Abstract
von Willebrand factor (VWF) is a multimeric plasma protein that mediates platelet adhesion to sites of vascular injury. The hemostatic function of VWF depends upon the formation of disulfide-linked multimers, which requires the VWF propeptide (D1D2 domains) and adjacent D′D3 domains. VWF multimer assembly occurs in the trans-Golgi at pH ∼6.2 but not at pH 7.4, which suggests that protonation of one or more His residues (pKa ∼6.0) mediates the pH dependence of multimerization. Alignment of 30 vertebrate VWF sequences identified 13 highly conserved His residues in the D1D2D′D3 domains, and His-to-Ala mutagenesis identified His395 and His460 in the D2 domain as critical for VWF multimerization. Replacement of His395 with Lys or Arg prevented multimer assembly, suggesting that reversible protonation of this His residue is essential. In contrast, replacement of His460 with Lys or Arg preserved normal multimer assembly, whereas Leu, Met, and Gln did not, indicating that the function of His460 depends primarily upon the presence of a positive charge. These results suggest that pH sensing by evolutionarily conserved His residues facilitates the assembly and packaging of VWF multimers upon arrival in the trans-Golgi.
Keywords: Endothelium, Evolution, Gene Structure, Hemostasis, Histidine, Intracellular Processing, Protein Assembly, von Willebrand Factor
Introduction
von Willebrand factor (VWF)2 is a multimeric hemostatic protein that mediates platelet adhesion to sites of vascular injury. VWF is assembled from identical 350-kDa precursors consisting of multiple structural domains in the order D1-D2-D′-D3-A1-A2-A3-D4-B1-B2-B3-C1-C2-CK (where CK indicates the cystine knot domain). The hemostatic function of VWF depends upon its assembly into multimers, and defects in VWF multimer formation are common causes of von Willebrand disease, the most prevalent inherited bleeding disorder (1).
The assembly of VWF multimers begins in the endoplasmic reticulum (ER) of endothelial cells, where pro-VWF monomers form homodimers through “tail-to-tail” intersubunit disulfide bonds between C-terminal cystine knot domains. After transport to the Golgi, pro-VWF dimers assemble into large multimers through “head-to-head” N-terminal interchain disulfide bonds between D3 domains. Subsequent cleavage of VWF propeptide (D1D2) by furin yields mature, multimeric VWF that is packed into tubular arrays and stored in rod-shaped granules called Weibel-Palade bodies (2).
The completion of VWF multimer assembly in the Golgi apparatus poses several challenges to the cell. Disulfide bonds typically form in the ER, where disulfide oxidoreductases and chaperones are devoted to this function. However, the Golgi is relatively hostile to disulfide rearrangement because the lower pH inhibits deprotonation of the sulfhydryl moiety of Cys residues, which is necessary for disulfide rearrangement. In addition, suitable oxidoreductases and chaperones are thought to be absent from the Golgi. To facilitate disulfide bond formation under these circumstances, the N-terminal D1D2D′D3 domains of the VWF precursor act as an endogenous pH-dependent oxidoreductase.
Deletion of the VWF propeptide prevents multimerization, but expression of the propeptide and mature subunit on separate plasmids (in trans) restores efficient multimer assembly (3). In the ER, the VWF propeptide forms a transient intrachain disulfide-linked intermediate with the D3 domain that rearranges in the Golgi to yield interchain disulfide bonds linking VWF subunits into multimers (4). In addition, the D1 and D2 domains both contain vicinal cysteines in a CGLC motif similar to the active site of disulfide isomerases, and insertion of an additional Gly into either motif inhibits VWF multimerization (5). These findings suggest that the VWF propeptide and protein disulfide isomerases employ similar mechanisms to facilitate disulfide bond formation.
VWF multimer assembly and packaging into Weibel-Palade bodies are both regulated by pH differences between the ER and relatively acidic Golgi. Neutralizing the pH of the Golgi by treating cells with ammonium chloride or chloroquine blocks VWF multimer assembly (6). In a cell-free system, VWF multimer assembly does not occur at pH 7.4, which is typical of the ER, but proceeds to some extent at pH 6.2 and is optimal at pH 5.8, conditions typical of the trans-Golgi and Weibel-Palade body, respectively (7, 8). In addition, neutralizing the intracellular pH disrupts the tubular packing of VWF multimers and causes rod-shaped Weibel-Palade bodies to become spherical, which prevents the orderly secretion of VWF filaments in response to secretagogues (9). In vitro, the VWF propeptide forms noncovalent homodimers that bind tightly to multimeric VWF at pH 6.4 but dissociate at pH 7.4 (10). Purified VWF propeptide and dimers of N-terminal D′D3 fragments self-assemble into tubules at pH 6.2 and disassemble at pH 7.4 (10). Thus, low pH is necessary to form intersubunit disulfide bonds during multimer assembly, after which low pH is also necessary for multimers to condense into tubules. These findings suggest that the two processes, multimer assembly and tubular packing, may be regulated by distinct sets of pH-sensing residues.
