Abstract
Binding of class I MHC molecules (MHCI) to an inhibitory receptor, PIR-B, expressed on B cells and myeloid cells provides constitutive cellular inhibition, thus ensuring peripheral tolerance. Recent unexpected findings pointed to a novel inhibitory role of PIR-B in neurite regeneration through binding to three axonal outgrowth inhibitors of myelin, including Nogo. Thus, it becomes interesting to determine whether the actions of the inhibitory myelin proteins and MHCI could coexist independently or be mutually exclusive as to the PIR-B-mediated immune and neural cell inhibition. Here, we present data supporting the competition of Nogo- and MHCI-mediated inhibition where they coexist. Kinetic analyses of Nogo and MHCI binding to the whole or a part of the recombinant PIR-B ectodomain revealed that PIR-B binds with higher affinity to Nogo than MHCI and that the MHCI binding only occurred with the N-terminal domains of PIR-B, whereas Nogo binding occurred with either the N- or C-terminal ectodomains. Importantly, kinetic tests indicated that the binding to PIR-B of Nogo and MHCI was competitive. Both endogenous and exogenous Nogo intensified the PIR-B-mediated suppression of interleukin-6 release from lipopolysaccharide-stimulated wild-type, but not PIR-B-deficient, cultured mast cells, indicating that PIR-B mediates Nogo-induced inhibition. Thus, we propose a novel mechanism by which PIR-B-mediated regulation is achieved differentially but competitively via MHCI and Nogo in cells of the immune system.
Keywords: Immunosupressor, Mast Cell, Neurobiology, Protein Domains, Receptor Structure-Function, Dual Regulation, Major Histocompatibility Complex Class I, Neurite Outgrowth Inhibitor, Paired Receptor
Introduction
PIR-B (paired Ig-like receptor-B), expressed on murine myeloid lineage cells, B cells, and prethymic stage T cells is an immunoregulatory cell-surface receptor (1–5), whose physiological ligand in the immune system has been identified as class I MHC molecules (MHCI4; also known as class I H-2 and HLA in mice and humans, respectively) (6). Because MHCI are expressed ubiquitously, the binding to MHCI causes PIR-B to deliver a constitutive inhibitory signal via recruitment of tyrosine phosphatase, Src homology 2 domain-containing tyrosine phosphatase-1 (7–9), whose substrates are critical cytosolic signaling molecules such as Bruton's tyrosine kinase (10). Similar to other stimulatory and inhibitory receptor pairs with shared ligand-binding specificities (11), PIR-B expression usually takes place concomitantly with that of the activating-type isoform PIR-A although the physiological role of PIR-A in the immune system remains largely unknown.
Recent unexpected findings have revealed additional inhibitory roles of PIR-B in synaptic plasticity (12) and axonal regeneration (13) in the neuronal system. In mice lacking functional PIR-B, the cortical ocular dominance plasticity is more robust, indicating that PIR-B functions to limit the extent of experience-dependent plasticity in the visual cortex and suggesting that it may function broadly to stabilize neuronal circuits (12). Surprisingly, it has also been reported (13) that PIR-B can bind to three axonal outgrowth inhibitory proteins in oligodendrocytes, i.e. neurite outgrowth inhibitor protein (Nogo) (14), myelin-associated glycoprotein (MAG) (15, 16), and oligodendrocyte myelin glycoprotein (OMgp) (17), like the Nogo receptor does (18, 19). Interfering with PIR-B activity, either with antibodies or by genetic deletion of the cytoplasmic portion of PIR-B, partially reverses the neurite inhibition by Nogo, MAG, and OMgp, implying that PIR-B mediates the regeneration blocking of axons (13).
Given the two kinds of physiological ligands for PIR-B, i.e. MHCI and neurite outgrowth inhibitor proteins, we were immediately interested in determining how differently PIR-B binds to these immuno- and neuromodulators, examining simultaneously whether or not PIR-A can also bind to the myelin-associated inhibitor proteins in addition to MHCI.
EXPERIMENTAL PROCEDURES
Preparation of Recombinant PIR Domains
PIR-B, like PIR-A, is a type I transmembrane glycoprotein with a ligand-binding ectodomain structure composed of six Ig-like domains (D1 to D6, from the N to the C terminus). Recombinant proteins for D1–D6, D1D2, and D3–D6 of PIR-B and PIR-A were prepared according to the following procedures. PIR-B domain fragment DNAs were amplified from a C57BL/10-derived whole-length PIR-B cDNA inserted into the pcEXV-3 vector as a template by using following primers: PIR-B (D1–D6), 5′-CAGGTACCGGGTCCCTCCCTAAG-3′ and 5′-ATGATATCTGAGACTGTGAGCTC-3′; PIR-B (D1D2), 5′-AGGATATCTGAGACCAGGAGCTC-3′ and 5′-CAGGTACCGGGTCCCTCCCTAAG-3′; and PIR-B (D3-D6), 5′-TGGGTACCGGTAATCTCCAAAAA-3′ and 5′- ATGATATCTGAGACTGTGAGCTC-3′. Each DNA fragment was inserted into pHLsec vector or pSegTag2-B harboring Fc portion DNA of BALB/c IgG3. The resultant plasmids were transfected to 293-F (Invitrogen) as a host cell to express the recombinant proteins. PIR-A4 D1–D6 fragment DNAs were amplified from C57BL/6 spleen cDNA with the primers 5′-TCTAGAGAGATGCCATGTCCTGCACC-3′ and 5′-TCTAGAAGATTAGTTCCCAGCTGCAGG-3′. The fragment was inserted into pcDNA3.1 Zeo(+) (Invitrogen). The resultant plasmid was further digested with BglII and NgoMIV to obtain CMV promoter-insert-poly(A) site fragment and ligated into pSV2-dhfr (ATCC) as described previously (20). PIR-A4 D1D2 and D3–D6 DNA fragments were amplified from PIR-A4 D1–D6 DNA-inserted pcDNA3.1 Zeo(+) vector as a template with following primers: PIR-A4 (D1D2), 5′-CCATGGCCCTCCCTAAGCCTATCCTC-3′ and 5′-CCATGGGGAGCTCCACGGATTCAC-3′; PIR-A4 (D3–D6), 5′-GATATCAGGTAATCTCCAAAAACCAACCAT-3′ and 5′-GATATCGCTGAGACTGTGAGCTCCAC-3′. The DNA fragments were inserted into pFUSE-hIgG1e3-Fc2 vector (InvivoGen, San Diego, CA). For the expression of PIR-A4 D1–D6 monomer, the plasmid was transfected to the DG44 cell (Invitrogen) and applied for gene amplification with methotrexate (Sigma) as described previously (20). For the expression of PIR-A4 D1D2 and D3–D6 Fc fusion protein, the plasmids were transfected to CHO-K1 cells. Purification of recombinant proteins was performed by affinity purification with HiTrap Protein A (GE Healthcare) or D1D2-specific anti-PIR-A/B (6C1, a generous gift from Dr. H. Kubagawa, University of Alabama at Birmingham, and Dr. M. D. Cooper, Emory University, Atlanta, GA)-immobilized affinity column.
