Abstract
Xanthomonas axonopodis pv. citri (Xac) causes citrus canker, provoking defoliation and premature fruit drop with concomitant economical damage. In plant pathogenic bacteria, lipopolysaccharides are important virulence factors, and they are being increasingly recognized as major pathogen-associated molecular patterns for plants. In general, three domains are recognized in a lipopolysaccharide: the hydrophobic lipid A, the hydrophilic O-antigen polysaccharide, and the core oligosaccharide, connecting lipid A and O-antigen. In this work, we have determined the structure of purified lipopolysaccharides obtained from Xanthomonas axonopodis pv. citri wild type and a mutant of the O-antigen ABC transporter encoded by the wzt gene. High pH anion exchange chromatography and matrix-assisted laser desorption/ionization mass spectrum analysis were performed, enabling determination of the structure not only of the released oligosaccharides and lipid A moieties but also the intact lipopolysaccharides. The results demonstrate that Xac wild type and Xacwzt LPSs are composed mainly of a penta- or tetra-acylated diglucosamine backbone attached to either two pyrophosphorylethanolamine groups or to one pyrophosphorylethanolamine group and one phosphorylethanolamine group. The core region consists of a branched oligosaccharide formed by Kdo2Hex6GalA3Fuc3NAcRha4 and two phosphate groups. As expected, the presence of a rhamnose homo-oligosaccharide as O-antigen was determined only in the Xac wild type lipopolysaccharide. In addition, we have examined how lipopolysaccharides from Xac function in the pathogenesis process. We analyzed the response of the different lipopolysaccharides during the stomata aperture closure cycle, the callose deposition, the expression of defense-related genes, and reactive oxygen species production in citrus leaves, suggesting a functional role of the O-antigen from Xac lipopolysaccharides in the basal response.
Keywords: Carbohydrate Structure, Glycolipid Structure, Lipopolysaccharide (LPS), Mass Spectrometry (MS), Pathogen-associated Molecular Pattern (PAMP), UV-MALDI-TOF MS, Xanthomonas axonopodis pv. citri
Introduction
Citrus canker, one of the most devastating citrus diseases in the world, is caused by the Gram-negative bacterium Xanthomonas axonopodis pv. citri (Xac).6 The disease is characterized by the formation of circular and water-soaked spots on the abaxial surface of leaves. Then the bacteria colonize the apoplast, and the leaf epidermis is broken due to cell hyperplasia induced by the pathogen. Finally, the lesions raise and then darken and thicken into a light tan to brown corky canker. On heavily infected trees, citrus canker causes defoliation and premature fruit drop (1, 2).
Plants have basal perception systems for characteristic molecules from different classes of microorganisms, named pathogen-associated molecular patterns (PAMPs). Several components of the bacterial surfaces have been demonstrated as PAMPs, including flagellin, the lipopolysaccharides from Gram-negative bacteria, fungal cell wall-derived glucans, chitins, mannans, or proteins and peptidoglycans from Gram-positive bacteria (3).
In plant pathogenic bacteria, lipopolysaccharides (LPSs) are important virulence factors, and they are being increasingly recognized as major PAMPs for plants. LPSs share a common structure for all Gram-negative bacteria composed of a membrane-anchored phosphorylated and acylated 1–6-linked glucosamine (GlcN) disaccharide, named lipid A, to which a carbohydrate moiety of varying size is attached. The latter may be divided into a lipid A proximal core oligosaccharide and a distal O-antigen, whose presence or absence determines the smooth or rough appearance of the bacterial colony. Accordingly, LPSs are named smooth (S-) or rough (R-) respectively, with R-LPSs also known as lipo-oligosaccharides (LOS) (4, 5). Due to their amphipathic nature, structural characterization of LPSs is not an easy task, and molecular weight measurements are not straightforward because LPSs form aggregates and present intermolecular cross-linking of negative charges via divalent cations (6). Therefore, exquisitely sensitive analytical methods are required to determine the whole structure. Matrix-assisted laser desorption/ionization (MALDI) mass spectra have been widely used to gain knowledge about lipid A heterogeneity (7–9). Although much of the work on LPSs was performed after chemical degradation, some intact smaller structures have been obtained, particularly when only short O-specific chains are present or when the O-chain is missing (10).
LPSs apparently play diverse roles in bacterial pathogenesis of plants. As major components of the outer membrane, they are involved in the protection of bacterial cells, contributing to reduce the membrane permeability and thus allowing growth of bacteria in the unfavorable conditions of the plant environment (11). Moreover, LPSs can be recognized by plants to elicit or potentiate plant defense-related responses (12, 13). One of the most widely studied effects of LPSs on plant cells is their ability to prevent the hypersensitive response (HR) induced in plants by avirulent bacteria. HR is a rapid and localized response characterized by reactive oxygen species (ROS) production and programmed cell death that is often associated with plant host resistance (14, 15). In comparison with animal and human cells, where the role of LPS has been established, little is known about the mechanisms of LPS perception by plants and the cognate signal transduction pathway. Recent findings have suggested that the lipid A moiety may be at least partially responsible for LPS perception by Arabidopsis thaliana, leading to a rapid burst of NO, a hallmark of innate immunity in animals (13). Using synthetic O-antigen polysaccharides (oligorhamnans), it has been shown that the O-chain of LPS is recognized by Arabidopsis and that this recognition leads to elicitation of a specific gene transcription response associated with defense. These observations, together with the established influence of the core region on lipid A toxicity in animals, indicate that the elucidation of the structure and of the biological activity of both lipid A and core region are of high importance for a better understanding of LPS action in plants (16).
Regarding Xanthomonas campestris pv. campestris (Xcc), several studies on LPS structure or substructure have been reported in an attempt to understand its activity as a PAMP (14, 17, 18). In this work, we have determined the structure of purified LPSs obtained from X. axonopodis pv. citri wild type and a mutant of the O-antigen ABC transporter encoded by wzt gene named Xacwzt. High pH anion exchange chromatography (HPAEC) and MALDI mass spectrometry analysis under different conditions and with different matrices were performed, enabling structure determination of not only the released oligosaccharides and lipid A moieties but also the intact LPSs. Interestingly, striking differences from other Xanthomonas LPSs described before were evidenced. Moreover, we have also examined the LPS function from Xac in the pathogenesis process. In this context, we analyzed the response of the different LPSs during the stomata aperture closure cycle, the callose deposition, the expression of defense-related genes, and ROS production in citrus leaves, suggesting a functional role of the O-antigen from Xac LPS in the basal response.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Culture Growth Conditions, and Media
Escherichia coli cells were cultivated at 37 °C in Luria-Bertani (LB) medium. X. axonopodis pv. citri were grown at 28 °C in Silva-Buddenhagen (SB) medium (19). Antibiotics were used at the following final concentrations: ampicillin, 100 μg/ml for E. coli and 25 μg/ml for Xac; kanamycin, 40 μg/ml. All Xac strains were derivatives of the strain Xcc99-1330 kindly provided by Blanca I. Canteros.
Mating and Mutagenesis
All DNA manipulations, including plasmid purification, restriction enzyme digestion, DNA ligation, and agarose gel electrophoresis, were performed with standard techniques (20). Total bacterial genomic DNA from Xac was isolated using the cetyltrimethylammonium bromide procedure (21) and was used for the wzt fragment PCR amplification using the following primers: wzt-F (5′-ATGCAAGCTTCCTCTCAAGCGTCTATTCTCGT-3′) and wzt-R (5′-CGCGGATCCAGGCCCTTATCG GTAAAAAGAC-3′). Underlined letters correspond to restriction sites. The PCR-amplified product wzt-1031bp, digested with BamHI and HindIII, was cloned into the suicide vector pK18mobGII (22), yielding the plasmid pK/wzt. Plasmid DNA was transferred to Xac by biparental mating from the broad host-range-mobilizing E. coli strain S17–1 (23). Bacterial mixtures were spotted onto Hybond-C membranes, placed on SB agar, and incubated for 48 h at 28 °C. The membranes were then washed with 0.9% NaCl, and the bacteria were transferred to selective medium as described previously (24). Xac mutant strain was selected by the vector-encoded antibiotic resistance (kanamycin) and confirmed by PCR hybridization analysis (data not shown). Mutants were complemented by cloning the wzt amplified fragments in pBBR1MCS-5 (25).
LPS Extraction and Purification
Bacterial cultures of Xac wild type and Xacwzt were grown in liquid SB medium to stationary phase (A600 ∼3) and centrifuged for 20 min at 10,000 × g. LPSs from harvested cells were extracted with a 50% phenol/water mixture (26). The aqueous phases after three extractions were pooled and exhaustively dialyzed (membrane cut-off, 12 kDa) against distilled water at 4 °C. The LPS preparations were evaluated by SDS-PAGE and silver staining analysis as described by Marolda et al. (27) and Tsai and Frasch (28), respectively.
Isolation and Purification of Lipid A
LPS was hydrolyzed with 1% acetic acid for 3 h at 100 °C (17). Precipitated lipid A was recovered by ultracentrifugation at 4 °C, 8000 × g for 60 min. The solution containing the sugar moiety (called oligosaccharide) was separated and lyophilized. The solid was washed and further extracted with CHCl3/MeOH 3:1 (v/v). After drying the organic phase, purified lipid A was obtained.
Characterization of Lipids A by TLC
Lipids A obtained from Xac wild type and Xacwzt LPSs were analyzed by thin layer chromatography (TLC) on silica gel (Merck) developed with CHCl3/MeOH/H2O/NEt3, 12: 6: 1: 0.04 (v/v/v/v) and revealed with 5% H2SO4 in ethanol and heat.