Several proteins employ His residues as pH sensors to regulate their function upon transport to the Golgi. For example, His69 in the autoinhibitory propeptide of furin becomes protonated in the trans-Golgi, after which the propeptide dissociates and furin can cleave other substrates (11). The recycling mannose lectin ERGIC-53 binds cargo in the ER and releases it in the slightly more acidic ER-Golgi intermediate compartment (ERGIC), and cargo release requires protonation of conserved His178 (12). A recent His-to-Ala mutagenesis study of the ligand-receptor binding interface showed that the evolutionarily conserved residues His180 in human prolactin and His188 in the prolactin receptor are critical for pH-dependent ligand binding at the cell surface and release in acidic endosomes (13). We have used phylogenetic comparisons to identify candidate pH-sensing His residues in VWF and homologous gel-forming mucins, which were functionally evaluated by mutagenesis. The results indicate that at least two His residues in the D2 domain of the VWF propeptide are critical for pH-dependent multimer assembly in the Golgi.
EXPERIMENTAL PROCEDURES
Sequences
Vertebrate VWF sequences were identified by using human VWF cDNA (14–17), genomic DNA (18, 19), and encoded protein sequences for iterative BLASTN and TBLASTN (National Center for Biotechnology Information (NCBI) version 2.2.24) searches of GenBankTM sequences including genome assemblies, high throughput genome sequences, genome survey sequences, whole genome shotgun sequences, expressed sequence tags, nonredundant nucleotide sequences, trace archives, and short read archives. Exons were identified based on sequence similarity using the programs SeqMan, MegAlign, and EditSeq (version 8.1.5, DNASTAR, Inc., Madison, WI) and assembled into cDNA sequences. Splice site prediction with the program SplicePort (20) was used to support the identification of exon 2, which encodes a cleaved signal peptide. Gaps were assigned lengths based on the conserved length of the corresponding exons in VWF genes of other species. Sequences encoding the D1D2D′D3 segment of the mucins MUC2, MUC5AC, MUC5B, MUC6, MUC19, and FIMB1 were identified by methods similar to those used for VWF. The sequences, supporting evidence, and methods for phylogenetic analyses are described in detail in the supplemental material. All sequences have been scanned against these databases, and accession numbers for sequences with significant relatedness to the deposited sequences are included.
Constructs
Mutations were constructed utilizing a QuikChange XL kit (Stratagene, La Jolla, CA) with oligonucleotides from Integrated DNA Technologies (Coralville, IA). Mutations were prepared in pSVHVWF1.1, which encodes full-length human VWF (21), in plasmid pSVH-propeptide-FLAGNT, which encodes the VWF propeptide (amino acid residues 1–763) with an N-terminal FLAG tag between the signal peptide and domain D1 (4), or in plasmid pSVH-D3-FLAGNT/cMycCT, which encodes the VWF D1D2D′D3 domains (residues 1–1241) with an N-terminal FLAG tag and a C-terminal c-Myc tag (4). Plasmid pSVH-KSDR-D3-FLAGNT/cMycCT is similar to pSVH-D3-FLAGNT/cMycCT except that amino acid residues 760RSKR763 were mutated to 760KSDR763, which prevents cleavage by furin (22). Plasmid pSVH-D′D3Δpro-c-Myc encodes the VWF signal peptide (residues 1–22) followed by domains D′D3 (residues 764–1220) and a c-Myc tag (4). Plasmid pSVH-Δpro encodes the VWF signal peptide (residues 1–22) followed by the mature VWF subunit (residues 764–2813).
Cell Culture
BHK-fur4 cells that express human furin (23) were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum and 2 mm glutamine. BHK-fur4-D1D2D′D3-FLAGNT-c-MycCT cells were described previously (4). BHK-fur4-D1D2-KSDR-D′D3-FLAGNT-c-MycCT cells, stably transfected with pSVH-KSDR-D3-FLAGNT/cMycCT, were prepared similarly.
BHK cells were transiently transfected with pSVH-propeptide-FLAGNT and pSVH-D′D3Δpro-c-Myc constructs, pSVH-VWF constructs, or pSVH-D3-FLAGNT/cMycCT constructs, using Lipofectamine Plus reagent (Invitrogen) in Opti-MEM (Invitrogen) according to the manufacturer's instructions. Conditioned medium was collected after 48 h and treated with 40 mm N-ethylmaleimide and 144 μm phenylmethylsulfonyl fluoride.
Protein Analysis
The concentration of VWF was determined by ELISA (24). Falcon Pro-bind ELISA plates (BD Biosciences) were coated with a 1:1000 dilution of rabbit polyclonal anti-VWF antibody (082; DAKO, Carpinteria, CA). Bound VWF was detected with a 1:5000 dilution of HRP-conjugated rabbit polyclonal anti-VWF antibody (P226; DAKO) and tetramethyl-benzidine (Pierce). Plates were read by a Spectra MaxPLUS microplate reader with the SoftMaxPro 4.7.1 software (Molecular Devices, Silicon Valley, CA).
