Abstract
In mammals, viral infections are detected by innate immune receptors, including Toll-like receptor and retinoic acid inducible gene I (RIG-I)-like receptor (RLR), which activate the type I interferon (IFN) system. IFN essentially activates genes encoding antiviral proteins that inhibit various steps of viral replication as well as facilitate the subsequent activation of acquired immune responses. In this study, we investigated the expression of non-coding RNA upon viral infection or RLR activation. Using a microarray, we identified several microRNAs (miRNA) specifically induced to express by RLR signaling. As suggested by Bioinformatics (miRBase Target Data base), one of the RLR-inducible miRNAs, miR-23b, actually knocked down the expression of very low density lipoprotein receptor (VLDLR) and LDLR-related protein 5 (LRP5). Transfection of miR-23b specifically inhibited infection of rhinovirus 1B (RV1B), which utilizes the low density lipoprotein receptor (LDLR) family for viral entry. Conversely, introduction of anti-miRNA-23b enhanced the viral yield. Knockdown experiments using small interfering RNA (siRNA) revealed that VLDLR, but not LRP5, is critical for an efficient infection by RV1B. Furthermore, experiments with the transfection of infectious viral RNA revealed that miR-23b did not affect post-entry viral replication. Our results strongly suggest that RIG-I signaling results in the inhibitions of infections of RV1B through the miR-23b-mediated down-regulation of its receptor VLDLR.
Keywords: Innate Immunity, Interferon, MicroRNA, RNA Helicase, Virus Entry, RLR, Rhinovirus
Introduction
Among the viral components that trigger the antiviral responses of a host, nucleic acids have been considered critical. In invertebrates and plants, the RNA interference (RNAi) system, which destroys non-self RNA in a sequence-dependent manner, is the major antiviral mechanism (1–3). In mammals, certain RNA structures rather than primary sequences are sensed as non-self to initiate a series of antiviral programs including the activation of type I IFN genes. There are at least two receptor systems functioning as sensors to detect viral RNA signatures: the endosomal Toll-like receptor (TLR)2 3 and TLR7/8, which interact with extracellular viral RNA (4), and the cytosolic RIG-I, melanoma differentiation associated gene 5 and laboratory of genetics and physiology 2, collectively termed the RLR (5, 6), which sense intracellular viral RNA with a double-stranded and/or 5′-triphosphate structure (7). Signaling generated by the innate immune receptors is transduced through networks with various adaptor molecules and results in the activation of IFN genes (8–10). IFN is secreted, thus expanding the antiviral signal to other cells through physical interaction with cell surface receptors thereby activating the IFN-stimulated genes (ISGs) (11). Several ISGs execute antiviral activity by inhibiting viral transcription, translation, viral assembly, and release of viral particles from the cells. Another feature of the IFN system is that it is tightly regulated by positive and negative feedback loops (12).
Although the RNAi system is operative in mammals, the IFN system appears to dominate the antiviral immune response. Recently, non-coding RNA known as miRNA has received much attention for its post-transcriptional regulation of gene expression (13, 14). Over 500 miRNA-encoding genes, which are exclusively transcribed by RNA polymerase II, have been identified in mammals. These primary miRNAs are processed by the enzyme Drosha into hairpin loop-containing pre-miRNAs, which are then subjected to export from the nucleus to cytoplasm via exportin 5 (15). Further enzymatic processing of the pre-miRNAs by Dicer leads to generation of a mature miRNA duplex that is loaded into the RNA-induced silencing complex, which is then guided by the miRNA to complementary messenger RNAs. Notably, the sequence complementary in the 6–8-base pair “seed region” at the 5′ end of the miRNA seems to determine the specificity of miRNA-target mRNA interaction (16, 17).
Accumulated information has revealed that some miRNAs are involved in immune regulation. Expression profiling showed that stimulation of monocytes with lipopolysaccharide (LPS) induced the expression of miR-146. miR-146 targets tumor necrosis factor receptor-associated factor 6 (Traf6) and interleukin-1 receptor-associated kinase 1(Irak1), encoding components of the TLR signaling pathway, suggesting a negative feedback loop (18). Furthermore, liver-specific miR-122 contributes to the liver tropism of the hepatitis C virus by accelerating the binding of ribosomes to the viral RNA and hence aiding translation (19–21). These reports highlight the importance of understanding the function of miRNAs.
In this report, we examined miRNA expression upon RIG-I signaling and identified 37 miRNAs. Among them, miR-23b exhibited antiviral activity to rhinovirus (RV) 1B. RV, a member of the family Picornaviridae, causes an extensive range of human respiratory disorders including the common cold, viral bronchiolitis, and exacerbations of asthma and chronic obstructive pulmonary disease (22–26). Recently, primary human bronchial epithelial cells from asthmatics were found to be defective in IFN-β and IFN-λ mRNA and protein, (27, 28), providing a likely explanation for the increased vulnerability to virus-induced asthma exacerbations. Furthermore, it was revealed that TLR3, RIG-I, and melanoma differentiation associated gene 5 were important for RV-inducing innate responses (29–31). Our analyses revealed that miR-23b blocks infections of RV1B through down-regulation of its receptor, VLDLR. This is a novel antiviral mechanism activated by RIG-I signaling.
EXPERIMENTAL PROCEDURES
Cell Culture
HeLa cells were maintained in Dulbecco's modified Eagle's medium with fetal bovine serum and penicillin-streptomycin (100 units/ml and 100 μg/ml, respectively). L929 cells were maintained in minimum essential medium with fetal bovine serum and penicillin-streptomycin (50 units/ml and 100 μg/ml, respectively).
miRNA Microarray
Total RNA was isolated using TRIzol (Invitrogen) and miRNA was purified with a PureLink miRNA Isolation Kit (Invitrogen). The purified miRNA was labeled using Label IT miRNA Labeling Kits (Mirus). Hybridization was performed, using a three-dimensional gene miRNA Oligo Chip (TORAY).
