Abstract
Cerebral cavernous malformation (CCM) is a disease that affects between 0.1 and 0.5% of the human population, with mutations in CCM3 accounting for ∼15% of the autosomal dominant form of the disease. We recently reported that CCM3 contains an N-terminal dimerization domain (CCM3D) and a C-terminal focal adhesion targeting (FAT) homology domain. Intermolecular protein-protein interactions of CCM3 are mediated by a highly conserved surface on the FAT homology domain and are affected by CCM3 truncations in the human disease. Here we report the crystal structures of CCM3 in complex with three different leucine-aspartate repeat (LD) motifs (LD1, LD2, and LD4) from the scaffolding protein paxillin, at 2.8, 2.7, and 2.5 Å resolution. We show that CCM3 binds LD motifs using the highly conserved hydrophobic patch 1 (HP1) and that this binding is similar to the binding of focal adhesion kinase and Pyk2 FAT domains to paxillin LD motifs. We further show by surface plasmon resonance that CCM3 binds paxillin LD motifs with affinities in the micromolar range, similar to FAK family FAT domains. Finally, we show that endogenous CCM3 and paxillin co-localize in mouse cerebral pericytes. These studies provide a molecular-level framework to investigate the protein-protein interactions of CCM3.
Keywords: Brain, Crystal Structure, Extracellular Matrix, Protein Domains, Protein Structure, Protein-Protein Interactions, Cerebral Cavernous Malformation, FAT Homology Domain, LD Motif, Vasculature
Introduction
Mutations in three genes, CCM1, CCM2, and CCM3, are implicated in the inherited autosomal dominant form of cerebral cavernous malformation (CCM)2 (1, 2). CCM is a dysplasia that affects the central nervous system, resulting in thin-walled, dilated blood vessels. Complications resulting from CCM include hemorrhagic stroke, seizure, epilepsy, and other focal neurological outcomes (3–5). Approximately 15% of inherited CCM cases are associated with truncations or mutations in CCM3, the protein product of which is cerebral cavernous malformation 3 (CCM3, also called PDCD10 (programmed cell death 10), or TFAR15 (TF-1 cell apoptosis-related protein 15) (6, 7). The function of this protein is not fully described, and its protein binding partners are still being determined; however, both global and endothelial-specific deletions of CCM3 are incompatible with life (8). This protein seems to have a scaffolding function and has been shown to directly interact with CCM2 (9–11), germinal center kinase III family members STK24, STK25, and MST4 (10, 12–17), the STRIPAK complex (18), and paxillin (11) and can associate the cytoplasmic tail of VEGFR2 (8). The atomic-level mechanisms of CCM3 interactions with its binding partners have not been described.
Paxillin is a scaffolding protein important for localization of many proteins to the intracellular side of cell adhesions (19–21). Importantly, the localization of protein kinases, phosphatases, and GTPase regulating proteins (guanine nucleotide exchange factors, GTPase-activating proteins, and effectors) to the actin cytoskeleton and the spatial-temporal dynamics of cellular adhesion are impacted by paxillin (22–24). Examples of proteins that paxillin recruits include FAK, Pyk2, PTP-PEST, CrkII, DOCK180, p210RasGAP, and GIT1/2 (21). The domain architecture of paxillin facilitates this role as a scaffolding protein; the N-terminal region contains multiple phosphorylation sites and five leucine-rich LD (LDXLLXXL) motifs, and the C-terminal region contains four LIM domains (21, 25). The LD motifs have been shown to directly interact with multiple proteins, including FAK and Pyk2 (26–28), vinculin (26–28), GIT1 (29), and α-parvin (30) and to allow regulation of paxillin association by phosphorylation within the LD motif region (21).
LD motifs are named after the first two amino acids of their consensus sequence. These 8-residue α-helical sequences are important sites of protein-protein interaction and are found in the paxillin superfamily (including paxillin, Hic-5, leupaxin, and PaxB) and also in other unrelated proteins such as E6-AP and ERC-55 (21). Multiple proteins are targeted to focal adhesions by interaction with LD motifs, including FAK, Pyk2, GIT1, and the parvins (21). The mechanism of localization of FAK, Pyk2, and GIT1 is by a focal adhesion targeting (FAT) domain. This domain comprises an up-down-up-down four-helix bundle that directly interacts with the LD motifs of paxillin on two diametrically opposite faces, named hydrophobic patches 1 and 2 (HP1 and HP2) (31–35).
In our previous structural study we discovered that CCM3 is an all-α-helical, two-domain protein (11). The N-terminal domain is a homodimerization domain with a novel fold, the CCM3 dimerization fold, and buries a large surface area of ∼3700 Å2. The C-terminal domain is a four-helix bundle that we termed helices αF through αI. Structural similarity between CCM3 and FAT domain-containing proteins was not predicted, as there is poor sequence homology (14 and 10% identity between CCM3 C-terminal domain and the FAT domains of FAK and Pyk2, respectively); however, Dali searches yield Z-scores higher than 14 and r.m.s.d. over 110 residues of less than 2 Å between CCM3 C-terminal domain and the FAT domains of Pyk2 and FAK (11). Interestingly, our sequence conservation analysis of CCM3 revealed that the C-terminal domain contains a stunningly well conserved surface on helices αG and αH (11). The center of this surface is aliphatic and surrounded by multiple conserved lysine residues. The molecular surface displays significant structural similarity to the LD motif binding surfaces of FAK and Pyk2 (11). We, therefore, conducted biochemical and cell biological experiments and showed that CCM3 can directly bind paxillin LD motifs LD1, LD2, and LD4 but not LD3 or LD5 in a specific manner and that point mutations in CCM3 or paxillin could abrogate this interaction (11). These discoveries led us to term the CCM3 C-terminal domain a “FAT homology” domain.
We have now conducted a structural study to investigate the direct interaction of CCM3 with paxillin LD motifs. We have determined the co-crystal structures of CCM3 with paxillin LD motifs LD1, LD2, and LD4 to 2.8, 2.7, and 2.5 Å, respectively. We find that overall CCM3 is capable of binding of LD motif proteins in a manner analogous to that seen for the FAT domain proteins, FAK and Pyk2, and show potentially significant differences in the binding pocket between CCM3 and these other FAT domain-containing proteins. We also find by SPR that CCM3 can bind paxillin LD motifs with affinities in the range observed for FAK, Pyk2, GIT1, and α-parvin. Finally, we show that endogenous CCM3 and paxillin co-localize in mouse cerebral pericytes. We, therefore, provide the first molecular-level description, binding affinity, and endogenous co-localization study of CCM3 interactions with an LD motif-containing protein by the CCM3 FAT homology domain.
EXPERIMENTAL PROCEDURES
Protein Purification and Crystallization
Full-length human CCM3 (Uniprot ID Q9BUL8, residues 1–212) was expressed, purified and crystallized as previously described (11) and the supplemental material.
