Abstract
Mass spectrometry has emerged as a powerful tool for the analysis of all lipids. Lipidomic analysis of biological systems using various approaches is now possible with a quantitative measurement of hundreds of lipid molecular species. Although availability of reference and internal standards lags behind the field, approaches using stable isotope-labeled derivative tagging permit precise determination of specific phospholipids in an experimental series. The use of reactivity of ozone has enabled assessment of double bond positions in fatty acyl groups even when species remain in complex lipid mixtures. Rapid scanning tandem mass spectrometers are capable of quantitative analysis of hundreds of targeted lipids at high sensitivity in a single on-line chromatographic separation. Imaging mass spectrometry of lipids in tissues has opened new insights into the distribution of lipid molecular species with promising application to study pathophysiological events and diseases.
Keywords: Lipids, Lipid Ether, Lipid Structure, Mass Spectrometry (MS), Steroid, MALDI Imaging, MBOAT, Lipidomics, Stable Isotope Tagging
Introduction
Mass spectrometry has been applied to the analysis of lipids since its early origins as an analytical tool for organic chemistry. Fundamental studies of fatty acid esters proved that MS not only could reveal detailed structural information from known compounds (1) but also was highly useful for the structural elucidation of unknown lipids such as the prostaglandins (2). Lipids also were used as molecules to probe the basic mechanisms of ion fragmentation following electron ionization (3). However, very few lipids were amenable for direct MS analysis because of the absolute requirement that the molecule must have a sufficient vapor pressure to enter as a gas into the ion source of the mass spectrometer. This remarkably changed when fast atom bombardment ionization was first introduced (4), and nonvolatile lipids such as phospholipids (5) could be directly analyzed; yet it was the development of electrospray ionization (ESI)2 by Fenn et al. (6) and MALDI by Karas and Hillenkamp (7) that truly opened the vast array of lipids found in biology (Table 1) to direct analysis by MS. The situation today is that all lipids are amenable to MS and the numerous ancillary techniques engaged in current practice.
TABLE 1.
Lipid categories and examples of molecular species
A complete list of lipids and classification numbers can be found at the Nature/LIPID MAPS website. SM, sphingomyelin.
| Category | Specific lipid species |
|---|---|
| Fatty acyls | Fatty acids, eicosanoids, endocannabinoids |
| Glycerolipids | TAGs, diacylglycerols |
| Glycerophospholipids | PC, PE, PI, PS, PG, PA, cardiolipin |
| Sphingolipids | SM, sulfatides, sphingosine, ceramides, ganglioside |
| Sterol lipids | Cholesterol, estradiol, testosterone, bile acids |
| Prenol lipids | Farnesol, dolichols, vitamin K |
| Saccharolipids | Lipid A, acyltrehaloses |
| Polyketides | Aflatoxins, tetracyclines, erythromycin |
These desorption and spray ionization techniques surprisingly impart very little energy to the ionized lipid, and thus, protonated molecular ions ([M + H]+) or adducted molecular ions (such as [M + NH4]+, [M + Na]+, and [M + K]+) are the usually observed cations. Deprotonated molecular anions ([M − H]−) and adducted anions ([M + Cl]− and [M + acetate]−) are observed if the molecule preferentially forms negative ions. At the same time that these remarkable ionization techniques were being developed, tools were emerging to impart energy to break covalent bonds within ions using techniques such as collisional activation and collision-induced dissociation (CID) and to transmit product ions using efficient collision cells (radio frequency-only quadrupole fields) and then reanalyze the product ions by a second mass spectrometer (MS/MS). The triple quadrupole mass spectrometer encompassed these advances of CID and was found to be well suited for the analysis of lipids through its many modes of MS/MS operation, including product ion scanning, precursor ion scanning, neutral loss scanning, and selected reaction monitoring (SRM; also termed multiple reaction monitoring). High resolution mass analysis of molecular ion species and product ions after CID became routinely possible with the second generation time-of-flight analyzers (8) and ion-trapping technology of the ion cyclotron resonance cell and the orbitrap mass spectrometer (9). Much of this instrumental development occurred in the commercial manufacturing sector because of the large market for these tools in proteomic research. Nevertheless, these tools were well suited for lipid analysis, and application of MS and MS/MS to solve challenging problems was now possible.
Lipids are somewhat different from biomolecules such as peptides, oligonucleic acids, and oligosaccharides from many standpoints of MS analysis. Lipids are hydrophobic as well as hydrophilic molecules (10), and the hydrophobicity typically means a large number of –CH2 groups or a large number of hydrogens in the molecule that impart a significant mass defect observed as the fraction of exact mass following the integer mass of the molecular ion species. Information related to chemical structure can often be obtained by CID, but this information is encoded in the gas-phase ion chemistry of the lipid. As a result of these developments in MS, which were largely realized a decade or more ago, recent applications of MS to lipid analysis have resulted in remarkable advances in our understanding of lipid biochemistry.
