Abstract
Neural adhesion molecule NB-3 plays an important role in the apical dendrite development of layer V pyramidal neurons in the visual cortex, and receptor-like protein-tyrosine phosphatase α (PTPα) mediates NB-3 signaling in this process. Here we investigated the role of PTPα in regulating cell surface expression of NB-3. We found that cortical neurons from PTPα knock-out mice exhibited a lower level of NB-3 at the cell surface. When expressed in COS1 cells, NB-3 was enriched in the Golgi apparatus with a low level of cell surface expression. However, co-expression of PTPα increased the cell surface distribution of NB-3. Further analysis showed that PTPα facilitated Golgi exit of NB-3 and stabilized NB-3 protein at the cell surface by preventing its release from the plasma membrane. The extracellular region of PTPα but not its catalytic activity is necessary for its effect on NB-3 expression. Thus, the PTPα-mediated increase of NB-3 level at the cell surface represents a novel function of PTPα in NB-3 signaling in neural development.
Keywords: Cell Adhesion, Cell Surface, Golgi, Phosphatase, Protein-Protein Interactions, GPI Anchor, NB-3, PTPα
Introduction
Protein-tyrosine phosphatase α (PTPα)4 is a receptor-like protein phosphatase with two cytoplasmic protein-tyrosine phosphatase domains (D1 and D2) and a relatively short and highly glycosylated extracellular domain. PTPα is widely expressed in many tissues, including the nervous system, the immune system, and many cancer cells (1). As an essential component of several Src family kinase-dependent signaling pathways, PTPα acts in conjunction with ligand-activated receptors, ion channels, and cell adhesion molecules to dephosphorylate and activate Src family kinases (2–9). In this manner PTPα affects many fundamental cellular processes, including mitosis (10–12), migration (13–15), proliferation (16, 17), and transformation and tumorigenesis (5, 18). PTPα is particularly highly expressed in the brain, and it affects many aspects of neural development and function, such as neurite outgrowth, neuronal differentiation and migration, ion channel activity, oligodendrocyte differentiation and myelination, hippocampal long term potentiation, and spatial memory (2, 6, 8, 9, 13, 19–24). In particular, PTPα mediates neural adhesion molecules NB-3 and Close Homolog of L1 (CHL1) signaling in developing layer V pyramidal neurons in the caudal cortex and is required for correct apical dendrite projection of these neurons in vivo (25).
Dendrite development is an important process in neural development. Apical dendrites of cortical pyramidal neurons, the major sites for these neurons to receive excitatory inputs, exhibit a stereotypic orientation toward the pial surface. Neural adhesion molecules NB-3 and CHL1 regulate apical dendrite orientation in the mouse visual cortex (25, 26). NB-3 belongs to the contactin subgroup of the immunoglobulin (Ig) superfamily (27). Like other contactin family members, NB-3 contains six Ig-like domains and four fibronectin type III (FNIII) repeats. It lacks a transmembrane and intracellular domain and is anchored at the cell surface via a glycosylphosphatidylinositol (GPI) link. NB-3 forms a co-receptor complex with CHL1, an L1 family cell adhesion molecule, in developing neurons. Knocking out either Nb-3 or Chl1 genes in mice leads to abnormal apical dendrite orientation in layer V of the caudal cortex, indicating that both are important for apical dendrite development (25, 26). Besides regulating dendrite development, NB-3 has also been shown to regulate synaptic formation. It is located at the presynaptic site of glutamatergic synapses between parallel fibers and Purkinje cells in the cerebellum. In Nb-3−/− mice vesicular glutamate transporter 1-positive synaptic density is reduced (28), which may explain the motor coordination impairment observed in these mice (29). In the human genome, both NB-3 and CHL1 genes are located on chromosome 3p26-p25. This region is associated with the human 3p syndrome, a disease characterized by mental retardation or low IQ and delayed speech and motor development (30, 31). Involvement of NB-3 and CHL1 in dendrite development and synaptogenesis may explain some aspects of 3p syndrome. Although CHL1 gene deletion has been found in some patients with 3p syndrome (32, 33), the association of NB-3 gene and this disease needs to be determined.
To function as a receptor in developing neurons, NB-3 needs to present at the cell surface at a sufficient level. However, our previous study suggested that other proteins might play a role in the optimal cell surface expression of NB-3 (25). In the present study, we examined the role of PTPα in regulating NB-3 cell surface expression. We found that Ptpα−/− cortical neurons exhibited a lower level of cell surface NB-3 as compared with wild-type neurons. In transfected COS1 cells NB-3 was mainly retained in the Golgi apparatus during its synthesis. PTPα facilitated Golgi exit of NB-3 and increased NB-3 surface level by inhibiting NB-3 release from the plasma membrane. The extracellular domain but not the catalytic activity is necessary for this effect of PTPα. These results indicate that PTPα not only transduces NB-3 signals but also increases its surface presentation in developing neurons, thus revealing a dual function of PTPα in this signaling pathway.
EXPERIMENTAL PROCEDURES
Expression Constructs
Expression constructs for VSVG-PTPα (7) and CHL1-HA (25) were described. Briefly, a VSVG epitope was inserted at a PacI site (Ile-16) in the extracellular region of full-length human PTPα in the pXJ41-neo vector. An HA tag was linked to the C terminus of full-length mouse CHL1 during the construction. Various NB-3 and PTPα constructs used in this study are illustrated in the figures, and the methods for construction are presented in the supplemental Materials and Methods.
Antibodies
For immunostaining, immunoprecipitation, and immunoblotting, we used anti-VSVG (mAb from Roche Applied Science), anti-Myc (mAb and pAb from Cell Signaling), anti-HA (mAb from Cell Signaling), and anti-α-tubulin (mAb from Sigma). To visualize various intracellular organelles, anti-PDIA4 (pAb from Proteintech Group) for endoplasmic reticulum, anti-GORASP2 (pAb from Proteintech Group) for medial Golgi apparatus, and LysoTracker DND-99 (from Invitrogen) for lysosomes were used. To detect endogenous NB-3 and PTPα protein levels in immunoblotting in cultured cortical neurons, rabbit pAb anti-NB-3 (25, 29) and rabbit pAb anti-PTPα (7) were used. Mouse mAb anti-NB-3 (25, 29) was used for whole cell and cell surface staining of NB-3 in cultured cortical neurons. Horseradish peroxidase-conjugated (from GE Healthcare) or Alexa Fluor 488- or 594-conjugated immunoglobulins (from Invitrogen) were used as secondary antibodies for immunoblotting and immunofluorescent staining.