Compositional Analysis of Fatty Acids
Fatty acids were determined by gas chromatography-mass spectrometry (GC-MS) analysis as methyl ester derivatives. GC-MS was performed on a Shimadzu QP 5000-GC 17A. Electron impact mass spectra were recorded at 70 eV. Spectra were acquired from m/z 40–700. Fatty acids were obtained from the lipid A by method 1) acid hydrolysis of lipid A with 4 n HCl (100 °C, 18 h) (29), in which fatty acids were extracted with chloroform and after drying were methylated with 1 n HCl/MeOH (65 °C, 2h), and method 2) basic hydrolysis with 4 n NaOH (100 °C, 5 h), neutralization, and extraction with chloroform (30). The dried sample was dissolved in anhydrous methanol, and immediately before injection, it was treated with methanolic 0.5 m phenyltrimethylammonium hydroxide solution (Fluka) in the presence of molecular sieves. Analysis was performed on a capillary column (Ultra-1, 25 m × 0.20 mm). The temperature program was initially 80 °C for 2 min and then rose to 290 °C at a rate of 10 °C/min, final time 30 min. Injector temperature was 260 °C for method 1 producing in situ pyrolytic derivatization.
Compositional Analysis of Sugar Moieties
Determination of the sugar residues was carried out by HPAEC analysis. The oligosaccharides were hydrolyzed with 2 m trifluoroacetic acid (100 °C, 2 h), lyophilized, and analyzed in a DX-3000 Dionex BioLC system with pulsed amperometric detection (HPAEC-PAD; Dionex Corp., Sunnyvale, CA). Separation of carbohydrates was carried out on a CarboPac PA-10 column (25 × 0.4 cm) with a PA-10 precolumn and a 20-μl injection loop. The following conditions were employed. For neutral sugar analysis, an isocratic elution with 16 mm NaOH aqueous solution at 1 ml/min was used; for acidic sugar analysis, an isocratic elution with 50 mm NaOH and 0.1 m sodium acetate aqueous solution at 1 ml/min was used.
Additionally, 3N-acetylfucosamine (Fuc3NAc) was detected by GC-MS as the methyl peracetylated derivative on an Rxi-5 ms column (30 m × 0.25 mm; Thermo Scientific), temperature program 140 °C (5 min) to 290 °C, rate 5.
UV-MALDI-TOF Mass Spectrometry Analysis
Matrices and calibrating chemicals were purchased from Sigma-Aldrich. Measurements were performed using an Ultraflex II TOF/TOF mass spectrometer equipped with a high performance solid-state laser (λ = 355 nm) and a reflector. The system is operated by the Flexcontrol 2.4 software package (Bruker Daltonics GmbH, Bremen, Germany).
The mass spectra reported are the result of 1000 laser shots. All samples were measured in the linear and the reflectron mode and as routine in both positive and negative polarity.
The samples were loaded onto a ground steel sample plate (MTP 384 ground steel, Bruker Daltonics GmbH) using the sandwich method or by the classic dried drop method: a sample/matrix solution mixture (1 μl, 1:1 (v/v)) was deposited on the target plate and allowed to dry at room temperature.
Different matrices were assayed to obtain adequate spectra. For lipid A analysis, nor-harmane was the best; for oligosaccharides and complete LPSs, 2,4,6-trihydroxyacetophenone alone or with the addition of 20 mm ammonium citrate was used.
Callose Staining
Orange (Citrus sinensis cv. valencia late) was used as the host plant for Xac. All plants were grown in a growth chamber in incandescent light at 28 °C with a photoperiod of 16 h. Leaves were harvested 24 h after Xac wild type LPS (100 μg/ml), Xacwzt LPS (100 μg/ml), Xac wild type (107 colony-forming units (cfu)/ml), and Xacwzt (107 cfu/ml) inoculations, and thin sections (10 μm) were cut with a cryostat. The leaf sections were mounted on glass slides and stained with 0.05% (w/v) aqueous aniline blue solution in 100 mm phosphate buffer, pH 8.0, and coverslips were placed over them (19). The sections were examined with a UV fluorescence microscope (Nikon Eclipse E800).
Stomatal Guard Cell Assay
Segments of abaxial epidermis from orange leaves kept in darkness overnight were floated on buffer A (10 mm MES, 10 mm KOH, pH 6.15) for 2 h at 25 °C in darkness. Epidermis was transferred to LPS solutions (50 μg/ml) or buffer A containing 50 mm KCl and 100 μm CaCl2 with 1 μm naphtaleneacetic acid (NAA) or 50 μm abscisic acid (ABA) and exposed under incandescent light (λ = 430 nm at 35 watts/m2) at 25 °C for additional time (31). Pictures were taken at random regions under the microscope. Pore widths of >20 stomata from three separate segments for each treatment were measured using the software Image-Pro version 4.5 for Windows (Cybernetics, Inc.) (32). Results were assessed by the Kruskal-Wallis non-parametric analysis of variance.
RNA Preparation and Reverse Transcription-PCR (RT-PCR) Assays
Orange leaves were inoculated with 100 μg/ml LPS isolated from Xac wild type and Xacwzt. Leaves were harvested at 2, 6, and 24 h postinoculation (hpi). Plant total RNA was isolated using TRIzol® reagent (Invitrogen) according to the manufacturer's recommendations. After extraction, the RNA was treated with RNase-free DNase (Promega), and its integrity was checked by agarose gel electrophoresis. cDNA first strand was synthesized from 1 μg of total RNA as template using 200 units of Moloney murine leukemia virus reverse transcriptase (Promega), 0.5 mm dNTP mixture, 2.5 μg of oligonucleotide dT22 and incubating for 60 min at 42 °C. PCRs were performed using specific primers for pathogenesis-related 1 (PR1), phenylalanine ammonia-lyase (PAL), and mitogen-activated protein kinase kinase 4 (MKK4) genes (supplemental Table S1).
Control reactions, where RT was omitted, were done in parallel for all of the samples to rule out the possibility of amplification from contaminating DNA. PCRs were carried out with 2 μl of cDNA template under the following conditions: 25 cycles of denaturation at 94 °C for 1 min, annealing at 59 °C for 1 min, and extension at 72 °C for 1 min with a final extension step at 72 °C for 5 min. The number of cycles to be used, avoiding reaching the plateau of the PCRs, was previously determined by taking samples at a different number of cycles during the PCR amplification step and analyzing the products obtained by agarose gel electrophoresis. As a constitutive control, a 329-bp fragment of the actin gene was amplified using the same PCR conditions. RT-PCR products were resolved on 1.5% (w/v) agarose gels.
Peroxide Determination in Plant Tissue Extracts
A modified version of the FOX II assay was used to quantify the presence of peroxides in plant tissue extracts (33). Tobacco (Nicotina tabacum cv. Petit Havana), as non-host plant, and orange leaves were inoculated with Xac wild type and Xacwzt LPSs and H2O as control and harvested 2 hpi. The tissue was immediately frozen in liquid nitrogen, ground to a fine powder, and extracted in 300 μl of ethanol/H2O (80:20, v/v) with 0.01% butylated hydroxytoluene added. After centrifugation at 3000 × g for 10 min, 30 μl of the supernatant was combined with 30 μl of 10 mm TPP (a peroxide reducing agent) in methanol and incubated for 30 min in a 96-well microplate. Samples without TPP were treated in the same way. FOX reagent (90% MeOH (v/v), 25 mm H2SO4, 4 mm butylated hydroxytoluene, 250 μm ferrous ammonium sulfate hexahydrate, and 100 μm xylenol orange) was then added to each sample. Absorbance at 560 nm was recorded 10 min after FOX addition in a multimode microplate reader (Synergy 2, BioTek). The absorbance differences between equivalent samples with and without TPP indicate the levels of peroxides, which were calculated using a 0–20 μm H2O2 standard curve. The results were expressed as μm H2O2/g fresh weight. Statistical analysis was carried out by means of analysis of variance and LSD Fisher multiple comparisons.
RESULTS
Characterization of Xac LPS Cluster Involved in LPS Biosynthesis
Xanthomonas is a sizable genus of plant pathogenic bacteria that possess a diverse range of host plants rendering different types of diseases: vascular and non-vascular or localized disease (34). The LPS clusters of several Xanthomonas species were completely analyzed, and it was found that the LPS locus is flanked by two conserved genes: the cystathionine γ-lyase gene (metB) at one end and the electron transport flavoprotein gene (etfA) at the other end. The locus presents a size of 19.9 kb and contains 17 open reading frames (35). The Xac genome has been completely sequenced, and the genes involved in LPS biosynthesis have been identified (36, 37). Based on the report by Patil et al. (35), we have identified the Xac wzt gene (ABC type transporter O-antigen) located between the metB and etfA genes. This gene corresponds to 1.NP_643907.1 in GenBankTM. We have constructed a Xac mutant in the wzt gene responsible for the transport of the O-antigen region, in order to determine the structure of the Xac mutant LPS and evaluate the role of this region in the virulence process during citrus canker disease.
Isolation of LPSs
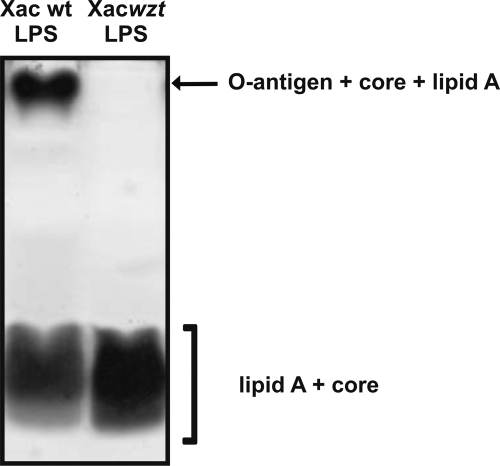
LPSs from Xac wild type and Xacwzt were purified by the hot phenol method (26) and analyzed by SDS-PAGE (Fig. 1). The LPS from Xac wild type showed one well defined slowly migrating band that corresponds to the entire LPS, composed of the O-antigen + core + lipid A moiety and a diffuse and broad faster migrating band. The upper part of the faster migrating bands represents the lipid A + core, and the lower part represents the lipid A + inner core of the LPS (38). The Xacwzt LPS lacked the slowly migrating band, corresponding to the high molecular weight LPS molecule containing a polymeric O-antigen, when compared with Xac wild type LPS. The electrophoretic mobility of the rapidly migrating bands representing the lipid A + core structure was not influenced in this mutant. This finding strongly suggested that Xacwzt LPS bears no O-chain at all and that only the lipid A + core portions are present. The pattern observed in the LPS from Xac wild type is similar to other Xanthomonas spp. (14, 39).