Surface Plasmon Resonance (SPR) Analysis of PIR Domain Binding to MHCI, Nogo-66, and MAG
For the SPR analysis of PIR, the following recombinant proteins were used. Biotinylated H-2Kb monomer (loaded with a tyrosinase-related protein 2 peptide, SVYDFFVWL and coupled with mouse β2m-B) (Sanquin, Amsterdam, Netherlands), H-2Db monomer (loaded with an influenza virus nucleoprotein F5 peptide, ASNENMDAM, and coupled with mouse β2m-B) (Sanquin), HLA-G monomer (loaded with a histone H2A derived peptide, RIIPRHLQL; kind gift from Dr. Maenaka, Kyushu University), Nogo-66-Fc (recombinant rat Nogo-A/Fc amino acids 1026–1090, R&D Systems, Minneapolis, MN) and MAG-Fc (recombinant rat MAG/Fc, R&D Systems). SPR analysis was performed with a BIAcore 2000 (BIAcore, Uppsala, Sweden) as essentially described elsewhere (6). In brief, ligand proteins were immobilized at 1500–2500 response units on research-grade CM5 chips (BIAcore) through covalently coupled streptavidin. Analyte proteins, after buffer exchange to HBS-EP buffer comprising 10 mm HEPES, pH 7.4, 150 mm NaCl, 3.4 mm EDTA, and 0.005% Surfactant P20 (BIAcore), were injected over the immobilized flow cells at 25 °C. The binding interactions were assayed at 20 μl/min. The binding responses with various concentrations were subtracted from the nonspecific responses to an empty flow cell. Kinetic constants were calculated from sensorgram data with the BIA evaluation program (version 3.0.2; BIAcore). KD values were obtained using the bivalent analyte model for Fc fusion proteins or determined from the equilibrium binding data fit to a 1:1 binding model for monomer proteins.
Mice
C57BL/6 mice were purchased from Charles River Japan Inc. (Yokohama, Japan). Pirb−/− mice were generated (21) and back-crossed with C57BL/6 for twelve generations. β2-microglobulin-deficient (B2m−/−) mice were purchased from The Jackson Laboratory (Bar Harbor, MA). NogoA/B-deficient mice were developed by Stephen M. Strittmatter (Yale University) (22). Mice were maintained and bred in the Animal Facility of The Institute of Development, Aging and Cancer, Tohoku University, an environmentally controlled and specific pathogen-free facility, according to the guidelines for experimental animals defined by the university, and the animal protocols were reviewed and approved by the Animal Studies Committee of the university. All experiments were performed on 8-to-14-week-old age-matched male and female mice.
Mast Cell Culture
For preparation of bone marrow (BM)-derived cultured mast cells (BMMCs), bone marrow cells were obtained from both hind limbs (femur and tibia) of 8-week-old female C57BL/6 mice. After lysing the erythrocytes with lysis buffer (0.144 m ammonium chloride solution), BM cells were suspended at a density of 5 × 105 cells/ml in RPMI 1640 medium (Sigma-Aldrich), supplemented with 10% FCS, 50 international units/ml penicillin, 50 μg/ml streptomycin, 50 μm 2-mercaptoethanol, 1 mm sodium pyruvate, 0.1 mm minimum essential medium nonessential amino acids solution, and 5 ng/ml mouse rIL-3 (R&D Systems), and then incubated at 37 °C under a humidified atmosphere of 5% CO2 and 95% air. The BM-derived nonadherent cells were transferred every 6–7 days to new culture flasks at a density of 3 × 105 cells/ml over a 4-to-10-week culture period. Over 90% of the cells were identified as immature mast cells at 4 weeks after initiation of the culture (23) and were designated as BMMCs.
Confocal Microscopy Analysis
Confocal laser-scanning microscopy of BMMCs was performed with Alexa Fluor®-546-conjugated (Invitrogen) Nogo-66-Fc (R&D Systems), Alexa 488-conjugated anti-PIR-A/B (6C1), and Alexa 647-conjugated anti-H-2Kb/Db (clone 28-6-8). All of these mAbs were directly labeled by an Alexa Fluor 488, 546, or 647 monoclonal labeling kit (Invitrogen). Alexa Fluor 488, 546, or 647-positive signals were collected by a confocal laser-scanning microscope (Fluoview FV1000, Olympus, Tokyo, Japan).
Linearization Analysis and Calculation of Correlation Coefficient
Linearization analysis determines the location of the receptors observed in one plane of the cell versus the whole cell. Scans of the BMMC surface were made by drawing ring-shaped area at the mid focal plane of each cell (20, 24). The fluorescence intensity of the two target molecules was normalized relative to the highest peak (y axis). The correlation coefficient was calculated on a pixel-by-pixel basis according to Equation 1,
where x and y are the intensities of the green and red channels in a pixel, and x and y are the average intensities of the green and red channels within the region of interest. The correlation coefficient theoretically ranges from −1 to 1, where 1 indicates perfect overlap, 0 indicates random distribution, and −1 indicates avoidance.
