Abstract
Aberrant up-regulation of P-Rex1 expression plays important roles in cancer progression and metastasis. The present study investigated the regulatory mechanism underlying P-Rex1 gene expression in prostate cancer cells. We showed that P-Rex1 expression was much higher in metastatic prostate cancer cells than in prostate epithelial cells and non-metastatic prostate cancer cells. Histone deacetylase (HDAC) inhibitors or silence of endogenous HDAC1 and HDAC2 markedly elevated P-Rex1 transcription in non-metastatic prostate cancer cells, whereas overexpression of recombinant HDAC1 in metastatic prostate cancer cells suppressed P-Rex1 expression. HDAC inhibitor trichostatin A (TSA) also significantly increased P-Rex1 promoter activity and caused acetylated histones to accumulate and associate with the P-Rex1 promoter. One Sp1 site, essential for basal promoter activity, was identified as critical for the TSA effect. TSA treatment did not alter the DNA-binding activity of Sp1 toward the P-Rex1 promoter; however, it facilitated the dissociation of the repressive HDAC1 and HDAC2 from the Sp1 binding region. Interestingly, HDAC1 association with Sp1 and with the P-Rex1 promoter were much weaker in metastatic prostate cancer PC-3 cells than in non-metastatic prostate cancer cells, and HDAC inhibitors only had very modest stimulatory effects on P-Rex1 promoter activity and P-Rex1 expression in PC-3 cells. Altogether, our studies demonstrate that HDACs could regulate P-Rex1 gene transcription by interaction with Sp1 and by region-specific changes in histone acetylation within the P-Rex1 promoter. Disassociation of HDACs from Sp1 on the P-Rex1 promoter may contribute to aberrant up-regulation of P-Rex1 in cancer.
Keywords: Epigenetics, G-Protein-coupled Receptors, Histone Deacetylase, Receptor Tyrosine Kinase, Sp1, Tumor Metastases, P-Rex1, Prostate Cancer
Introduction
Rac is a member of the Rho family of small G-proteins that are important regulators of the cell cytoskeleton during the establishment of cell polarity and cell processes such as migration, vesicle trafficking, and cell division (1, 2). Hyperactivation of Rac has been found in some cancers and may control tumor cell movement (3, 4). P-Rex13 is a guanine nucleotide exchange factor that specifically activates Rac by catalyzing exchange of GDP for GTP bound to Rac (5). It is among the few Rac-guanine nucleotide exchange factors that are known to be synergistically activated by both receptor tyrosine kinases and G-protein-coupled receptors (5–7). In addition, the mammalian target of rapamycin, which integrates nutrient and growth factor signals (8), also activates P-Rex1 (9). Thus, P-Rex1 simultaneously integrates signals from several input pathways and plays important roles in both normal physiology and pathological conditions. For example, P-Rex1 has been implicated in chemotactic migration of neutrophils (5, 10), neurite differentiation (11), migration of cortical neurons (12), cerebellar long term potentiation (13), prostate cancer metastasis (14), the angiogenic responses of microvascular endothelial cells (15), and breast cancer progression (16).
In humans, P-Rex1 has a very limited distribution. It is expressed in cells of hematopoietic lineage and in neurons (5, 12), suggesting that the P-Rex1 gene is normally repressed in most human tissues. We recently found that the expression levels of P-Rex1 were very low in normal human primary prostate epithelial cells and in non-metastatic prostate cancer cells, whereas levels were highly elevated in metastatic prostate cancer cells (14). Immunohistochemical staining of P-Rex1 protein in human prostate cancer specimens also suggests that P-Rex1 expression levels are an index of metastasis, the major cause of prostate cancer death. Indeed, using a mouse xenograft model, we demonstrated that up-regulated P-Rex1 promotes the spontaneous metastasis of human prostate cancer cells to mouse lymph nodes (14). In addition, P-Rex1 is also overexpressed in estrogen receptor-positive and/or ErbB2-positive breast cancers (16), and increased expression of P-Rex1 correlates with poor patient outcome in breast cancer (17). Thus, it has been suggested that up-regulated P-Rex1 could be a novel therapeutic target in cancers and other pathological conditions (9, 14, 16, 17). Understanding fundamental mechanisms controlling expression of P-Rex1 gene is crucial for introducing effective therapies.
P-Rex1 is located at 20q13.13, a chromosomal region that is often relatively spared from amplifications and somatic mutations in prostate cancer cell lines (18), implying that other mechanisms may be involved in P-Rex1 up-regulation in metastatic prostate cancer cells. Recent evidence has suggested that DNA-change-independent or epigenetic events, such as aberrant histone modifications and DNA methylation, play an important role in transcriptional regulation of a number of target genes, critical for prostate tumor progression (19–22). P-Rex1 expression is normally repressed in most cells, suggesting that epigenetic mechanisms may be responsible for its low levels, whereas a reversal of epigenetic inhibition could contribute to the elevated expression of P-Rex1 during the later phases of prostate cancer progression. However, the exact molecular mechanism for P-Rex1 regulation is entirely unknown. In the present study, we demonstrate for the first time that HDACs have a major role in suppressing Sp1-driven P-Rex1 gene transcription. Disassociation of HDACs from Sp1 on the P-Rex1 promoter may contribute to aberrant up-regulation of P-Rex1 in metastatic prostate cancer cells.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
All cell lines were from the American Type Culture Collection (Manassas, VA). PC-3 cells were cultured in RPMI 1640 medium with 5% fetal bovine serum (FBS). Non-metastatic prostate cancer LNCaP and CWR22Rv1 (22Rv1) cells were grown in RPMI 1640 with 10% FBS, 10 mm HEPES, 4.5 g/liter glucose, and 1.0 mm sodium pyruvate. Immortalized human prostate epithelial RWPE-1 cells were cultured in keratinocyte medium supplemented with 5 ng/ml human recombinant epidermal growth factor and 0.05 mg/ml bovine pituitary extract. HEK293 cells were maintained in DMEM with 10% FBS. Pre-immune rabbit IgG, anti-HDAC1, anti-HDAC2, anti-histone H4, anti-acetyl-H4 (Ac-H4) antibodies, and sodium butyrate (NaB) were from Millipore (Billerica, MA). Sp1 and β-actin antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-hemagglutinin (HA) antibody was from Covance (Princeton, NJ). Sp1 IRdye-labeled oligonucleotide and IRdye700 and IRdye800 secondary antibodies were from LI-COR Biosciences (Lincoln, NE). Lipofectamine LTX and Plus reagent, Lipofectamine 2000, and a Dynabeads® Protein A Immunoprecipitation Kit were from Invitrogen (Carlsbad, CA). ON-TARGETplus SMARTpools targeting human Sp1, HDAC1, and HDAC2 were purchased from Thermo Scientific Dharmacon (Lafayette, CO). P-Rex1 monoclonal antibody 6F12 was a gift from Dr. Marcus Thelen (Institute for Research in Biomedicine, Switzerland). P-Rex1 polyclonal antibody, TSA, 5-aza-2′-deoxycytidine (5-Aza-dC), and mithramycin A (MMA) were from Sigma-Aldrich. Other reagents were from either Sigma-Aldrich or ThermoFisher Scientific (Waltham, MA).