FIGURE 1.
Electrophoretic analysis of LPS from Xac. SDS-PAGE analysis of LPSs prepared from Xac wild type (wt) (lane 1) and Xacwzt (lane 2) strains. LPSs were isolated by the hot phenol method. The polyacrylamide gel (14% acrylamide in the separating gel and 5% in the stacking gel) was run with a Tricine buffer system and subsequently silver-stained. The arrows on the right indicate the different regions of the LPS molecules.
Structural Analysis of Lipids A
First, to elucidate the structures of lipids A and oligosaccharides of the Xac wild type and Xacwzt LPSs, acid hydrolysis with acetic acid was performed on the corresponding LPSs to split the lipid A from the oligosaccharide (17). The corresponding lipids A were purified and analyzed by TLC (Fig. 2A). Notably, several spots were detected in both samples, probably due to the variability of the degree of acylation and the phosphorylation pattern.
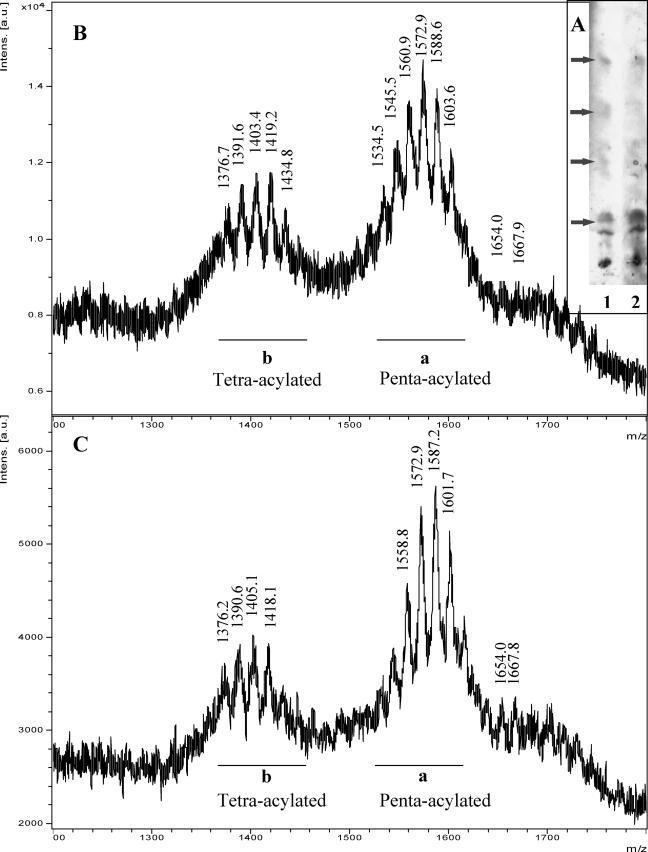
FIGURE 2.
A, analysis by TLC of lipid A obtained from Xac wild type (lane 1) and from Xacwzt (lane 2) in CHCl3/MeOH/H2O/NEt3 (12:6:1.0:0.04) (v/v/v/v). The arrows indicate species with different degree of acylation/phosphorylation. B, UV-MALDI-TOF spectrum in the positive ion mode of Xac wild type lipid A. C, UV-MALDI-TOF spectrum in the positive ion mode of the Xacwzt lipid A. a and b, penta- and tetra-acylated species respectively. Mass numbers correspond to [M + Na]+ ions. The mass numbers given are those of monoisotopic peaks. For the assignment of the structures, see Table 1 and “Results.”
UV-MALDI-TOF mass spectra of lipids A from Xac wild type and Xacwzt LPSs using nor-harmane as matrix were similar (Fig. 2, B and C, respectively). Analysis in the positive ion mode showed two main clusters of ions. One of them, the most abundant (Fig. 2B, a), ranging from m/z 1530 to 1605, with the most intense peaks at m/z 1560.9, 1572.9, and 1588.6, indicates the presence of [M + Na]+ adducts corresponding to a penta-acylated diglucosamine backbone attached to one phosphorylethanolamine (P-EtNH2) and one pyrophosphorylethanolamine (PP-EtNH2) group. The other cluster, ranging from m/z 1375 to 1435 (Fig. 2B, b), with the most intense peaks at m/z 1391.6, 1403.4, and 1419.2, corresponds to [M + Na]+ ions of tetra-acylated species. Each family consisted of several signals with mass differences of Δm/z 14 or 16, indicating different lengths of fatty acid residues and/or the presence of an additional hydroxyl group. In addition, low intensity signals at m/z 1667.9 and 1654.0 consistent with a diglucosamine backbone penta-acylated and carrying two PP-EtNH2 groups were observed.
GC-MS analysis of fatty acids (obtained by method 2) from lipid A (supplemental Fig. S1) revealed mainly the presence of saturated C8:0, C9:0, C10:0, C11:0, C14:0, and C15:0 and three peaks showing characteristic signals at m/z 75, M-15, M-30, M-73, and M-89 corresponding to the linear 3-methoxy-methylesters C11:0, C12:0, and C13:0. In accordance, fatty acid analysis after derivatization by method 1 showed a group of signals with diagnostic base peak at m/z 103 of 3-hydroxymethylesters of C11:0, C12:0, and C13:0. (supplemental Fig. S2) Therefore, 3-hydroxyl group resulted, methylated by phenyltrimethylammonium hydroxide. Table 1 summarizes the proposed structures for lipid A MALDI-TOF main ions, taking into account the results of fatty acid analysis.
TABLE 1.
Proposed structures for lipid A obtained from Xac wild type LPS after MALDI-TOF mass spectrometry analysis in the positive ion mode (sugar backbone: [GlcNH2-GlcNH2])
| Masscalc.a [M + Na]+ | Massmeas.b [M + Na]+ | Proposed structures | PP-EtNH2 | P-EtNH2 | |
|---|---|---|---|---|---|
| Penta-acylated (a) | 1667.9 | 1667.9 | C10:0 C13:0(OH) C11:0 C15:0 C9:0 | 2 | |
| 1653.9 | 1653.2 | C10:0 C13:0(OH) C11:0 C14:0 C9:0 | 2 | ||
| 1603.9 | 1603.6 | C11:0 C13:0(OH) C11:0 C13:0 (OH) C10:0 | 1 | 1 | |
| 1587.9 | 1588.6 | C10:0 C13:0(OH) C11:0 C14:0 C10:0 | 1 | 1 | |
| 1573.9 | 1572.9 | C10:0 C13:0(OH) C11:0 C14:0 C9:0 | 1 | 1 | |
| 1559.9 | 1560.9 | C10:0 C13:0(OH) C11:0 C14:0 C8:0 | 1 | 1 | |
| 1545.8 | 1545.2 | C10:0 C12:0(OH) C11:0 C14:0 C8:0 | 1 | 1 | |
| 1533.8 | 1534.5 | C 8:0 C11:0(OH) C11:0 C12:0(OH) C9:0 | 1 | 1 | |
| Tetra-acylated (b) | 1433.8 | 1434.6 | C13:0(OH) C11:0 C14:0 C10:0 | 1 | 1 |
| 1419.8 | 1419.2 | C13:0(OH) C11:0 C14:0 C9:0 | 1 | 1 | |
| 1405.8 | 1403.4 | C13:0(OH) C11:0 C14:0 C8:0 | 1 | 1 | |
| 1391.7 | 1391.6 | C12:0(OH) C11:0 C14:0 C8:0 | 1 | 1 | |
| 1377.7 | 1376.7 | C11:0(OH) C11:0 C14:0 C8:0 | 1 | 1 |
a calc., calculated monoisotopic masses.
b meas. measured monoisotopic masses.
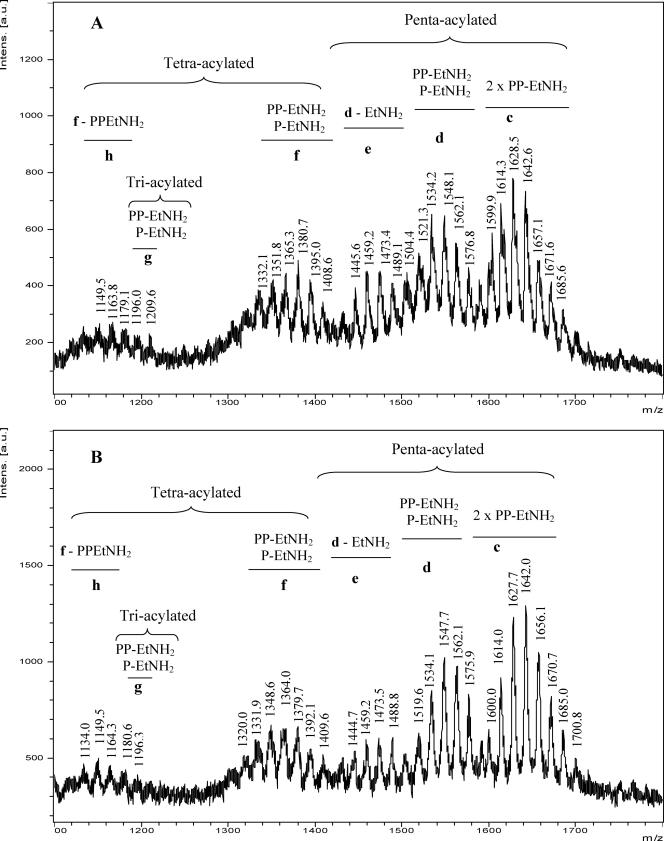
When the MALDI-TOF MS analysis of the lipid A was performed in the negative ion mode, a similar pattern for both LPSs was also observed, although five cluster ions were detected (Fig. 3, A and B). Cluster d ranging from m/z 1580 to 1520 in the form [M − H]− correlate with the penta-acylated species detected in the spectra recorded in the positive polarity (Fig. 2B, a). In addition, group e, ranging from m/z 1450 to 1510, was consistent with the loss of ethanolamine (60 mass units) from penta-acylated species. Ions ranging from m/z 1410 to 1350 (f) confirm the tetra-acylated structures detected in the positive ion mode (Fig. 2B, b). Group h ranging from m/z 1140 to 1200, present in a very low amount, corresponds to the loss of 204 mass units (PP-EtNH2) from tetra-acylated species (group f). Furthermore, ions m/z 1209.9 and 1196.0 are consistent with triacylated species yielded by the loss of the C10–C13 (OH) acyloxy group from the penta-acylated species. Moreover, ions in the highest mass range (Fig. 3, A and B, c) ranging from m/z 1690 to 1600, with 80 mass units more than d, correspond to penta-acylated lipid A moieties carrying two PP-EtNH2 groups. Interestingly, ions m/z 1657.1, 1671.6, and 1685.6 were not detected as [M + Na]+ in the positive ion mode spectra. Table 2 summarizes the proposed structures for the main ions differing in the phosphorylation pattern.