Western Blotting
D′D3 samples were heated to 100 °C for 5 min in Laemmli sample buffer and subjected to SDS-PAGE on 4–15% gradient gels (Bio-Rad). VWF multimer gel electrophoresis was performed as described previously (25). Multimer gels were incubated in 1.34 mm mercaptoethanol in PBS for 15 min to increase the efficiency of protein transfer by electroblotting onto polyvinylidene (PVDF) membranes (Whatman). Blots were blocked for 1 h in 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.1% Tween 20, and 0.5% casein and then incubated overnight at 4 °C in a 1:1000 dilution of HRP-conjugated rabbit polyclonal anti-VWF (P226; DAKO) and developed using the ECL Plus kit (GE Healthcare).
RESULTS AND DISCUSSION
VWF Structure
Coding sequences for VWF were assembled for 30 vertebrates including 20 placental mammals, a marsupial, two birds, a reptile, an amphibian, and five fish (supplemental material and supplemental Fig. S1). Exon 2 encodes a signal peptide and was identified or could be proposed with a high degree of confidence for all amniotes except dolphin because of a gap in the dolphin genome assembly. The proposed second exons for frog and fish VWF are relatively uncertain because they lack support from cDNA sequences or from similarity to signal peptides for VWF genes of other species. Exon 1 is noncoding and could not be identified for 10 species. The remaining coding sequences are complete except for portions of six exons (0.4% of 1530 total exons) encoding 199 amino acid residues (0.08% of total amino acids).
The VWF gene structure varies relatively little among the species studied. All 52 exons present in human VWF are conserved, and exon length is invariant for 23 exons. Among 1504 total exons (excluding exons 1–2), only 113 (0.7%) deviate from the length of the corresponding human exon, and in 63 instances, the difference is one codon. All splice junctions are standard except for eight (0.53%) that are predicted to employ a GC splice donor (supplemental Table S1), which is similar to the 0.56% prevalence of noncanonical GC-AG splice sites reported for a dataset of more than 22,000 mammalian introns (26). The use of these noncanonical splice sites in anole VWF intron 26 and zebrafish VWF intron 44 was verified by comparison of genomic DNA and cDNA sequences (supplemental Table S1).
Exon 28 exhibits more extreme variation in structure. In tetrapods and zebrafish, exon 28 varies in length from 1346 to 1436 nucleotides and encodes VWF domains A1 and A2. However, exon 28 is split into exons 28a (304–310 nucleotides) and 28b (1066–1162 nucleotides) in the three-spined stickleback, Japanese medaka, spotted green pufferfish, and fugu. These four fish are closely related when compared with zebrafish (supplemental Fig. S1), which suggests that a single exon 28 is ancestral and the split exon 28 is a derived character.
The amino acid sequence of VWF is also highly conserved. Human VWF (amino acid residues 23–2813) is at least 78% identical to VWF of other placental mammals, 73% identical to opossum VWF, 55–56% identical to bird, reptile, and amphibian VWF, and 45–46% identical to fish VWF (supplemental Fig. S2). Human VWF has 233 Cys residues (not including one Cys in the signal peptide), and all are conserved in other vertebrates with two kinds of exceptions that involve four Cys residues.
VWF of the Western clawed frog lacks two Cys residues corresponding to Cys418 and Cys521 in the D2 domain of human VWF. The simultaneous absence of these two Cys residues suggests that they form a disulfide bond in frog VWF, which is supported by the identification of a Cys898–Cys993 disulfide bond between the corresponding residues in the homologous D3 domain of human VWF (19, 27).
VWF of all five fish studied is missing two Cys residues that correspond to Cys1669 and Cys1670 of human VWF and are conserved in other species. These residues are located at the C-terminal end of the VWF A2 domain, where they form an unusual vicinal disulfide bond (27). The A2 domain unfolds in response to hydrodynamic shear stress to expose a cleavage site for ADAMTS13, a regulatory metalloprotease that is specific for VWF. The rigid Cys1669–Cys1670 disulfide bond is tightly buried in a hydrophobic pocket and resists the force-dependent unfolding of the A2 domain (28). The absence of the Cys1669–Cys1670 disulfide bond would be expected to decrease the stability of the A2 domain and decrease the force required to initiate unfolding. Therefore, the lack of this disulfide bond in fish VWF may reflect adaptation to distinct hemostatic requirements. For example, blood circulates in fish at relatively low velocities that are likely to generate low shear forces, which may require a reduced threshold for shear-induced unfolding of VWF to allow proteolytic cleavage by ADAMTS13. The relationship between structure and force sensing could be evaluated directly by comparing the unfolding of fish and mammalian A2 domains with laser tweezers (29).
