Abstract
Genome integrity is maintained by a network of DNA damage response pathways, including checkpoints and DNA repair processes. In Saccharomyces cerevisiae, the BRCT domain-containing protein Rtt107/Esc4 is required for the restart of DNA replication after successful repair of DNA damage and for cellular resistance to DNA-damaging agents. In addition to its well characterized interaction with the endonuclease Slx4, Rtt107 interacts with a number of other DNA repair and recombination proteins. These include the evolutionarily conserved SMC5/6 complex, which is involved in numerous chromosome maintenance activities, such as DNA repair, chromosome segregation, and telomere function. The interaction between Rtt107 and the SMC5/6 complex was mediated through the N-terminal BRCT domains of Rtt107 and the Nse6 subunit of SMC5/6 and was independent of methyl methane sulfonate-induced damage and Slx4. Supporting a shared function in the DNA damage response, Rtt107 was required for recruitment of SMC5/6 to DNA double strand breaks. However, this functional relationship did not extend to other types of DNA lesions such as protein-bound nicks. Interestingly, Rtt107 was phosphorylated when SMC5/6 function was compromised in the absence of DNA-damaging agents, indicating a connection beyond the DNA damage response. Genetic analyses revealed that, although a subset of Rtt107 and SMC5/6 functions was shared, these proteins also contributed independently to maintenance of genome integrity.
Keywords: Chromatin Immunoprecipitation (ChIP), Chromosomes, DNA Damage, Genetics, Yeast, DNA Double Strand Breaks
Introduction
Eukaryotic cells have evolved complex mechanisms to maintain genome integrity, which is essential for genetic inheritance and cell viability. One of the central causes of genome instability is the failure to repair damaged DNA resulting from the constant assault of chemicals, radiation, or biological processes. Many different types of DNA lesions can occur, the most severe being double strand breaks (DSBs).4 In Saccharomyces cerevisiae, DSB ends are immediately sensed and bound by the MRX (Mre11-Rad50-Xrs2) complex (1). A signaling cascade is triggered, leading to the activation of the kinases Mec1 and Tel1, the yeast homologues of mammalian ATR (ATM and Rad 3-related) and ATM (ataxia-telangiectasia mutated), and recruitment of a whole host of DNA damage response proteins to the DSB (2, 3).
One of the downstream phosphorylation targets of Mec1 is Rtt107/Esc4, which is required for reinitiating replication after repair of alkylating DNA damage (4, 5). Deletion of the RTT107 gene results in hypersensitivity to DNA-damaging agents such as the DNA-alkylating agent methyl methane sulfonate (MMS), the nucleotide reductase inhibitor hydroxyurea, and the topoisomerase I poison camptothecin (4–6). However, the requirement of Rtt107 for resistance to DNA-damaging agents is alleviated when the chromatin regulatory pathway leading to H3 K79 trimethylation is inhibited (7). Nevertheless, even in the absence of DNA-damaging agents, rtt107Δ mutants exhibit chromosome instability and an increased incidence of Rad52 foci, indicative of a failure to properly process DNA damage or stalled DNA replication forks (8, 9).
Rtt107 contains several BRCT (BRCA1 C-terminal) homology domains, which often serve as phospho-binding modules to recruit signaling complexes and repair factors to DNA damage-induced lesions (4, 10). Consistent with a role as a scaffold for protein-protein interactions during the DNA damage response, Rtt107 interacts with a number of DNA repair and recombination proteins (5, 11–13). The best characterized Rtt107-interacting partner is the replication-specific endonuclease Slx4, which interacts with the N-terminal BRCT domains of Rtt107 (5). Slx4 is required for Mec1-dependent phosphorylation of Rtt107 and, like Rtt107, facilitates resumption of DNA replication after DNA damage (5). However, it has become clear over the last few years that Rtt107 also has Slx4-independent functions, and vice versa. Consistent with this, the defects in DNA damage response are generally more severe in rtt107Δ mutants than those of slx4Δ mutants, and the rtt107Δ slx4Δ double mutants are more sensitive to MMS than either of the single mutants (5).
Although the Rtt107-Slx4 interaction is well characterized, the Slx4-independent functions of Rtt107 and the proteins associated with these have yet to be elucidated. As an example, Rtt107 was recently identified to associate with the evolutionarily conserved SMC5/6 complex (11). The latter is a large multisubunit complex comprising the Smc5-Smc6 heterodimer and six non-Smc elements (Nse1–6), which are all encoded by essential genes in budding yeast (14–16). Smc5 and Smc6 are members of the structural maintenance of chromosome (SMC) proteins, a group that includes condensin (Smc1–3) and cohesin (Smc2–4) (15). The SMC5/6 complex is important for numerous chromosome maintenance activities, including DNA repair, chromosome segregation, and telomere function (15). Genome-wide mapping of the budding yeast SMC5/6 complex revealed that it localizes to centromeric regions, long chromosome arms, and rDNA arrays in unchallenged cells, as well as stalled DNA replication forks and DSBs (17). Interestingly, the recruitment of the SMC5/6 complex to these diverse chromosomal regions is differentially regulated, suggesting the involvement of multiple mechanisms and protein interaction partners (17).
In this study we characterized the interaction between Rtt107 and the SMC5/6 complex, and found that this association was mediated via the N-terminal BRCT domain of Rtt107 and the Nse6 subunit of the SMC5/6 complex. Here, we determined that the function underlying this interaction was a requirement of Rtt107 for the recruitment of the SMC5/6 complex to DNA DSBs, although not to certain DNA lesions such as protein-bound nicks. Moreover, this interaction was independent of Slx4 and DNA damage, consistent with this being a Slx4-independent role for Rtt107. Our results suggest that Rtt107 and the SMC5/6 complex cooperate together at DSBs, but they also have distinct functions in the DNA damage response.
