Abstract
Clumping factor B (ClfB) from Staphylococcus aureus is a bifunctional protein that binds to human cytokeratin 10 (K10) and fibrinogen (Fg). ClfB has been implicated in S. aureus colonization of nasal epithelium and is therefore a key virulence factor. People colonized with S. aureus are at an increased risk for invasive staphylococcal disease. In this study, we have determined the crystal structures of the ligand-binding region of ClfB in an apo-form and in complex with human K10 and Fg α-chain-derived peptides, respectively. We have determined the structures of MSCRAMM binding to two ligands with different sequences in the same site showing the versatile nature of the ligand recognition mode of microbial surface components recognizing adhesive matrix molecules. Both ligands bind ClfB by parallel β-sheet complementation as observed for the clumping factor A·γ-chain peptide complex. The β-sheet complementation is shorter in the ClfB·Fg α-chain peptide complex. The structures show that several residues in ClfB are important for binding to both ligands, whereas others only make contact with one of the ligands. A common motif GSSGXG found in both ligands is part of the ClfB-binding site. This motif is found in many human proteins thus raising the possibility that ClfB recognizes additional ligands.
Keywords: Bacteria, Cell Surface Protein, Crystal Structure, Extracellular Matrix Proteins, Fibrinogen, MSCRAMM, Cytokeratin
Introduction
The Gram-positive bacterium Staphylococcus aureus is in a commensal relationship with humans and is an important opportunistic pathogen that causes infections in both hospital and community settings (1). It can cause mild superficial skin infections as well as invasive life-threatening infections such as endocarditis and necrotizing pneumonia (2). S. aureus infections are aggravated by the ability of the organism to acquire resistance to antimicrobial drugs (3). Approximately 20% of the human population persistently carry S. aureus in the moist squamous epithelium of their anterior nares, and the remainder of the population can carry the organism intermittently (4). Subjects carrying S. aureus are at increased risk of infection compared with the non-carriers (5).
Understanding the bacterial surface factors that mediate colonization of the nares is of fundamental importance. A number of staphylococcal cell wall-anchored proteins may participate in this process. The surface protein clumping factor B (ClfB)4 and iron-regulated surface determinant protein A (IsdA) promote adhesion of bacteria to desquamated nasal epithelial cells ex vivo (6, 7). This correlates with preclinical in vivo studies in rodents where expression of ClfB and IsdA by S. aureus is associated with increased colonization of the nares (8, 9). Clinically, it has also been demonstrated that ClfB deletion strains are cleared more rapidly from humans compared with isogenic wild type strains (10). The serine aspartate repeat proteins SdrC and SdrD and the S. aureus surface protein SasG also promote bacterial adhesion to squamous cells in vitro (11, 12), but a role for these proteins in colonization of the nares has not yet been reported.
Prevention of nasal carriage by topical administration of the antibiotic mupirocin reduces the risk of infection (13). Active immunization of mice with recombinant ClfB protein or passive immunization with a monoclonal anti-ClfB antibody reduced experimental colonization (9) suggesting that reduction of colonization should be a desirable end point for any human anti-S. aureus vaccination program. Understanding the molecular details of bacterial adhesion to squames and colonization of the nares could lead to novel strategies to reduce carriage and the risk of infection.
ClfB is a member of a family of proteins that are covalently anchored to the cell wall peptidoglycan. Staphylococci can express up to 22 such proteins on their cell surface (14). Some of the surface proteins promote adhesion to components of the extracellular matrix (e.g. collagen, fibrinogen, and fibronectin) during infection and are called microbial surface components recognizing adhesive matrix molecules (MSCRAMMS) (14). Thus, the MSCRAMM, SdrG of Staphylococcus epidermidis, binds to the fibrinogen (Fg) β-chain (15, 16), and ClfA of S. aureus binds to the Fg γ-chain (17, 18). Cna binds to the collagen triple helix (19, 20). Functional redundancy is a common theme with several surface proteins capable of binding to fibrinogen. Also, dual or multiple binding specificities are often observed. The aforementioned ClfB binds to the α-chain of fibrinogen (21) as well as to cytokeratin 10 (22). Fibronectin-binding proteins FnBPA and FnBPB bind to fibrinogen (23) and elastin (24) as well as to the ligand after which they are named, fibronectin (25, 26).
Members of the ClfA-Sdr family of MSCRAMMS have a similar domain organization and protein folding characteristics (Fig. 1A) (14). A signal sequence is located at the N terminus, which is cleaved during protein secretion. The N terminus of the mature surface-exposed protein includes the ligand-binding A region that is composed of the three subdomains N1, N2, and N3. Following the A region are one or two repeated regions. Some members have between 2 and 5 separately folded B repeat units (27), which form rigid rod-like structures. This is followed by the flexible repeats of the di-peptide serine-aspartate. Both repeat segments are believed to facilitate extension of the ligand-binding region from the cell surface. The C terminus includes a cell wall-spanning region, an LPXTG sequence, which is cleaved by the trans-peptidase sortase during covalent anchorage of the protein to the cell wall peptidoglycan (28), a hydrophobic membrane-spanning domain, and positively charged residues at the extreme C terminus. The last two segments are cleaved from the mature protein during secretion and anchoring.
FIGURE 1.
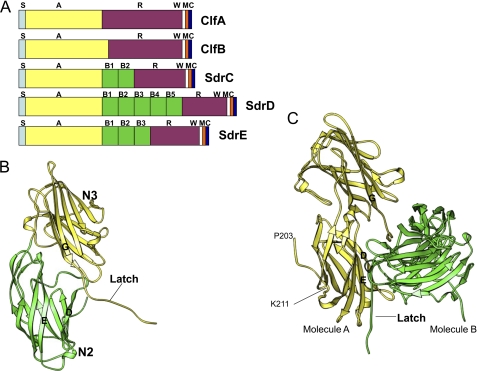
Representation of apoClfBN2N3(199–542). A, domain organization of Clf-Sdr proteins. S, signal peptide; A, ligand binding region; B, homologous repeats; R, SD repeats; W, wall-spanning region; M, membrane anchor; C, cytoplasmic tail. B, ribbon representation of structure of the apo-form of ClfB. The N2 and N3 subdomains are shown in green and yellow, respectively. C, representation of the two symmetry-related molecules in the unit cell. The latch of the molecule A (yellow) is inserted in the latching trench of molecule B (green) and vice versa.