Luciferase Reporter Constructs and HA-tagged Sp1 Construct
All primers are shown in supplemental Table S1. The P-Rex1 promoter fragments (−2024/+3) and (−576/+3) were amplified from human genomic DNA by PCR with BglII- and HindIII-flanked primers and were cloned into the pGL3-Basic luciferase reporter vector (Promega, Madison, WI). Progressive deletion mutants of the P-Rex1 promoter-luciferase construct were created by inverse PCR with promoter-specific primers, using the P-Rex1 promoter luciferase construct (−576/+3) as a template, followed by self-ligation. The P-Rex1 luciferase construct (−190/+3) with mutations in Sp1 sites were also generated by inverse PCR. Nucleotide changes are indicated underlined for each primer. The HA-tagged Sp1 construct was generate by amplifying from the pBS-Sp1 plasmid (Addgene, Cambridge, MA) using the indicated primers and subsequently subcloning into pcDNA3.1-HA vector (Invitrogen) via XhoI/KpnI sites. All constructs were validated by DNA sequencing.
Conventional RT-PCR and Quantitative Real-time RT-PCR
Conventional and real-time RT-PCR analyses were done as described previously (14) for P-Rex1 (28 cycles) and β-actin (22 cycles). The conventional PCR products were separated by 2% agarose gel electrophoresis and confirmed by DNA sequencing analysis. The P-Rex1 and β-actin primers are listed in supplemental Table S1.
RNA Interference
PC-3 cells in suspension were transfected with 50 nm control siRNA (negative control #1 siRNA, Ambion) or siRNAs targeting Sp1 or HDAC1 and HDAC2 using Lipofectamine 2000. Cells were seeded on six-well plates, and 90% of the transfection medium was replaced with fresh culture medium after 5 h of incubation. The following day, adherent PC-3 cells were re-transfected with the same siRNAs. Two days later, cells were harvested for Western blot analysis of Sp1, HDAC1, HDAC2, and P-Rex1 expression.
Western Blot
Protein was extracted from exponentially growing cells using 1× radioimmune precipitation assay lysis buffer (Santa Cruz Biotechnology). Protein samples (40 μg) were loaded on SDS-PAGE, electrophoresed, and transferred to an Immobilon-FL membrane (Millipore). Primary antibodies were used to identify the relevant protein of interest and loading control (β-actin). IRdye700- or IRdye800-labeled secondary antibodies were used for protein band detection. The images were captured with a LI-COR Odyssey infrared imaging system (LI-COR Biosciences) at wavelengths of 700 or 800 nm.
Transient Transfection and Luciferase Assay
To analyze the effect of recombinant HDAC1 on endogenous P-Rex1 expression, PC-3 cells (100-mm dish) were transfected with 10 μg of pEGFP-N1 or 20 μg of HDAC1-GFP plasmids (Addgene) using Lipofectamine LTX with Plus reagent. 18 h post-transfection, the cells were treated with or without 500 nm TSA for 24 h. GFP+ cells were then separated from GFP− cells by fluorescence-activated cell sorting using a FACSAria (BD Immunocytometry Systems) at the Creighton University Flow Cytometry Core Facility. Alteration of P-Rex1 mRNA and protein expression in GFP+ cells was analyzed by quantitative RT-PCR and Western blot analysis, respectively.
For luciferase reporter assays, cells were seeded in 24-well plates, and transfections were performed when the cells were grown to 90% confluence. 22Rv1 cells or HEK293 cells were transfected with 500 ng of pGL3-Basic vector or P-Rex1 promoter reporter constructs with 10 ng of TK-Renilla plasmid (pRL-tk), used to normalize for transfection efficiency. For Sp1 overexpression experiments, HEK293 cells were co-transfected with P-Rex1 promoter reporter constructs (100 ng), pRL-tk (10 ng), and 1000 ng of pcDNA3.1 empty vector or vector encoding HA-tagged Sp1. For HDAC1 overexpression experiments, HEK293 cells were co-transfected with the P-Rex1 promoter reporter construct, pRL-tk, and GFP-tagged HDAC1 or control pEGFP-N1 plasmid. To examine P-Rex1 promoter activities in various cell lines, 100 ng of pRL-tk and 3 μg of the P-Rex1 promoter luciferase construct (−576/+3), cloned from the genomic DNA of RWPE-1 cells, were transfected into RWPE-1, 22Rv1, or PC-3 cells (2 × 106) with a Cell Line Nucleofector Kit V using the Amaxa Nucleofector System (Lonza Inc., Walkersville, MD), and transfected cells were seeded onto 24-well plates. After 24 h of culture, cells were harvested and subjected to luciferase assays. The luciferase activities were measured using the dual luciferase assay kits (Promega) (23). The data presented are averages of at least three independent experiments.
Nuclear Extracts and Electrophoretic Mobility Shift Assays
Nuclear extracts from PC-3 cells were prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents (ThermoFisher Scientific) according to the manufacturer's instructions. EMSA was preformed as previously described (23), using the Sp1 IRdye-labeled oligonucleotide as a probe. Briefly, PC-3 nuclear extract (5 μg) was incubated with the IRdye-labeled double-stranded Sp1 consensus binding motif (50 fmol) in 20-μl solution containing 10 mm Tris, pH 7.5, 50 mm KCl, 1 mm dithiothreitol, 0.25% Tween 20, 1 mm EDTA, and 100 μg/ml poly(deoxyinosinic-deoxycytidylic acid) for 30 min on ice. For competition assays, the competitive oligonucleotide (2.5 pmol) was preincubated with nuclear extracts for 5 min before adding the Sp1 IRdye-labeled oligonucleotide. The protein-DNA complexes were resolved on a 4% non-denaturing polyacrylamide gel containing 2.5% glycerol. Gel imaging was carried out using the Odyssey infrared imaging system at 700 nm.
Chromatin Immunoprecipitation Assay
ChIP assay was carried out using the ChIP-IT express kit (Reactive Motif, Carlsbad, CA) according to the manufacturer's instructions. Briefly, cells (80% confluence) were cross-linked with 1% formaldehyde for 10 min at room temperature. The cell lysates were centrifuged to pellet the nuclei at 5000 rpm for 10 min in 4 °C. DNA was sheared into 200- to 800-bp fragments by sonications, followed by centrifugation to remove debris. The chromatin fraction was incubated for 4 h at 4 °C in ChIP buffer containing protein G magnetic beads and 5 μg of the following antibodies: anti-Sp1, anti-HDAC1, anti-HADC2, anti-Ac-H4, or control rabbit IgG. The chromatin-protein complexes were eluted from magnetic beads, reverse-cross-linked, and then treated with proteinase K at 37 °C for 1 h. The final DNA products were used as PCR templates for amplification using the P-Rex1 proximal promoter-specific primers (supplemental Table S1).
Co-immunoprecipitation of Sp1 and HDAC1
Nuclear extracts of PC-3 and 22Rv1 cells were prepared using a Nuclear Complex Co-IP Kit (Active Motif, Carlsbad, CA) in the presence of phosphatase inhibitors (Sigma) following the manufacture's protocol. Co-immunoprecipitation was performed using a Dynabeads Protein A Immunoprecipitation Kit (Invitrogen). In brief, 3 μg of polyclonal Sp1 antibody or pre-immune rabbit IgG was incubated with 40 μl of Dynabeads Protein A suspension. Cell nuclear extracts were incubated with IgG-Dynabeads or Sp1-Dynabeads for 5 h at 4 °C. Immunoprecipitated complexes were washed four times with immunoprecipitation buffer and eluted in SDS sample buffer and detected by Western blot using monoclonal HDAC1 antibody and polyclonal Sp1 antibody.
Statistical Analysis
Results are the mean ± S.E. of at least three determinations. Statistical comparisons used a Student's t test, or a two-way analysis of variance with the Bonferroni correction or Newman-Keuls test where there were multiple comparisons. A probability (p) value of <0.05 was considered significant.
