Abstract
Inactivating mutations in the transcription factor hepatocyte nuclear factor (HNF) 1A cause HNF1A–maturity-onset diabetes of the young (HNF1A-MODY), the most common monogenic form of diabetes. To examine HNF1A-MODY-induced defects in gene expression, we performed a microarray analysis of the transcriptome of rat INS-1 cells inducibly expressing the common hot spot HNF1A frameshift mutation, Pro291fsinsC-HNF1A. Real-time quantitative PCR (qPCR), Western blotting, immunohistochemistry, reporter assays, and chromatin immunoprecipitation (ChIP) were used to validate alterations in gene expression and to explore biological activities of target genes. Twenty-four hours after induction of the mutant HNF1A protein, we identified a prominent down-regulation of the bone morphogenetic protein 3 gene (Bmp-3) mRNA expression. Reporter assays, qPCR, and Western blot analysis validated these results. In contrast, inducible expression of wild-type HNF1A led to a time-dependent increase in Bmp-3 mRNA and protein levels. Moreover, reduced protein levels of BMP-3 and insulin were detected in islets of transgenic HNF1A-MODY mice. Interestingly, treatment of naïve INS-1 cells or murine organotypic islet cultures with recombinant human BMP-3 potently increased their insulin levels and restored the decrease in SMAD2 phosphorylation and insulin gene expression induced by the HNF1A frameshift mutation. Our study suggests a critical link between HNF1A-MODY-induced alterations in Bmp-3 expression and insulin gene levels in INS-1 cells and indicates that the reduced expression of growth factors involved in tissue differentiation may play an important role in the pathophysiology of HNF1A-MODY.
Keywords: Apoptosis, Bone Morphogenetic Protein (BMP), Diabetes, Insulin, Pancreatic Islets, Hepatocyte Nuclear Factor 1A, Maturity-onset Diabetes of the Young
Introduction
Maturity-onset diabetes of the young (MODY)2 is a familial form of non-insulin-dependent diabetes with autosomal dominant inheritance (1). Most forms of MODY are caused by decreased expression of functional transcription factors leading to a decrease in beta cell mass and islet architecture (2). Inactivating mutations in the transcription factor hepatocyte nuclear factor (HNF) 1A cause HNF1A-MODY (3), the most common monogenic form of diabetes (4). Pancreatic beta cell dysfunction is a hallmark feature of the pathogenesis of HNF1A-MODY and type 2 diabetes (5–7). The control of pancreatic differentiation by extracellular signaling molecules and growth factors during development is an area of intensive research (8). However, our knowledge on the role of extracellular signaling molecules or growth factors in the pathogenesis of HNF1A-MODY or type 2 diabetes is currently limited.
Cytokines of the transforming growth factor-β (TGF-β) superfamily, which includes the TGF-β isoforms, activins, and the bone morphogenetic proteins (BMPs), regulate gene expression in diverse cell types and are involved in numerous processes including lineage determination, tissue differentiation, cell proliferation, and apoptosis (9–11). TGF-β/activin and BMP signaling have been shown to play a critical role in pancreatic development (12–14). Using an unbiased transcriptomics approach in INS-1 cells inducibly expressing the common human hot spot HNF1A frameshift mutation, Pro291fsinsC-HNF1A, we demonstrate here that HNF1A controls the expression of the TGF-β superfamily member, BMP-3. We further demonstrate a role for BMP-3 in the control of insulin expression with potential implications for HNF1A-MODY pathophysiology.
EXPERIMENTAL PROCEDURES
Cell Culture: INS-1 Cells Overexpressing HNF1A in an Inducible System
Rat INS-1 insulinoma cells overexpressing wild-type HNF1A (WT-HNF1A), the HNF1A-MODY-associated frameshift mutant Pro291fsinsC-HNF1A, carrying a C nucleotide insertion in a poly(C) tract mutation hot spot, or a dominant negative mutant of HNF1A (DN-HNF1A/SM6) all under the control of a doxycycline-dependent transcriptional activator have been described previously (15–17). Cells were cultured in RPMI 1640 medium at 6 mm glucose supplemented with 10% fetal bovine serum (FBS) (PAA, Cölbe, Germany), 2 mm l-glutamine, 1 mm pyruvate, penicillin (100 units/ml), streptomycin (100 μg/ml), 10 mm HEPES (pH 7.4), and 50 μm 2-mercaptoethanol (Sigma) (18). For treatments with recombinant BMP-3 (rBMP-3), cells were cultured in 2% serum overnight and then treated with vehicle or 500 ng/ml doxycycline and 10 ng/ml rBMP-3 protein made up in 0.1% bovine serum albumin (BSA) (R&D Systems Europe Ltd.) in 0.05% serum-containing medium for 24 h.
