Abstract
MALDI-imaging MS is a new molecular imaging technology for direct in situ analysis of thin tissue sections. Multiple analytes can be monitored simultaneously without prior knowledge of their identities and without the need for target-specific reagents such as antibodies. Imaging MS provides important insights into biological processes because the native distributions of molecules are minimally disturbed, and histological features remain intact throughout the analysis. A wide variety of molecules can be imaged, including proteins, peptides, lipids, drugs, and metabolites. Several specific examples are presented to highlight the utility of the technology.
Keywords: Lipids, Mass Spectrometry (MS), Peptides, Proteomics, Tissue, Drugs, Imaging Mass Spectrometry, Metabolites, Proteins
Introduction
MALDI imaging MS (IMS)4 is an emerging new tool for the analysis of biological and clinical tissue samples. It has been shown to be amenable for the analysis of proteins, peptides (both endogenous and enzymatically produced), lipids, and small molecules (such as drugs and endogenous metabolites). Spatial relationships of molecules within a specimen are preserved because intact tissue is directly analyzed without homogenization. In this way, molecules can be interrogated in their native environments, providing new insights into the biological processes involved.
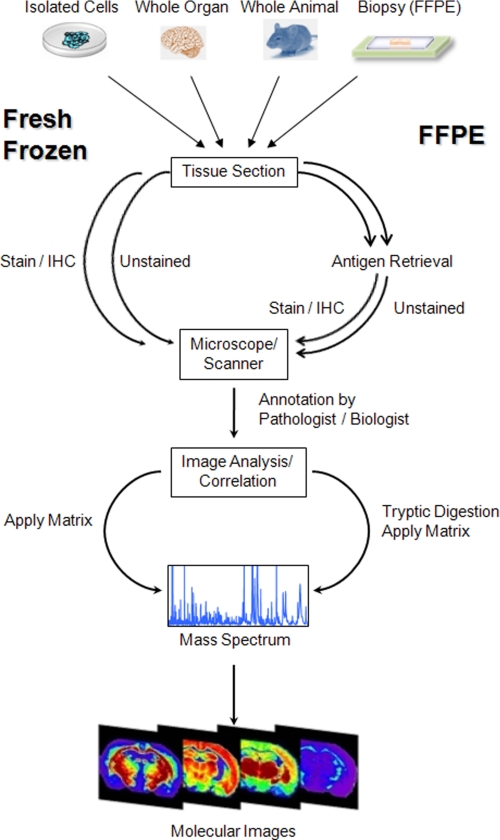
IMS requires minimal sample preparation for analysis. Thin sections of tissue samples (typically, 5–10 μm thick) are obtained from frozen or formalin-fixed paraffin-embedded (FFPE) tissue blocks and collected on conductive MALDI targets. A matrix compound (typically, a small organic acid as well as a proteolytic enzyme when necessary) is applied to the surface of the tissue sample. Mass spectra are subsequently collected by firing a laser in an ordered pattern of thousands of ablated spots on the tissue section, and a discrete spectrum is collected from each location on the sample. Each ablated spot is analogous to a pixel in a digital photograph. Each pixel (spectrum) contains many analytes that can be individually displayed as a function of their position and relative intensity within the tissue section. In this way, hundreds of images from specific molecular species can be generated simultaneously from a single tissue section without prior knowledge of their identities. Specific reagents such as antibodies are not needed. Alternatively, IMS can be carried out in a profiling mode in which only relatively small selected areas from each tissue section are targeted for analysis. A stained serial section is typically used to guide the analysis. The sample preparation and analysis process of fresh-frozen and FFPE tissues are summarized in Fig. 1.
FIGURE 1.
Schematic outline of a typical workflow for tissue samples. Sample pretreatment steps include cutting and mounting the tissue section on a conductive target as well as paraffin removal and antigen retrieval for FFPE sections. A frozen serial section or the FFPE section to be analyzed is stained, a digital image is taken, and areas of interest are selected by a pathologist. Trypsin and/or matrix is applied to the tissue section, and mass spectra are generated at each x,y coordinate. IHC, immunohistochemistry.
IMS can be very high-throughput in nature. Often, 10–20 tissue sections can be collected on a single target plate and analyzed concurrently. With currently available high repetition rate lasers (1 kHz or greater), the entire target plate can be analyzed in a matter of a few minutes to a few hours, depending on the desired spatial resolution. It has recently been shown that, through the use of a 5 kHz laser and continuous laser raster, a rat brain measuring 185 mm2 can be imaged at 100 μm resolution in <10 min (1). Commercial lasers are capable of achieving spot diameters on tissue of tens of microns. Nevertheless, the spatial resolution used in a given image analysis is dependent primarily on the biological question of interest. High-resolution spatial analysis necessarily leads to quite large data files and increases analysis times, unnecessary attributes if only a low-resolution question is being asked.
Mass resolution is also an important factor for IMS. TOF mass analyzers, commonly used for IMS, routinely have a resolving power on tissue of 1 part in 10,000. If higher mass resolution and mass accuracy are necessary such as for lipids and small metabolites, a Fourier transform ion cyclotron resonance or an orbitrap mass spectrometer can be used, which are capable of <1 ppm mass accuracy and mass resolution (m/Δm) of >1,000,000. These instruments allow for accurate determination of elemental composition as well as base-line separation of analytes differing in mass by <0.01 Da for low-molecular-mass species.