RESULTS
Stimulation of P-Rex1 Expression by HDAC Inhibitors in Human Prostate Epithelial Cells and Prostate Cancer Cells
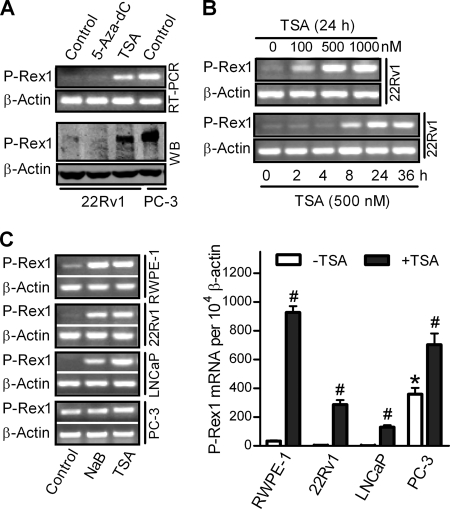
To understand the epigenetic mechanisms underlying regulation of P-Rex1 expression, non-metastatic prostate cancer 22Rv1 cells were treated with a DNA methyltransferase inhibitor 5-Aza-dC or a histone deacetylase inhibitor TSA (24). P-Rex1 mRNA expression (Fig. 1A, top section) increased dramatically after TSA treatment, whereas 5-Aza-dC treatment had no effect. Western blot analysis using a monoclonal P-Rex1 antibody confirmed that P-Rex1 protein was also significantly increased in TSA-treated but not in 5-Aza-dC-treated 22Rv1 cells (Fig. 1A, lower section). In contrast, the expression of the β-actin gene was not affected. As previously reported (14), human metastatic prostate cancer PC-3 cells express high levels of P-Rex1 mRNA and protein (Fig. 1A, lane 4).
FIGURE 1.
Stimulation of P-Rex1 expression by HDAC inhibitors in human prostate epithelial cells and prostate cancer cells. A, 22Rv1 cells were treated without (control) or with 5-Aza-dC (10 μm) for 2 weeks or TSA (500 nm) for 24 h. PC-3 cells, which express higher levels of endogenous P-Rex1, were used as a positive control. P-Rex1 mRNA and protein expression were determined by conventional RT-PCR (top section) and Western blot (WB) analysis using P-Rex1 monoclonal antibody 6F12 (bottom section), respectively. β-Actin was used as an internal control. B, concentration-dependent (top section) and time-dependent (bottom section) stimulation of P-Rex1 mRNA expression in 22Rv1 cells following TSA treatment. C, P-Rex1 mRNA expression in different prostate cell lines treated with HDAC inhibitors (10 mm NaB or 500 nm TSA) or vehicle (Control). Left section, representative images of conventional RT-PCR results; right section, quantitative RT-PCR results. Data shown are means ± S.E. of at least three independent experiments. In the absence of TSA, P-Rex1 expression in PC-3 cells was significantly higher than that in RWPE-1, 22Rv1, and LNCaP cells (*, p < 0.01). #, p < 0.05 indicated that TSA treatment induced a significant increase of P-Rex1 expression.
TSA treatment significantly increased P-Rex1 transcription in 22Rv1 cells in a time-dependent and concentration-dependent manner (Fig. 1B). To further confirm that transcription of P-Rex1 gene was subject to regulation by HDACs through chromatin modifications, NaB, another HDAC inhibitor (25), was also used. In human immortalized prostate epithelial RWPE-1 cells and non-metastatic prostate cancer 22Rv1 and LNCaP cells, both NaB and TSA significantly increased P-Rex1 expression by over 30-fold (Fig. 1C). In contrast, the basal level of P-Rex1 expression in PC-3 cells was significantly higher than that in RWPE-1, 22Rv1, or LNCaP cells, and NaB or TSA only increased P-Rex1 expression by <2-fold in PC-3 cells (Fig. 1C).
HDACs Suppress P-Rex1 Expression in Human Prostate Cancer Cells
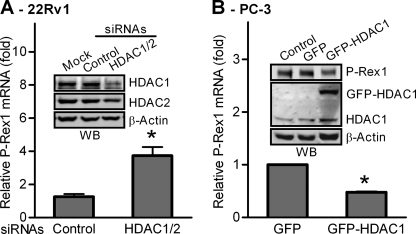
The functions of HDACs, especially HDAC1 and HDAC2, in gene transcriptional regulation have been extensively studied. Thus, we next examined the effects of knocking down endogenous HDAC1 and HDAC2 on P-Rex1 expression in 22Rv1 cells. As shown in Fig. 2A, a combination of siRNAs against HDAC1 and HDAC2 reduced expression of HDAC1 and HDAC2 in 22Rv1 cells by ∼70 and 50%, respectively. Consequently, P-Rex1 gene expression in 22Rv1 cells was significantly increased by ∼4-fold. We further investigated the effect of overexpression of HDAC1 on endogenous P-Rex1 expression in PC-3 cells. As shown in Fig. 2B, the endogenous P-Rex1 mRNA expression in PC-3 cells transfected with GFP-tagged HDAC1 was ∼60% lower than that in cells transfected with GFP control. Western blot analysis confirmed that P-Rex1 protein was also significantly decreased by ∼50% in PC-3 cells expressing GFP-HDAC1 as compared with cells expressing GFP alone (Fig. 2B, inset). Together, our results demonstrate that HDACs such as HDAC1 and HDAC2 act as a negative regulator of the transcription of the P-Rex1 gene.
FIGURE 2.
HDACs inhibit P-Rex1 gene expression in prostate cancer cells. A, silence of HDAC1 and HDAC2 by siRNAs increased P-Rex1 transcription in 22Rv1 cells, determined by quantitative RT-PCR. The P-Rex1 mRNA level in untransfected 22Rv1 cells was set as 1. Data shown are means ± S.E. of three independent experiments with *, p < 0.01 compared with cells transfected with scramble siRNA. Inset, Western blot analysis of HDAC1, HDAC2, and β-actin. B, PC-3 cells were transfected with either GFP or GFP-HDAC1. Alteration of P-Rex1 mRNA expression in GFP+ cells was analyzed by quantitative RT-PCR. The P-Rex1 mRNA level in PC-3 cells transfected with GFP was set as 1. Data are mean ± S.E. of three experiments performed in duplicate. *, p < 0.01 compared with cells transfected with GFP. Inset, Western blot analysis of protein levels of P-Rex1, GFP-HDAC1, endogenous HDAC1, and β-actin in GFP+ cells. Images shown are representative of two independent experiments.
Identification of the Promoter Region Regulating P-Rex1 Expression
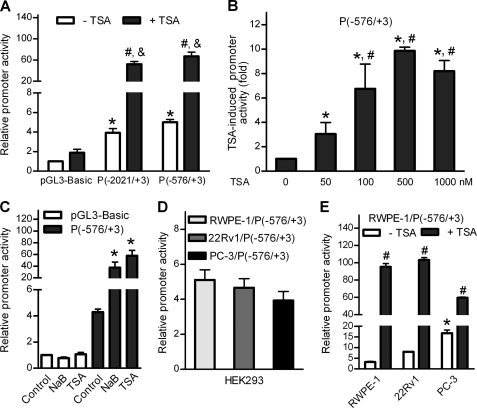
It is generally accepted that HDACs suppress the transcription of a specific gene by binding to its promoter region. To identify the putative promoter that regulates P-Rex1 expression, a 2024-bp DNA fragment upstream of the human P-Rex1 coding region was amplified using genomic DNA of human prostate tissue as a template and was then cloned into the luciferase reporter vector pGL3-Basic, designated as P(−2021/+3) (the “+3” represents the third base from the putative transcription start site). 22Rv1 cells transiently transfected with the P(−2021/+3) or the control pGL3-Basic vector were treated with or without 500 nm TSA. As shown in Fig. 3A, the P(−2021/+3) construct displayed a 4-fold basal activity over the pGL3-Basic vector, and a further 13.8-fold induction after TSA treatment. In contrast, TSA treatment only increased the activity of pGL3-Basic vector by <2-fold. This result demonstrates that the fragment cloned is both transcriptionally active and TSA-inducible. Analysis of this fragment by PromoterScan software identified a putative promoter region within the 579-bp fragment upstream of the P-Rex1 gene. The P(−576/+3) plasmid containing this region displayed similar basal and TSA-induced promoter activities as compared with P(−2021/+3) (Fig. 3A), and the induction of its promoter activity by TSA was concentration-dependent (Fig. 3B). In 22Rv1 cells, NaB also stimulated the P(−576/+3) promoter activity by 9-fold (Fig. 3C).