Viral Infection
Cells were treated with the culture medium (“mock-treated”) or infected with SeV, NDV, VSV, EMCV, RV16, or RV1B in serum-free and antibiotic-free medium. After adsorption for 1 h at 37 °C, the medium was changed and infection was continued for 24 h in the presence of serum-containing DMEM.
Treatment with Interferon-β (IFN-β)
HeLa cells were maintained in 12-well plates in the presence of serum-containing DMEM. Recombinant human IFN-β (1000 units/ml) was added to each well.
Plaque Assay
L929 cells were seeded in 24-well plates (2.5 × 105 cells/well) in minimal essential medium with 5% FBS for 24 h at 37 °C. After cell propagation, the growth medium was removed and serial dilutions of viral supernatants in serum-free and antibiotic-free minimal essential medium were added to the wells. The inoculated cells were further incubated to allow adsorption of the virus for 1 h at 37 °C. Subsequently, a mixture of agar overlay was added and the plates were incubated at 37 °C for 24 h. The plaques were visualized by staining with a 0.02% neutral red solution. The viral titer is expressed as plaque forming units.
Transfection of miR-23b and Anti-miR-23b
Double-stranded RNA oligonucleotides representing mature sequences that mimic endogenous miR-23b and anti-miR-23b (Ambion) were transfected into HeLa cells with RNAi MAX (Invitrogen) according to the manufacturer's instructions. Silencer negative control #2 small interfering RNA (Ambion) at the same concentration as miR-23b and anti-miR-23b was used in each experiment.
Real-time PCR
Total RNA was isolated using TRIzol (Invitrogen). Real-time PCR for miR-23b, -24, and -27b was performed using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer's protocol. Normalization was performed by using RNU48 primers and probes. Real-time PCR was performed using TaqMan Gene Expression Assay probes for IFN-β and IL-6. The 18S rRNA gene was used as an internal control to normalize differences in each sample. The expression levels for each gene were assessed relative to the expression of 18S rRNA. Real-time PCR for RV RNA was performed using the following primers and probe: RV forward, 5-GTGAAGAGCCSCRTGTGCT-3, RV reverse, 5-GCTSCAGGGTTAAGGTTAGCC-3, RV probe, 5-TGAGTCCTCCGGCCCCTGAATG-3.
Amido Black Staining
Cells were washed in PBS three times and fixed with methanol. 0.5% Amido Black solution was added and incubated 20 min at RT. After 20 min, the Amido Black solution was removed and eluted by 0.1 m NaOH. 630 nm absorption was measured.
TCID50
The RV1B titer was measured by the Reed-Muench method (43).
Small Interfering RNA (siRNA) Knockdown of VLDLR and LRP5
siRNA for VLDLR (Digital Biology) and siRNA for LRP5 (IDT) were transfected into HeLa cells with RNAi MAX (Invitrogen) according to the manufacturer's instructions. Negative control siRNA (Digital Biology) at the same concentration as siVLDLR and siLRP5 was used in each experiment.
Immunoblotting
Cells were lysed with Nonidet P-40 lysis buffer (50 mm Tris (pH 8), 150 mm NaCl, and 1% Nonidet P-40). The lysate was resolved by SDS-PAGE (30 μg of protein/lane). Proteins were transferred to a nitrocellulose membrane. The membrane was blocked in 5% milk for 30 min at room temperature and probed with mouse anti-SeV Nucleocapsid protein, mouse anti-NDV Nucleocapsid protein, mouse anti-VLDLR (Santa Cruz sc-18824), or rabbit anti-LRP5 (Cell Signaling D23F7). Ab binding was detected with alkaline phosphatase-conjugated anti-mouse or anti-rabbit IgG.
RV Genomic RNA Isolation and Transfection
RV-containing medium was centrifuged at 10,000 × g for 24 h. The pellet was suspended in TRIzol (Invitrogen). The RV genomic RNA (0.5 μg) and miR-23b were transfected into HeLa cells with Lipofectamine 2000 (Invitrogen).
Statistical Analysis
Statistical analysis were conducted with an unpaired t test, with values of p < 0.05 considered statistically significant. Each point in a graph represents the mean ± S.E. for at least three independent experiments.
RESULTS
Induction of miRNA Expression by RIG-I Signaling
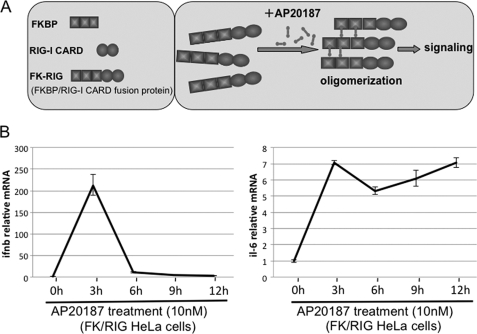
To directly examine the impact of the activation of RIG-I on the gene expression profile, we developed an artificial RIG-I activation system (details will be reported elsewhere).3 Briefly, the N-terminal signaling domain of RIG-I, caspase recruitment domain (CARD), was introduced into the ARGENTTM Regulated Homodimerization Kit (ARIAD). The resultant FKBP/RIG-I CARD fusion protein (FK/RIG) allowed CARD to oligomerize by the chemical compound, AP20187 (AP), and to introduce CARD-mediated signaling into the cells (Fig. 1A). As shown in Fig. 1B, the endogenous mRNA levels of IFN-β and IL-6 in HeLa cells stably expressing FK/RIG (HeLa FK/RIG) were significantly increased by the enforced oligomerization of RIG-I CARD (Fig. 1B). Furthermore, microarray analyses of the transcripts of these cells revealed transient expression of IFN genes and ISGs with a concomitant induction of IFN regulatory factor (IRF-3) dimerization and NF-κB DNA-binding activity (data not shown). Thus we concluded that the FK/RIG system could mimic the virus-induced activation of RIG-I signaling.