EXPERIMENTAL PROCEDURES
Yeast Strains and Plasmids
All yeast strains used in this study are listed in supplemental Table S1 and created using standard yeast genetic techniques (18). Complete gene deletions and integration of TAP, FLAG, HA, or VSV tags at the 3′ end of genes were achieved using one-step gene integration of PCR-amplified modules (19–22). The mec1Δ mutants were created by complete deletion of MEC1 in sml1Δ strains. The nse5-R103G allele was created in Katsuhiko Shirahige's laboratory using standard procedures. The yeast strains containing the smc6–9 or nse3-SB1 allele were generous gifts from Luis Aragon (Imperial College London) and Philip Hieter (University of British Columbia), respectively. The GFP::NATMX6 plasmid and the pJ69–4a/α yeast strains were obtained from Elizabeth Conibear (University of British Columbia). The yeast two-hybrid plasmids were constructed by first cloning the genes into the pCR8GW-Topo Gateway Entry vector (Invitrogen) and subsequent cloning into either the pGal4 DBD-Dest or pGal4 AD-Dest gateway destination vectors obtained from Stefan Taubert (University of British Columbia).
Growth and DNA Damage Sensitivity Assays
Overnight cultures grown in YPD were diluted to 0.3 A600 and grown in YPD (Bacto Yeast extract, Bacto Peptone, 2% dextrose) to ∼1–2 A600. The cells were 10-fold serially diluted and spotted onto solid YPD plates or plates with MMS, camptothecin, or hydroxyurea (Sigma) at various concentrations. The plates were then incubated at the indicated temperature for 2–3 days.
Analytical-scale Interaction Assays, Immunoprecipitation, and Phosphatase Treatment
Overnight cultures were diluted to 0.3 A600 and grown in YPD to 0.8 A600, and cells were collected for immunoprecipitation. The procedure for analytical-scale immunoprecipitation of the epitope-tagged proteins was adapted from a previous report (23). Briefly, yeast cells were harvested and lysed in TAP-IP buffer (50 mm Tris (pH 7.8), 150 mm NaCl, 1.5 mm MgAc, 0.15% Nonidet P-40, 1 mm DTT, 10 mm NaPPi, 5 mm EGTA, 5 mm EDTA, 0.1 mm Na3VO4, 5 mm NaF, CompleteTM Protease inhibitor mixture (Roche)) using acid-washed glass beads and mechanically disrupted using a bead beater (BioSpec Products). Epitope-tagged fusion proteins were captured using IgG-Sepharose beads (Amersham Biosciences), anti-FLAG M2-agarose beads (Sigma), anti-VSV-agarose beads (Abcam), or anti-HA-agarose beads (Sigma) and subsequently washed in TAP-IP buffer. Captured and co-purifying proteins were detected by immunoblotting with anti-FLAG M2 (Sigma), anti-HA (Applied Biological Materials), or anti-VSV (Bethyl) antibodies and visualized using the Odyssey Infrared Imaging System (Licor).
The protocol for phosphatase treatment was adapted from a previous report (24). Briefly, analytical-scale immunoprecipitation was performed as described above and washed with TAP-IP buffer lacking NaPPi, EGTA, EDTA, Na3VO4, and NaF. Beads were resuspended in 40 μl of 1× NEbuffer for protein metallophosphatases (50 mm HEPES, 100 mm NaCl, 2 mm DTT, 0.01% Brij 35, 1 mm MnCl2) with or without the addition of 200 units of λ-phosphatase (New England Biolabs) in the presence or absence of EDTA (100 mm) and incubated for 30 min at 30 °C. After the incubation, beads were washed twice with the modified TAP-IP buffer, and analyzed as above.
Rtt107-TAP complexes were purified from extracts obtained from 2-liter cultures that were harvested in late logarithmic phase. Our protocol was modified from published protocols to allow immunoprecipitation of the TAP tag using calmodulin beads (23). Briefly, cells were disrupted with a coffee grinder in the presence of dry ice pellets and resuspended in 0.8 volumes/weight of lysis buffer (40 mm Hepes-KOH (pH 7.3), 200 mm NaCl, 0.1% Tween 20, 10% glycerol, 5 mm β-mercaptoethanol, Complete Protease inhibitor mixture (Roche Applied Science)). Crude extracts were prepared by centrifugation in an SS-34 rotor for 20 min at 14,000 rpm. These were then incubated with 250 μl of calmodulin beads (Stratagene) for 120 min at 4 °C. Beads were washed with 2 × 200 μl of lysis buffer with 2 mm CaCl2 followed by 200 μl of lysis buffer with 0.5 mm CaCl2. Finally, the proteins were eluted by adding 2 × 300 μl of elution buffer (50 mm Tris-Cl (pH 8.0), 100 mm NaCl, 5 mm EGTA, 10% glycerol, 1 mm DTT) to the beads and incubating for 30 min at 4 °C. A fraction of the protein eluates was then mixed with SDS sample buffer, and the rest were incubated with either anti-FLAG M2-agarose beads or anti-HA-agarose beads (Sigma), and then washed with wash buffer (10 mm HEPES-KOH (pH 7.3), 200 mm NaCl, 0.1% Tween 20, 10% glycerol, 1 mm EDTA, 1 mm DTT). The beads were resuspended in SDS sample buffer and analyzed as described above.
