Abstract
LC-MS-based quantitative proteomics has become increasingly applied to a wide range of biological applications due to growing capabilities for broad proteome coverage and good accuracy and precision in quantification. Herein, we review the current LC-MS-based quantification methods with respect to their advantages and limitations and highlight their potential applications.
Keywords: Chemical Modification, Mass Spectrometry (MS), Peptides, Protein Chemistry, Proteomics, LC-MS, MS/MS, Quantitation, Stable Isotope Labeling
Introduction
LC-MS-based quantitative proteomic approaches have become increasingly popular over the past decade (1–7). In general, discovery-based proteomic efforts lend themselves to global analyses whereby a broad survey of the proteome is performed across various samples, and the quantitative differences among them are estimated. In discovery-based efforts, the breadth of analysis is emphasized more than the precision and accuracy of quantification. For studies in which these qualities are crucial such as verification efforts, the tactic switches to sensitive, precise, and accurate analysis of a few targeted proteins in relatively large set of samples and internal standards are often used.
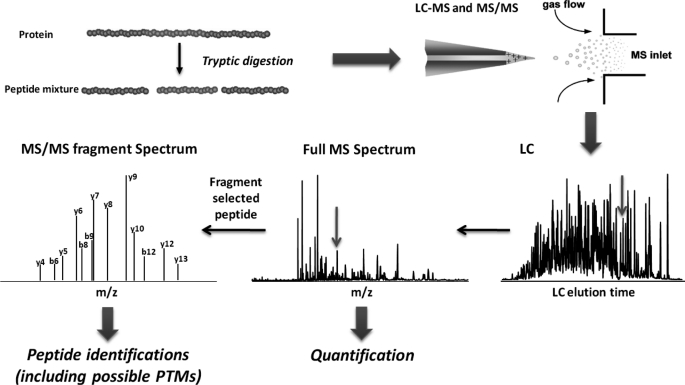
Fig. 1 illustrates an LC-MS-based global proteomic workflow in which proteins are converted into peptides for identification and quantification (i.e. “bottom-up” proteomics). Typically, methods are applied in conjunction with enzymatic digestion of proteins and subsequent measurement of one or more peptides from each protein that serve as effective measurement surrogates. We note that direct measurement of intact proteins (i.e. “top-down” proteomics) is another analytical option but is beyond the scope of this minireview and therefore not discussed herein. In global analyses, relative quantification of peptides usually involves either label-free or stable isotope labeling techniques (1) to discern differences in protein abundances among different biological conditions, and results are often expressed as “-fold changes.” Overall, label-free approaches have wider dynamic range and broader proteome coverage, whereas stable isotope labeling approaches offer higher quantification precision and accuracy (8). Another common approach is absolute quantification, which determines the exact amount or concentration of a peptide/protein in a given sample and requires the use of an appropriate “internal” standard. All of these approaches have considerably different pros and cons that must be weighed before deciding which one is best for a specific course of study.
FIGURE 1.
General workflow for LC-MS-based global proteomics. Proteins in complex biological samples are first converted into peptides by proteolytic digestion (e.g. tryptic digestion). The resulting peptide mixture is then separated by LC and ionized by electrospray before entering the mass spectrometer. In a typical data-dependent acquisition operation mode, a full MS spectrum is acquired for the peptides that are eluting from the LC column at any given time; one of the most intensive ion species (i.e. peptides) is then isolated and fragmented to obtain the MS pattern of its fragments (i.e. MS/MS spectrum). Because peptide bonds are prone to fragmentation under collision-induced dissociation conditions in the MS/MS analysis and produce predominantly b- or y-type ions (N- or C-terminal fragments carrying charge, respectively), the peptide sequence can be readily deduced from the MS/MS spectrum. This process is fully automated by searching the MS/MS spectra against protein sequence databases. Possible post-translational modifications can also be identified by including dynamic modification on certain amino acid residues (e.g. Ser, Thr, or Tyr for phosphorylation) in the database search. Quantification of each peptide is typically performed at the extracted ion chromatogram level; however, for isobaric tagging approaches (e.g. iTRAQ), quantification is carried out at the MS/MS spectrum level. PTMs, post-translational modifications.
Challenges affecting quantification in a bottom-up proteomic workflow stem from the wide range of peptide and protein physicochemical properties that give rise to large differences in MS responses (8). Sample handling, digestion efficiency, and separation also can have an impact on results. As such, relative peptide intensities may not directly reflect the relative abundances of different proteins. A major factor that influences LC-MS-based quantification via electrospray ionization is ion suppression (9). Peptide intensity depends on the quantity of the peptide being ionized as well as on ionization efficiency and, under some conditions, on the properties of co-eluting peptides. The use of lower flow rates (e.g. <100 nl/min) (9, 10) or internal standards (11, 12) can help alleviate ion suppression. Other issues in LC-MS-based quantification include the separation peak capacity and reproducibility of the chromatography and the mass measurement accuracy and resolving power of the mass spectrometer. Significant technological advances such as the development and commercialization of ultra-performance LC and high-mass accuracy/resolution mass spectrometers have substantially overcome these issues, making LC-MS-based quantification more reliable and accessible to biologists.