FIGURE 3.
Negative ion UV-MALDI-TOF analysis of the lipid A released from Xac LPSs. Spectra obtained from Xac wild type (A) and Xacwzt (B) LPSs are shown. Major ions corresponding to the penta-acylated diglucosamine backbone carrying two PP-EtNH2 (c); one P-EtNH2 and one PP-EtNH2 (d); d species after loss of EtNH2 (e); tetra-acylated diglucosamine backbone carrying one P-EtNH2 and one PP-EtNH2 (f); triacylated species carrying one P-EtNH2 and one PP-EtNH2 (g); and f species after loss of PP-EtNH2 (h) are indicated. Mass numbers correspond to [M − H]− ions. The mass numbers given are those of monoisotopic peaks. For the assignment of the structures, see Table 2 and “Results.”
TABLE 2.
Assignment of the main ions obtained by MALDI-TOF MS in the negative ion mode of lipid A from Xac wild type LPS (sugar backbone: [GlcNH2-GlcNH2])
| Acylation | Masscalc.a [M − H]− | Massmeas.b [M − H]− | PP-EtNH2 | P-EtNH2 | M-HOEtnH2 or M-PPEtNH2 |
|---|---|---|---|---|---|
| Penta-acylated | |||||
| c | 1685.9 | 1685.6 | 2 | ||
| 1671.9 | 1671.6 | 2 | |||
| 1657.8 | 1657.1 | 2 | |||
| 1643.9 | 1642.6 | 2 | |||
| 1629.9 | 1628.5 | 2 | |||
| 1615.9 | 1614.3 | 2 | |||
| 1601.9 | 1599.9 | 2 | |||
| d | 1577.8 | 1576.8 | 1 | 1 | |
| 1563.8 | 1562.1 | 1 | 1 | ||
| 1549.9 | 1548.1 | 1 | 1 | ||
| 1535.9 | 1534.2 | 1 | 1 | ||
| 1521.9 | 1521.3 | 1 | 1 | ||
| e | 1503.9 | 1504.4 | 1563.8–60 | ||
| 1490.8 | 1489.1 | 1549.9–60 | |||
| 1474.2 | 1473.4 | 1534.2–60 | |||
| 1461.3 | 1459.2 | 1521.3–60 | |||
| Tetra-acylated | |||||
| f | 1409.8 | 1408.6 | 1 | 1 | |
| 1395.8 | 1395.0 | 1 | 1 | ||
| 1381.8 | 1380.7 | 1 | 1 | ||
| 1367.7 | 1365.3 | 1 | 1 | ||
| 1353.7 | 1351.8 | 1 | 1 | ||
| h | 1177.8 | 1179.1 | 1 | 1381.8–204 | |
| 1163.7 | 1163.8 | 1 | 1367.7–204 | ||
| 1149.7 | 1149.5 | 1 | 1353.7–204 | ||
| Tri-acylated | |||||
| g | 1210.6 | 1209.6 | 1 | 1 | |
| 1196.6 | 1196.0 | 1 | 1 |
a calc., calculated monoisotopic masses.
b meas., measured monoisotopic masses.
Sugar Analysis of the Oligosaccharides Released from LPSs
The oligosaccharides from both LPSs were subjected to acid hydrolysis and further analysis by HPAEC-PAD (supplemental Fig. S3, A and B). Analysis of the neutral sugars showed main peaks corresponding to rhamnose (Rha), glucose (Glc), and mannose (Man). A minor component eluting faster than Rha was identified as Fuc3NAc by comparison with an authentic sample and by GC-MS. When the acidic components were analyzed by HPAEC-PAD, galacturonic acid (GalA) and 3-deoxy-d-manno-octulosonic acid (Kdo) were detected in both LPSs (supplemental Fig. S3C). The differences observed between the Glc/Man/Rha relationship in the Xac wild type LPS (1:4.3:11.2) and the Xacwzt LPS (1:0.2:1.3) support the fact that the latter lacks the O-antigen. Furthermore, these differences also suggest that rhamnose is the main component of the O-antigen moiety.
Structural Characterization of the Oligosaccharides
In order to perform a structural characterization of the oligosaccharides present in both LPSs, a detailed MALDI-TOF MS analysis was carried out (Fig. 4).
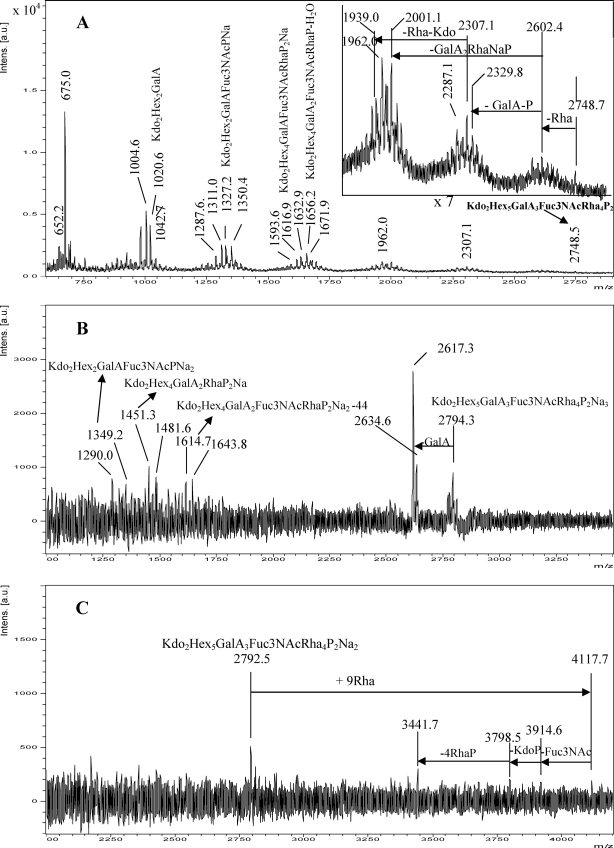
FIGURE 4.
UV-MALDI-TOF MS analysis of the oligosaccharides released from Xac LPSs. A, spectrum obtained from the Xacwzt LPS analyzed in the negative ion mode. The upper inset shows in detail the high mass region of the spectrum. Spectra were obtained from the Xacwzt LPS in positive ion mode (B) and from Xac LPS in the positive ion mode (C). Schematic structures of the most representative ions are shown. The mass numbers given are those of monoisotopic peaks. For the assignment of the structures, see Scheme 1 and “Results.”
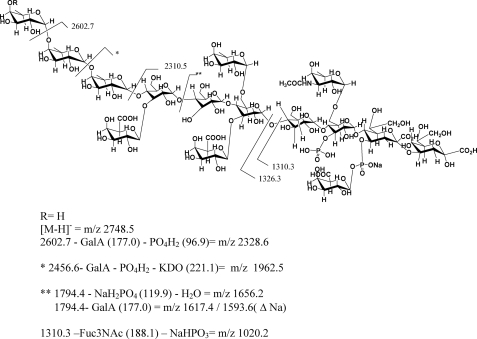
Regarding the oligosaccharide obtained from the Xacwzt LPS analyzed in the linear negative ion mode, the ion peak at m/z 2748.7 (calc. m/z 2748.5 [M − H]−) corresponds to the highest signal detected that matches with an oligosaccharide consisting of two Kdo, three GalA, five hexoses, four Rha, and one Fuc3NAc residue, bearing two phosphate groups (Fig. 4A and Scheme 1). Loss of one Rha unit (146 mass units) renders m/z 2602.4 (calc. m/z 2602.7). Subsequent loss of one GalA residue (177 mass units) and one phosphate group (97 mass units) renders ion peak m/z 2329.8 (calc. m/z 2328.6) and m/z 2307.1 (calc. m/z 2306.6) (ΔNa). Further loss of one Rha unit (146 mass units) and one Kdo (221 mass units) gives m/z 1962.0 (calc. m/z 1962.5) and m/z 1939.0 (calc. m/z 1939.5) (ΔNa). On the other hand, the signal at m/z 2001.1 (calc. m/z 2000.7) may be ascribed to loss of one GalA (177 mass units), one Rha (146 mass units), and NaPO3GalA (279 mass units) from m/z 2602.4.
SCHEME 1.
Major cleavages detected in the MALDI MS analysis of the oligosaccharidic moiety obtained from the Xacwzt LPS.
It is well known that in MALDI MS, “in source” fragmentation major cleavages, termed glycosidic cleavages, involve breaking a single bond between adjacent sugar rings (40). Then signals at m/z 1794.4 and 1810.4 (Δ16 mass units) were expected. However, signals at m/z 1656.2 (calc. m/z 1656.4) and 1671.9 (calc. m/z 1672.4) were detected, indicating further loss of water and NaH2PO4 from them, respectively (see Scheme 1). Similarly, the loss of a GalA residue from m/z 1794.4 and 1810.4 (Δ16 mass units) gave rise to m/z 1616.9 (calc. m/z 1617.4) and 1632.9 (calc. m/z 1633.4), respectively. Ions at m/z 1327.2 (calc. m/z 1326.3), 1311.0 (calc. m/z 1310.3) and m/z 1287.6 (ΔNa) correspond to the hexasaccharide-reducing moiety consisting in two Kdo, two hexoses, one Fuc3NAc, one GalA, and one phosphate group. Ion peaks at m/z 1020.6 (calc. m/z 1020.2) and 1004.6 (calc. m/z 1004.2) (Δ16 mass units) are consistent with the loss of Fuc3NAc and NaH2PO3 from m/z 1311.0. Furthermore, at low molecular masses, signals at m/z 675.0 (calc. m/z 675.1) and 652.2 (calc. m/z 652.1) (ΔNa) would correspond to the loss of 44 mass units from the Kdo-Kdo disaccharide linked to a GalA-phosphate residue.