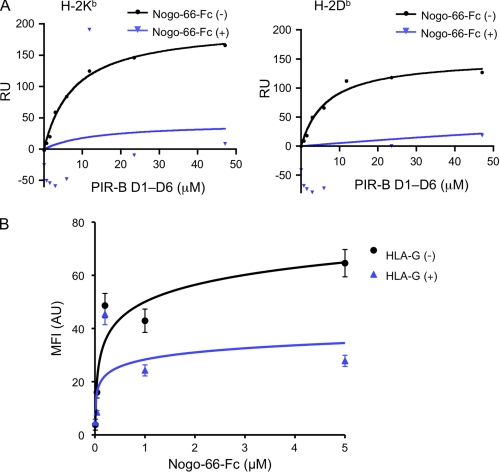
Competition Assay of Nogo-66-Fc/HLA-G Binding to PIR-B
5 × 105 cells/sample of PIR-B-transfected E.G7 cells (E.G7-PirB) (20) were stained with graded concentration (0 to 5 μm) of Alexa Fluor 546-conjugated Nogo-66-Fc in the presence or absence of a fixed concentration (7 μm) of recombinant HLA-G monomer for 40 min at 37 °C. The samples were then washed three times with PBS and suspended in 100 μl of PBS. Ten μl aliquot of the suspensions was mounted on a water-repellent and aminopropylsilane-coated grass slide (TF2404A, Matsunami Glass Ind., Ltd.), incubated for 15 min at room temperature, fixed with 4% paraformaldehyde, washed three times with PBS, and mounted with Fluoromount® (Diagnostic BioSystems, Pleasanton, CA) and coverglass. The slides were scanned and fluorescence intensity of the images was measured with Fluoview FV1000 confocal laser scanning microscopy (Olympus). Mean fluorescence intensity of each concentration was calculated from 16 images of two replicate wells.
Measurement of Cytokine IL-6
BMMCs cultured at 2 × 106 cells/ml in complete RPMI medium containing 10% FCS supplemented with 5 ng/ml IL-3 (PeproTech, Hamburg, Germany) in 96-well flat plates (Corning, Lowell, MA) were stimulated with 0–1000 ng/ml LPS (K12; InvivoGen) in the presence of 5 μg/ml NogoA-Fc chimeric protein (R&D Systems) or murine recombinant IgG2a-Fc (R&D Systems). Culture supernatants were collected 16 h after stimulation. The amount of IL-6 produced was determined with a mouse IL-6 ELISA Max Set standard (BioLegend, San Diego, CA).
Reverse Transcription-PCR
For RT-PCR analysis of mRNAs for Nogo isoforms (NogoA, B1, and B2), MAG, and OMgp in immune cells, total RNA was extracted with an RNeasy Mini Kit (Qiagen, Valencia, CA) from brain samples, and splenic CD19+ B cells and Thy-1+ T cells isolated from splenocytes with a magnetic-activated cell sorter (Miltenyi Biotec, Bergisch Gladbach, Germany), BM-derived GM-CSF-induced cultured DCs, BM-derived M-CSF-induced cultured macrophages, and BM-derived IL-3-induced BMMCs: all samples were from C57BL/6 mice. First-strand cDNA was synthesized with olido(dT) primer (TaKaRa Biotechnology, Ohtsu, Japan), and the cDNA was amplified with the following target-specific primers: NogoA (forward), 5′-GCAGAGCTGAATAAAACTTCAG-3′; NogoB1 (forward), 5′-GCAGGGGCTCGGGCTCAGTG-3′; NogoB2 (forward), 5′-GCTCTTCCTGCTGCATCTGAG-3′ and the common reverse primer for NogoA/B1/B2, 5′-GTTCACATGACCAAGAGCAG-3′; MAG, 5′-CCATGGGCCACTGGGGTGCCTG-3′ and 5′-CCATGGCAGGACCGATTTTGGCCCAC-3′; OMgp, 5′-ACCTCAAGCTTATTTACTATGAG-3′ and 5′-AGTGTTTCCATTTGCAGTGGTT-3′; and hypoxanthine phosphoribosyltransferase, 5′-TGATCAGTCAACGGGGGACA-3′ and 5′-TTCGAGAGGTCCTTTTCACCA-3′. Thirty cycles of PCR were carried out with Ex-TaqDNA polymerase (TaKaRa Biotechnology) with denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s, and elongation at 72 °C for 60 s.
Statistical Analysis
Statistical analyses were performed using two-way analysis of variance or Student's t test. p < 0.05 was considered as being statistically significant.
RESULTS
PIR-B D1D2 Binds to H-2 Molecules
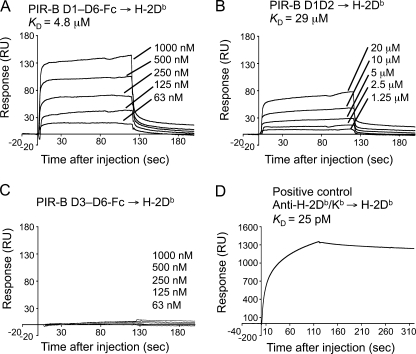
We prepared recombinant ectodomains of PIR-B, i.e. the whole D1–D6 protein, or the N-terminal D1D2 and C-terminal D3–D6 separately (supplemental “Results” and Fig. S1). These recombinant proteins, at various concentrations, were examined for their binding to a recombinant H-2 α chain·β2m complex fixed on a sensor chip for SPR analysis (Fig. 1). As shown in the sensorgrams for D1-D6 (Fig. 1A), D1D2 (Fig. 1B), and D3-D6 (Fig. 1C), binding of the PIR-B domains to H-2Db was detected for D1-D6 (KD = 4.0 ± 1.9 μm, Table 1) and D1D2 (KD = 29 ± 5 μm), but not for D3-D6. Thus, we concluded that the binding site for the H-2 α chain·β2m complex resides exclusively in D1D2 of PIR-B.
FIGURE 1.