Although the applications of IMS are multifold, here we will briefly discuss four common types of molecular analysis amenable to this technology. This article is not intended to be a review of the field of IMS but rather is a perspective on the current status of MALDI-IMS.
Protein Analysis
Protein IMS requires relatively simple sample preparation in that no prior knowledge of the proteins in the sample or specialized reagents are need. A section of a frozen tissue block is collected onto a conductive MALDI target. A serial section is usually collected at this time for histological analysis. The section for MS analysis is fixed in graded alcohol to remove lipids and biological salt, thereby enhancing signal quality (2). The analysis of proteins can be approached in two different ways: whole section imaging and histology-directed profiling.
In the imaging mode, a matrix is applied uniformly over the entire tissue section using either a robotic reagent spotter (3) or a spray nebulization device (4). Spectra are acquired from a defined pattern by moving the sample target plate beneath a fixed laser, and spectra are collected. Each ion within the spectrum can then be viewed for its spatial distribution and relative intensity across the entire section. Molecular correlations with spatial distinctions can be clearly observed and compared in normal and diseased tissues (5, 6).
In the profiling mode, spectra are acquired only from distinct locations within the section, usually carried out in a histology-directed manner (7). Briefly, two serial sections of a tissue sample are collected: one on a MALDI target and one on a standard microscope slide for histological staining. A digital photomicrograph of the stained section is taken and is then reviewed by a biologist or pathologist. The areas of interest are annotated for analysis, and individual cell types of interest are marked with a color-coded circle (which is typically 100–200 μm in diameter). The resulting selective image is then superimposed on an image of the unstained section, and the coordinates of the annotations are determined and transferred to a robotic spotter. A matrix is deposited at the desired location, and mass spectra are acquired. For protein analysis, spectra are generally acquired in the linear positive ion mode on a TOF mass spectrometer. This allows for high sensitivity over a relatively broad mass range. Histology-directed profiling is very conducive to high-throughput analysis and biostatistical evaluation for determination of proteins that discriminate disease states.
Imaging and profiling of proteins have been applied to a wide variety of tissues and diseases, including brain (8, 9), breast (10–12), gastric (13), lung (14), kidney (5), and prostate (15) cancer; embryonic development (16); inflammatory bowel diseases (17); and Alzheimer disease (18). Several of these studies are described in some detail below.
A recent study employing IMS technology examined 106 gastric biopsy samples from 63 cancer patients and 43 healthy volunteers to determine a molecular signature of gastric cancer (13). A histology-directed protein profiling approach was used to target areas enriched (>75%) in either carcinoma or normal epithelial cells. The samples were split into a training set (31 cancer and 21 normal) and a testing set (32 cancer and 22 normal). A support vector machine algorithm was applied to the training set to determine a classifier composed of 73 signals that produced a cross-validation accuracy of 96.6%. When applied to the independent validation set, the classification resulted in a sensitivity of 93.8% and a specificity of 95.5%. Several of the proteins that were part of the classifier were identified through traditional proteomic approaches. These included α-defensin-1 and α-defensin-2 (m/z 3439 and 3368), calgranulin A (m/z 10840), and two forms of calgranulin B (m/z 13158 and 12694) that exhibited higher expression in the tumor samples, as well as lysozyme C (m/z 14697), a C-terminal fragment of anterior gradient protein 2 homolog (m/z 2968), and an N-terminal fragment of histone H2B (m/z 7767) that exhibited higher expression in the normal samples. Proteomic expression in these samples could also be correlated with disease stage, with 17 peaks differentiating stage Ia from stage Ib and higher. These findings can potentially be used to differentiate gastric cancer patients who are candidates for surgical intervention (aggressive disease) and those who can be treated nonsurgically. An additional proteomic signature could be used to differentiate between Lauren intestinal and diffuse types of gastric cancer.
Another report utilized IMS for the analysis and classification of myxoid sarcomas (19). In this study, a total of 20 myxofibrosarcomas (MFS) and 20 myxoid liposarcomas (MLS) were analyzed. Each type of tumor was further histologically subdivided into 10 high-grade and 10 low-grade cases. Spectra from histologically relevant areas were extracted and subjected to statistical analysis using ClinProTools software. Principal component analysis showed separation of high-grade tumors from low-grade tumors when using the first two principal components but not a distinction between MFS and MLS. The four different sample types could be distinguished from each other when using the second and third principal components, indicating that there is a greater difference between grades of tumors than between types. From the imaging experiments, specific m/z values were found that were unique to each diagnosis: m/z 4320 was found only in low-grade MFS, m/z 6639 only in high-grade MFS, m/z 5592 only in low-grade MLS, and m/z 11483 only in high-grade MLS. Proteins that could specifically discriminate between low- and high-grade sarcomas included calgizzarin, calcyclin, and histones H2A, H2B, H3, and H4 (previously identified in Ref. 20). A support vector machine classification algorithm was built from the data from the high- and low-grade MFS samples and applied to a set of five histologically diagnosed intermediate-grade MFS samples. Class imaging showed that these samples contained separate nodules of low and high grade-like mass spectral profiles. Hierarchical clustering of all spectra showed separate branches for high- and low-grade MFS samples, with a subset of intermediate-grade spectra clustering as subbranches on the high- and low-grade arms. These samples contained separate foci of different grades (high and low) as determined by molecular analysis as opposed to a true “intermediate” grade of samples.
