Abstract
Structural studies of ribosome complexes with bound tRNAs and release factors show considerable contacts between these factors and helix 69 (H69) of 23 S rRNA. Although biochemical and genetic studies have provided some general insights into the role of H69 in tRNA and RF selection, a detailed understanding of these contributions remains elusive. Here, we present a pre- steady-state kinetic analysis establishing that two distinct regions of H69 make critical contributions to substrate selection. The loop of H69 (A1913) forms contacts necessary for the efficient accommodation of a subset of natural tRNA species, whereas the base of the stem (G1922) is specifically critical for UGA codon recognition by the class 1 release factor RF2. These data define a broad and critical role for this centrally located intersubunit helix (H69) in accurate and efficient substrate recognition by the ribosome.
Keywords: Kinetics, Ribosome Function, Transfer RNA (tRNA), Translation, Translation Release Factors
Introduction
The ribosome is the ribonucleoprotein machine responsible for the faithful translation of the genetic material into the encoded polypeptide. The core RNA components of the ribosome (the 16 S and 23 S rRNA) that reflect its earliest evolution play a key role in interactions that specify the selection of tRNAs and release factors on recognition of sense and stop codons, respectively. During translation elongation, the selection of a cognate ternary complex (composed of aa-tRNA, EFTu, and GTP) from solution is specified primarily in the small ribosomal subunit where three nucleotides in 16 S rRNA directly evaluate the geometry of the codon-anticodon interaction (1). Though different in molecular detail, the selection of class 1 release factors takes place in the same decoding center of the small ribosomal subunit where conserved protein features of the RF engage the stop codon. Thus, for both decoding events, molecular interactions in the small subunit “decoding center” are early and critical contributors to the selection of cognate substrates during the translational cycle.
In addition to these interactions in the decoding center, it is believed that other molecular interactions between the ribosome and these substrates contribute to binding and specificity. Structural studies of tRNA (2–5), release factors (6–8), and ribosome recycling factor (9, 10) bound to the ribosome reveal that H692 of the large subunit rRNA makes extensive contacts with all of these factors. This universally conserved helix forms an intersubunit bridge that is proximal to the substrate binding cleft and directly contacts the functionally critical h44 of 16 S rRNA which contains two of the three nucleotides known to directly interact with the decoding helix (codon-anticodon) during tRNA selection (A1492 and A1493).
What is the specific nature of these molecular interactions, and what do they suggest about the role of H69 in ribosome function? H69 contacts incoming tRNA in both the pre- and postaccommodation steps near positions 25/26 of the D stem and position 38, located near the anticodon stem; tRNA mutations at these positions are linked to miscoding phenotypes (11, 12). Likewise, the class 1 RFs make extensive contacts with H69 during stop-codon recognition to extend the GGQ domain into the large ribosomal subunit to promote catalysis. A particular metastable helical element referred to as the “switch helix” appears to occupy a cavity in the ribosome that is normally occupied by H69 but that is displaced during stop codon recognition. H69 is heavily modified by the synthase RluD, which pseudouridylates positions 1911, 1915, and 1917 (13). Recent studies have shown that ribosomes isolated from a strain lacking RluD show defects in the ability of RF2 (but not RF1) to catalyze the release of a peptide on UAA-programmed ribosome complexes (14). Finally, other studies observed large conformational changes in H69 in ribosome structures where ribosome recycling factor is bound, consistent with the idea that movement of the helix plays a critical role in the dissociation reaction that ribosome recycling factor catalyzes (9). Simple positioning makes H69 an excellent candidate for participation in substrate selection events and overall ribosome function.
In an effort to define the contribution of H69 to ribosome function, the properties of ribosomes from which the helix had been deleted were characterized (15). Although the ΔH69 ribosomes displayed substantial defects in subunit association (and even exhibit ribosome recycling factor-independent recycling) and RF1-mediated catalysis, the particles were surprisingly active in poly-Phe synthesis and displayed only modest defects in accuracy during tRNA selection (the variant ribosomes incorporated ∼2-fold less leucine on the poly-Uridine template). These results were somewhat at odds with the expectation that H69 would be critical to both tRNA and RF selection events mediated at the subunit interface.
Genetic screens (16–18) using in vivo-based reporters for stop codon read-through and frame-shifting had also previously identified several positions of interest in H69. In particular, these studies identified “restrictive” mutations in the terminal loop of the helix (C1914, Ψ1911) based on observed decreases in UGA read-through, and other “ribosomal ambiguity” mutations in the stem (G1922) based on observed increases in UGA read-through. Although these studies highlighted a potential role for H69 in substrate selection events, their genetic nature did not allow for direct evaluation of ribosome function, especially because the read-through assay cannot distinguish between defects in the tRNA and RF selection steps.