Phylogenetic Analysis of Candidate pH Sensors
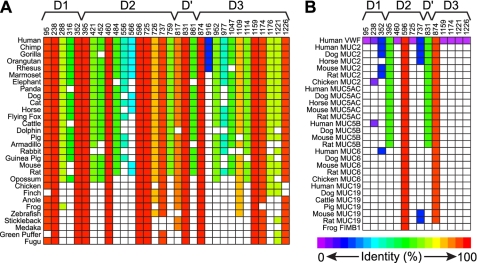
Histidine has a pKa value suitable for detecting the difference in pH between ER (pH 7.4) and the trans-Golgi (pH 6.2) and is likely to perform this function during the assembly and storage of VWF multimers in Weibel-Palade bodies. Intracellular multimerization and storage of VWF are conserved among vertebrates, which suggests that pH-sensing His residues could be identified by phylogenetic comparison of VWF sequences. The D1D2D′D3 segment of VWF is sufficient for pH-dependent oligomerization and tubular storage, and relevant His residues should reside in these domains. Of the 32 His residues in the D1D2D′D3 domains of human VWF, 9 are conserved in all 30 species studied, and 4 others are conserved in at least 28 of 30 species (supplemental Fig. S3 and Fig. 1A).
FIGURE 1.
Conservation of His residues in VWF and gel-forming mucins. Human VWF was aligned with VWF (A) or representative gel-forming mucins MUC2, MUC5AC, MUC5B, MUC6, MUC19, and FIMB1 (B) from the vertebrate species at the left of each graph. The positions of His residues in human VWF and their locations with domains D1, D2, D′, and D3 are indicated at the top. A square in the grid is colored when the aligned amino acid is His, as in human VWF, and white when it is not His. The color key indicates the percentage of sequence identity at that position from purple (0%) to red (100%).
VWF and gel-forming mucins belong to a family of evolutionarily related vertebrate proteins that form large polymers (30). Like VWF, the homologous gel-forming mucins MUC2, MUC5AC, MUC5B, MUC6, MUC19, and FIMB1 have N-terminal D1D2D′D3 domains (31). MUC2 (32), MUC5AC (33), and MUC19 (34) have been shown to form interchain disulfide bonds between N-terminal D′D3 domains. As in the case of VWF, these mucins form dimers in the ER that assemble into disulfide-linked multimers in the Golgi (31), suggesting a conserved role for mucin D1D2D′D3 domains in the pH-dependent assembly of multimers.
Alignment of human VWF with representative gel-forming mucins (supplemental Fig. S4) shows that a subset of His residues conserved in VWF is also conserved in mucins (Fig. 1B). His596 is present in all mucins, and His874 is present in all mucins except FIMB1. His395 and His831 occur in all MUC2, MUC5AC, and MUC5B sequences, but not in MUC6, MUC19, or FIMB1.
Conservation across VWF and gel-forming mucins suggests that some of these His residues might participate in pH-sensitive biosynthetic steps that are shared by all of these proteins. However, VWF and mucins have structural differences that may reflect significant differences in the mechanism of biosynthesis and intracellular storage. For example, the D1D2 domains of VWF comprise a propeptide that is separated from the D′D3 domains by a conserved furin cleavage site, but this cleavage site is not present in gel-forming mucins (supplemental Fig. S4). Consequently, the D1D2 domains are likely to remain covalently attached to multimeric mucins, as demonstrated for recombinant MUC19 (35). The pH-dependent binding of the VWF propeptide to the mature VWF subunit is important for storage in Weibel-Palade bodies, but any His residues that mediate this noncovalent binding in VWF may be unnecessary for mucins if the D1D2 domains and D′D3 domains remain covalently linked.
Dependence of D′D3 Dimerization on Intracellular pH
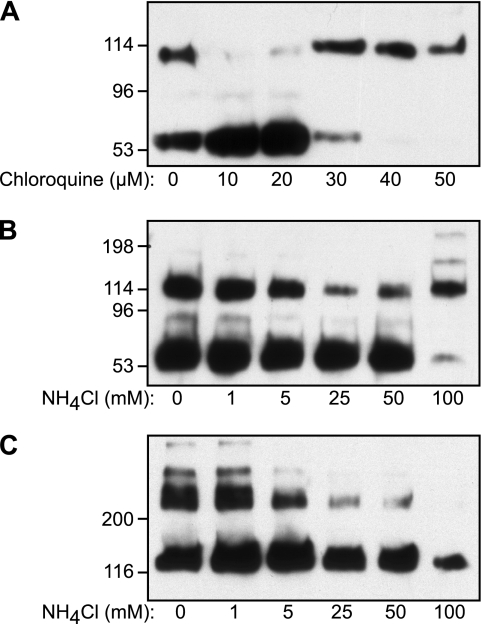
The formation of intersubunit disulfide bonds between D3 domains of VWF subunits occurs in the Golgi and is blocked by raising the intracellular pH with ammonium chloride, chloroquine, or monensin (6). This pH-dependent assembly process can be studied conveniently with constructs of VWF that are truncated after the D3 domain (4, 36). For example, the expression of VWF D1D2D′D3 domains in BHK cells results in the secretion of D′D3 55-kDa monomers and 110-kDa dimers, as well as cleaved 85-kDa VWF propeptide (domains D1D2) (4), and treatment of cells expressing this construct with chloroquine prevented D′D3 dimerization (Fig. 2A). Chloroquine concentrations of 30 μm or greater also inhibited cleavage of VWF propeptide, probably because furin is inactive at elevated pH (11), which resulted in secretion of monomeric 140-kDa D1D2D′D3 that has the same electrophoretic mobility as D′D3 dimers.