Basically, there is no recognized “one-size-fits-all” method that fulfills every quantitative need, and available options for quantification can make it difficult for an investigator to choose the most appropriate approach to answer particular biological questions. This minireview presents the advantages and limitations of commonly used LC-MS-based protein quantification approaches and provides guidelines for researchers who may not be familiar with but would like to benefit from quantitative proteomic measurements.
Label-free Quantification
Straightforward and inexpensive, label-free quantification is being increasingly applied to proteomic measurements. Without the need to modify peptides/proteins with stable isotope-containing compounds or to add heavy isotope-labeled internal standards to the sample, label-free approaches require minimal manipulation of the sample and can be used on any type of biological materials. Conceptually, label-free quantification allows for the comparison of an unlimited number of samples; however, each sample has to be analyzed individually (no sample multiplexing). This type of approach usually offers wide dynamic range, which is especially advantageous when relatively large-abundance changes are expected. The two major label-free quantification approaches are spectral counting and MS ion intensity (or peak area) measurement.
Spectral counting, i.e. counting the number of MS/MS spectra identifying a given peptide or protein, represents a simple approach for relative quantification without stable isotope labeling (13, 14). The rationale behind this method, which has been experimentally validated (13), is that the frequency for which a peptide is selected for MS/MS fragmentation is positively correlated to its quantity in the data-dependent acquisition operation mode. In this mode, a survey MS scan is acquired, followed by selection of typically ≤10 of the most abundant ions in the survey MS scan for subsequent MS/MS analysis (i.e. undersampling). Spectral counts for different peptides from a given protein can be summed up for relative quantification of the protein; however, the linearity and the number of quantified proteins will depend on the data-dependent acquisition setting details (15). Although spectral counting is straightforward, small numbers of spectral counts for proteins present in low abundance provide less robust quantitative measurements of these proteins due to statistical limitations and other factors such as the need to limit the false discovery rate so as not to “count” lower quality spectra (16).
Label-free quantification can also be performed based on MS ion intensity (or peak area) for the peptides in a given sample. These LC-MS-based approaches alleviate the undersampling issue inherent in typical LC-MS/MS analysis (17). An example of this type of approach is the accurate mass and time tag strategy (18–20), which utilizes high-resolution high-mass accuracy LC-MS to analyze individual samples. Peptides are identified by matching accurate mass and normalized elution time features to those stored in a previously established reference database of peptides. Following accurate alignment of detected LC-MS features (i.e. LC retention time and m/z values) across different analyses, the areas under chromatographic elution profiles of the identified peptides can be compared among different samples for relative quantification. Similar quantitative approaches that rely on direct LC-MS measurements, feature alignment, and peak identifications have also been reported (21, 22).
In label-free intensity-based quantification, any variations in sample preparation, LC-MS reproducibility, ionization efficiency, and other sources of “instrument drift” can lead to increased measurement error. Therefore, it is important to normalize the data to correct (as much as possible) for systematic variations (23). Software tools for performing LC-MS feature alignments, peak matching, data normalization, and statistical analyses of label-free quantification data are available (20, 22, 24). Although label-free intensity-based quantification has been broadly applied, its accuracy and reliability for quantification are inherently limited by the reproducibility related to sample processing, LC-MS platform, etc. These factors should be considered and minimized when applying label-free quantification approaches.
Stable Isotope Labeling Approaches for Quantification
Stable isotope labeling approaches enable accurate quantification based on a stable isotope dilution concept (1). Because a stable isotope-labeled peptide has the same chemical properties as its native equivalent, the two peptides within a mixture should exhibit identical behaviors during LC and electrospray ionization processes and be separated by their differences in mass, thus enabling accurate peptide (but not necessarily protein) quantification. The relative abundance differences measured by MS between the two peptide forms are taken to quantitatively reflect true differences in abundance within the mixture. 13C, 15N, and 18O are commonly used stable isotopes (25) that can be incorporated into proteins or peptides metabolically, enzymatically, or chemically. Although sometimes used, 2H is less desirable because it changes the physicochemical properties of peptides such that the heavy form elutes slightly earlier than the light form in reverse-phase LC (26). In stable isotope labeling methods, samples are combined after labeling and analyzed by LC-MS or LC-MS/MS, which essentially avoids the uncertainty induced by variations in instrument performance between measurements. As a result, quantification precision and accuracy are markedly improved compared with label-free approaches.