Here, we examine the role of previously identified key residues in H69 using transient kinetic approaches to isolate and quantify defects in specific steps in the tRNA and RF selection processes. We conclude that although contacts between the loop of H69 (A1913) are not essential for tRNA selection for a majority of tRNA substrates, these contacts are nevertheless critical for acceptance of a subset of natural tRNAs (specifically, tRNAk2CAUIle on the AUA codon and tRNAICGArg on the CGA codon). These tRNAs depend on noncanonical pairings in the third codon position, and we argue that this inherent instability necessitates additional contacts with H69. In other experiments, we find that although contacts between the stem of H69 (G1922) and class 1 RFs are generally not critical to selection, these contacts are specifically essential for facilitating the recognition of UGA codons by RF2. These data argue for a critical role for H69 in the overall fidelity of translation.
EXPERIMENTAL PROCEDURES
Buffers and Reagents
All experiments were performed in HiFi buffer (50 mm Tris-HCl (pH 7.5) with 70 mm NH4Cl, 30 mm KCl, 3.5 mm MgCl2, 0.5 mm spermidine, 8 mm putrescine, and 2 mm DTT). Accommodation and GTPase activation measurements were performed at 20 °C and release factor assays at 37 °C. mRNA was prepared by PCR and T7 transcription and features an epsilon sequence, a Shine-Dalgarno sequence, and a seven-nucleotide spacer followed by the coding sequence AUG XXX UUU, where XXX is the codon specified in individual experiments. EFTu and aaRS were purified as described previously (11). tRNATrp and tRNATrp24/27/59 were expressed, isolated, and purified from the MY87 strain as described previously (11). Other WT tRNA species were isolated from Escherichia coli MRE 600 total tRNA (Roche Applied Science) by aminoacylation using the corresponding aaRS and incubation with EFTu prior to mixing with ribosomal complexes. To account for differences in tRNA abundance, the amount of total tRNA used to form ternary complexes was adjusted to ensure that a sufficient ternary complex was provided for the reaction to proceed to completion (with 1.0 μm ribosome complexes); RF1 and RF2 were purified as described previously (19). Ribosomes (WT and mutants) were purified as described previously (20).
Kinetic Analysis
Accommodation and GTPase activation measurements were carried out essentially as described previously (11, 12, 21). Peptide release experiments were carried out as described previously (19), where 0.25 μm initiation complexes were typically mixed with 5 μm purified release factor protein in a rapid quench apparatus. To account for any subunit association defects of variant ribosomes, all initiation complexes were supplemented with purified WT 30 S ribosomal subunits (21).
RESULTS
Variant Ribosomes and Experimental Background
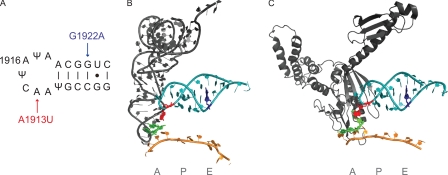
Structural studies of the ribosome and its substrates have made the targeted study of individual regions of potential importance easy to accomplish, once the most interesting targets have been identified. As the ternary complex encounters the ribosomal A site (2, 4), the incoming tRNA is distorted into an A/T state. This strained conformation is stabilized not only by interactions with the decoding center residues (A1492, A1493, and G530) but also by interactions with the 16 S rRNA shoulder, protein S12, the L11 region of 23 S rRNA, and the terminal loop of H69. After GTP hydrolysis and dissociation of EFTu, the tRNA accommodates and adopts the A/A state (Fig. 1B). We note that although there many interactions outside the decoding center that appear to stabilize the A/T state, the interaction between H69 and the tRNA core (between residue A1913 of H69 and residues 25/26 and 38 of the tRNA) is one of a few that appears to be maintained as the tRNA enters the A/A state. When a stop codon is present in the A site and a class 1 release factor binds to the ribosome, A1913 also appears to be involved, forming a stacking interaction with the displaced decoding helix residue A1493 (Fig. 1C). As such, A1913 does not directly contact the release factor in the accommodated state but may still be important for function during this stage or earlier in the process during codon recognition (though we have no structural information on a potential “preaccommodation” release factor). Interestingly, the terminal loop of H69 has been implicated in substrate selection on the ribosome because C1914A/U mutant ribosomes yield phenotypes of increased frame-shifting and decreased UGA read-through in reporter assays. Another potential site of interest found in H69 is G1922 (Fig. 1A). This residue, though somewhat remote from the ribosomal A site and involved in a canonical Watson-Crick interaction in the H69 stem, has been singled out by genetic screens because mutation of this residue results in substantial UGA stop codon read-through (16).
FIGURE 1.
Positioning of Helix 69 of 23 S rRNA. A, schematic representation of H69 residues including pseudouridines (Ψ) at positions 1911, 1915, and 1917. Mutations introduced in this study are highlighted in red (A1913U) and blue (G1922A). B, structure of a post-accomodation tRNA in the A/A state. Regions corresponding to ribosomal tRNA binding sites are indicated underneath the mRNA (gold). Decoding center residues 1492 and 1493 (green) evaluate the geometry of the codon-anticodon interaction. H69 (teal) is shown with residues highlighted as in A. C, structure of RF2 in the ribosomal A site with the GGQ domain extended into the peptidyl transferase center. Decoding center residues 1492 and 1493 are seen to adopt a different conformation than in B, and a stacking interaction is observed between 1493 of 16 S rRNA and A1913 of H69 of 23 S rRNA. Figures were made from Protein Data Bank codes 2WDK/2WDL and 2WH1/2WH2 (8, 35) using PyMOL software.