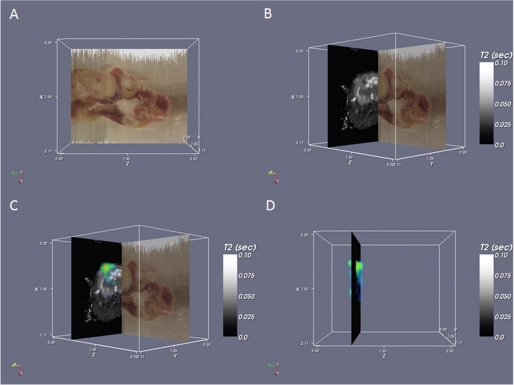
An emerging application of IMS is three-dimensional analysis of organ structures (21, 22) and co-registration of MS data with other three-dimensional imaging modalities such as magnetic resonance imaging (MRI) (23) and CAT scans. IMS allows the determination of molecular changes within or surrounding a tumor or other structure. These studies have allowed a more in-depth picture of the biological disease process and provide molecular information for spatial correlation to non-invasive three-dimensional imaging modalities such as MRI. Fig. 2 shows an example of this type of correlation: the brown data plane (A) is the (optical) block face image acquired during sectioning, the black background plane (B) shown perpendicular to the block face is a single image from the MRI scan, and the blue/green image (C and D) superimposed on the magnetic resonance image is the MS image of astrocytic phosphoprotein PEA, shown as a volume.
FIGURE 2.
Three-dimensional IMS and co-registration. A, block face imaging of a rat head. Sections were collected axially but can be viewed in any orientation such as the sagittal view shown here. B, block face image (sagittal) co-registered with the T2 magnetic resonance image (axial). Brain structure, including the corpus callosum and a tumor, can be observed in the MRI data. C, MS data for a tumor-specific protein, astrocytic phosphoprotein PEA, are superimposed on the MRI data. D, three-dimensional rendered volume of the MS data (sagittal) on a single magnetic resonance plane (axial).
Peptide Analysis
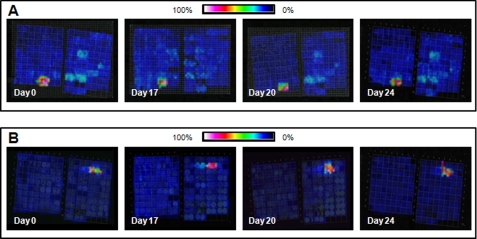
The analysis of peptides by IMS has involved studies of endogenous peptides as well as peptides produced after on-tissue protease digestion. The latter has become an important approach for the analysis of proteins in FFPE tissue specimens. The vast majority of samples stored in tissue banks worldwide are FFPE specimens (24), as this type of fixation is standard practice in pathology because it allows samples to be stored at room temperature for many years. These samples are often well documented, and furthermore, samples from rare diseases are available where fresh-frozen samples are scarce (25). Many protocols have been described within recent years to attain peptide analysis of FFPE tissue samples, usually combining heat-induced antigen retrieval techniques and enzymatic digestion (26–31). Even though only partial reversal of formalin-induced modifications is achieved, several studies have substantiated the potential of this approach (29). These protocols have proven to be highly reproducible, and Fig. 3 shows selected peptide images collected from a tissue microarray (TMA) constructed from normal and cancerous FFPE tissue and cell lines on 4 different days (days 0, 17, 20, and 24) from the same TMA block. This example shows the reproducibility of on-tissue tryptic digestion in revealing unique molecular features from the tissue cores of several organs and tumors and underscores the robustness of the imaging technology over an extended period of time, an essential aspect in the analysis of patient tissue biopsies.
FIGURE 3.
Reproducibility of on-tissue tryptic digestion from a TMA comprising samples from multiple different organ sites and cancers. A, a feature at m/z 1650.0 was found only in the leiomyosarcoma samples. B, a feature at m/z 1177.7 was present only in the astrocytoma samples. Both features were reproducible when comparing serial sections that were analyzed on 4 different days.
MALDI-IMS has been utilized to map the distribution of low-molecular-mass peptides derived from the breakdown of crystallins, the major eye lens proteins (32). To identify peptides from the water-insoluble fraction, frozen equatorially sectioned lens samples were thoroughly washed in graded ethanol solutions to remove interfering salts, lipids, and water-soluble proteins. Spray-coated sections from human lenses at different ages (20–86 years) were compared with one another at a spatial resolution of 200 μm. Peptides were identified in lens extracts using MALDI-MS and nanoflow LC-electrospray ionization MS/MS. In the 37-year-old lens, the C-terminal βA3 peptides (m/z 3197, 3253, and 3390) represented the major species, whereas in the 42-year-old lens, additional N-terminal peptides of αB-crystallins (m/z 2187 and 2359) and γS-crystallins (m/z 2598) could be detected in the nuclear region of the lens. A 58-year-old lens showed an evident increase in the αB2–18 peptide (m/z 2187) in the nuclear region and a lesser increase in the other peptides. In the case of an 80-year-old, all peptides could be found in the cortex and the nucleus. The oldest lens (age 86) additionally showed a significant increase in the αA66–80-H2O peptide (m/z 1848). This lens was the only one with an age-related cataract. These findings were consistent with previously published work (33).