FIGURE 1.
Artificial oligomerization of the CARD of RIG-I results in signaling to activate the IFN-β and IL-6 genes. A, schematic representation of the FK/RIG fusion protein and its oligomerization by a cross-linking reagent, AP20187. B, HeLa cells stably expressing FK/RIG were treated with AP for the period indicated and cells were harvested to examine endogenous IFN-β and IL-6 mRNA levels by real-time PCR.
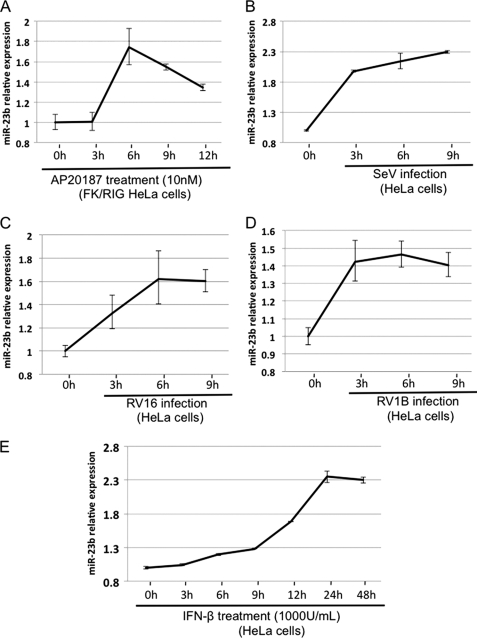
To analyze the miRNA expression profile in HeLaFK/RIG cells, RNA fractions were prepared at 3, 6, 9, and 12 h after the addition of AP and subjected to analysis with a miRNA microarray (TORAY) that covers 900 miRNAs. The expression level of several miRNAs was up- or down-regulated after the addition of AP (supplemental Fig. S1), the miRNAs exhibiting elevated levels of expression are listed in Table 1. For up-regulated miRNA, the induction kinetics appear to be different. This is commonly observed for different ISGs, presumably the induction is regulated by transcription and post-transcriptional mechanisms. Certainly this issue requires further investigation. Among these RIG-I-inducible miRNA, we focused on miR-23b because (i) it has been reported that miR-23b is regulated by NF-κB (32) and (ii) a data base search revealed that miR-23b has several target sites on mRNA encoding VLDLR (see below). To confirm the results obtained with the miRNA microarray, we monitored expression of miR-23b by real-time PCR. AP treatment resulted in a 1.7-fold increase in miR-23b at 6 h in HeLaFK/RIG with similar kinetics as in the microarray assay (Fig. 2A). The Sendai virus (SeV) and two strains of rhinovirus (RV16 and RV1B) also induced miR-23b accumulation (Fig. 2, B–D) with similar kinetics to the endogenous IFN-β mRNA expression (supplemental Fig. S2, A–C). miR-23b is reported to be derived from the polycistronic miRNA cluster that consists of miR-23b-27b-24 in the human gene, C9orf3 (supplemental Fig. S3A) (32). As expected, the expression of miR-24 and miR-27b was also induced by RV16 and RV1B infection (supplemental Fig. S3, B and C), suggesting that these miRNA are regulated by a common mechanism. On the other hand, treatment of cells with IFN-β also induced miR-23b expression with a peak at 24 h, suggesting that in addition to RIG-I-mediated signaling, IFN receptor-mediated signaling induces miR-23b expression albeit with slow kinetics (Fig. 2E). These results suggest that, although the expression of miR-23b is up-regulated by both RIG-I- and IFN-mediated signaling, the former is mainly responsible for transient induction after oligomerization of RIG-I CARD.
TABLE 1.
miRNA induced by RIG-I-mediated signaling
HeLa cells stably expressing FK/RIG were treated with AP for the periods indicated and miRNA expression was determined by miRNA microarray (TORAY). The relative expression (fold-increase compared to the baseline miRNA level at 0 h) of representative miRNA whose expression was up-regulated more than 2-fold is shown. The opposite strand of miRNA is denoted with an asterisk (*).