FIGURE 2.
Effects of chloroquine and ammonium chloride on D′D3 dimerization. BHK-fur4-D1D2D′D3-FLAGNT-c-MycCT cells (A and B) or BHK-fur4-D1D2-KSDR-D′D3-FLAGNT-c-MycCT cells (C) were treated with the indicated concentration of chloroquine (A) for 48 h or NH4Cl for 72 h (B and C). Conditioned medium was analyzed by SDS-PAGE (4–15%) and Western blotting with HRP-conjugated rabbit anti-human VWF antibody, which preferentially detects products containing D′D3 domains. The VWF propeptide, domains D1D2, can be seen as a weakly reactive 95-kDa band in some lanes. The mass in kDa of marker proteins is indicated at the left.
Similar effects were observed upon treatment of cells with increasing concentrations of ammonium chloride, which progressively inhibited the dimerization of D′D3. At 100 mm ammonium chloride, propeptide cleavage was inhibited, and the predominant secreted product appeared to be monomeric D1D2D′D3 (Fig. 2B). This interpretation was confirmed by expressing a variant of D1D2D′D3 in which the furin recognition site was mutated to KSDR to prevent cleavage. For this construct, dimerization was completely inhibited at 100 mm ammonium chloride, and the secreted product consisted entirely of monomeric 140-kDa D1D2D′D3 (Fig. 2C). These results show that the formation of intersubunit disulfide bonds by VWF D1D2D′D3 exhibits the expected dependence on Golgi pH. This system was employed to evaluate the role of conserved His residues on pH-dependent disulfide bond formation.
Effects of His Mutagenesis on D′D3 Dimerization
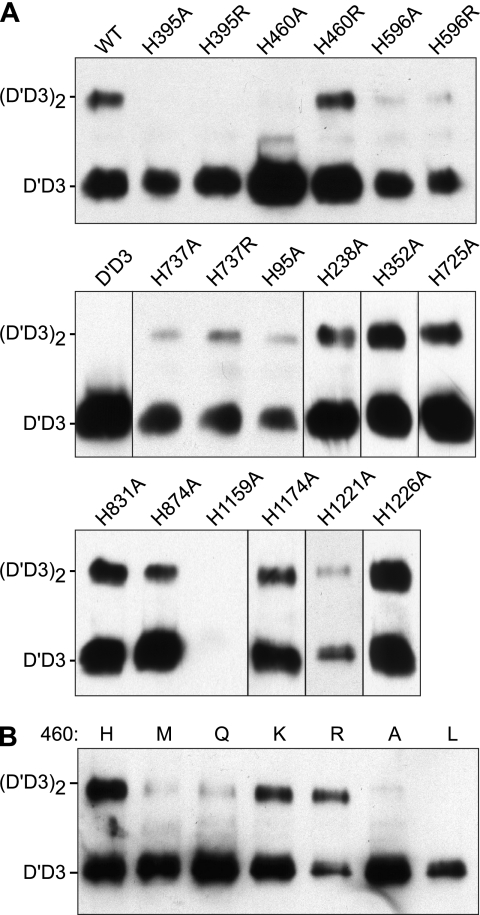
The decrease in pH between ER and Golgi would facilitate the reversible protonation of a pH-sensing His residue. To prevent the acquisition of positive charge at acidic pH, His residues were replaced by Ala, and the effect of charge removal on D′D3 dimerization was assessed in BHK-fur4 cells transiently transfected with the corresponding D1D2D′D3 constructs (Fig. 3A). Similar results were obtained by expressing D1D2 and D′D3 domains on separate plasmids (supplemental Fig. S5).
FIGURE 3.
Expression of truncated VWF D1D2D′D3 constructs with His mutations. BHK-fur4 cells were transiently transfected with wild type (WT) or mutant D1D2D′D3 constructs having the indicated amino acid substitutions (A) or with D1D2D′D3 constructs that encode the indicated amino acids at position 460 (B). Conditioned medium was analyzed by SDS-PAGE and Western blotting with HRP-conjugated anti-VWF. Positions corresponding to monomeric D′D3 and dimeric (D′D3)2 are indicated at the left.
All constructs were expressed efficiently except H1159A, for which secretion was severely impaired. Dimerization was prevented by the substitutions H395A and H460A, whereas decreased but detectable dimerization was observed for H95A, H596A, and H737A. The remaining constructs assembled dimers with approximately normal efficiency, indicating that the corresponding His residues are not essential for intersubunit disulfide bond formation.
Replacement by Ala at five of eight conserved His residues in the VWF propeptide impaired D′D3 dimerization; one is in domain D1 (His95), and four are in domain D2 (His395, His460, His596, His737). These results suggest that the VWF propeptide contains pH-sensing His residues, which is consistent with the observation that recombinant D1D2 exhibits increased noncovalent self-association at pH 6.2 when compared with pH 7.4 (10). Conversely, none of five evaluable His residues in the D′D3 domains was required to form D′D3 dimers.