The classification of subtypes of non-small cell lung cancer was achieved by MALDI-IMS in a high-throughput manner using FFPE TMA samples with high accuracy (29). TMA sections containing 22 squamous cell carcinoma cores from 14 different patients and 18 adenocarcinoma cores from 12 different patients were subjected to heat-induced antigen retrieval, on-tissue tryptic digestion, and matrix deposition. Spectra generated from duplicate samples were highly reproducible, with no statistically significant differences observed in peak intensities between duplicates as calculated using a t test and minimum 1.6-fold intensity difference comparison for the 200 most intense signals. The correct classification of all patients could be achieved by combining 73 peaks in a support vector machine algorithm-based model. Direct on-tissue identification by MALDI-MS/MS of numerous peptides from ∼50 proteins was performed. For example, three tryptic peptides (m/z 987.60, 1163.62, and 1905.99), almost exclusively expressed in squamous cell carcinomas, were identified as originating from heat shock protein β1. A selective distribution in a subset of the squamous cell carcinoma samples could be found for a tryptic peptide from keratin type II cytoskeletal 5 (m/z 1410.70).
Other studies on FFPE samples include the analysis of samples from gastric cancer (34) and pancreatic adenocarcinoma using MALDI ion mobility IMS (31, 35). In the latter, multiple tryptic peptides could be identified directly from tissue sections.
Lipid Analysis
Lipids are an important target for mass spectral analysis because they have been implicated in a variety of biological and disease processes, including signal transduction (36), kinase pathways (37), cancer (38, 39), Alzheimer disease (40), and embryo implantation (41), and have been found to have differential expression in different parts of the brain (42). With respect to MALDI-MS, dry matrix applications such as sublimation (43) and dry coating (44) can be employed. This provides advantages over wet application techniques in that analyte delocalization cannot occur, and higher spatial resolution imaging can be achieved. Typically, 2,5-dihydroxybenzoic acid (DHB) or 2,6-dihydroxyacetophenone is used as a matrix for lipid imaging. DHB is used for positive ion mode analysis, and 2,6-dihydroxyacetophenone can be used for either positive or negative ion mode analysis. More recently, liquid ionic matrices have been used in IMS experiments, combining DHB with aniline, pyridine, or 3-acetylpyridine (45). These combinations have led to increased vacuum stability during imaging, decreased total matrix volume requirements, and the ability to work in both positive and negative ionization modes. However, these matrices have been demonstrated only via microspotting, thus limiting the achievable spatial resolution to 100–200 μm. For lipid analysis, spectra are generally collected using a reflectron TOF analyzer for increased mass resolution and differentiation of molecular species. Lipid classes such as phosphatidylcholines (PCs) and sphingomyelins are observed in positive mode, whereas classes such as phosphatidylethanolamines (PEs), phosphatidylinositols (PIs), and phosphatidylserines (PSs) are observed in negative mode. Lipid analyses are complex because as many as 40 different lipids along with sodiated and potassiated lipid species and lipid-matrix adducts can have molecular masses within 1 Da of each other (LIPID MAPS). Some of these mass overlap issues can be circumvented through the use of a higher mass resolution mass spectrometer such as an orbitrap or Fourier transform ion cyclotron resonance mass spectrometer or through MS/MS analysis, which can provide structural information on the composition of the side chains (46). Additionally, the matrix can be doped with lithium to produce lithiated ionic species, reducing the spectral complexity and aiding in lipid fragmentation (47). However, the exact structure of side chains (i.e. positions of unsaturations) cannot be readily determined without further studies.
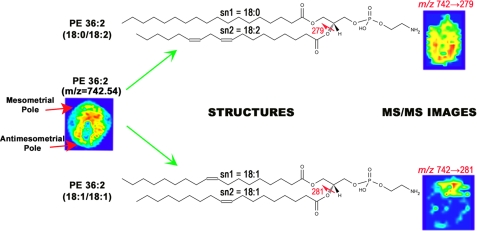
A recent study has examined the role that lipids play in the embryo implantation process (41). Thin sections of mouse embryos were imaged at days 4–8 post-fertilization in both positive and negative modes. Substantial increases in expression of most phospholipids were observed in the stromal area after implantation (day 5 and later) compared with before implantation (day 4). Subsequent to implantation, there was a restructuring of lipid species to either the mesometrial (vascularized) pole or antimesometrial pole. On day 6, PC (16:0/18:1), PE (16:0/18:1), and PI (18:1/20:4) were observed to increase at the mesometrial pole, whereas higher expression of PC (18:0/18:2) and PI (16:0/18:2) was observed at the antimesometrial pole. By day 8, PCs, PEs, phosphatidylglycerols, and PSs (18:1) and PIs and PEs (20:4) along with PE plasmalogen (16:0/22:4) showed higher expression levels at the mesometrial pole, and PCs, PIs, and PSs (18:2), PCs (20:4), and PC, PE plasmalogen, and PI (22:6) demonstrated higher intensity at the antimesometrial pole. Quantitation results were validated through microdissection of the embryo sections into top and bottom regions and subjecting them to LC-MS/MS analysis. IMS results were consistent with LC-MS/MS quantitation results. It was also determined through MS/MS imaging that isobaric species showed very different localization patterns (Fig. 4). For example, PE (36:2; 18:0/18:2) was expressed throughout the embryo, with higher expression at the antimesometrial pole (bottom) of the implantation site, whereas PE (36:2; 18:1/18:1) was observed only at the mesometrial pole (top) of the implantation site.