The interaction between the N2N3 subdomains of the A region of SdrG and its ligand has been studied in detail at the submolecular level (16). Crystal structures of the apo-form of SdrG and of a complex of the interacting Fg β-chain peptide bound to SdrG revealed the so-called “dock, lock, latch” mechanism of binding (16). A schematic describing the dock, lock, latch mechanism is shown in supplemental Fig. S1. The Fg β-chain peptide docks into a ligand-binding trench located between subdomains N2 and N3 through an anti-parallel β-strand complementation of the G-strand of N3. Subsequently, the peptide is covered and locked in place by redirection of the C-terminal extension of N3. The C terminus of the lock segment of the N3 extension forms a latch by β-strand complementation with a β-strand exposed in a trench within subdomain N2. A closed structure of the apo-form of the SdrG N2N3 segment, containing an engineered di-sulfide bond to covalently keep the latch in place, failed to bind the ligand peptide (29). Thus, the ligand can only bind to an open form of the apoprotein where the latching sequence is free. More recent analysis (30) showed that ClfA binds the Fg γ-chain peptide by a variation of the dock, lock, latch mechanism whereby the ligand peptide forms a parallel β-strand with the G-strand of the N3 module of the MSCRAMM and slides into the binding trench of a stabilized locked form.
The A domain of ClfB binds to two seemingly disparate ligands, K10 and Fg (21, 31). The binding site within K10 was mapped to the C-terminal Tyr-(Gly-Ser)n Ω loop region (22). One Ω loop YGGGSSGGGSSGGGH segment was studied in detail. The binding site for ClfB in the α-chain of Fg was mapped to repeat 5 of the flexible region of the αC domain (GSWNSGSSGTGSTGNQ), a segment that might form a loop similar to those in the K10 tail region (31).
In this study, we have investigated the structural basis for the interaction of ClfB with synthetic peptides corresponding to the identified binding sites in Fg α-chain and K10, respectively. We show that the N2N3 segment of ClfB folds into two subdomains containing DEv-IgG folds (32), similar to SdrG and ClfA, and that the two ligands bind to the same region of the binding trench. Both ligand peptides form parallel β-strand complementations with a β-strand (strand G) in N3. We show that several residues in ClfB are important for binding to both ligands and identify an interactive motif GSSGXG found in both ligands. However, the MSCRAMM residues engaged by residues in the common motif differ somewhat between the two ligands. In addition, ClfB interacts with several Fg-specific residues outside the common motif suggesting that the identified motif constitutes only part of the ClfB-binding sites.
EXPERIMENTAL PROCEDURES
Bacteria and Growth Conditions
Escherichia coli strains XL-1 Blue and TOPP3 (Stratagene, La Jolla, CA) were used as hosts for cloning and expression, respectively. Cells were grown in LB broth containing 100 μg/ml ampicillin.
DNA Manipulation
DNA restriction enzymes were purchased from New England Biolabs and used according to the manufacturer's instructions. DNA segments encoding the N2N3 domain of ClfB (accession number YP_001333563.1) or its corresponding truncated versions were amplified by PCR using S. aureus Newman chromosomal DNA as template. Enzyme restriction sites BamHI and HindIII were incorporated at the 5′ ends of the primers to facilitate cloning in pQE30 expression plasmid (Qiagen, Chatsworth, CA).
Site-directed mutagenesis was performed using the pQE30 ClfB(212–542) plasmid as template. The introduction of specific mutations was performed using primers listed in Table 1. The resulting PCR mixtures were digested using DpnI for 1 h at 37 °C to eliminate parental DNA and transformed into E. coli XL-1 Blue competent cells. The presence of appropriate DNA sequences was confirmed by Sanger sequencing using specific forward and reverse pQE30 sequencing primers at the Baylor College of Medicine DNA Core Facility, Houston, TX.
TABLE 1.
Oligonucleotide primers used to amplify gene fragments
F is forward and R is reverse.
| ClfB(212–542)_F | 5′cacgggatccggtacaaatgtaaatgataaagt |
| ClfB(212–542)_R | 5′cacgaagcttttaatttactgctgaatcacca |
| ClfB(212–526)_F | 5′cacgggatccggtacaaatgtaaatgataaagtt |
| ClfB(212–526)_R | 5′cacgaagcttttaattctcattattccaaccg |
| ClfB_Q235A_F | 5′aatttcaagttagaaaagactacatttgaccctaatgcaagtggtaacacatttatg |
| ClfB_Q235A_R | 5′cataaatgtgttaccacttgcattagggtcaaatgtagtcttttctaacttgaaatt |
| ClfB_S236Q_F | 5′tagaaaagactacatttgaccctaatcaacagggtaacacatttatggcggcaaatt |
| ClfB_S236Q_R | 5′aatttgccgccataaatgtgttaccctgttgattagggtcaaatgtagtcttttcta |
| ClfB_N238A_F | 5′acatttgaccctaatcaaagtggtgccacatttatggcggcaaattttac |
| ClfB_N238A_R | 5′gtaaaatttgccgccataaatgtggcaccactttgattagggtcaaatgt |
| ClfB_D270E_F | 5′cagatagtttaactggtaatggagaggtggattattctaattcaaataata |
| ClfB_D270E_R | 5′tattatttgaattagaataatccacctctccattaccagttaaactatctg |
| ClfB_F328A_F | 5′agaaaatattaacggacaattttcattacctttagctacagaccgagcaaaggc |
| ClfB_F328A_R | 5′gcctttgctcggtctgtagctaaaggtaatgaaaattgtccgttaatattttct |
| ClfB_Q377A_F | 5′caaatggcgcgaacatttcttctgcaattattggtgtagatacagctt |
| ClfB_Q377A_R | 5′aagctgtatctacaccaataattgcagaagaaatgttcgcgccatttg |
| ClfB_T383A_F | 5′acatttcttctcaaattattggtgtagatgcagcttcaggtcaaaac |
| ClfB_T383A_R | 5′gttttgacctgaagctgcatctacaccaataatttgagaagaaatgt |
| ClfB_W522A_F | 5′caaatagagactacagtattttcggtgcgaataatgagaatgttgtacgttatg |
| ClfB_W522A_R | 5′cataacgtacaacattctcattattcgcaccgaaaatactgtagtctctatttg |
| ClfB_N526Q_F | 5′gtattttcggttggaataatgagcaggttgtacgttatggtggtggaag |
| ClfB_N526Q_R | 5′cttccaccaccataacgtacaacctgctcattattccaaccgaaaatac |
| ClfB_Y530A_F | 5′ggttggaataatgagaatgttgtacgtgctggtggtggaagtg |
| ClfB_Y530A_R | 5′cacttccaccaccagcacgtacaacattctcattattccaac |
Expression and Purification of Recombinant Proteins
Expression and purification of recombinant MSCRAMM proteins were performed as described previously for SdrC (33). Briefly, recombinant proteins were expressed from appropriate plasmids in E. coli TOPP3. Overnight starter cultures were diluted 1:50 in LB-containing ampicillin (100 μg/ml) and incubated with shaking until the culture reached A600 0.6–0.8. Recombinant protein expression was induced by addition of 0.1 mm isopropyl 1-thio-β-d-galactopyranoside (final concentration) and continued during a 4-h incubation. Bacterial cells were harvested by centrifugation, resuspended in PBS, and frozen at −80 °C. Recombinant proteins were purified from cell lysates by Ni2+ affinity and ion exchange chromatography.