To study the contributions of H69 to tRNA selection and release factor function, we mutated these two separate sites (A1913U and G1922A) on H69 and evaluated the biochemical properties of the resulting large subunit variants (Fig. 1A). Both ribosome variants were generated by site-directed mutagenesis and purified as described previously (20). Because we anticipated that both might have defects in ribosome association, all assays were performed in the presence of additional WT 30 S subunits (see “Experimental Procedures”).
Transient Kinetic Analysis of Effects of H69 Mutations on Sense Codon Recognition
To decipher the role of H69 in tRNA selection, we began by evaluating kinetic parameters according to the previously established kinetic and thermodynamic framework (22). Based on earlier studies, we anticipated that there were two steps of primary interest, GTPase activation and accommodation, which are key determinants of the fidelity of tRNA selection. The first of these steps, GTPase activation, occurs when the incoming ternary complex is poised in the A/T state and sampling the codon. Codon-anticodon interactions trigger conformational rearrangements in EFTu that lead to the hydrolysis of GTP and subsequent dissociation of EFTu from the ribosome complex. Because GTPase activation (k3) limits the rate of GTP hydrolysis (kGTP), we measure kGTP as a proxy for this process as reported previously (11, 22). For this assay, ribosomes (WT and variant) were programmed with an mRNA template with coding region AUG XXX UUU (where XXX is the indicated mRNA codon) and were subjected to initiation and elongation reactions that yield a peptidyl-tRNA (fMet-AA-tRNAAA) in the P site. Using these ribosome complexes, we measured the rates of GTP hydrolysis by EFTu stimulated by a chosen single concentration of WT, A1913U, and G1922A ribosome complexes. We found no substantial differences between ribosomal species in the rates of kGTP triggered by interactions between tRNAGAUIle or tRNAk2CAUIle ternary complex and ribosomes (WT, A1913U, or G1922A) carrying either the AUU or AUA codons in the A site (supplemental Fig. S1). These data suggest that H69 is minimally involved in this early stage of tRNA selection.
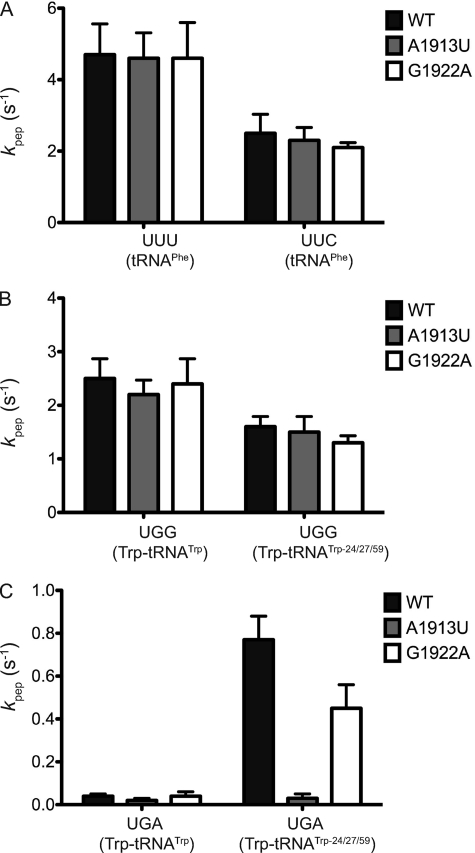
The second key step in the tRNA selection framework represents the “accommodation” of the tRNA into the A/A state once it is released from EFTu. As this conformational change is rate-limiting for the subsequent peptide bond forming reaction (kpep) (23), the rate of peptidyl transfer can be used as a proxy for measuring accommodation (k5). For this measurement, WT and variant ribosome complexes were prepared with fMet-tRNAfMet in the P site, and several distinct codons in the A site and the rate of dipeptide formation was measured on addition of ternary complexes carrying distinct tRNA isoacceptors corresponding to those codons (24). When we followed tRNAPhe interactions with UUU and UUC (both Phe codons) programmed complexes, we found the rates of accommodation to be indistinguishable on WT, A1913U, and G1922A ribosomes (Fig. 2A and Table 1). These results are broadly consistent with previous reports indicating that deletion of H69 has little effect on poly-Phe synthesis (15). Similar results were obtained with tRNATyr deciphering UAU and UAC codons and tRNAcmo5UGCAla or tRNAGGCAla deciphering GCU, GCC, GCA, and GCG codons (supplemental Table S1).
FIGURE 2.
Effects of H69 mutations on tRNA accommodation. Observed rates of peptide bond formation measured by following the formation of fMet-AA dipeptide for WT and H69 variant ribosomes for tRNAPhe on UUU and UUC codons (A), tRNATrp and a miscoding mutant (tRNATrp-24/27/59) on UGG (B) and UGA (C) codons. Error bars represent S.E. Figures reported are from at least three (n = 3) independent experiments.
