Abstract
The IκB kinase (IKK) complex acts as a gatekeeper of canonical NF-κB signaling in response to upstream stimulation. IKK activation requires sensing of ubiquitin chains by the essential IKK regulatory subunit IKKγ/NEMO. However, it has remained enigmatic whether NEMO binding to Lys-63-linked or linear ubiquitin chains is critical for triggering IKK activation. We show here that the NEMO C terminus, comprising the ubiquitin binding region and a zinc finger, has a high preference for binding to linear ubiquitin chains. However, immobilization of NEMO, which may be reminiscent of cellular oligomerization, facilitates the interaction with Lys-63 ubiquitin chains. Moreover, selective mutations in NEMO that abolish association with linear ubiquitin but do not affect binding to Lys-63 ubiquitin are only partially compromising NF-κB signaling in response to TNFα stimulation in fibroblasts and T cells. In line with this, TNFα-triggered expression of NF-κB target genes and induction of apoptosis was partially compromised by NEMO mutations that selectively impair the binding to linear ubiquitin chains. Thus, in vivo NEMO interaction with linear and Lys-63 ubiquitin chains is required for optimal IKK activation, suggesting that both type of chains are cooperating in triggering canonical NF-κB signaling.
Keywords: NF-kappa B (NF-KB), Signal Transduction, T-cell Receptor, Tumor Necrosis Factor (TNF), Ubiquitin, IKK, NEMO, Thermophoresis
Introduction
Induction of gene expression by the transcription factor NF-κB controls many physiological processes, including immunity, differentiation, or apoptosis. The IκB kinase (IKK)3 complex acts as the gatekeeper of canonical NF-κB signaling. In response to extracellular stimuli, IKKs become activated and catalyze the phosphorylation of NF-κB inhibitors (IκBs), leading to their ubiquitination and proteasomal degradation (1, 2). The IKK complex consists of the two catalytic domains IKKα and IKKβ and the regulatory subunit IKKγ or NF-κB essential modulator (NEMO). NEMO mutations scattered throughout the entire gene are causing severe pathologies such as anhidrotic ectodermal dysplasia with immunodeficiency and incontinentia pigmenti (3–6).
NEMO serves as a critical integrating platform coupling upstream receptor signaling to the canonical NF-κB pathway. Biochemical and genetic studies have highlighted a pivotal function of polyubiquitination for IKK/NF-κB activation. Upon TNFα or T cell receptor/CD28 stimulation, signaling adaptors RIP1 or MALT1 are modified by covalent attachment of Lys-63-linked ubiquitin (Ub) chains to recruit NEMO and thereby promote IKK activation (7–9). Recently, the assembly of linear Ub chains by the linear Ub chain assembly complex was shown to promote cytokine-triggered IKK activation (10). The Ub binding surface in NEMO called UBAN (Ub binding in ABIN and NEMO) or NOA (NEMO-optineurin-ABIN) is required for signal induced IKK activation. NEMO UBAN has been co-crystallized with linear as well as with Lys-63 Ub chains (11, 12). The crystal structures reveal a bipartite Ub binding region from amino acids 290 to 330, distinguishing between a proximal and a distal moiety of bound di-Ub.
In solution, linear di-Ub was found to bind with ∼100 fold higher affinity to the UBAN than Lys-63 Ub (13). However, the C-terminal zinc finger (ZF) of NEMO may contribute a second Ub binding site (14), and it was reported that the UBAN in conjunction with ZF (NEMO UBAN-ZF) displays similar affinities for linear or Lys-63 tetra-Ub (15). Thus, the efficiency of linear versus Lys-63 Ub chains to bind to NEMO in vitro as well as the physiological relevance of these associations for IKK activation has remained unresolved.
Using in vitro association studies, we provide evidence that the entire NEMO C terminus displays a high preference for interacting with linear Ub chains. However, depending on the assay, also Lys-63 Ub chains can associate weakly with NEMO through the UBAN domain. By analyzing NEMO UBAN mutations that selectively interfere with binding to linear but not Lys-63 Ub chains, we show that canonical IKK/NF-κB activation is partially compromised upon loss of NEMO binding to linear Ub chains. Hence, in vivo, different Ub chains can contribute to optimal NF-κB activation.
EXPERIMENTAL PROCEDURES
Prokaryotic Expression Plasmids
The vector system pASK-IBA3plus (IBA GmbH, Göttingen, Germany) was used for production of bacterial recombinant proteins. This vector is inducible with anhydrotetracycline (IBA GmbH). Using SacII and NcoI restriction sites, the NEMO sequences were cloned into the multiple cloning site leading to the in-frame fusion of the StrepTagII sequence to the C terminus. The N-terminal StrepTagII was introduced using the 5′-primer. With the 3′-primer, stop codons were introduced before the StrepTagII sequence to prevent fusion of the StrepTagII to the C terminus. His-tagged ubiquitin was also cloned into pASK-IBA3plus using the same strategy like N-terminally Strep-tagged NEMO.
Eucaryotic Expression Plasmids
In the pPHAGE plasmid, a third generation lentiviral SIN nonreplicative vector, the truncated human CD2 (hΔCD2) and human NEMO cDNA were linked by the 18 amino acids T2A sequence from Thosea asigna. The T2A linker co-translationally prevents the formation of its last peptide bond, which allows concomitant expression of two genes under the control of one promoter, in this case the phosphoglycerate kinase (PGK) promoter.
The empty PGK-PHAGE plasmid was supplied with a NotI-SalI-BamHI linker. Subsequently, hΔCD2 (without STOP codon) and the T2A sequence were inserted in-frame, each with an EagI/NotI restriction site deletion strategy. Thus, the only remaining NotI site was situated behind PGK-hΔCD2-T2A and could be used to insert NotI-FLAG-hNEMO-NotI WT or mutated fragments in-frame.
Antibodies and Reagents
Cells were treated with polybrene (Sigma), TNFα (Biomol), and phorbol 12-myristate 13-acetate (PMA)/ionomycin (Calbiochem). The following antibodies were used: allophycocyanin-labeled anti-human CD2 (eBioscience), anti-IκBα (sc-371), anti-NEMO (sc-8330), anti-β-actin (sc-1616; all Santa Cruz Biotechnology), anti-p-IκBα (Cell Signaling), anti-IKKα (05–536; Millipore), anti-IKKβ (05–535; Millipore), anti-FLAG M2 (F3165, Sigma). Anti-His-tagged DELFIA Europium-N1 (AD0108; PerkinElmer Life Sciences).
Protein expression and purification
Recombinant proteins were produced in Escherichia coli strain BL21-CodonPlus (DE3) RILP (Stratagene) using the pIBA3plus expression system (IBA GmbH, Göttingen, Germany). Bacteria were grown to a density of an A600 value of 0.8–1.0. Protein expression was induced by addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside and 200 ng/ml anhydrotetracycline (IBA GmbH), and bacteria were cultured overnight at 21 °C. Cultures were harvested by centrifugation at 3000 × g, and pellets were resuspended in lysis buffer (StrepTagII purification (100 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.5 mm DTT, 0.5 mg/ml lysozyme, and protease inhibitors); His tag purification (75 mm phosphate buffer, pH 7.4, 400 mm NaCl, 10 mm imidazole, 0.5 mg/ml lysozyme, and protease inhibitors)) at 4 °C. After 20 min at room temperature, the culture suspension was sonicated on ice. 20 min centrifugation at 20,000 × g (4 °C) led to clarification of the lysate from cellular debris. Finally, the supernatant was again centrifuged at 20,000 × g (4 °C).