Peptide Synthesis and Crystal Soaking
Paxillin LD motif-derived peptides were soaked into CCM3 crystals as described in the Supplemental Experimental Procedures. For clarity, paxillin residues specifically discussed in this manuscript are notated with superscript PAX; additionally, FAK and Pyk2 residues are notated with superscripts FAK and Pyk2.
Data Collection and Structure Determination
X-ray diffraction data for the CCM3 in complex with paxillin LD motifs LD1, LD2, and LD4 were collected to 2.8, 2.7, and 2.5 Å resolution, respectively, and structures were determined as described in the Supplemental Experimental Procedures.
LD motif peptides were first modeled into the unbiased positive difference density map as a poly-alanine α-helix using the COOT automatic helix placement function. For LD1 and LD4 peptides, automatic helix placement returned only one peptide direction. Opposite directions for LD1 and LD4 peptides were tested in parallel by refinement of a poly-alanine peptide built in the reverse direction. The reverse direction poly-alanine peptide raised Rfree by more than 1%. For the LD2 peptide complex, we built into the unbiased positive difference density map in two opposite directions in parallel. Refinement of these poly-alanine peptides showed a 0.5% difference in R and Rfree. The correct direction of LD2 was confirmed by careful analysis of electron density maps throughout refinement. The unbiased positive difference maps were consulted throughout refinement. Each crystal contained clear difference density for one LD motif peptide.
The final models for CCM3 complexed with LD motif peptides were validated using MolProbity (36) and Procheck (37). Final refined R and Rfree values for CCM3-LD1 motif were 24.0/29.5% between 50.0 and 2.8 Å, for CCM3-LD2 motif were 23.7/29.1% between 50.0 and 2.7 Å, and CCM3-LD4 motif were 24.1/29.1% between 50.0 and 2.5 Å. Structure analyses were performed using PISA (38), Dali (39), and PDBsum (40). Superpositions were performed using TOPP (41). The refined models have been deposited in the PDB with accession codes 3RQE, 3RQF, and 3RQG.
Surface Plasmon Resonance
CCM3 wild-type proteins were purified as described above. A CCM3 LD motif binding-deficient mutant, CCM3–4KE (quadruple lysine to glutamate mutation K132E,K139E,K172E,K179E) was purified by nickel-nitrilotriacetic acid and size-exclusion chromatography. Human FAK FAT domain (residues Ser-892 to His-1052) was subcloned into pGEX6p-1. Purification was by glutathione-Sepharose affinity and size-exclusion chromatography. GST-LD motif constructs were provided by Christopher Turner and encoded paxillin LD-motifs 1, 2, and 4 (30, 42). GST-LD1, GST-LD2, and GST-LD4 fusion proteins were purified by glutathione-Sepharose affinity and size-exclusion chromatography. All the proteins were further buffer-exchanged by additional size exclusion in the Biacore buffer used for all experiments (10 mm Hepes, pH 7.5, 150 mm NaCl, 3 mm EDTA). Binding studies were performed at 25 °C using a Biacore T100 optical biosensor equipped with a CM5 sensor chip coated with anti-GST antibodies from GST Capture kit (Biacore BR-1002-23). Fusion proteins GST-LD1, GST-LD2, and GST-LD4 were captured on the anti-GST antibody surface. Recombinant GST was captured on the reference cell. Analytes were delivered over the four cells from two- and three-fold dilution series in a range of concentrations. Binding of CCM3 proteins was tested in a concentration range of 0.26–50 μm for CCM3 wild-type protein, 2.5–200 μm for CCM3–4KE mutant, and 0.51–32.5 μm for the FAK FAT domain. The lowest concentration was duplicated in experiments with CCM3–4KE mutant and FAK FAT domain; for CCM3, wild-type protein multiple duplicates were included within each single experiment. We attempted to measure the affinity of CCM3 FAT homology domain alone for GST-LD motifs but found that nonspecific binding to the antibody prevented analysis. Sensorgrams were double-referenced by subtracting binding to the GST control surface and correcting for buffer injections. Data were analyzed using the Biacore T100 Evaluation software.
Isolation of Mouse Cerebral Pericytes
Mouse cerebral pericytes were isolated as described in the Supplemental Experimental Procedures.
Transfection of CCM3 siRNA
CCM3 siRNA knockdown in pericyte was performed as described previously for vascular endothelial cells (8). CCM3 siRNA (Ambion) was resolved to the concentration of 20 μm. 2 μl of CCM3 siRNA and 8 μl of Oligofectamine were mixed in Opti-MEM I (Invitrogen) and were incubated for 30 min at room temperature. The siRNA-Oligofectamine mixture were then added to 90% confluence pericyte culture in a 6-well plate and incubated in a 37 °C 95% O2, 5% CO2 incubator for 12 h. pericytes were then replaced with the regular culture medium and incubated for additional 36 h before harvest for immunostaining.
Indirect Immunofluorescence Confocal Microscopy
Fixation, permeabilization, and staining of cultured endothelial cell were performed as described previously (8). The primary antibodies were used as follows. Rabbit polyclonal anti-CCM3 was generated from our laboratory; total paxillin was from BD Transduction Laboratories (mouse). Alexa Fluor 488 (green) and 594 (red) conjugated-secondary antibodies (Molecular Probes, Eugene, OR) were used. Confocal immunofluorescence microscopy was performed using an Olympus confocal microscope (La Jolla, CA), and the acquired images were transferred to Photoshop 6.0 (Adobe Systems, San Jose, CA) to generate the final figures.
RESULTS
Overall Structure of CCM3 in Complex with Paxillin LD Motifs LD1, LD2, and LD4
We previously reported the crystal structure of CCM3 in two different crystal forms: a tetragonal crystal form in space group P4122 with one CCM3 chain dimerizing with a crystallographic symmetry mate, and an orthorhombic crystal form in space group P212121 with two CCM3 dimers per asymmetric unit. We revealed that CCM3 is a multidomain protein composed of an N-terminal CCM3 dimerization domain and C-terminal FAT homology domain. Our structural analysis discovered that the CCM3 FAT homology domain contains an exquisitely conserved surface analogous to the HP1 of FAK and Pyk2. We, therefore, conducted structure-directed biochemical and cell biological analysis to investigate whether CCM3 can specifically bind LD motif-containing proteins in a manner similar to FAK and Pyk2. Our analyses found that CCM3 can bind to the well studied cytoskeleton protein paxillin, which contains five LD motifs, and that this binding was specific to LD1, LD2, and LD4. We, therefore, proposed that CCM3 binds LD motifs in a manner similar to FAT domain proteins. Here we show that CCM3 is indeed capable of binding LD motifs in a FAK-analogous fashion and provide a molecular level description, affinities of the interaction, and evidence of endogenous co-localization.
In the previously determined CCM3 crystal structures, the HP1 binding pocket is located on the outer face of the CCM3 dimer and is solvent-exposed, suggesting that these crystal forms may be amenable to peptide soaking. We, therefore, conducted peptide soaking experiments of the orthorhombic CCM3 crystal form using paxillin-derived peptides for LD motifs LD1, LD2, and LD4. We successfully obtained co-crystals of CCM3 with each of these LD motifs by soaking peptide into unliganded CCM3 crystals. We determined the complex structures of CCM3 with LD1 to 2.8 Å, CCM3 with LD2 to 2.7 Å, and CCM3 with LD4 to 2.5 Å (Fig. 1; Table 1).