TABLE 1.
Peptidyl transfer rates of rRNA mutants
S.E. are calculated from at least three independent determinations.
| Codon (tRNA) | WT (kPEP) | A1913U (kPEP) | G1922A (kPEP) | % Codona |
|---|---|---|---|---|
| s−1b | s−1b | s−1b | ||
| UUU (Phe) | 4.7 (0.86) | 4.6 (0.71) | 4.6 (1.0) | 2.2 |
| UUC (Phe) | 2.5 (0.53) | 2.3 (0.36) | 2.1 (0.14) | 1.7 |
| UGG (WT-Trp) | 2.5 (0.37) | 2.2 (0.27) | 2.4 (0.47) | 1.5 |
| UGG (Mut-Trp) | 1.6 (0.19) | 1.5 (0.29) | 1.3 (0.13) | 1.5 |
| UGA (WT-Trp) | .04 (0.01) | 0.02 (0.01) | 0.04 (0.02) | 0.09 |
| UGA (Mut-Trp) | .77 (0.11) | 0.03 (0.02) | 0.45 (0.11) | 0.09 |
| CGU (Arg-ICG) | 3.0 (0.50) | 2.9 (0.35) | 3.2 (0.48) | 2.1 |
| CGA (Arg-ICG) | 2.4 (0.23) | 0.37 (0.04) | 1.9 (0.16) | 0.40 |
| CGC (Arg-ICG) | 2.2 (0.31) | 2.1 (0.31) | 1.9 (0.38) | 2.2 |
| AUU (Ile-GAU) | 2.1 (0.47) | 2.1 (0.42) | 1.9 (0.41) | 3.0 |
| AUA (Ile-k2CAU) | 1.3 (0.28) | 0.20 (0.04) | 0.79 (0.23) | 0.43 |
a % Codon values calculated for E. coli K12 from the Genomic tRNA Database.
b kPEP is calculated at 1 μm ribosomes and 0.15 μm ternary complex. Values presented do not include a calculation for the rate of rejection (k7) seen in the CGA measurements.
In our previous work, we found that tRNATrp-miscoding tRNAs exploit an interaction with residue A1913 of H69 to effectively accommodate on certain near-cognate codon ribosome complexes (21). These results are extended in Fig. 2 where a potent miscoding tRNATrp variant (tRNATrp-24/27/59) exhibits no defects in cognate codon recognition on any ribosome that we have characterized (Fig. 2B), but exhibits clear defects in accommodation on the near-cognate codon UGA in the A1913U ribosome background (Fig. 2C). Notably, on the near-cognate UGA codon, none of the ribosome variants significantly stimulates accommodation by WT tRNATrp, ruling out miscoding as causative for the previously described increases in stop codon read-through associated with the G1922A variant (Fig. 2C) (16). Indeed, while the variant tRNATrp-24/27/59 substantially promotes accommodation on the UGA codon (0.77 s−1 versus 0.03 s−1, a 25-fold effect, Fig. 2c), if anything, the G1922A ribosomes slightly attenuate this elevated rate (0.77 s−1 versus 0.45 s−1).
So far, taken together, the A1913U ribosomes appear to behave normally with cognate codon-anticodon pairing interactions, only exhibiting alterations in the tRNA selection profile when miscoding by a variant tRNATrp is specifically probed. These data suggest that A1913 plays some specialized role in stabilizing more compromised interactions between tRNA and the ribosome. To further explore the sensitivity (or hyperaccuracy) of the A1913U ribosomes, we looked at the decoding parameters of several codons that specifically depend on noncanonical wobble position interactions. When comparing two different isoleucine codons (AUU and AUA) (Fig. 3A) and three different arginine codons (CGU, CGA, and CGC) (Fig. 3B), we noticed that accommodation rates on several of these codons (AUU, CGU, and CGC) were unaffected by the different H69 mutations, whereas they were sensitive to substitution of A1913 for the AUA (2.4 s−1 versus 0.37 s−1) and CGA (1.3 s−1 versus 0.20 s−1) codons.
FIGURE 3.
A1913 is critical for tRNA accommodation. Observed rates (kpep) of peptide bond formation measured by following the formation of fMet-AA dipeptide for wild-type (WT) and H69 variant ribosomes for tRNAIle on AUU (decoded by tRNAGAUIle) and AUA (decoded by tRNAk2CAUIle) codons (A) and tRNAIGCArg on CGU, CGA, and CGC codons (B). Shown is the time course of peptide bond formation (C) for tRNAIGCArg on CGU (circles) and CGA (squares) codons with wild-type (WT, black lines) and A1913U (1913, red lines) ribosomes. Fraction dipeptide values are normalized to the plateau of the CGU measurements. Error bars represent S.E.