FIGURE 3.
HDAC inhibitors increased P-Rex1 promoter activity. Promoter activities were measured using the dual luciferase assay kits. Data shown are means ± S.E. of at least three independent experiments. A, identification of a putative proximal promoter of the human P-Rex1 gene. 22Rv1 cells transfected with pGL3-Basic vector or P-Rex1 promoter reporter constructs were treated without or with 500 nm of TSA for 18 h. Relative promoter activities are indicated as -fold increases over pGL3-Basic vector. *, p < 0.001 and #, p < 0.001 compared with pGL3-Basic in the absence and presence of TSA, respectively. &, p < 0.01 compared with the promoter activity in the absence of TSA. B, concentration-dependent induction of promoter activity of the P(−576/+3) construct in 22Rv1 cells by TSA was determined by analysis of variance and a Newman-Keuls multiple comparison test. TSA-induced promoter activities were shown as -fold induction with *, p < 0.05 and #, p < 0.01 compared with that in the absence of TSA or presence of 50 nm TSA, respectively. C, HDAC inhibitors stimulated P-Rex1 promoter activities. 22Rv1 cells transfected with pGL3-Basic vector or the P(−576/+3) construct were treated with 10 mm NaB or 500 nm TSA for 18 h before luciferase assays. *, p < 0.01 compared with untreated cells (control). D, the P(−576/+3) constructs, cloned from the genomic DNA of RWPE-1, 22Rv1, or PC-3 cells, displayed similar promoter activities in HEK293 cells. E, the P(−576/+3) construct, cloned from the genomic DNA of RWPE-1 cells, displayed different basal promoter activities and TSA-stimulation in RWPE-1, 22Rv1, and PC-3 cells. *, p < 0.01 compared with the basal promoter activity in RWPE-1 and 22Rv1 cells. #, p < 0.001 indicated that TSA induced a significant increase of P-Rex1 promoter activity.
Interestingly, the P(−576/+3) constructs cloned from RWPE-1, 22Rv1, or PC-3 cells displayed similar promoter activities in HEK293 cells (Fig. 3D). DNA sequence analysis of the 579-bp fragments cloned from these cell lines confirmed the identical sequences (data not shown). We then compared the promoter activity of P(−576/+3) construct cloned from RWPE-1 cells in various prostate cell lines. The data shown in Fig. 3E indicated that the basal promoter of the P(−576/+3) activity was 5-fold and 2-fold higher in PC-3 cells than that in RWPE-1 cells and 22Rv1 cells, respectively. In contrast, TSA-mediated stimulation of the P(−576/+3) promoter activity was only 3.5-fold in PC-3 cells but 30-fold and 13-fold in RWPE-1 cells and 22Rv1 cells, respectively. Thus, the P-Rex1 promoter has higher basal activity with less TSA response in PC-3 cells than that in RWPE-1 cells or 22Rv1 cells, similar to the endogenous P-Rex1 expression patterns and TSA-response in these cell lines.
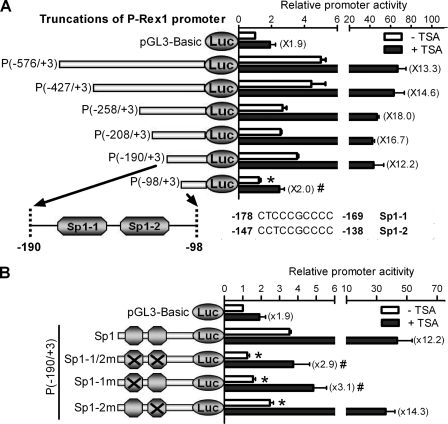
To further determine the cis-element responsible for TSA stimulation within the P-Rex1 promoter, luciferase reporter gene constructs containing the various 5′ flanking regions of the P-Rex1 promoter were transiently transfected into 22Rv1 cells, and their promoter activities were determined in the presence or absence of 500 nm TSA. As shown in Fig. 4A, following progressive deletion, the P(−190/+3) plasmid still retains the basal promoter activity, similar to that of the P(−576/+3) plasmid (3.5- versus 5.0-fold, respectively). Moreover, P(−190/+3) displayed a similar TSA induction compared with P(−576/+3) (12.2- versus 13.3-fold, respectively). However, a further 92-bp deletion of P(−190/+3) resulted in a significant loss of the basal promoter activity (1.2- versus 3.5-fold) and TSA induction (2.0- versus 12.2-fold) (Fig. 4A), suggesting that this deleted 92-bp fragment (−190/−98) contains key elements of the P-Rex1 promoter.
FIGURE 4.
Identification of the Sp1 binding domain as critical for TSA-mediated activation/derepression of P-Rex1 gene transcription. The promoter activity and TSA response of pGL3 vectors containing various lengths of the P-Rex1 promoter (A) or its Sp1 binding site mutants (B) were assayed in 22Rv1 cells. Data shown are means ± S.E. of at least three independent experiments. A, identification of TSA response region in the P-Rex1 promoter. Upper section: serial deletion constructs from the 5′ site were created as shown on the left, and their relative promoter activities are shown on the right. Lower section: schematic diagram depicting the putative Sp1 binding domains in the −190/−98 region of the P-Rex1 promoter. *, p < 0.05 and #, p < 0.01 compared with P(−576/+3) in the absence and presence of TSA, respectively. B, mutation of a putative Sp1 site at −178/−169 caused the significant loss of both basal promoter activity and TSA induction of the P(−190/+3) construct. The two putative Sp1 sites located at the −190/−98 region were double or single mutated as depicted on the left, and relative promoter activities are shown on the right. *, p < 0.05 and #, p < 0.01 compared with the P(−190/+3) construct with wild-type Sp1 sites in the absence and presence of TSA, respectively.
Identification of the Sp1 Binding Domain as Critical for TSA-mediated Activation/Derepression of P-Rex1 Gene Transcription
Using TFSEARCH, a tool for searching transcription factor binding sites, we identified two consensus Sp1 binding domains at −178/−169 bp (Sp1-1) and −147/−138 bp (Sp1-2) in this 92-bp fragment (Fig. 4A, lower section). Simultaneous mutation of these two consensus Sp1 binding domains in the P(−190/+3) construct (Sp1-1/2m) significantly reduced both basal promoter activity and TSA induction by ∼85% (Fig. 4B). Interestingly, mutation of the first Sp1 binding domain (Sp1-1m) had similar inhibitory effects on the basal promoter activity and TSA induction as the mutations of both Sp1 binding domains, whereas mutation of the second Sp1 binding domain (Sp1-2m) only reduced basal promoter activity by ∼25% (p < 0.05) without any significant effect on TSA induction (Fig. 4B). Taken together, these studies demonstrate that the first Sp1 binding domain (−178/−169) is critical for both basal promoter activity and activation/derepression of P-Rex1 gene transcription by the HDAC inhibitor TSA.