Accordingly, the mass spectra recorded in the linear positive mode of the oligosaccharide coming from the Xacwzt LPS using the same matrix showed, in the highest mass range, main peaks at m/z 2794.3 (calc. m/z 2794.7) and 2617.3 (calc. m/z 2617.7) due to the loss of a GalA unit from the total structure assigned above bearing three sodium atoms (Fig. 4B). Other peaks between m/z 1644 to 1290 due to in source fragmentation, reinforce the proposed structure. For example, the ion at m/z 1349.2 would correspond to the addition of sodium to Kdo2Hex2 GalAFuc3NAcPNa (m/z 1327.2 in Fig. 4A); m/z 1451.3 is consistent with the composition Kdo2Hex4GalA2RhaP2Na; and m/z 1614.7 would correspond to the loss of CO2 (44 mass units) from Kdo2Hex4GalA2Fuc3NAcRhaP2Na.
On the other hand, regarding the oligosaccharide obtained from the Xac LPS, analysis in the negative ion mode showed similar signals up to m/z 2748.0 (supplemental Fig. S4). However, it was interesting to note that when the analysis was performed in the linear positive ion mode, in addition to signal at m/z 2792.5 corresponding to the core moiety, the highest mass signal corresponded to m/z 4117.7 (calc. m/z 4118.7). This ion may be assigned to the addition of nine Rha (9 × 146 mass units) to the core oligosaccharide (Fig. 4C). Furthermore, although of lower intensity, signals at m/z 3914.6, m/z 3798.5, and m/z 3441.7 are in agreement with m/z 4117.7 lacking a Fuc3NAc (204 mass units), one Kdo plus one phosphate group (221 mass units + 97 mass units), and four Rha and a phosphate group (4 × 146 mass units + 80 mass units), respectively (Fig. 4C). These signals confirm that a rhamnose homopolysaccharide constitutes the O-antigen of the Xac wild type LPS. It is worth noting that although this method is useful to determine sugar sequence, it provides no information about the configuration and type of linkages of the sugar components.
Structural Analysis of Intact LPS
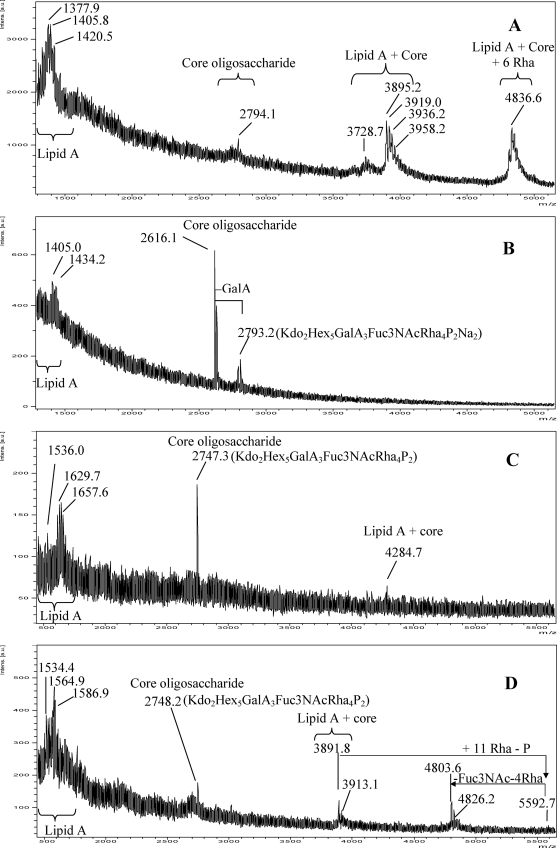
When the MS analysis of the intact Xac LPS was performed in the linear positive ion mode, signals in five regions were clearly detected: a region between m/z 1332 and 1650 corresponding to the lipid A signals, a region between m/z 2700 and 2800 corresponding to the core, a region between m/z 3700 and 3750 and a region between m/z 3850 and 4000 corresponding to lipid A with different degrees of acylation linked to the core, and finally the region between m/z 4700 and 4900 corresponding to the LPS structure (Fig. 5A).
FIGURE 5.
UV-MALDI-TOF MS analysis of the LPSs isolated from Xac. Xac wild type LPS in the positive ion mode (A), Xacwzt LPS in the positive ion mode (B), Xacwzt LPS in the negative ion mode (C), and Xac wild type LPS in the negative ion mode (D) spectra are shown. The mass numbers given are those of monoisotopic peaks. Main regions corresponding to the lipid A moiety, core, core + lipid A, and lipid A + core + O-antigen are indicated. Schematic structures of the most representative ions are shown. For the assignment of the structures, see Fig. 9B and “Results.”
In the lower mass range, major peaks m/z 1377.9, 1405.8, and 1420.5 are in accordance with the tetra-acylated sugar backbone of the lipid A (see Table 1). In the core region, peak at m/z 2794.1 (calc. m/z 2793.5) correlates with the oligosaccharide formed by two Kdo, five hexoses, three GalA, one Fuc3NAc, four Rha units, and two phosphate groups. Moreover, the signal at m/z 3919.0 (calc. m/z 3919.9) correlates with the oligosaccharide linked to the lipid A with the characteristic multiple signals due to fatty acid microheterogeneity. In the highest mass region, the signal at m/z 4836.6 (calc. m/z 4836.7) may be assigned to the addition of six Rha units to the core + lipid A moiety.
Noticeably, the spectrum of the intact Xacwzt LPS presents signals only at m/z 1405.0 and 1434.2 due to the lipid A moiety and at m/z 2793.2 and 2616.1 (ΔGalA) ascribed to the core region, reinforcing the fact that the O-antigen is truncated (Fig. 5B).
Accordingly, when the spectra were recorded in the negative polarity, the Xacwzt LPS presents only peaks corresponding to the lipid A moiety (m/z 1536.0, 1629.7, and 1657.6), to the core oligosaccharide (m/z 2747.3), and to the core + lipid A moiety (m/z 4284.7) (Fig. 5C). Finally, the Xac wild type LPS shows in the high mass range peaks ascribed to the lipid A + core region at m/z 3891.8 and 3913.1. The signal at m/z 5592.7 corresponds to the addition of 11 Rha units to m/z 3891.8 with the concomitant loss of a phosphate group (80 mass units). Also, signals at m/z 4826.2, 4803.6 (ΔNa), ascribed to the loss of one Fuc3NAc and four Rha units from m/z 5592.7, confirm the structure of the LPS (Fig. 5D).
Callose Deposition
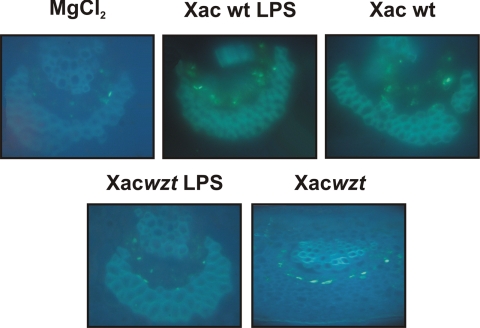
Cell wall alterations like the formation of minute papillae directly beneath the sites of pathogen penetration constitute a basal form of resistance to bacterial colonization (41). These papillae, which are primarily composed of callose (a β-1,3-glucan) and lignin (a highly complex phenolic polymer), are thought to act as a physical barrier, blocking pathogen colonization of plant cells. Callose deposition may be determined by staining with aniline blue. This selective staining of callose depends upon the structure of this polymer; in callose, the 1,3-glycosidic linkages of the glucose units cause the polymer to arrange in a helix that binds aniline blue, and the resulting complex leads to yellow fluorescence (19). As is shown in Fig. 6, callose deposition in orange leaves was higher and very similar in response to Xac wild type strain and Xac wild type LPS compared with the negative control MgCl2. On the other hand, callose deposition was less pronounced for the Xacwzt LPS compared with both bacterial strains and the LPS from Xac wild type. These results suggest that LPS from Xac probably has a role in the basal response during biotic stress.
FIGURE 6.
Callose deposition in orange leaves inoculated with Xac wild type, Xacwzt, and the different LPSs. The leaves were infiltrated with 10 mm MgCl2 as a negative control, with Xac wild type LPS and Xacwzt LPS solutions (100 μg/ml), and with 107 cfu/ml of the different bacterial suspension. The leaves were stained to visualize callose deposits 24 hpi and observed by fluorescence microscopy.
Lipopolysaccharide Role in Stomatal Aperture
Microbial entry into host tissue is a critical first step in the pathogenic process causing infection in animals and plants. In plants, it has been assumed that microscopic surface openings, such as stomata, serve as passive ports of bacterial entry during infection. Recent research has shown that stomatal closure by guard cells is part of a plant innate immune response to restrict bacterial invasion (32). In this work, we have analyzed the role of Xac LPS in the early stages of plant-pathogen interactions.
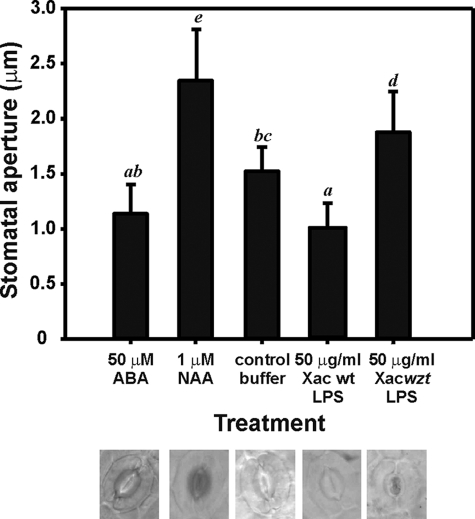
Inoculation procedures that we normally used artificially deliver bacteria directly underneath the epidermis, jumping the first barrier that bacteria encounter naturally during the plant colonization. For that reason, stomatal aperture was evaluated in orange leaves that were incubated in the presence of Xac wild type and Xacwzt LPSs for 2 h. As a positive control for opening the auxin analog, NAA was used, and for stomatal closure, ABA was employed (Fig. 7). LPS from Xac wild type did not induce stomatal opening in the studied conditions, presenting values similar to ABA and significantly lower than buffer (p < 0.01). On the other hand, LPS from Xacwzt did not manage to maintain closed the stomata, producing a significant opening in comparison with buffer, ABA, and LPS from Xac wild type (p < 0.0001). NAA control presented significant opening differences with all treatments but lower compared with LPS from Xacwzt. These results suggest that LPS from Xac probably has a role as PAMP, closing stomata at early stages of the infection. Furthermore, this result suggests that the O-antigen region from Xac LPS could be important for basal response during the plant-pathogen interaction.