| AP20187 treatment (FK/RIG-HeLa cells) |
|||||
|---|---|---|---|---|---|
| 0 h | 3 h | 6 h | 9 h | 12 h | |
| miR-423–3p | 1.00 | 2.31 | 2.77 | 4.34 | 4.59 |
| miR-301b | 1.00 | 2.31 | UDa | UD | UD |
| miR-923 | 1.00 | 3.63 | 1.34 | 1.50 | 0.84 |
| miR-181a* | 1.00 | UD | 7.16 | UD | UD |
| miR-23b | 1.00 | 1.47 | 2.32 | 1.03 | 1.27 |
| miR-125b | 1.00 | 1.94 | 2.73 | 1.16 | 1.38 |
| miR-505* | 1.00 | UD | 2.41 | UD | UD |
| miR-940 | 1.00 | UD | 2.11 | UD | UD |
| miR-1226 | 1.00 | 0.90 | 2.05 | UD | UD |
| miR-1229 | 1.00 | UD | 2.80 | UD | UD |
| miR-1281 | 1.00 | UD | 2.21 | UD | UD |
| miR-149 | 1.00 | UD | 0.99 | 3.36 | UD |
| miR-188–5p | 1.00 | 1.85 | 1.36 | 3.70 | UD |
| miR-320 | 1.00 | 1.22 | 1.31 | 2.62 | 1.71 |
| miR-200c | 1.00 | 1.20 | 1.03 | 2.63 | UD |
| miR-362–5p | 1.00 | 1.09 | 0.94 | 5.65 | UD |
| miR-425* | 1.00 | UD | UD | 8.81 | UD |
| miR-769–3p | 1.00 | 1.00 | 1.01 | 2.65 | UD |
| miR-801 | 1.00 | 1.09 | 0.92 | 9.18 | 1.27 |
| miR-29b-1* | 1.00 | 1.43 | 1.27 | 2.00 | UD |
| miR-92b* | 1.00 | 0.70 | 0.88 | 2.29 | UD |
| miR-1225–3p | 1.00 | UD | UD | 7.16 | UD |
| miR-1228 | 1.00 | UD | 1.24 | 5.77 | UD |
| miR-1249 | 1.00 | UD | UD | 5.89 | UD |
| miR-1280 | 1.00 | 0.63 | 1.19 | 8.00 | 1.74 |
| miR-664 | 1.00 | 1.55 | UD | 23.75 | UD |
| miR-664* | 1.00 | 1.55 | UD | 22.62 | UD |
| miR-19b | 1.00 | 1.26 | 1.40 | 1.14 | 2.05 |
| miR-7i | 1.00 | 1.42 | 1.50 | 1.68 | 2.92 |
| miR-484 | 1.00 | 0.84 | 1.38 | 1.67 | 2.04 |
| miR-500* | 1.00 | 1.58 | 1.76 | 1.73 | 3.18 |
| miR-421 | 1.00 | 1.61 | 1.90 | 0.96 | 2.11 |
| miR-671–5p | 1.00 | UD | UD | UD | 5.38 |
| miR-768–5p | 1.00 | 0.61 | 0.57 | UD | 2.94 |
| miR-939 | 1.00 | 0.79 | 0.71 | UD | 2.75 |
| miR-320b | 1.00 | 1.39 | 1.44 | 1.92 | 2.20 |
| miR-1304 | 1.00 | 1.69 | 1.9 | UD | 31.12 |
a UD, undetermined.
FIGURE 2.
Induction of miR-23b by RIG-I signaling, SeV and RV infection, or IFN-β treatment. HeLa cells stably expressing FK/RIG were treated with AP (A). B–E, HeLa cells were infected with SeV (B), infected with RV16 (C), RV1B (D), or treated with IFN-β (1000 units/ml) (E). The miRNA fraction was extracted at the time points indicated and the amount of miR-23b was determined by real-time PCR.
Selective Effect of miR-23b on Rhinovirus 1B Replication
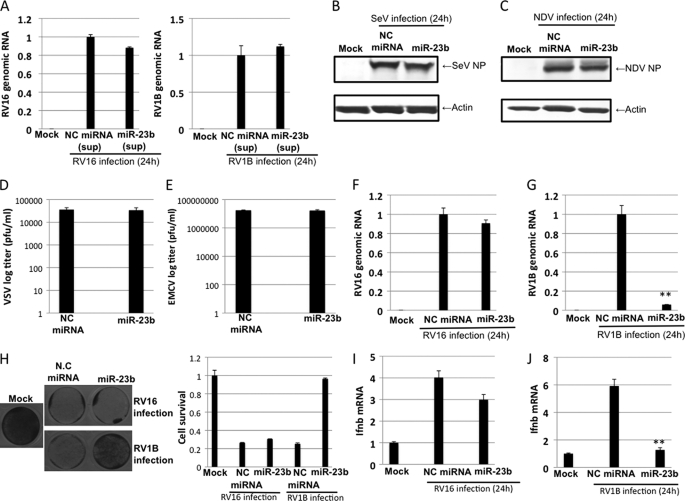
To examine whether miR-23b has an antiviral effect, HeLa cells were transiently transfected with synthetic miR-23b (Ambion), which is a chemically modified double-stranded RNA and mimics endogenous miR-23b, or control miRNA (NC miRNA) for 48 h and infected with SeV, Newcastle disease virus (NDV), vesicular stomatitis virus (VSV), encephalomyocarditis virus (EMCV), and two strains of rhinovirus (RV16 and RV1B) for 24 h. RV16 and RV1B are representative of major and minor group RVs, respectively, and utilize distinct cellular receptors for viral entry (33–35). First, to exclude the possibility that miRNA induces production of antiviral humoral factors such as IFNs, we examined antiviral activity in the culture supernatant of HeLa cells transfected with either control miRNA or miR23b. The supernatant did not affect viral RNA yields from RV-infected HeLa cells, indicating that miR23b does not induce production of antiviral cytokines (Fig. 3A). Next, we tried to examine the effect of miR-23b on viral growth. For SeV and NDV infections, nucleocapsid proteins (NPs), which are known to interact with viral genome in infected cells, were detected by Western blotting with specific antibodies (Fig. 3, B and C). Transfection with miR-23b did not influence the accumulation of NPs in SeV- or NDV-infected cells, suggesting no effect on growth of these viruses. In the case of VSV and EMCV, we determined viral titer from the infected cells by plaque assays (Fig. 3, D and E). Transfection with miR-23b did not influence yields of VSV or EMCV. For RV16 and RV1B, the accumulation of viral RNA was examined by real-time PCR. The introduction of miR-23b moderately reduced the amount of RV16 RNA, however, a dramatic reduction of RV1B viral RNA was observed (Fig. 3, F and G). Furthermore, RV1B titer was severely reduced by miR-23b treatment (supplemental Fig. S4A). Consistent with this, the cell survival rate was significantly restored by miR-23b transfection after RV1B infection but not RV16 infection (Fig. 3H). Again, to verify whether the effect of miR-23b on viral RNA yield of RV1B is indirectly mediated by the secreted IFNs, endogenous IFN-β mRNA levels in miR-23b-transfected and RV-infected cells were determined (Fig. 3, I and J). In cells infected with RV16 or RV1B, IFN-β mRNA levels were closely correlated to the viral yields (compare Fig. 3, F and G to I and J, respectively), suggesting that the decrease of RV1B RNA caused by miR-23b is unlikely due to enhanced production of IFN-β. Taken together, these results suggest that miR-23b has an antiviral effect specific to RV1B.