Solid Phase Assay
Immulon 4BH plates were coated with 1 μg of Fg (obtained from ChromogenixAB) or recombinant K10 (produced as described previously (22). Coated wells were blocked for 1 h at room temperature with 3% BSA in 0.5% Tween/TBS buffer. Increasing concentrations of ClfB recombinant proteins were added to the wells and incubated for 1 h at room temperature. The wells were washed, and bound ClfB protein was detected with a polyclonal antibody (1:3000) against ClfBN2N3 (H.T.I. BioProducts) followed by an anti-rabbit HRP-labeled antibody (1:5000) (Bio-Rad). HRP-dependent color development using SigmaFast OPD (Sigma) as a substrate was monitored using a microtiter plate reader (Molecular Devices) at 450 nm. Data presented represent the mean ± S.D. of three independent experiments performed in triplicate. The binding was analyzed by nonlinear regression for one binding site (GraphPad Prism). The relative binding was calculated as percent of the wild type protein binding which was considered 100%.
Crystallization
Apo-ClfB(199–542)
Initially, a droplet of 2 μl of rClfBN123 at 9 mg/ml and 2 μl of 0.7 m sodium formate was equilibrated against a reservoir containing 1 ml of 1.44 m sodium citrate. The crystals belong to tetragonal P43212 space group. Electron spray ionization-mass spectrometry measurements on the dissolved crystals revealed that the rClfBN123 (59 kDa) had broken down to a molecular mass of 38 kDa, and the N-terminal sequencing revealed Ala-199 as the N terminus. Subsequently, for the structural and biochemical ligand binding studies, recombinant ClfB(212–542) was used.
ClfB·Ligand Peptide Complexes
Crystals of the ClfB·K10 peptide complex were generated by soaking apo-crystals of ClfB(212–542) in the peptide solution. Apo-crystals of ClfB(212–542) were first grown by mixing 1.5 μl of protein solution with 1 μl of reservoir solution containing 0.9 m sodium citrate, 90 mm imidazole, pH 8.0.
These apo-ClfB(212–542) crystals were soaked in 1 mm of K10 peptide containing the reservoir solution for 5 h at 20 °C. Large chunky apo-ClfB(212–542) crystals cracked after a few minutes of soaking, but a large piece of crystal was used for the x-ray data collection. A similar soaking method was adopted to obtain the co-crystal of 16 residues Fg α-chain peptide with ClfB(212–542).
X-ray Data Collection, Structure Solution, and Refinement
Apo-ClfB(199–542)
Diffraction data to a 2.45-Å resolution was collected on a R-axis IV image plate detector mounted on a Rigaku rotating-anode generator. Data were collected over 120° at a distance of 200 mm with 1° oscillations. Data statistics are reported in Table 2. The crystal structure was solved by the multiple isomorphous replacement method. Four derivatives, trimethyl lead acetate, ethyl mercurithiosalicylate, uranyl acetate, and mersalyl acid were identified by difference Patterson analysis using the program XTALVIEW (34). The trimethyl lead acetate and uranyl acetate had two sites each, and ethyl mercurithiosalicylate and mersalyl acid had one. Refinement of heavy atom derivatives and subsequent phase calculations were performed using the program SOLVE (35). An initial 3.5-Å electron density map clearly defined the solvent and revealed well connected main-chain densities. The initial CA trace was made using the baton mode in O. Side chains were incorporated into the model, and further refinements were done using CNS (36). The initial R-factor/Rfree of the fully built model with side chains was 33.9/41.0%. The model building was performed using O in conjunctions with Oops. Maximum likelihood refinement and bulk solvent correction were done with CNS (36). The final model has 99% of the residues in the allowed region of the Ramachandran plot. Three residues, Val-204, Asn-207, and Thr-213, were in the disallowed region. However, all three with higher temperature factors and weak electron density were in the flexible region of the N terminus of the protein.
TABLE 2.
Crystallographic data measurement and refinement data
| ClfB(199–542) | ClfB((212–542) )·K10 peptide | ClfB((212–542) )·Fg α-chain peptide | |
|---|---|---|---|
| Cell dimensions | |||
| a, b, c | 96.4, 96.4, 84.1 Å | 85.6, 85.6, 84.8 Å | 86.2, 86.2, 83.9 Å |
| α, β, γ | 90° | 90° | 90° |
| Space group | P43 21 2 | P43 | P43 |
| Max resolution | 2.45 Å | 2.60 Å | 2.50 Å |
| Reflections unique | 15,061 | 18,830 | 21,349 |
| Completeness | 98.9% (96.0%) | 99.6% (99.8%) | 99.8% (100%) |
| Rmergea | 0.061 | 0.080 | 0.061 |
| No. of molecules in the asymmetric unit | 1 | 1 | 1 |
| R-factor/Rfreeb | 0.204/0.281 | 0.182/0.219 | 0.178/0.207 |
| Average B value | 31.3 Å | 38.2 Å | 25.8 Å |
| No. of non-hydrogen atoms | 2821 | 2653 | 2733 |
| Protein | 2642 | 2463 | 2463 |
| Peptide | 56 | 66 | |
| Water | 179 | 134 | 204 |
| Root mean square deviations from ideal values | |||
| Bond lengths | 0.006 Å | 0.019 Å | 0.016 Å |
| Bond angles | 1.20° | 1.64° | 1.55° |
a Rmerge = Σ|Ij − 〈I〉|/Σ Ii; where Ij is the measured and 〈I〉 is the mean intensity of reflection hkl.
b +Rfree is calculated over 5% of randomly selected reflections not included in the refinement.