Bacterial lysates containing the Strep-tagged proteins of interest were applied on the StrepTrap columns (GE Healthcare), and bacterial proteins were washed away using washing buffer (100 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.5 mm DTT). Target proteins were eluted with an elution buffer (100 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.5 mm DTT, 2.5 mm d-desthiobiotin). His-tagged proteins were applied on HisTrap columns (GE Healthcare), and bacterial proteins were washed away with washing buffer (20 mm phosphate buffer, pH 7.4, 500 mm NaCl, 30 mm imidazole), and His-tagged proteins were eluted using a gradient of elution buffer (20 mm phosphate buffer, pH 7.4, 500 mm NaCl, 500 mm imidazole). Elution fractions containing the proteins of interest were pooled and concentrated using Amicon Ultra-15 spin columns (Millipore) with an exclusion size of 3 kDa.
Lys-63 di-Ub was generated by in vitro ligation of His-Ub K63R and Ub Asp-77 (16). By using these two mutations, directed synthesis of di-Ub is feasible. To generate the Lys-63 di-Ub, the ubiquitin conjugation kit (Boston Biochem) was used in combination with His-Ub K63R, Ub Asp-77, the activating E1 enzyme UBE1 (Boston Biochem) and the Lys-63-specific E2 complex UbcH13/Uev1a (Boston Biochem). The ligation reaction was incubated for 4 h at 37 °C. Excess of nonligated monoubiquitin was removed by subsequent gel filtration (see supplemental Fig. 1).
Microscale Thermophoresis (MST)
The labeling was performed with a reactive dye (NT-647) using N-hydroxy succinimide-ester chemistry, which reacts efficiently with the primary amines of proteins to form highly stable dye-protein conjugates. The labeling procedure was performed as described previously (17). MST assays were mainly carried out as described in Wienken et al. (17). Serial dilutions of unlabeled NEMO proteins were mixed with 200 nm of NT-647-labeled Ub proteins in MST buffer (25 mm Tris-HCl, pH 8.0, 100 mm NaCl, 0.1% BSA, 0.1% Tween 20, 0.5 mm DTT) and incubated for 30 min. MST assays were measured in a NanoTemper Monolith NT.015T. By plotting NEMO concentration to % changes of normalized fluorescence (ΔFnorm [%]), curve fitting was performed using GraphPad prism software, and KD values were determined.
DELFIA Assay (ELISA-based Plate Assay)
DELFIA ELISA assays (dissociation-enhanced lanthanide fluorescence immunoassay) were performed using the manufactory's protocol (PerkinElmer Life Sciences). 20 pmol of Strep-tagged NEMO proteins in DELFIA assay buffer (Perkin Elmer) were coupled to Strep-Tactin-coated 96-well plates (IBA GmbH). After 2 h of incubation, the unbound fraction was washed away using DELFIA washing buffer (PerkinElmer Life Sciences), and bound NEMO proteins were incubated with 250 pmol of His-tagged Ub proteins in DELFIA assay buffer. Again, after 2 h of incubation, an excess of Ub proteins was washed away (with DELFIA washing buffer) and 500 ng/ml europium-labeled anti-His-tagged antibody in DELFIA assay buffer was added to the reaction for 1 h. After extensive washing (with DELFIA washing buffer), enhancer solution (PerkinElmer Life Sciences) was added to activate europium fluorescence, and this was measured in a BioTek Synergy 2 fluorescence plate reader (excitation, 340 nm; emission, 615 nm).
Cell Culture, Lentiviral Infection, EMSA, Western Blot, and Co-IP
Mouse embryonic fibroblasts (MEF) and HEK 293T cells were grown in DMEM, and Jurkat T cells were cultured in RPMI supplemented with 10% FCS according to standard procedures. Lentiviruses were produced according to Refs. 18, 19. In short, proviral plasmids (pHAGE-PGK-hΔCD2-T2A-NEMO, pMDL, pVSVG, and pREV) were transfected in HEK 293T cells by calcium chloride precipitation. NEMO−/Y MEF were incubated with the sterile-filtrated viral supernatant harvested from transfected HEK 293T cells in the presence of 8 μg/ml polybrene for 72 h. Infected cells expressing hΔCD2 were stained with allophycocyanin-labeled anti-human CD2 and enriched by FACS sorting. A hΔCD2-low population was sorted to yield exogenous NEMO expression at the level of endogenous NEMO from WT MEF. Reconstitution of NEMO-deficient Jurkat T cells was performed accordingly. Cells were subsequently expanded and stimulated with 8 ng/ml or 20 ng/ml TNFα (MEF or Jurkat cells, respectively), 400 ng/ml PMA, and 600 ng/ml ionomycin (P/I). EMSA and Western blotting was carried out as described previously (20). NEMO-IKKα/β interaction was investigated after FLAG IP of FLAG-NEMO constructs and subsequent detection of IKKα and IKKβ by Western blotting. Co-IP and Western blotting was done as described (21).
Quantitative Real-time PCR
Reconstituted MEF were treated with 8 ng/ml TNFα for 1 h. mRNA was isolated with Qiagen RNeasy Mini Kit and subsequently DNase-treated (Promega). cDNA was synthesized using the Invitrogen SuperScript II kit with poly-dT primer. Quantitative RT-PCR was basically performed as described previously (21). For data analysis, Cp values were first normalized to the levels of the murine housekeeping gene porphobilinogen deaminase, and afterward, mRNA levels of stimulated samples were related to the values of the corresponding unstimulated control. Finally, levels of WT-reconstituted cells were set to 1 and all other data related to this. The Following primers were used in this study: A20, 5′-GCTCAACTGGTGTCGTGAAG-3′ and 5′-ATGAGGCAGTTTCCATCACC-3′; CXCL2, 5′-AGTGAACTGCGCTGTCAATG-3′ and 5′-CTTCAGGGTCAAGGCAAACT-3′; ICAM-1, 5′-GGAGACGCAGAGGACCTTAAC-3′ and 5′-CGCTCAGAAGAACCACCTTC-3′; and porphobilinogen deaminase, 5′-GCGCTAACTGGTCTGTAGGG-3′ and 5′-TGAGGGAAAGGCAGATATGG-3′.
Apoptosis Assay
Rate of apoptosis was determined after 22 h of 8 ng/ml TNFα stimulation in MEF by phycoerythrin-annexin V and 7-aminoactinomycin D staining using the BD Pharmingen apoptosis kit. Phycoerythrin-annexin V stains preapoptotic cells and 7-aminoactinomycin D dead cells. FACS assays were measured on an LSRII flow cytometer (BD Biosciences), and data evaluation was carried out using FlowJo software (Treestar).