FIGURE 1.
Overall structure of CCM3 in complex with paxillin LD motifs. A, a schematic diagram shows one CCM3 dimer in complex with paxillin LD4 motif. For one monomer of CCM3 the molecular surface is shown; for the other, α-helices are shown in green as cylinders and labeled. The bound LD4 motif is colored salmon. The open and closed ends of the CCM3 FAT homology domain and the N and C termini of CCM3 are indicated. B, shown is a close-up of LD4 motif binding to CCM3 FAT homology domain. The surface of CCM3 is colored by conservation as per Li et al. (11). Dark blue indicates complete conservation, gray indicates poor conservation. LD motifs bind CCM3 in the extremely well conserved HP1 site. Paxillin LD4 is colored salmon. C, structure-based sequence alignment of the CCM3 FAT homology domain with the FAT domains of FAK and Pyk2 is shown. Pyk2 residues 868–1009 (SwissProt Q14289), FAK residues 912–1052 (SwissProt Q05397), and CCM3 residues, 98–212 (SwissProt Q9BUL8) are shown. Green or yellow boxes indicate the extent of α-helices. CCM3 residues that contact LD motifs are shown in red. CCM3 residues discussed, Ala-135 and Lys-172, are boxed in red. Residue numbers for each protein are indicated. All structural figures made using CCM4MG (41).
TABLE 1.
Diffraction data collection and refinement statistics for CCM3 in complex with paxillin LD motifs LD1, LD2, and LD4
Values in parentheses are for the highest resolution shell. A single crystal was used for each dataset.
| CCM3-LD1 | CCM3-LD2 | CCM3-LD4 | |
|---|---|---|---|
| Data collection | |||
| Space group | P212121 | P212121 | P212121 |
| X-ray source | NSLS X29 | NSLS X6A | NSLS X6A |
| Wavelength (Å) | 1.0750 | 1.0781 | 1.0781 |
| Cell | |||
| a, b, c (Å) | 63.2, 118.0, 123.7 | 62.9, 115.5, 124.4 | 63.0, 116.2, 124.6 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution range (Å) | 50.0-2.8 (2.9-2.8) | 50.0-2.7 (2.8-2.7) | 50.0-2.5 (2.59-2.50) |
| No. of unique reflections | 23,488 | 25,432 | 32,672 |
| Completeness (%) | 99.9 (100) | 100 (100) | 100 (100) |
| Rsym (%) | 10.4 (86.0) | 11.1 (82.0) | 11.1 (98.4) |
| Mn2+I/σ(I) | 17.6 (2.5) | 18.4 (2.3) | 18.9 (2.4) |
| Redundancy | 7.2 (7.4) | 8.2 (7.3) | 8.3 (8.2) |
| Wilson B-factor (Å2) | 81.5 | 78.5 | 70.2 |
| Refinement statistics | |||
| Resolution range (Å) | 50.0-2.8 (2.80-2.87) | 50.0-2.7 (2.70-2.77) | 50.0-2.5 (2.50-2.56) |
| R-factor (%) | |||
| Working set | 24.0 (28.8) | 23.7 (32.2) | 24.1 (29.1) |
| Test set | 29.5 (36.2) | 29.1 (38.0) | 29.1 (35.6) |
| Free R reflections (%) | 5.1 (5.0) | 5.1 (4.8) | 5.0 (4.2) |
| Free R reflections, no. | 1192 (82) | 1292 (81) | 1617 (95) |
| Residues built | A: 15–90, 97–210 | A: 16–212 | A: 16–212 |
| B: 13–152, 157–208 | B: 10–152, 157–210 | B: 10–210 | |
| C: −1–148, 158–210 | C: −1–87, 97–152, 158–209 | C: −1–210 | |
| D: 12–91, 97–152, 158–208 | D: 9–87, 97–209 | D: 8–210 | |
| LD1: 5–14 | LD2: 143–153 | LD4: 262–273 | |
| Non-H atoms; protein/LD motif | 6279/72 | 6379/91 | 6648/94 |
| No. water molecules | 0 | 4 | 15 |
| Mean B-factor (Å2) | 86/60/101/194 | 85/63/98/143 | 84/59/98/138 |
| CCM3/CCM3D/FAT-homology/LD peptide | |||
| Model statistics | |||
| r.m.s.d. bond lengths (Å) | 0.011 | 0.006 | 0.007 |
| r.m.s.d. bond angles (°) | 1.316 | 0.897 | 0.985 |
| Ramachandran plot (%), favored/allowed/disallowed | 91.7/8.3/0 | 94.5/5.5/0 | 92.0/8.0/0 |
To avoid model bias, the models were built and refined to convergence without the addition of the LD motif peptides. We found that one of the HP1 binding pockets in each crystal was occupied by unattributed continuous electron density that was clearly α-helical, the expected secondary structure of these LD motif peptides. We then used an unbiased automated placement routine in the program COOT to place a poly-alanine α-helix in each of these positive difference densities and discovered a predominant direction in two of the three LD motif peptides built. Careful analysis of the electron density and refined R-factors for each of these helices allowed us to accurately build the specific LD motif. We confirmed the direction of each peptide by analysis of poly-alanine models built in parallel in the opposite direction and found lower Rfree values for the final built orientation by between 0.5 and 1%. Detailed description of model building is provided under “Experimental Procedures.” Each of the final built LD motif helices show the hydrophobic stripe to be oriented into the HP1 hydrophobic cleft of the CCM3 FAT homology domain and allow salt-bridge formation between the acidic residues of the LD motif and the highly conserved lysine residues on the edge of HP1 (Fig. 1B). This mode of interaction is highly similar to that seen previously for the LD motif binding FAT domains of FAK and Pyk2 (Fig. 1C).
In the orthorhombic crystal form of CCM3, four protein chains form two dimers per asymmetric unit. In this crystal form the HP1 pocket of chains A, C, and D are well exposed to solvent. For the co-crystal structure of CCM3 in complex with paxillin LD1 motif, we built one 10-residue LD1 motif (human paxillin residues 5–14, DALLADLEST) bound in the HP1 site of chain C (Fig. 2, A and B, and supplemental Fig. 1) and observed continuous density around the HP1 pocket of chain D and scattered unattributed density in the HP1 pocket of chain A that we could not build. For the co-crystal structure of CCM3 in complex with paxillin LD2 motif, we built 11 residues (human paxillin residues 143–153, SELDRLLLELN) bound to HP1 of chain A (Fig. 2, A and B, and supplemental Fig. 1). We also observed other unattributed densities in the HP1 pocket of chains C and D but were unable to build LD2 into this density. For the co-crystal structure of CCM3 in complex with paxillin LD4 motif, we built one paxillin/LD4 motif of 12 residues (human paxillin residues 262–273, ATRELDELMASL) bound to HP1 of chain C (Fig. 2, A and B and supplemental Fig. 1) and also observed unbuildable continuous electron density in the HP1 pocket of chain A. We hypothesize that the microenvironment of the different binding pockets, perhaps mediated by interactions on the dorsal side of the LD peptide chain, can account for the differences in quality of electron density bound to each chain. Interestingly, partial occupancy of FAT domain HP1 sites has previously been seen crystallographically for FAK (31). Overall, we built a single chain each of LD1, LD2, and LD4 bound to the HP1 binding site of CCM3 FAT homology domain.