FIGURE 3.
The effect of miR-23b transfection on viral growth. A, HeLa cells were transfected with negative control miRNA (NC miRNA) or miR-23b for 48 h, the culture supernatant (sup) was collected, and HeLa cells were treated with supernatant for 24 h and then infected with RV for 24 h. Levels of RV genomic RNA were determined by real-time PCR. B-G, HeLa cells transfected with NC miRNA or miR-23b for 48 h were infected with SeV (B), NDV (C), VSV (D), EMCV (E), RV16 (F), or RV1B (G) for an additional 24 h. The viral NP level was determined by Western blotting (B, SeV; C, NDV). Infectivity in the culture supernatant was determined by plaque assay (D, VSV; E, EMCV). Levels of RV genomic RNA were determined by real-time PCR (F, RV16; G, RV1B) using specific primer sets. H, Amido Black staining of miRNA-transfected and RV-infected cells. IFN-β mRNA levels of the RNA samples in F and G are shown in I and J, respectively. **, p < 0.005.
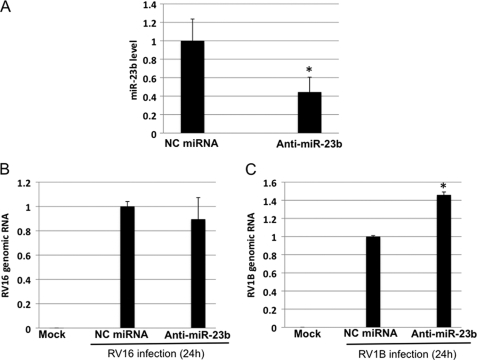
Down-regulation of miR-23b Expression Resulted in Increased RNA Yields of RV1B
To further investigate the mechanism of the antiviral effect of miR-23b on RV1B, we transiently expressed anti-miR-23b in HeLa cells. This anti-miR-23b (Ambion) is a chemically modified single-stranded RNA and specifically bind miR-23b, inhibiting the function of miR-23b. The transfection led to a 60% decrease of endogenous miR-23b production, compared with NC-miRNA transfection (Fig. 4A). Under these conditions, the RV16 RNA level was not changed by anti-miR-23b (Fig. 4B). On the other hand, the RV1B RNA level was significantly increased by anti-miR-23b (Fig. 4C). Consistent with this, RV1B titer was clearly increased by anti-miR-23b transfection (supplemental Fig. S4B). These results further confirm that the inhibition of RV1B RNA is mediated by miR-23b.
FIGURE 4.
Anti-miR-23b enhanced RNA yields of RV1B. HeLa cells were transfected with NC miRNA or anti-miR-23b for 48 h and infected with RV for 24 h. A, the expression of miR-23b after anti-miR-23b transfection was determined by real-time PCR. B and C, the yield of genomic RNA of RV16 (B) and RV1B (C) was determined by real-time PCR. *, p < 0.05.
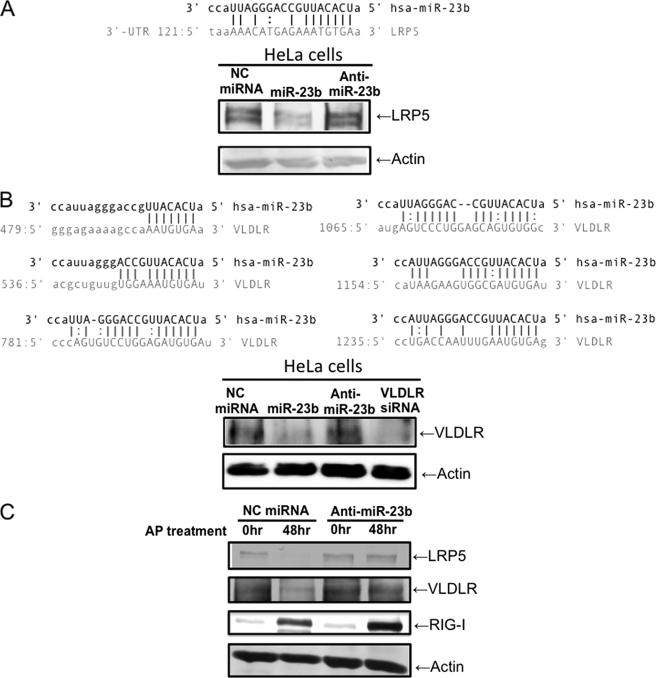
LRP5 and VLDLR Are Targets of miR-23b
A bioinformatic-based (microRNA.org) search of databases revealed 1 and 6 target sites on mRNA encoding LDLR-related protein 5 (LRP5) and VLDLR, respectively (Fig. 5, A and B). Because it has been reported that minor group RVs, including RV1B, use LDLR family proteins (LDLR, VLDLR, and LRPs) as cellular receptors and major group RVs utilize ICAM-1, we speculate that miR-23b specifically affects expression of RV1B receptors. To confirm this, miR-23b was introduced into HeLa cells and its effect on expression of LRP5 and VLDLR was determined by Western blotting (Fig. 5). The levels of LRP5 and VLDLR were significantly reduced by transfection of miR-23b, suggesting that miR-23b targets mRNA for these proteins. Furthermore, induction of endogenous miR-23b expression through the activation of FK/RIG by AP treatment in HeLaFK/RIG cells resulted in decreased levels of LRP5 and VLDLR (Fig. 5C). However, anti-miR-23b blocked this reduction. These results strongly suggest that endogenous miR-23b participate in the down-regulation of LRP5 and VLDLR expression via RIG-I-mediated signaling.