The most popular metabolic labeling approach is stable isotope labeling with amino acids in cell culture (SILAC)2 (27). Briefly, one or several amino acids (typically Arg or Lys) are labeled with “heavy” isotopes (e.g. 13C and/or 15N atoms) and added to the growth medium. Then, the heavy isotope-labeled amino acids are incorporated into all the proteins after several cell doublings. Equal amounts of the heavy isotope-labeled proteome and the normal-labeled proteome can be mixed at the level of either intact cells (i.e. equal numbers of heavy and normal isotope-labeled cells) or cell lysates (i.e. equal amounts of heavy and normal isotope-labeled protein contents) and analyzed. Peptide abundance ratios are determined in the MS mode by comparing the intensities of the labeled and unlabeled peptides within a survey mass spectrum. SILAC can be used to quantify in vivo changes (e.g. protein turnover) as the isotopes are introduced during the natural biosynthesis process. Another advantage is that multiplexing is possible due to the availability of several labels (e.g. 12C614N4-Arg, 13C614N4-Arg, and 13C615N4-Arg) (1, 12). A shortcoming is that SILAC is limited to cell cultures and cannot easily be applied to tissues or biofluids.
With enzymatic labeling, 18O atoms are incorporated into the C terminus of every proteolytic peptide during or after protein digestion catalyzed by trypsin (28), Glu-C (29), or some other proteases (30). Previously, two main issues, incomplete labeling and 18O back-exchange to 16O, limited the application of 18O labeling for global quantitative proteomics. Typically, 18O labeling is performed when a sample is digested in H218O solution with trypsin; however, different peptides often have different labeling efficiencies. We and others demonstrated more effective labeling by directly incubating peptides in 18O water with trypsin as a catalyst (31, 32), and a recently improved protocol that incorporates a boiling step after labeling now prevents oxygen back-exchange (33). 18O labeling coupled with the accurate mass and time tag approach has been applied in pairwise global quantitative proteomics (31, 34), and more large-scale applications have also been reported (35, 36). The main advantages of 18O labeling are its simplicity in labeling, relatively low cost, and applicability to all types of samples (e.g. tissues, cells, and biological fluids). Additionally, many different software tools are available to facilitate the 16O/18O-based quantification (31, 37, 38).
Another commonly applied stable isotope labeling strategy is based on incorporating isotope-containing tags into proteins or peptides via chemical reactions, a strategy that can be applied to any type of biological materials. The first reported chemical labeling proteomic approach was isotope-coded affinity tag (ICAT), which reacts specifically to the sulfhydryl group of Cys residues (39). Relative protein abundance is determined in the MS mode, and the mass shift is 9 Da per ICAT-labeled Cys (provided 13C is used in the heavy form) (40). More recently, novel chemically reactive tags have been developed for labeling peptides that facilitate quantitative analysis of multiple samples simultaneously (i.e. multiplexing), which is particularly useful for following a biological system over multiple time points (8, 41). The most commonly used tags for peptide labeling are isobaric tags for relative and absolute quantification (iTRAQ) (42–44) and tandem mass tags (TMT) (45, 46). iTRAQ and TMT labels react specifically with primary amine groups of tryptic peptides, e.g. N-termini and the side chains of Lys residues, and the tagging reactions are largely complete without major side reactions. The same peptides labeled with different isobaric tags have exactly the same mass and co-elute precisely in LC separations. Although tags remain indistinguishable in the MS scan, they fragment into reporter ions of different masses in the MS/MS scan. The intensities of the different reporter ions are then used to determine the relative abundance of the corresponding peptides and proteins in different samples. It is important to note that both iTRAQ- and TMT-based quantifications require the ability to observe low m/z fragment ions (i.e. the reporter ions), which limits the type of mass spectrometers that can be used. The iTRAQ reagent can be utilized to analyze up to four (4-plex iTRAQ) or eight (8-plex iTRAQ) samples simultaneously, whereas TMT can address two to six samples. Because iTRAQ and TMT label-based quantifications are measured at the MS/MS level, potentially higher signal/noise ratios may be obtained for quantification compared with those obtained at the MS level. Additionally, the accuracy of MS/MS level quantification depends on the isolation window for selected precursors in the first stage MS (8, 47), which is typically 3 Thomson, as all ions within that window will fragment, and potential interferences could skew the quantification results.
“Universal” Reference for Relative Quantification
A challenge for large-scale quantitative applications involving label-free and/or labeling-based proteomics is maintaining platform reproducibility for large studies that may extend over several months or years. The use of a stable isotope-labeled whole proteome as a universal internal standard offers a solution to this challenge. Briefly, a pooled reference sample can be generated, and the digested peptides can be labeled with 18O (as an example) to serve as the universal reference. The labeled reference sample can be added to each unlabeled biological sample so that each unlabeled peptide will have a corresponding 18O-labeled version of the peptide from the universal reference. Relative peptide and protein abundances in many different samples can be compared based on their ratios to the universal reference in each analysis. The concept is similar to the use of synthetic isotope-labeled peptides as internal standards (48) that are added to each biological sample. The 18O-labeled universal reference can be generated from any type of biological sample and requires only a single step to label a digest of the pooled sample. This strategy has been applied to two-dimensional proteome mapping of mouse brain (49) and in human plasma proteome studies to discover biomarkers (36, 50).