D′D3 monomers do not self-associate at low pH (10), which might suggest that D′D3 domains lack pH-sensing His residues. However, low pH does promote the binding of D′D3 monomers to D1D2. Low pH also induces D′D3 dimers to bind D1D2 domains and form large helical tubules similar to the VWF tubules within Weibel-Palade bodies of endothelial cells (10). Thus, it remains possible that pH-sensing residues on D′D3 domains mediate interactions with D1D2 that contribute to intersubunit disulfide bond formation, tubular packing in Weibel-Palade bodies, or both processes.
For selected His-to-Ala substitutions that impaired dimerization, the His residue was replaced by Arg to prevent the loss of positive charge at neutral pH, and the effect of this charge stabilization was assessed (Fig. 3A). D′D3 dimerization was not preserved in constructs H395R, H596R, or H737R. However, H460R formed dimers approximately as efficiently as the wild type construct.
Mutagenesis of His460 was extended to include replacement with nonpolar amino acids of varying sizes (Ala, Leu, Met), positively charged amino acids (Arg, Lys), and the polar amino acid Gln. Only Lys and Arg restored D′D3 dimerization, suggesting that formation of disulfide bonds between D3 domains depends on a positive charge at this position (Fig. 3B).
Conserved His and VWF Multimer Assembly
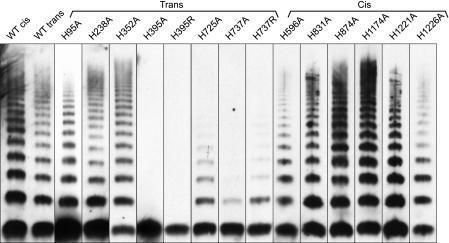
Conserved His residues within the D1D2D′D3 domains were also changed to Ala in the context of full-length VWF. When expressed in BHK cells, VWF assembles into disulfide-linked multimers with more than 20 bands visible upon SDS-agarose gel electrophoresis. Bands correspond to multimers with even numbers of subunits ranging from dimers to larger than 40-mers. Whether VWF is encoded by a single plasmid (cis) or by separate plasmids for the VWF propeptide and mature subunit (trans), multimers assemble with similar efficiency (Fig. 4). We took advantage of this property by constructing most D1D2 mutations in the smaller propeptide construct, which was coexpressed with the mature subunit. Mutations located in the D′D3 domains were constructed and expressed in a single full-length VWF plasmid.
FIGURE 4.
Multimer assembly by full-length VWF constructs with His mutations. BHK-fur4 cells were transiently transfected to express wild type VWF from a single plasmid (WT cis) or to express the VWF propeptide and mature subunit on separate plasmids (WT trans) as described under “Experimental Procedures.” Trans indicates mutations at His residues in D1D2 domains that were constructed in the VWF propeptide and cotransfected with the mature VWF subunit. Cis indicates mutations that were constructed in full-length VWF. Conditioned medium was analyzed by SDS-agarose gel electrophoresis and Western blotting with HRP-conjugated rabbit anti-human VWF antibody.
In most cases, mutations at His residues behaved similarly whether expressed in truncated D1D2D′D3 (Fig. 3) or in full-length VWF (Fig. 4). Multimers composed of VWF H1159A were not secreted (data not shown), which is comparable with the results obtained for the same mutation in the truncated D1D2D′D3 construct. As expected, His residues that were not required for dimerization of D1D2D′D3 (Fig. 3) were not essential in full-length VWF (Fig. 4). In particular, none of five evaluable His-to-Ala substitutions in the D′D3 domains had a pronounced effect upon multimer assembly: H831A, H874A, H1174A, H1221A, and H1226A.
Three His-to-Ala mutations produced somewhat different results when expressed in truncated or full-length VWF. H95A and H596A allowed the secretion of trace amounts of D′D3 dimer (Fig. 3A) but were compatible with the secretion of a range of VWF multimers, although the size distribution was skewed toward smaller species (Fig. 4). Conversely, the mutation H725A did not reduce the production of D′D3 dimer (Fig. 3A) but was associated with the secretion of relatively small VWF multimers when compared with wild type VWF (Fig. 4).
This variation suggests that sequences C-terminal to the critical D1D2D′D3 region can influence the efficiency of VWF multimer assembly. Alternatively, the intermediate and variable phenotype caused by the substitution H725A may reflect the presence of the adjacent His726, which is found in VWF from 24 of 30 species sequenced (supplemental Fig. S3). If both His725 and His726 contribute to pH-dependent changes in the D2 domain, then loss of one may cause a partial loss of function.
Ala substitutions at His395 and His737 markedly decreased D′D3 dimer formation (Fig. 3A) and VWF multimer assembly (Fig. 4). In addition, neither H395A nor H395R formed multimers when expressed in full-length VWF. H737A and H737R also had similar defects in D′D3 dimer formation (Fig. 3A) and multimer assembly (Fig. 4), although less severe than associated with mutations at His395. These results indicate that the fixed positive charge of a guanidinium group in Arg cannot substitute functionally for His395 or His737.