FIGURE 5.
miR-23b targets LRP5 and VLDLR. A and B, the results of the search for target sequences of miR-23b (miRBase Target Data base). Candidate sequences in LRP5 (A) and VLDLR (B) mRNA are shown. HeLa cells were transfected with NC miRNA, miR-23b, or anti-miR-23b for 48 h and LRP5 (A) and VLDLR (B) were detected by Western blotting. C, HeLa FK/RIG cells were transfected with either NC miRNA or anti-miR-23b for 24 h and AP20187 was treated for 48 h. LRP5 and VLDLR were detected by Western blotting.
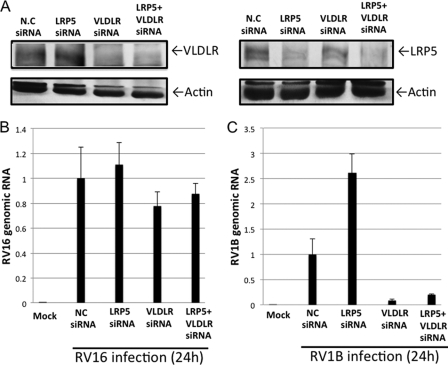
Knockdown of VLDLR Reduced RV1B RNA Yields
To clarify which of the miR-23b-targeted LDLR family proteins are critical for RV1B infections, we individually knocked down LRP5 and VLDLR using specific siRNA (Fig. 6A) and the RV viral RNA yield was compared (Fig. 6, B and C). As expected, the viral RNA of RV16, which utilizes ICAM-1 as a receptor (36), was not affected by either siRNA treatment. However, knockdown of VLDLR dramatically reduced RV1B RNA yields. Knockdown of LRP5 enhanced RV1B RNA yields, however, this effect was not observed in cells with the double knockdown of VLDLR and LRP5. The protein level of VLDLR is slightly increased in cells transfected with LRP5 siRNA by an unknown mechanism (Fig. 6A). The higher RV1B replication in cells transfected with LRP5 siRNA (Fig. 6C) could be explained by the increased expression of its receptor, VLDLR. These results suggest that VLDLR but not LRP5 is critical for viral growth of RV1B.
FIGURE 6.
Knockdown of VLDLR blocks accumulation of viral RNA in RV1B-infected cells. HeLa cells were transfected with control siRNA or siRNA targeting VLDLR or LRP5 as indicated. A–C, at 48 h after the transfection, cells were mock infected or infected with RV16 or RV1B for an additional 24 h. VLDLR and LRP5 were detected by Western blotting in mock-infected cells (A). RNA levels of RV16 (B) or RV1B (C) were determined by real-time PCR.
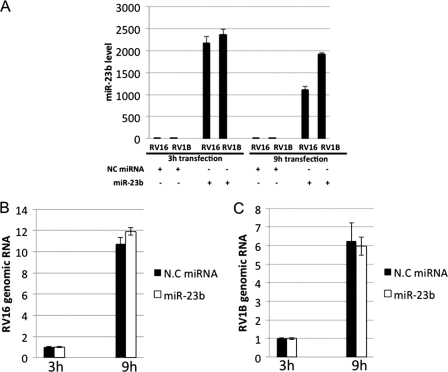
miR-23b Did Not Influence Intracellular Replication of RV1B
The above results suggest that miR-23b blocks entry of RV1B by inhibiting expression of the receptor VLDLR. Because there are several reports that viral genomic RNA is directly targeted by host miRNA (37, 38), we investigated whether RV1B replication initiated by transfection of infectious viral genomic RNA is affected by miR-23b. HeLa cells were co-transfected with viral RNA and miRNA (Fig. 7), and the cells were harvested to examine the level of miR-23b and viral RNA at 3 and 9 h post-infection. High levels of miR-23b were detected in miR-23b-transfected but not NC miRNA-transfected cells, suggesting that the miR-23b was efficiently incorporated, and remained in the cells at 9 h after transfection (Fig. 7A). In the cells transfected with RV16 or RV1B genomic RNAs, although increasing amounts of viral RNAs were detected, no inhibitory effect of miR-23b was observed (Fig. 7, B and C). These results suggest that miR-23b inhibits infection of RV1B by down-regulating the expression of its major receptor, VLDLR, rather than impairment of the viral replication.
FIGURE 7.
miR-23b did not influence intracellular replication of RV1B. A–C, HeLa cells were co-transfected with RV genomic RNA and NC miRNA or miR-23b for 3 and 9 h. Levels of miR-23b (A), RV16 RNA (B), and RV1B RNA (C) were determined by real-time PCR.
DISCUSSION
It has been well established that IFN treatment activates a Janus kinase-signal transducer activator of transcription (STAT) pathway resulting in activation of a variety of ISGs (39–41). Some ISGs encode proteins, collectively known as antiviral proteins, which directly inhibit viral replication. In this report, we demonstrated an alternative mechanism of inducing an antiviral state, that is, reducing the level of a protein essential for viral infection via activating a gene encoding miRNA. Thus innate immune responses restrict viral replication either by adding antiviral proteins or by removing proteins necessary for viral infection.
In addition to the regulation of host genes, miRNAs targeting viral RNA genomes have been reported. Pedersen and colleagues (38) reported that treatment of hepatic cells with IFN-β resulted in the production of at least eight miRNAs (miR-1, miR-30, miR-128, miR-196, miR-296, miR-351, miR-431, and miR-448) that perfectly complement hepatitis C virus mRNAs. These findings suggest that the mammalian immune system utilizes miRNA to combat viral infections via multiple mechanisms.