FIGURE 2.
Comparison of LD motif binding to CCM3. A, stereoviews show refined 2Fo −Fc electron density for CCM3 in complex with paxillin LD motifs, LD1, LD2, and LD4. Maps are contoured at 2 σ (light blue) and 0.6 σ (blue). B, shown is a close-up view of the LD motif peptides. All peptides are shown in an identical orientation with respect to the CCM3 FAT homology domain, and the N and C termini are labeled. LD1 is colored gray, LD2 is green, and LD4 is salmon. Residues are labeled, and side chains are shown. C, superposition of the CCM3 FAT homology domains bound to paxillin LD motifs is shown. The LD motifs bind CCM3 at the same location in the HP1 binding site. CCM3-LD1 is colored gray, CCM3-LD2 is colored dark green, and CCM3-LD4 is colored light green (CCM3) and salmon (LD4). α-Helical regions of CCM3 are shown as cylinders. D, sequence alignment of the paxillin LD motifs is shown. Residues built in the complex structures are shaded, and the consensus eight-residue LD motif is shown below. ELD or DLE tri-peptide unit is colored red.
The overall topology of CCM3 in complex with paxillin LD motifs is extremely similar to that of native CCM3 and shows that association of CCM3 with LD motifs does not alter the conformation of the protein. Overall r.m.s.d. for the whole asymmetric unit of CCM3 in the complex structures with paxillin LD1, LD2, and LD4 are 0.7 Å over 762 Cα residues, 0.6 Å over 780 Cα residues, and 0.7 Å over 807 Cα residues, respectively, when compared with the native orthorhombic CCM3 structure (PDB ID 3L8I). For the LD motif peptide binding FAT homology domain of CCM3, the r.m.s.d. for all determined CCM3 structures (PDB IDs 3L8I, 3L8J, 3AJM) (11, 43) range from 0.4 to 0.6 Å. All three bound LD motif peptides can be superposed together and locate to the same position of the HP1 binding pocket composed of helices αG and αH (Fig. 2C).
LD Motifs Bind to the HP1 Pocket of CCM3
The crystal structures of CCM3 in complex with the LD motif peptides for paxillin LD1, LD2, and LD4 all display a broadly similar mode of binding (Fig. 2, C and D). In each of the CCM3-LD motif complex structures, the LD motif forms a 2–3 turn α-helix that presents a hydrophobic stripe to the HP1 pocket of the CCM3 FAT homology domain. This hydrophobic stripe presentation is reminiscent of paxillin LD motifs binding to the HP1 site of FAK and Pyk2 FAT domains. In CCM3 the HP1 binding site is situated between helices αG and αH of the four-helical bundle FAT homology domain. This surface is composed of a hydrophobic cleft bounded by multiple lysine residues and is exquisitely conserved through evolution (11). Detailed description of the binding is provided in the Supplemental Results. In the CCM3-LD1 motif structure, LD1 buries a total of 880 Å2 and provides the first reported crystallographic study of a FAT homology domain in complex with paxillin LD1 motif. In the structure of CCM3 in complex with the paxillin LD2 motif, the bound LD2 peptide buries a total of 998 Å2. In the highest resolution complex of an LD motif with CCM3 (2.5 Å), paxillin LD4 is found in complex with CCM3 and buries a surface area totaling 972 Å2.
Comparison of LD Motif Binding for CCM3, FAK, and Pyk2
The interactions of the FAK FAT domain with paxillin LD motifs LD2 and LD4 have been shown by both x-ray crystallography and NMR to be mediated by two hydrophobic patches on opposite faces of the FAT domain (31–35). HP1 is at the interface of helix 2 and 3, and HP2 is at the interface of helix 1 and 4. These surfaces can interact with two paxillin LD motifs, which adopt an α-helical secondary structure. As LD2 preferentially binds HP2 and LD4 preferentially binds HP1, the current model suggests that the paxillin LD repeat region wraps around one FAT domain and utilizes two LD motifs to bind each hydrophobic patch (34). In the structures of the FAK FAT domain in complex with LD motifs, both LD binding sites share similar architecture, characterized by a hydrophobic patch between two helices, surrounded by conserved basic residues that interact with the negative charges of the LD peptides. Pyk2 binds the LD motifs of paxillin in a very similar fashion to that previously seen for FAK (32). Interestingly, for CCM3, LD motif binding is observed only for HP1 as the location of the HP2 site is occluded (discussed below).
Superposition of the CCM3-LD motif complexes with all of the previously determined FAT domain crystal structures in complex with LD motifs shows that at the HP1 site CCM3 binds LD motifs in a highly similar location to FAT domains. This HP1 binding site is found juxtaposed between helices 2 and 3 in FAK and Pyk2 and the analogous helices αG (residues 131–142) and αH (residues 171–178) in CCM3. In the structures of LD2 and LD4 bound to the FAK family (FAK and Pyk2), the three residue Glu-Leu-Asp (ELD) tri-peptide unit at the N terminus of the LD motifs points toward the open end of the FAT four-helix bundle. In contrast, for the LD2 and LD4 complexes with CCM3, the ELD unit points toward the closed end of the four-helix bundle of the FAT homology domain (Figs. 2D and 3). Interestingly, LD1 does not contain an N-terminal ELD unit but instead contains a C-terminal DLE unit. In our structure of CCM3 in complex with paxillin LD1 there is a direction reversal compared with LD2 and LD4 (Fig. 2C and supplemental Fig. 2).
FIGURE 3.
CCM3 binds LD motifs in an analogous manner to FAK and Pyk2 FAT domains. A, a surface diagram of the CCM3 FAT homology domain (helices αF to αI) is colored by electrostatic potential (calculated in CCP4MG). Paxillin LD4 is colored salmon. Selected residues discussed in the text are labeled. B, shown is a surface diagram of the FAK FAT domain colored by electrostatic potential. Paxillin LD4 is shown in yellow. Selected residues discussed in the text are labeled. The PDB accession code for FAK/LD4 is 1OW7 (31). C, shown is a surface diagram of the Pyk2 FAT domain colored by electrostatic potential. Paxilln LD4 is shown in cyan. Selected residues discussed in the text are labeled. The PDB accession code for Pyk2/LD4 is 3GM1 (32). D, superposition of CCM3-LD4, FAK-LD4, and Pyk2-LD4 is shown. FAT and FAT homology domains are shown as cylinders, and LD4 is in schematic format. FAK-LD4 is colored yellow, Pyk2/LD4 is colored blue, and CCM3-LD4 is colored light green and salmon. E, Cα superposition of the C-terminal 15 residues of CCM3 helix αG (salmon), FAK helix 2 (yellow), and Pyk2 helix 2 (cyan) illustrates the hinge that occurs at residues Ala-135, Gly-995FAK, and Gly-914Pyk2. Residues superposed are indicated by a black bar. The PDB accession code for FAK is 1OW6 (31) and for Pyk2 is 3GM1 (32).