Immunostaining
For whole cell staining, cells were fixed using 4% paraformaldehyde in PBS and incubated in the blocking solution (5% normal goat serum, 0.3% Triton X-100 in PBS), then with the primary antibody overnight at 4 °C, followed by incubation with corresponding secondary antibodies. Staining of cell surface Myc-tagged NB-3 in transfected COS1 cells or endogenous NB-3 in cultured cortical neurons was described previously (25). Briefly, live cells were incubated with rabbit anti-Myc antibody (from Cell Signaling) or mouse anti-NB-3 antibody in normal growth medium for 2 h at 37 °C. Cells were then fixed and stained with fluorescent anti-rabbit IgG or permeabilized and double-stained with monoclonal anti-VSVG antibody for VSVG-PTPα. For double labeling of NB-3-Myc and lysosome, live COS1 cells were incubated with LysoTracker DND-99 (1:1000 in normal growth medium) at 37 °C for 30 min, followed by wash, fixation, and immunostaining using the mAb anti-Myc antibody. Images were taken using an Olympus FV1000 confocal microscope and analyzed using the FV10-ASW software.
Cell Surface Protein Biotinylation Assay
The assay to use biotin to label cell surface proteins was described before (25). Briefly, COS1 cells (36–48 h after transfection) or cortical neurons from embryonic stage 17.5 (E17.5) mice (7 days in culture) were washed three times in cold PBS and incubated with the membrane-impermeable biotin derivative sulfo-NHS-SS-biotin (0.5 mg/ml, from Pierce) in PBS for 15 min at 4 °C. After three washes in cold PBS (first wash with 25 mm Tris-HCl included), cells were lysed in radioimmune precipitation assay (RIPA) buffer. Biotin-labeled proteins were precipitated using UltraLink® Immobilized NeutrAvidinTM Gel (from Pierce) at 4 °C for 2 h. Pulldown samples were analyzed using SDS-PAGE and immunoblotting. Membranes were scanned using Bio-Rad Molecular Imager® FX, and the intensity of signal was analyzed using Quantity One software.
Removal of GPI-anchored Protein from Cell Surface
24 h after transfection, cells were treated with phosphatidylinositol-phospholipase C (PI-PLC, 10−4 units/ml, from Sigma) in the normal growth medium for 16 h at 37 °C followed by extensive wash with cold PBS (25, 34). Cells were then proceeded for immunostaining or cell surface biotinylation assay.
Knock-out Mice and Primary Cortical Neuronal Culture
PTPα-deficient mice as described before (35) were on the 129J background. Heterozygous mice were mated to generate wild-type and knock-out pairs for primary cortical neuronal culture. At E17.5, mothers were euthanized. Cortices from embryos were dissected and dissociated after trypsin digestion. Cells from individual embryos were plated separately on poly-l-lysine-coated plates. Embryos were then genotyped using a PCR procedure, and cells from Ptpα+/− embryos were discarded. Wild-type and Ptpα−/− cells were cultured in Neural Basal medium with B27 supplement (from Invitrogen) for 7 days before immunostaining or cell surface biotinylation assay were carried out. All procedures were approved and monitored by the institutional animal care and use committee in the Biological Resource Center of Singapore and the institutional animal care and use committee in the Institute of Biophysics, China.
RESULTS
PTPα Affects NB-3 Cell Surface Expression in Cultured Mouse Cortical Neurons
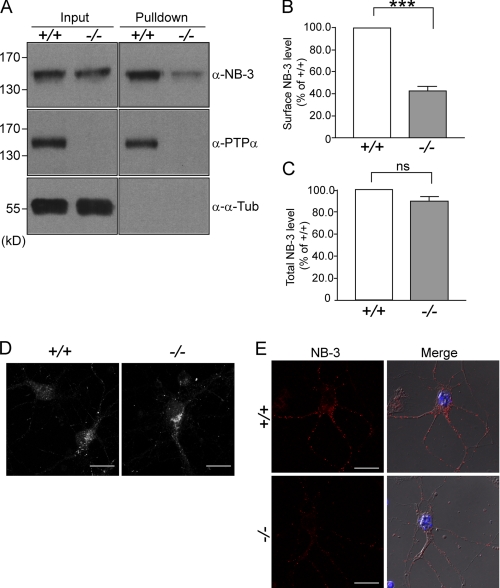
In search of proteins that enhance cell surface expression of NB-3, we found that PTPα, which interacts with and mediates signaling of NB-3 (25), also modulates its cell surface expression in neurons. Cortical neurons from E17.5 wild-type and Ptpα−/− mouse littermates were separately isolated and cultured for 7 days before cell surface protein was covalently labeled with biotin. Unlabeled biotin was removed, and neurons were lysed using RIPA buffer. Biotin-labeled cell surface proteins were precipitated using a NeutrAvidinTM gel. Probing of precipitates with a rabbit anti-NB-3 antibody revealed the amount of cell surface NB-3 protein. We detected a significant reduction of cell surface NB-3 in the Ptpα−/− cortical neurons (43.3 ± 3.3% of wild type), whereas the whole cell NB-3 level in the Ptpα−/− neurons was not significantly different from that in the wild-type neurons (Fig. 1, A–C). Consistently, when cell surface NB-3 was clustered and stained using a mouse anti-NB-3 antibody, Ptpα−/− neurons exhibited fewer clusters with lower signal intensity (Fig. 1E). Together, these observations indicate that PTPα positively regulates NB-3 cell surface expression in neurons and led us to investigate whether PTPα itself enhances NB-3 cell surface expression and the possible mechanism.
FIGURE 1.