One advance for lipid MS, perhaps not fully appreciated, has been the increased availability of sophisticated mass spectrometers in biochemical laboratories to study lipid biochemistry. A specific example was the use of MS to refine our understanding of the biological role of the gene product SRD5A3, which was previously annotated as steroid 5α-reductase type 3, and the involvement of this protein in a severe congenital glycosylation genetic disorder affecting humans (11). These investigators were able to identify increased levels of polyprenol lipids relative to dolichols in the plasma of affected humans that lacked this gene relative to normal subjects and were able to go on and define SRD5A3 as an NADPH-dependent reductase that saturates the α-isoprene unit of polyprenols to yield the dolichol structure. Because dolichols are required for N-linked glycosylation in the Golgi apparatus, the loss of this rate-limiting enzyme reduced overall glycosylation of proteins critical for proper function. A MS method for the analysis of dolichols was a recent result of having advanced instruments available for lipidomic studies in laboratories of lipid biochemists (12).
Lipidomic Studies
One of the significant changes that have occurred has been an understanding of lipids as they exist as complex mixtures within the cellular environment. This has naturally led to the concept that revealing the nature of this mixture is an important goal in understanding cell biology or system biology. This has resulted in the widespread use of the term “lipidomics,” which unfortunately means different things to different individuals. On one hand, some have a “global” view of lipidomics as the identification and quantitation of all lipids in a cell (tissue or organism) and studies of how they vary with state/challenge of the cell. Others use the term to define quantitative analysis of a restricted group such as the major lipids present in a biological sample. The sheer number of actual lipid substances within living cells is enormous (13). The identified and quantitated lipids within the macrophage have been reported to be as high as 1000 individual molecular species with caveats that more species were detected than could be quantitated (14). The dynamic range in which lipid concentrations can vary in a tissue may be 106 or more (from nanomolar fatty acids to attomolar eicosanoid lipid mediators), and this range challenges the capability of any single approach of analysis. As noted above, molecular weight alone is insufficient to absolutely define a lipid structure because this single measure introduces assumptions as to identification. Identification by chromatographic retention time, measurement at high resolution (elemental composition at submillimass unit accuracy), gas-phase chemical behavior, and elution with an isotope-labeled internal standard may still be insufficient to absolutely identify a complex lipid because of ambiguities arising from regio- or stereoisomerism (sn-substitution for phospholipids or double bond position and geometry for any fatty acyl substituent). Perhaps most insidious is that the criteria employed to identify a lipid are often not stated, which can also lead to misunderstandings on the part of the reader. Nevertheless, practicality demands that assumptions have to be made because even state-of-the-art MS is insufficiently powerful to deal with the complex mixture of lipids in a lipidomics study, yet one must balance the need to achieve results.
One aspect of cellular lipid mixture complexity is that families of closely related lipids are present that differ by the number of fatty acyl carbons and number of double bonds (as well as position of double bonds) as well as minor variants in structure such as ether substitution for esters. These closely related families comprise molecular species that have very similar structure but often different molecular weights. Molecular species that differ by common chain elongation and the number of double bonds in the fatty acyl chain appear at intervals 24–28 Da higher or lower in the mass spectrum of the mixture. Many of the lipids presented in Table 1 exist within cells as mixtures of individual molecular species, and the MS experiment allows one to readily discover those molecular ions species present in an extract by either positive and/or negative ion MS. There have been several reviews on the behavior of each of the lipid classes and the individual molecular species in terms of their CID ion chemistry after ionization by either electrospray or MALDI (15–18). The challenge has been to analyze the nature of the complex mixture and understand whether information collected about different molecular species reveals unique biochemical events or possibly pathophysiology.
There have been several approaches to deal with complex mixtures of molecular species. The approaches taken can be divided into two general methodologies. The first involves little pre-separation of the lipids other than by simple solvent extraction, followed by direct analysis as the complex mixture of many different lipid classes. This approach is called the “shotgun” method and certainly has specific advantages as well as some disadvantages. The second approach has been to use chromatographic separation of crude lipid extracts to isolate specific lipid classes prior to analysis (Table 1) with or without additional separation of the individual molecular species. For example, normal-phase chromatography (which is a separation method based on analyte molecular polarity) is well suited to separate each of the lipid classes such as glycerophospholipids from glycerolipids (triacylglycerols (TAGs) or diacylglycerols). However, molecular species are not typically separated by this technique, and to achieve this, reversed-phase HPLC is employed. If individual classes of lipids are not pre-separated through a normal phase-type approach, then separation only by reversed-phase chromatography separates by lipophilicity and is largely not influenced by the polar headgroup and the overall polar character of the lipid.