RESULTS
NEMO C Terminus Is Selectively Binding to Linear Ub Chains in Solution
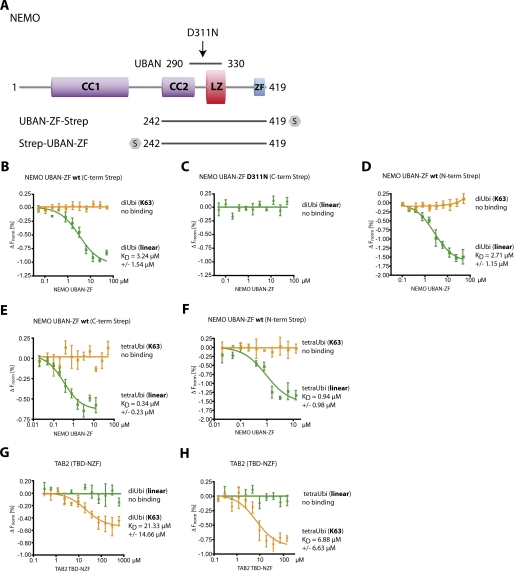
We used MST, a sensitive type of protein interaction assay, to investigate the association of the NEMO C terminus (amino acids 242–419) with fluorescence-labeled di-Ub or tetra-Ub in solution. Binding of an interaction partner influences the thermal migration behavior of ubiquitin, and the fluorescence depletion in a heated spot of the protein solution was measured in dependence of increasing interactor concentration. KD values were derived from the depletion curves. All NEMO UBAN-ZF fragments were C- or N-terminally fused to StrepTagII and purified from E. coli (Fig. 1A and supplemental Fig. 1, A and C). Binding of linear di-Ub to NEMO UBAN-ZF is evident from the shift in thermal migration of linear di-Ub induced upon incubation with increasing amounts of NEMO UBAN-ZF (supplemental Fig. 2A). For the binding of linear di-Ub to UBAN-ZF, we determined a KD of ∼3.2 μm (Fig. 1B), which is in the range measured by isothermal titration calorimetry for linear di-Ub and the NEMO UBAN alone (13, 22). We were unable to detect an association between Lys-63 di-Ub and the NEMO UBAN-ZF (supplemental Fig. 2C and Fig. 1B). As expected, mutation of Asp-311 to Asn (D311N), which prevents formation of two critical hydrogen bonds and a salt bridge between the UBAN and the distal Ub moiety (11) and is known to cause severe pathological defects in vivo, completely abolished interaction with NEMO UBAN-ZF (supplemental Fig. 2B and Fig. 1C). Similar results were obtained with N-terminally StrepTagII-tagged NEMO UBAN-ZF and linear or Lys-63 di-Ub chains (Fig. 1D), excluding the possibility that the C-terminal tag interferes with the function of the ZF.
FIGURE 1.
In solution, recombinant NEMO UBAN-ZF WT selectively binds linear Ub. A, scheme of the NEMO construct. NEMO UBAN-ZF with the UBAN domain and the mutation site Asp-311 is indicated. CC1/CC2, coiled coil domains 1/2; LZ, leucin zipper; ZF, zinc finger; UBAN, ubiquitin binding domain; S, StrepTagII. B, C-terminal (C-term) Strep NEMO UBAN-ZF WT bound linear di-Ub with a KD of 3.24 μm, but not Lys-63-linked di-Ub. C, MST assays of NEMO UBAN-ZF D311N mutation, which is not able to bind linear Ub. D, MST experiments were carried out with N-terminally Strep-tagged NEMO UBAN-ZF WT as described in B. It bound linear di-Ub with a KD of 2.71 μm, but not Lys-63-linked di-Ub. E, linear tetra-Ub chains bound to C-terminal Strep NEMO UBAN-ZF WT with a KD of 0.34 μm, whereas no binding was detectable with Lys-63-linked tetra-Ub. F, linear tetra-Ub chains also bound to N-terminally (N-term) Strep-tagged NEMO UBAN-ZF WT with a KD of 0.94 μm, whereas no binding was detectable with Lys-63-linked tetra-Ub. G, the TBD-NFZ domain of the Lys-63-linked ubiquitin recognizing protein TAB2 served as an independent control for the validity of the assay. TAB2 did not recognize linear di-Ub, whereas Lys-63-linked di-Ub was bound with a KD of 21.33 μm. H, linear tetra-Ub also did not bind TAB2 TBD-NFZ, whereas Lys-63-linked tetra-Ub was bound by TAB2 TBD-NZF with a KD of 6.88 μm. All data represent the mean from three independent experiments, and error bars indicate S.D. ΔFnorm, change normalized fluorescence.
As longer Lys-63 Ub chains were suggested to enhance the affinity to the C terminus of NEMO (15), we performed MST using linear or Lys-63 tetra-Ub. Whereas the affinity of linear tetra-Ub toward C- or N-terminally StrepTagII-tagged UBAN-ZF was even enhanced compared with linear di-Ub (KD ∼ 0.34 or 0.94 μm, respectively), a Lys-63 tetra-Ub did not bind to NEMO independent of the position of the StrepTagII (Fig. 1, E and F). These data suggest that longer Ub chains are not per se enhancing the affinity between Lys-63 Ub chains and NEMO in solution. To verify that Lys-63 di-Ub and tetra-Ub are functional, we determined their association to the NZF of TAB2 (TAK1 binding domain–Npl4 zinc finger), which was shown to present a selective surface for the interaction with Lys-63 Ub chains (23). As expected, TBD-NZF bound to Lys-63 di-Ub and tetra-Ub with a KD of ∼21.3 μm and ∼6.9 μm, respectively. However, no association to linear Ub was observed (Fig. 1, G and H).
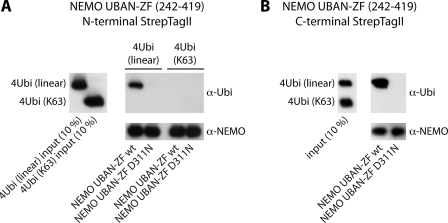
We also determined association of either linear or Lys-63 tetra-Ub to NEMO UBAN-ZF in StrepTagII pulldown experiments and confirmed that the C terminus of NEMO is exclusively precipitating linear tetra-Ub after incubation of individual chains or in a mixed chain reaction (Fig. 2, A and B). Taken together, MST data and pulldown experiments provide evidence that in solution the NEMO C terminus is associating to linear Ub chains with KD in the low micromolar range, whereas a putative association to Lys-63 Ub chains is beyond the limit of detection.
FIGURE 2.
Pulldown experiments of N- or C-terminally Strep-tagged NEMO-UBAN-ZF with linear and Lys-63-linked tetra-Ub. A, Western blot of the StrepTagII pulldown experiment with N-terminal Strep NEMO-UBAN-ZF. Left panel, 10% input of linear and Lys-63-linked tetra-Ub chains. Right panel, N-terminal Strep NEMO-UBAN-ZF WT was only capable of recognizing linear tetra-Ub but not Lys-63-linked tetra-Ub. D311N mutation was unable to bind any of the investigated tetra-Ub chain. B, in an approach where a mixture of both chain types (linear and Lys-63) was provided for recognition, C-terminal Strep NEMO-UBAN-ZF WT only pulled down linear tetra-Ub.