A “super-SILAC” approach has been applied to quantify proteins in human tumor tissues (51). In this approach, equal amounts of SILAC-labeled proteins from several previously established cancer-derived cell lines were combined (i.e. a super-SILAC mixture). This mixture served as a “global internal standard” to quantify relative protein abundance from multiple tissue samples of the same tumor type. Unlike the 18O-labeled universal reference, the super-SILAC strategy is limited by the availability of appropriate cell lines for generating SILAC-labeled samples and cannot be applied to biofluids.
Although the universal reference or global internal standard approach offers great flexibility for large-scale relative quantification studies, it is difficult to produce a “true” universal reference that contains all the proteins and protein forms of interest and that can be reproducibly generated. The nature of adding a reference/standard makes it inevitable to dilute every sample 1:1 with the standard and thus makes it more difficult to detect and quantify lower abundance proteins.
Targeted Quantification
Global quantitative proteomics inherently suffers some limitations as a result of missing data in individual analyses, false identifications, reproducibility issues, and computational challenges. Targeted quantification approaches using selected reaction monitoring (SRM) or multiple reaction monitoring for accurate quantification of selected analytes are gaining in popularity as a means of overcoming these limitations. SRM is typically performed using a “triple quadrupole” tandem mass spectrometer that consists of a selection quadruple for the precursor ion, a collision quadrupole, and another selection quadrupole for the fragments. Specifically, an SRM assay defines a list of precursor m/z values associated with specific retention times. The critical rules for precursor selection are defined in a comprehensive review (52). For each m/z value, the assay defines one or several fragment ions that are predicted to have good MS response and are readily distinguished from interference. By setting the MS platform to exclusively monitor predefined precursor-to-fragment ion transitions in rapid succession, a specific ion can be detected and quantified when it has the expected m/z value and produces fragments of the expected m/z. Recently, an interesting variant, intelligent SRM, was developed to confirm the precursor identity without significantly perturbing the SRM quantification (53). In this method, additional transitions are acquired in a data-dependent fashion (triggered when all the primary transitions exceed a predefined threshold), which increases the specificity of the analysis.
Among the benefits of SRM-based quantification is the excellent reproducibility attained by using labeled synthetic peptides as internal standards (54). Additionally, the approach is less affected by sample complexity, as noise signals are filtered out at both the precursor and fragment levels (55). As a result, SRM has the highest sensitivity and a wide dynamic range that extends 4–5 orders of magnitude, which makes SRM well suited for targeted quantification experiments such as biomarker verification. SRM can also be applied to quantify particular peptide modifications (e.g. ubiquitination and phosphorylation) (41, 56).
An important application of SRM is absolute quantification, which is achieved by using known concentrations of synthetic isotope-labeled peptides as internal standards (48). In this method, a target peptide is synthesized in an isotope-labeled form and added to the protein digest at a known concentration. The concentration of the native peptide is determined by comparing the ion intensities between the labeled and unlabeled forms. The internal standard can correct for ion suppression and matrix effects (11). For an experiment with multiple samples, the use of an internal standard ensures a fair comparison across all samples. However, the protein amount determined by absolute quantification may not reflect the true expression levels in the original sample because the internal standard is added after digestion and thus cannot correct for the variable losses or enrichments that occur during sample preparation (57), i.e. not true “absolute quantification.” Using an isotope-labeled protein as the internal standard may alleviate this problem because the surrogate protein can be combined with the target protein at the very beginning of the analytical process. The standard can be either a full-length isotope-labeled equivalent of a target protein (58) or an isotope-labeled artificial concatamer of proteotypic peptides from several proteins of interest, which allows multiplexing (59). Regardless, identifying and synthesizing suitable internal standards are not trivial tasks, which limits the absolute quantification approach to a restricted set of prescreened proteins (1), making it suitable for applications such as validating potential protein candidates of interest or quantifying particular post-translational modifications such as ubiquitination (60).
Factors to Consider When Choosing a Quantification Strategy
When it comes to quantitative proteomics, many options are available, each with their own set of advantages and disadvantages. Some factors to be considered when making a choice include the types of samples, the number of samples to be compared, the biological sources and complexity of the samples, the analytical needs (e.g. quantification precision, accuracy and whether an absolute concentration is necessary) of the biological problem, and the cost of the experiment (12).