Yeast Two-hybrid Assays
Two-hybrid assays were performed as described previously (25). In brief, two-hybrid strain pJ69–4a was transformed with Gal4 DNA-binding domain (GBD) vector or GBD fusion plasmids; transformants were selected and grown on SC-Leu medium. Similarly, two-hybrid strain pJ69–4α was transformed with Gal4 activation domain (GAD) vector or GAD fusion plasmids, selected, and grown on SC-Trp medium. The resulting transformants were then mated, and the diploid cells containing both the GAD and GBD constructs were selected on SC-Leu-Trp plates. Overnight cultures of the diploid strains grown in SC-Leu-Trp were diluted to ∼0.3 A600 and grown in SC-Leu-Trp to ∼1–2 A600. The cells were 10-fold serially diluted and spotted onto solid SC-Leu-Trp plates, SC-Leu-Trp-His plates, SC-Leu-Trp-Ade plates, or SC-Leu-Trp-His plates containing 1 mm 3-amino-1,2,4-triazole (Fisher). The plates were then incubated at 30 °C for 5 days. Positive interactions were indicated by cell growth on SC-Leu-Trp-Ade and/or SC-Leu-Trp-His (plus 1 mm 3-amino-1,2,4-triazole). Each construct was tested with vector alone as a control.
Chromatin Immunoprecipitation (ChIP)
ChIP experiments were performed as described previously (26). In brief, yeast cells (250 ml) were grown in a rich medium to an A600 of 0.5–0.6 and were cross-linked with 1% formaldehyde for 20 min before chromatin was extracted. The chromatin was sonicated (Bioruptor, Diagenode, Sparta, NJ: 10 cycles, 30 s on/off, high setting) to yield an average DNA fragment of 500 bp. Anti-FLAG antibody (Sigma, 4 μl) was coupled to 60 μl of protein A magnetic beads (Invitrogen). After reversal of the cross-linking and DNA purification, the immunoprecipitated and input DNA were analyzed by quantitative real-time PCR (qPCR) using Rotor-Gene 6000 (Qiagen). Samples were analyzed in triplicate for three independent ChIP experiments. Primer sequences are listed in supplemental Table S2.
Fluorescence Microscopy
To visualize Rad52-GFP foci, cells were grown at 25 °C in SC-complete medium to logarithmic phase, briefly sonicated to loosen cell aggregates, and then immobilized on a glass slide with a solution of 1.0%-agarose in ddH2O. Multiple images were obtained at 0.3-μm intervals along the z axis, and Rad52-GFP foci were counted by inspection of all focal planes. The cells were categorized as G1 (unbudded), S (small bud), or G2/M (medium to large bud). At least 400 cells were counted for each strain. All imaging was done with the Zeiss Axioplan 2 fluorescence microscope using Metamorph software. Statistical significance was assessed using Student's t test.
RESULTS
Rtt107 Physically Interact with the SMC5/6 Complex
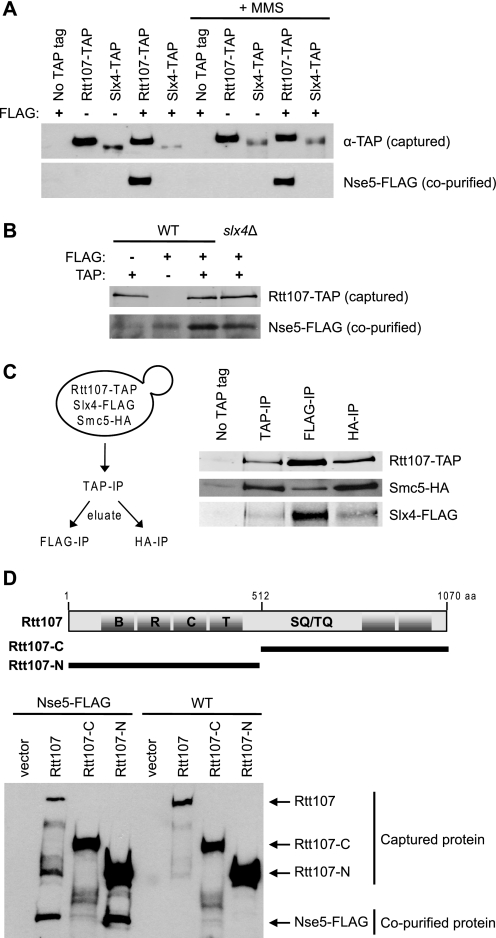
Having previously characterized the interaction of Rtt107 with Slx4 (5), we wished to expand our understanding of Rtt107 function by studying its interaction with the SMC5/6 complex, recently identified by a mass spectrometry approach (11). We first confirmed the physical interaction between Rtt107 and the SMC5/6 complex found by Ohouo et al. by analytical-scale interaction assays, testing immunoprecipitated epitope-tagged Rtt107 for co-purifying SMC5/6 subunits (Fig. 1A, supplemental Fig. S1, and data not shown). Because Rtt107 interacted with all the subunits tested, we chose Nse5 as a representative subunit to further characterize the interaction. The interaction of Nse5-FLAG and Rtt107-TAP was independent of DNA damage induced by MMS (Fig. 1A). Interestingly, Slx4 did not associate with SMC5/6, suggesting that only a fraction of cellular Rtt107 associated with SMC5/6 (Fig. 1A). Furthermore, Slx4 was not required for the interaction between Rtt107 and SMC5/6, nor did its absence affect the relative level of Nse5 interacting with Rtt107, supporting the model that separate pools of Rtt107 interacted with either Slx4 or SMC5/6 (Fig. 1B). To measure this more directly, Slx4-FLAG or Smc5-HA was immunoprecipitated from purified Rtt107-TAP complexes. In the FLAG immunoprecipitation fraction, Smc5-HA was depleted, whereas the opposite trend was observed in the HA immunoprecipitation fraction (Fig. 1C). This suggested that the majority of Rtt107 interacted with Slx4 or SMC5/6 separately, although there may be a minor population that interacts with both.