ClfB(212–542)·K10 Peptide and ClfB(212–542)·Fg α-Chain Peptide
Crystals soaked in the peptide solution were flash-frozen with a stabilizing solution containing 25% glycerol. Diffraction data were measured on Rigaku R-Axis IV2+ detector. A total of 120 frames was collected at a detector distance of 150 mm with 1° oscillation for the ClfB·K10 peptide complex and 180 frames for the ClfB·Fg α-chain peptide complex. Data were indexed, integrated, and scaled using HKL2000 (37). The co-crystals of K10 and Fg α-chain peptides diffracted to 2.6 and 2.5 Å resolution, respectively, and the data statistics are listed in Table 2. The structure was solved by molecular replacement with the program PHASER (38) using apo-ClfB as the search model. After obtaining the structure solution of ClfB using molecular replacement, it was subjected to simulated annealing at 1000 K using CNS (36). Electron density maps calculated at this point clearly showed electron density for peptide molecules in the binding trench between the N2 and N3 domains. Model building and refinement were performed using the program COOT (39). After several cycles of refinement using CNS (36), rebuilding and adding water using COOT (39), a few cycles of additional refinement was performed using Refmac5 (40). In the ClfB·K10 peptide complex, electron density was not observed for the N-terminal peptide residues Tyr-473, Gly-474, Gly-475, and Gly-476. Electron density for the side chain of Ser-477 was not observed, but backbone atoms of Ser-477 could be modeled. The numbering refers to residues in the human K10. The C-terminal residues of ClfB from 532 to 542 in the ClfB·K10 peptide complex and residues 529 to 542 in the ClfB·Fg α-chain peptide complex were not modeled due to lack of detected electron density. In the ClfB·Fg α-chain complex, additional density was observed for the three modeled terminal residues 526–528 and were therefore modeled as an alternative conformation. The ClfB·K10 peptide and ClfB·Fg α-chain peptide structures, respectively, were refined to final R-factors of 18.1 and 17.8% and Rfree values of 21.8 and 20.7%. Stereochemical quality of the model was validated using PROCHECK (41), and figures were made using RIBBONS (42) and PyMOL.
RESULTS
Crystal Structure of Apo-ClfBN2N3(199–542) Domain
Crystals of ClfB(199–542) were obtained by the hanging drop method using the conditions described under “Experimental Procedures.” The crystal structure was determined at 2.45 Å resolution by the multiple isomorphous replacement method. The recombinant ClfB(199–542) crystallized in the tetragonal space group P43212 consists of eight asymmetric units. The crystallographic and refinement statistics are presented in Table 2. ClfB(199–542) folded into two domains, each adopting the DE variant of the IgG fold in a similar fashion to ClfA and SdrG (Fig. 1B) (16, 32).
The structure of the N2 and N3 subdomains superposes with each other with a root mean square deviation of 0.95 Å. The inter-domain orientation of the N2-N3 subdomains is similar to that observed in the SdrG apoprotein and in the ClfA·Fg γ-peptide complex (16, 18). During model building, the multiple isomorphous replacement map at 3 Å resolution showed a section of continuous electron density between the D and E strands of the N2 domain that could not be connected to any other part of the molecule. Similarly, an additional electron density present adjacent to the strand G of the N3 domain could not be linked to any part of the structural model. During the final stages of refinement, the two additional densities were identified as extensions of the N and the C termini, respectively, of the symmetry-related molecules. The C terminus of one molecule extends out and interacts with the N2 domain of a crystallographic 2-fold related molecule (Fig. 1C). The structure revealed that the N-terminal residues 199–211 do not belong to the N2 subdomain thus refining the ligand-binding region to residues 212–542 (Fig. 1C). Subsequent studies were performed using recombinant ClfBN2N3(212–542).
Structure of ClfBN2N3(212–542) in Complex with a Cytokeratin 10-derived Peptide
We determined the structure of the ClfB·K10 peptide complex at 2.6 Å resolution by the molecular replacement method using the ClfB apo-structure as the search model. The co-crystal was obtained by soaking newly obtained apo-ClfB(212–542) crystals in a stabilizing solution containing a synthetic peptide corresponding to the identified ClfB-binding site in K10. Similar to SdrG·ligand and ClfA·ligand complexes (16, 18), the K10 peptide binds in the trench formed between the N2 and N3 subdomains of the A region (Fig. 2A). The 15-amino acid peptide corresponds to residues Tyr-473 to His-487 of human K10. The final refined model contained 11 of the 15 amino acids of the peptide ligand. Electron densities were not observed for amino acids Tyr-473, Gly-474, Gly-475, and Gly-476 of the peptide and the C-terminal residues 532–542 of ClfB.
FIGURE 2.
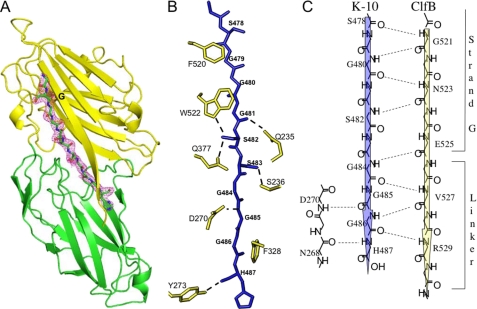
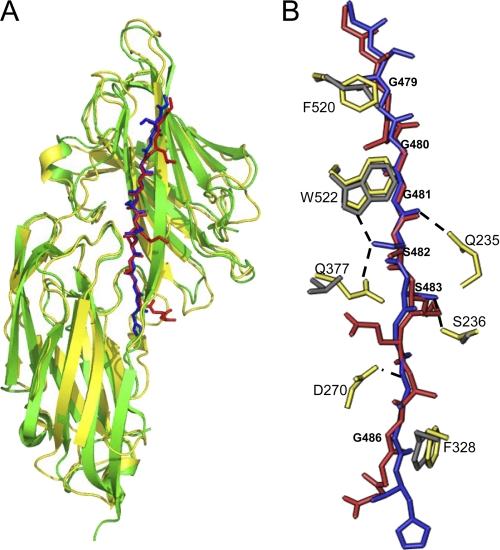
Crystal structure of the ClfBN2N3·K10 peptide complex. A, ribbon representation of the ClfB·K10 peptide complex. The peptide is shown in stick format and is colored by atom type: carbon, green; nitrogen, blue; and oxygen, red. 2Fo − Fc electron density map around the peptide contoured at 1σ is shown in purple. B, representation of the key side-chain interactions of ClfB and the K10 peptide. The K10 peptide is shown in blue stick format. Side-chain atoms of interacting residues in ClfB are shown in yellow. Hydrogen bonds are shown as dotted lines. C, schematic representation of the backbone hydrogen bonding interactions between ClfB and the K10 peptide. Hydrogen bonds are shown as dotted lines. The K10 peptide makes a parallel β-sheet complementation with ClfB.