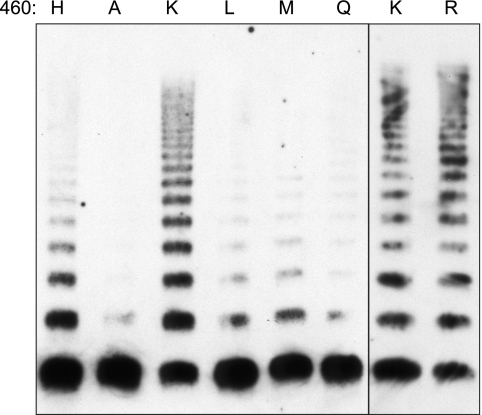
Amino acid substitutions at His460 had congruent effects on D′D3 dimerization (Fig. 3) and VWF multimer assembly (Fig. 5). As observed for the truncated D1D2D′D3 construct, mutation of His460 to Ala, Leu, Met, or Gln profoundly impaired multimerization, but replacement by Arg or Lys restored multimerization at least to the level of wild type VWF. H460K and H460R constructs formed multimers with equal efficiency (Fig. 5), indicating that intersubunit disulfide bond formation depends on the presence of a positive charge at this position.
FIGURE 5.
Mutational analysis of His460 and VWF multimer assembly. BHK-fur4 cells were transiently transfected to express full-length VWF with the indicated amino acid residues at position 460. Conditioned medium was analyzed by SDS-agarose gel electrophoresis and Western blotting with HRP-conjugated rabbit anti-human VWF antibody.
Results were consistent with the proposed function of conserved His residues as pH sensors. The conserved His395 and His460 located in the propeptide play an essential role in VWF multimerization.
Among the residues examined, replacement of His395 or His460 by Ala (that is H395A or H460A, respectively) caused the most profound defect in intersubunit disulfide bond formation, which suggests that these residues contribute to the pH-dependent regulation of VWF multimer assembly. Both residues are highly conserved in VWF (Fig. 1A), but His460 is not conserved in gel-forming mucins (Fig. 1B). The significance of this distinction is unknown at present but may be related to the absence of a furin site between the D1D2 and D′D3 domains of mucins (supplemental Fig. S4); His460 may be required for pH-dependent interactions between the D1D2 propeptide and mature VWF subunit after cleavage by furin but dispensable for mucins. In addition, VWF is stored intracellularly in densely packed helical tubules, whereas gel-forming mucins do not appear to undergo a similar process of highly ordered condensation in secretory granules. Therefore, the pH-dependent tubular packing of VWF may well depend on His residues that would not need to be conserved in mucins.
Multimer assembly and tubular packing are critical for the hemostatic function of VWF. Both events depend on low pH in the trans-Golgi but can be dissociated by appropriately targeted mutations. For example, the von Willebrand disease mutation Y87S impairs both processes, preventing multimer assembly and disrupting tubular packing; VWF Y87S dimers are stored in spherical, disorganized Weibel-Palade bodies (9, 37). However, insertion of an extra Gly into the CGLC motif of domain D1 blocks multimer assembly without affecting storage in rod-shaped Weibel-Palade bodies (5). Whether specific pH-sensing His residues participate in one or both of these processes will be established by additional analyses that discriminate between the noncovalent dimerization of the VWF propeptide (D1D2), binding of propeptide to D′D3 domains, disulfide-mediated multimerization, and packing of VWF into tubules (4, 10).
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant HL72917 (to J. E. S.) and by American Heart Association Midwest Affiliate Postdoctoral Fellowship Award 0825817G (to R. H. H.).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental methods, sequences for Gorilla gorilla and Tursiops truncatus, Table S1, and Figs. S1–S5.
- VWF
- von Willebrand factor
- ER
- endoplasmic reticulum
- BHK
- baby hamster kidney.