SPR analysis of the binding of recombinant PIR-B and PIR-A domains to H-2. A–D, SPR analysis of PIR-B domain binding to H-2Db immobilized on biosensor chips. The H-2Db protein was immobilized on a sensor chip at ≤2200 response units (RU). Representative SPR profiles are shown for various concentrations of the D1–D6 (A), D1D2 (B), and D3–D6 proteins (C) bound to H-2Db, as well as for anti-H-2 antibodies bound to H-2Db as a positive control (D). Control Fc protein did not exhibit any detectable binding to H-2Db (not shown). The low response unit values of PIR-B D1–D6 and D1D2 binding to MHCI (maximally 6% of immobilized H-2 has been reacted with PIR-B D1–D6 in (A)) may represent instability of the steric structure of PIR recombinant proteins in solution, which might also cause an insufficient recovery of the proteins from culture supernatants.
TABLE 1.
Binding affinities of PIR ectodomains to various ligands
| Recombinant ectodomaina |
KDb |
||
|---|---|---|---|
| H-2Db | HLA-G | Nogo-66 | |
| μm | |||
| PIR-B | |||
| D1–D6 | 4.0 ± 1.9c (4) | 2.4 ± 0.7 (3) | 0.57d (1) |
| D1D2 | 29 ± 5 (3) | 24 ± 4 (3) | 8.5 (1) |
| D3–D6 | NBe (1) | NDf | 0.80 (1) |
| PIR-A | |||
| D1–D6 | +g (1) | ND | 6.9 (2) |
| D1D2 | ND | ND | NB (1) |
| D3–D6 | ND | ND | 4.3 (2) |
a Recombinant extracellular Ig-like domains of PIR (see supplemental Fig. S1A).
b Affinity was determined from the binding data fit to a bivalent analyte model for Fc fusion proteins or to a 1:1 binding model for fusion proteins without Fc.
c Affinity values were determined from the numbers of response units with various concentrations of analytes. Data are presented as the value(s) for either a single determination, the means for two different experiments, or the means ± S.D. for three to four different experiments, the number of experiments are given in parentheses. The KD value of PIR-B–H-2Db binding was larger than that reported previously (6), possibly due partly to the fact that in our previous SPR analysis, we employed the H-2 α chain complexed with human β2m to ensure the stability of the complex, and we calculated the KD at lower concentrations of anlayte than the current analysis.
d The KD value is larger than that reported previously (13), possibly due to the difference in the assay methods: Atwal et al. (13) employed an enzyme-linked immunosorbent assay.
e NB, no binding detected.
f ND, not done.
g Binding was detected with a fixed concentration of an analyte.
PIR-B Can also Bind to Either β2m-free H-2 or to Human HLA-G
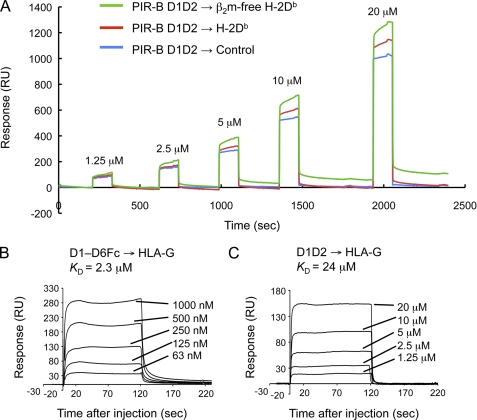
Recent reports have demonstrated that a human PIR-B ortholog, LILRB1 (leukocyte Ig-like receptor), does not recognize the β2m-free form of HLA-B27 or HLA-G, but that LILRB2 does (25, 26). This arises from differences in a limited number of amino acid residues (25, 26). Also, in the HLA-B27 transgenic rodent model, HLA-B27 heavy chain homodimers become ligands for PIR (27). These preceding observations suggest that PIR-B could also bind to the β2m-free form of MHC-I. Thus, we attempted to directly measure the PIR-B binding to the β2m-free α chain of H-2Db, prepared by acid treatment of the whole H-2 complex immobilized on a sensor chip. Successful removal of β2m probably along with a peptide from the H-2 heavy chain was confirmed by the fact that the H-2 on the sensor chip was not recognizable with anti-β2m polyclonal antibodies (data not shown). Our SPR analysis revealed that PIR-B D1D2 was able to bind to the β2m-free α chain of H-2Db (Fig. 2A) and even to a human nonclassical (or less polymorphic) MHCI, HLA-G (Fig. 2, B and C), indicating that PIR-B possesses broad binding specificity, binding to the whole MHCI complex, β2m (6), and the MHCI heavy chain lacking β2m and a peptide, and even to human MHCI.
FIGURE 2.
SPR analysis of the binding of recombinant PIR-B domains to β2m-free H-2Db and HLA-G. A, representative SPR profiles of the binding of D1D2, at various concentrations, to the acid-treated β2m-free H-2Db (green), nontreated H-2Db (red), and to an empty chip (blue). The H-2 protein was immobilized on sensor chips. B and C, representative SPR profiles of the binding of D1–D6 (B) and D1D2 (C), at various concentrations, to the HLA-G protein. RU, response units.
PIR-B D1-D6 Is Flexible
Although PIR-B D1D2 plays a major role in MHCI binding, PIR-B D3-D6 may play a role by supporting the MHCI binding by D1D2, such stabilization of the steric structure of D1D2. Also, the D3–D6 region together with D1D2 may be flexible under physiological conditions, which might make the receptor easily accessible to MHCI expressed on the same cell (in cis) (24) or on different cells (in trans) (20, 28). Binding of MHCI to the terminal ectodomain of PIR-B and a flexible nature of the ectodomain were supported by our data from protein tomography analysis of PIR-B ectodomain and MHCI in aqueous solution (supplemental “Results” and Fig. S2).