FIGURE 1.
Rtt107 physically interacted with the SMC5/6 complex. A, Rtt107-TAP but not Slx4-TAP co-immunoprecipitated with Nse5-FLAG independently of exposure to 0.025% MMS for 2 h. Analytical-scale TAP purifications were performed on whole cell extracts of the indicated strains. Immunoblotting was performed using anti-rabbit IgG or anti-FLAG antibodies. B, Rtt107-TAP co-immunoprecipitated with Nse5-FLAG independently of Slx4. C, separate pools of Rtt107 interacted with Slx4 or SMC5/6. Eluates from large-scale TAP purifications were subsequently immunoprecipitated with anti-FLAG or anti-HA-agarose beads. Immunoblotting was performed using anti-rabbit IgG, anti-HA, or anti-FLAG antibodies. D, the N-terminal portion of Rtt107 was responsible for the interaction with Nse5-FLAG. The truncation mutants of Rtt107 were from Roberts et al. (5).
BRCT domains are known to mediate protein-protein interactions among a variety of DNA damage repair proteins, including the Rtt107-Slx4 interaction (5). Rtt107 contains four BRCT domains in the N terminus and two BRCT domains in the C terminus (Fig. 1D). We expressed and purified TAP-tagged truncation mutants comprising the N- or C-terminal portion of Rtt107 from a low copy plasmid (5) in an rtt107Δ NSE5-FLAG strain and tested their ability to co-purify with Nse5-FLAG. In contrast to the C-terminal fragment, the N-terminal fragment of Rtt107 co-purified with Nse5-FLAG, suggesting that Nse5 interacted with the N-terminal BRCT domains of Rtt107 (Fig. 1D).
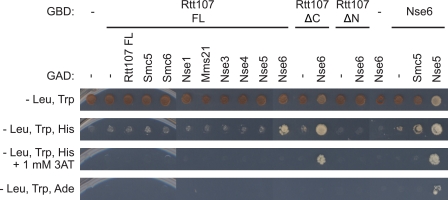
To decipher which subunit within the SMC5/6 complex was mediating the contact with Rtt107, we utilized a pairwise yeast two-hybrid interaction matrix between Rtt107 fused to the GBD and the individual SMC5/6 subunits fused to the GAD. As expected, Nse6 displayed a weak two-hybrid interaction with Smc5 and a strong interaction with Nse5, consistent with previous reports (Fig. 2) (27). Cells expressing both full-length GBD-Rtt107 (FL) and GAD-Nse6 were able to grow better on SC-His medium than the GAD-vector control, but not on SC-Ade medium, indicating that the full-length GBD-Rtt107 (FL) exhibited a weak two-hybrid interaction with Nse6, but not with the other subunits of the SMC5/6 complex. In addition, GAD-Nse6 displayed a strong yeast two-hybrid interaction with the N-terminal fragment of Rtt107 and very little interaction with the C-terminal fragment of Rtt107, consistent with our co-immunoprecipitation data. Together, these results suggested that Rtt107 interacted with the SMC5/6 complex via its N-terminal BRCT domains and that this interaction was likely mediated by the Nse6 subunit.
FIGURE 2.
Rtt107 interacted with the SMC5/6 complex via the Nse6 subunit in a yeast two-hybrid analysis. Rtt107 was fused to the Gal4 DNA binding domain (GBD), and the individual SMC5/6 subunits were fused to the Gal4 activation domain (GAD). Physical interaction was indicated by growth of the yeast on SC-His and/or SC-Ade selection media. The N-terminal and C-terminal fragments of Rtt107 used here were equivalent to the constructs in Fig. 1D.
Rtt107 Is Required for Recruitment of the SMC5/6 Complex to a DNA Double-stranded Break
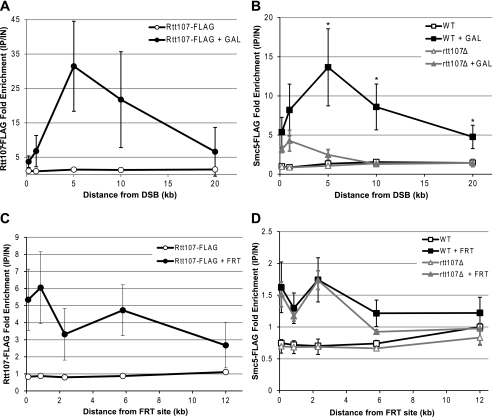
To identify a function for the interaction between Rtt107 and the SMC5/6 complex, we first examined the DNA damage response, because both partners are involved in this process (4, 5, 15). Given that the SMC5/6 complex is recruited to a DSB (17, 28), we tested whether Rtt107 physically associated with this genomic lesion as well. To this end, we FLAG-tagged Rtt107 in a strain that contained the HO endonuclease cut site in the mating locus and the HO endonuclease under control of the galactose promoter, allowing for creation of a single DSB at a specific locus (29). After 2 h of galactose induction, we performed ChIP and measured Rtt107-FLAG enrichment at various distances from the HO cut site by qPCR. Interestingly, Rtt107 was significantly enriched at regions near the DSB created by the HO endonuclease after galactose induction (Fig. 3A). Although Rtt107 was recruited at low levels at the DSB itself, its enrichment slowly increased up to a maximum at 5 kb from the DSB, and then slowly decreased with increasing distance from the DSB, up to 20 kb. Because Rtt107 physically interacted with the SMC5/6 complex, we next tested whether Rtt107 was required for recruitment of the SMC5/6 complex to the DSB. As expected, Smc5-FLAG was recruited to regions near the HO cut site after galactose induction, and this enrichment reached a maximum at 5 kb from the DSB, thus mirroring the Rtt107 binding pattern (Fig. 3B). However, Smc5-FLAG enrichment was dramatically reduced in the absence of Rtt107, particularly in the region 5–20 kb away from the DSB where the majority of Smc5-FLAG was observed (Fig. 3B).