The shotgun technique has been significantly refined to capitalize on the anionic and weakly anionic nature of many lipid classes to optimize the analysis of highly different lipid classes present in an extract (19). Direct analysis of the negative ions formed by electrospray can reveal the presence of the very acidic lipids such as phosphatidylinositol (PI), phosphatidylserine (PS), phosphatidylglycerol (PG), phosphatidic acid (PA), cardiolipin, and sulfatides (sphingolipids) by the addition of a base such as LiOH. Negative ion ESI can also yield signals from the phosphatidylethanolamine (PE) molecular species. Positive ion ESI-MS yields abundant [M + H]+ (or [M + Li]+ if LiOH is added) for glycerophosphatidylcholine (PC) and glycerolipid molecular species present in a complex mixture. Among the potential confounding features of shotgun lipidomics is ion suppression during the electrospray process that limits the ability to detect minor lipid species that may be present (20) and the fact that any observed m/z may contain multiple molecular ion species. The shotgun method has been used for the quantitation of specific lipids using techniques such as neutral loss and precursor ion scanning (MS/MS) and internal standard/reference standard calibration protocols to generate standard curves to convert ratios of target lipid ion abundance/internal standard ion abundance to absolute concentration of lipids in the electrospray solvent system (21). The major advantages of the shotgun method are the ease of implementation and the broad coverage of major lipid molecular species that can be quantitated in a relatively short period of time by a single laboratory.
Pre-separation of a lipid extract using specific chromatographic protocols developed for each lipid class can enable the identification of far more lipid species that differ in subtle ways. The analysis of the neutral lipids from RAW 264.7 cells by normal-phase chromatography followed by LC-MS enabled the detection of ether-linked triradylglycerols because they were separated by polarity from the TAGs (22). An ether-linked triradylglycerol such as 16:0 ether/18:0/18:1 differs only slightly (0.027 thomson units) in the mass/charge ratio of the observed [M + NH4]+ from an odd-chained analog such as 15:0/18:0/18:1 TAG but substantially differs in mobility under the normal-phase chromatographic separation employed. These ether-linked glycerides could have easily been misidentified without a separation step. As with shotgun lipidomics, this LC-MS and LC-MS/MS approach for lipid analysis can be set up for quantitative analysis using internal standards and calibration curves established with reference standards (22). The chromatographic pre-separation approach was used to identify >1000 different molecular species recently measured in the macrophage lipidome (14). To achieve this quantitative measure for this large number of molecular species, it was necessary to separate and to employ a different separation protocol that was ideal for each individual class of lipid. Lipidomic analyses of yeast (23), Caenorhabditis elegans (24), tuberculosis mycobacteria (25), human plasma (26), and others (27) have emerged.
Challenges still remain in the lipidomics area, in large part because it is very tedious to establish and carry out the quantitative analysis method. However, more importantly, there are insufficient internal as well as few reference standards commercially available that can be used to generate the appropriate calibration curves to convert abundance of ions into a quantitative measure of lipid concentration for many lipid classes. The abundance of any molecular ion species, e.g. [M + H]+, typically carries information on concentration, but ion abundance is confounded by a number of features, including instrument response factors, ionization efficiency of the molecule, stability of the molecular ion species, and the presence of other molecules that could cause ion suppression of the analyte of interest. Quantitative MS has evolved to the highly specific and sensitive “gold standard” level of acceptance because of measures to avoid many of these problems. The central strategy of isotope dilution has been to use a stable isotope-labeled analog of the analyte as an internal standard and to measure the ratio of signal intensities of the analyte and internal standard rather than any absolute intensity. The ratio is then converted to analyte concentration by a calibration curve generated using reference standards. However, this approach is not available for most lipidomic studies because of the absence of internal standards for each and every lipid molecular species and, practically, the potential overlap of m/z between internal standards and endogenous lipids. The absence of reference standards for the different variants of structure observed in a molecular species series, e.g. number and position of double bonds in fatty acyl chains, variety of fatty acyl chain length, ether analogs, even positional isomers for phospholipids and glycerolipids, further complicates analysis because one cannot easily measure parameters such as ionization efficiency, ion stability, and response factors that are critical for the accuracy of the quantitative analysis. Often, quantitation is based on an “average standard curve” or, worse, just a single reference standard for many different structural variants in a series. Nevertheless, given the problems with accuracy, these measurements remain precise and useful when comparing changes of the same lipid molecular species within an experimental series.
An alternative approach to absolute quantitation has been to express the abundance of molecular species within a single class of lipids as mole fraction using the abundance ratios of all molecular species in that class to normalize the data. Relevant information can be readily gleaned as exemplified in an extensive report of phospholipid molecular specie measurements in a study of lysophosphatidic acid acyltransferases that supply polyunsaturated fatty acids to phospholipids that become incorporated into cellular membranes (28).