Another feature of tRNA selection that is commonly evaluated is the overall rate of rejection of aa-tRNAs post-GTP hydrolysis; increases in rejection at this late stage show up as end point defects in the single turnover assay utilized to evaluate accommodation rates. Interestingly, although we generally see no difference in the rates of rejection between the ribosome species tested on other cognate codons, there is a striking rejection of tRNAICGArg on the CGA codon, both with the WT and the A1913U variant ribosomes (Fig. 3C); we do not see end point defects for the other potentially challenging decoding event on the AUA isoleucine codon. These increased rejection rates must be accounted for when calculating the accommodation rate (k5) specifically on the CGA codon (22).
Transient Kinetic Analysis of Effects of H69 Mutations on Stop Codon Recognition
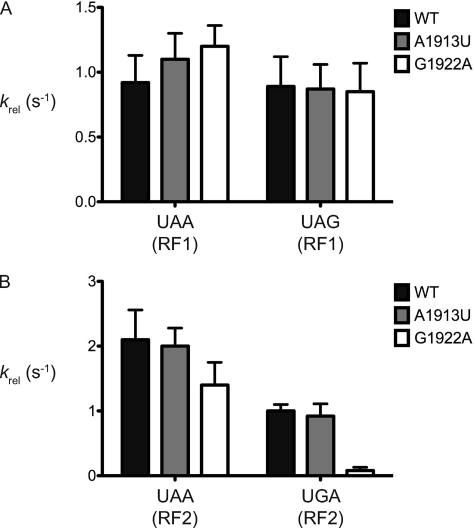
In light of the many similarities between recognition of sense and stop codons by tRNAs and RFs, respectively, we set out to evaluate the properties of the same ribosome variants in RF recognition and function. To determine the contributions of specific regions of H69 to termination, we tested the ability of our ribosome variants to promote peptide release by both RF1 and RF2 on all three stop codons. As for the tRNA selection assays, ribosome complexes were programmed with an mRNA directing the initiator [35S]fMet-tRNAfMet to the P site and the appropriate codons to the A site (UAA, UAG, or UGA). These complexes where subsequently mixed with saturating amounts of the appropriate release factor, RF1 or RF2, and the rates of peptide release were determined (19). The activity of RF1, which normally recognizes UAA and UAG codons, appears to be completely unaffected by the introduced changes in H69 (Fig. 4A); the rates of peptide release on both codons are similar for all ribosomes tested (Table 2). The relatively subtle changes to H69 studied here likely explain the apparent discrepancy with earlier studies where defects in RF1 function were detected when the entire helix (H69) was deleted (15). Interestingly, the activity of RF2 is differentially affected on its two cognate codons, UAA and UGA (Fig. 4B); whereas there were no appreciable defects for the variant ribosomes in recognition by RF2 of UAA codons, there was a notable defect (krel reduced by ∼10-fold) on UGA recognition for the G1922A ribosomes (Table 2). To test whether this decrease in function is a result of diminished binding by RF2 to the G1922A ribosome, we measured the same rates with 3-fold more RF2 and found the observed rates to be unaffected (supplemental Fig. S2), indicating that the values measured reflect kcat for the release reaction.
FIGURE 4.
G1922 is critical for RF2 recognition of UGA. Observed rates of peptide release (krel) measured by following the hydrolysis of fMet from fMet-tRNAfMet present in the ribosomal P site. A, rates of peptide release for release factor 1 (RF1) on UAA and UAG codons for WT and H69 variant ribosomes. B, rates of peptide release for release factor 2 (RF2) on UAA and UGA codons for WT and H69 variant ribosomes. Error bars represent S.E.
TABLE 2.
Peptide release rates of rRNA mutants
S.E. are calculated from at least three independent determinations.
| Codon (RF) | WT (krel) | A1913U (krel) | G1922A (krel) | % Codona |
|---|---|---|---|---|
| s−1b | s−1b | s−1b | ||
| UAA (RF1) | 0.92 (0.21) | 1.1 (0.20) | 1.2 (0.16) | 0.21 |
| UAG (RF1) | 0.89 (0.23) | 0.87 (0.19) | 0.85 (0.22) | 0.03 |
| UAA (RF2) | 2.1 (0.46) | 2.0 (0.28) | 1.4 (0.35) | 0.21 |
| UGA (RF2) | 1.0 (0.10) | 0.92 (0.19) | 0.08 (0.05) | 0.09 |
a % Codon values calculated for E. coli K12 from the Genomic tRNA Database.
b krel is calculated at 0.25 μm ribosomes and 5 μm release factor.
The defects associated with stop codon recognition by H69 variant ribosomes agree broadly with genetic screens that identified positions in H69 that affected stop codon read-through (16). Although earlier interpretation of the genetic results was ambiguous in defining the affected step, our results clearly indicate that stop codon read-through results from specific defects in UGA decoding by RF2 on the G1922A variant ribosomes; we note that read-through was only observed on UGA, but not on a UAA or UAG codon at the same position (16).