On superposition of the CCM3 FAT homology domain with FAK family FAT domains there is a structural deviation in helix 2. Glycines are highly flexible residues that tend to disrupt α-helices and so are not preferred; however, in the FAK family, at approximately the mid-point of helix 2 there is a highly conserved glycine residue, Gly958FAK or Gly914Pyk2. For the FAK family, helix 2 is kinked at the location of this glycine. In CCM3 the corresponding helix, αG, has a completely conserved alanine (Ala-135) at the corresponding position, and as a result, the helix is straighter when compared with helix 2 of FAK or Pyk2 (Fig. 3E). The difference in linearity of this helix between CCM3, FAK, and Pyk2 results in a broader HP1 site in CCM3 at the closed end of the four-helix bundle (Fig. 3D). Additionally, residues corresponding to conserved Lys-172 in CCM3 are small Gly-995FAK and Ala-951Pyk2 (Fig. 1C); thus, the charge and hydrophobic surface of HP1 are different in CCM3 around Lys-172. This combined with the extended 3–4 loop in FAK/Pyk2 compared with CCM3, gives the surface of FAK/Pyk2 a more torpedo-like shape compared with the stubbier CCM3. Overall, these differences between FAK/Pyk2 and CCM3 potentially indicate that the closed end of the HP1 binding site could be important for altered CCM3 ligand binding specificity when compared with the canonical FAT domains of FAK and Pyk2.
Overall, however, the mode of binding for LD motifs of paxillin to CCM3 is analogous to that previously seen for paxillin LD motifs to FAK family members utilizing mostly conserved residues and resulting in an orientation of the LD helix broadly parallel to the FAT or FAT homology domain axis. It is, therefore, interesting to note that our findings for the orientation of LD motif binding to CCM3 contrast with crystallographic studies of the FAK FAT domain in complex with an LD-like motif (IXXLL) of CD4, which binds the FAT domain in an almost orthogonal direction with a KD of 5–16 μm (44).
Directionality of LD Motif Binding to CCM3
On determination of these structures, we were surprised to discover that CCM3 binds paxillin LD1 in the opposite direction when compared with LD2 and LD4. Interestingly, bidirectionality of binding for LD motif peptides has been described previously. For the crystal structures of α-parvin in complex with LD1, LD2, and LD4 peptides (45), the orientation of LD1 was found to be reversed when compared with LD2 and LD4. This bidirectionality was ascribed to the “pseudo-palindromic character of the LD consensus” (45). There are other examples of directionality being imposed on protein binding to highly similar peptides; for example, the binding direction of PXXP motifs to SH3 domains is determined by the presence of an arginine residue N or C terminus to the PXXP motif (46). The reason for divergence in paxillin LD1 binding may arise from the spacing of charged residues that decorate the perimeter of the helical LD motif hydrophobic stripe. Interestingly, for all three LD motifs of paxillin (LD1, LD2, and LD4) bound to CCM3, a three-residue ELD or DLE motif places the side chains of an aspartate and a glutamate proximal to Lys-179 and Lys-132 and places a leucine residue in the hydrophobic patch of HP1. In the three LD motifs studied here the glutamate residue interacts with Lys-132 (Glu12PAX, Glu144PAX, or Glu265PAX) and the aspartate with Lys-179 (Asp10PAX, Asp146PAX, or Asp267PAX). In LD2 and LD4 this three-residue arrangement (LD144E and LD265E) occurs at the N terminus of the peptide, but in LD1 it occurs at the C terminus (D10LE) (supplemental Fig. 2). Interestingly, in the complex crystal structures of α-parvin, the direction of LD1 is reversed when compared with LD2 and LD4 (45), with this reversal placing the DLE tripeptide motif of LD1 in the same location with respect to α-parvin as the ELD tri-peptide of LD2 and LD4. We hypothesize that the orientation of this tripeptide may be important for directionality of LD motif binding to both CCM3 and α-parvin. An alternative explanation for the differing directionality of LD1 is that a longer peptide could bind in the opposite direction; however, the previous studies for α-parvin (45) show that bidirectional binding can occur with longer peptides than those utilized in this study, suggesting peptide length may not impact directionality (supplemental Table 1), and an NMR study of the interaction between α-parvin and LD1 showed the same orientation as the crystal structure (47). Overall, our structure-based analysis, therefore, implies that bidirectionality of LD motif binding might be a general theme for LD motif recognition that is utilized for other LD-protein interactions.
A Pocket Analogous to FAK HP2 Is Conserved but Occluded in CCM3
The FAT domains of FAK and Pyk2 bind paxillin LD motifs in two sites, termed HP1 and HP2. The HP1 site is located between helices 2 and 3 (equivalent to HP1 helices αG and αH in CCM3), and the HP2 site is located on the opposite face of FAK and Pyk2 between helices 1 and 4 (equivalent to helices αF and αI in CCM3) (31, 32). Superposition of CCM3 with Pyk2 and FAK structures in complex with paxillin LD motif peptides shows that the location of the EF loop between helices αE and αF in CCM3 overlaps well with the location of the LD repeats that bind HP2 of FAK family and occludes the corresponding location of the LD motif-binding HP2 site on CCM3 (supplemental Fig. 3A). In CCM3, completely conserved Leu-81 (11) from helix αE inserts into a hydrophobic patch bounded by Leu-114, Pro-118, Phe-191, and Asn-195 from helices αF and αI (supplemental Fig. 3B), helping occlude the HP2 site for the CCM3 FAT homology domain.