Stable Isotope Tagging
An alternative approach for aminophospholipid quantitation within a complex mixture of molecular species had its origins in the growing need for quantitation in peptide analysis. In this case, phospholipids such as PE and PS are derivatized with stable isotope-labeled reagents that tag the molecule within a specific treatment series. For example, identical aliquots of cells are treated under conditions A, B, C, and D. Each cell treatment aliquot is derivatized with a different stable isotope internal standard to tag each incubation condition. The tag is then used to decode the lipid molecular species as to each experimental condition, and the ratios of isotopically distinct ion signal intensities allow determination of changes in concentration. This multiplexed quantitative approach was first demonstrated with commercial derivatization iTRAQ (isobaric tag for relative and absolute quantitation) reagents, in which product ions of derivatized aminophospholipids were analyzed but required MS3 to decode (29). An alternative derivative was developed using four different stable isotope-labeled dimethylaminobenzoic acid N-hydroxysuccinimide esters and precursor ion scanning to decode modified or newly formed aminophospholipids in cells treated with ozone (30). These studies readily revealed the differences in concentrations of PE molecular species because the ionization phenomenon was virtually identical for all of the products under analysis, and in effect, a stable isotope variant was made for each aminophospholipid molecular species.
Fatty Acyl Double Bond Positional Analysis
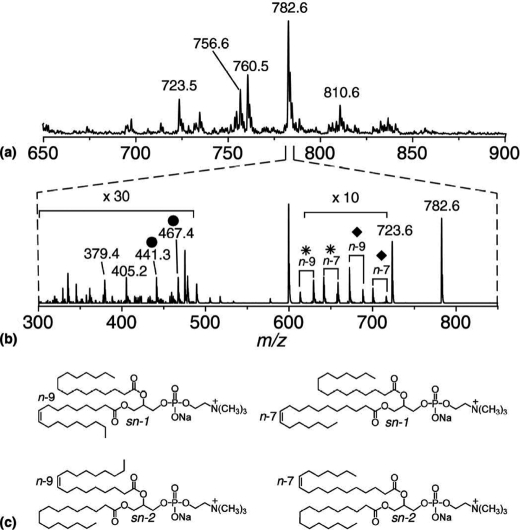
The exact position of a double bond in a fatty acyl chain remains rather difficult to determine by MS alone, yet important information can be ignored if such determinations are not made. For example, vaccenic acid (18:1(n-7)) is a common trans-fatty acid in cow's milk, but it is isomeric in terms of double bond position to oleic acid (18:1(n-9)), the more common monounsaturated fatty acyl group found in phospholipids, sphingolipids, and glycerolipids. Thus, the identification of an 18:1 fatty acyl group, for example, by neutral loss scanning or product ion scanning in lipidomic analyses is ambiguous with respect to positional isomers. Recently, a method using ozone to effect carbon bond cleavage in phospholipids was introduced and revealed that often this fatty acyl group is a mixture of n-7 and n-9 fatty acids (31, 32). This ozone approach has been expanded to include collisional activation of ions in the presence of ozone (combined CID-ozone identification), which can lead to product aldehyde ions that reveal the double bond position (Fig. 1).
FIGURE 1.
Ozone reaction with phospholipids in the collision cell of a linear ion trap mass spectrometer. a, ESI mass spectrum obtained by direct infusion of a crude lipid extract from cow brain. b, combined CID-ozone identification mass spectrum acquired by applying collision energy to the mass-selected precursor ion at m/z 782.6 with ozone vapor present in the collision cell (q2). c, molecular structure of four regioisomeric lipids that could give rise to the combined spectral features observed in b. This figure has been reprinted with permission from the American Chemical Society (31).
Targeted Lipid Quantitation
The quantitation of eicosanoids and other lipid mediators by MS was well established even when only GC-MS and negative ion chemical ionization were available (33). The analysis of these lipids has always been a challenge because of the low quantities made within tissues or cells. LC-MS/MS has emerged as the technique of choice to study these molecules because no derivatization is required, and this class of arachidonic acid metabolite has a free carboxylic acid moiety that renders efficient negative ion formation (carboxylate anion) by ESI. The recent advances in MS instrumentation include rapid scanning; for example, in the single reaction monitoring mode of MS/MS operation, it is now quite possible to monitor 20–50 selected ion transitions with a duty cycle of less than a second. The recent advances in chromatography at high pressure (ultra-HPLC) have greatly reduced retention times, and the combination of ultra-HPLC with rapid SRM has enabled development of methods to target not one eicosanoid in a single LC-MS/MS assay but 100 eicosanoids at high sensitivity and specificity (34).