DISCUSSION
Early cross-linking studies, structural work on the ribosome, and genetic screens have implicated H69 as playing an important role in tRNA selection during elongation by the ribosome. However, the details of the molecular mechanisms by which H69 influences this process remain poorly understood. Here, we establish a critical role for H69 in tRNA selection and termination by measuring the presteady-state kinetic parameters of several variant ribosomes carrying point mutations in this helix. In particular, the terminal loop of H69 appears to be critical for the recognition of certain tRNA species that depend on noncanonical third position pairing interactions, whereas the stem of H69 appears to be critical specifically for recognition of UGA stop codons by RF2. We suggest that the sensitivity of these particular decoding events (on typically rare codons in E. coli) results from nonoptimal interactions in the decoding center that are supplemented by additional more remote interactions between the ribosome and the substrate.
Despite an abundance of structural data placing H69 in a functionally critical region of the ribosome and genetic studies implicating the helix in core ribosome function, the most thorough biochemical study to date found that although H69 contributed substantially to subunit association (and thus recycling) and to termination by RF1, it contributed only modestly to the fidelity of tRNA selection. In this earlier study, using a processive poly-Phe synthesis assay, the authors found that ribosomes lacking H69 are only modestly restrictive in decoding with ∼2-fold decreases in leucine misincorporation. Because this steady-state assay does not isolate the critical steps of tRNA selection, more substantial effects on individual steps in tRNA selection could have been masked by rate-limiting processes that are not affected by the selected ribosomal mutations. Another potential problem with the assay is that it only measures the encounter of the ribosome with two different tRNA substrates (tRNAPhe and tRNALeu) on a single codon (UUU) and, as such, ignores other tRNA-codon interactions that may have different requirements.
Other key studies implicated H69 in tRNA selection, largely through genetic screens that identified mutations in the ribosome that affected UGA read-through or frame-shifting on mRNA transcripts that contain a stop codon in the middle of the gene (16–18). One class of mutations discovered in these studies clustered in the terminal loop of H69 (C1914, Ψ1911) and generally exhibited slightly decreased rates of UGA read-through; as a representative of this class of mutation, we mutated residue A1913 as it also appears to be located in a functionally critical location. Another class of H69 mutation uncovered by genetic screens was located in the stem of H69. Mutations in this region (G1922, G1921) exhibit very strong UGA read-through phenotypes; as a representative of this class of mutation, we mutated G1922 (Fig. 1). The importance of H69 as a critical component of ribosome function is hinted at in these studies as they find that a number of point mutations in the region exhibit lethal and severe slow growth phenotypes. By measuring rates of tRNA and RF selection under varied conditions with multiple substrates, we define the underlying ribosome defect that results in distinct differences in UGA read-through for the chosen simple base substitutions (A1913U, G1922A) in H69. In particular, we were interested in knowing whether a strong UGA read-through phenotype in reporter assays in vivo arises because of increases in the rate of incorporation of tRNATrp or because of decreases in the activity of RF2.
Although our earlier studies had suggested that an intact H69 provided additional stabilization to certain near-cognate miscoding events during tRNA selection (21), our results here indicate that H69 plays a key role in facilitating “normal” codon-anticodon interactions during translation. It is well known that the first two mRNA codon positions are deciphered by the ribosome through strict Watson-Crick RNA base pairs. Decoding of the third position is known to be less stringent, allowing a certain breadth of noncanonical interactions. These noncanonical interactions principally include so called “wobble” pairing with G-U and U-G base pairs being the most common example (exemplified in this study by tRNAPhe on the UUC and UUU codons, respectively). Other wobble interactions include more heavily modified bases such as queosine (a modified G) (as in tRNATyr, which decodes UAU and UAC), or uridine-5-oxyacetic acid (as in tRNAcmo5UGCAla, which decodes GCA and GCG). The ribosome variants characterized in this study (A1913U and G1922A) did not distinguish between these particular noncanonical third position pairing interactions and other more canonical WC pairing interactions (Table 1 and supplemental Table S1).
Other known third position pairing interactions, however, appear to rely more heavily on the identity of A1913 of H69, as indicated by the defects reported on in Fig. 3, A and B. The well characterized wobble interactions with the post-transcriptionally modified nucleotide inosine that decodes C, U, and A residues at the third codon position is one such example (25). Although the third position C and U (CGC and CGU) are decoded through the purine-pyrimidine interactions I-C and I-U with tRNAICGArg, the third position A (CGA) is decoded through a purine-purine interaction, I-A, which results in a significantly wide base pair in the decoding helix (26). Another example is the AUA codon in E. coli, decoded by tRNAk2CAUIle, which contains a lysidine (k2C) residue in its anticodon (5′-k2CAU-3′). The addition of this lysine moiety to the cytidine residue in the first position of the anticodon changes the specificity of tRNAk2CAUIle from AUG to AUA (27). Though little work has focused on this particular modification, a similar change also appears to be critical for the proper decoding of AUA in archaea (28). For both of these examples, A1913U variant ribosomes are compromised in their ability to select these tRNA species for accommodation into the ribosome.