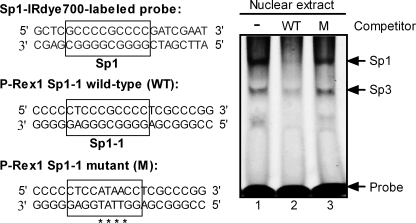
We further investigated whether oligonucleotide containing this putative Sp1 binding domain can attenuate the binding of Sp1 from PC-3 nuclear extracts to the Sp1 IRdye-labeled oligonucleotide. The electrophoretic mobility shift assay (EMSA) results shown in Fig. 5 indicate that transcriptional factors in PC-3 nuclear extracts bind to this Sp1 probe. As previously reported (26, 27), the bands with lower mobility and higher mobility are Sp1 binding and Sp3 binding, respectively (Fig. 5, lane 1). Preincubation with 50-fold excess of non-labeled P-Rex1 Sp1-1 wild-type oligonucleotide almost completely abolished Sp1 binding to its probe and partially reduced the Sp3 binding (Fig. 5, lane 2 versus lane 1). In contrast, a 50-fold excess of non-labeled P-Rex1 Sp1-1 mutant competitor had only slight inhibitory effects on Sp1 or Sp3 binding to the Sp1 IRdye-labeled oligonucleotide (lane 3 versus lane 1). These results further confirm the existence of the Sp1 binding domain within the P-Rex1 promoter.
FIGURE 5.
EMSA demonstrated that P-Rex1 Sp1-1 oligonucleotide (−182/−161) blocked Sp1 binding to the IRdye-labeled Sp1 probe. Left section: sequences of IRdye 700-labeled Sp1 probe, P-Rex1 Sp1-1 wild-type (WT), and its mutant (M) oligonucleotide. The sequences of putative Sp1 binding sites are boxed, and mutations are denoted by asterisks. Right section: EMSA. PC-3 nuclear extracts (5 μg) were preincubated without (lane 1) or with 2.5 pmol of P-Rex1 Sp1-1 WT (lane 2) or its mutant oligonucleotide (lane 3) for 5 min before adding the IRdye 700-labeled Sp1 probe (50 fmol) to the mixture. The image shown is representative of four independent experiments.
Sp1 Dependence of Human P-Rex1 Promoter Activity
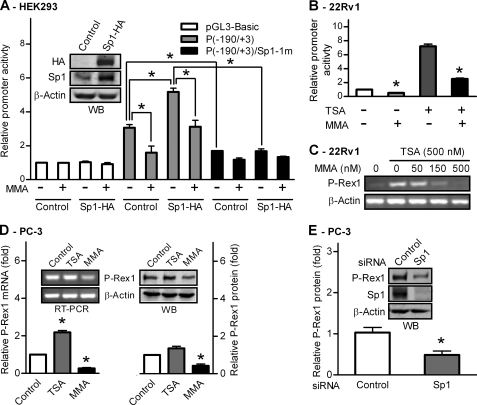
To evaluate the functional importance of Sp1 on human P-Rex1 promoter activity, an HA-tagged human Sp1-expressing plasmid was co-transfected with P(−190/+3) plasmids containing the wild-type Sp1 binding domain or the Sp1-1m mutant, into HEK293 cells. Western blot analysis using anti-HA and anti-Sp1 antibodies confirmed ectopic expression of HA-tagged Sp1 (Sp1-HA) in HEK293 cells (Fig. 6A, inset). Ectopic expression of Sp1-HA enhanced the promoter activity of P(−190/+3) by 1.7-fold, which was significantly reduced by an Sp1-specific inhibitor, MMA (500 nm) (Fig. 6A). In addition, MMA treatment also inhibited the promoter activity of P(−190/+3) by >50% in control cells (Fig. 6A), suggesting endogenous Sp1 in HEK293 cells has a significant role in regulating P-Rex1 promoter activities. Mutation of the first Sp1 site (Sp1-1m) of P(−190/+3) resulted in significant loss of both basal and Sp1-HA-enhanced P-Rex1 promoter activities, and MMA treatment had no additional inhibitory effect. As a control, neither expression of Sp1-HA nor MMA treatment had any significant effect on pGL3-Basic activities (Fig. 6A). Thus, Sp1 has a positive regulatory function on the P-Rex1 promoter, and the Sp1 binding site located at −178/−169 is required for its activation.
FIGURE 6.
Sp1 activity is required for P-Rex1 gene expression in prostate cancer cells. A, Sp1 stimulates P-Rex1 promoter activity. HEK293 cells were transfected with pGL3-Basic vector, P(−190/+3) wild type, or its Sp1-1m mutant along with pcDNA3.1-Sp1-HA or vector control. Cells were treated without or with 500 nm of the Sp1 inhibitor MMA for 24 h, and promoter activities were measured using the dual luciferase assay. *, p < 0.01 indicates a significant difference. Inset: Western blot analysis of ectopic expression of HA-tagged Sp1 using anti-HA or anti-Sp1 antibodies. B, MMA suppresses both basal and TSA-induced promoter activity of the P(−190/+3) construct in 22Rv1 cells. The basal activity of the P(−190/+3) construct in the absence of MMA was set as 1, and data shown are mean ± S.E. of three experiments performed in duplicate. *, p < 0.01 compared with untreated control cells. C, MMA inhibits 500 nm TSA-induced expression of P-Rex1 mRNA in 22Rv1 cells. D, MMA inhibits endogenous P-Rex1 expression in PC-3 cells. Relative P-Rex1 mRNA (left) and protein expression (right) in control PC-3 cells and cells treated with 500 nm of TSA or MMA for 24 h. Inset: representative images of conventional RT-PCR analysis (left) and Western blot analysis (right) of P-Rex1 expression. E, silence of Sp1 by siRNAs suppressed P-Rex1 expression in PC-3 cells. P-Rex1 protein levels in mock transfected cells were set as 1. Data shown are means ± S.E. of three independent experiments with *, p < 0.01 compared with cells transfected with control siRNA. Inset: representative images of WB analysis of P-Rex1, Sp1, and β-actin expression.
Sp1 Activity Is Required for P-Rex1 Gene Expression in Prostate Cancer Cells
We also found that 500 nm MMA treatment suppressed both basal and TSA-induced P-Rex1 promoter activity in 22Rv1 cells by ∼80% (Fig. 6B). Thus, we further investigated whether the transcription factor Sp1 functionally regulates endogenous P-Rex1 expression in prostate cancer cells. As shown in Fig. 6C, TSA stimulation of P-Rex1 expression in 22Rv1 cells was significantly suppressed by MMA treatment in a concentration-dependent manner. Treatment with 500 nm MMA completely abolished TSA-induced P-Rex1 expression in 22Rv1 cells. MMA treatment also blocked NaB-stimulated P-Rex1 promoter activity and endogenous P-Rex1 expression in 22Rv1 (data not shown).
Interestingly, the expression levels of endogenous P-Rex1 mRNA and protein in PC-3 cells were only modestly increased by TSA treatment, but were reduced by >50% following 500 nm MMA treatment, as determined by quantitative RT-PCR (Fig. 6D, left) and Western blot (Fig. 6D, right), respectively. In addition, when endogenous Sp1 expression in PC-3 cells was knocked down by over 90%, endogenous P-Rex1 protein expression was also reduced by ∼50% (Fig. 6E). These results demonstrate that Sp1 plays an important role in up-regulated P-Rex1 gene expression in PC-3 cells.