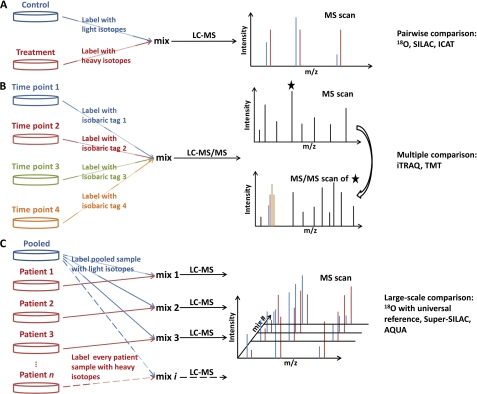
Table 1 summarizes the performance characteristics for each of the common LC-MS-based proteomic quantification strategies. Among the stable isotope labeling techniques, SILAC offers the best accuracy for quantification because it labels at the cell culture level. Both 18O and isobaric labeling (e.g. iTRAQ or TMT), which label at the peptide level, offer similar levels of accuracy. Fig. 2 further exemplifies application of the distinctive strategies for different types of studies. Note that advances in the field continue to broaden the spectrum of applications.
TABLE 1.
Overview of the characteristics of different LC-MS-based quantification approaches
| Proteome coverage | Sample preparation workflow complexity | Quantification precision | Quantification dynamic range (log10) | Quantification level | No. of samples to compare | Cost per sample | |
|---|---|---|---|---|---|---|---|
| Spectral counting | High | Low | Low (>30% RSDa) | 2–3 | MS/MS | Unlimited | Low |
| AUC/ion intensity | High | Low | Medium (10–30% RSD) | 2–3 | MS | Unlimited | Low |
| SILAC | High | High | High (<10% RSD) | 1–2 | MS | 2–3 | High |
| 18O | Medium | Medium | Medium (10–20% RSD) | 1–2 | MS | 2 | Low |
| ICAT | Low | Medium | High (<10% RSD) | 1–2 | MS | 2 | Medium |
| iTRAQ/TMT | Medium to highb | Medium | High (<10% RSD) | 1–2 | MS/MS | 2–8 | High |
| 18O universal reference | Medium | Medium | Medium (10–20% RSD) | 1–2 | MS | Unlimited | Low |
| Super-SILAC | Medium | High | High (<10% RSD) | 1–2 | MS | Unlimited | High |
| SRM | Low | High | High (<10% RSD) | 4–5 | MS/MS | Unlimited | Highc |
a RSD, relative standard deviation; AUC, area under the curve.
b Prefractionation can lead to high proteome coverage in the iTRAQ or TMT approach.
c Synthesis of standard peptides or proteins is complex and can be very expensive.
FIGURE 2.
Schematic diagrams of three major strategies in quantitative proteomics using stable isotope labeling. A, pairwise comparison is used to compare two samples; 18O labeling, SILAC, and ICAT fall into this category. B, multiple comparison is used to compare up to four, six, or eight samples depending the isobaric tags used (i.e. 4-plex iTRAQ, 6-plex TMT, or 8-plex iTRAQ). C, large-scale comparison employs an internal standard: 18O-labeled universal reference, super-SILAC mixture, or synthetic isotope-labeled peptides. AQUA, absolute quantification.
Deciding which is the best quantitative approach to use is made more difficult by the fact that the modification stoichiometry may also change within a protein. For instance, many identified phosphoproteins have more than one phosphorylation site that is differentially regulated with individual functions (61). Therefore, it is crucial to distinguish whether the abundance change comes from site-specific phosphorylation or from the whole protein. Several quantitative phosphoproteomic studies have employed SILAC or iTRAQ-labeled SRM techniques to map the phosphorylation signaling network upon epidermal growth factor stimulation in different samples (41, 62, 63).
Conclusion and Perspective
In conclusion, proteomic quantification is a multifaceted term encompassing global and targeted measurements that can involve relative and/or absolute abundance determinations across large sets of proteins. No single “gold standard method” can resolve all of the analytical problems associated with proteomic quantification; biologists must choose the most appropriate method for particular biological applications.
Although LC-MS-based quantitative proteomics has made and is continuing to make large strides toward better understanding biological systems, its potential has not been fully realized. Bottom-up proteomic approaches rely on the assumption that proteins are completely digested into peptides that are all reproducibly detectable in MS analysis (12); this is rarely true in practice. Moreover, precise and accurate quantification of a specific protein is only achievable when the peptides are exclusively derived from a particular protein, so-called proteotypic peptides (64). “Missing values” still present a formidable challenge in proteomic data analysis, and effective quantification of most post-translational modifications is still in its infancy.
To realize true proteome-wide quantification, higher performance platforms providing better separations and linear responses over a wider dynamic range are required. Higher mass resolution and measurement accuracy can help differentiate target peptides from co-eluting molecules with similar mass; higher sensitivity will facilitate quantification on low-abundant proteins; and faster scanning rates will assist in quantifying proteins on the basis of more peptides (1). In addition, automated and more complete/uniform protein digestion, more efficient and higher throughput labeling, and more intelligent platform control software are also required to promote the wide application of quantitative proteomics.