FIGURE 7.
Stomatal guard cell assay. Shown is quantification of stomatal apertures in epidermal peels of orange leaves incubated with 50 μm ABA (bar 1), 1 μm NAA (bar 2), control buffer (bar 3), 50 μg/ml Xac LPS (bar 4), and Xacwzt LPS (bar 5). Below, representative images of stomatal apertures of each treatment are shown. Data are expressed as the mean of apertures of >60 stomata ± S.D. (error bars) of three independent experiments. Statistically significant differences are identified by lowercase letters (p < 0.01).
Expression of Defense-related Genes in Orange Leaves
During plant-pathogen interactions, including bacteria, the expression levels of many different genes are modified in the plants, allowing the activation/inhibition of defense mechanisms, depending on the response triggered (42, 43). Although special emphasis has been placed on gene expression during incompatible interactions, defense-related genes are also activated in compatible interactions, usually at later time points (44). PAL, PR1, and MKK4 are well known plant-inducible defense genes (45, 46).
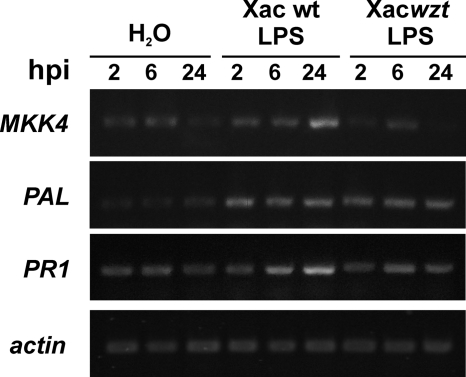
We examined the expression of the defense-related genes MKK4, PAL, and PR1 by RT-PCR using as a control for orange expression a 329-bp fragment of the actin gene. The products obtained in the PCRs are shown in Fig. 8. Transcript levels in response to Xac wild type LPS showed a later substantial accumulation at 24 hpi for MKK4 and PR1 genes compared with Xacwzt LPS- and control (carrier H2O)-inoculated leaves. On the other hand, a relatively weak increase in accumulation of the PAL transcripts was observed for Xac wild type- and Xacwzt LPS-inoculated leaves at all of the times assayed.
FIGURE 8.
Expression analysis of C. sinensis defense-related genes. Shown are amplified products of the PAL, PR1, and MKK4 genes by RT-PCR from orange leaves infiltrated with Xac wild type and Xacwzt LPSs and H2O as a control. Leaves were harvested at 2, 6, and 24 hpi. Equal cDNA amounts were controlled by amplification of the constitutively expressed actin gene (bottom gel). Three independent biological repetitions of the experiment were performed with similar results. The data shown from one experiment are therefore representative.
Peroxide Determination in Plant Tissue Extracts
ROS are important components of the host and non-host innate immunity during plant-pathogen interaction (19, 47). In previous reports, it was observed that the inoculation of several PAMPs like flagellin and EF-Tu also induced the ROS production in plants (48). In addition, it has been demonstrated that LPSs from various bacterial sources trigger H2O2 production in tobacco cell cultures (49).
In this context, we analyzed the ROS production in tobacco and orange leaves inoculated with LPS from Xac wild type and Xacwzt. We evaluated total peroxide accumulation by the FOX II assay. As is shown in Table 3, peroxide accumulation was significant higher in tobacco and orange leaves inoculated with Xac wild type LPS than in leaves inoculated with Xacwzt LPS and H2O-infiltrated leaves (p < 0.05). On the other hand, no differences were observed between Xacwzt LPS- and H2O-inoculated leaves.
TABLE 3.
Determination of peroxides by FOX assay in N. tabacum and C. sinensis leaves inoculated with Xac wild type and Xacwzt LPSs, at 2 hpi
Peroxide concentrations are expressed as μm H2O2/g fresh weight (fw).
| Tobacco | Orange | |
|---|---|---|
| H2O | 89.54 ± 7.06a | 123.66 ± 15.49a |
| Xac WT LPS | 138.51 ± 16.59b | 173.04 ± 18.42b |
| Xacwzt LPS | 92.64 ± 11.78a | 131.27 ± 14.35a |
a,b Statistically significant differences (p < 0.05).
DISCUSSION
The knowledge of how X. axonopodis pv. citri interacts with the host plant is a requisite to diminish the economic damage that citrus canker causes worldwide. This paper reports for the first time the characterization of the LPS molecule with a deep rough phenotype from the plant pathogenic bacterium X. axonopodis pv. citri and a wzt mutant strain. UV-MALDI-TOF MS analysis allowed the characterization of not only the lipid A moiety but also the core region plus the O-antigen. In order to preserve labile substituents, neither deacylation by hydrazinolysis nor deacylation by harsh alkaline treatment was used.
In the present work, we provide evidence that X. axonopodis pv. citri LPS shows striking differences with other bacterial LPS or Xanthomonas LPSs. In this sense, five major aspects may be pointed out: 1) the presence of two PP-EtNH2 groups in the lipid A moiety; 2) lipid A mainly penta-acylated; 3) the presence of two Kdo units linking lipid A to the core region; 4) the presence of a Fuc3NAc unit in the core; 5) a rhamnose homo-oligosaccharide as the O-antigen.
Xac lipids A could be isolated from the LPSs after mild acid hydrolysis and ultracentrifugation, indicating that the core-O-specific polysaccharide moieties are linked to the lipid A by an acid-labile constituent (i.e. 3-deoxy-d-manno-octulosonic acid) as is the case in most LPSs (50)).
Regarding aspect 1, UV-MALDI-TOF MS analysis of the chemically released lipid A showed a bis-pyrophosphorylethanolamine substituted diglucosamine disaccharide structure. As expected, signals corresponding to structures carrying the two pyrophosphorylethanolamine groups were more intense in the negative ion mode, due to the anionic nature of these species. The lipids A isolated from the Xac wild type and Xacwzt LPSs did not show differences in the degree of acylation and phosphorylation, as was evident in the mass spectrum analysis.
Regarding Xanthomonas genus, lipids A from LPSs of X. campestris pv. pruni and X. fragariae showed the same backbone structure, P-4-β-d-GlcN-(1–6)-α-d-GlcN-1-P acylated with fatty acids with a remarkable variability in chain lengths. Slight structural differences were identified between them: an additional P-EtNH2 residue in lipid A of the LPS from X. campestris pv. pruni and, on average, shorter acyl chains in the lipid A of the LPS from X. fragariae (51). In the lipid A moiety of the LOS from the mutant Xcc strain 8530, derived from the wild type strain 8004 of Xcc, a non-stoichiometric substitution of both polar heads by phosphorylethanolamine groups was reported (18). This feature also has been described in Neisseria meningitidis lipid A (52). Furthermore, the MALDI mass spectra of Xac lipid A show that structures carrying one pyrophosphoryethanolamine group and one phosphorylethanolamine group were also important. Interestingly, the substitution with ethanolamine groups would amend the net charge of the lipid A with possible consequences for binding of the molecule to putative plant receptors.
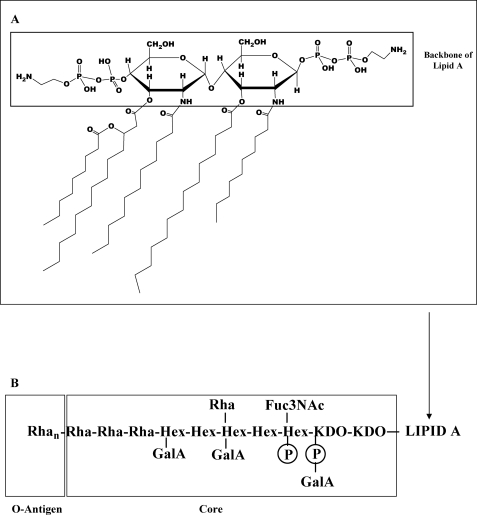
The second structural difference may be ascribed to the presence of mainly penta-acylated structures in the lipid A Xac moiety. Mainly saturated fatty acids that vary among C8:0 to C15:0 and 3-hydroxylated C11:0, C12:0, and C13:0 were determined in the LPS from Xac. Minor signals corresponding to tetra- and triacylated species were observed. By contrast, whereas X. campestris pv. pruni wild type lipid A was mainly hexa-acylated (51), the Xcc 8530 mutant's lipid A species were mainly penta-acylated (18). In the Fig. 9A, a schematic representation of lipid A from Xac is shown, taking into account the type and configuration of linkages determined for other Xanthomonas lipids A. The chemical peculiarities in the Xac LPS could play an important role in the pathogen infection in the host plant, and maybe the variability of the fatty acid could in turn help the bacterium to evade the host's response (53).
FIGURE 9.
Structure of LPS from Xac. A, chemical structure of the most representative penta-acylated lipid A from Xac LPS intensity. The box contains the diglucosamine + two pyrophosphorylethanolamine groups backbone of lipid A. B, schematic representation of the LPS from a Xac wild type. Boxes are drawn around the lipid A moiety, core region, and O-antigen.