Immobilization of NEMO UBAN-ZF Enhances Binding to Lys-63 Ub
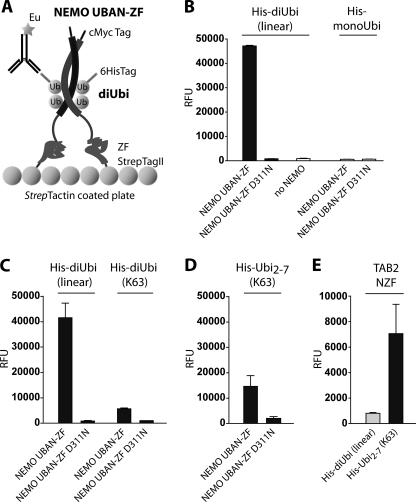
Despite the apparent inability of Lys-63 di-Ub to associate with the NEMO C terminus in solution, Lys-63 di-Ub and the NEMO UBAN have been co-crystallized and were found to bind when NEMO UBAN was coated on a biosensor (12, 24). We hypothesized that a more static orientation of the NEMO C terminus might facilitate the interaction of the UBAN with Lys-63 di-Ub. Therefore, we established DELFIA assays as a very sensitive sandwich-based ELISA method to measure interaction of plate bound NEMO UBAN-ZF with di-Ub (Fig. 3A). For this purpose, StrepTagII-tagged NEMO UBAN-ZF was attached to StrepTactin-coated plates and incubated with His-tagged mono-Ub, linear di-Ub, or Lys-63 di-Ub (supplemental Fig. 1A and B). Binding of NEMO UBAN-ZF to His-Ub proteins was detected by time-resolved fluorescence after extensive washing, using europium-labeled anti-His antibody. Interaction of linear His-di-Ub with NEMO UBAN-ZF was evident from a strong time-resolved fluorescence signal (Fig. 3B). Again, the substitution D311N in the NEMO UBAN completely prevented binding to linear His-di-Ub. To validate the assay, we tested binding of His-mono-Ub to NEMO UBAN-ZF. MonoUbi did not interact with NEMO UBAN-ZF, providing evidence for the reliability of the assay (Fig. 3B).
FIGURE 3.
Immobilized NEMO UBAN-ZF binds to linear and more weakly to Lys-63-linked Ub chains. A, experimental setup of the DELFIA plate-based binding assay. Recombinant c-Myc-NEMO-UBAN-ZF-StrepTagII dimer is bound to a StrepTactin-coated plate. His-labeled di-Ub binds to NEMO and can be detected by a europium-labeled anti-His antibody. B, NEMO-UBAN-ZF WT showed a strong and robust signal when incubated with linear His-di-Ub compared with background level (no NEMO) or the NEMO UBAN-ZF D311N mutation. Also, no interaction of NEMO-UBAN-ZF and mono-Ub was detected. C, NEMO UBAN-ZF WT showed a weak interaction with Lys-63 His-di-Ub compared with binding to linear His-di-Ub. D, the binding of NEMO-UBAN-ZF to Lys-63 Ub was enhanced by use of a mixture of His-Ub2–7 chains. E, the functionality of Lys-63 Ub chains for DELFIA plate assays was validated by the use of TAB2 NZF domain, which specifically recognized Lys-63 His-Ub2–7. All data represent the mean from three independent experiments, and error bars indicate S.D. RFU, relative fluorescence unit.
Next, we compared the interaction between linear His-di-Ub and Lys-63 His-di-Ub with the NEMO UBAN-ZF (Fig. 3C). In this setup, we detected binding of the NEMO UBAN-ZF to Lys-63 His-di-Ub, even though the time-resolved fluorescence signal was ∼7–8-fold decreased when compared with linear His-di-Ub. This reduced affinity of Lys-63 di-Ub compared with linear di-Ub is in the range that has been observed in a biosensor assay where the NEMO UBAN alone was immobilized (24). The substitution D311N completely prevented association to Lys-63 His-di-Ub, proving that this interaction critically depends on the binding of the distal Ub moiety to the N-terminal part of the UBAN (12). To determine whether longer Lys-63 Ub chains could enhance the interaction, we used a mixture of Lys-63 Ub chains ranging from 2–7 Ub (Lys-63 His-Ubi2–7) in length (Fig. 3D). Association of Lys-63 His-Ubi2–7 was slightly enhanced when compared with Lys-63 di-Ub. To ascertain that the DELFIA assay did not generally prefer detection of linear versus Lys-63 di-Ub, we compared binding of Lys-63 His-Ubi2–7 and linear His-di-Ub to TAB2 NZF fused to StrepTagII. Also in the plate bound assay the NZF domain of TAB2 bound with high preference to Lys-63 Ub chains when compared with linear di-Ub (Fig. 3E).
C-terminal part of UBAN Domain Is Selectively Required for Association to Linear Di-Ub
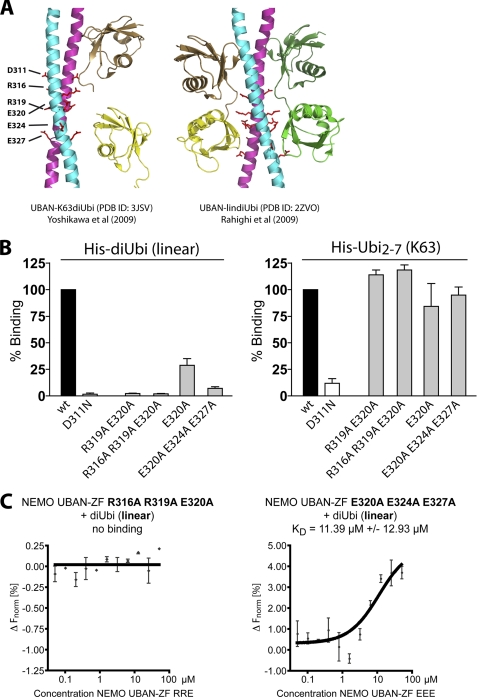
The crystal structures of the NEMO UBAN domain bound to linear or Lys-63 di-Ub reveal that the distal Ub moiety is associating with the N-terminal part of the UBAN domain that contains Asp-311 (11, 12). Due to the different positioning of the proximal Ub in linear and Lys-63 di-Ub, the C-terminal part of the UBAN domain binds to the proximal Ub moiety only in the context of linear di-Ub (Fig. 4A). We purified NEMO UBAN-ZF proteins where residues in the C-terminal part of the UBAN are mutated that are exclusively contacting the distal moiety of linear di-Ub (supplemental Fig. 1B). Because Ub binding requires NEMO dimerization through the surrounding CC2-LZ region (25), we first verified by gel filtration that NEMO UBAN-ZF WT and mutations were predominantly forming solution dimers, ruling out that the mutations are distorting the overall structure of the NEMO UBAN-ZF (supplemental Fig. 3). DELFIA assays revealed that the point mutation of E320A decreased the interaction of NEMO UBAN-ZF with linear di-Ub by ∼75% and that the combined mutation of all glutamates (E320A/E324A/E327A) led to 90% reduced binding (Fig. 4B). Moreover, mutations R319A/E320A and R316A/R319A/E320A abolished the association to linear di-Ub to the same extent as the D311N mutation in the DELFIA assays. Importantly, all of the distal NEMO UBAN-ZF mutations are still binding to the Lys-63 Ub, confirming that the C-terminal UBAN domain is exclusively responsible for contacting the linear Ub (Fig. 4B). To verify the results, we also determined the affinity of the triple NEMO UBAN-ZF mutations E320A/E324A/E327A and R316A/R319A/E320A to linear di-Ub in solution by MST (Fig. 4C). Whereas the triple glutamate mutation still bound linear di-Ub, even though with a decreased affinity when compared with NEMO UBAN-ZF WT (KD ∼11.4 μm versus ∼3.2 μm) (compare Fig. 1C), mutation of the contact residues R316A/R319A/E320A completely abolished association of the NEMO C terminus and linear di-Ub in solution (Fig. 4C).