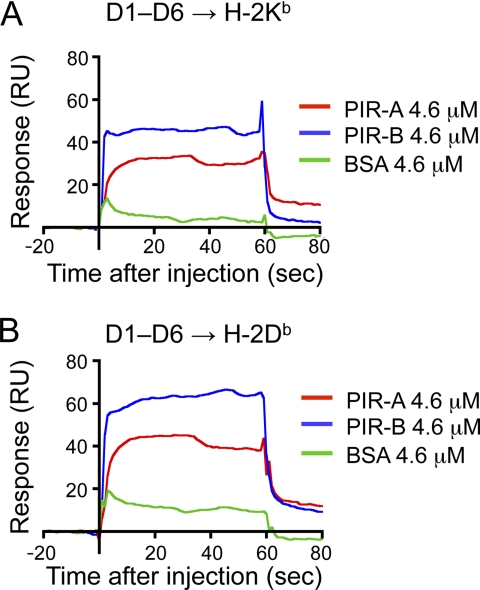
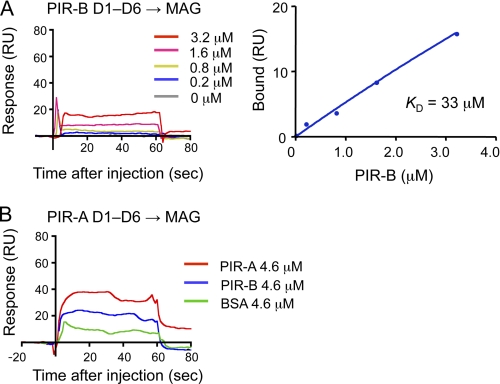
PIR-A Binds to MHCI with Less Efficiency than PIR-B
Regarding PIR-A binding to MHCI, although our previous study demonstrated that MHCI tetramer binds to PIR-A on Pirb−/− macrophages (6), SPR measurement of the binding of PIR-A to MHCI remains to be determined. Therefore, we also generated recombinant PIR-A D1D2, D3–D6, and D1–D6 (supplemental Fig. S1). Due to an unknown reason, we could not obtain enough amounts of the recombinant PIR-A proteins to determine the affinity values. Instead, we compared the binding response to MHCI between PIR-A and PIR-B. As shown in Fig. 3, PIR-A D1-D6 binds to the whole H-2 complex with a lesser efficiency than PIR-B D1–D6.
FIGURE 3.
SPR analysis of PIR-A binding to H-2. A and B, SPR analysis of PIR-A D1–D6 domain binding to H-2Kb (A) or H-2Db (B) immobilized on a biosensor chip. As positive and negative controls, PIR-B D1–D6 and BSA, respectively, are analyzed at the same concentration as that of PIR-A (4.6 μm). RU, response units.
PIR-B and PIR-A Bind to Nogo and MAG
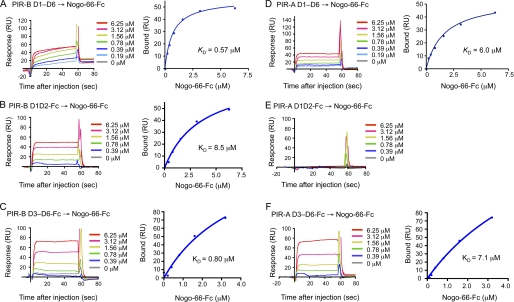
We next attempted to determine the binding kinetics of Nogo and MAG as to PIR-B. We also examined whether PIR-A binds to these axonal outgrowth inhibitors. Our SPR analysis of PIR-B D1–D6 binding to Nogo-66, a 66-amino acid peptide of Nogo essential for binding to the Nogo receptor (18, 19), revealed that the binding affinity was higher (KD = 0.57 μm, Fig. 4A) than that in the case of MHCI (Fig. 1A and Table 1). Interestingly, similar analysis revealed that PIR-A D1–D6 can also bind to Nogo-66 (Fig. 4D), although its affinity is much lower (KD = 6.9 μm, mean of two separate determinations, Table 1) than that of PIR-B. Additionally, we detected the binding of PIR-B D1-D6 to MAG (Fig. 5A), but the affinity was much lower (KD = 33 μm) than that of PIR-B as to Nogo-66. PIR-A D1-D6 binding to MAG was detected, and the affinity seemed to be higher than that in the case of PIR-B (Fig. 5B).
FIGURE 4.
SPR analysis of the binding of recombinant PIR-B and PIR-A domains to Nogo-66. A and D, SPR profiles of the binding of D1–D6 of PIR-B (A) or PIR-A (D), at various concentrations, to Nogo-66 protein immobilized on sensor chips. Negative control profiles (0 μm D1–D6) were utilized for subtraction from each profile using the Blank Run Subtraction program. B and C, SPR profiles of the binding of D1D2 (B) or D3-D6 (C) of PIR-B, at various concentrations, to Nogo-66 immobilized on sensor chips. Negative control profiles (0 μm D1D2) were utilized for subtraction from each profile using the Blank Run Subtraction program in A. E and F, SPR profiles of the binding of PIR-A D1D2 (E) or D3-D6 (F), at various concentrations, to Nogo-66 immobilized on sensor chips. RU, response units.
FIGURE 5.
SPR analysis of the binding of recombinant PIR-B and PIR-A D1-D6 domains to MAG. SPR profiles for various concentrations of PIR-B D1-D6 (A) or a fixed concentration (4.6 μm) of PIR-A D1–D6 (B) to MAG immobilized on sensor chips. In B, as positive and negative controls, PIR-B D1–D6 and BSA, respectively, are analyzed at the same concentration as that of PIR-A. RU, response units.
Both D1D2 and D3–D6 of PIR-B Bind to Nogo
Given that PIR-B and PIR-A bind to Nogo-66, we were interested in dissecting the binding domains. Again, D1D2 and D3–D6 of PIR-B and PIR-A were subjected to SPR analysis of binding to Nogo-66. We found that both D1D2 and D3-D6 of PIR-B can bind to Nogo-66 (Fig. 4B, C). The affinity of D1D2 or D3–D6 to Nogo-66 (KD = 8.5 μm or 0.80 μm, respectively) was lower than, or roughly comparable with, that of D1–D6 (0.57 μm, Fig. 4A and Table 1), respectively. On the other hand, PIR-A D3–D6 was found to bind to Nogo-66 with affinity (KD = 4.3 μm, mean of two separate determinations, Fig. 4F and Table 1) comparable with that of the D1–D6 of PIR-A (KD = 6.9 μm, mean of two separate determinations, Fig. 4D and Table 1). We failed to detect, however, binding activity of PIR-A D1D2 as to Nogo-66 (Fig. 4E). These results indicate that, unlike MHCI binding, both D1D2 and D3–D6 of PIR-B can bind to Nogo-66, and that, unlike PIR-B binding, PIR-A binding to Nogo-66 only occurs with D3–D6.