Cell surface expression of NB-3 is reduced in Ptpα−/− cortical neurons. A–C, analysis of steady-state cell surface level of endogenous NB-3 protein in cultured cortical neurons from wild-type (+/+) and Ptpα−/− (−/−) mice. Cell surface proteins of E17.5 cortical neuronal cultures (7 days in culture) were labeled with biotin and were subsequently pulled down using NeutrAvidinTM gel. Input and pulldown samples were resolved in SDS-PAGE, and the endogenous proteins were detected in immunoblotting. For quantification in B, levels of cell surface NB-3 were normalized to input NB-3 levels. Relative surface NB-3 protein levels in Ptpα−/− neurons were calculated as a percentage of that in the wild-type neurons. Note that the total NB-3 level is not significantly changed in the Ptpα−/− neurons (C). Results from five independent experiments (n = 5) are presented as mean ± S.E. ***, p < 0.001; ns, not significantly different; one-sample t test. α-α-Tub, anti-α-tubulin antibody. D, immunostaining of whole cell endogenous NB-3 in wild-type (+/+) and Ptpα−/− (−/−) cortical neurons using a mouse monoclonal anti-NB-3 antibody. E, cell surface staining of NB-3 in wild-type (+/+) and Ptpα−/− (−/−) cortical neurons. Live mouse cortical neurons (7 days in culture) were incubated with a mouse monoclonal anti-NB-3 antibody at 37 °C for 2 h, followed by fixation and incubation with Alexa Fluor 594-conjugated anti-mouse IgG antibody. Scale bars, 20 μm.
PTPα Enhances Cell Surface Expression of NB-3 in Transfected COS1 Cells
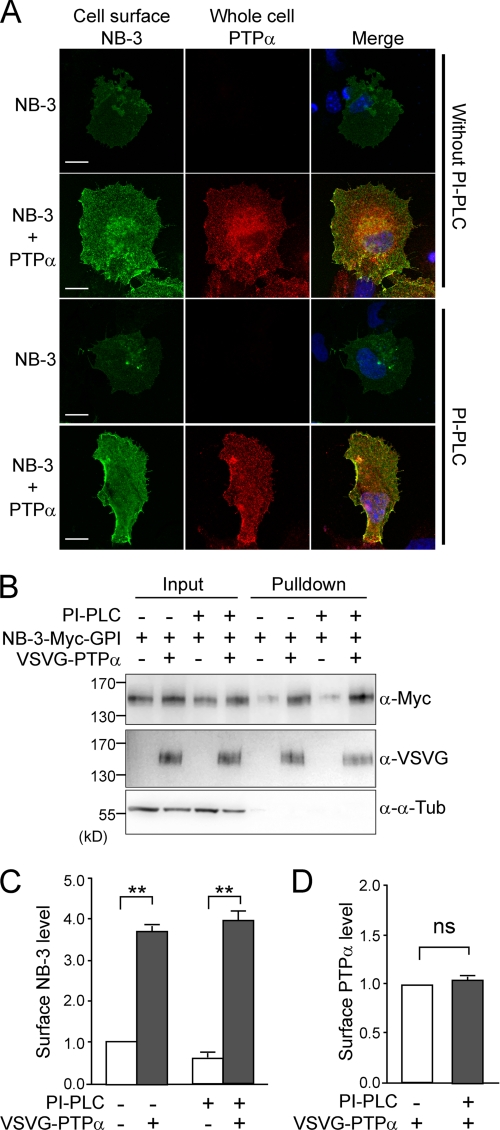
We used COS1 cells to examine the effects of PTPα on NB-3 in more detail. Full-length mouse NB-3 cDNA fused with a Myc tag between the fourth FNIII repeat and the consensus GPI site (NB-3-Myc-GPI, Fig. 2A) and human PTPα cDNA fused with a VSVG tag (VSVG-PTPα) were transfected into the COS1 cells. To visualize only the cell surface NB-3-Myc protein in the transfected cells, live cells were incubated with a rabbit polyclonal anti-Myc antibody in normal growth medium, followed by fixation and incubation with fluorescent secondary antibody. Cells co-transfected with NB-3-Myc-GPI and VSVG-PTPα exhibited a stronger cell surface staining for NB-3-Myc than the cells transfected with NB-3-Myc-GPI cDNA alone (Fig. 2B). Double immunostaining of cell surface NB-3-Myc and whole cell VSVG-PTPα revealed that cells with a strong cell surface signal for NB-3-Myc also expressed VSVG-PTPα (Fig. 2C).
FIGURE 2.
PTPα enhances cell surface expression of NB-3 in transfected COS1 cells. A, schematic structure of the NB-3 constructs. The NB-3-Myc-GPI construct has a Myc tag between the fourth FNIII repeat and the consensus GPI site. The sNB-3-Myc has a Myc tag at the C terminus, and the GPI consensus site is disrupted. The NB-3-Ig-Myc construct has four FNIII repeats removed. The NB-3-FN-Myc construct has six Ig domains removed. B and C, increase of cell surface NB-3-Myc-GPI when co-expressed with VSVG-PTPα in COS1 cells. After transfection with NB-3-Myc-GPI alone (B, −PTPα) or with both NB-3-Myc-GPI and VSVG-PTPα cDNAs (B, +PTPα), live COS1 cells were incubated with a rabbit anti-Myc antibody followed by fixation and staining with Alexa Fluor 488-conjugated anti-rabbit IgG antibody for visualization of cell surface NB-3-Myc protein. Images were taken using the same confocal settings. In C, COS1 cells were transfected with both NB-3-Myc-GPI and VSVG-PTPα. Cell surface NB-3-Myc-GPI was stained using the same procedure as in B, followed by permeabilization of cell membrane and staining of VSVG-PTPα using a mouse anti-VSVG antibody. Scale bars, 20 μm. D–F, analysis of steady-state cell surface level of NB-3-Myc-GPI and VSVG-PTPα in single- and double-transfected COS1 cells. Cell surface proteins were labeled with biotin and precipitated using NeutrAvidinTM gel. Samples were blotted with anti-Myc or anti-VSVG antibodies to assess the level of NB-3-Myc-GPI and VSVG-PTPα proteins. For quantification in E and F, levels of cell surface proteins were normalized to corresponding input proteins. Readings for single-transfected cells were given the value of 1.0. Relative surface protein levels in double-transfected cells were calculated accordingly. Results from three independent experiments (n = 3) are presented as mean ± S.E. for each panel. **, p < 0.01; ns, not significantly different; one-sample t test.