Regarding the oligosaccharide moiety, released chains from the Xac wild type and Xacwzt LPS share a common mass spectra pattern ascribed to the core region. In addition, mass spectra of the Xac LPS released oligosaccharide showed signals at higher masses due to the presence of the O-antigen, confirming its absence in the mutant LPS. As expected, spectra from the intact LPSs from both strains also showed a common mass spectra pattern corresponding to the lipid A region, core region, and lipid A + core region, but only in the spectra of the Xac wild type LPS were significant signals at m/z >4500 shown. Thus, a signal at m/z 5592.7 correlates with a sugar chain carrying nine rhamnose units more than the core oligosaccharide (m/z 4284.7). As far as we know, only mass spectra of rough LPS or LOS have been reported, and our approach indicates that smooth LPS mass spectrum analysis is also valuable.
As in many LPSs, the inner core region carries anionic substituents; Kdo is linked to a galacturonyl phosphate substituent, and a hexose is linked to a phosphate group. The presence of negatively charged substituents in close proximity of the lipid A + core region is functionally important for intermolecular associations by cross-linking of divalent cations (54). In contrast with the core region described for Xcc, in Xac two Kdo units and one Fuc3NAc are also present. The latter has been reported as mainly being part of the O-antigen synthesized by most Xanthomonas species, such as Xcc strain 8004 (17) and X. hortorum pv. vitians (55). Fig. 9B shows a schematic representation of the LPS from Xac wild type.
Although some O-antigens from Xanthomonas species bear different sugars (rhamnose and xylose (56), rhamnose and fucose (55), and rhamnose and 3N-acetylfucosamine (57)), Xac shares with Xcc (14) a characteristic rhamnose homo-oligosaccharide as O-antigen. Bedini et al. (58) have constructed synthetic oligorhamnans showing a pronounced chain length-dependent induction of the PR1 gene expression, and prevention of the HR in A. thaliana. They demonstrated that short oligosaccharides can induce the expression of defense-related genes and prevent HR in a size-dependent manner. Therefore, they proposed that the coiled structure exposed from dimeric and trimeric oligomers of the repeating unit [α-l-Rha-(1–3)-α-l-Rha-(1–3)-α-l-Rha-(1–2)]n, peculiar for many O-chains of phytopathogenic bacteria, will act as a PAMP, being recognized by plants and consequently eliciting specific responses, including HR suppression and PR1 gene expression (58).
Callose deposition in plants is characteristic of the basal response being induced by different bacterial molecules (59). The callose deposition was detected when LPS isolated from Xac wild type was inoculated in orange plants, demonstrating that the LPS of Xac acts in the basal response. Braun et al. (14) showed that a truncated O-chain of the LPS produced a reduced induction of the oxidative burst in tobacco cell cultures. By comparing the elicitor activity of the different LPS species in tobacco cell cultures, they suggested that the O-antigen as well as the outer core region seem to play no role in the recognition process. On the other hand, the LPS of deep rough mutant Xcc H14 showed a decreased but still measurable activity, whereas the lipid A seems to be inactive (14).
The stomata are natural points of pathogen entry into plant leaves. Melotto and colleagues (32, 60) demonstrated that plant stomata are closed in response to the invasion of a plant pathogen, Pseudomonas syringae pv. tomato DC3000, and a human pathogen, E. coli O157:H7, concluding that both phytopathogenic and non-phytopathogenic living bacteria can also promote stomatal closure through the PAMPs like flagellin and lipopolysaccharide (32, 60). Furthermore, this response was also triggered by Flg22 and LPS (61, 62), supporting that the stomata closure is part of the mechanisms of immune response against bacterial invasion. These results strongly suggest that LPS is capable of triggering the stomatal closure during the plant basal response. In this context, we tested both LPSs with stomata from orange plant epidermis. In the presence of the Xac LPS, the stomata from the orange plant remained closed, while the Xacwzt LPS without O-antigen enabled stomata opening.
Newman et al. (63) demonstrated that purified lipopolysaccharide from X. campestris pv. campestris induced accumulation of transcripts, some of them defense-related genes. PAL, PR1, and MKK4 are some of the most studied defense-related genes. Phenylalanine ammonia-lyase (PAL) is a key enzyme in the phenylpropanoid pathway, which leads to the production of many phytoalexins or reactive compounds during HR development after treatment with various elicitors (64). Pathogenesis-related proteins (PRs) are assigned an important role in plant defense against pathogenic constraints and in general adaptation to stressful environment (65). In addition, the recognition of microbial PAMPs by a host plant cell activates downstream signaling mechanisms and cellular responses and includes the activation of mitogen-activated protein kinase (MAPK) cascades. A complete MAPK cascade (MEKK1, MKK4/MKK5, and MPK3/MPK6) and WRKY22/WRKY29 transcription factors were identified in A. thaliana downstream after the fagellin receptor FLS2 recognition. The activation of this MAPK cascade leads to resistance to both bacterial and fungal pathogens, suggesting that signaling events initiated by diverse pathogens converge into a conserved MAPK cascade. MKK4 is among the genes that are rapidly induced at the transcriptional level after the perception and transmission of the Flg22 signal in A. thaliana (45, 46). We evaluated the expression levels of the PAL, PR1, and MKK4 genes in orange leaves at 2, 6, and 24 hpi with Xac wild type and Xacwzt LPSs. PR1 and MKK4 genes were induced only by LPS from Xac wild type, whereas the LPS from Xacwzt rendered a similar pattern to the control. On the other hand, a low induction of PAL was observed with both LPS treatments. These results demonstrated that LPS from Xac is capable of inducing defense-related genes.
Plant basal defenses include a rapid accumulation of ROS. Furthermore, it has also been observed that PAMP inoculation elicits various defense responses, including H2O2 production (66). More specifically, Meyer et al. (49) have demonstrated that the addition of purified LPS from Xcc to tobacco cell cultures results in the induction of an oxidative burst reaction similar to that induced by already known elicitors. We observed an induction of the peroxide levels in orange and tobacco leaves inoculated with Xac wild type LPS compared with Xacwzt LPS and the control.
Altogether, these results suggest that the O-antigen region of X. axonopodis pv. citri LPS could be involved in the innate immunity. Our future aims are oriented to study further the interaction between Xac wild type and Xacwzt strains with citrus plants.
Supplementary Material
Acknowledgments
We thank Dr. Göran Widmalm (Stockholm University) for an authentic sample of d-Fuc3NAc. We thank Dr. Lucas Daurelio (IBR-CONICET-UNR) for statistical analyses.
This work was supported by a grant from the Agencia Nacional de Promoción Cientifica y Tecnológica (ANPCyT PICT 01-12783) (to E. G. O.), the University of Buenos Aires, and CONICET. The Ultraflex II (Bruker) TOF/TOF mass spectrometer was supported by an ANPCyT (Grant PME 125).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. S1–S4.
- Xac
- X. axonopodis pv. citri
- Xcc
- X. campestris pv. campestris
- HPAEC
- high pH anion exchange chromatography
- PAD
- pulsed amperometric detection
- HR
- hypersensitive response
- Kdo
- 3-deoxy-d-manno-octulosonic acid
- LOS
- lipo-oligosaccharides
- Man
- mannose
- Fuc3NAc
- 3N-acetylfucosamine
- PAMP
- pathogen-associated molecular pattern
- P-EtNH2
- phosphorylethanolamine
- PP-EtNH2
- pyrophosphorylethanolamine
- Rha
- rhamnose.