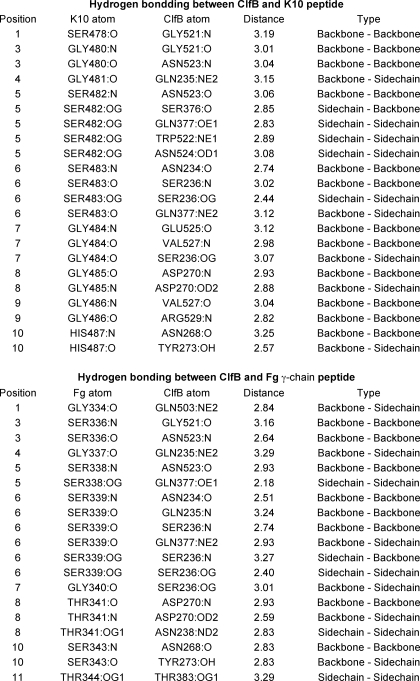
The interaction between the ClfB(212–542) and the K10 peptide buries a total accessible surface area of 1463Å2. Six of the 11 K10 residues that interact with ClfB are glycines anchored by the aromatic residues Phe-520 and Trp-522 in the N3 subdomain as well as Phe-328 in the N2 subdomain (Fig. 2B). The ClfB Phe-520 and Phe-328 anchor Gly-479 and Gly-486, respectively, and both Gly-480 and Gly-481 are anchored by Trp-522. Tryptophan fluorescence experiments performed in an earlier study (22) indicated that Trp-522 and/or Trp-405 were involved in binding to K10. The co-crystal structure confirms that Trp-522, but not Trp-405, is involved in anchoring the K10 peptide. The stretch of residues between Ser-478 and Gly-484 in the K10 peptide are involved in backbone hydrogen bonding interactions (β-sheet complementation) with strand G of the N3 subdomain of ClfB (Fig. 2, A and C, and Table 3). The backbone atoms of Asn-268 and Asp-270 from the N2 domain interact with the backbone of the K10 peptide residues Gly-485 and His-487 (Fig. 2C and Table 3). In addition to the β-sheet complementation, several side-chain to side-chain/main-chain hydrogen bonding interactions are involved in anchoring the ligand peptide. Side chains of Gln-377 and Ser-236 of ClfB are involved in hydrogen bonding interactions with the Oγ atoms of Ser-482 and Ser-483 of the K10 peptide (Fig. 2B). Hydrogen bonding interactions were also observed between Asp-270 and the main-chain nitrogen of Gly-485, Gln-235, and the main-chain carbonyl oxygen of Gly-481 and between Tyr-273 and the main-chain carbonyl oxygen of His-487 (Fig. 2B).
TABLE 3.
Hydrogen bonds between ClfB and ligand in the complexes
Hydrogen bonds were detected by Discovery Studio (Accelrys Inc.) using the criterion distance <3.3 Å between donor and acceptor atoms.
Structure of ClfB in Complex with a Fg α-Chain Peptide and Comparison with the ClfB·K10 Peptide Complex
Similar to the ClfB·K10 peptide complex, the ClfB·Fg α-chain peptide complex was obtained by soaking the apo-ClfB(212–542) crystal in a solution of ligand peptide. Electron densities were not observed for C-terminal residues 529–542 of ClfB. The Fg α-chain peptide bound in the trench formed between the N2 and N3 subdomains of ClfB with the same orientation as that of the K10 peptide (Fig. 3A). ClfB binds to its ligands by making parallel β-sheet interactions with the N3 subdomain. The 16-mer peptide contains residues from Gly-332 to Gln-347 (GSWNSGSSGTGSTGNQ) of the α-chain of human Fg (Fig. 4A). The final refined model contains electron densities only for the last 12 C-terminal peptide residues. Unlike other MSCRAMM·ligand complexes for which the structures have been determined, the N-terminal residues of the Fg α-chain peptide Trp-334, Asn-335, and Ser-336 do not participate in β-sheet complementation with the G-strand of the N3 domain but rather swing away from the G-strand (Fig. 3A). As a result, the β-strand complementation observed in the ClfB·Fg peptide complex is shorter compared with the ClfB·K10 peptide complex. The interaction between the ClfB(212–542) and the Fg α-chain peptide buries a total accessible surface area of 1579Å2.
FIGURE 3.
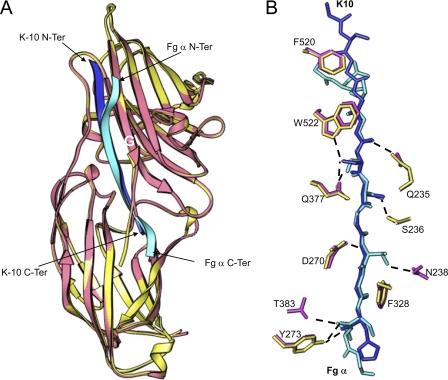
Comparison of ClfB·K10 and ClfB·Fg α-chain peptide. A, ribbon representation of the superimposition of ClfB·K10 and ClfB·Fg α-chain complexes. ClfB from the K10 and Fg α-chain complexes is shown in yellow and pink, respectively. The K10 peptide is shown in blue, and the Fg α-chain is shown in cyan. B, comparison of the key interactions of Fg α-chain and K10 with ClfB. The ClfB molecules of the two complexes were superimposed and are not shown for clarity. K10 peptide is shown in blue and the Fg α-chain peptide is shown in cyan. The side chains of ClfB from the K10 complex are shown in yellow, and those from Fg α-chain complex are shown in pink.
FIGURE 4.
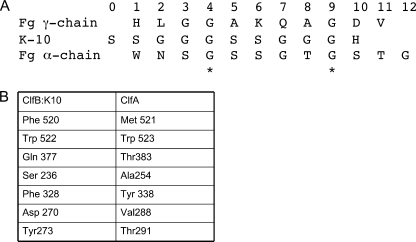
Structure-based alignment of the sequences of ClfB·ligand complexes and the ClfA·Fg γ-chain peptide complex. A, alignment of the amino acid sequences of the ligand molecules based on the structure of their complexes. Each of these amino acid-binding subsites within the binding site were designated 0–12 starting from the N terminus. The ClfB·K10 and ClfB·Fg α-chain peptide complexes and the ClfA·Fg γ-chain peptide complex were superimposed, and the aligned sequences are listed. B, table of the key ligand-binding residues in ClfB and the corresponding residues in ClfA.
An alignment of the structure-based superposition of the ligand sequence of ClfB is shown in Fig. 4. The positions of the residues occupying the binding sites in the MSCRAMMS were designated position 0–12 from the N terminus. Both ligand peptides K10 and Fg have an identical segment (GSSGXG) at positions 4–7 and a Gly in position 9, and these residues largely engage the same residues in ClfB. However, there are significant differences (Figs. 2–5). Gly-337 and Ser-338 at positions 4 and 5 (Fig. 3B and Table 3) in the ClfB-Fg α-chain peptide complex deviate from the corresponding positions in the ClfB·K10 peptide complex due to a kink observed in the N terminus of the Fg α-chain peptide (Figs. 3B and 4). This deviation in the relative positions of the three N-terminal residues of the peptide results in a weaker interaction with the backbone atoms at position 2 of the Fg α-chain peptide (Fig. 3, A and B). In addition, in the ClfB·Fg α-chain peptide complex, Ser-338 at position 5 makes contact only with Gln-377 of ClfB compared with the bifurcated hydrogen bond with both Trp-522 and Gln-377 by Ser-482 of K10 (Fig. 3B and Table 3). Similar to the ClfB·K10 peptide complex, the conserved Gly-343 residue at position 9 of the Fg α-chain peptide (Figs. 4 and 3B) interacts with the hydrophobic side chain of Phe-328. The hydrogen bonds between the residues Asp-270, Gln-377, and Tyr-273 of ClfB with the backbone atoms of the two ligand peptides are also a common feature observed in both the complexes (Fig. 3B and Table 3). In addition to these interactions, Thr residues at positions 8 and 11 interact in unique ways with Asn-238 and Thr-383 in the ClfB·Fg α-chain peptide complex (Fig. 3B and Table 3).