To quantitatively compare the cell surface level of NB-3-Myc protein in single- and double-transfected COS1 cells, cell surface protein biotinylation was performed on live cells 36–48 h after transfection. The biotin-labeled cell surface proteins were precipitated, resolved by SDS-PAGE, and blotted with anti-Myc antibody to reveal the amount of cell surface NB-3-Myc-GPI. We found an average 3.5 ± 0.2-fold increase in the amount of cell surface NB-3-Myc in co-transfected cells, compared with that in the cells transfected with NB-3-Myc-GPI alone (Fig. 2, D and E), whereas the cell surface VSVG-PTPα level was relatively high and not significantly affected by the co-expression of NB-3-Myc-GPI (1.2 ± 0.1-fold change as compared with cells transfected with VSVG-PTPα alone (Fig. 2, D and F)). Together, these results indicate that co-expression with PTPα is sufficient to enhance NB-3 cell surface expression in COS1 cells.
PTPα Facilitates Golgi Exit of NB-3
We next investigated how PTPα increased cell surface expression of NB-3. The low level of NB-3-Myc surface expression in COS1 cells could result from either its intracellular retention in the endoplasmic reticulum (ER) or Golgi apparatus during synthesis, its intense degradation in lysosomes, or cleavage of its GPI link at the cell surface and subsequent release from the cell membrane. To examine the cellular distribution of NB-3 protein in detail, COS1 cells transfected with NB-3-Myc-GPI were co-stained with anti-Myc antibody and various markers for intracellular organelles. NB-3-Myc exhibited a strong and focused signal at a perinuclear location and relatively weak signal at the cell plasma and periphery location. This strong signal was co-localized with a marker for medial Golgi apparatus, to a lesser extent with the marker for ER, but not with lysosomes (Fig. 3). These results indicate that a significant amount of NB-3-Myc protein is retained in the Golgi apparatus during synthesis.
FIGURE 3.
Analysis of the intracellular distribution of NB-3-Myc-GPI in COS1 cells. COS1 cells were transfected with NB-3-Myc-GPI. Thirty-six hours after transfection, cells were co-stained with anti-Myc (for NB-3-Myc-GPI) and markers for medial Golgi apparatus (anti-GORASP2), ER (anti-PDIA4), and lysosomes (LysoTracker), respectively. NB-3-Myc-GPI was mainly co-localized with the medial Golgi apparatus. Lys, lysosomes. Scale bars, 20 μm.
When co-expressed with VSVG-PTPα, however, the perinuclear signal of NB-3-Myc was not obvious anymore, whereas the NB-3-Myc signal at the cell periphery was increased (Fig. 4A). Indeed, the Pearson's coefficient for NB-3-Myc and the Golgi marker co-localization was significantly decreased in co-transfected cells as compared with that in the cells expressing NB-3-Myc-GPI alone, whereas co-localization of NB-3-Myc with the ER marker remained the same (Fig. 4B). The reduction of co-localization of NB-3-Myc with the medial Golgi marker and the increase of NB-3-Myc signal at the cell periphery in co-transfected cells suggest that PTPα facilitates the Golgi exit of NB-3, which reduces intracellular retention of NB-3 during synthesis and increases its protein level at the cell surface. Consistent with this finding, we also observed an increase of co-localization of endogenous NB-3 with the Golgi apparatus in cultured Ptpα−/− neurons compared with the wild-type neurons (Fig. 4, C and D).
FIGURE 4.
PTPα facilitates Golgi exit of NB-3. A, co-expression with VSVG-PTPα reduced NB-3-Myc-GPI in the medial Golgi and increased its signal at the cell periphery (arrowheads). Transfected COS1 cells were immunostained with anti-Myc antibody and markers for medial Golgi apparatus (anti-GORASP2) or ER (anti- PDIA4). Scale bars, 20 μm. B, change of co-localization of NB-3-Myc-GPI with medial Golgi and ER measured as Pearson's coefficient. Results (presented as mean ± S.E.) are from three independent experiments with over 100 cells measured in each experiment. ***, p < 0.001; ns, not significantly different; two-way analysis of variance followed by Bonferroni post test. C, Ptpα−/− cortical neurons exhibit a more enriched NB-3 signal in the Golgi apparatus. Cultured cortical neurons from wild-type (+/+) and Ptpα−/− (−/−) mice (7 days in culture) were fixed and immunostained with mouse anti-NB-3 antibody and rabbit anti-GORASP2 antibody. Scale bars, 10 μm. D, co-localization of endogenous NB-3 with medial Golgi measured as Pearson's coefficient. Results (presented as mean ± S.E.) are from three independent experiments with over 50 cells measured in each experiment. ***, p < 0.001; t test.
PTPα Stabilizes NB-3 at the Cell Surface
NB-3 protein lacks a transmembrane domain and is anchored at the cell surface via a GPI link. The GPI link can be cleaved by extracellular phospholipase activity and leads to subsequent release of NB-3 protein from the cell membrane. Indeed, secreted NB-3 protein can function as a ligand for other receptors, such as Notch1 (36). Thus, we hypothesized that PTPα might increase the surface level of NB-3 via inhibition of NB-3 release from the cell surface. To test this possibility, we used PI-PLC to cleave the GPI link at the cell surface. After a 16-h treatment with PI-PLC, cell surface NB-3-Myc level was not significantly changed in cells co-expressing NB-3-Myc and VSVG-PTPα, compared with cells without PI-PLC treatment, as shown by both immunostaining (Fig. 5A) and cell surface protein biotinylation assay (Fig. 5, B and C). PI-PLC treatment resulted in a decrease of cell surface NB-3-Myc in cells expressing NB-3-Myc alone; however, the change was not statistically significant (Fig. 5, B and C). Thus, PTPα is capable of increasing the cell surface level of NB-3 independent of the GPI link.
FIGURE 5.