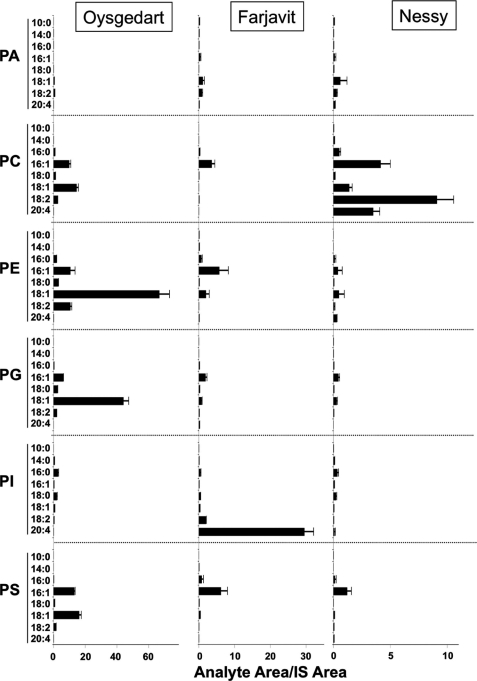
Rapid SRM scanning was also used in the development of an enzyme choice assay to characterize newly discovered lysophospholipid acyltransferases (LATs) (35). Enzymatic properties such as substrate specificity have hitherto often been determined using radiolabeled tracers and Michaelis-Menten kinetic parameters. An alternative approach was recently taken that relied on the ability to rapidly determine the reaction products when mixtures of potential substrates were mixed with cloned enzymatic proteins. In this particular case, four different LATs were studied by the addition of a mixture of six lysophospholipid subclasses (choline, ethanolamine, inositol, glycerol, serine, and lyso-PA) with eight different CoA esters. This resulted in 48 potential products, requiring (after inclusion of controls) 60 SRM ion transitions to monitor during an LC-MS/MS run. Remarkable substrate specificity was observed in this competition assay for each of the different LATs of the MBOAT (membrane-bound O-acyltransferases) family (35) as well as three unique LATs from Drosophila (Fig. 2) (36). A similar mixed substrate approach to assess enzymatic activity was also used in studies of LPAAT3 (lysophosphatidic acid acyltransferase 3) (28).
FIGURE 2.
Dual choice LAT enzyme assay by selected ion recording and LC-MS/MS. The fly MBOAT proteins Oys, Frj, and Nes were expressed in the acyltransferase-deficient ale1Δ yeast strain. Isolated microsomes containing the fly proteins were incubated with a mixture of eight acyl-CoA species and six lysophospholipids, and the products formed by each enzyme were separated and quantified by LC-MS/MS. Scales were adjusted to highlight the substrate preferences of each enzyme. Results given are the mean ± S.E. of three experiments. Acyl-CoAs are abbreviated as x:y, where x is the number of carbon atoms in the chain and y is the number of double bonds. This figure has been reprinted with permission from the American Society for Cell Biology (36).
Imaging MS (IMS) of Lipids
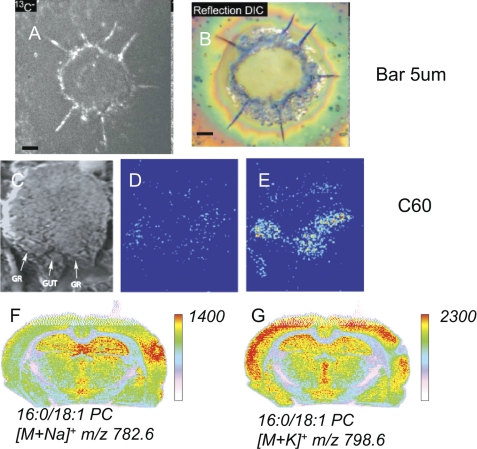
An area of very rapid growth has been the imaging of tissues by MS. Studies of lipid biochemistry have always suffered from the inability to determine the location of specific lipid substances at or near the cellular level. IMS partially bridges this gap but has the additional advantage of MS specificity. Several different ionization methods have been explored to generate images of the distribution of lipid species as they occur in tissues. Secondary ion MS (SIMS) has historically been the method used to generate lipid-derived ions, but the energy of this technique often results in lipid structural damage and loss of molecular species information (37). Nonetheless, this technique has the best lateral resolution (<50 nm is possible) and is well adapted to specific lipid experiments such as tracing the transport of 13C-labeled oleate in an adipocyte (Fig. 3, A and B), where the ion observed is 13C− (38). Recent advances using caged C60+ (buckyballs) as projectile ions have enabled intact lipids (cholesterol) to be imaged in the SIMS experiments and suggest that it may be possible to image abundant lipids at cellular resolution (Fig. 3, C–E) (39).
FIGURE 3.
Imaging of lipids using MS. A, SIMS imaging of fatty acid transport in cultured adipocytes after unwashed 3T3F442A adipocytes were incubated with [13C]oleate. Images are of 13C−. Scale bar = 5 μm. This figure has been reprinted with permission as open access from Ref. 38. B, optical image using a reflection differential interference contrast microscope of the same cells before analysis with SIMS. Reflection differential interference contrast (DIC) images (magnification ×500) were obtained using a Nikon Eclipse E800 upright microscope. Scale bar = 5 μm. This figure has been reprinted with permission as open access from Ref. 38. C, scanning ion image of an axial slice from a 9-day-old mouse embryo. The gut and genital ridge (GR) are identified by white arrows. D and E, SIMS images of cholesterol (m/z 366–370) from the tissue in the SIMS image. The image in E was taken before a sputter dose of 1 × 1013 C60+60+/cm2, and the image in C was taken after nanotome sputtering. This figure has been reprinted with permission from Elsevier (39). F and G, rat brain sections from a traumatic brain injury model and imaging by MALDI-IMS corresponding to 16:0/18:1 PC ([M + Na]+, m/z 782.6) and 16:0/18:1 PC ([M + K]+, m/z 798.6), respectively (51).