FIGURE 4.
MS/MS lipid imaging. MS imaging of m/z 742.54 shows a ubiquitous pattern from top to bottom of the implantation site. However, MS/MS imaging of this ion shows that it is made up of two distinct isobaric structures that have very different localization patterns. PE (36:2; 18:0/18:2) is localized to the antimesometrial pole (lower), whereas PE (36:2; 18:1/18:1) is localized to the mesometrial pole (upper).
A second study looked at expression of lipids and, in particular, sulfatides (STs) in human ovarian cancer (38). In this study, 12 ovarian cancer samples along with 12 normal ovaries were collected and cryopreserved. Sections were prepared through thin sectioning and spray nebulization of 2-mercaptobenzothiazole over the entire section. Three ions, ST d18:1/C16:0 (m/z 778.6), d18:1/C24:1 (m/z 888.6), and d18:1/C24:0 (m/z 890.6), were observed at high expression levels in the epithelial carcinoma regions but were absent from neighboring stromal areas. Image analysis of normal ovarian tissue was negative for these three ions. Statistical analysis of extracted regions from the IMS experiments showed significantly higher expression of an ion at m/z 778.6, corresponding to ST d18/C16:0 in epithelial carcinoma compared with stromal areas. Two different stroma regions had ion intensities of 153 ± 34 and 204 ± 44, whereas, the carcinoma region had an ion intensity of 507 ± 135 (p = 0.0016 and 0.0066, respectively, versus the two stromal regions). ST identification was confirmed through extraction of lipids from the tissues and analysis by LC-MS/MS by multiple reaction monitoring. ST species can be confirmed by precursor scans for m/z 96.9, which corresponds to HSO4 in the negative ionization mode.
Drug Analysis
Data on drug absorption, distribution, metabolism, and excretion are critical to the development process. MALDI-IMS has been used in pharmaceutical research to provide information on the distribution and metabolism of drugs within targeted organ sections, including whole body tissue sections (48, 49). Drug analyses are performed using MS/MS to monitor the drug itself using structure-specific fragments, increasing the sensitivity and providing identification with high confidence. To accomplish the analysis in a high-throughput manner, single or multiple reaction monitoring can be utilized to monitor the structure-related composition of the drug (precursor or parent ion) in the mass spectrometer to form one (single reaction monitoring) or multiple (multiple reaction monitoring) specific fragment ions (50, 51). Compared with whole body autoradioluminography, a traditional method to study the spatial distribution of drug candidates in tissue, MALDI-IMS has two advantages: it does not require radiolabeling of the drug of interest and can distinguish between the parent drug and its metabolites (49, 52, 53). Other commonly used methods in drug analysis such as LC-MS require tissue homogenization and therefore preclude the acquisition of high-resolution spatial and histological information (54, 55).
Tissue handling and sample pretreatment steps in drug analysis vary from those commonly implemented in protein analysis in that washing steps should be avoided because they can degrade or compromise the level of the drug in the tissue. Because most drugs have a molecular mass below 1000 Da, the matrix should be carefully selected to avoid interferences from spectral noise generated from the matrix (56).
Recently, investigators studied the correlation of skin blanching and percutaneous absorption of glucocorticoid receptor (GR) agonists to assess their potency and clinical anti-inflammatory efficacy (57). MALDI-IMS was utilized to directly analyze the distribution of three GR agonists in porcine skin tissue. Pig ear samples (∼2 cm2) were placed in Petri dishes and incubated with a mixture of all three compounds together for 16 h. Images were acquired by monitoring a selected fragment of each GR agonist in MS/MS mode across the tissue sections (spatial resolution of 200 μm) at 0- and 16-h time points after incubation. The results showed that the arylpyrazole penetrated through the epidermis layer into the dermis, whereas the arylpyrazolopyrimidine showed only some penetration and the arylindazole only limited penetration into the dermis. None of the three compounds could be detected in the adjacent connective tissue. These results were in accordance with those obtained from skin blanching responses wherein the arylpyrazole showed the greatest response, peaking 2–3 h after dressing removal.
In another study, investigators tracked and quantified the distribution of tiotropium bromide (TTP) within the lungs of dosed rats (58). An inhaled bronchodilator, TTP is used for the treatment of asthma and chronic obstructive pulmonary disease. Rats were dosed for 15 min in a two-stage flow-past inhalation chamber (total delivered lung dose of 50 μg) and killed 15 min after administration. The distribution of TTP within the lung tissue was analyzed by MALDI-IMS in both MS and MS/MS modes (spatial resolution of 200 μm). Parent and fragment ion signals from serial sections correlated well and revealed the drug to be present in the alveolar beds in the upper and lower parenchyma. Utilizing a linear regression concentration curve obtained from drug standards spotted on lung tissue sections, the amount of TTP within areas of tissue sections from dosed animals was calculated. Those results were in accordance with results from quantification analysis of tissue extracts by LC-MS/MS.