We examined 900 miRNAs using a microarray and found that 37 and 28 miRNAs were up- and down-regulated, respectively (Table 1). The results show a marked regulation of miRNA expression upon viral infection (3–4%). In this report, we focused on miR-23b, because its possible target genes encode cell-surface proteins that are known to be viral receptors. Overexpression of miR-23b and anti-miR-23b resulted in repressed and enhanced production of RV1B, respectively. Although miR-23b targets both LRP5 and VLDLR, our analyses revealed the down-regulation of VLDLR to be responsible for the inhibition of RV1B.
Among the minor group rhinovirus there are 12 types of RV (RV1A, RV1B, RV2, RV44, RV47, RV49, RV23, RV25, RV29, RV30, RV31, and RV62). Considering that these minor group RVs commonly utilize VLDLR for their entry, miR-23b should exhibit an antiviral effect on these 12 types RVs. Importantly, the minor group RVs are shown to cause disease more often than the major group RV (42), suggesting that down-regulation of VLDLR by miR-23b is of significance for host defense to the minor group of RVs. Because the binding of viruses to the host cell is the initial step for viral entry, transient down-regulation of cell surface molecules could be an effective strategy to avoid viral transmission.
Artificial activation of RIG-I or infection by SeV and RV resulted in the accumulation of miR-23b with a peak at 6 and 9 h, respectively (Fig. 2). IFN-β treatment also induced the accumulation of miR-23b albeit with slower kinetics and a peak around 24 h. These results suggest that the expression of miR-23b is regulated by similar mechanisms to that of some ISGs, which are stimulated by both IRF-3/IRF-7 and IFN-stimulated gene factor 3 (ISGF3), the trimeric complex of STAT1, STAT2, and IRF-9. As reported previously, NF-κB may participate in the production of miR-23b inducted by viral infection or RIG-I stimulation (32). Because type I IFN is produced and secreted in the initial stages of viral infection, IFN induces the accumulation of miR-23b in uninfected cells, thereby protecting them from initial infection.
In summary, our study presents evidence that RIG-I-mediated signaling up-regulates the expression of 37 miRNAs, one of which, miR-23b, strongly inhibits minor group rhinoviruses through down-regulation of VLDLR, which functions as a receptor for entry into the cell. This finding provides a novel perspective for RIG-I-mediated antiviral effects.
Supplementary Material
Acknowledgments
We thank Prof. T. Sakaguchi for anti-NDV NP antibody and Prof. K. Atsushi for anti-SeV NP antibody.
This work was supported in part by a Grant-in-aid from the Ministry of Education, Science, Sports and Culture in Japan, the Ministry of Health, Labor, and Welfare, the PRESTO Japan Science and Technology Agency, the Uehara Memorial Foundation, the Mochida Memorial Foundation for Medical and Pharmaceutical Research, and Nippon Boehringer Ingelheim.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
K. Onomoto, M. Yoneyama, and T. Fujita, unpublished data.
- TLR
- Toll-like receptor
- RIG-I
- retinoic acid-inducible gene I
- LGP2
- laboratory of genetics and physiology 2
- VLDLR
- very low density lipoprotein receptor
- LRP
- LDLR-related protein
- SeV
- Sendai virus
- NDV
- Newcastle disease virus
- VSV
- vesicular stomatitis virus
- EMCV
- encephalomyocarditis virus
- RV
- rhinovirus
- ISG
- IFN-stimulated gene
- CARD
- caspase recruitment domain
- IRF
- IFN regulatory factor
- NP
- nucleocapsid protein
- ICAM-1
- intercellular adhesion molecule 1.
REFERENCES
- 1. Vaucheret H., Béclin C., Fagard M. (2001) J. Cell Sci. 114, 3083–3091 [DOI] [PubMed] [Google Scholar]
- 2. Takaoka A., Yanai H. (2006) Cell. Microbiol. 8, 907–922 [DOI] [PubMed] [Google Scholar]
- 3. Baulcombe D. (2004) Nature 431, 356–363 [DOI] [PubMed] [Google Scholar]
- 4. Akira S., Takeda K. (2004) Nat. Rev. Immunol. 4, 499–511 [DOI] [PubMed] [Google Scholar]
- 5. Yoneyama M., Kikuchi M., Matsumoto K., Imaizumi T., Miyagishi M., Taira K., Foy E., Loo Y. M., Gale M., Jr., Akira S., Yonehara S., Kato A., Fujita T. (2005) J. Immunol. 175, 2851–2858 [DOI] [PubMed] [Google Scholar]
- 6. Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. (2004) Nat. Immunol. 5, 730–737 [DOI] [PubMed] [Google Scholar]
- 7. Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., Hiiragi A., Dermody T. S., Fujita T., Akira S. (2008) J. Exp. Med. 205, 1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K. J., Takeuchi O., Akira S. (2005) Nat. Immunol. 6, 981–988 [DOI] [PubMed] [Google Scholar]
- 9. Kumar H., Kawai T., Kato H., Sato S., Takahashi K., Coban C., Yamamoto M., Uematsu S., Ishii K. J., Takeuchi O., Akira S. (2006) J. Exp. Med. 203, 1795–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Potter J. A., Randall R. E., Taylor G. L. (2008) BMC Struct. Biol. 8, 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shingai M., Ebihara T., Begum N. A., Kato A., Honma T., Matsumoto K., Saito H., Ogura H., Matsumoto M., Seya T. (2007) J. Immunol. 179, 6123–6133 [DOI] [PubMed] [Google Scholar]
- 12. Dalpke A., Heeg K., Bartz H., Baetz A. (2008) Immunobiology 213, 225–235 [DOI] [PubMed] [Google Scholar]