Cleavage of the GPI-anchor of NB-3 does not abolish the effect of PTPα on the NB-3 cell surface expression. A, COS1 cells transfected with NB-3-Myc-GPI alone or both with VSVG-PTPα were treated with PI-PLC (10−4 units/ml) for 16 h before cells were immunostained for cell surface NB-3-Myc and whole cell VSVG-PTPα. PI-PLC treatment did not exhibit obvious effect on the cell surface Myc signals in double-transfected cells. Images were taken using the same confocal settings. Scale bars, 20 μm. B, transfected COS1 cells were treated with or without PI-PLC (10−4 units/ml) for 16 h before cell surface proteins were labeled with biotin and pulled down using NeutrAvidinTM gel. Input and pulldown samples were analyzed using immunoblotting. For quantification in C and D, levels of cell surface proteins were normalized to corresponding input proteins. Readings for single-transfected cells without PI-PLC treatment were given the value of 1.0. Relative surface protein levels in double-transfected cells were calculated accordingly. Results from three independent experiments (n = 3) are presented as mean ± S.E. for each panel. **, p < 0.01; ns, not significantly different; two-way analysis of variance with repeated measures followed by Bonferroni post test for C and one-sample t test for D.
Treatment with PI-PLC has been shown to modulate the expression of GDNF, a GPI-anchored molecule whose synthesis is auto-regulated (37). To avoid possible effects of PI-PLC on NB-3 synthesis, we performed another experiment to test if PTPα can stabilize NB-3 at the cell surface independent of the GPI link. A Myc tag was added to the C terminus of the full-length mouse NB-3 cDNA, which disrupts the GPI consensus site and results in a secreted form of NB-3 (sNB-3-Myc, Fig. 2A). Indeed, the sNB-3-Myc signal was barely detectable at the plasma membrane of cells transfected with sNB-3-Myc alone; however, the cell surface sNB-3-Myc signal was much higher in cells co-transfected with VSVG-PTPα (Fig. 6A). After performing double staining of cell surface sNB-3-Myc and whole cell VSVG-PTPα, we found that the cells expressing a higher level of VSVG-PTPα also exhibited a higher level of cell surface sNB-3-Myc (Fig. 6B). In addition, a cell surface biotinylation assay revealed that the cell surface sNB-3-Myc was increased over 10-fold in cells co-expressing VSVG-PTPα, compared with cells expressing sNB-3-Myc alone (Fig. 6, C–E). Consistent with these findings, the level of sNB-3-Myc in the conditioned medium of PTPα co-transfected COS1 was significantly lower than that of the cells expressing sNB-3-Myc alone (21.3 ± 0.7%, Fig. 6, F and G). Together, these results indicate that protein-protein interaction of PTPα with NB-3 may stabilize NB-3 at the cell surface and leads to increased retention of NB-3 protein at the plasma membrane.
FIGURE 6.
PTPα stabilizes a secreted form of NB-3 at the cell surface. A and B, increase of cell surface sNB-3-Myc when co-expressed with VSVG-PTPα in COS1 cells. After transfection with a secreted form of NB-3 (sNB-3-Myc) alone (A, −PTPα) or with both sNB-3-Myc and VSVG-PTPα cDNAs (A, +PTPα), live COS1 cells were stained with rabbit anti-Myc antibody for cell surface NB-3-Myc protein. Images were taken using the same confocal settings. In B, COS1 cells co-transfected with both sNB-3-Myc and VSVG-PTPα were stained for cell surface sNB-3-Myc and whole cell VSVG-PTPα. Note that cells expressing high level of VSVG-PTPα also exhibit high level of cell surface sNB-3-Myc (arrowhead). Scale bars, 20 μm. C–E, analysis of steady-state cell surface level of sNB-3-Myc and VSVG-PTPα in single- and double-transfected COS1 cells. Cell surface proteins were labeled with biotin and precipitated using NeutrAvidinTM Gel. Samples were blotted with anti-Myc or anti-VSVG antibodies to assess the level of sNB-3-Myc and VSVG-PTPα proteins. For quantification in D and E, levels of cell surface proteins were normalized to corresponding input proteins. Readings for single-transfected cells were given the value of 1.0. Relative surface protein levels in double-transfected cells were calculated accordingly. Results from four independent experiments (n = 4) are presented as mean ± S.E. for each panel. *, p < 0.05; ns, not significantly different; one-sample t test. α-α-Tub, anti-α-tubulin antibody. F, co-expression with VSVG-PTPα significantly reduced the sNB-3-Myc released into the culture medium. Forty-eight hours after transfection, culture medium was collected and the free sNB-3-Myc protein level in the conditioned medium was analyzed. For quantification in G, levels of sNB-3-Myc in the conditioned medium were normalized to that in the whole cell lysates. Relative protein levels in the medium of double-transfected cells were calculated as a percentage of that of the single-transfected cells. Results from four independent experiments (n = 4) are presented as mean ± S.E. ***, p < 0.001; one-sample t test.
Both Ig Domains and FNIII Repeats of NB-3 Interact with PTPα
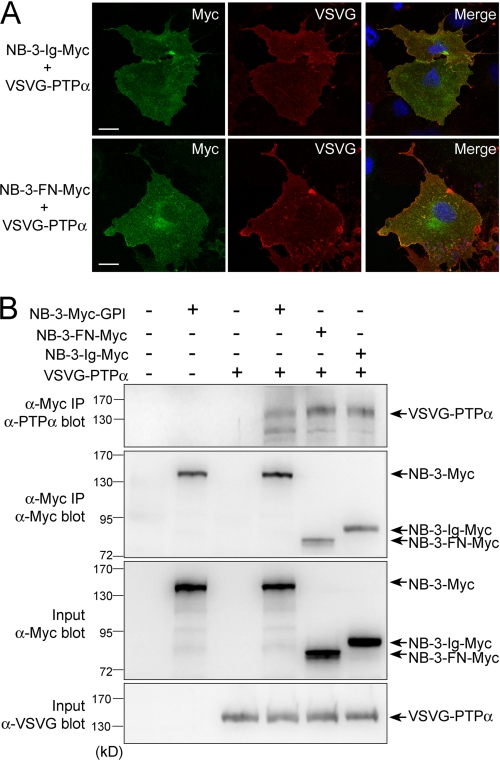
Our previous study showed that PTPα interacts with NB-3 and mediates its signaling to p59fyn (25). To determine the region in NB-3 that mediated its interaction with PTPα, we made truncated forms of NB-3 lacking either FNIII repeats (NB-3-Ig-Myc) or Ig-like domains (NB-3-FN-Myc) (Fig. 2A). In transfected COS1 cells, both NB-3-Ig-Myc and NB-3-FN-Myc were co-localized with VSVG-PTPα intracellularly and at the cell periphery (Fig. 7A). Thirty-six hours after transfection, transfected COS1 cells were lysed using RIPA buffer and immunoprecipitated with mouse monoclonal anti-Myc antibody. The immune precipitates were analyzed by immunoblotting with rabbit polyclonal anti-PTPα antibody. VSVG-PTPα co-transfected with either NB-3-Ig-Myc or NB-3-FN-Myc was recovered in the anti-Myc immune precipitates (Fig. 7B), indicating that both Ig domains and the FNIII repeats of NB-3 can interact with PTPα. However, shorter truncated forms of NB-3 (Ig1–4, Ig4–6, FN1–2, and FN3–4) were not able to co-immunoprecipitate VSVG-PTPα (supplemental Fig. S1), indicating that all six Ig-like domains or four FNIII repeats are necessary for NB-3-PTPα interaction.