This work was supported, in whole or in part, by National Institutes of Health Grant RR018522 from the National Center for Research Resources for Integrative Biology (to R. D. S.). This work was also supported by a United States Department of Energy early career research award (to W.-J. Q.). Experimental work was performed in the Environmental Molecular Sciences Laboratory, a Department of Energy/Biological and Environmental Research national scientific user facility on the Pacific Northwest National Laboratory campus in Richland, WA. Pacific Northwest National Laboratory is a multiprogram national laboratory operated by Battelle for the Department of Energy under Contract DE-AC05-76RLO 1830. This is the fourth article in the Thematic Minireview Series on Biological Applications of Mass Spectrometry. This minireview will be reprinted in the 2011 Minireview Compendium, which will be available in January, 2012.
- SILAC
- stable isotope labeling with amino acids in cell culture
- ICAT
- isotope-coded affinity tag
- iTRAQ
- isobaric tag for relative and absolute quantification
- TMT
- tandem mass tag
- SRM
- selected reaction monitoring.
REFERENCES
- 1. Ong S. E., Mann M. (2005) Nat. Chem. Biol. 1, 252–262 [DOI] [PubMed] [Google Scholar]
- 2. Domon B., Aebersold R. (2010) Nat. Biotechnol. 28, 710–721 [DOI] [PubMed] [Google Scholar]
- 3. Qian W. J., Jacobs J. M., Liu T., Camp D. G., 2nd, Smith R. D. (2006) Mol. Cell. Proteomics 5, 1727–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Domon B., Aebersold R. (2006) Science 312, 212–217 [DOI] [PubMed] [Google Scholar]
- 5. Cravatt B. F., Simon G. M., Yates J. R., 3rd (2007) Nature 450, 991–1000 [DOI] [PubMed] [Google Scholar]
- 6. Choudhary C., Mann M. (2010) Nat. Rev. Mol. Cell Biol. 11, 427–439 [DOI] [PubMed] [Google Scholar]
- 7. Wilm M. (2009) Proteomics 9, 4590–4605 [DOI] [PubMed] [Google Scholar]
- 8. Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B. (2007) Anal. Bioanal. Chem. 389, 1017–1031 [DOI] [PubMed] [Google Scholar]
- 9. Tang K., Page J. S., Smith R. D. (2004) J. Am. Soc. Mass Spectrom. 15, 1416–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schmidt A., Karas M., Dülcks T. (2003) J. Am. Soc. Mass Spectrom. 14, 492–500 [DOI] [PubMed] [Google Scholar]
- 11. Annesley T. M. (2003) Clin. Chem. 49, 1041–1044 [DOI] [PubMed] [Google Scholar]
- 12. Elliott M. H., Smith D. S., Parker C. E., Borchers C. (2009) J. Mass Spectrom. 44, 1637–1660 [DOI] [PubMed] [Google Scholar]
- 13. Liu H., Sadygov R. G., Yates J. R., 3rd (2004) Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 14. Qian W. J., Jacobs J. M., Camp D. G., 2nd, Monroe M. E., Moore R. J., Gritsenko M. A., Calvano S. E., Lowry S. F., Xiao W., Moldawer L. L., Davis R. W., Tompkins R. G., Smith R. D. (2005) Proteomics 5, 572–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Y., Wen Z., Washburn M. P., Florens L. (2009) Anal. Chem. 81, 6317–6326 [DOI] [PubMed] [Google Scholar]
- 16. Zhou J. Y., Schepmoes A. A., Zhang X., Moore R. J., Monroe M. E., Lee J. H., Camp D. G., Smith R. D., Qian W. J. (2010) J. Proteome Res. 9, 5698–5704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tabb D. L., MacCoss M. J., Wu C. C., Anderson S. D., Yates J. R., 3rd (2003) Anal. Chem. 75, 2470–2477 [DOI] [PubMed] [Google Scholar]
- 18. Smith R. D., Anderson G. A., Lipton M. S., Pasa-Tolic L., Shen Y., Conrads T. P., Veenstra T. D., Udseth H. R. (2002) Proteomics 2, 513–523 [DOI] [PubMed] [Google Scholar]
- 19. Strittmatter E. F., Ferguson P. L., Tang K., Smith R. D. (2003) J. Am. Soc. Mass Spectrom. 14, 980–991 [DOI] [PubMed] [Google Scholar]
- 20. Monroe M. E., Tolić N., Jaitly N., Shaw J. L., Adkins J. N., Smith R. D. (2007) Bioinformatics 23, 2021–2023 [DOI] [PubMed] [Google Scholar]