The differential binding of MHCI and Nogo to PIR-B shown above suggests that PIR-B harbors only one binding region for MHCI, D1D2, whereas it harbors two or more separate binding regions for Nogo-66. MHCI is a 57-kDa protein complex. On the other hand, a dimeric Nogo-66, which is a critical portion of Nogo-66-Fc chimera employed as a ligand for our SPR analysis, is 16 kDa. Because the molecular masses of PIR-B D1D2 and D3–D6 are 22 and 44 kDa, respectively, each of these portions of PIR-B is sufficiently large enough to accept one or more dimeric Nogo-66 peptides. Therefore, our next question was how much the binding site(s) for Nogo-66 in PIR-B overlap(s) with that of MHCI, or if these two ligands compete with each other for binding.
Binding of Nogo and MHCI to PIR-B Is Competitive
We initially anticipated that PIR-B is able to bind simultaneously to MHCI and Nogo-66. In partial support of this, our confocal laser microscopic analysis of intrinsically expressed PIR-B and MHCI on BMMCs and the extrinsically added Nogo-66-Fc recombinant protein bound to the cells revealed that these three molecules co-localized partly, but not solely, on the same cell surface (supplemental Fig. S3). However, this result does not verify a formation of a ternary complex of Nogo-PIR-B-MHCI. Therefore, we next examined a potential competition between Nogo and MHCI to PIR-B by SPR analysis (Fig. 6A). H-2Kb- or H-2Db-immobilized sensor chip was subjected for binding to PIR-B D1-D6 in the presence or absence of fixed concentration of Nogo-66-Fc chimera. We found that in the presence of Nogo-66-Fc, MHCI binding to PIR-B was markedly reduced, indicating that Nogo-66 inhibits MHCI binding efficiently in our experimental setting. We then intended to verify the competitive nature of Nogo-66 and MHCI in a cell-based assay. To this end, PIR-B-transfected E.G7 cells were stained with graded concentration of fluorochrome-conjugated Nogo-66-Fc in the presence or absence of fixed concentration of recombinant HLA-G protein (Fig. 6B). We found that in the presence of HLA-G, Nogo-66 binding to the cells was inhibited. These results indicate that Nogo-66 and MHCI are competitive in their binding to PIR-B. Our results also suggest that PIR-B on cell surface can exert its regulatory function differentially but competitively through endogenous Nogo and MHCI. This notion is examined below.
FIGURE 6.
Competitive binding of MHCI and Nogo-66 to PIR-B. A, SPR analysis of the competitive binding to PIR-B of H-2 and Nogo-66-Fc. Various concentrations of PIR-B D1-D6 was flowed over an H-2Kb-immobilized (2200 response units (RU)) or H-2Db-immobilized (1700 response units) sensor chip in the presence or absence of a fixed concentration (5 μm) of Nogo-66-Fc. B, cell-based competitive binding assay of PIR-B and Nogo-66-Fc in the presence or absence of HLA-G. PIR-B-transfected E.G7 cells were stained with graded concentration (0 to 5 μm) of Alexa Fluor 546-conjugated Nogo-66-Fc in the presence or absence of a fixed concentration (7 μm) of recombinant HLA-G. Acquisition of the image and measurement of the fluorescent intensity were performed under a Fluoview FV1000 confocal laser scanning microscope. Each point represents the mean ± S.E. from 16 images of two replicate wells. MFI, mean fluorescence intensity; AU, arbitrary unit; RU, response units.
Endogenous Nogo and MHCI Induce PIR-B-mediated Inhibition of IL-6 Release from LPS-stimulated Mast Cells
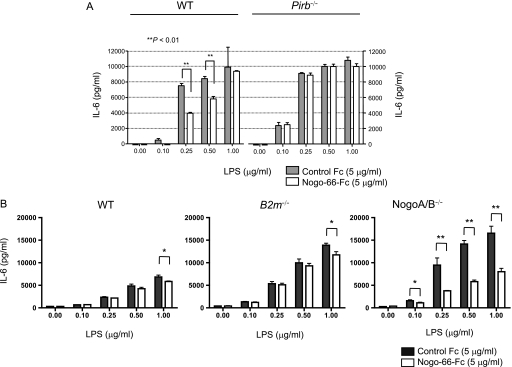
We stimulated BMMCs from wild-type C57BL/6 or Pirb−/− mice with various concentrations of LPS, a ligand for Toll-like receptor 4, in the presence of plate-bound Nogo-66-Fc fusion protein or control Fc. As already shown in our previous study (24), the IL-6 release from LPS-stimulated C57BL/6 BMMCs was lower than that from Pirb−/− cells, suggesting that Toll-like receptor 4-mediated BMMC activation is inhibited by PIR-B (Fig. 7A). Interestingly, this attenuated release of IL-6 from C57BL/6 BMMCs decreased to some extent in the presence of plate-bound Nogo-66, whereas the IL-6 release from Pirb−/− BMMCs was not modulated significantly by Nogo-66. These observations suggest that exogenous Nogo-66 induces an inhibitory response via PIR-B in BMMCs, and that constitutive PIR-B-mediated inhibition could further be strengthened to some extent by exogenous Nogo-66. To verify our notion further, we additionally employed BMMCs from B2m−/− and NogoA/B-deficient mice and tested them for IL-6 release upon LPS stimulation in the presence or absence of extrinsic Nogo-66-Fc. As shown in Fig. 7B, endogenous B2m and NogoA/B deficiencies both resulted in the increased IL-6 release compared with wild-type BMMCs, suggesting the inhibitory function of these molecules working as the endogenous ligands for PIR-B. Of note, addition of Nogo-66 in the culture reduced the IL-6 release response of wild-type and B2m−/− BMMCs only marginally but significantly (Fig. 7B, left and middle). Interestingly, in the absence of intrinsic Nogo, the suppression by extrinsic Nogo-66 was more evident (Fig. 7B, right), suggesting that intrinsic NogoA/B in BMMCs play a role in the suppression.