FIGURE 7.
Co-immunoprecipitation of NB-3-Ig-Myc and NB-3-FN-Myc with VSVG-PTPα in transfected COS1 cells. A, co-localization of NB-3-Ig-Myc and NB-3-FN-Myc with VSVG-PTPα in transfected COS1 cells. Scale bars, 20 μm. B, COS1 cells transfected with VSVG-PTPα and various NB-3 constructs were lysed with RIPA buffer and immunoprecipitated using a mouse anti-Myc antibody. Immunoprecipitates were analyzed in immunoblotting using a rabbit anti-PTPα antibody. VSVG-PTPα was detected in both NB-3-Ig-Myc and NB-3-FN-Myc immune precipitates.
The Extracellular Domain of PTPα, but Not Its Catalytic Activity, Is Necessary for Enhancing NB-3 Cell Surface Level
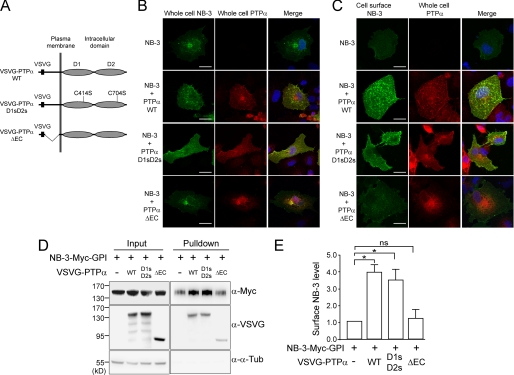
Because contactin family members lack transmembrane and intracellular domains, the extracellular region of PTPα is required for its association with F3/contactin (3). To determine whether the extracellular region of PTPα is required for enhancing NB-3 cell surface expression, we co-transfected COS1 cells with NB-3-Myc-GPI and a PTPα mutant construct, VSVG-PTPα-ΔEC, which has the majority of the extracellular region deleted (amino acid residues Ser-18 to Ser-119, Fig. 8A). Similar to cells transfected with NB-3-Myc-GPI alone, co-transfected cells exhibited a strong and focused NB-3-Myc signal at a perinuclear location and relatively weak signals at the cell surface (Fig. 8, B and C). Cell surface biotinylation assay revealed a similar cell surface NB-3-Myc level in cells transfected with or without VSVG-PTPα-ΔEC, suggesting that removal of the extracellular region of PTPα abolishes the effect of PTPα on NB-3 cell surface expression.
FIGURE 8.
The extracellular domain of PTPα, but not its catalytic activity, is necessary for enhancing cell surface expression of NB-3. A, schematic structure of the PTPα constructs. The VSVG-PTPα-D1sD2s construct has two essential cysteine residues (Cys-414 and Cys-704 in the D1 and D2 catalytic domains, respectively) mutated to serine residues, which abolishes its catalytic activity. The VSVG-PTPα-ΔEC constructs has the majority of its extracellular domain removed. WT, wild type. B and C, COS1 cells transfected with NB-3-Myc-GPI and various VSVG-PTPα constructs were stained for whole cell NB-3-Myc (B) or cell surface NB-3-Myc (C) and whole cell VSVG-PTPα. Images were taken using the same confocal settings. Scale bars, 20 μm. D and E, analysis of cell surface NB-3-Myc-GPI in cells co-transfected with various VSVG-PTPα constructs. Cell surface proteins in transfected COS1 cells were labeled with biotin and precipitated using NeutrAvidinTM gel, followed by analysis using immunoblotting. For quantification in E, levels of cell surface NB-3-Myc were normalized to corresponding input samples. Readings for single-transfected cells were given the value of 1.0. Relative surface protein levels in double-transfected cells were calculated accordingly. Results from three independent experiments (n = 3) are presented as mean ± S.E. *, p < 0.05; ns, not significantly different; one-way analysis of variance with repeated measures followed by Tukey's post test.
The intracellular region of PTPα contains two catalytic domains (D1 and D2), both of which are necessary for its optimal phosphatase activity to activate downstream Src family members (38). Mutation of two essential cysteine residues (Cys-414 and Cys-704) to serine residues in the D1 and D2 catalytic domains, respectively, abolishes its catalytic activity (38). To test whether catalytic activity of PTPα and its downstream signaling is necessary for enhancing NB-3 cell surface expression, we co-transfected COS1 cells with a PTPα construct harboring these two mutations (VSVG-PTPα-D1sD2s, Fig. 8A). The strong perinuclear NB-3-Myc signal was not obvious (Fig. 8B), and the cell surface NB-3-Myc level was similar to that of cells expressing the wild-type PTPα (Fig. 8, C–E). Together, these results indicate that the extracellular region but not the catalytic activity of PTPα is necessary for its role in enhancing NB-3 cell surface expression.
DISCUSSION
NB-3 is a neural adhesion molecule that functions in the developing nervous system, including the neocortex and cerebellum. Upon stimulation, cell surface NB-3 activates PTPα, which transduces signals inside the cells via dephosphorylation and activation of p59fyn. This signaling pathway is required for proper apical dendrite orientation of deep layer pyramidal neurons (25, 39). In this study, we have uncovered a previous unknown role of PTPα in the NB-3 signaling pathway: increasing the surface expression level of NB-3 protein in both neurons and transfected COS1 cells (Figs. 1 and 2). PTPα executes this function mainly through two mechanisms: facilitating Golgi exit of NB-3 and stabilizing NB-3 protein at the plasma membrane.