REFERENCES
- 1. Brunings A. M., Gabriel D. W. (2003) Mol. Plant Pathol. 4, 141–157 [DOI] [PubMed] [Google Scholar]
- 2. Graham J. H., Gottwald T. R., Cubero J., Achor D. S. (2004) Mol. Plant Pathol. 5, 1–15 [DOI] [PubMed] [Google Scholar]
- 3. Gust A. A., Biswas R., Lenz H. D., Rauhut T., Ranf S., Kemmerling B., Götz F., Glawischnig E., Lee J., Felix G., Nürnberger T. (2007) J. Biol. Chem. 282, 32338–32348 [DOI] [PubMed] [Google Scholar]
- 4. Holst O., Molinaro A. (2009) in Microbial Glycobiology: Structures, Relevance, and Applications (Moran A., Brennan P., Holst O., von Itszstein M. eds) pp. 29–56, Elsevier, San Diego, CA [Google Scholar]
- 5. Knirel Y. A., Senchenkova S. N., Shashkov A. S., Esteve C., Alcaide E., Merino S., Tomás J. M. (2009) Carbohydr. Res. 344, 479–483 [DOI] [PubMed] [Google Scholar]
- 6. John C. M., Liu M., Jarvis G. A. (2009) J. Lipid Res. 50, 424–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lindner B. (2000) Methods Mol. Biol. 145, 311–325 [DOI] [PubMed] [Google Scholar]
- 8. Therisod H., Labas V., Caroff M. (2001) Anal. Chem. 73, 3804–3807 [DOI] [PubMed] [Google Scholar]
- 9. Worrall T. A., Lin S., Cotter R. J., Woods A. S. (2000) J. Mass Spectrom. 35, 647–650 [DOI] [PubMed] [Google Scholar]
- 10. Harvey D. J. (2009) Mass Spectrom. Rev. 28, 273–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dow J. M., Osbourn A. E., Wilson T. J., Daniels M. J. (1995) Mol. Plant Microbe Interact. 8, 768–777 [DOI] [PubMed] [Google Scholar]
- 12. Conrath U., Thulke O., Katz V., Schwindling S., Kohler A. (2001) Eur. J. Plant Pathol. 107, 113–119 [Google Scholar]
- 13. Zeidler D., Zähringer U., Gerber I., Dubery I., Hartung T., Bors W., Hutzler P., Durner J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15811–15816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Braun S. G., Meyer A., Holst O., Pühler A., Niehaus K. (2005) Mol. Plant Microbe Interact. 18, 674–681 [DOI] [PubMed] [Google Scholar]
- 15. Newman M. A., von Roepenack E., Daniels M., Dow M. (2000) Mol. Plant Pathol. 1, 25–31 [DOI] [PubMed] [Google Scholar]
- 16. Silipo A., Molinaro A., Sturiale L., Dow J. M., Erbs G., Lanzetta R., Newman M. A., Parrilli M. (2005) J. Biol. Chem. 280, 33660–33668 [DOI] [PubMed] [Google Scholar]
- 17. Molinaro A., Silipo A., Lanzetta R., Newman M. A., Dow J. M., Parrilli M. (2003) Carbohydr. Res. 338, 277–281 [DOI] [PubMed] [Google Scholar]
- 18. Silipo A., Sturiale L., Garozzo D., Erbs G., Jensen T. T., Lanzetta R., Dow J. M., Parrilli M., Newman M. A., Molinaro A. (2008) Chembiochem 9, 896–904 [DOI] [PubMed] [Google Scholar]
- 19. Daurelio L. D., Tondo M. L., Dunger G., Gottig N., Ottado J., Orellano E. G. (2009) in Book on Plant Bioassays (Narwal S. S., Catalán A. N., Sampietro D. A., Vattuone M. A., Polyticka B. eds) Chapter 10, pp. 187–206, Studium Press, LLC, Houston, TX [Google Scholar]
- 20. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 21. Murray M. G., Thompson W. F. (1980) Nucleic Acids Res. 8, 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Katzen F., Becker A., Ielmini M. V., Oddo C. G., Ielpi L. (1999) Appl. Environ. Microbiol. 65, 278–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Simon R., Priefer U., Puhler A. (1983) Nat. Biotechnol. 1, 784–791 [Google Scholar]
- 24. Dunger G., Arabolaza A. L., Gottig N., Orellano E. G., Ottado J. (2005) Plant Pathol. 54, 781–788 [Google Scholar]
- 25. Kovach M. E., Elzer P. H., Hill D. S., Robertson G. T., Farris M. A., Roop R. M., 2nd, Peterson K. M. (1995) Gene 166, 175–176 [DOI] [PubMed] [Google Scholar]
- 26. Westphal O., Jann K. (1965) in Methods in Carbohydrate Chemistry (Whistler R. L. ed) pp. 83–91, Academic Press, Inc., New York [Google Scholar]
- 27. Marolda C. L., Lahiry P., Vinés E., Saldías S., Valvano M. A. (2006) Methods Mol. Biol. 347, 237–252 [DOI] [PubMed] [Google Scholar]
- 28. Tsai C. M., Frasch C. E. (1982) Anal. Biochem. 119, 115–119 [DOI] [PubMed] [Google Scholar]
- 29. Qureshi N., Takayama K., Ribi E. (1982) J. Biol. Chem. 257, 11808–11815 [PubMed] [Google Scholar]
- 30. Rietschel E. T., Lüderitz O., Volk W. A. (1975) J. Bacteriol. 122, 1180–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gottig N., Garavaglia B. S., Daurelio L. D., Valentine A., Gehring C., Orellano E. G., Ottado J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 18631–18636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Melotto M., Underwood W., Koczan J., Nomura K., He S. Y. (2006) Cell 126, 969–980 [DOI] [PubMed] [Google Scholar]
- 33. DeLong J. M., Prange R. K., Hodges D. M., Forney C. F., Bishop M. C., Quilliam M. (2002) J. Agric. Food Chem. 50, 248–254 [DOI] [PubMed] [Google Scholar]
- 34. Büttner D., Bonas U. (2010) FEMS Microbiol. Rev. 34, 107–133 [DOI] [PubMed] [Google Scholar]
- 35. Patil P. B., Bogdanove A. J., Sonti R. V. (2007) BMC Evol. Biol. 7, 243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. da Silva A. C., Ferro J. A., Reinach F. C., Farah C. S., Furlan L. R., Quaggio R. B., Monteiro-Vitorello C. B., Van Sluys M. A., Almeida N. F., Alves L. M., do Amaral A. M., Bertolini M. C., Camargo L. E., Camarotte G., Cannavan F., Cardozo J., Chambergo F., Ciapina L. P., Cicarelli R. M., Coutinho L. L., Cursino-Santos J. R., El-Dorry H., Faria J. B., Ferreira A. J., Ferreira R. C., Ferro M. I., Formighieri E. F., Franco M. C., Greggio C. C., Gruber A., Katsuyama A. M., Kishi L. T., Leite R. P., Lemos E. G., Lemos M. V., Locali E. C., Machado M. A., Madeira A. M., Martinez-Rossi N. M., Martins E. C., Meidanis J., Menck C. F., Miyaki C. Y., Moon D. H., Moreira L. M., Novo M. T., Okura V. K., Oliveira M. C., Oliveira V. R., Pereira H. A., Rossi A., Sena J. A., Silva C., de Souza R. F., Spinola L. A., Takita M. A., Tamura R. E., Teixeira E. C., Tezza R. I., Trindade, dos Santos M., Truffi D., Tsai S. M., White F. F., Setubal J. C., Kitajima J. P. (2002) Nature 417, 459–463 [DOI] [PubMed] [Google Scholar]
- 37. Van Sluys M. A., Monteiro-Vitorello C. B., Camargo L. E., Menck C. F., Da Silva A. C., Ferro J. A., Oliveira M. C., Setubal J. C., Kitajima J. P., Simpson A. J. (2002) Annu. Rev. Phytopathol. 40, 169–189 [DOI] [PubMed] [Google Scholar]
- 38. Vorhölter F. J., Niehaus K., Pühler A. (2001) Mol. Genet. Genomics 266, 79–95 [DOI] [PubMed] [Google Scholar]
- 39. Sidhu V. K., Vorhölter F. J., Niehaus K., Watt S. A. (2008) BMC Microbiol. 8, 87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Harvey D. J. (2003) Int. J. Mass Spectrom. 226, 1–35 [Google Scholar]
- 41. Keshavarzi M., Soylu S., Brown I., Bonas U., Nicole M., Rossiter J., Mansfield J. (2004) Mol. Plant Microbe Interact. 17, 805–815 [DOI] [PubMed] [Google Scholar]
- 42. Kazan K., Schenk P. M., Wilson I., Manners J. M. (2001) Mol. Plant Pathol. 2, 177–185 [DOI] [PubMed] [Google Scholar]
- 43. Daurelio L. D., Petrocelli S., Blanco F., Holuigue L., Ottado J., Orellano E. G. (2011) J. Plant Physiol. 168, 382–391 [DOI] [PubMed] [Google Scholar]
- 44. Dong X., Mindrinos M., Davis K. R., Ausubel F. M. (1991) Plant Cell 3, 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Asai T., Tena G., Plotnikova J., Willmann M. R., Chiu W. L., Gomez-Gomez L., Boller T., Ausubel F. M., Sheen J. (2002) Nature 415, 977–983 [DOI] [PubMed] [Google Scholar]
- 46. Zipfel C., Robatzek S., Navarro L., Oakeley E. J., Jones J. D., Felix G., Boller T. (2004) Nature 428, 764–767 [DOI] [PubMed] [Google Scholar]
- 47. Green J., Paget M. S. (2004) Nat. Rev. Microbiol. 2, 954–966 [DOI] [PubMed] [Google Scholar]
- 48. Ingle R. A., Carstens M., Denby K. J. (2006) BioEssays 28, 880–889 [DOI] [PubMed] [Google Scholar]
- 49. Meyer A., Pühler A., Niehaus K. (2001) Planta 213, 214–222 [DOI] [PubMed] [Google Scholar]
- 50. Zähringer U., Lindner B., Rietschel E. T. (1999) in Endotoxin in Health and Disease (Morrison D. C., Brade H., Opal S., Vogel S. eds) Marcel Dekker, Inc., New York [Google Scholar]
- 51. Silipo A., Molinaro A., Lanzetta R., Parrilli M., Lindner B., Holst O. (2004) Eur. J. Org. Chem. 2004, 1336–1343 [Google Scholar]
- 52. Kulshin V. A., Zähringer U., Lindner B., Frasch C. E., Tsai C. M., Dmitriev B. A., Rietschel E. T. (1992) J. Bacteriol. 174, 1793–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dow M., Newman M. A., von Roepenack E. (2000) Annu. Rev. Phytopathol. 38, 241–261 [DOI] [PubMed] [Google Scholar]
- 54. Raetz C. R., Whitfield C. (2002) Annu. Rev. Biochem. 71, 635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Molinaro A., Lanzetta R., Evidente A., Parrilli M., Holst O. (1999) FEMS Microbiol. Lett. 181, 49–53 [DOI] [PubMed] [Google Scholar]
- 56. Shashkov A. S., Senchenkova S. N., Laux P., Ahohuendo B. C., Kecskés M. L., Rudolph K., Knirel Y. A. (2000) Carbohydr. Res. 323, 235–239 [DOI] [PubMed] [Google Scholar]
- 57. Molinaro A., De Castro C., Lanzetta R., Parrilli M., Petersen B. O., Broberg A., Duus J. Ø. (2002) Eur. J Biochem. 269, 4185–4193 [DOI] [PubMed] [Google Scholar]
- 58. Bedini E., De Castro C., Erbs G., Mangoni L., Dow J. M., Newman M. A., Parrilli M., Unverzagt C. (2005) J. Am. Chem. Soc. 127, 2414–2416 [DOI] [PubMed] [Google Scholar]
- 59. Brown I., Trethowan J., Kerry M., Mansfield J., Bolwell G. P. (1998) Plant J. 15, 333–343 [Google Scholar]
- 60. Zeng W., Melotto M., He S. Y. (2010) Curr. Opin. Biotechnol. 21, 599–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gómez-Gómez L. (2004) Mol. Immunol. 41, 1055–1062 [DOI] [PubMed] [Google Scholar]
- 62. Felix G., Duran J. D., Volko S., Boller T. (1999) Plant J. 18, 265–276 [DOI] [PubMed] [Google Scholar]
- 63. Newman M. A., Daniels M. J., Dow J. M. (1995) Mol. Plant Microbe Interact. 8, 778–780 [DOI] [PubMed] [Google Scholar]
- 64. Fujikawa T., Ishihara H., Leach J. E., Tsuyumu S. (2006) Mol. Plant Microbe Interact. 19, 342–349 [DOI] [PubMed] [Google Scholar]
- 65. Edreva A. (2005) Gen. Appl. Plant Physiol. 31, 105–124 [Google Scholar]
- 66. Mishina T. E., Zeier J. (2007) Plant J. 50, 500–513 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.