FIGURE 5.
Superimposition of ClfB·K10 and ClfA·Fg γ-chain peptide complexes. A, ribbon representation of the superimposition of the ClfB·10 and ClfA·Fg γ-chain-peptide complexes. ClfA and ClfB molecules are shown in green and yellow, respectively. The K10 peptide is shown in blue, and the Fg γ-chain peptide is shown in red. B, comparison of the key side-chain interactions of the ClfB·K10 peptide and ClfA·Fg γ-chain peptide complexes. The K10 peptide is shown as a blue stick object. The Fg γ-chain peptide from the ClfA·γ-chain complex (18) has been overlaid on the K10 peptide and is shown as a red stick object. Side-chain atoms of interacting residues in ClfB are shown in yellow, and the corresponding residues in ClfA are shown in gray. Hydrogen bonds are shown as dotted lines.
Comparison of MSCRAMM·Ligand Complexes
The N2N3 subdomains of ClfB and ClfA as found in their respective peptide complexes superimpose with a root mean square deviation of 1.8 Å and bind their corresponding peptide ligands in the same orientation (Fig. 5A). In contrast to the S. epidermidis SdrG Fg β-chain peptide, which makes an anti-parallel β-sheet complementation with the N3 subdomain, the ClfA and ClfB ligands adopt a parallel β-sheet complementation. The key difference between ClfA and ClfB is that ClfA binds to the C terminus of the Fg γ-chain, whereas ClfB binds to the Fg α-chain at a site that is flanked by many residues on either side of the binding region (16). Because of the internal location of the MSCRAMM-binding site in Fg, it is likely that ClfB (like SdrG) needs to be in an open conformation for ligand binding This open conformation does not allow the N3 extension to be deposited as a latch in the N2 subdomain. In contrast, both the open and closed forms of ClfA are able to bind Fg (18).
The identified ligands of ClfA and ClfB show some intriguing similarities even though the two MSCRAMMs exhibit only 25% sequence identity and bind different segments in different polypeptides of Fg, and only ClfB binds CK-10. Three Gly residues in K10 and the Fg γ-chain are conserved at positions 3, 4, and 9 and make similar interactions with ClfB and ClfA, respectively. The Gly residues in position 4 and 9 are also conserved in the α-chain peptide bound by ClfB. The corresponding amino acids in the MSCRAMMs that anchor these ligand residues are conserved in ClfA and ClfB. The aromatic amino acid Trp-523 (ClfA) that anchors the conserved Gly ligand residue at positions 3 and 4 is also found in ClfB as Trp-522. Phe-328 in ClfB plays the role of ClfA Tyr-338 in anchoring the conserved Gly at position 9 (Figs. 4 and 5B). The polar residue Ser-482 at position 5 in the K10 peptide is replaced by a hydrophobic residue, Ala-405, in the Fg γ-chain-derived peptide. This amino acid difference is accommodated by Gln-377 (Thr-383 in ClfA), which swings toward the ligand and makes a hydrogen bonding interaction with Ser-482 at position 5 (Figs. 4 and 5B). Similarly, at position 6 the Ser-483 residue in the K10 peptide is replaced by Lys-406 in the Fg γ-chain peptide, and its binding is accommodated by the Ser-236 of ClfB (Figs. 4 and 5B). Comparison of the two structures suggests that a few amino acid variations in the MSCRAMM-binding site could alter the binding specificity of the ligand.
Investigation of ClfB-binding Mechanisms by Analyzing Mutants
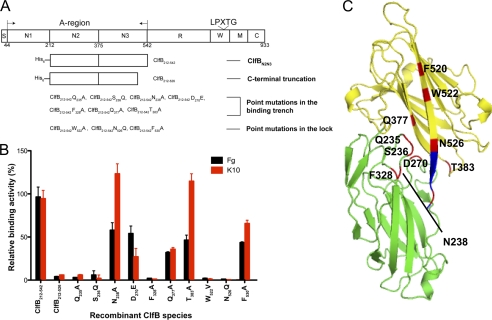
The structure of ClfB(212–542) in complex with synthetic peptides mimicking the MSCRAMM-binding sites in Fg α-chain and K10 showed that both ligands dock in the binding trench formed between the N2 and N3 subdomains. However, the complexes analyzed were obtained by soaking crystals of the apo-form of ClfB(212–542) in solutions containing short peptides from the two ligands. To evaluate if binding of the intact ligand proteins involved the identified interactions with the peptides, we used a mutational approach. Mutated ClfB proteins containing truncations or selected amino acid substitutions were generated, purified, and tested for binding to immobilized intact Fg and K10 in solid phase binding assays. The C-terminal residues following the G-strand of MSCRAMMs makes direct interaction with the ligand and locks the ligand in place. Truncation of amino acid residues 526–542 ablated ClfB binding to both Fg and K10 suggesting that one or more of the residues in positions Asn-526, Val-527, and Val-528 are critical for ClfB interaction with both ligands (Fig. 6, A and B). These residues belong to the Lock region that makes direct interaction with the main-chain atoms of the ligand and therefore are crucial for ligand binding. The N2 and N3 subdomains and the reoriented lock region in the complex almost completely bury the C-terminal region of the ligand peptide. The ClfB Asn-526 residue points inward and lines the binding pocket. Replacing Asn with a polar but bulkier Gln residue would potentially hinder the proper locking event. This hypothesis was tested by substituting Asn-526 in ClfB with Gln, which resulted in a greatly reduced binding of the MSCRAMM to both ligands. In addition, substitution of four other residues that participate in the interaction of ClfB with both ligands (Q235A, S236Q, F328A, and W522V) completely eliminated the binding of ClfB to both Fg and K10, whereas the D270E, Q377A, and F520A substitutions reduced the binding to both ligands by 50%. Amino acid residues Asn-238 and Thr-383 contact only the Fg peptide (Fig. 3B). Substitution of the two residues with Ala (N238A and T383A) results in reduced binding to Fg, whereas these replacements did not affect K10 binding to ClfB (Fig. 6B).