FIGURE 3.
Rtt107 was required for recruitment of the SMC5/6 complex to a double-stranded break but not to a protein-bound nick. Fold enrichment of Rtt107-FLAG or Smc5-FLAG was determined by ChIP-qPCR. Enrichment at the target loci were normalized to the PRP8 reference locus and the corresponding input DNA. Graphs show averages from three independent experiments, and error bars represent standard deviations. A, Rtt107 was recruited to regions near the DSB after 2 h of galactose induction in asynchronous cells. B, Smc5 recruitment to the DSB was dependent on Rtt107. *, p < 0.05 comparing WT to rtt107Δ after galactose induction. C, Rtt107 was recruited to regions near the protein-bound nick in the presence of the FRT target site 2 h after release into S phase. D, Smc5 was similarly recruited to regions near the protein-bound nick independently of Rtt107.
Because the above experiment represented only one type of DNA lesion that can be encountered by cells, we turned to another system that allows interrogation of a protein-bound nick to further expand our understanding of the roles of Rtt107 and the SMC5/6 complex in the DNA damage response. In this system, a protein-bound nick is introduced at a specific locus in the genome by expressing a ligation-defective Flp recombinase that remains covalently bound to the DNA after forming a nick at its recognition target site (termed the “FRT site”) (30). During S phase, DNA replication forks run into the protein-bound nick and become stalled or collapsed, thus mimicking DNA damage produced by camptothecin. We expressed either Rtt107-FLAG or Smc5-FLAG in this strain and measured its enrichment near the site of DNA damage by ChIP-qPCR. Cells were arrested in G1 with alpha factor, treated with galactose to induce expression of the mutant Flp recombinase, released into S phase, and collected after 2 h. Rtt107-FLAG was recruited to this type of DNA lesion as well, albeit at lower levels than at a DSB, and was enriched up to 12 kb from the protein-bound nick (Fig. 3C). Similarly, Smc5-FLAG was recruited up to 6 kb away from the protein-bound nick (Fig. 3D). Although the relative Smc5-FLAG enrichment was lower at a protein-bound nick compared with a DSB, the levels were comparable to the fold enrichment measured by qPCR at other chromosomal loci enriched for the SMC5/6 complex identified in ChIP-on-chip studies (data not shown (17)). Intriguingly, the absence of Rtt107 had no significant effect on Smc5-FLAG recruitment to a protein-bound nick, thus starkly contrasting with the situation at the HO-induced DSB (Fig. 3D). Taken together, this data suggested that Rtt107 and the SMC5/6 complex cooperate only in response to specific DNA lesions, although they both may have broad roles in the DNA damage response.
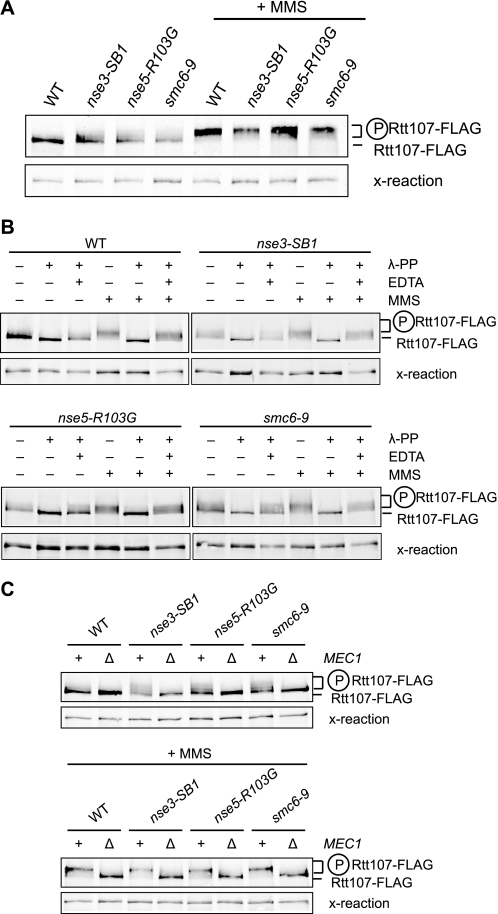
Mutations in the SMC5/6 Complex Resulted in Phosphorylation of Rtt107 in the Absence of DNA Damage Agents
Rtt107 is phosphorylated upon exposure to DNA damage agents, and this modification is important for its role in the DNA damage response (4, 5). To explore a potential regulatory relationship between Rtt107 and the SMC5/6 complex, we tested whether Rtt107 phosphorylation was dependent on the SMC5/6 complex. We FLAG-tagged Rtt107 in strains containing the nse3-SB1, nse5-R103G, or the smc6–9 hypomorph alleles (31, 32) and exposed them to 0.03% MMS for 1 h. Surprisingly, in the cultures not exposed to MMS, Rtt107-FLAG from the strains with an altered SMC5/6 complex exhibited partially retarded migration through the gel when compared with the wild-type strain (Fig. 4A, compare lanes 2–4 to lane 1). This result suggested that Rtt107 was modified in the absence of MMS, most likely by phosphorylation, although not to the same extent as the phosphorylation induced by DNA-damaging agents. After exposure to MMS, Rtt107 was phosphorylated regardless of whether the genes encoding for subunits of the SMC5/6 complex were mutated (Fig. 4A). To confirm that the retarded migration of Rtt107 was caused by phosphorylation, we treated the protein extracts with λ-phosphatase alone or in the presence of the phosphatase inhibitor EDTA. The retarded migration was completely abolished in the presence of λ-phosphatase, an effect that was reversed by the addition of EDTA (Fig. 4B). Moreover, this phosphorylation was dependent on the checkpoint kinase Mec1, because the retarded migration was eliminated in strains lacking MEC1 (Fig. 4C).