The dependence of these two inherently weak codon-anticodon interactions on the terminal loop of H69 for efficient acceptance into the ribosome leads to interesting questions about the mechanism by which the ribosome achieves uniformity of tRNA selection (29). When a rare AUA or CGA codon (Table 1) is encountered by a translating ribosome, interactions other than those determined by decoding center residues (A1493, A1492, G530) are apparently necessary for efficient accommodation of the incoming tRNA. Indeed, even when the H69 interactions are intact (with a WT ribosome), we measure increased rates of rejection (k7) of tRNAICGArg on the CGA codon, an observation consistent with recent reports in Saccharomyces cerevisiae identifying the inherently weak nature of the CGA codon (30). Overall, as the accommodation defects that we observe in A1913U ribosomes are of a magnitude comparable to those observed when the small subunit decoding residues (1493, 1492, 530) are themselves mutated (31), we propose that the terminal loop of H69 functions as a critical core component in tRNA selection on the ribosome.
Another interesting observation emerging from our studies is that the variant H69 ribosomes appear to have no defects in GTPase activation, though it is generally thought that increased rates of GTPase activation and accommodation are triggered by common structural signals in the decoding center (22). Although there are numerous examples in the literature where both GTPase activation and accommodation are similarly affected by a miscoding tRNA (11, 12), we have recently shown that a mutation in the elbow region of tRNATrp-G59A stimulates accommodation, but not GTPase activation, on a near-cognate codon (21). Interestingly, this variant tRNA also depends on H69 residue A1913 to stimulate this accommodation step. Recent structural work on tRNATrp miscoding variants in the A/T state has argued that miscoding depends on the formation of hydrogen bonding interactions in the tRNA core and not on altered contacts with the ribosome (32). These data argue for a H69-specific contribution to accommodation (but not to GTPase activation) with special sensitivity to the third position discrimination.
RF selection is in many ways similar to tRNA selection wherein large scale conformational rearrangements triggered by interactions in the decoding center of the small subunit lead to optimal catalysis in the large subunit (19, 33). Nevertheless, the signals that lead to these outcomes are clearly somewhat different because they depend on protein-RNA rather than RNA-RNA interactions in the decoding center (6, 8, 34). One likely critical difference is the stacking interaction that forms between A1913 of H69 and A1493 of H44 when cognate interactions in the decoding center are sensed. Interestingly, we show here that A1913U ribosomes have no apparent defect in interactions with RF1 or RF2 on authentic stop codons, suggesting that the requisite stacking interaction may not depend on the identity of the nucleotide found at position 1913 but only on the presence of some nucleotide that can stack. Earlier studies similarly showed that mutation of A1493 had no effect on RF recognition and catalysis on authentic stop codons (19).
How then do we rationalize the defects in RF recognition by G1922A ribosomes where the mutation of interest appears to be more structurally remote from presumed critical contacts at the decoding center? We notice that for A1913 to stack with residue A1493 of the decoding center, H69 must undergo a relatively large conformational change which results in the formation of a new pocket within the ribosome core that cradles (and undoubtedly stabilizes) the so-called “switch” loop that forms in an extended conformation of the RF. Because of the proximity of G1922 to this switch loop pocket, we speculate that G1922A ribosomes are compromised in their ability to stabilize the extended conformation of RF2 bound to the ribosome (Fig. 4B). The specificity of the defects, observed only with RF2-UGA interactions, can perhaps be attributed to subtle differences observed on comparing the switch loop regions of RF1 and RF2 (34). For example, RF2 is seen to make additional contacts with H69 residues 1914 and 1915 (8), and these might be somewhat destabilized on interaction with the UGA-programmed ribosome complex. Whatever the cause, the specific deficiencies that we observe in the G1922A ribosomes are consistent with multiple earlier reports of RF-related deficiencies with H69 variant ribosomes (14–16).
A thorough biochemical examination of specific ribosome variants and the defects associated with recognition of certain “cognate” interactions by tRNAs and RFs has allowed us to identify a role for H69 in both tRNA accommodation and RF2 function. This study highlights how individual mRNA codons are distinctly dealt with by the translational machinery and discovers a new mechanism that the ribosome employs to ensure uniformity in tRNA selection. Perturbation of this mechanism slows down the rates of incorporation of natural tRNA substrates on the AUA and CGA codons, at a minimum, and the rate of peptide release on the UGA codon. These findings not only shed light on the mechanism of ribosomal codon recognition but may also prove useful in manipulating the ribosomal machinery to accept foreign amino acids into codons that were deemed previously inaccessible or inefficient.
Supplementary Material
Acknowledgments
We thank B. Rogers and S. He for providing reagents and technical expertise for ribosome tagging and purification steps and H. Zaher for comments and suggestions on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM059425 and the Howard Hughes Medical Institute (to R. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
- H69
- helix 69
- RF
- release factor.