FIGURE 6.
Illustration of ClfB domains and recombinant ClfB proteins affinities for K10 and Fg. A, schematic representation of the domain structure of ClfB. S, signal sequence; A-region, containing N1, N2, and N3; R, serine-aspartic acid repeat region; W, wall-spanning fragment; M, transmembrane domain; C, cytoplasmic tail; LPXTG, cell wall anchoring motif. Also shown are the His-tagged truncated proteins used in this study. B, relative binding affinities of different ClfB recombinant protein species for Fg and K10. Fg and recombinant K10 were immobilized on Immulon 4BH plates, and increasing concentrations of ClfB recombinant protein species were added to the wells. Bound protein was detected with rabbit anti-ClfBN2N3 followed by an anti-rabbit HRP-labeled antibody. The binding constants were analyzed by nonlinear regression for one binding site (GraphPad Prism). The relative binding activity of the mutant recombinant ClfB species were calculated as percent of the wild type protein binding. C, positions of the amino acid mutations carried out for characterization of the ligands binding to ClfB are plotted on the Ribbon model of ClfB. The N2 and the N3 subdomains of ClfB are shown in green and blue, respectively. The lock residues are shown in blue, and the positions of the point mutation are shown in red and labeled.
DISCUSSION
The ability of S. aureus to colonize the host by adhering to desquamated epithelial cells in the nasal vestibule, to bind to plasma proteins, and to adhere to the extracellular matrix during invasive infection is mediated by proteins located on the cell surface. A family of structurally related MSCRAMMs forming the Sdr family is involved in these processes.
The interaction of these proteins with their specific ligands follows a similar pattern. The ligand docks into a binding cleft formed between two IgG-like folded subdomains located in the A region of the MSCRAMM. Known ligands for ClfA and SdrG are locked in place, and the complex is stabilized by a C-terminal extension of the last IgG-like module through direct interaction and a β-complementation process involving a strand in the first module.
Clumping factor B is a bifunctional adhesin, which has a major role in nasal colonization as well as being able to bind Fg and contribute to the disease process. Because nasal carriage is a risk factor for bloodstream infection, the role of ClfB role in nasal colonization may be of particular importance. Expression of ClfB promotes nasal colonization by binding to K10 molecules that are exposed on the surface of desquamated epithelial cells. Specifically, ClfB binds to the repeated Gly/Ser-rich Ω loops found in the C-terminal region of the K10 molecule. Similarly, ClfB binds another Gly/Ser-rich sequence located in the C-terminal part of the Fg α-chain. This study presents the crystal structure of apo-ClfBN2N3 as well as ClfBN2N3 in complex with either the Fg α-chain-derived peptide (GSWNSGSSGTGSTGNQ) or the K10-derived peptide (YGGGSSGGGSSGGGH).
In its apo-form, the ligand-binding domain of ClfB folds into two IgG-like domains like other crystallized members of the family, SdrG and ClfA. In the apoprotein structure, the C-terminal extension of the protein interacts with another ClfB molecule in the crystal (Fig. 1B).
In addition, we determined the crystal structures of ClfBN2N3 in complex with synthetic peptides corresponding to the identified ClfB-binding sites in the Fg α-chain and K10, respectively. The crystals of the ClfB·Fg α-chain peptide and ClfB·K10 peptide diffracted with 2.4 and 2.6 Å, respectively. Although a 15-residue K10-peptide was used, electron densities were observed only for 11 residues. Similarly, only 12 residues of the 16-residue Fg peptide diffracted in the ClfB·Fg peptide complex. Further analysis revealed that both ligand peptides dock in the binding cleft located between the N2 and N3 subdomains, like the previously described SdrG·ligand and ClfA·ligand complexes.
We identified a common motif GSSGXG present in the two ligand peptides that interact with ClfB. This motif is fairly common and is found in many mammalian proteins. Thus, it is possible that ClfB recognizes additional ligands. The proteins dermokine and plakopilin are particularly interesting as potential ligands because they represent extracellular host proteins and could serve as targets for bacterial adhesins mediating attachment to skin or epithelial surfaces. However, preliminary studies indicate that the affinities of ClfB N2N3 for intact Fg or K10 are significantly higher compared with the corresponding binding site peptides (data not shown), suggesting that residues outside the identified ligand-binding motifs in Fg and K10 contribute to the interaction.
To explore if the residues in ClfB shown to interact with the ligand peptides are also important for the binding of the intact protein ligands, we used a mutational strategy. The results of these experiments showed that Gln-235, Trp-522, Ser-236, and Phe-328 in ClfB that are predicted to make contact with the conserved GSSGXG motif in the ligands are indeed critical for MSCRAMM binding to the intact ligand proteins (Fig. 6C). Significantly, all ClfB residues that bind the K10 and/or Fg α-chain peptides are conserved in a large collection of S. aureus strains representing 48 unique protein variants (43). Asn-238 and Thr-383 interact with Fg but not with K10 due to unique residues at the C-terminal region of the Fg ligand peptide. In addition, we identified that the backbone atoms of Gly-521 in the G strand of the ClfB make contact only with the K10-derived peptide due to a kink in the Fg α-chain-derived peptide, which alters the orientation of the N-terminal part of this ligand. Thus residues in ClfB shown in the crystal structure to engage the two ligand peptides are also important in binding the intact ligand proteins.
Supplementary Material
This work was supported in part by a grant from Pfizer Vaccine Research (to V. K. G. and M. H.) and by Science Foundation of Ireland (to T. J. F.). A. S. A., Y. V. M., and S. L. L. are employees of Pfizer. M. H. and T. J. F. are inventors on a United States patent claiming the use of ClfB as an S. aureus vaccine component and licensed to Pfizer through Inhibitex Inc.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
The atomic coordinates and structure factors (codes 3AU0, 3ASW, and 3AT0) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- ClfB
- clumping factor B
- K10
- cytokeratin 10
- MSCRAMMS
- microbial surface components recognizing adhesive matrix molecules
- Fg
- fibrinogen.