REFERENCES
- 1. Sadler J. E. (1998) Annu. Rev. Biochem. 67, 395–424 [DOI] [PubMed] [Google Scholar]
- 2. Wagner D. D. (1990) Annu. Rev. Cell Biol. 6, 217–246 [DOI] [PubMed] [Google Scholar]
- 3. Wise R. J., Pittman D. D., Handin R. I., Kaufman R. J., Orkin S. H. (1988) Cell 52, 229–236 [DOI] [PubMed] [Google Scholar]
- 4. Purvis A. R., Sadler J. E. (2004) J. Biol. Chem. 279, 49982–49988 [DOI] [PubMed] [Google Scholar]
- 5. Mayadas T. N., Wagner D. D. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 3531–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wagner D. D., Mayadas T., Marder V. J. (1986) J. Cell Biol. 102, 1320–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mayadas T. N., Wagner D. D. (1989) J. Biol. Chem. 264, 13497–13503 [PubMed] [Google Scholar]
- 8. Erent M., Meli A., Moisoi N., Babich V., Hannah M. J., Skehel P., Knipe L., Zupancic G., Ogden D., Carter T. (2007) J. Physiol. 583, 195–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Michaux G., Abbitt K. B., Collinson L. M., Haberichter S. L., Norman K. E., Cutler D. F. (2006) Dev. Cell 10, 223–232 [DOI] [PubMed] [Google Scholar]
- 10. Huang R. H., Wang Y., Roth R., Yu X., Purvis A. R., Heuser J. E., Egelman E. H., Sadler J. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 482–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Feliciangeli S. F., Thomas L., Scott G. K., Subbian E., Hung C. H., Molloy S. S., Jean F., Shinde U., Thomas G. (2006) J. Biol. Chem. 281, 16108–16116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Appenzeller-Herzog C., Roche A. C., Nufer O., Hauri H. P. (2004) J. Biol. Chem. 279, 12943–12950 [DOI] [PubMed] [Google Scholar]
- 13. Kulkarni M. V., Tettamanzi M. C., Murphy J. W., Keeler C., Myszka D. G., Chayen N. E., Lolis E. J., Hodsdon M. E. (2010) J. Biol. Chem. 285, 38524–38533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sadler J. E., Shelton-Inloes B. B., Sorace J. M., Harlan J. M., Titani K., Davie E. W. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 6394–6398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shelton-Inloes B. B., Titani K., Sadler J. E. (1986) Biochemistry 25, 3164–3171 [DOI] [PubMed] [Google Scholar]
- 16. Shelton-Inloes B. B., Broze G. J., Jr., Miletich J. P., Sadler J. E. (1987) Biochem. Biophys. Res. Commun. 144, 657–665 [DOI] [PubMed] [Google Scholar]
- 17. Bonthron D., Orr E. C., Mitsock L. M., Ginsburg D., Handin R. I., Orkin S. H. (1986) Nucleic Acids Res. 14, 7125–7127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mancuso D. J., Tuley E. A., Westfield L. A., Lester-Mancuso T. L., Le Beau M. M., Sorace J. M., Sadler J. E. (1991) Biochemistry 30, 253–269 [DOI] [PubMed] [Google Scholar]
- 19. Mancuso D. J., Tuley E. A., Westfield L. A., Worrall N. K., Shelton-Inloes B. B., Sorace J. M., Alevy Y. G., Sadler J. E. (1989) J. Biol. Chem. 264, 19514–19527 [PubMed] [Google Scholar]
- 20. Dogan R. I., Getoor L., Wilbur W. J., Mount S. M. (2007) Nucleic Acids Res. 35, W285–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matsushita T., Sadler J. E. (1995) J. Biol. Chem. 270, 13406–13414 [DOI] [PubMed] [Google Scholar]
- 22. Rehemtulla A., Kaufman R. J. (1992) Blood 79, 2349–2355 [PubMed] [Google Scholar]
- 23. Bodó I., Katsumi A., Tuley E. A., Eikenboom J. C., Dong Z., Sadler J. E. (2001) Blood 98, 2973–2979 [DOI] [PubMed] [Google Scholar]
- 24. Tuley E. A., Gaucher C., Jorieux S., Worrall N. K., Sadler J. E., Mazurier C. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 6377–6381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Raines G., Aumann H., Sykes S., Street A. (1990) Thromb. Res. 60, 201–212 [DOI] [PubMed] [Google Scholar]
- 26. Burset M., Seledtsov I. A., Solovyev V. V. (2001) Nucleic Acids Res. 29, 255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marti T., Rösselet S. J., Titani K., Walsh K. A. (1987) Biochemistry 26, 8099–8109 [DOI] [PubMed] [Google Scholar]
- 28. Zhang Q., Zhou Y. F., Zhang C. Z., Zhang X., Lu C., Springer T. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9226–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang X., Halvorsen K., Zhang C. Z., Wong W. P., Springer T. A. (2009) Science 324, 1330–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lang T., Hansson G. C., Samuelsson T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 16209–16214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Perez-Vilar J., Hill R. L. (1999) J. Biol. Chem. 274, 31751–31754 [DOI] [PubMed] [Google Scholar]
- 32. Godl K., Johansson M. E., Lidell M. E., Mörgelin M., Karlsson H., Olson F. J., Gum J. R., Jr., Kim Y. S., Hansson G. C. (2002) J. Biol. Chem. 277, 47248–47256 [DOI] [PubMed] [Google Scholar]
- 33. Sheehan J. K., Kirkham S., Howard M., Woodman P., Kutay S., Brazeau C., Buckley J., Thornton D. J. (2004) J. Biol. Chem. 279, 15698–15705 [DOI] [PubMed] [Google Scholar]
- 34. Perez-Vilar J., Eckhardt A. E., Hill R. L. (1996) J. Biol. Chem. 271, 9845–9850 [DOI] [PubMed] [Google Scholar]
- 35. Perez-Vilar J., Eckhardt A. E., DeLuca A., Hill R. L. (1998) J. Biol. Chem. 273, 14442–14449 [DOI] [PubMed] [Google Scholar]
- 36. Voorberg J., Fontijn R., van Mourik J. A., Pannekoek H. (1990) EMBO J. 9, 797–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rosenberg J. B., Haberichter S. L., Jozwiak M. A., Vokac E. A., Kroner P. A., Fahs S. A., Kawai Y., Montgomery R. R. (2002) Blood 100, 1699–1706 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.