Binding Affinities of CCM3 for Paxillin-derived LD Motifs
To further study the binding properties of CCM3 with paxillin-derived LD motifs and to compare with the binding of the FAK FAT domain with these LD motifs, we measured the binding affinities of CCM3 and the FAK FAT domain with GST fusion LD motifs by SPR. We used GST fusion proteins of paxillin LD motifs, LD1, LD2, and LD4 (GST-LD1, GST-LD2, GST-LD4), as ligands captured by an anti-GST antibody immobilized on the activated surface of a CM5 sensor chip. We found that the KD of CCM3 binding to GST-LD1 is 17 μm, and the KD of CCM3 binding to LD2 and LD4 are 39 and 23 μm, respectively (Table 2 and supplemental Fig. 4). As a control we measured the affinity of the FAK FAT domain, which has previously been shown to bind paxillin LD motifs with KD in the range of ∼4–11 μm (33, 48). We observed FAK FAT domain binding affinities to GST-LD1 at 9 μm, GST-LD2 at 2 μm, and LD4 at 1 μm. The binding affinity of FAK FAT domain with paxillin LD1 has not previously been reported. Our SPR data indicate that FAK FAT domain binds LD1; however, this interaction is of lower affinity than binding of LD2 or LD4 (∼5- and 9-fold weaker binding, respectively). We then tested whether the quadruple lysine to glutamic acid mutant CCM3 (CCM3–4KE) could bind paxillin GST fusion LD motifs. We found that this CCM3 HP1 mutant protein could not bind paxillin GST fusion LD motifs (Table 2 and supplemental Fig. 4). These SPR data are consistent with the co-crystal structures presented above in which the CCM3 FAT homology HP1 binding site interacts with LD motifs. This result is also consistent with the structural similarities of the HP1 binding pockets for CCM3 FAT homology and FAK family FAT domains. Although we were unable to purify a paxillin fragment including multiple LD motifs to investigate the biophysical properties of the interactions with CCM3, it would be interesting in future experiments to investigate the interaction of CCM3 with an extended paxillin N-terminal fragment.
TABLE 2.
Binding affinities of CCM3 for paxillin LD motifs measured by SPR
The affinities of CCM3 for GST fusions of paxillin LD motifs, LD1, LD2, and LD4, were determined by SPR. Control experiments were conducted using the FAK FAT domain and the quadruple lysine to glutamic acid CCM3 mutant, CCM3–4KE. Binding affinities (KD) of CCM3 for the GST-LD fusion proteins, GST-LD1, GST-LD2, and GST-LD4, were derived from two independent SPR experiments with difference surface densities of GST-LDs. Binding affinities for the FAK FAT domain and CCM3–4KE are derived from one SPR experiment. The table shows the dissociation constant (KD) for each interaction (±S.D.). NA, not applicable.
| Paxillin LD motif |
KD Values |
||
|---|---|---|---|
| CCM3 | CCM3–4KE | FAK FAT domain | |
| (μm) | |||
| LD1 | 17 ± 3 | NA | 9 |
| LD2 | 39 ± 2 | NA | 2 |
| LD4 | 23 ± 6 | NA | 1 |
Endogenous CCM3 and Paxillin Co-localize
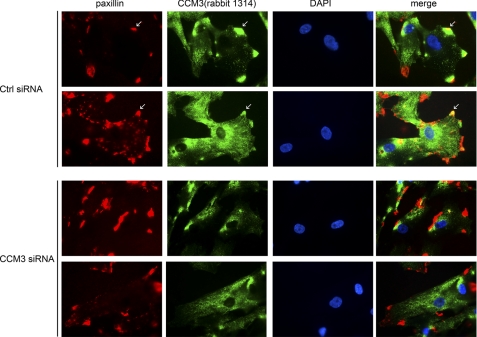
Previously we showed that CCM3 partially localizes with paxillin to leading edges in vascular endothelial cells in an overexpression system (11). Co-localization of endogenous CCM3 and paxillin and the effects of CCM3 on paxillin function have, however, not been determined. To address these questions, we examined several cell lines for endogenous CCM3/paxillin staining. We found that pericytes produced the strongest co-localization of CCM3 and paxillin. As shown in Fig. 4, CCM3 was co-localized with paxillin in the leading edges (arrows). Only partial knockdown was achieved in pericytes by CCM3 siRNA due to the long half-life of CCM3 protein (8); however, this partial knockdown dramatically reduced co-staining of CCM3 with paxillin in the leading edge (Fig. 4, bottom).
FIGURE 4.
Co-localization of endogenous CCM3 and paxillin in pericytes. Pericytes were transfected with control (Ctrl) or CCM3 siRNA. 48 h post-transfection, cells were immunostained with anti-CCM3 and paxillin followed by counterstaining with DAPI (blue) for nuclei. Merged images are shown on the right. Representative images from 10 cells in each group are shown. Scale bar, 20 μm.
DISCUSSION
In this study we have determined the crystal structures of full-length human CCM3 in complex with the LD1, LD2, and LD4 motifs of human paxillin. We show that CCM3 binds these LD motifs using the highly conserved HP1 surface of its FAT homology domain, that CCM3 can bind LD motifs in a bidirectional fashion, and that the mode of CCM3 binding to LD motifs is similar to that previously seen for FAK and Pyk2. We also found that CCM3 binds paxillin LD motifs with affinities in the range seen for FAT domain proteins and that endogenous CCM3 and paxillin co-localize.
LD motifs encode an eight-residue consensus sequence (LDXLLXXL) that folds as short α-helices with a central hydrophobic stripe and are decorated by surrounding residues often capable of hydrogen-bond or salt-bridge formation. These motifs are observed in many proteins including paxillin, leupaxin Hic5, and unrelated proteins such as E6-AP and ERC-55 (21, 49). The best-studied LD motif protein is paxillin, which binds directly to the FAT domains of FAK and Pyk2, the most structurally homologous proteins to CCM3 (11). The interactions of FAK and Pyk2 with paxillin LD motifs occur via their FAT domain HP1 site; the region of CCM3 that is structurally analogous to this site is extremely well conserved (11) (Fig. 1B). We, therefore, targeted our studies to investigate the binding of CCM3 with the LD motif containing protein paxillin and previously demonstrated the interaction by pulldown assay, co-immunoprecipitation, and partial cellular co-localization of CCM3 with paxillin in an overexpression system (11). In the present study we show that CCM3 can directly bind the paxillin LD motifs LD1, LD2, and LD4 in co-crystals using the FAK-homologous HP1 binding site of the CCM3 FAT homology domain. We also determined that the affinity of CCM3 for paxillin LD motifs is in the same range as the affinities of the paxillin LD motif binding proteins, FAK, Pyk2, GIT1, and α-parvin. For example, KD values previously reported for paxillin LD motif binding of FAK FAT, Pyk2 FAT, GIT1 FAT, and α-parvin CH2 are 4–11, 45, 7–25, and ∼100 μm, respectively (29, 33, 45, 48, 50). We further confirmed the binding of CCM3 to paxillin LD motifs by a quadruple lysine to glutamate mutation that abrogated binding. This provides direct evidence for the mode of intermolecular binding of LD motifs by CCM3. We go on to show that endogenous CCM3 and paxillin can co-localize in leading edges of mouse cerebral pericytes, indicating that CCM3 and paxillin may be direct binding partners in vivo. This conclusion is not without caveats as there are many LD motif-containing proteins; therefore, it is not unreasonable to suggest that CCM3 can bind multiples of these proteins utilizing a FAT homology/LD motif interaction similar to those described in this study. It also remains to be investigated whether the LD motif is the exclusive driver of protein-protein interaction for CCM3, whether the dimeric CCM3 binds two different LD motifs by both FAT homology domains in a bi-dentate fashion analogous to HP1 and HP2 of FAK binding paxillin LD2 and LD4, or whether dimeric CCM3 binds two copies of the same protein by the HP1 sites of the dimers FAT homology domains.