REFERENCES
- 1. Diekema D. J., Pfaller M. A., Schmitz F. J., Smayevsky J., Bell J., Jones R. N., Beach M. (2001) Clin. Infect. Dis. 32, S114–S132 [DOI] [PubMed] [Google Scholar]
- 2. Lowy F. D. (1998) N. Engl. J. Med. 339, 520–532 [DOI] [PubMed] [Google Scholar]
- 3. Chambers H. F. (2009) J. Infect. Dis. 199, 291–293 [DOI] [PubMed] [Google Scholar]
- 4. Kluytmans J., van Belkum A., Verbrugh H. (1997) Clin. Microbiol. Rev. 10, 505–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. von Eiff C., Becker K., Machka K., Stammer H., Peters G. (2001) N. Engl. J. Med. 344, 11–16 [DOI] [PubMed] [Google Scholar]
- 6. O'Brien L. M., Walsh E. J., Massey R. C., Peacock S. J., Foster T. J. (2002) Cell. Microbiol. 4, 759–770 [DOI] [PubMed] [Google Scholar]
- 7. Clarke S. R., Brummell K. J., Horsburgh M. J., McDowell P. W., Mohamad S. A., Stapleton M. R., Acevedo J., Read R. C., Day N. P., Peacock S. J., Mond J. J., Kokai-Kun J. F., Foster S. J. (2006) J. Infect. Dis. 193, 1098–1108 [DOI] [PubMed] [Google Scholar]
- 8. Clarke S. R., Wiltshire M. D., Foster S. J. (2004) Mol. Microbiol. 51, 1509–1519 [DOI] [PubMed] [Google Scholar]
- 9. Schaffer A. C., Solinga R. M., Cocchiaro J., Portoles M., Kiser K. B., Risley A., Randall S. M., Valtulina V., Speziale P., Walsh E., Foster T., Lee J. C. (2006) Infect. Immun. 74, 2145–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wertheim H. F., Walsh E., Choudhurry R., Melles D. C., Boelens H. A., Miajlovic H., Verbrugh H. A., Foster T., van Belkum A. (2008) PLoS Med. 5, e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Corrigan R. M., Miajlovic H., Foster T. J. (2009) BMC Microbiol. 9, 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Roche F. M., Meehan M., Foster T. J. (2003) Microbiology 149, 2759–2767 [DOI] [PubMed] [Google Scholar]
- 13. Talon D., Rouget C., Cailleaux V., Bailly P., Thouverez M., Barale F., Michel-Briand Y. (1995) J. Hosp. Infect. 30, 39–49 [DOI] [PubMed] [Google Scholar]
- 14. Foster T. J., Höök M. (1998) Trends Microbiol. 6, 484–488 [DOI] [PubMed] [Google Scholar]
- 15. Davis S. L., Gurusiddappa S., McCrea K. W., Perkins S., Höök M. (2001) J. Biol. Chem. 276, 27799–27805 [DOI] [PubMed] [Google Scholar]
- 16. Ponnuraj K., Bowden M. G., Davis S., Gurusiddappa S., Moore D., Choe D., Xu Y., Hook M., Narayana S. V. (2003) Cell 115, 217–228 [DOI] [PubMed] [Google Scholar]
- 17. McDevitt D., Francois P., Vaudaux P., Foster T. J. (1994) Mol. Microbiol. 11, 237–248 [DOI] [PubMed] [Google Scholar]
- 18. Ganesh V. K., Rivera J. J., Smeds E., Ko Y. P., Bowden M. G., Wann E. R., Gurusiddappa S., Fitzgerald J. R., Höök M. (2008) PLoS Pathog. 4, e1000226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Patti J. M., Jonsson H., Guss B., Switalski L. M., Wiberg K., Lindberg M., Höök M. (1992) J. Biol. Chem. 267, 4766–4772 [PubMed] [Google Scholar]
- 20. Zong Y., Xu Y., Liang X., Keene D. R., Höök A., Gurusiddappa S., Höök M., Narayana S. V. (2005) EMBO J. 24, 4224–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ní Eidhin D., Perkins S., Francois P., Vaudaux P., Höök M., Foster T. J. (1998) Mol. Microbiol. 30, 245–257 [DOI] [PubMed] [Google Scholar]
- 22. Walsh E. J., O'Brien L. M., Liang X., Hook M., Foster T. J. (2004) J. Biol. Chem. 279, 50691–50699 [DOI] [PubMed] [Google Scholar]
- 23. Wann E. R., Gurusiddappa S., Hook M. (2000) J. Biol. Chem. 275, 13863–13871 [DOI] [PubMed] [Google Scholar]
- 24. Roche F. M., Downer R., Keane F., Speziale P., Park P. W., Foster T. J. (2004) J. Biol. Chem. 279, 38433–38440 [DOI] [PubMed] [Google Scholar]
- 25. Flock J. I., Fröman G., Jönsson K., Guss B., Signäs C., Nilsson B., Raucci G., Höök M., Wadström T., Lindberg M. (1987) EMBO J. 6, 2351–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jönsson K., Signäs C., Müller H. P., Lindberg M. (1991) Eur. J. Biochem. 202, 1041–1048 [DOI] [PubMed] [Google Scholar]
- 27. Josefsson E., McCrea K. W., Ní Eidhin D., O'Connell D., Cox J., Höök M., Foster T. J. (1998) Microbiology 144, 3387–3395 [DOI] [PubMed] [Google Scholar]
- 28. Mazmanian S. K., Liu G., Ton-That H., Schneewind O. (1999) Science 285, 760–763 [DOI] [PubMed] [Google Scholar]
- 29. Bowden M. G., Heuck A. P., Ponnuraj K., Kolosova E., Choe D., Gurusiddappa S., Narayana S. V., Johnson A. E., Höök M. (2008) J. Biol. Chem. 283, 638–647 [DOI] [PubMed] [Google Scholar]
- 30. Ganesh V. K., Rivera J. J., Smeds E., Ko Y. P., Bowden M. G., Wann E. R., Gurusiddappa S., Fitzgerald J. R., Höök M. (2008) PLoS Pathog. 4, e1000226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Walsh E. J., Miajlovic H., Gorkun O. V., Foster T. J. (2008) Microbiology 154, 550–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Deivanayagam C. C., Wann E. R., Chen W., Carson M., Rajashankar K. R., Höök M., Narayana S. V. (2002) EMBO J. 21, 6660–6672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Barbu E. M., Ganesh V. K., Gurusiddappa S., Mackenzie R. C., Foster T. J., Sudhof T. C., Höök M. (2010) PLoS Pathog. 6, e1000726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McRee D. E. (1999) J. Struct. Biol. 125, 156–165 [DOI] [PubMed] [Google Scholar]
- 35. Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 37. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 38. McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 39. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 40. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 41. Laskowski R. A., Moss D. S., Thornton J. M. (1993) J. Mol. Biol. 231, 1049–1067 [DOI] [PubMed] [Google Scholar]
- 42. Carson M. J. (1997) J. Mol. Graph. 5, 103–106 [Google Scholar]
- 43. Murphy E., Lin S. L., Nunez L., Andrew L., Fink P. S., Dilts D. A., Hoiseth S. K., Jansen K. U., Anderson A. S. (2011) Hum. Vaccines 7, 51–59 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.