FIGURE 4.
In the absence of MMS, Rtt107 was phosphorylated in mutants expressing a compromised SMC5/6 complex. A, cells expressing Rtt107-FLAG were untreated or treated with 0.03% MMS for 1 h. Analytical-scale immunoprecipitations of Rtt107-FLAG were performed and analyzed by immunoblotting with anti-FLAG antibodies. The reduced mobility of Rtt107-FLAG indicated phosphorylation of the protein. Cross-reaction bands were used as a loading control. B, as in A, except immunoprecipitates were left untreated or incubated with λ-phosphatase (λ-PP; 200 units) for 30 min at 30 °C in the presence or absence of EDTA (100 mm). C, cells expressing Rtt107-FLAG with and without MEC1 were treated as in A. All strains contained sml1Δ to suppress the lethality of mec1Δ mutants.
Rtt107 and the SMC5/6 Complex Have Independent Functions in the DNA Damage Response
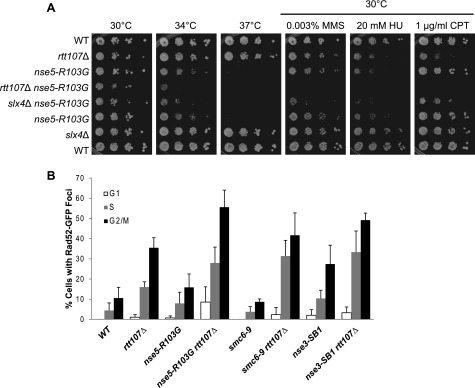
To further understand the functional relationship between Rtt107 and the SMC5/6 complex in the DNA damage response, we examined the effect of deleting RTT107 on cell growth during continuous exposure to DNA-damaging agents when SMC5/6 function was compromised. The nse5-R103G mutant alone was sensitive to high temperature and to DNA-damaging agents. In comparison, the rtt107Δ nse5-R103G double mutant grew much slower at the semi-permissive temperature (34 °C) and on low concentrations of the DNA-damaging agents MMS, hydroxyurea, and camptothecin (Fig. 5A). We then tested the effect of deleting SLX4 to compare its involvement in SMC5/6 functions to that of RTT107. The slx4Δ nse5-R103G double mutant also exhibited slower growth than either single mutant in all conditions tested (Fig. 5A). A similar pattern of growth phenotypes was observed when rtt107Δ or slx4Δ was combined with hypomorph alleles encoding for other SMC5/6 subunits (supplemental Fig. S2 and data not shown). The overall poorer growth of all double mutants suggested that both RTT107 and SLX4 contributed to other DNA damage response pathways that were partially redundant with the functions of the SMC5/6 complex.
FIGURE 5.
Rtt107 and the SMC5/6 complex had independent functions. A, double mutants containing nse5-R103G and rtt107Δ or slx4Δ grew significantly more slowly than the respective single mutants. 10-fold serial dilutions of the indicated strains were plated onto media containing various DNA-damaging agents and incubated at the indicated temperatures. B, rtt107Δ mutants exhibited an increased level of Rad52-GFP foci, whereas mutants expressing hypomorph alleles of genes encoding SMC5/6 exhibited a range of phenotypes. The percentage of cells in G1, S, or G2/M phase containing Rad52-GFP foci was calculated by dividing the number of cells in G1, S, or G2/M phase containing Rad52-GFP foci by the total number of cells in that cell cycle phase. At least 150 cells were counted in a minimum of three independent experiments. Error bars represent standard deviations of the values.
As an additional indicator of involvement in the DNA damage response, we measured Rad52-GFP foci to determine whether Rtt107 and the SMC5/6 complex cooperate in limiting spontaneous DNA damage. In the absence of exogenous DNA-damaging agents, rtt107Δ mutants exhibited an increased fraction of cells with Rad52-GFP foci, as previously reported (Fig. 5B) (7, 8). The strains containing a compromised SMC5/6 complex exhibited a range of phenotypes, from wild-type levels to increased levels of spontaneous Rad52-GFP foci, due to the hypomorphic nature of the alleles (Fig. 5B). The double mutants displayed a tendency for higher levels of Rad52-GFP foci than the respective single mutants, suggesting that Rtt107 and SMC5/6 contribute independently to controlling spontaneous DNA damage (Fig. 5B). Taken together, these data indicated that Rtt107 and SMC5/6 functioned separately in the DNA damage response in addition to their shared roles as one complex.
DISCUSSION
In this study, we characterized the interaction between Rtt107 and the SMC5/6 complex and documented that this interaction was independent of DNA damage and of Slx4, the best characterized Rtt107 interaction partner. We revealed that Rtt107 was required for recruitment of the SMC5/6 complex to DNA DSBs but not at a protein-bound nick. The relationship of these proteins extended to the phosphorylation status of Rtt107, because mutating genes encoding for SMC5/6 resulted in Rtt107 phosphorylation in the absence of DNA-damaging agents. Although Rtt107 and the SMC5/6 complex clearly worked together in some aspects of the DNA damage response, we demonstrated that they also had independent functions.