MALDI can be readily coupled with imaging of lipids in tissues because the most abundant ions released in this MALDI-IMS experiment are the lipids that make up the cellular membranes and are in lipid droplets. Remarkable images are appearing as to the regional distribution of lipids in tissues such as brain (40) and kidney (41) as well as large biological structures such as embryos (42) and entire organisms such a mouse (43). The secondary lipid ions that are observed do correlate fairly well with local concentrations of specific lipids that are readily desorbed and ionized by this technique. This correlation is particularly high for PC in the positive ion mode (44) and for PI and sulfatides in the negative ion mode. However, there is bias in this experiment in that some lipids known to be present are not easily observed, perhaps due to ion suppression. For example, PE molecular species are not readily observed in the positive ion mode in some instruments, possibly due to their rather facile loss of the polar headgroup (141 Da) to yield positively charged diglyceride-like ions. Another curious feature is that the phospholipids and glycerolipids are observed as alkali metal adducts (Na+ and K+) as well as the protonated species. Recent reports (51) have suggested that there is information in the relative abundance of these ions that reflects edema and loss of the ATPases that maintain the cellular cation gradient (Fig. 3, F and G). Another ionization technique, desorption ESI, has also been successfully used to image lipids in various tissues, including brain (45, 46).
Combined Ion Mobility-MS (IM-MS) of Lipids
The coupling of ion mobility separation with either single stage or tandem MS provides an additional dimension of analysis on a far more rapid time scale than achieved with conventional pre-separation of lipid classes using condensed-phase chromatographic methods. Woods and co-workers (47) reported a systematic study of several polar lipid classes, illustrating distinct behaviors in the relationship between ion mobility and m/z ratio. The combination of MALDI and IM-MS was exploited by Kliman et al. (48) in examining intact brain tissue from Drosophila; quantitative comparisons of different tissue samples were followed by IM-MS/MS analyses to allow identification of those molecular species present in differing amounts. Further studies by Trimpin et al. (49), this time using ESI, provided evidence for phospholipid aggregates in the gas phase, interpreted as inverted micelles, which were separated by IM-MS prior to disaggregation and further mobility analysis. Three stages of IM-MS interspersed with activated decomposition also allowed distinction between sn-1 and sn-2 isomers, illustrated using PGs. Such sophisticated analyses, employing tandem ion mobility coupled with MS, are unlikely to become routine, but the improved separation power of combining single stage mobility separation and MS or MS/MS is such that widespread application is likely.
Conclusion
Over the past decade, significant advances have been made in MS instrumentation that significantly impact the analysis of lipids in biological samples. Sensitivity for the LC-MS detection of lipids has increased perhaps 50–100-fold with the development of advanced tandem quadrupole mass spectrometers as well as advanced LC technology. Along with this increased in sensitivity has come increased scanning speed, which permits almost unlimited selected ion recording of ion transitions when coupled with computer-driven timing of the SRM experiments. Remarkable advances in IMS of lipids will likely drive the development of improvements in rastering laser beams for the MALDI mass spectral data acquisition stage. The expected application of ion mobility technology to real problem solving in lipid biochemistry will likely prove the value of this MS technology, which is still in its infancy. The utility of ion-trapping instruments and, in particular, the newer technologies of high energy collision in the orbitrap promise the re-emergence of capability to carry out charge-remote decomposition experiments useful for positional analysis of double bonds in phospholipids and glycerolipids (50). In the midst of these technological advances are the advances in lipid biochemistry that can result.
Acknowledgments
R. C. M. thanks the Queen Mary University of London and the William Harvey Institute for access to resources used in the preparation of this minireview.
This work was supported, in whole or in part, by National Institutes of Health Grant HL25785 and LIPID MAPS Consortium Grant GM06338. This is the second article in the Thematic Minireview Series on Biological Applications of Mass Spectrometry. This minireview will be reprinted in the 2011 Minireview Compendium, which will be available in January, 2012.
- ESI
- electrospray ionization
- CID
- collision-induced dissociation
- SRM
- selected reaction monitoring
- TAG
- triacylglycerol
- PI
- phosphatidylinositol
- PS
- phosphatidylserine
- PG
- phosphatidylglycerol
- PA
- phosphatidic acid
- PE
- phosphatidylethanolamine
- PC
- glycerophosphatidylcholine
- LAT
- lysophospholipid acyltransferase
- IMS
- imaging MS
- SIMS
- secondary ion MS
- IM-MS
- ion mobility-MS.