FIGURE 4.
Directed mutagenesis of NEMO selectively impairs binding to linear di-Ub. A, crystal structures of NEMO UBAN dimers co-crystallized with either Lys-63 di-Ub (left panel) or linear di-Ub (right panel). Asp-311 contacting the distal Ub of Lys-63 and linear di-Ub as well as contact residues solely involved in the recognition of the proximal Ub in linear di-Ub are indicated. B, DELFIA plate assay of NEMO-UBAN-ZF WT and mutations targeting the linear ubiquitin specific recognition site. Mutation of the C-terminal UBAN domain selectively impedes binding of linear Ub. Association of WT and mutations D311N, R319A/E320A, R316A/R319A/E320A, E320A, or E320A/E324A/E327A to linear His-di-Ub (left) and Lys-63 HisUb2–7 (right) was measured by plate-coupled DELFIA assays. For comparison, binding of WT was set to 100%. C, in MST, the NEMO UBAN-ZF R316A/R319A/E320A was not binding to linear di-Ub, whereas NEMO UBAN-ZF E320A/E324A/E327A interacted with linear di-Ub with a lower affinity of 11.39 μm compared with NEMO UBAN-ZF WT (compare with Fig. 1A). All data represent the mean from three independent experiments, and error bars indicate S.D. ΔFnorm, change normalized fluorescence.
Partial Impairment of NF-κB Signaling by Selective Interference with NEMO Binding to Linear Ub
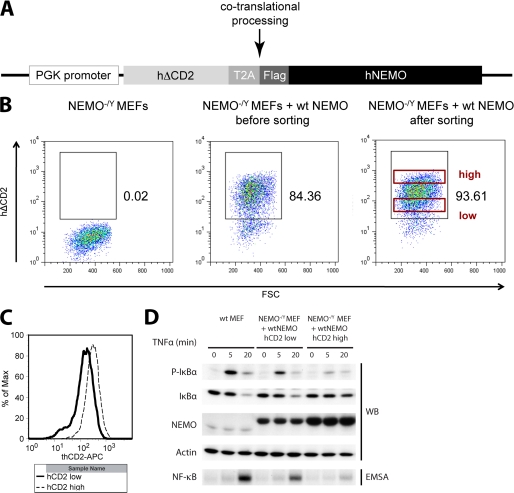
To analyze the functional impact of NEMO UBAN mutations on TNFα-induced NF-κB activation, we reconstituted NEMO-deficient (NEMO−/Y) MEF. Overexpression of NEMO was shown to inhibit NF-κB activation (26, 27). Thus, we chose a lentiviral expression system to co-express FLAG-NEMO together with the cell surface marker hΔCD2 to be able to sort for cells expressing different NEMO constructs at equivalent levels in the range of the endogenous protein (Fig. 5). FLAG-NEMO and hΔCD2 were separated by the co-translational processing sequence T2A (Fig. 5A) (28). This allowed us to FACS sort for cells expressing equal NEMO amounts.
FIGURE 5.
Expression levels of reconstituted NEMO in MEF. A, schematic of the lentiviral construct used to reconstitute MEF or Jurkat T cells. Promoter, PGK; surface infection marker, human truncated CD2 (hΔCD2). The T2A sequence mediates interruption of its own translation, thus creating two polypeptides under the control of one promoter. Triple FLAG-tagged full-length wild type human NEMO was used for reconstitution. B, FACS analysis of infection marker (hΔCD2) levels in the course of the NEMO reconstitution experiment (staining with anti-hCD2-allophycocyanin). Left panel, uninfected NEMO-deficient MEF. Middle panel, infected cells before sorting. Right panel, CD2-positive cell populations after sorting. Two populations (NEMO high and low) are indicated. C, restaining of “high” and “low” cells from the right panel (B) Western blot (WB) and EMSA analysis (D) of reconstituted NF-κB signaling in WT, low, and high MEF. Quantification of NEMO levels yielded WT:low:high = 1:8:20. In the case of the NEMO high cells, the NF-κB signaling is severely impaired. The NEMO levels of the cells actually used for experimentation matched the endogenous NEMO levels of WT MEF much closer. See Fig. 6, A and B, left panels.
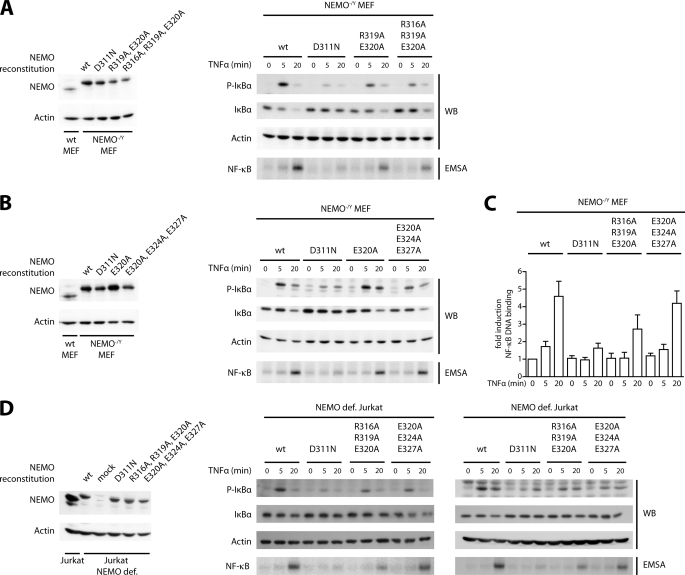
Restaining of sorted MEF (Fig. 5B) revealed that surface amounts of hΔCD2 directly correlated with intracellular NEMO amounts (Fig. 5, C and D). Comparison of the TNFα-induced NF-κB signaling in reconstituted hΔCD2/NEMO high and low cells showed that increasing amounts of NEMO indeed strongly impaired NF-κB activation (Fig. 5D). Thus, all subsequent reconstitution analyses were carried out with MEF expressing equivalent amounts of WT and mutated NEMO protein at levels slightly above the endogenous NEMO levels in WT MEF (Fig. 6, A and B, left panels). NF-κB signaling was determined by IκBα and phospho-IκBα Western blotting and measurement of NF-κB DNA binding by EMSA (Fig. 6, A and B, and supplemental Fig. 4A). Quantification of EMSA results is shown in Fig. 6C. As expected, UBAN mutation D311N, which abolished association of linear and Lys-63 Ub in vitro, is strongly impaired in TNFα-dependent NF-κB activation. NEMO carrying the UBAN mutations E320A or E320A/E324A/E327A, which displayed a diminished binding to linear Ub, efficiently mediated TNFα induced NF-κB activation similar to NEMO WT (Fig. 6, B and C). Mutations R319A/E320A or R316A/R319A/E320A, which completely abrogated association of linear but not Lys-63 Ub, only partially impaired reconstitution of NF-κB signaling (Fig. 6, A and C). To exclude that mutations in the NEMO UBAN affect IKK complex assembly, we immunoprecipitated FLAG-NEMO from WT, D311N, E320A/E324A/E327A, and R316A/R319A/E320A reconstituted MEF and probed for IKKα and IKKβ. As expected, the mutations had no influence on IKK complex composition (supplemental Fig. 4B). Taken together, our results suggest that NEMO binding to linear Ub chains contributes but is not exclusively responsible for optimal NF-κB activation.