Association of Sp1 with the P-Rex1 Promoter in both 22Rv1 and PC-3 Cells
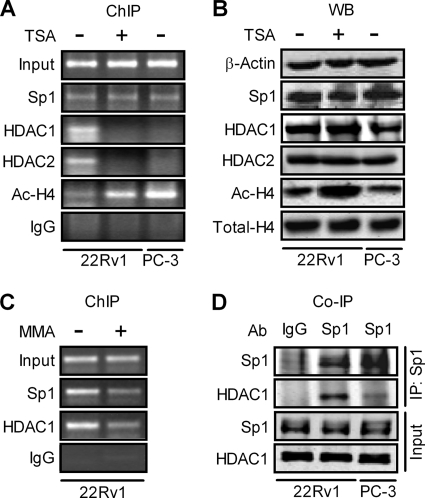
To directly assess whether the Sp1 transcription factor is associated with the proximal region of the P-Rex1 promoter, ChIP assays were carried out using an antibody against Sp1. The genomic DNA that was co-immunoprecipitated with the Sp1 transcription factor was amplified with the specific primers for the P-Rex1 promoter fragment, which harbors the previously identified TSA-responsive Sp1 binding domain (see Fig. 4). As shown in Fig. 7A, Sp1 binding to the P-Rex1 promoter proximal region was comparable between 22Rv1 and PC-3 cells, and was not altered by TSA treatment. In addition, Western blot analysis indicated that the expression level of Sp1 protein in 22Rv1 cells was also similar to that in PC-3 cells and was not affected by TSA treatment (Fig. 7B). These results suggest that up-regulation of P-Rex1 expression in PC-3 cells and TSA induction of P-Rex1 expression in 22Rv1 cells are not due to altered Sp1 levels or accessibility of Sp1 to the P-Rex1 promoter. It should be noted that a negative control IgG failed to immunoprecipitate the P-Rex1 promoter DNA in either 22Rv1 or PC-3 cells (Fig. 7A).
FIGURE 7.
Association of HDAC1 with the P-Rex1 promoter via interaction with Sp1 in 22Rv1 cells but not in PC-3 cells or TSA-treated 22Rv1 cells. A, ChIP analysis of association of Sp1, HDAC1, HDAC2, and Ac-H4 with the P-Rex1 promoter in PC-3 cells and 22Rv1 cells treated with or without 500 nm TSA. Results of amplification of soluble chromatin before immunoprecipitation are shown as Input, whereas normal rabbit IgG was used a negative control. B, whole cell extracts prepared from PC-3 cells and 22Rv1 cells were subjected to Western blot (WB) analyses using antibodies against β-actin, Sp1, HDAC1, HDAC2, Ac-H4, or total H4. C, Sp1 inhibitor MMA abolishes the association of both Sp1 and HDAC1 with the P-Rex1 promoter in 22Rv1 cells. ChIP assays were performed using soluble chromatin prepared from 22Rv1 cells treated without or with 500 nm MMA for 12 h. Images shown are representative of at least three independent experiments. D, determination of Sp1 and HDAC1 interactions. Nuclear extracts prepared from 22Rv1 cells and PC-3 cells were subject to co-immunoprecipitation (Co-IP) assay with rabbit polyclonal Sp1 antibody, followed by Western blot analysis using monoclonal HDAC1 antibody and polyclonal Sp1 antibody (top section). The bottom section shows the image of Western blot analysis of Sp1 and HDAC1 in 10% of input.
Association of HDAC1 and HDAC2 with the P-Rex1 Promoter in 22Rv1 but Not in PC-3 Cells
We next investigated association of HDAC1 and HDAC2 with the P-Rex1 promoter in 22Rv1 cells and PC-3 cells. As shown in Fig. 7A, binding of both HDAC1 and HDAC2 to the P-Rex1 promoter was detected in 22Rv1 cells but was diminished after TSA treatment. In contrast, little association of HDAC1 and HDAC2 with the P-Rex1 promoter was found in PC-3 cells. These results suggest that association of HDAC1 and HDAC2 with the P-Rex1 promoter may be involved in the repression of P-Rex1 gene transcription in 22Rv1 cells. It should be noted that reduced association of HDAC1 and HDAC2 with the P-Rex1 promoter in TSA-treated 22Rv1 cells was not due to TSA-induced degradation of HDACs, because Western blot analysis showed that there were no differences in the levels of HDAC1 and HDAC2 proteins in control and TSA-treated 22Rv1 cells (Fig. 7B).
The HDAC family of proteins suppresses the transcription of a specific gene by binding to its promoter region, leading to the deacetylation of core histones in the chromosomal context (28, 29). Indeed, more Ac-H4 was found to be associated with the P-Rex1 promoter in PC-3 cells than in 22Rv1 cells (Fig. 7A). TSA treatment substantially increased binding of Ac-H4 to the P-Rex1 promoter in 22Rv1 cells (Fig. 7A). Interestingly, Western blot analysis indicated that the total levels of Ac-H4 between 22Rv1 cells and PC-3 cells are comparable even though TSA treatment could remarkably increase levels of Ac-H4 in 22Rv1 cells (Fig. 7B). Total histone H4 protein levels in 22Rv1 cells and PC-3 cells are also comparable, which was not affected by TSA.
Association of HDAC1 With the P-Rex1 Promoter Is Dependent on Sp1
Sp1 was originally identified as a positive transcription factor, but recent studies show that Sp1 can also suppress gene expression, especially as a component of an inhibitory complex that contains HDAC1 (30–32). Thus, we further examined whether the binding of HDAC1 to the P-Rex1 promoter is via the transcription factor Sp1. As shown in Fig. 7C, treatment of 22Rv1 cells with the Sp1-specific inhibitor MMA reduced the association of both Sp1 and HDAC1 with the P-Rex1 promoter. In contrast, TSA treatment only reduced the binding of HDAC1 to the P-Rex1 promoter without effects on Sp1 association with the P-Rex1 promoter (see Fig. 7A). Therefore, these results suggest that Sp1 binding to the P-Rex1 promoter is critical for HDAC1 association with the P-Rex1 promoter.
Association of HDAC1 with Sp1 in PC-3 Cells Was Much Weaker Than That in 22Rv1 Cells
We further examined the potential interaction between Sp1 and HDAC1 in 22Rv1 and PC-3 cells. The nuclear protein extracts derived from 22Rv1 and PC-3 cells were immunoprecipitated with the Sp1-specific antibody and analyzed for the presence of HDAC1 by Western blot using the antibody against HDAC1. As shown in Fig. 7D, HDAC1 was present in the immunoprecipitates from 22Rv1 cells with much less HDAC1 in the immunoprecipitates from PC-3 cells. It should be noted that, in the presence of phosphatase inhibitors, Sp1 appears in double bands, as compared with a single band in the absence of phosphatase inhibitors (see Fig. 7B).
DISCUSSION
In humans, P-Rex1 has a very limited distribution (5, 12), suggesting that the P-Rex1 gene is normally repressed in most human tissues. Up-regulation of P-Rex1 gene expression has been linked with cancer progression and metastasis (14, 16, 17). Thus the one or more mechanisms regulating P-Rex1 gene expression could be a novel therapeutic target in cancers. However, how this gene is controlled has not been elucidated. In the present study, using prostate cells as a model, we have presented the first evidence that histone deacetylation, but not DNA methylation, is involved in the suppression of P-Rex1 expression. More importantly, our studies suggest that disassociation of HDACs from Sp1 on the P-Rex1 promoter may contribute to aberrant up-regulation of P-Rex1 in cancers.