The N-terminal domain of CCM3 is a homodimerization domain with a novel fold, the CCM3 dimerization fold, and buries a high surface area of ∼3700 Å2. This domain has been shown to be critical for CCM3 interactions with the germinal center kinase III kinases, MST3, STK24, and STK25, by interaction with a conserved ∼100-residue C-terminal tail in these kinases (12). These kinases are important for cell migration (51, 52) and apoptosis (53–55). MST3 has been proposed to play a role in paxillin phosphorylation and PTP-PEST regulation; however, the mechanism of localization for this protein to focal adhesions is not understood (51). We, therefore, propose that CCM3 could target germinal center III kinases to specific locations within the cell using an LD motif guidance system.
Cerebral cavernous malformations are vascular dysplasias that affect the central nervous system and result in dilated blood vessels with thin walls (3–5). These dysplasias are associated with focal neurological defects including hemorrhagic stroke, seizure, and epilepsy. The three genes mutated in CCM, CCM1, CCM2, and CCM3, encode three proteins, CCM1 (Krit1), CCM2 (malcavernin), and CCM3, that do not have inherent enzymatic activity but instead seem to have roles as regulators of signal transduction and as scaffolding proteins. The molecular level basis for the function and interactions of these proteins has not previously been investigated. In this study we provide the first structural description and affinity measurements of intermolecular interactions by one of these CCM-associated proteins, CCM3. We also show that endogenous CCM3 and paxillin can co-localize. Together these data provide a framework for further investigations into the functional roles of protein-protein interactions by CCM3 and indicate that disease-associated CCM3 truncations (11) will result in loss of these protein-protein interactions by the CCM3 FAT homology domain.
Supplementary Material
Acknowledgments
We thank Joseph Schlessinger, Jae-Hyun Bae, and James Lulo for peptides and cDNA. GST-LD constructs were provided by Christopher Turner. We thank David Calderwood, Yong Zhang, Amy Stiegler, and Nilda Alicea-Velázquez for helpful discussions and Ya Ha and Yi Xue for data collection of CCM3-LD1 at NSLS beamline X29. We also thank Vivian Stojanoff and Jean Jakoncic of NSLS beamline X6A. Modified pET-32 vector was designed by Florence Poy.
This work was supported, in whole or in part, by National Institutes of Health Grants HL077357 and HL085789 (to W. M.), RR026992 (to E. F.-S.), and AI075133, GM088240, and DK087844 (to T. J. B.). This work was also supported by startup funds to T. J. B. from Yale University.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table and Figs. 1–4.
The atomic coordinates and structure factors (codes 3RQE, 3RQF, and 3RQG) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- CCM
- cerebral cavernous malformation
- FAK
- focal adhesion kinase
- FAT
- focal adhesion targeting
- HP
- hydrophobic patch
- r.m.s.d.
- root mean square deviation
- SPR
- surface plasmon resonance
- NSLS
- Brookhaven National Synchrotron Light Source
- LD
- leucine-aspartate repeat.
REFERENCES
- 1. Revencu N., Vikkula M. (2006) J. Med. Genet. 43, 716–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Labauge P., Denier C., Bergametti F., Tournier-Lasserve E. (2007) Lancet Neurol. 6, 237–244 [DOI] [PubMed] [Google Scholar]
- 3. Rigamonti D., Hadley M. N., Drayer B. P., Johnson P. C., Hoenig-Rigamonti K., Knight J. T., Spetzler R. F. (1988) N. Engl. J. Med. 319, 343–347 [DOI] [PubMed] [Google Scholar]
- 4. Pozzati E., Acciarri N., Tognetti F., Marliani F., Giangaspero F. (1996) Neurosurgery 38, 662–670 [PubMed] [Google Scholar]
- 5. Davenport W. J., Siegel A. M., Dichgans J., Drigo P., Mammi I., Pereda P., Wood N. W., Rouleau G. A. (2001) Neurology. 56, 540–543 [DOI] [PubMed] [Google Scholar]
- 6. Bergametti F., Denier C., Labauge P., Arnoult M., Boetto S., Clanet M., Coubes P., Echenne B., Ibrahim R., Irthum B., Jacquet G., Lonjon M., Moreau J. J., Neau J. P., Parker F., Tremoulet M., Tournier-Lasserve E. (2005) Am. J. Hum. Genet. 76, 42–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guclu B., Ozturk A. K., Pricola K. L., Bilguvar K., Shin D., O'Roak B. J., Gunel M. (2005) Neurosurgery 57, 1008–1013 [DOI] [PubMed] [Google Scholar]
- 8. He Y., Zhang H., Yu L., Gunel M., Boggon T. J., Chen H., Min W. (2010) Sci. Signal. 3, ra26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liquori C. L., Berg M. J., Siegel A. M., Huang E., Zawistowski J. S., Stoffer T., Verlaan D., Balogun F., Hughes L., Leedom T. P., Plummer N. W., Cannella M., Maglione V., Squitieri F., Johnson E. W., Rouleau G. A., Ptacek L., Marchuk D. A. (2003) Am. J. Hum. Genet. 73, 1459–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Voss K., Stahl S., Schleider E., Ullrich S., Nickel J., Mueller T. D., Felbor U. (2007) Neurogenetics 8, 249–256 [DOI] [PubMed] [Google Scholar]
- 11. Li X., Zhang R., Zhang H., He Y., Ji W., Min W., Boggon T. J. (2010) J. Biol. Chem. 285, 24099–24107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Voss K., Stahl S., Hogan B. M., Reinders J., Schleider E., Schulte-Merker S., Felbor U. (2009) Hum. Mutat. 30, 1003–1011 [DOI] [PubMed] [Google Scholar]
- 13. Ma X., Zhao H., Shan J., Long F., Chen Y., Chen Y., Zhang Y., Han X., Ma D. (2007) Mol. Biol. Cell 18, 1965–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rual J. F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G. F., Gibbons F. D., Dreze M., Ayivi-Guedehoussou N., Klitgord N., Simon C., Boxem M., Milstein S., Rosenberg J., Goldberg D. S., Zhang L. V., Wong S. L., Franklin G., Li S., Albala J. S., Lim J., Fraughton C., Llamosas E., Cevik S., Bex C., Lamesch P., Sikorski R. S., Vandenhaute J., Zoghbi H. Y., Smolyar A., Bosak S., Sequerra R., Doucette-Stamm L., Cusick M. E., Hill D. E., Roth F. P., Vidal M. (2005) Nature. 437, 1173–1178 [DOI] [PubMed] [Google Scholar]