NB-3, as a GPI-anchored cell surface protein, is processed in the ER-Golgi-plasma membrane secretary pathway during synthesis. Post-translational modification, including GPI-anchor formation mainly takes place in the ER, whereas the Golgi apparatus is the major sorting station that directs newly synthesized NB-3 protein to the plasma membrane. We found that NB-3 protein is enriched in the medial Golgi in transfected COS1 cells (Fig. 3), resulting in a low cell surface expression of NB-3, whereas surface expression of transfected PTPα is relatively higher (Fig. 2). According to a rapid-partitioning model of intra-Golgi progression, proteins entering the Golgi are quickly partitioned between two phases, the processing domain enriched in Golgi enzymes and the export domain capable of budding transport intermediates for their final target sites (40). Enrichment of NB-3 in the medial Golgi suggests that most NB-3 protein may be partitioned into the processing domain, which leads to a low Golgi exit rate for NB-3 protein. PTPα is expressed at the cell surface at a relatively high level, suggesting that more PTPα is partitioned into the export domain to translocate to the plasma membrane. As an NB-3-interacting protein, co-expression with PTPα would probably help partition of NB-3 into the export domain, and facilitate Golgi exit of NB-3. Indeed, co-expression with PTPα significantly reduced the co-localization of NB-3 with the Golgi marker, and in Ptpα−/− neurons more endogenous NB-3 protein was co-localized with the Golgi marker (Fig. 4). Moreover, cargo proteins exit the Golgi at an exponential rate proportional to total Golgi load (40). Co-expression with PTPα would increase the total Golgi abundance, which may also increase NB-3 Golgi exit rate to a certain extent.
NB-3 belongs to the F3/contactin family. Adhesion molecules in this family lack a transmembrane domain and are anchored at the cell surface via a GPI link, which is frequently cleaved by extracellular phospholipase activity and leads to the release of the proteins. Indeed, released F3 or NB-3 can function as a ligand for other cell surface receptors, such as Notch1 (36, 41). However, to function as a receptor instead, NB-3 needs to be present at the plasma membrane. We found that cleavage of the GPI link using PI-PLC did not reduce the cell surface level of NB-3 in COS1 cells co-expressing PTPα (Fig. 5). Also, PTPα significantly increased the cell surface level of a secreted form of NB-3 and reduced its release into the culture medium (Fig. 6), suggesting that PTPα is able to stabilize NB-3 at the plasma membrane independent of the GPI link. PTPα most likely exerts this effect through direct interaction with both Ig domains and FNIII repeats of NB-3 protein at the plasma membrane (Fig. 7). The extracellular region but not the catalytic activity of PTPα is necessary for its effects on NB-3 cell surface expression (Fig. 8), suggesting that the physical interaction of PTPα and NB-3 protein is sufficient to facilitate NB-3 intracellular processing and stabilization at the cell surface.
In summary, our present study demonstrated a novel function of PTPα in NB-3/PTPα signaling. In addition to transducing the NB-3 signal inside neurons and eventually leading to cytoskeleton rearrangement and dendritic growth, PTPα also helped Golgi exit of NB-3 and stabilized NB-3 at the plasma membrane, thus increasing the cell surface level of NB-3. In this way, PTPα, though a downstream effector, may also be able to enhance NB-3 signaling strength and efficiency at the receptor level in developing neurons. It would be interesting to investigate whether PTPα also regulates cell surface expression of other membrane-bound molecules and whether this signaling mechanism also applies to other GPI-anchored proteins to enhance and facilitate their functions as receptors.
Supplementary Material
Acknowledgments
We thank Dr. Kazutada Watanabe from Nagaoka University of Technology, Japan, for kindly providing the anti-NB-3 antibodies. We thank Dr. Jane Y. Wu from Northwestern University, Dr. Li Zhu from the Institute of Biophysics, Chinese Academy of Sciences, China, and Dr. Shi-Qiang Wang from Peking University, China, for helpful comments on the manuscript.
This work was supported in part by Ministry of Science and Technology Grants 2009CB825402 and 2010CB529603 and National Science Foundation Grant 30900845 in China (to H. Y.) and by Canadian Institutes of Health Research Grant MOP-62759 (to C. J. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Materials and Methods.
- PTPα
- protein-tyrosine phosphatase α
- SFK
- src family kinase
- CHL1
- Close Homolog of L1
- FNIII
- fibronectin type III
- GPI
- glycosylphosphatidylinositol
- E17.5
- embryonic day 17.5
- mAb and pAb
- mouse monoclonal and rabbit polyclonal antibodies
- RIPA
- radioimmune precipitation assay
- ER
- endoplasmic reticulum
- PI-PLC
- phosphatidylinositol-phospholipase C.