The activation/derepression of endogenous P-Rex1 gene expression in three different cell lines by HDAC inhibitors, in contrast to the unchanged level of the β-actin gene mRNA, indicates that histone deacetylation is a general mechanism underlying repression of P-Rex1 and that the HDAC inhibitor-mediated up-regulation is a gene-selective effect. Inhibition of HDAC activity affects a very limited portion of genes (∼2%) (33), but the P-Rex1 gene may belong to the small group whose expression is highly sensitive to the degree of histone acetylation in chromatin. When compared with non-metastatic prostate cancer 22Rv1 cells, acetylated histone H4 associated with the P-Rex1 promoter was significantly higher in metastatic prostate cancer PC-3 cells that have high basal level of P-Rex1 expression, and HDAC inhibitors only had very modest stimulatory effects (<2-fold) on its P-Rex1 expression. In contrast, HDAC inhibitor significantly increased levels of acetylated histone H4 associated with the P-Rex1 promoter in 22Rv1 cells and markedly increased P-Rex1 gene expression by over 30-fold. Thus, HDACs could repress P-Rex1 gene transcription via deacetylation of histones associated with the P-Rex1 promoter.
Four classes of HDACs with 18 members have been identified (34). The functions of class 1 HDACs, especially HDAC1 and HDAC2, in gene transcriptional regulation have been extensively studied (35–37). In 22Rv1 cells, the P-Rex1 promoter is occupied with HDAC1 and HDAC2, whereas little association of HDAC1 and HDAC2 with the P-Rex1 promoter was found in PC-3 cells. More importantly, after TSA treatment, the binding of HDAC1 and HDAC2 to the P-Rex1 promoter in 22Rv1 cells was significantly decreased, which correlates with the drastic induction of P-Rex1 promoter activity and gene expression. These results highlight the possibility that HDAC1 and HDAC2 may be involved in the histone deacetylation-related repression of P-Rex1 gene expression. Indeed, reduction of endogenous HDAC1 and HDAC2 by siRNAs significantly enhanced P-Rex1 gene expression in 22Rv1 cells, whereas ectopic expression of HDAC1 reduced P-Rex1 gene expression in PC-3 cells.
To identify the regions involved in the HDAC-dependent suppression of P-Rex1 expression, luciferase reporter constructs containing various lengths of the P-Rex1 promoter were generated and promoter activities were analyzed. The P-Rex1 promoter cloned from different prostate cell lines displayed similar promoter activities in HEK293 cells. In contrast, the same P-Rex1 promoter had different basal activity and TSA response in different prostate cell lines, similar to the endogenous P-Rex1 expression patterns and TSA response in these cell lines. These results suggest that the differences in P-Rex1 expression levels among different prostate cell lines are largely due to epigenetic modifications of the P-Rex1 promoter. In addition, a 92-bp minimal region that is essential for both basal and TSA-induced P-Rex1 promoter activity was identified. Results obtained from mutagenesis and EMSA suggest that this 92-bp region contains a Sp1 binding site and may act as a regulatory cis element of P-Rex1 gene transcription. Indeed, ectopic overexpression of Sp1 significantly increased P-Rex1 promoter activity unless this Sp1 binding site was mutated. Conversely, the Sp1 inhibitor MMA attenuated P-Rex1 promoter activity and abolished TSA-induced P-Rex1 expression in 22Rv1 cells. MMA treatment or silence of Sp1 also decreased endogenous P-Rex1 expression in PC-3 cells by 50%. Together, our study identified the minimal promoter regulating P-Rex1 expression and shows that the Sp1 transcription factor is critical for P-Rex1 expression.
Interestingly, this Sp1 binding domain is also responsible for mediating the TSA effect, because mutation of this site totally abolished TSA-induced P-Rex1 promoter activity. We had verified that Sp1 is associated with the P-Rex1 promoter in both 22Rv1 and PC-3 cells. The unchanged binding pattern of Sp1 to the Sp1 binding domain of the P-Rex1 promoter in PC-3 cells and 22Rv1 cells in the presence or absence of TSA indicates that activation of P-Rex1 promoter activity is not related to increased DNA-binding activity of Sp1. In addition, in 22Rv1 cells, treatment with MMA released both Sp1 and HDAC1 from the P-Rex1 promoter, whereas TSA treatment only reduced the HDAC1 occupancy without effect on Sp1 association with the P-Rex1 promoter. These results suggest that Sp1 may be directly bound to the Sp1 site in the P-Rex1 promoter, which is indispensable for HDAC1 occupancy. Our data are therefore consistent with previous studies suggesting that Sp1 could serve as a docking site for HDACs, enabling HDACs to suppress gene transcription through modulating chromatin structure (30–32). Indeed, HDAC1 associates with Sp1 in 22Rv1 cells. Thus, the Sp1 binding domain in the P-Rex1 promoter may also be involved in HDAC-mediated repression of P-Rex1 gene expression in addition to its central contribution to the basal P-Rex1 gene promoter activity. However, HDAC1 association with Sp1 and with the P-Rex1 promoter were barely detected in metastatic prostate cancer PC-3 cells, and HDAC inhibitors had very modest stimulatory effects on P-Rex1 promoter activity and P-Rex1 expression in PC-3 cells. Thus, disassociation of HDAC1 from Sp1 may be one of the mechanisms underlying up-regulation of P-Rex1 expression in metastatic prostate cancer.
Histone acetylation/deacetylation is commonly known to be involved in the regulation of gene transcription through modulating chromatin structure, so it is most likely that low expression of P-Rex1 in 22Rv1 cells is attributable to the condensed and repressive chromatin structure in its promoter region due to histone deacetylation. The higher expression of P-Rex1 in PC-3 cells could possibly be caused by a more open and active chromatin structure due to histone acetylation. Although the total amount of acetylated H4 was similar between PC-3 cells and 22Rv1 cells, the association of acetylated H4 with the P-Rex1 promoter is much higher in PC-3 cells than that in 22Rv1 cells. Thus, up-regulation of P-Rex1 in metastatic prostate cancer cells is not due to a global increase in histone acetylation but rather region-specific changes in histone acetylation within the P-Rex1 promoter. Because histone deacetylation regulates a set of genes that are actively involved in prostate cancer development and progression (19, 21), HDAC inhibitors are considered promising anti-cancer drugs due to their restoration of the expression of silenced tumor suppressor genes. However, our data caution that these inhibitors could have severe side-effects by derepressing genes, such as P-Rex1, that promote tumor progression and metastasis. Thus, further identifying specific signaling pathways involved in the disassociation of HDACs from Sp1 on the P-Rex1 gene promoter may provide an alternative and effective strategy to suppress cancer progression and metastasis.
It should be noted that TSA treatment significantly increased P-Rex1 mRNA expression in 22Rv1 cells, to levels comparable to that in PC-3 cells. However, P-Rex1 protein level in TSA-treated 22Rv1 cells was still much lower than that in PC-3 cells. Thus, although promoter repression or activation is required for P-Rex1 repression or expression, post-transcriptional regulatory events also play critical roles in aberrant up-regulation of P-Rex1 protein in metastatic prostate cancer.
In summary, our study has identified the P-Rex1 gene promoter as a target for regulatory repression of P-Rex1 gene transcription by a Sp1-HDACs complex. P-Rex1 was postulated to function as a molecule that simultaneously integrates signals from several input pathways as the cells rearrange their cytoskeleton and migrate in response to those signals. The finding that P-Rex1 gene expression is subject to HDAC-mediated regulation extends our understanding of the essential role of P-Rex1 in both physiological and pathological situations. More importantly, our data suggest that disassociation of HDACs from Sp1 on the P-Rex1 promoter may contribute to aberrant up-regulation of P-Rex1 in metastatic prostate cancer.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants CA125661 (to Y. T.) and CA88184 (to M. F. L.). This work was also supported by the U.S. Dept. of Defense Prostate Cancer Research Program (Grant W81XWH-07-1-0189 to Y. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- P-Rex1
- phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1
- HDAC
- histone deacetylase
- TSA
- trichostatin A
- MMA
- mithramycin A
- pRL-tk
- TK-Renilla plasmid
- 5-Aza-dC
- 5-aza-2′-deoxycytidine
- NaB
- sodium butyrate.