The localization of oxaliplatin and its derivatives in heated intraoperative chemotherapy-like treated rat kidneys was recently analyzed using MALDI-IMS (59). Oxaliplatin was detectable down to 0.23 ± 0.05 mg of total oxaliplatin/g of tissue in 15-μm-thick tissue sections. In the kidney, oxaliplatin and its derivatives were present in the cortex, whereas in the medulla, little or no drug penetration was detected by MALDI-IMS. Similarly, the distribution of a small-molecule EGF receptor inhibitor (erlotinib) and its metabolites was analyzed by MALDI-IMS in tissue sections from rat liver, spleen, and muscle (60). After oral administration of the drug, the highest concentrations of the drug and its major metabolite were found in the liver. Comparing the direct quantitative analysis of the drug by MALDI-MS and LC-MS/MS on tissue homogenates gave similar results for the liver and spleen.
Limitations
Although limitations have been noted throughout this minireview, they fall mainly in the area of detection limits due to the difficulty in desorbing higher molecular mass analytes (more than ∼30 kDa) and to ion interference/suppression effects experienced with some samples. Image resolution has improved markedly, so 20 μm spatial resolution can be achieved with many commercial instruments. Similarly, the speed of imaging has now improved, so an image that required many hours to acquire can now be obtained in minutes (1).
Perspectives
MALDI-IMS has been shown to be of significant benefit in the analysis of tissue specimens for monitoring changes in proteins, peptides, lipids, and drugs. Recent technological developments have allowed the analysis of FFPE tissues, providing a new source of samples that will greatly expand the number and types of samples that can be analyzed by MS. The use of human as well as animal samples will provide considerable insight into many disease and biological processes. As the technology advances, the applications of IMS in the clinic will continue to expand, enabling it to play a central role in the diagnosis and prognosis of disease and in the evaluation of patient therapy.
Acknowledgments
We thank Kristin Burnum, Sheerin (Khatib-Shahidi) Latham, and Tuhin Sinha for data used in the figures.
This work was supported, in whole or in part, by National Institutes of Health Grant 5R01 GM58008 from NIGMS. This work was also supported by United States Department of Defense Grant W81XWH-05-1-0179 and Vanderbilt Ingram Cancer Center Core Support Grant P30 CA68485. This is the sixth article in the Thematic Minireview Series on Biological Applications of Mass Spectrometry. This minireview will be reprinted in the 2011 Minireview Compendium, which will be available in January, 2012.
- IMS
- imaging MS
- FFPE
- formalin-fixed paraffin-embedded
- MFS
- myxofibrosarcoma(s)
- MLS
- myxoliposarcoma(s)
- MRI
- magnetic resonance imaging
- TMA
- tissue microarray
- DHB
- 2,5-dihydroxybenzoic acid
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PI
- phosphatidylinositol
- PS
- phosphatidylserine
- ST
- sulfatide
- GR
- glucocorticoid receptor
- TTP
- tiotropium bromide.
REFERENCES
- 1. Spraggins J. M., Caprioli R. M. (2011) J. Am. Soc. Mass Spectrom. 22, 1022–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Seeley E. H., Oppenheimer S. R., Mi D., Chaurand P., Caprioli R. M. (2008) J. Am. Soc. Mass Spectrom. 19, 1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aerni H. R., Cornett D. S., Caprioli R. M. (2006) Anal. Chem. 78, 827–834 [DOI] [PubMed] [Google Scholar]
- 4. Walch A., Rauser S., Deininger S. O., Höfler H. (2008) Histochem. Cell Biol. 130, 421–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oppenheimer S. R., Mi D., Sanders M. E., Caprioli R. M. (2010) J. Proteome Res. 9, 2182–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Han E. C., Lee Y. S., Liao W. S., Liu Y. C., Liao H. Y., Jeng L. B. (2011) Clin. Chim. Acta 412, 230–239 [DOI] [PubMed] [Google Scholar]
- 7. Cornett D. S., Mobley J. A., Dias E. C., Andersson M., Arteaga C. L., Sanders M. E., Caprioli R. M. (2006) Mol. Cell. Proteomics 5, 1975–1983 [DOI] [PubMed] [Google Scholar]