FIGURE 6.
Reconstitution of NEMO-deficient MEF and Jurkat T cells. NEMO-deficient MEF or Jurkat T cells were lentivirally reconstituted with NEMO WT or mutation constructs. A and B, expression of NEMO WT and NEMO in NEMO−/Y MEF was analyzed by Western blotting (WB; upper panels), and the extent of NF-κB signaling was evaluated by determining p-IκBα and IκBα amounts by Western blotting and NF-κB DNA binding by EMSA (lower panels). Whereas mutation D311N almost completely prevented NF-κB signaling, mutation R319A/E320A and R316A/R319A/E320A led to reduced NF-κB signaling and mutations E320A and E320A/E324A/E327A had almost no discernible effect on NF-κB activation. C, quantification of NF-κB DNA binding after TNFα stimulation from three independent EMSA experiments with S.D. Actin was used as an internal control. D, expression of NEMO WT and mutations in NEMO-deficient (NEMO def.) Jurkat T cells was analyzed by Western blotting (left panels). NF-κB signaling was determined after TNFα (middle panel) or P/I (right panel) stimulation according to A and B. P/I re-enacts T cell receptor stimulation and serves as a TNF-independent control for downstream signaling involving NEMO. In T cells, mutations R316A/R319A/E320A showed impaired NF-κB activation after TNFα and P/I stimulation, whereas mutations E320A/E324A/E327A are only slightly reduced.
To verify the results in an independent cellular system, we infected NEMO-deficient Jurkat T cells by the same lentiviral approach (Fig. 6D). FACS-sorted hΔCD2-positive cells expressed NEMO at comparable levels slightly below the endogenous level of Jurkat T cells (Fig. 6D, left panel). Next, we compared NF-κB activation in response to TNFα (Fig. 6D, middle panel) or PMA/ionomycin (P/I; Fig. 6D, right panel) stimulation after reconstitution. Similar to the results obtained in MEF, the mutation of D311N in NEMO nearly abolished NF-κB activation in response to both stimuli. Whereas NEMO E320A/E324A/E327A was almost fully functional in rescuing TNFα- or P/I-induced NF-κB activation, the triple R316A/R319A/E320A exchange impaired but did not completely block NF-κB signaling. Thus, the C-terminal part of the UBAN that contacts the proximal Ub moiety in linear Ub not only contributes to canonical NF-κB in response to TNFα stimulation, but linear chains are also involved in PKCθ-dependent IKK/NF-κB activation in T cells.
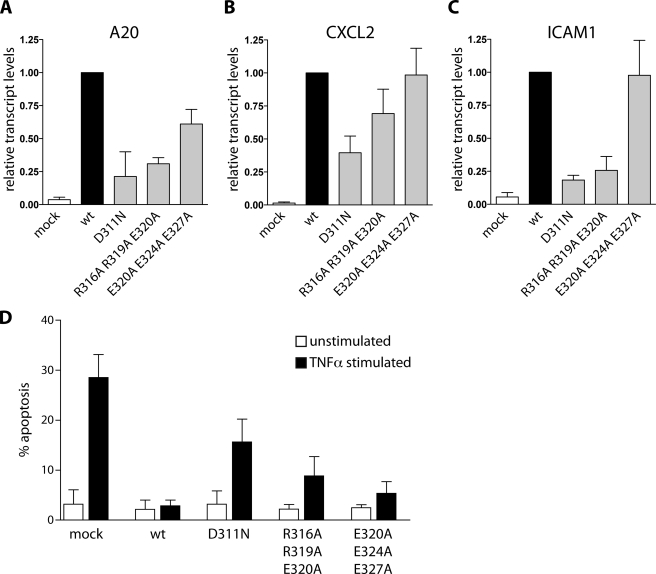
NEMO Binding to Linear Ub Partially Impairs Induction of NF-κB Target Genes and Prevention of Apoptosis
To gain more insight into the functional impact of selective disruption of NEMO-linear Ub binding, we monitored TNFα-induced expression of the NF-κB target genes A20 (TNFα-induced protein 3, TNFAIP3), CXCL2 (chemokine (C-X-C motif) ligand 2), and ICAM-1 (intercellular adhesion molecule 1) by quantitative RT-PCR in MEF reconstituted with NEMO mutations (Fig. 7A). Congruent with the result obtained for NF-κB activation, the D311N mutation had the most severe effect on the expression of all three genes, whereas the E320A/E324A/E327A mutation showed almost no influence compared with wild type NEMO. NF-κB target gene expression was reduced in NEMO R316A/R319A/E320A reconstituted cells. However, particularly in the case of CXCL2, the effect of NEMO R316A/R319A/E320A was not as severe as seen with NEMO D311N, which again reflected the partial rescue with the selectively linear Ub binding mutant.
FIGURE 7.
TNFα induced NF-κB target gene transcription and susceptibility to apoptosis in NEMO-reconstituted MEF. A–C, NEMO-reconstituted MEF were stimulated with TNFα for 1 h, and NF-κB target gene mRNA levels (A20, CXCL2, and ICAM-1) were determined by quantitative PCR. Bars indicate mRNA levels of TNFα-stimulated cells versus unstimulated control cells. For comparison, all calculated values were related to levels of WT-reconstituted cells, which were set to 1. In accordance with all previous results, mutations D311N and R316A/R319A/E320A led to reduced NF-κB target gene transcript levels, whereas mutation E320A/E324A/E327A had only mild effects. D, NEMO-reconstituted MEF were assayed for early apoptosis after 22 h of TNFα treatment. Late apoptotic cells were excluded in the FACS analysis, and levels of annexin V-positive cells are depicted in the graph. NEMO WT reconstitution is able to completely rescue TNFα-induced apoptosis of knock-out cells. NEMO D311N shows elevated annexin V levels, and thus is only partially able to prevent apoptosis. The mutation R316A/R319A/E320A shows decreased apoptosis levels in comparison with D311N, and E320A/E324A/E327A is even more capable to rescue.
We determined apoptosis induction in fibroblasts as a functional readout for the effectiveness of the NEMO constructs to rescue NF-κB-mediated prosurvival signaling after TNFα stimulation (Fig. 7B). Whereas expression of NEMO WT clearly counteracted TNFα-induced apoptosis when compared with mock-reconstituted fibroblasts, NEMO D311N-reconstituted cells showed a significant increase in the level of apoptotic cells. Again, the NEMO E320A/E324A/E327A mutant acted highly similar to NEMO WT and almost completely prevented TNFα-induced apoptosis. In contrast, NEMO R316A/R319A/E320A reconstitution displayed an intermediate phenotype that had less severe effects on the ability of NEMO to prevent apoptosis than the NEMO D311N mutation. Altogether, our data suggest that selective interference with NEMO-linear Ub binding is not sufficient to completely abrogate NF-κB functional responses, which is in line with the differential capacity of the NEMO mutations in triggering NF-κB activation after TNFα stimulation.