REFERENCES
- 1. Yamazaki D., Kurisu S., Takenawa T. (2005) Cancer Sci. 96, 379–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H., Borisy G., Parsons J. T., Horwitz A. R. (2003) Science 302, 1704–1709 [DOI] [PubMed] [Google Scholar]
- 3. Knight-Krajewski S., Welsh C. F., Liu Y., Lyons L. S., Faysal J. M., Yang E. S., Burnstein K. L. (2004) Oncogene 23, 5513–5522 [DOI] [PubMed] [Google Scholar]
- 4. Sanz-Moreno V., Gadea G., Ahn J., Paterson H., Marra P., Pinner S., Sahai E., Marshall C. J. (2008) Cell 135, 510–523 [DOI] [PubMed] [Google Scholar]
- 5. Welch H. C., Coadwell W. J., Ellson C. D., Ferguson G. J., Andrews S. R., Erdjument-Bromage H., Tempst P., Hawkins P. T., Stephens L. R. (2002) Cell 108, 809–821 [DOI] [PubMed] [Google Scholar]
- 6. Hill K., Krugmann S., Andrews S. R., Coadwell W. J., Finan P., Welch H. C., Hawkins P. T., Stephens L. R. (2005) J. Biol. Chem. 280, 4166–4173 [DOI] [PubMed] [Google Scholar]
- 7. Barber M. A., Donald S., Thelen S., Anderson K. E., Thelen M., Welch H. C. (2007) J. Biol. Chem. 282, 29967–29976 [DOI] [PubMed] [Google Scholar]
- 8. Fingar D. C., Blenis J. (2004) Oncogene 23, 3151–3171 [DOI] [PubMed] [Google Scholar]
- 9. Hernández-Negrete I., Carretero-Ortega J., Rosenfeldt H., Hernández-García R., Calderón-Salinas J. V., Reyes-Cruz G., Gutkind J. S., Vázquez-Prado J. (2007) J. Biol. Chem. 282, 23708–23715 [DOI] [PubMed] [Google Scholar]
- 10. Zhao T., Nalbant P., Hoshino M., Dong X., Wu D., Bokoch G. M. (2007) J. Leukoc. Biol. 81, 1127–1136 [DOI] [PubMed] [Google Scholar]
- 11. Waters J. E., Astle M. V., Ooms L. M., Balamatsias D., Gurung R., Mitchell C. A. (2008) J. Cell Sci. 121, 2892–2903 [DOI] [PubMed] [Google Scholar]
- 12. Yoshizawa M., Kawauchi T., Sone M., Nishimura Y. V., Terao M., Chihama K., Nabeshima Y., Hoshino M. (2005) J. Neurosci. 25, 4406–4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jackson C., Welch H. C., Bellamy T. C. (2010) PLoS One 5, e11962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qin J., Xie Y., Wang B., Hoshino M., Wolff D. W., Zhao J., Scofield M. A., Dowd F. J., Lin M. F., Tu Y. (2009) Oncogene 28, 1853–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carretero-Ortega J., Walsh C. T., Hernández-García R., Reyes-Cruz G., Brown J. H., Vázquez-Prado J. (2010) Mol. Pharmacol. 77, 435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sosa M. S., Lopez-Haber C., Yang C., Wang H., Lemmon M. A., Busillo J. M., Luo J., Benovic J. L., Klein-Szanto A., Yagi H., Gutkind J. S., Parsons R. E., Kazanietz M. G. (2010) Mol. Cell 40, 877–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Montero J. C., Seoane S., Ocaña A., Pandiella A. (2011) Oncogene 30, 1059–1071 [DOI] [PubMed] [Google Scholar]
- 18. van Bokhoven A., Varella-Garcia M., Korch C., Johannes W. U., Smith E. E., Miller H. L., Nordeen S. K., Miller G. J., Lucia M. S. (2003) Prostate 57, 205–225 [DOI] [PubMed] [Google Scholar]
- 19. Murillo H., Schmidt L. J., Karter M., Hafner K. A., Kondo Y., Ballman K. V., Vasmatzis G., Jenkins R. B., Tindall D. J. (2006) Genes Chromosomes Cancer 45, 702–716 [DOI] [PubMed] [Google Scholar]
- 20. Richiardi L., Fiano V., Vizzini L., De Marco L., Delsedime L., Akre O., Tos A. G., Merletti F. (2009) J. Clin. Oncol. 27, 3161–3168 [DOI] [PubMed] [Google Scholar]
- 21. Nelson W. G., De Marzo A. M., Yegnasubramanian S. (2009) Endocrinology 150, 3991–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wolff D. W., Xie Y., Deng C., Gatalica Z., Yang M., Wang B., Wang J., Lin M. F., Abel P. W., Tu Y. (2011) Int. J. Cancer, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xie Y., Wolff D. W., Lin M. F., Tu Y. (2007) Mol. Pharmacol. 72, 73–85 [DOI] [PubMed] [Google Scholar]
- 24. Yoshida M., Kijima M., Akita M., Beppu T. (1990) J. Biol. Chem. 265, 17174–17179 [PubMed] [Google Scholar]
- 25. Marks P. A., Richon V. M., Rifkind R. A. (2000) J. Natl. Cancer Inst. 92, 1210–1216 [DOI] [PubMed] [Google Scholar]
- 26. Sellak H., Yang X., Cao X., Cornwell T., Soff G. A., Lincoln T. (2002) Circ. Res. 90, 405–412 [DOI] [PubMed] [Google Scholar]
- 27. Camarero N., Nadal A., Barrero M. J., Haro D., Marrero P. F. (2003) Nucleic Acids Res. 31, 1693–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wolffe A. P. (1997) Nature 387, 16–17 [DOI] [PubMed] [Google Scholar]
- 29. Struhl K. (1998) Genes Dev. 12, 599–606 [DOI] [PubMed] [Google Scholar]
- 30. Doetzlhofer A., Rotheneder H., Lagger G., Koranda M., Kurtev V., Brosch G., Wintersberger E., Seiser C. (1999) Mol. Cell. Biol. 19, 5504–5511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bu Y., Gelman I. H. (2007) J. Biol. Chem. 282, 26725–26739 [DOI] [PubMed] [Google Scholar]
- 32. Kang J. E., Kim M. H., Lee J. A., Park H., Min-Nyung L., Auh C. K., Hur M. W. (2005) Cell Physiol. Biochem. 16, 23–30 [DOI] [PubMed] [Google Scholar]
- 33. Van Lint C., Emiliani S., Verdin E. (1996) Gene Expr. 5, 245–253 [PMC free article] [PubMed] [Google Scholar]
- 34. Witt O., Deubzer H. E., Milde T., Oehme I. (2009) Cancer Lett. 277, 8–21 [DOI] [PubMed] [Google Scholar]
- 35. Gui C. Y., Ngo L., Xu W. S., Richon V. M., Marks P. A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 1241–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Biçaku E., Marchion D. C., Schmitt M. L., Münster P. N. (2008) Cancer Res. 68, 1513–1519 [DOI] [PubMed] [Google Scholar]
- 37. Ma P., Schultz R. M. (2008) Dev. Biol. 319, 110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.