REFERENCES
- 1. Ogle J. M., Brodersen D. E., Clemons W. M., Jr., Tarry M. J., Carter A. P., Ramakrishnan V. (2001) Science 292, 897–902 [DOI] [PubMed] [Google Scholar]
- 2. Schmeing T. M., Voorhees R. M., Kelley A. C., Gao Y. G., Murphy F. V., 4th, Weir J. R., Ramakrishnan V. (2009) Science 326, 688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Korostelev A., Trakhanov S., Laurberg M., Noller H. F. (2006) Cell 126, 1065–1077 [DOI] [PubMed] [Google Scholar]
- 4. Li W., Agirrezabala X., Lei J., Bouakaz L., Brunelle J. L., Ortiz-Meoz R. F., Green R., Sanyal S., Ehrenberg M., Frank J. (2008) EMBO J. 27, 3322–3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Valle M., Sengupta J., Swami N. K., Grassucci R. A., Burkhardt N., Nierhaus K. H., Agrawal R. K., Frank J. (2002) EMBO J. 21, 3557–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Korostelev A., Zhu J., Asahara H., Noller H. F. (2010) EMBO J. 29, 2577–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Laurberg M., Asahara H., Korostelev A., Zhu J., Trakhanov S., Noller H. F. (2008) Nature 454, 852–857 [DOI] [PubMed] [Google Scholar]
- 8. Weixlbaumer A., Jin H., Neubauer C., Voorhees R. M., Petry S., Kelley A. C., Ramakrishnan V. (2008) Science 322, 953–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pai R. D., Zhang W., Schuwirth B. S., Hirokawa G., Kaji H., Kaji A., Cate J. H. (2008) J. Mol. Biol. 376, 1334–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weixlbaumer A., Petry S., Dunham C. M., Selmer M., Kelley A. C., Ramakrishnan V. (2007) Nat. Struct. Mol. Biol. 14, 733–737 [DOI] [PubMed] [Google Scholar]
- 11. Cochella L., Green R. (2005) Science 308, 1178–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ledoux S., Olejniczak M., Uhlenbeck O. C. (2009) Nat. Struct. Mol. Biol. 16, 359–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ejby M., Sørensen M. A., Pedersen S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19410–19415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kipper K., Sild S., Hetényi C., Remme J., Liiv A. (2011) Biochimie 93, 834–844 [DOI] [PubMed] [Google Scholar]
- 15. Ali I. K., Lancaster L., Feinberg J., Joseph S., Noller H. F. (2006) Mol. Cell 23, 865–874 [DOI] [PubMed] [Google Scholar]
- 16. O'Connor M. (2009) Mol. Genet. Genomics 282, 371–380 [DOI] [PubMed] [Google Scholar]
- 17. O'Connor M., Dahlberg A. E. (1995) J. Mol. Biol. 254, 838–847 [DOI] [PubMed] [Google Scholar]
- 18. Hirabayashi N., Sato N. S., Suzuki T. (2006) J. Biol. Chem. 281, 17203–17211 [DOI] [PubMed] [Google Scholar]
- 19. Youngman E. M., He S. L., Nikstad L. J., Green R. (2007) Mol. Cell 28, 533–543 [DOI] [PubMed] [Google Scholar]
- 20. Youngman E. M., Green R. (2005) Methods 36, 305–312 [DOI] [PubMed] [Google Scholar]
- 21. Ortiz-Meoz R. F., Green R. (2010) RNA 16, 2002–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gromadski K. B., Rodnina M. V. (2004) Mol. Cell 13, 191–200 [DOI] [PubMed] [Google Scholar]
- 23. Pape T., Wintermeyer W., Rodnina M. (1999) EMBO J. 18, 3800–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Agris P. F. (2004) Nucleic Acids Res. 32, 223–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Curran J. F. (1995) Nucleic Acids Res. 23, 683–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murphy F. V., 4th, Ramakrishnan V. (2004) Nat. Struct. Mol. Biol. 11, 1251–1252 [DOI] [PubMed] [Google Scholar]
- 27. Muramatsu T., Nishikawa K., Nemoto F., Kuchino Y., Nishimura S., Miyazawa T., Yokoyama S. (1988) Nature 336, 179–181 [DOI] [PubMed] [Google Scholar]
- 28. Ikeuchi Y., Kimura S., Numata T., Nakamura D., Yokogawa T., Ogata T., Wada T., Suzuki T., Suzuki T. (2010) Nat. Chem. Biol. 6, 277–282 [DOI] [PubMed] [Google Scholar]
- 29. Ledoux S., Uhlenbeck O. C. (2008) Mol. Cell 31, 114–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Letzring D. P., Dean K. M., Grayhack E. J. (2010) RNA 16, 2516–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cochella L., Brunelle J. L., Green R. (2007) Nat. Struct. Mol. Biol. 14, 30–36 [DOI] [PubMed] [Google Scholar]
- 32. Schmeing T. M., Voorhees R. M., Kelley A. C., Ramakrishnan V. (2011) Nat. Struct. Mol. Biol. 18, 432–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. He S. L., Green R. (2010) Nat. Struct. Mol. Biol. 17, 465–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Korostelev A., Asahara H., Lancaster L., Laurberg M., Hirschi A., Zhu J., Trakhanov S., Scott W. G., Noller H. F. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19684–19689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Voorhees R. M., Weixlbaumer A., Loakes D., Kelley A. C., Ramakrishnan V. (2009) Nat. Struct. Mol. Biol. 16, 528–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.