REFERENCES
- 1. Pallen C. J. (2003) Curr. Top Med. Chem. 3, 821–835 [DOI] [PubMed] [Google Scholar]
- 2. Bodrikov V., Leshchyns'ka I., Sytnyk V., Overvoorde J., den Hertog J., Schachner M. (2005) J. Cell Biol. 168, 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zeng L., D'Alessandri L., Kalousek M. B., Vaughan L., Pallen C. J. (1999) J. Cell Biol. 147, 707–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. von Wichert G., Jiang G., Kostic A., De Vos K., Sap J., Sheetz M. P. (2003) J. Cell Biol. 161, 143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zheng X. M., Wang Y., Pallen C. J. (1992) Nature 359, 336–339 [DOI] [PubMed] [Google Scholar]
- 6. den Hertog J., Pals C. E., Peppelenbosch M. P., Tertoolen L. G., de Laat S. W., Kruijer W. (1993) EMBO J. 12, 3789–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bhandari V., Lim K. L., Pallen C. J. (1998) J. Biol. Chem. 273, 8691–8698 [DOI] [PubMed] [Google Scholar]
- 8. Tsai W., Morielli A. D., Cachero T. G., Peralta E. G. (1999) EMBO J. 18, 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lei G., Xue S., Chéry N., Liu Q., Xu J., Kwan C. L., Fu Y. P., Lu Y. M., Liu M., Harder K. W., Yu X. M. (2002) EMBO J. 21, 2977–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zheng X. M., Shalloway D. (2001) EMBO J. 20, 6037–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zheng X. M., Resnick R. J., Shalloway D. (2002) J. Biol. Chem. 277, 21922–21929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vacaru A. M., den Hertog J. (2010) Mol. Cell. Biol. 30, 2850–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Petrone A., Battaglia F., Wang C., Dusa A., Su J., Zagzag D., Bianchi R., Casaccia-Bonnefil P., Arancio O., Sap J. (2003) EMBO J. 22, 4121–4131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zeng L., Si X., Yu W. P., Le H. T., Ng K. P., Teng R. M., Ryan K., Wang D. Z., Ponniah S., Pallen C. J. (2003) J. Cell Biol. 160, 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krndija D., Schmid H., Eismann J. L., Lother U., Adler G., Oswald F., Seufferlein T., von Wichert G. (2010) Oncogene 29, 2724–2738 [DOI] [PubMed] [Google Scholar]
- 16. Møller N. P., Møller K. B., Lammers R., Kharitonenkov A., Hoppe E., Wiberg F. C., Sures I., Ullrich A. (1995) J. Biol. Chem. 270, 23126–23131 [DOI] [PubMed] [Google Scholar]
- 17. Steták A., Csermely P., Ullrich A., Kéri G. (2001) Biochem. Biophys. Res. Commun. 288, 564–572 [DOI] [PubMed] [Google Scholar]
- 18. Zhang X. Q., Kondrikov D., Yuan T. C., Lin F. F., Hansen J., Lin M. F. (2003) Oncogene 22, 6704–6716 [DOI] [PubMed] [Google Scholar]
- 19. Kostic A., Sap J., Sheetz M. P. (2007) J. Cell Sci. 120, 3895–3904 [DOI] [PubMed] [Google Scholar]
- 20. Sap J., D'Eustachio P., Givol D., Schlessinger J. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 6112–6116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Skelton M. R., Ponniah S., Wang D. Z., Doetschman T., Vorhees C. V., Pallen C. J. (2003) Brain Res. 984, 1–10 [DOI] [PubMed] [Google Scholar]
- 22. Su J., Yang L. T., Sap J. (1996) J. Biol. Chem. 271, 28086–28096 [DOI] [PubMed] [Google Scholar]
- 23. Wang P. S., Wang J., Xiao Z. C., Pallen C. J. (2009) J. Biol. Chem. 284, 33692–33702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang L. T., Alexandropoulos K., Sap J. (2002) J. Biol. Chem. 277, 17406–17414 [DOI] [PubMed] [Google Scholar]
- 25. Ye H., Tan Y. L., Ponniah S., Takeda Y., Wang S. Q., Schachner M., Watanabe K., Pallen C. J., Xiao Z. C. (2008) EMBO J. 27, 188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Demyanenko G. P., Schachner M., Anton E., Schmid R., Feng G., Sanes J., Maness P. F. (2004) Neuron 44, 423–437 [DOI] [PubMed] [Google Scholar]
- 27. Ogawa J., Kaneko H., Masuda T., Nagata S., Hosoya H., Watanabe K. (1996) Neurosci. Lett. 218, 173–176 [DOI] [PubMed] [Google Scholar]
- 28. Sakurai K., Toyoshima M., Ueda H., Matsubara K., Takeda Y., Karagogeos D., Shimoda Y., Watanabe K. (2009) Dev. Neurobiol. 69, 811–824 [DOI] [PubMed] [Google Scholar]
- 29. Takeda Y., Akasaka K., Lee S., Kobayashi S., Kawano H., Murayama S., Takahashi N., Hashimoto K., Kano M., Asano M., Sudo K., Iwakura Y., Watanabe K. (2003) J. Neurobiol. 56, 252–265 [DOI] [PubMed] [Google Scholar]
- 30. Drumheller T., McGillivray B. C., Behrner D., MacLeod P., McFadden D. E., Roberson J., Venditti C., Chorney K., Chorney M., Smith D. I. (1996) J. Med. Genet. 33, 842–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Verjaal M., De Nef M. B. (1978) Am. J. Dis. Child 132, 43–45 [DOI] [PubMed] [Google Scholar]
- 32. Cuoco C., Ronchetto P., Gimelli S., Béna F., Divizia M. T., Lerone M., Mirabelli-Badenier M., Mascaretti M., Gimelli G. (2011) Orphanet. J. Rare Dis. 6, 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pohjola P., de Leeuw N., Penttinen M., Kääriäinen H. (2010) Am. J. Med. Genet. A. 152A, 441–446 [DOI] [PubMed] [Google Scholar]
- 34. Faivre-Sarrailh C., Gauthier F., Denisenko-Nehrbass N., Le Bivic A., Rougon G., Girault J. A. (2000) J. Cell Biol. 149, 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ponniah S., Wang D. Z., Lim K. L., Pallen C. J. (1999) Curr. Biol. 9, 535–538 [DOI] [PubMed] [Google Scholar]
- 36. Cui X. Y., Hu Q. D., Tekaya M., Shimoda Y., Ang B. T., Nie D. Y., Sun L., Hu W. P., Karsak M., Duka T., Takeda Y., Ou L. Y., Dawe G. S., Yu F. G., Ahmed S., Jin L. H., Schachner M., Watanabe K., Arsenijevic Y., Xiao Z. C. (2004) J. Biol. Chem. 279, 25858–25865 [DOI] [PubMed] [Google Scholar]
- 37. He D. Y., Ron D. (2006) FASEB J. 20, 2420–2422 [DOI] [PubMed] [Google Scholar]
- 38. Lim K. L., Lai D. S., Kalousek M. B., Wang Y., Pallen C. J. (1997) Eur. J. Biochem. 245, 693–700 [DOI] [PubMed] [Google Scholar]
- 39. Schmid R. S., Maness P. F. (2008) Curr. Opin. Neurobiol. 18, 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Patterson G. H., Hirschberg K., Polishchuk R. S., Gerlich D., Phair R. D., Lippincott-Schwartz J. (2008) Cell 133, 1055–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hu Q. D., Ang B. T., Karsak M., Hu W. P., Cui X. Y., Duka T., Takeda Y., Chia W., Sankar N., Ng Y. K., Ling E. A., Maciag T., Small D., Trifonova R., Kopan R., Okano H., Nakafuku M., Chiba S., Hirai H., Aster J. C., Schachner M., Pallen C. J., Watanabe K., Xiao Z. C. (2003) Cell 115, 163–175 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.