We have confirmed the physical interaction between Rtt107 and the SMC5/6 complex first identified by mass spectrometry (11) and found that it was mediated by the N-terminal BRCT domains of Rtt107. A unique feature of the interaction between Rtt107 and the SMC5/6 complex was its independence from Slx4, in contrast to the Slx4-dependent interaction between Rtt107 and the DNA replication and repair protein Dpb11 (11). Rtt107 forms a complex with Slx4 to provide resistance to DNA-alkylating agents and promote recovery from DNA damage, and the loss of their function can be suppressed by inhibiting the pathway leading to H3 K79 trimethylation (5, 7). Whereas many of the established functions of Rtt107 involve Slx4, the interaction between Rtt107 and the SMC5/6 complex did not, suggesting that this interaction represented a distinct role for Rtt107. Our data were consistent with a model in which separate pools of Rtt107 in the cell interact with either Slx4 and Dpb11 or the SMC5/6 complex. Interestingly, both Slx4 and the SMC5/6 complex bound the N terminus of Rtt107 (5). The functional relationship of Rtt107 and SMC5/6 was further supported by the finding that Rtt107 was phosphorylated by Mec1 when the SMC5/6 complex was compromised, even in the absence of DNA damage induced by exogenous agents. This suggested that Rtt107 acted directly or indirectly as an indicator of malfunction of the SMC5/6 complex.
At least one of the functions of the Rtt107-SMC5/6 complex appears to be at the site of DSBs, because both partners were recruited there upon induction of a DSB and Rtt107 was required for recruitment of the SMC5/6 complex. However this does not represent a sequential recruitment to the DSB, because Rtt107 interacted with the SMC5/6 complex constitutively, in the absence of DNA-damaging agents. Therefore, they must be recruited to the DSB together as one complex upon receiving a signal from the DNA damage site. Because Rtt107 was required for the recruitment of the SMC5/6 complex, it is likely that the triggering signal occurs via Rtt107. As such, we propose a model whereby Rtt107 is recruited to the DSB by binding to phosphorylated H2A S129, a histone modification that occurs immediately after DSB formation (33). In support of this model, Brc1, the Schizosaccharomyces pombe homologue of Rtt107, binds specifically to a phosphorylated H2A peptide in vitro via the phospho-binding BRCT domains of Brc1 (34). In addition, given that Mre11, one of the early sensors of DSBs, is also required for the recruitment of the SMC5/6 complex to DSBs, it is tempting to speculate that Mre11 is responsible for recruiting Rtt107, and consequently the SMC5/6 complex, to the site of DNA damage (17, 35). Although we have determined that Rtt107 was required for the recruitment of the SMC5/6 complex to DSBs, the question remains whether Rtt107 has subsequent functions in regulating the function of the SMC5/6 complex at DSBs, because both rtt107Δ and smc5/6 mutants have defects in sister chromatid recombination (4, 28).
Using the recently established Flp-nick system (30), we demonstrated that both Rtt107 and the SMC5/6 complex were recruited to a protein-bound nick. This was consistent with previous studies showing that both Rtt107 and the SMC5/6 complex are recruited to stalled replication forks (13, 17), which may occur at the protein-bound nick introduced by the mutant Flp recombinase. Interestingly, Smc5-FLAG recruitment was not affected by the absence of Rtt107, in contrast to the situation at DSBs. This result supported the model that SMC5/6 is involved in multiple DNA repair pathways that are differentially regulated, consistent with previous studies (17). Taken together, this data suggested that Rtt107 and the SMC5/6 complex cooperate only in response to specific DNA lesions, although they both may have broad roles in the DNA damage response.
Although our data strongly suggested a shared function of Rtt107 and the SMC5/6 complex at DNA DSBs, their distinct biochemical associations and the phenotypes of mutants in either RTT107 or genes encoding SMC5/6 subunits point toward differing additional functions. From a genetic perspective, all the subunits of the SMC5/6 complex are encoded by essential genes, whereas RTT107 is non-essential, indicating that one or more crucial functions carried out by SMC5/6 do not require Rtt107. The genetic interactions between RTT107 and the genes encoding for SMC5/6 support independent functions, because the double mutants grew much more slowly than the respective single mutants. Furthermore, they exhibit contrasting genetic interactions with other genes in the DNA damage response. Whereas deletion of DOT1 suppresses the DNA damage sensitivity of rtt107Δ mutants, it had no effect on mutants with a compromised SMC5/6 complex (Ref. 7 and data not shown). Conversely, deletion of RAD52 partially suppresses the temperature sensitivity of mutants expressing the hypomorphic alleles encoding for SMC5/6 subunits (32), but rad52Δ rtt107Δ double mutants have increased sensitivity to DNA-damaging agents (4, 12).
Our work revealed that a subset of Rtt107 and SMC5/6 functions is shared, most likely at the site of DSBs. However, the details of the regulatory network surrounding this relationship are yet to be discovered. The challenge of future research will be to fully elucidate the functional connection between these two important players in genome integrity.
Supplementary Material
Acknowledgments
We thank L. Aragon, E. Conibear, P. Hieter, J. Haber, L. Bjergbaek, and S. Taubert for yeast strains and plasmids and A. Wang and M. Aristizabal for critical reading of the manuscript.
This work was supported in part by Canadian Institutes of Health Research Grant MOP-79442 (to M. S. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2, Tables S1 and S2, and references.