REFERENCES
- 1. Ryhage R., Stenhagen E. (1960) J. Lipid Res. 1, 361–390 [PubMed] [Google Scholar]
- 2. Granström E., Samuelsson B. (1969) J. Am. Chem. Soc. 91, 3398–3400 [DOI] [PubMed] [Google Scholar]
- 3. VanLear G. E., McLafferty F. W. (1969) Annu. Rev. Biochem. 38, 289–322 [DOI] [PubMed] [Google Scholar]
- 4. Barber M., Bordoli R. S., Sedgwick R. D., Tyler A. M. (1981) Nature 293, 270–275 [Google Scholar]
- 5. Murphy R. C., Mathews W. R., Rokach J., Fenselau C. (1982) Prostaglandins 23, 201–206 [DOI] [PubMed] [Google Scholar]
- 6. Fenn J. B., Mann M., Meng C. K., Wong S. F., Whitehouse C. M. (1989) Science 246, 64–71 [DOI] [PubMed] [Google Scholar]
- 7. Karas M., Hillenkamp F. (1988) Anal. Chem. 60, 2299–2301 [DOI] [PubMed] [Google Scholar]
- 8. Vestal M. L., Campbell J. M. (2005) Methods Enzymol. 402, 79–108 [DOI] [PubMed] [Google Scholar]
- 9. Perry R. H., Cooks R. G., Noll R. J. (2008) Mass Spectrom. Rev. 27, 661–699 [DOI] [PubMed] [Google Scholar]
- 10. Fahy E., Subramaniam S., Brown H. A., Glass C. K., Merrill A. H., Jr., Murphy R. C., Raetz C. R., Russell D. W., Seyama Y., Shaw W., Shimizu T., Spener F., van Meer G., VanNieuwenhze M. S., White S. H., Witztum J. L., Dennis E. A. (2005) J. Lipid Res. 46, 839–861 [DOI] [PubMed] [Google Scholar]
- 11. Cantagrel V., Lefeber D. J., Ng B. G., Guan Z., Silhavy J. L., Bielas S. L., Lehle L., Hombauer H., Adamowicz M., Swiezewska E., De Brouwer A. P., Blümel P., Sykut-Cegielska J., Houliston S., Swistun D., Ali B. R., Dobyns W. B., Babovic-Vuksanovic D., van Bokhoven H., Wevers R. A., Raetz C. R., Freeze H. H., Morava E., Al-Gazali L., Gleeson J. G. (2010) Cell 142, 203–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Garrett T. A., Guan Z., Raetz C. R. H. (2007) Methods Enzymol. 432, 117–143 [DOI] [PubMed] [Google Scholar]
- 13. Wenk M. R. (2010) Cell 143, 888–895 [DOI] [PubMed] [Google Scholar]
- 14. Dennis E. A., Deems R. A., Harkewicz R., Quehenberger O., Brown H. A., Milne S. B., Myers D. S., Glass C. K., Hardiman G., Reichart D., Merrill A. H., Jr., Sullards M. C., Wang E., Murphy R. C., Raetz C. R., Garrett T. A., Guan Z., Ryan A. C., Russell D. W., McDonald J. G., Thompson B. M., Shaw W. A., Sud M., Zhao Y., Gupta S., Maurya M. R., Fahy E., Subramaniam S. (2010) J. Biol. Chem. 285, 39976–39985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Murphy R. C., Axelsen P. H. (2011) Mass Spectrom. Rev. 30, 579–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Murphy R. C., Fiedler J., Hevko J. (2001) Chem. Rev. 101, 479–526 [DOI] [PubMed] [Google Scholar]
- 17. Griffiths W. J. (2003) Mass Spectrom. Rev. 22, 81–152 [DOI] [PubMed] [Google Scholar]
- 18. Hsu F. F., Turk J., Stewart M. E., Downing D. T. (2002) J. Am. Soc. Mass Spectrom. 13, 680–695 [DOI] [PubMed] [Google Scholar]
- 19. Han X., Gross R. W. (2005) Mass Spectrom. Rev. 24, 367–412 [DOI] [PubMed] [Google Scholar]
- 20. Nakanishi H., Ogiso H., Taguchi R. (2009) Methods Mol. Biol. 579, 287–313 [DOI] [PubMed] [Google Scholar]
- 21. Shaner R. L., Allegood J. C., Park H., Wang E., Kelly S., Haynes C. A., Sullards M. C., Merrill A. H., Jr. (2009) J. Lipid Res. 50, 1692–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hutchins P. M., Barkley R. M., Murphy R. C. (2008) J. Lipid Res. 49, 804–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ejsing C. S., Sampaio J. L., Surendranath V., Duchoslav E., Ekroos K., Klemm R. W., Simons K., Shevchenko A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2136–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schwudke D., Hannich J. T., Surendranath V., Grimard V., Moehring T., Burton L., Kurzchalia T., Shevchenko A. (2007) Anal. Chem. 79, 4083–4093 [DOI] [PubMed] [Google Scholar]
- 25. Jain M., Petzold C. J., Schelle M. W., Leavell M. D., Mougous J. D., Bertozzi C. R., Leary J. A., Cox J. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5133–5138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Quehenberger O., Armando A. M., Brown A. H., Milne S. B., Myers D. S., Merrill A. H., Bandyopadhyay S., Jones K. N., Kelly S., Shaner R. L., Sullards C. M., Wang E., Murphy R. C., Barkley R. M., Leiker T. J., Raetz C. R., Guan Z., Laird G. M., Six D. A., Russell D. W., McDonald J. G., Subramaniam S., Fahy E., Dennis E. A. (2010) J. Lipid Res. 51, 3299–3305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Esch S. W., Tamura P., Sparks A. A., Roth M. R., Devaiah S. P., Heinz E., Wang X., Williams T. D., Welti R. (2007) J. Lipid Res. 48, 235–241 [DOI] [PubMed] [Google Scholar]