DISCUSSION
We found that the entire NEMO C terminus displays in vitro a very high preference for binding to linear over Lys-63 Ub chains. This is consistent with previous data showing that the NEMO UBAN alone represents a selective surface for interaction with linear di-Ub (11, 13, 29). Even though the NEMO ZF was suggested to bind mono-Ub and to enhance interaction with longer Lys-63 Ub chains (14, 15), KD measurements by MST did not confirm that the ZF augments interaction of the UBAN with Lys-63 Ub chains. Nevertheless, in plate-bound assays, Lys-63 Ub can also associate with NEMO, and this interaction critically depends on residue Asp-311, which contacts the distal Ub moiety in either linear or Lys-63 di-Ub (11, 12). We conclude that the binding of NEMO to distinct Ub chains is highly dependent on the type of assay. As we used a truncated NEMO mutant that comprises the UBAN-linker-ZF domains (amino acids 242–419) for all in vitro assays, it is well conceivable that the N terminus may also contribute to the association of distinct Ub chains in the full-length NEMO.
At a first glance, the enhanced binding of Lys-63 Ub to plate-bound NEMO seems to represent an artificial scenario. However, cellular oligomerization of NEMO is critical for NF-κB signaling (30); thus, higher order oligomers of NEMO may promote a more static orientation that could facilitate Lys-63 Ub binding. Interestingly, the crystal structure also revealed that one Lys-63 di-Ub couples to the UBAN of two NEMO dimers that are aligned in parallel (12). In addition, our plate bound analyses suggest that a dual binding of one Lys-63 di-Ub to the distal UBAN regions of two NEMO dimers may enhance the affinity. This may become relevant, as our reconstitutions of MEF and Jurkat T cells clearly indicate that selective destruction of linear Ub binding to the NEMO UBAN is not sufficient to abrogate NF-κB signaling. Thus, in vivo only the N-terminal part of the NEMO UBAN surrounding Asp-311 that contacts the distal moiety of linear and Lys-63 Ub is absolutely essential for triggering activation of canonical NF-κB signaling. Interestingly, three recent reports highlight SHANK-associated RH domain interacting protein as a new member of the linear Ub chain assembly complex, including heme-oxidized IRP2 ubiquitin ligase-1 and HOIL-1 interacting protein, which catalyzes assembly of linear chains to upstream components of the TNF receptor pathway (31–33). These studies show that TNF signaling is highly dependent on the linear ubiquitin chain, but in accordance with our data, the signaling is not completely abolished in SHARPIN knock-out cells.
Overexpression of NEMO has been suggested to inhibit optimal activation of NF-κB (26, 27). By utilizing a tightly controlled lentiviral expression system, we confirmed that overexpression of NEMO is in fact strongly interfering with NF-κB activation and thus a critical issue for analyzing the physiological impact of NEMO mutations. By sorting cell populations that express homogenous NEMO amounts, we can show quantitative differences in the capacity of distinct UBAN mutations to reconstitute NF-κB signaling. By this approach, we were able to demonstrate in two different cell types (fibroblasts and Jurkat T cells) that only the combined interruption of linear and Lys-63 Ub binding to the UBAN abrogates TNFα-mediated NF-κB activation. These differential levels of NF-κB activation have biological consequences as NF-κB-dependent target gene expression and NF-κB-induced antiapoptotic pathways in fibroblasts are also affected. Moreover, we show for the first time that besides Lys-63 Ub, the binding of linear Ub to NEMO UBAN also controls NF-κB signaling in response to T cell activation. Thus, the combined requirement for linear and Lys-63 Ub binding is apparently a common mechanism to integrate specific upstream signaling pathways to the IKK complex.
Our comprehensive analysis suggests that the NEMO UBAN domain is bifunctional, because it binds with low affinity to Lys-63 and with high affinity to linear Ub chains, and both types of chains can contribute to NF-κB activation in response to TNFα or P/I stimulation. Thus, in cells, the NEMO oligomerization and local concentrations of distinct Ub chains will determine the occupancy of the UBAN. It is conceivable that consecutive binding of Lys-63 and linear Ub chains to the UBAN could cooperate in optimal IKK activation. Possibly, low affinity binding to Lys-63 Ub chains mediates NEMO recruitment to the TNFR, where the linear ubiquitin assembly complex increases the local concentrations of linear Ub (10, 34). Within the TNFR complex, an exchange of Lys-63 to linear Ub chain binding to UBAN could be required to promote full IKK activation. Such a cooperative model is in agreement with our observation that selective destruction of linear Ub binding to the NEMO UBAN is not sufficient to completely prevent NF-κB signaling. Recent findings demonstrate that cIAP1/UbcH5 mediate the attachment of Lys-11 Ub chains to RIP1, thereby facilitating the recruitment of NEMO (24). In fact, this is in line with the observation that neither the lack of Lys-63 Ub chains nor linear chains were sufficient to completely abrogate NF-κB signaling (35). Hence, a detailed analysis of the NEMO-K11 Ub binding mode will be necessary to fully understand the role of Lys-11-modified RIP1 in the TNFα receptor pathway. In light of the recent reports regarding the significant role of linear Ub chain assembly complex and linear ubiquitin chains in the TNF pathway (31–33), future studies must further reveal how different types of Ub chains cooperate to activate NF-κB.
Supplementary Material
Acknowledgments
We thank K. Demski for excellent technical assistance. Lentiviral vectors were a kind gift of G. Mostoslavsky and NEMO-deficient Jurkat T cells were kindly provided by S. C. Sun.
This work was funded by Deutsche Forschungsgemeinschaft Priority Program SPP1365 (to D. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- IKK
- IκB kinase
- DELFIA
- dissociation-enhanced lanthanide fluorescence immunoassay
- NEMO
- NF-κB essential modulator
- ZF
- zinc finger
- PGK
- phosphoglycerate kinase
- MST
- microscale thermophoresis
- MEF
- mouse embryonic fibroblast(s)
- P/I
- phorbol 12-myristate 13-acetate/ionomycin
- TBD-NZF
- TAK1 binding domain–Npl4 zinc finger
- Ub
- ubiquitin.