- 21. Li X. J., Yi E. C., Kemp C. J., Zhang H., Aebersold R. (2005) Mol. Cell. Proteomics 4, 1328–1340 [DOI] [PubMed] [Google Scholar]
- 22. Jaffe J. D., Mani D. R., Leptos K. C., Church G. M., Gillette M. A., Carr S. A. (2006) Mol. Cell. Proteomics 5, 1927–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Callister S. J., Barry R. C., Adkins J. N., Johnson E. T., Qian W. J., Webb-Robertson B. J., Smith R. D., Lipton M. S. (2006) J. Proteome Res. 5, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Polpitiya A. D., Qian W. J., Jaitly N., Petyuk V. A., Adkins J. N., Camp D. G., 2nd, Anderson G. A., Smith R. D. (2008) Bioinformatics 24, 1556–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang R., Regnier F. E. (2002) J. Proteome Res. 1, 139–147 [DOI] [PubMed] [Google Scholar]
- 26. Zhang R., Sioma C. S., Wang S., Regnier F. E. (2001) Anal. Chem. 73, 5142–5149 [DOI] [PubMed] [Google Scholar]
- 27. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 28. Stewart I. I., Thomson T., Figeys D. (2001) Rapid Commun. Mass Spectrom. 15, 2456–2465 [DOI] [PubMed] [Google Scholar]
- 29. Reynolds K. J., Yao X., Fenselau C. (2002) J. Proteome Res. 1, 27–33 [DOI] [PubMed] [Google Scholar]
- 30. Yao X., Afonso C., Fenselau C. (2003) J. Proteome Res 2, 147–152 [DOI] [PubMed] [Google Scholar]
- 31. Qian W. J., Monroe M. E., Liu T., Jacobs J. M., Anderson G. A., Shen Y., Moore R. J., Anderson D. J., Zhang R., Calvano S. E., Lowry S. F., Xiao W., Moldawer L. L., Davis R. W., Tompkins R. G., Camp D. G., 2nd, Smith R. D. (2005) Mol. Cell. Proteomics 4, 700–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fenselau C., Yao X. (2009) J. Proteome Res. 8, 2140–2143 [DOI] [PubMed] [Google Scholar]
- 33. Petritis B. O., Qian W. J., Camp D. G., 2nd, Smith R. D. (2009) J. Proteome Res. 8, 2157–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu T., Qian W. J., Strittmatter E. F., Camp D. G., 2nd, Anderson G. A., Thrall B. D., Smith R. D. (2004) Anal. Chem. 76, 5345–5353 [DOI] [PubMed] [Google Scholar]
- 35. Petyuk V. A., Qian W. J., Smith R. D., Smith D. J. (2010) Methods 50, 77–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Qian W. J., Liu T., Petyuk V. A., Gritsenko M. A., Petritis B. O., Polpitiya A. D., Kaushal A., Xiao W., Finnerty C. C., Jeschke M. G., Jaitly N., Monroe M. E., Moore R. J., Moldawer L. L., Davis R. W., Tompkins R. G., Herndon D. N., Camp D. G., Smith R. D. (2009) J. Proteome Res. 8, 290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mason C. J., Therneau T. M., Eckel-Passow J. E., Johnson K. L., Oberg A. L., Olson J. E., Nair K. S., Muddiman D. C., Bergen H. R., 3rd (2007) Mol. Cell. Proteomics 6, 305–318 [DOI] [PubMed] [Google Scholar]
- 38. Ramos-Fernández A., López-Ferrer D., Vázquez J. (2007) Mol. Cell. Proteomics 6, 1274–1286 [DOI] [PubMed] [Google Scholar]
- 39. Gygi S. P., Rist B., Gerber S. A., Turecek F., Gelb M. H., Aebersold R. (1999) Nat. Biotechnol. 17, 994–999 [DOI] [PubMed] [Google Scholar]
- 40. Yi E. C., Li X. J., Cooke K., Lee H., Raught B., Page A., Aneliunas V., Hieter P., Goodlett D. R., Aebersold R. (2005) Proteomics 5, 380–387 [DOI] [PubMed] [Google Scholar]
- 41. Wolf-Yadlin A., Hautaniemi S., Lauffenburger D. A., White F. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 43. Choe L., D'Ascenzo M., Relkin N. R., Pappin D., Ross P., Williamson B., Guertin S., Pribil P., Lee K. H. (2007) Proteomics 7, 3651–3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pierce A., Unwin R. D., Evans C. A., Griffiths S., Carney L., Zhang L., Jaworska E., Lee C. F., Blinco D., Okoniewski M. J., Miller C. J., Bitton D. A., Spooncer E., Whetton A. D. (2008) Mol. Cell. Proteomics 7, 853–863 [DOI] [PubMed] [Google Scholar]