FIGURE 7.
Nogo augments PIR-B-mediated suppression of IL-6 release from cultured mast cells stimulated with LPS. Bone marrow-derived cultured mast cells from C57BL/6 (wild type), Pirb−/−, B2m−/−, or NogoA/B-deficient mice were stimulated with various concentrations of LPS in the presence of Nogo-66-Fc or control Fc proteins. After 16 h (A) or 10 h (B), the culture supernatants were collected and subjected to IL-6 measurement by ELISA. Data are shown as mean of triplicate samples ± S.D. Representative data for two experiments with similar results are shown (*, p < 0.05; **, p < 0.01). In A, the IL-6 release from wild-type mast cells was slightly but significantly inhibited by Nogo-66 at some LPS concentrations, whereas such inhibition was not observed in the absence of PIR-B. Note that with PIR-B deficiency, the IL-6 release was higher than from the cells with PIR-B, indicating that PIR-B inhibits the mast cell response. In B, although the observed Nogo-66-mediated suppression in wild type and B2m−/− cells was statistically significant only at 1 μg/ml LPS, its suppression was much more significant in cells with no intrinsic NogoA/B. Note again that with B2m and NogoA/B deficiencies, the IL-6 release was higher than from wild type cells, indicating that endogenous β2m and NogoA/B inhibit mast cell response.
DISCUSSION
In this study, we have shown for the first time that MHCI and Nogo bind to PIR-B differentially in terms of the binding affinities and the binding site(s). Interestingly, the binding was competitive when MHCI and Nogo co-exist. Additionally, we have found that PIR-A can bind to Nogo and MAG and that its affinity of the Nogo binding to PIR-A was ∼10 times lower than that of Nogo-PIR-B binding. Moreover, we have found that Nogo can augment the PIR-B-mediated inhibition of mast cell activation through Toll-like receptor 4 in vitro. Based on our observation, in which Nogo binds PIR-B with a higher affinity than MHCI does, one may speculate that the formation of Nogo·PIR-B complex dominates than that of MHCI·PIR-B complex. We have to, however, make a precise titration of MHCI and Nogo on cell surface before we fully understand the nature of PIR-B-mediated inhibition by these two endogenous ligands. Nevertheless, our observations postulate a novel mechanism in which Nogo and MHC-I competitively modulate the PIR-B-mediated regulation of the immune cells.
Can Neuronal PIR-B also Be Regulated Differentially via Nogo and MHCI?
Nogo splicing isoforms, NogoA and NogoB, are implicated in the regulation of neural and cardiovascular functions through the Nogo receptor, such as axonal regeneration, vascular remodeling, and chemotaxis (14, 29, 30). We detected several mRNAs for Nogo isoforms, including NogoA, NogoB1, and NogoB2, and two other axonal outgrowth inhibitory proteins, MAG and OMgp, in T cells, B cells, dendritic cells, macrophages, and BMMCs at different levels (supplemental Fig. S4). Our present observation of a Nogo-mediated inhibitory effect on mast cell function, together with the RT-PCR detection of Nogo/MAG/OMgp mRNAs, suggests that Nogo, and potentially MAG and OMgp as well, have a physiological impact on the immune system. Conversely, in the neuronal system, one may immediately speculate that PIR-B-mediated inhibition of neurite regeneration (13) and neural plasticity (12) could also be mediated synergistically by Nogo/MAG/OMgp and MHCI (31, 32). This scenario, particularly a potential co-expression and co-operation of Nogo/MAG/OMgp and MHCI in the cells, should be examined in detail in the next step toward a full understanding of the mechanism of PIR-B-mediated neuronal regulation.
Implication of PIR-A Binding to MHC-I and Nogo/MAG
In this study, we performed parallel analysis of PIR-A as a reference for PIR-B and found that it also binds to Nogo, but unlike in the case of PIR-B, the binding was restricted to D3–D6 with much lower affinity than that for PIR-B (Fig. 4 and Table 1). Also, PIR-A D1–D6 binding to MAG was detected, like for PIR-B (Fig. 5). Although it has not been examined whether neuronal cells express PIR-A, the possibility of its expression should not necessarily be excluded, because transcriptional activation of Pirb and Pira occurs in parallel at least in B cells (3, 9). However, cell-surface expression of PIR-A requires its association with the Fc receptor common γ chain (3, 33, 34), whose expression in neural cells is yet to be defined. If these are the cases, PIR-A can also have an opposite impact on axonal regeneration to that of PIR-B, namely as an accelerator of neurite outgrowth or synaptic plasticity. Enhancing such an activating function of PIR-A and blocking PIR-B-mediated inhibition might become important for therapeutic use for neural regeneration after injury.
Supplementary Material
Acknowledgments
We thank H. Furukawa, H. Kubagawa, and M. D. Cooper for providing the reagents and N. Halewood for editorial assistance.
This work was supported in part by the Core Research for Evolutional Science and Technology (CREST) Program of the Japan Science and Technology Agency, a grant-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and a grant from the 21st Century Center of Excellence (COE) Program “Center for Innovative Therapeutic Development Towards the Conquest of Signal Transduction Diseases” and Global COE Program “Innovative Therapeutic Development Towards the Conquest of Signal Transduction Diseases with Network Medicine”.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” “Results,” Figs. S1–S4, and additional references.
- MHCI
- major histocompatibility complex class I molecule(s)
- HLA
- human leukocyte antigen
- β2m
- β2 microglobulin
- BMMC
- bone marrow-derived cultured mast cell
- MAG
- myelin-associated glycoprotein
- Nogo-66
- neurite outgrowth inhibitor protein 66 amino-acid peptide
- OMgp
- oligodendrocyte-myelin glycoprotein
- SPR
- surface plasmon resonance.