- 28. Koeberle A., Shindou H., Harayama T., Shimizu T. (2010) FASEB J. 24, 4929–4938 [DOI] [PubMed] [Google Scholar]
- 29. Zemski Berry K. A., Murphy R. C. (2006) Anal. Biochem. 349, 118–128 [DOI] [PubMed] [Google Scholar]
- 30. Zemski Berry K. A., Turner W. W., VanNieuwenhze M. S., Murphy R. C. (2009) Anal. Chem. 81, 6633–6640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thomas M. C., Mitchell T. W., Harman D. G., Deeley J. M., Murphy R. C., Blanksby S. J. (2007) Anal. Chem. 79, 5013–5022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Poad B. L., Pham H. T., Thomas M. C., Nealon J. R., Campbell J. L., Mitchell T. W., Blanksby S. J. (2010) J. Am. Soc. Mass Spectrom. 21, 1989–1999 [DOI] [PubMed] [Google Scholar]
- 33. Blair I. A. (1990) Methods Enzymol. 187, 13–23 [DOI] [PubMed] [Google Scholar]
- 34. Masoodi M., Eiden M., Koulman A., Spaner D., Volmer D. A. (2010) Anal. Chem. 82, 8176–8185 [DOI] [PubMed] [Google Scholar]
- 35. Gijón M. A., Riekhof W. R., Zarini S., Murphy R. C., Voelker D. R. (2008) J. Biol. Chem. 283, 30235–30245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Steinhauer J., Gijón M. A., Riekhof W. R., Voelker D. R., Murphy R. C., Treisman J. E. (2009) Mol. Biol. Cell 20, 5224–5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fletcher J. S., Lockyer N. P., Vickerman J. C. (2011) Mass Spectrom. Rev. 30, 142–174 [DOI] [PubMed] [Google Scholar]
- 38. Lechene C., Hillion F., McMahon G., Benson D., Kleinfeld A. M., Kampf J. P., Distel D., Luyten Y., Bonventre J., Hentschel D., Park K. M., Ito S., Schwartz M., Benichou G., Slodzian G. (2006) J. Biol. 5, 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kurczy M. E., Piehowsky P. D., Willingham D., Molyneaux K. A., Heien M. L., Winograd N., Ewing A. G. (2010) J. Am. Soc. Mass Spectrom. 21, 833–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Koizumi S., Yamamoto S., Hayasaka T., Konishi Y., Yamaguchi-Okada M., Goto-Inoue N., Sugiura Y., Setou M., Namba H. (2010) Neuroscience 168, 219–225 [DOI] [PubMed] [Google Scholar]
- 41. Murphy R. C., Hankin J. A., Barkley R. M. (2009) J. Lipid Res. 50, S317–S322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Burnum K. E., Cornett D. S., Puolitaival S. M., Milne S. B., Myers D. S., Tranguch S., Brown H. A., Dey S. K., Caprioli R. M. (2009) J. Lipid Res. 50, 2290–2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reyzer M. L., Chaurand P., Angel P. M., Caprioli R. M. (2010) Methods Mol. Biol. 656, 285–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hankin J. A., Murphy R. C. (2010) Anal. Chem. 82, 8476–8484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Girod M., Shi Y., Cheng J. X., Cooks R. G. (2010) J. Am. Soc. Mass Spectrom. 21, 1177–1189 [DOI] [PubMed] [Google Scholar]
- 46. Manicke N. E., Nefliu M., Wu C., Woods J. W., Reiser V., Hendrickson R. C., Cooks R. G. (2009) Anal. Chem. 81, 8702–8707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jackson S. N., Ugarov M., Post J. D., Egan T., Langlais D., Schultz J. A., Woods A. S. (2008) J. Am. Soc. Mass Spectrom. 19, 1655–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kliman M., Vijayakrishnan N., Wang L., Tapp J. T., Broadie K., McLean J. A. (2010) Mol. BioSyst. 6, 958–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Trimpin S., Tan B., Bohrer B. C., O'Dell D. K., Merenbloom S. I., Pazos M. X., Clemmer D. E., Walker J. M. (2009) Int. J. Mass Spectrom. 287, 58–69 [Google Scholar]
- 50. Cheng C., Gross M. L. (2000) Mass Spectrom. Rev. 19, 398–420 [DOI] [PubMed] [Google Scholar]
- 51. Hankin J. A., Farias S., Barkley R. M., Heidenreich K., Frey L. C., Hamazaki K., Kim H. Y., Murphy R. C. (2011) J. Am. Soc. Mass Spectrom. 22, 1014–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]