- 45. Thompson A., Schäfer J., Kuhn K., Kienle S., Schwarz J., Schmidt G., Neumann T., Johnstone R., Mohammed A. K., Hamon C. (2003) Anal. Chem. 75, 1895–1904 [DOI] [PubMed] [Google Scholar]
- 46. Dayon L., Hainard A., Licker V., Turck N., Kuhn K., Hochstrasser D. F., Burkhard P. R., Sanchez J. C. (2008) Anal. Chem. 80, 2921–2931 [DOI] [PubMed] [Google Scholar]
- 47. Schulze W. X., Usadel B. (2010) Annu. Rev. Plant Biol. 61, 491–516 [DOI] [PubMed] [Google Scholar]
- 48. Gerber S. A., Rush J., Stemman O., Kirschner M. W., Gygi S. P. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Petyuk V. A., Qian W. J., Chin M. H., Wang H., Livesay E. A., Monroe M. E., Adkins J. N., Jaitly N., Anderson D. J., Camp D. G., 2nd, Smith D. J., Smith R. D. (2007) Genome Res. 17, 328–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Qian W. J., Petritis B. O., Kaushal A., Finnerty C. C., Jeschke M. G., Monroe M. E., Moore R. J., Schepmoes A. A., Xiao W., Moldawer L. L., Davis R. W., Tompkins R. G., Herndon D. N., Camp D. G., 2nd, Smith R. D. (2010) J. Proteome Res. 9, 4779–4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Geiger T., Cox J., Ostasiewicz P., Wisniewski J. R., Mann M. (2010) Nat. Methods 7, 383–385 [DOI] [PubMed] [Google Scholar]
- 52. Lange V., Picotti P., Domon B., Aebersold R. (2008) Mol. Syst. Biol. 4, 222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kiyonami R., Schoen A., Prakash A., Peterman S., Zabrouskov V., Picotti P., Aebersold R., Huhmer A., Domon B. (2011) Mol. Cell. Proteomics, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Addona T. A., Abbatiello S. E., Schilling B., Skates S. J., Mani D. R., Bunk D. M., Spiegelman C. H., Zimmerman L. J., Ham A. J., Keshishian H., Hall S. C., Allen S., Blackman R. K., Borchers C. H., Buck C., Cardasis H. L., Cusack M. P., Dodder N. G., Gibson B. W., Held J. M., Hiltke T., Jackson A., Johansen E. B., Kinsinger C. R., Li J., Mesri M., Neubert T. A., Niles R. K., Pulsipher T. C., Ransohoff D., Rodriguez H., Rudnick P. A., Smith D., Tabb D. L., Tegeler T. J., Variyath A. M., Vega-Montoto L. J., Wahlander A., Waldemarson S., Wang M., Whiteaker J. R., Zhao L., Anderson N. L., Fisher S. J., Liebler D. C., Paulovich A. G., Regnier F. E., Tempst P., Carr S. A. (2009) Nat. Biotechnol. 27, 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Duncan M. W., Aebersold R., Caprioli R. M. (2010) Nat. Biotechnol. 28, 659–664 [DOI] [PubMed] [Google Scholar]
- 56. Kirkpatrick D. S., Hathaway N. A., Hanna J., Elsasser S., Rush J., Finley D., King R. W., Gygi S. P. (2006) Nat. Cell Biol. 8, 700–710 [DOI] [PubMed] [Google Scholar]
- 57. Havlis J., Shevchenko A. (2004) Anal. Chem. 76, 3029–3036 [DOI] [PubMed] [Google Scholar]
- 58. Brun V., Dupuis A., Adrait A., Marcellin M., Thomas D., Court M., Vandenesch F., Garin J. (2007) Mol. Cell. Proteomics 6, 2139–2149 [DOI] [PubMed] [Google Scholar]
- 59. Pratt J. M., Simpson D. M., Doherty M. K., Rivers J., Gaskell S. J., Beynon R. J. (2006) Nat. Protoc. 1, 1029–1043 [DOI] [PubMed] [Google Scholar]
- 60. Kirkpatrick D. S., Gerber S. A., Gygi S. P. (2005) Methods 35, 265–273 [DOI] [PubMed] [Google Scholar]
- 61. Nita-Lazar A., Saito-Benz H., White F. M. (2008) Proteomics 8, 4433–4443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang Y., Wolf-Yadlin A., Ross P. L., Pappin D. J., Rush J., Lauffenburger D. A., White F. M. (2005) Mol. Cell. Proteomics 4, 1240–1250 [DOI] [PubMed] [Google Scholar]
- 63. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 64. Mallick P., Schirle M., Chen S. S., Flory M. R., Lee H., Martin D., Ranish J., Raught B., Schmitt R., Werner T., Kuster B., Aebersold R. (2007) Nat. Biotechnol. 25, 125–131 [DOI] [PubMed] [Google Scholar]