] [PubMed] [Google Scholar]
- 31. Hare S., Gupta S. S., Valkov E., Engelman A., Cherepanov P. (2010) Nature 464, 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lubkowski J., Yang F., Alexandratos J., Wlodawer A., Zhao H., Burke T. R., Jr., Neamati N., Pommier Y., Merkel G., Skalka A. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4831–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Asante-Appiah E., Skalka A. M. (1997) J. Biol. Chem. 272, 16196–16205 [DOI] [PubMed] [Google Scholar]
- 34. Espeseth A. S., Felock P., Wolfe A., Witmer M., Grobler J., Anthony N., Egbertson M., Melamed J. Y., Young S., Hamill T., Cole J. L., Hazuda D. J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 11244–11249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen A., Weber I. T., Harrison R. W., Leis J. (2006) J. Biol. Chem. 281, 4173–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Iqbal A., Arslan S., Okumus B., Wilson T. J., Giraud G., Norman D. G., Ha T., Lilley D. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 11176–11181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li M., Craigie R. (2005) J. Biol. Chem. 280, 29334–29339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vora A., Bera S., Grandgenett D. (2004) J. Biol. Chem. 279, 18670–18678 [DOI] [PubMed] [Google Scholar]
- 39. Canutescu A. A., Dunbrack R. L., Jr. (2005) Bioinformatics 21, 2914–2916 [DOI] [PubMed] [Google Scholar]
- 40. Hare S., Vos A. M., Clayton R. F., Thuring J. W., Cummings M. D., Cherepanov P. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 20057–20062 [DOI] [PMC free article] [PubMed] [Google Scholar]