- DSB
- double strand break
- MMS
- methyl methane sulfonate
- SMC
- structural maintenance of chromosome
- GAD
- Gal4 activation domain
- GBD
- Gal4 DNA-binding domain
- FL
- full-length
- ChIP
- chromatin immunoprecipitation
- qPCR
- quantitative real-time PCR
- FRT
- Flp recognition target site
- HO
- homothallic switching.
REFERENCES
- 1. Ogawa H., Johzuka K., Nakagawa T., Leem S. H., Hagihara A. H. (1995) Adv. Biophys. 31, 67–76 [DOI] [PubMed] [Google Scholar]
- 2. Harper J. W., Elledge S. J. (2007) Mol. Cell 28, 739–745 [DOI] [PubMed] [Google Scholar]
- 3. Lisby M., Rothstein R. (2009) DNA Repair 8, 1068–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rouse J. (2004) EMBO J. 23, 1188–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roberts T. M., Kobor M. S., Bastin-Shanower S. A., Ii M., Horte S. A., Gin J. W., Emili A., Rine J., Brill S. J., Brown G. W. (2006) Mol. Biol. Cell 17, 539–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chang M., Bellaoui M., Boone C., Brown G. W. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16934–16939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lévesque N., Leung G. P., Fok A. K., Schmidt T. I., Kobor M. S. (2010) J. Biol. Chem. 285, 35113–35122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alvaro D., Lisby M., Rothstein R. (2007) PLoS Genet. 3, e228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yuen K. W., Warren C. D., Chen O., Kwok T., Hieter P., Spencer F. A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3925–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mohammad D. H., Yaffe M. B. (2009) DNA Repair 8, 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ohouo P. Y., Bastos de Oliveira F. M., Almeida B. S., Smolka M. B. (2010) Mol. Cell 39, 300–306 [DOI] [PubMed] [Google Scholar]
- 12. Chin J. K., Bashkirov V. I., Heyer W. D., Romesberg F. E. (2006) DNA Repair 5, 618–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roberts T. M., Zaidi I. W., Vaisica J. A., Peter M., Brown G. W. (2008) Mol. Biol. Cell 19, 171–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao X., Blobel G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4777–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Piccoli G., Torres-Rosell J., Aragón L. (2009) Chromosome Res. 17, 251–263 [DOI] [PubMed] [Google Scholar]
- 16. Fujioka Y., Kimata Y., Nomaguchi K., Watanabe K., Kohno K. (2002) J. Biol. Chem. 277, 21585–21591 [DOI] [PubMed] [Google Scholar]
- 17. Lindroos H. B., Ström L., Itoh T., Katou Y., Shirahige K., Sjögren C. (2006) Mol. Cell 22, 755–767 [DOI] [PubMed] [Google Scholar]
- 18. Amberg D. C., Burke D. J., Strathern J. N. (2005) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, pp. 155–160, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 19. Funakoshi M., Hochstrasser M. (2009) Yeast 26, 185–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gelbart M. E., Rechsteiner T., Richmond T. J., Tsukiyama T. (2001) Mol. Cell Biol. 21, 2098–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. (1999) Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 22. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 23. Kobor M. S., Venkatasubrahmanyam S., Meneghini M. D., Gin J. W., Jennings J. L., Link A. J., Madhani H. D., Rine J. (2004) PLoS Biol. 2, E131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Flott S., Rouse J. (2005) Biochem. J. 391, 325–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. James P., Halladay J., Craig E. A. (1996) Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schulze J. M., Jackson J., Nakanishi S., Gardner J. M., Hentrich T., Haug J., Johnston M., Jaspersen S. L., Kobor M. S., Shilatifard A. (2009) Mol. Cell 35, 626–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Duan X., Yang Y., Chen Y. H., Arenz J., Rangi G. K., Zhao X., Ye H. (2009) J. Biol. Chem. 284, 8507–8515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. De, Piccoli G., Cortes-Ledesma F., Ira G., Torres-Rosell J., Uhle S., Farmer S., Hwang J. Y., Machin F., Ceschia A., McAleenan A., Cordon-Preciado V., Clemente-Blanco A., Vilella-Mitjana F., Ullal P., Jarmuz A., Leitao B., Bressan D., Dotiwala F., Papusha A., Zhao X., Myung K., Haber J. E., Aguilera A., Aragón L. (2006) Nat. Cell Biol. 8, 1032–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haber J. E. (2002) Methods Enzymol. 350, 141–164 [DOI] [PubMed] [Google Scholar]
- 30. Nielsen I., Bentsen I. B., Lisby M., Hansen S., Mundbjerg K., Andersen A. H., Bjergbaek L. (2009) Nat. Methods 6, 753–757 [DOI] [PubMed] [Google Scholar]
- 31. Ben-Aroya S., Coombes C., Kwok T., O'Donnell K. A., Boeke J. D., Hieter P. (2008) Mol. Cell 30, 248–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Torres-Rosell J., Machin F., Farmer S., Jarmuz A., Eydmann T., Dalgaard J. Z., Aragón L. (2005) Nat. Cell Biol. 7, 412–419 [DOI] [PubMed] [Google Scholar]
- 33. Downs J. A., Lowndes N. F., Jackson S. P. (2000) Nature 408, 1001–1004 [DOI] [PubMed] [Google Scholar]
- 34. Williams J. S., Williams R. S., Dovey C. L., Guenther G., Tainer J. A., Russell P. (2010) EMBO J. 29, 1136–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lisby M., Barlow J. H., Burgess R. C., Rothstein R. (2004) Cell 118, 699–713 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.