- 8. Schwartz S. A., Weil R. J., Johnson M. D., Toms S. A., Caprioli R. M. (2004) Clin. Cancer Res. 10, 981–987 [DOI] [PubMed] [Google Scholar]
- 9. Schwartz S. A., Weil R. J., Thompson R. C., Shyr Y., Moore J. H., Toms S. A., Johnson M. D., Caprioli R. M. (2005) Cancer Res. 65, 7674–7681 [DOI] [PubMed] [Google Scholar]
- 10. Bauer J. A., Chakravarthy A. B., Rosenbluth J. M., Mi D., Seeley E. H., De Matos Granja-Ingram N., Olivares M. G., Kelley M. C., Mayer I. A., Meszoely I. M., Means-Powell J. A., Johnson K. N., Tsai C. J., Ayers G. D., Sanders M. E., Schneider R. J., Formenti S. C., Caprioli R. M., Pietenpol J. A. (2010) Clin. Cancer Res. 16, 681–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reyzer M. L., Caldwell R. L., Dugger T. C., Forbes J. T., Ritter C. A., Guix M., Arteaga C. L., Caprioli R. M. (2004) Cancer Res. 64, 9093–9100 [DOI] [PubMed] [Google Scholar]
- 12. Rauser S., Marquardt C., Balluff B., Deininger S. O., Albers C., Belau E., Hartmer R., Suckau D., Specht K., Ebert M. P., Schmitt M., Aubele M., Höfler H., Walch A. (2010) J. Proteome Res. 9, 1854–1863 [DOI] [PubMed] [Google Scholar]
- 13. Kim H. K., Reyzer M. L., Choi I. J., Kim C. G., Kim H. S., Oshima A., Chertov O., Colantonio S., Fisher R. J., Allen J. L., Caprioli R. M., Green J. E. (2010) J. Proteome Res. 9, 4123–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rahman S. M., Shyr Y., Yildiz P. B., Gonzalez A. L., Li H., Zhang X., Chaurand P., Yanagisawa K., Slovis B. S., Miller R. F., Ninan M., Miller Y. E., Franklin W. A., Caprioli R. M., Carbone D. P., Massion P. P. (2005) Am. J. Respir. Crit. Care Med. 172, 1556–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schwamborn K., Krieg R. C., Reska M., Jakse G., Knuechel R., Wellmann A. (2007) Int. J. Mol. Med. 20, 155–159 [PubMed] [Google Scholar]
- 16. Burnum K. E., Tranguch S., Mi D., Daikoku T., Dey S. K., Caprioli R. M. (2008) Endocrinology 149, 3274–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. M'Koma A. E., Seeley E. H., Washington M. K., Schwartz D. A., Muldoon R. L., Herline A. J., Wise P. E., Caprioli R. M. (2011) Inflamm. Bowel Dis. 17, 875–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stoeckli M., Knochenmuss R., McCombie G., Mueller D., Rohner T., Staab D., Wiederhold K. H. (2006) Methods Enzymol. 412, 94–106 [DOI] [PubMed] [Google Scholar]
- 19. Willems S. M., van Remoortere A., van Zeijl R., Deelder A. M., McDonnell L. A., Hogendoorn P. C. (2010) J. Pathol. 222, 400–409 [DOI] [PubMed] [Google Scholar]
- 20. Caldwell R. L., Holt G. E., Caprioli R. M. (2005) Cancer Genomics Proteomics 2, 333–346 [PubMed] [Google Scholar]
- 21. Andersson M., Groseclose M. R., Deutch A. Y., Caprioli R. M. (2008) Nat. Methods 5, 101–108 [DOI] [PubMed] [Google Scholar]
- 22. Crecelius A. C., Cornett D. S., Caprioli R. M., Williams B., Dawant B. M., Bodenheimer B. (2005) J. Am. Soc. Mass Spectrom. 16, 1093–1099 [DOI] [PubMed] [Google Scholar]
- 23. Sinha T. K., Khatib-Shahidi S., Yankeelov T. E., Mapara K., Ehtesham M., Cornett D. S., Dawant B. M., Caprioli R. M., Gore J. C. (2008) Nat. Methods 5, 57–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wisztorski M., Franck J., Salzet M., Fournier I. (2010) Methods Mol. Biol. 656, 303–322 [DOI] [PubMed] [Google Scholar]
- 25. Becker K. F., Schott C., Hipp S., Metzger V., Porschewski P., Beck R., Nährig J., Becker I., Höfler H. (2007) J. Patho.l 211, 370–378 [DOI] [PubMed] [Google Scholar]
- 26. Gustafsson J. O., Oehler M. K., McColl S. R., Hoffmann P. (2010) J. Proteome Res. 9, 4315–4328 [DOI] [PubMed] [Google Scholar]
- 27. Xu H., Yang L., Wang W., Shi S. R., Liu C., Liu Y., Fang X., Taylor C. R., Lee C. S., Balgley B. M. (2008) J. Proteome Res. 7, 1098–1108 [DOI] [PubMed] [Google Scholar]
- 28. Ronci M., Bonanno E., Colantoni A., Pieroni L., Di Ilio C., Spagnoli L. G., Federici G., Urbani A. (2008) Proteomics 8, 3702–3714 [DOI] [PubMed] [Google Scholar]
- 29. Groseclose M. R., Massion P. P., Chaurand P., Caprioli R. M. (2008) Proteomics 8, 3715–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lemaire R., Desmons A., Tabet J. C., Day R., Salzet M., Fournier I. (2007) J. Proteome Res. 6, 1295–1305 [DOI] [PubMed] [Google Scholar]