REFERENCES
- 1. Hayami K., Fukuta D., Nishikawa Y., Yamashita Y., Inui M., Ohyama Y., Hikida M., Ohmori H., Takai T. (1997) J. Biol. Chem. 272, 7320–7327 [DOI] [PubMed] [Google Scholar]
- 2. Kubagawa H., Burrows P. D., Cooper M. D. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 5261–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kubagawa H., Chen C. C., Ho L. H., Shimada T. S., Gartland L., Mashburn C., Uehara T., Ravetch J. V., Cooper M. D. (1999) J. Exp. Med. 189, 309–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Masuda K., Kubagawa H., Ikawa T., Chen C. C., Kakugawa K., Hattori M., Kageyama R., Cooper M. D., Minato N., Katsura Y., Kawamoto H. (2005) EMBO J. 24, 4052–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Takai T. (2005) Immunology 115, 433–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nakamura A., Kobayashi E., Takai T. (2004) Nat. Immunol. 5, 623–629 [DOI] [PubMed] [Google Scholar]
- 7. Bléry M., Kubagawa H., Chen C. C., Vély F., Cooper M. D., Vivier E. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2446–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maeda A., Kurosaki M., Ono M., Takai T., Kurosaki T. (1998) J. Exp. Med. 187, 1355–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ho L. H., Uehara T., Chen C. C., Kubagawa H., Cooper M. D. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 15086–15090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maeda A., Scharenberg A. M., Tsukada S., Bolen J. B., Kinet J. P., Kurosaki T. (1999) Oncogene 18, 2291–2297 [DOI] [PubMed] [Google Scholar]
- 11. Colonna M., Samaridis J. (1995) Science 268, 405–408 [DOI] [PubMed] [Google Scholar]
- 12. Syken J., Grandpre T., Kanold P. O., Shatz C. J. (2006) Science 313, 1795–1800 [DOI] [PubMed] [Google Scholar]
- 13. Atwal J. K., Pinkston-Gosse J., Syken J., Stawicki S., Wu Y., Shatz C., Tessier-Lavigne M. (2008) Science 322, 967–970 [DOI] [PubMed] [Google Scholar]
- 14. Prinjha R., Moore S. E., Vinson M., Blake S., Morrow R., Christie G., Michalovich D., Simmons D. L., Walsh F. S. (2000) Nature 403, 383–384 [DOI] [PubMed] [Google Scholar]
- 15. Domeniconi M., Cao Z., Spencer T., Sivasankaran R., Wang K., Nikulina E., Kimura N., Cai H., Deng K., Gao Y., He Z., Filbin M. (2002) Neuron 35, 283–290 [DOI] [PubMed] [Google Scholar]
- 16. Liu B. P., Fournier A., GrandPré T., Strittmatter S. M. (2002) Science 297, 1190–1193 [DOI] [PubMed] [Google Scholar]
- 17. Wang K. C., Koprivica V., Kim J. A., Sivasankaran R., Guo Y., Neve R. L., He Z. (2002) Nature 417, 941–944 [DOI] [PubMed] [Google Scholar]
- 18. Fournier A. E., GrandPre T., Strittmatter S. M. (2001) Nature 409, 341–346 [DOI] [PubMed] [Google Scholar]
- 19. McGee A. W., Strittmatter S. M. (2003) Trends Neurosci. 26, 193–198 [DOI] [PubMed] [Google Scholar]
- 20. Endo S., Sakamoto Y., Kobayashi E., Nakamura A., Takai T. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14515–14520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ujike A., Takeda K., Nakamura A., Ebihara S., Akiyama K., Takai T. (2002) Nat. Immunol. 3, 542–548 [DOI] [PubMed] [Google Scholar]
- 22. Kim J. E., Li S., GrandPré T., Qiu D., Strittmatter S. M. (2003) Neuron 38, 187–199 [DOI] [PubMed] [Google Scholar]
- 23. Kanehira M., Kaifu T., Maya K., Kaji M., Nakamura A., Obinata M., Takai T. (2006) J. Biochem. 140, 211–220 [DOI] [PubMed] [Google Scholar]
- 24. Masuda A., Nakamura A., Maeda T., Sakamoto Y., Takai T. (2007) J. Exp. Med. 204, 907–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shiroishi M., Kuroki K., Rasubala L., Tsumoto K., Kumagai I., Kurimoto E., Kato K., Kohda D., Maenaka K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16412–16417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Allen R. L., Raine T., Haude A., Trowsdale J., Wilson M. J. (2001) J. Immunol. 167, 5543–5547 [DOI] [PubMed] [Google Scholar]
- 27. Kollnberger S., Bird L. A., Roddis M., Hacquard-Bouder C., Kubagawa H., Bodmer H. C., Breban M., McMichael A. J., Bowness P. (2004) J. Immunol. 173, 1699–1710 [DOI] [PubMed] [Google Scholar]
- 28. Shatz C. J. (2009) Neuron 64, 40–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Acevedo L., Yu J., Erdjument-Bromage H., Miao R. Q., Kim J. E., Fulton D., Tempst P., Strittmatter S. M., Sessa W. C. (2004) Nat. Med. 10, 382–388 [DOI] [PubMed] [Google Scholar]
- 30. Miao R. Q., Gao Y., Harrison K. D., Prendergast J., Acevedo L. M., Yu J., Hu F., Strittmatter S. M., Sessa W. C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10997–11002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Datwani A., McConnell M. J., Kanold P. O., Micheva K. D., Busse B., Shamloo M., Smith S. J., Shatz C. J. (2009) Neuron 64, 463–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boulanger L. M. (2009) Neuron 64, 93–109 [DOI] [PubMed] [Google Scholar]
- 33. Maeda A., Kurosaki M., Kurosaki T. (1998) J. Exp. Med. 188, 991–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Taylor L. S., McVicar D. W. (1999) Blood 94, 1790–1796 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.