REFERENCES
- 1. Scheidereit C. (2006) Oncogene 25, 6685–6705 [DOI] [PubMed] [Google Scholar]
- 2. Hayden M. S., Ghosh S. (2008) Cell 132, 344–362 [DOI] [PubMed] [Google Scholar]
- 3. Smahi A., Courtois G., Vabres P., Yamaoka S., Heuertz S., Munnich A., Israël A., Heiss N. S., Klauck S. M., Kioschis P., Wiemann S., Poustka A., Esposito T., Bardaro T., Gianfrancesco F., Ciccodicola A., D'Urso M., Woffendin H., Jakins T., Donnai D., Stewart H., Kenwrick S. J., Aradhya S., Yamagata T., Levy M., Lewis R. A., Nelson D. L. (2000) Nature 405, 466–472 [DOI] [PubMed] [Google Scholar]
- 4. Courtois G., Israël A. (2000) Sci. STKE 2000, pe1 [DOI] [PubMed] [Google Scholar]
- 5. Fusco F., Pescatore A., Bal E., Ghoul A., Paciolla M., Lioi M. B., D'Urso M., Rabia S. H., Bodemer C., Bonnefont J. P., Munnich A., Miano M. G., Smahi A., Ursini M. V. (2008) Hum. Mutat. 29, 595–604 [DOI] [PubMed] [Google Scholar]
- 6. Hanson E. P., Monaco-Shawver L., Solt L. A., Madge L. A., Banerjee P. P., May M. J., Orange J. S. (2008) J. Allergy Clin. Immunol. 122, 1169–1177.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ea C. K., Deng L., Xia Z. P., Pineda G., Chen Z. J. (2006) Mol. Cell 22, 245–257 [DOI] [PubMed] [Google Scholar]
- 8. Wu C. J., Conze D. B., Li T., Srinivasula S. M., Ashwell J. D. (2006) Nat. Cell Biol. 8, 398–406 [DOI] [PubMed] [Google Scholar]
- 9. Oeckinghaus A., Wegener E., Welteke V., Ferch U., Arslan S. C., Ruland J., Scheidereit C., Krappmann D. (2007) EMBO. J. 26, 4634–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tokunaga F., Sakata S., Saeki Y., Satomi Y., Kirisako T., Kamei K., Nakagawa T., Kato M., Murata S., Yamaoka S., Yamamoto M., Akira S., Takao T., Tanaka K., Iwai K. (2009) Nat. Cell Biol. 11, 123–132 [DOI] [PubMed] [Google Scholar]
- 11. Rahighi S., Ikeda F., Kawasaki M., Akutsu M., Suzuki N., Kato R., Kensche T., Uejima T., Bloor S., Komander D., Randow F., Wakatsuki S., Dikic I. (2009) Cell 136, 1098–1109 [DOI] [PubMed] [Google Scholar]
- 12. Yoshikawa A., Sato Y., Yamashita M., Mimura H., Yamagata A., Fukai S. (2009) FEBS Lett. 583, 3317–3322 [DOI] [PubMed] [Google Scholar]
- 13. Lo Y. C., Lin S. C., Rospigliosi C. C., Conze D. B., Wu C. J., Ashwell J. D., Eliezer D., Wu H. (2009) Mol. Cell 33, 602–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cordier F., Grubisha O., Traincard F., Véron M., Delepierre M., Agou F. (2009) J. Biol. Chem. 284, 2902–2907 [DOI] [PubMed] [Google Scholar]
- 15. Laplantine E., Fontan E., Chiaravalli J., Lopez T., Lakisic G., Véron M., Agou F., Israël A. (2009) EMBO. J. 28, 2885–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pickart C. M., Raasi S. (2005) Methods Enzymol. 399, 21–36 [DOI] [PubMed] [Google Scholar]
- 17. Wienken C. J., Baaske P., Rothbauer U., Braun D., Duhr S. (2010) Nat. Commun. 1, 100 [DOI] [PubMed] [Google Scholar]
- 18. Mostoslavsky G., Fabian A. J., Rooney S., Alt F. W., Mulligan R. C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16406–16411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mostoslavsky G., Kotton D. N., Fabian A. J., Gray J. T., Lee J. S., Mulligan R. C. (2005) Mol. Ther. 11, 932–940 [DOI] [PubMed] [Google Scholar]
- 20. Kloo B., Nagel D., Pfeifer M., Grau M., Düwel M., Vincendeau M., Dörken B., Lenz P., Lenz G., Krappmann D. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 272–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eitelhuber A. C., Warth S., Schimmack G., Düwel M., Hadian K., Demski K., Beisker W., Shinohara H., Kurosaki T., Heissmeyer V., Krappmann D. (2011) EMBO. J. 30, 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ivins F. J., Montgomery M. G., Smith S. J., Morris-Davies A. C., Taylor I. A., Rittinger K. (2009) Biochem. J. 421, 243–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kulathu Y., Akutsu M., Bremm A., Hofmann K., Komander D. (2009) Nat. Struct. Mol. Biol. 16, 1328–1330 [DOI] [PubMed] [Google Scholar]
- 24. Dynek J. N., Goncharov T., Dueber E. C., Fedorova A. V., Izrael-Tomasevic A., Phu L., Helgason E., Fairbrother W. J., Deshayes K., Kirkpatrick D. S., Vucic D. (2010) EMBO. J. 29, 4198–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grubisha O., Kaminska M., Duquerroy S., Fontan E., Cordier F., Haouz A., Raynal B., Chiaravalli J., Delepierre M., Israël A., Véron M., Agou F. (2010) J. Mol. Biol. 395, 89–104 [DOI] [PubMed] [Google Scholar]
- 26. Krappmann D., Hatada E. N., Tegethoff S., Li J., Klippel A., Giese K., Baeuerle P. A., Scheidereit C. (2000) J. Biol. Chem. 275, 29779–29787 [DOI] [PubMed] [Google Scholar]
- 27. Ye J., Xie X., Tarassishin L., Horwitz M. S. (2000) J. Biol. Chem. 275, 9882–9889 [DOI] [PubMed] [Google Scholar]
- 28. Ben-Dor I., Itsykson P., Goldenberg D., Galun E., Reubinoff B. E. (2006) Mol. Ther. 14, 255–267 [DOI] [PubMed] [Google Scholar]
- 29. Komander D., Reyes-Turcu F., Licchesi J. D., Odenwaelder P., Wilkinson K. D., Barford D. (2009) EMBO. Rep. 10, 466–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tegethoff S., Behlke J., Scheidereit C. (2003) Mol. Cell. Biol. 23, 2029–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gerlach B., Cordier S. M., Schmukle A. C., Emmerich C. H., Rieser E., Haas T. L., Webb A. I., Rickard J. A., Anderton H., Wong W. W., Nachbur U., Gangoda L., Warnken U., Purcell A. W., Silke J., Walczak H. (2011) Nature 471, 591–596 [DOI] [PubMed] [Google Scholar]
- 32. Ikeda F., Deribe Y. L., Skånland S. S., Stieglitz B., Grabbe C., Franz-Wachtel M., van Wijk S. J., Goswami P., Nagy V., Terzic J., Tokunaga F., Androulidaki A., Nakagawa T., Pasparakis M., Iwai K., Sundberg J. P., Schaefer L., Rittinger K., Macek B., Dikic I. (2011) Nature 471, 637–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tokunaga F., Nakagawa T., Nakahara M., Saeki Y., Taniguchi M., Sakata S., Tanaka K., Nakano H., Iwai K. (2011) Nature 471, 633–636 [DOI] [PubMed] [Google Scholar]
- 34. Haas T. L., Emmerich C. H., Gerlach B., Schmukle A. C., Cordier S. M., Rieser E., Feltham R., Vince J., Warnken U., Wenger T., Koschny R., Komander D., Silke J., Walczak H. (2009) Mol Cell 36, 831–844 [DOI] [PubMed] [Google Scholar]
- 35. Xu M., Skaug B., Zeng W., Chen Z. J. (2009) Mol. Cell 36, 302–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.