- 31. Djidja M. C., Claude E., Snel M. F., Scriven P., Francese S., Carolan V., Clench M. R. (2009) J. Proteome Res. 8, 4876–4884 [DOI] [PubMed] [Google Scholar]
- 32. Su S. P., McArthur J. D., Andrew Aquilina J. (2010) Exp. Eye Res. 91, 97–103 [DOI] [PubMed] [Google Scholar]
- 33. Grey A. C., Schey K. L. (2009) Invest. Ophthalmol. Vis. Sci. 50, 4319–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morita Y., Ikegami K., Goto-Inoue N., Hayasaka T., Zaima N., Tanaka H., Uehara T., Setoguchi T., Sakaguchi T., Igarashi H., Sugimura H., Setou M., Konno H. (2010) Cancer Sci. 101, 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Djidja M. C., Claude E., Snel M. F., Francese S., Scriven P., Carolan V., Clench M. R. (2010) Anal. Bioanal. Chem. 397, 587–601 [DOI] [PubMed] [Google Scholar]
- 36. Berridge M. J., Irvine R. F. (1984) Nature 312, 315–321 [DOI] [PubMed] [Google Scholar]
- 37. Khan W. A., Blobe G. C., Hannun Y. A. (1995) Cell. Signal. 7, 171–184 [DOI] [PubMed] [Google Scholar]
- 38. Liu Y., Chen Y., Momin A., Shaner R., Wang E., Bowen N. J., Matyunina L. V., Walker L. D., McDonald J. F., Sullards M. C., Merrill A. H., Jr. (2010) Mol. Cancer 9, 186–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Katz-Brull R., Seger D., Rivenson-Segal D., Rushkin E., Degani H. (2002) Cancer Res. 62, 1966–1970 [PubMed] [Google Scholar]
- 40. Han X., Cheng H., Fryer J. D., Fagan A. M., Holtzman D. M. (2003) J. Biol. Chem. 278, 8043–8051 [DOI] [PubMed] [Google Scholar]
- 41. Burnum K. E., Cornett D. S., Puolitaival S. M., Milne S. B., Myers D. S., Tranguch S., Brown H. A., Dey S. K., Caprioli R. M. (2009) J. Lipid Res. 50, 2290–2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Woods A. S., Jackson S. N. (2006) AAPS J. 8, E391–E395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hankin J. A., Barkley R. M., Murphy R. C. (2007) J. Am. Soc. Mass Spectrom. 18, 1646–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Puolitaival S. M., Burnum K. E., Cornett D. S., Caprioli R. M. (2008) J. Am. Soc. Mass Spectrom. 19, 882–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Meriaux C., Franck J., Wisztorski M., Salzet M., Fournier I. (2010) J. Proteomics 73, 1204–1218 [DOI] [PubMed] [Google Scholar]
- 46. Römpp A., Guenther S., Schober Y., Schulz O., Takats Z., Kummer W., Spengler B. (2010) Angew. Chem. Int. Ed. Engl. 49, 3834–3838 [DOI] [PubMed] [Google Scholar]
- 47. Jackson S. N., Wang H. Y., Woods A. S. (2005) J. Am. Soc. Mass Spectrom. 16, 2052–2056 [DOI] [PubMed] [Google Scholar]
- 48. Cornett D. S., Frappier S. L., Caprioli R. M. (2008) Anal. Chem. 80, 5648–5653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Prideaux B., Staab D., Stoeckli M. (2010) Methods Mol. Biol. 656, 405–413 [DOI] [PubMed] [Google Scholar]
- 50. Kitteringham N. R., Jenkins R. E., Lane C. S., Elliott V. L., Park B. K. (2009) J. Chromatogr. B 877, 1229–1239 [DOI] [PubMed] [Google Scholar]
- 51. Wagner M., Varesio E., Hopfgartner G. (2008) J. Chromatogr. B 872, 68–76 [DOI] [PubMed] [Google Scholar]
- 52. Stoeckli M., Staab D., Schweitzer A. (2007) Int. J. Mass Spectrom. 260, 195–202 [Google Scholar]
- 53. Khatib-Shahidi S., Andersson M., Herman J. L., Gillespie T. A., Caprioli R. M. (2006) Anal. Chem. 78, 6448–6456 [DOI] [PubMed] [Google Scholar]
- 54. Wang H. Y., Jackson S. N., McEuen J., Woods A. S. (2005) Anal. Chem. 77, 6682–6686 [DOI] [PubMed] [Google Scholar]
- 55. Reyzer M. L., Hsieh Y., Ng K., Korfmacher W. A., Caprioli R. M. (2003) J. Mass Spectrom. 38, 1081–1092 [DOI] [PubMed] [Google Scholar]
- 56. Krutchinsky A. N., Chait B. T. (2002) J. Am. Soc. Mass Spectrom. 13, 129–134 [DOI] [PubMed] [Google Scholar]
- 57. Marshall P., Toteu-Djomte V., Bareille P., Perry H., Brown G., Baumert M., Biggadike K. (2010) Anal. Chem. 82, 7787–7794 [DOI] [PubMed] [Google Scholar]
- 58. Nilsson A., Fehniger T. E., Gustavsson L., Andersson M., Kenne K., Marko-Varga G., Andrén P. E. (2010) PLoS One 5, e11411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bouslimani A., Bec N., Glueckmann M., Hirtz C., Larroque C. (2010) Rapid Commun. Mass Spectrom. 24, 415–421 [DOI] [PubMed] [Google Scholar]
- 60. Signor L., Varesio E., Staack R. F., Starke V., Richter W. F., Hopfgartner G. (2007) J. Mass Spectrom. 42, 900–909 [DOI] [PubMed] [Google Scholar]