- 13. Friedman R. C., Farh K. K., Burge C. B., Bartel D. P. (2009) Genome Res. 19, 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Molnár A., Schwach F., Studholme D. J., Thuenemann E. C., Baulcombe D. C. (2007) Nature 447, 1126–1129 [DOI] [PubMed] [Google Scholar]
- 15. Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Rådmark O., Kim S., Kim V. N. (2003) Nature 425, 415–419 [DOI] [PubMed] [Google Scholar]
- 16. Gregory R. I., Chendrimada T. P., Cooch N., Shiekhattar R. (2005) Cell 123, 631–640 [DOI] [PubMed] [Google Scholar]
- 17. Okamura K., Ishizuka A., Siomi H., Siomi M. C. (2004) Genes Dev. 18, 1655–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taganov K. D., Boldin M. P., Chang K. J., Baltimore D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12481–12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Henke J. I., Goergen D., Zheng J., Song Y., Schüttler C. G., Fehr C., Jünemann C., Niepmann M. (2008) EMBO J. 27, 3300–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jopling C. L. (2008) Biochem. Soc. Trans. 36, 1220–1223 [DOI] [PubMed] [Google Scholar]
- 21. Sarasin-Filipowicz M., Krol J., Markiewicz I., Heim M. H., Filipowicz W. (2009) Nat. Med. 15, 31–33 [DOI] [PubMed] [Google Scholar]
- 22. Vlasak M., Roivainen M., Reithmayer M., Goesler I., Laine P., Snyers L., Hovi T., Blaas D. (2005) J. Virol. 79, 7389–7395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Savolainen C., Blomqvist S., Hovi T. (2003) Pediatr. Respir. Rev. 4, 91–98 [DOI] [PubMed] [Google Scholar]
- 24. Message S. D., Laza-Stanca V., Mallia P., Parker H. L., Zhu J., Kebadze T., Contoli M., Sanderson G., Kon O. M., Papi A., Jeffery P. K., Stanciu L. A., Johnston S. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13562–13567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johnston S. L. (2005) Proc. Am. Thorac. Soc. 2, 150–156 [DOI] [PubMed] [Google Scholar]
- 26. Johnston S. L., Pattemore P. K., Sanderson G., Smith S., Lampe F., Josephs L., Symington P., O'Toole S., Myint S. H., Tyrrell D. A., et al. (1995) BMJ 310, 1225–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wark P. A., Johnston S. L., Bucchieri F., Powell R., Puddicombe S., Laza-Stanca V., Holgate S. T., Davies D. E. (2005) J. Exp. Med. 201, 937–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Contoli M., Message S. D., Laza-Stanca V., Edwards M. R., Wark P. A., Bartlett N. W., Kebadze T., Mallia P., Stanciu L. A., Parker H. L., Slater L., Lewis-Antes A., Kon O. M., Holgate S. T., Davies D. E., Kotenko S. V., Papi A., Johnston S. L. (2006) Nat. Med. 12, 1023–1026 [DOI] [PubMed] [Google Scholar]
- 29. Slater L., Bartlett N. W., Haas J. J., Zhu J., Message S. D., Walton R. P., Sykes A., Dahdaleh S., Clarke D. L., Belvisi M. G., Kon O. M., Fujita T., Jeffery P. K., Johnston S. L., Edwards M. R. (2010) PLoS Pathog. 6, e1001178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hewson C. A., Jardine A., Edwards M. R., Laza-Stanca V., Johnston S. L. (2005) J. Virol. 79, 12273–12279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang Q., Nagarkar D. R., Bowman E. R., Schneider D., Gosangi B., Lei J., Zhao Y., McHenry C. L., Burgens R. V., Miller D. J., Sajjan U., Hershenson M. B. (2009) J. Immunol. 183, 6989–6997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou R., Hu G., Liu J., Gong A. Y., Drescher K. M., Chen X. M. (2009) PLoS Pathog. 5, e1000681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marlovits T. C., Abrahamsberg C., Blaas D. (1998) J. Virol. 72, 10246–10250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nizet S., Wruss J., Landstetter N., Snyers L., Blaas D. (2005) J. Virol. 79, 14730–14736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Okun V. M., Moser R., Ronacher B., Kenndler E., Blaas D. (2001) J. Biol. Chem. 276, 1057–1062 [DOI] [PubMed] [Google Scholar]
- 36. Grünberg K., Sharon R. F., Hiltermann T. J., Brahim J. J., Dick E. C., Sterk P. J., Van Krieken J. H. (2000) Clin. Exp. Allergy 30, 1015–1023 [DOI] [PubMed] [Google Scholar]
- 37. Otsuka M., Jing Q., Georgel P., New L., Chen J., Mols J., Kang Y. J., Jiang Z., Du X., Cook R., Das S. C., Pattnaik A. K., Beutler B., Han J. (2007) Immunity 27, 123–134 [DOI] [PubMed] [Google Scholar]
- 38. Pedersen I. M., Cheng G., Wieland S., Volinia S., Croce C. M., Chisari F. V., David M. (2007) Nature 449, 919–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu A. J., Chen Z. J., Kan E. C., Baum B. J. (1997) J. Cell Physiol. 173, 110–114 [DOI] [PubMed] [Google Scholar]
- 40. Lehtonen A., Matikainen S., Julkunen I. (1997) J. Immunol. 159, 794–803 [PubMed] [Google Scholar]
- 41. Meraz M. A., White J. M., Sheehan K. C., Bach E. A., Rodig S. J., Dighe A. S., Kaplan D. H., Riley J. K., Greenlund A. C., Campbell D., Carver-Moore K., DuBois R. N., Clark R., Aguet M., Schreiber R. D. (1996) Cell 84, 431–442 [DOI] [PubMed] [Google Scholar]
- 42. Andries K., Dewindt B., Snoeks J., Wouters L., Moereels H., Lewi P. J., Janssen P. A. (1990) J. Virol. 64, 1117–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reed L. J., Muench H. (1938) Am. J. Hygiene 27, 493–497 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.