- 15. Ewing R. M., Chu P., Elisma F., Li H., Taylor P., Climie S., McBroom-Cerajewski L., Robinson M. D., O'Connor L., Li M., Taylor R., Dharsee M., Ho Y., Heilbut A., Moore L., Zhang S., Ornatsky O., Bukhman Y. V., Ethier M., Sheng Y., Vasilescu J., Abu-Farha M., Lambert J. P., Duewel H. S., Stewart II, Kuehl B., Hogue K., Colwill K., Gladwish K., Muskat B., Kinach R., Adams S. L., Moran M. F., Morin G. B., Topaloglou T., Figeys D. (2007) Mol. Syst. Biol. 3, 89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zheng X., Xu C., Di Lorenzo A., Kleaveland B., Zou Z., Seiler C., Chen M., Cheng L., Xiao J., He J., Pack M. A., Sessa W. C., Kahn M. L. (2010) J. Clin. Invest. 120, 2795–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fidalgo M., Fraile M., Pires A., Force T., Pombo C., Zalvide J. (2010) J. Cell Sci. 123, 1274–1284 [DOI] [PubMed] [Google Scholar]
- 18. Goudreault M., D'Ambrosio L. M., Kean M. J., Mullin M. J., Larsen B. G., Sanchez A., Chaudhry S., Chen G. I., Sicheri F., Nesvizhskii A. I., Aebersold R., Raught B., Gingras A. C. (2009) Mol. Cell. Proteomics 8, 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deakin N. O., Turner C. E. (2008) J. Cell Sci. 121, 2435–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schaller M. D. (2001) Oncogene 20, 6459–6472 [DOI] [PubMed] [Google Scholar]
- 21. Brown M. C., Turner C. E. (2004) Physiol. Rev. 84, 1315–1339 [DOI] [PubMed] [Google Scholar]
- 22. Raftopoulou M., Hall A. (2004) Dev. Biol. 265, 23–32 [DOI] [PubMed] [Google Scholar]
- 23. Ridley A. J. (2001) Trends Cell Biol. 11, 471–477 [DOI] [PubMed] [Google Scholar]
- 24. Ridley A. J. (2001) J. Cell Sci. 114, 2713–2722 [DOI] [PubMed] [Google Scholar]
- 25. Turner C. E., Glenney J. R., Jr., Burridge K. (1990) J. Cell Biol. 111, 1059–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brown M. C., Perrotta J. A., Turner C. E. (1996) J. Cell Biol. 135, 1109–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Turner C. E., Miller J. T. (1994) J. Cell Sci. 107, 1583–1591 [DOI] [PubMed] [Google Scholar]
- 28. Turner C. E., Brown M. C., Perrotta J. A., Riedy M. C., Nikolopoulos S. N., McDonald A. R., Bagrodia S., Thomas S., Leventhal P. S. (1999) J. Cell Biol. 145, 851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schmalzigaug R., Garron M. L., Roseman J. T., Xing Y., Davidson C. E., Arold S. T., Premont R. T. (2007) Cell. Signal. 19, 1733–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nikolopoulos S. N., Turner C. E. (2000) J. Cell Biol. 151, 1435–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hoellerer M. K., Noble M. E., Labesse G., Campbell I. D., Werner J. M., Arold S. T. (2003) Structure 11, 1207–1217 [DOI] [PubMed] [Google Scholar]
- 32. Lulo J., Yuzawa S., Schlessinger J. (2009) Biochem. Biophys. Res. Commun. 383, 347–352 [DOI] [PubMed] [Google Scholar]
- 33. Gao G., Prutzman K. C., King M. L., Scheswohl D. M., DeRose E. F., London R. E., Schaller M. D., Campbell S. L. (2004) J. Biol. Chem. 279, 8441–8451 [DOI] [PubMed] [Google Scholar]
- 34. Bertolucci C. M., Guibao C. D., Zheng J. (2005) Protein. Sci. 14, 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hayashi I., Vuori K., Liddington R. C. (2002) Nat. Struct. Biol. 9, 101–106 [DOI] [PubMed] [Google Scholar]
- 36. Davis I. W., Murray L. W., Richardson J. S., Richardson D. C. (2004) Nucleic Acids Res. 32, W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 38. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 39. Holm L., Sander C. (1995) Trends Biochem. Sci 20, 478–480 [DOI] [PubMed] [Google Scholar]
- 40. Laskowski R. A. (2001) Nucleic Acids Res. 29, 221–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. CCP4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 42. Nikolopoulos S. N., Turner C. E. (2002) J. Biol. Chem. 277, 1568–1575 [DOI] [PubMed] [Google Scholar]
- 43. Ding J., Wang X., Li D. F., Hu Y., Zhang Y., Wang D. C. (2010) Biochem. Biophys. Res. Commun. 399, 587–592 [DOI] [PubMed] [Google Scholar]
- 44. Garron M. L., Arthos J., Guichou J. F., McNally J., Cicala C., Arold S. T. (2008) J. Mol. Biol. 375, 1320–1328 [DOI] [PubMed] [Google Scholar]
- 45. Lorenz S., Vakonakis I., Lowe E. D., Campbell I. D., Noble M. E., Hoellerer M. K. (2008) Structure 16, 1521–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Feng S., Chen J. K., Yu H., Simon J. A., Schreiber S. L. (1994) Science 266, 1241–1247 [DOI] [PubMed] [Google Scholar]
- 47. Wang X., Fukuda K., Byeon I. J., Velyvis A., Wu C., Gronenborn A., Qin J. (2008) J. Biol. Chem. 283, 21113–21119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Thomas J. W., Cooley M. A., Broome J. M., Salgia R., Griffin J. D., Lombardo C. R., Schaller M. D. (1999) J. Biol. Chem. 274, 36684–36692 [DOI] [PubMed] [Google Scholar]
- 49. Brown M. C., Curtis M. S., Turner C. E. (1998) Nat. Struct. Biol. 5, 677–678 [DOI] [PubMed] [Google Scholar]
- 50. Zhang Z. M., Simmerman J. A., Guibao C. D., Zheng J. J. (2008) J. Biol. Chem. 283, 18685–18693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lu T. J., Lai W. Y., Huang C. Y., Hsieh W. J., Yu J. S., Hsieh Y. J., Chang W. T., Leu T. H., Chang W. C., Chuang W. J., Tang M. J., Chen T. Y., Lu T. L., Lai M. D. (2006) J. Biol. Chem. 281, 38405–38417 [DOI] [PubMed] [Google Scholar]
- 52. Preisinger C., Short B., De Corte V., Bruyneel E., Haas A., Kopajtich R., Gettemans J., Barr F. A. (2004) J. Cell Biol. 164, 1009–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dan I., Ong S. E., Watanabe N. M., Blagoev B., Nielsen M. M., Kajikawa E., Kristiansen T. Z., Mann M., Pandey A. (2002) J. Biol. Chem. 277, 5929–5939 [DOI] [PubMed] [Google Scholar]
- 54. Nogueira E., Fidalgo M., Molnar A., Kyriakis J., Force T., Zalvide J., Pombo C. M. (2008) J. Biol. Chem. 283, 16248–16258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huang C. Y., Wu Y. M., Hsu C. Y., Lee W. S., Lai M. D., Lu T. J., Huang C. L., Leu T. H., Shih H. M., Fang H. I., Robinson D. R., Kung H. J., Yuan C. J. (2002) J. Biol. Chem. 277, 34367–34374 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.