Preparation of Mouse Organotypic Pancreatic Slice Cultures
Animal experiments were carried out in accordance with the principles of the European Communities Council Directive (86/609/EEC) and the National Institute of Health Guide for the Care and Use of Laboratory Animals. Procedures were reviewed and approved by the research ethics committee of the Royal College of Surgeons in Ireland, under license from the Department of Health, Dublin, Ireland. Pancreatic slices were obtained from C57BL/6J background mice. Mice were sacrificed by abdominal injection of 40 mg/kg pentobarbital (Dolethal; Vétoquinol, Buckingham, UK). The abdominal cavity was opened, and warm (37 °C) low gelling temperature agarose (2% w/v, Seaplaque GTG-agarose, BMA Products, Augst, Switzerland) was injected into the distally (duodenal side) clamped bile duct. Prior to the injection a ligature was performed at the height of the hepatic bifurcation of the bile duct to prevent the injected agarose from flowing backward toward the liver. During slicing and after the preparation, the tissue was kept in an ice-cold extracellular solution (125 mm NaCl, 2.5 mm KCl, 26 mm NaHCO3, 1.25 mm Na2HPO4, 2 mm sodium pyruvate, 0.25 mm ascorbic acid, 3 mm myo-inositol, 6 mm lactic acid, 1 mm MgCl2, 2 mm CaCl2, and 3 mm glucose) adjusted to pH 7.3 by gassing with carbogen (95% O2, 5% CO2). The injected and hardened pancreas was then extracted, placed in ice-cold extracellular solution, and, if necessary, supported with subcapsular injections of agarose. A small cube was cut from the agarose-embedded pancreatic tissue and glued (Super Glue; ND Industries, Troy, MI) onto the sample block of the vibratome (Intracel Vibratome 1500, Herts, UK). The tissue was sliced at a speed of 0.05 mm s−1 and 75 Hz into 140-μm-thick slices of 1 × 1 cm. During slicing and for storage the tissue slices were kept in ice cold extracellular solution and were gassed continuously with carbogen. Slices were then placed on the air-medium interface of Millicell semiporous membranes (pore size of 0.4 μm; Millipore) and incubated at 37 °C with 5% CO2. Slices were left to recover in RPMI 1640 medium at 6 mm glucose (as above) for at least 24 h in the incubator before treatment. For experiments with BMP-3, cells were cultured in 2% serum overnight and then treated with either vehicle or 50 ng/ml rBMP-3 protein made up in 0.1% BSA (R&D Systems Europe Ltd.) in 0.05% serum-enriched media for 24 h.
Real-time Absolute RT-PCR
For the assessment of Hnf1a expression by real-time quantitative PCR (qPCR) using absolute copy numbers, an Hnf1a gene-specific PCR amplicon was prepared as a standard. The calibration curve was created by plotting the threshold cycle corresponding to each standard versus their corresponding log number of Hnf1a standard (expressed as cDNA copy number of the Hnf1a gene). Normalization was carried out using total RNA values (19).
Microarray Analyses
Total RNA of INS-1 insulinoma cells overexpressing the Pro291fsinsC-HNF1A mutant for 24 h versus non-induced controls was extracted using the RNeasy Midi kit (Qiagen) and reverse transcribed to cDNA using the Superscript Choice kit (Invitrogen) with 20–40 μg of total RNA as template. The cRNA was prepared and biotin-labeled by in vitro transcription (Enzo Biochemical, Farmingdale, NY) and subsequently fragmented by incubation at 94 °C for 35 min in the presence of fragmentation buffer (40 mm Tris acetate (pH 8.1), 100 mm potassium acetate, and 30 mm magnesium acetate). Fifteen micrograms of fragmented cRNA was hybridized for 16 h at 45 °C to a rat U34A array (Affymetrix, Santa Clara, CA), and the gene chips were automatically washed and stained with streptavidin-phycoerythrin by using a fluidics station. Finally, probe arrays were scanned at 3-mm resolution using the Genechip System confocal scanner (Aligent Technologies, Palo Alto, CA). The Affymetrix Microarray Suite 5.0 was used to scan and analyze the relative abundance of each gene from the average difference of intensities. For each individual gene, the signal ratio between perfect match and mismatch probe cells was used to determine its “Absolute Call,” indicating whether the corresponding transcript was present (P), absent (A), or marginal (M). For analysis of differential target gene expression, the “Difference Call Decision Matrix” was employed. Individual transcript levels were ranked as either increased (I), marginally increased (MI), decreased (D), marginally decreased (MD), or not changed (NC). To be considered as differentially regulated, the Absolute Call of individual transcripts had to be present or marginal, and the Difference Call increased/decreased or marginally increased/marginally decreased. The gene identity of the obtained expressed sequence tag sequences represented on the chips refers to the assignment given on the Affymetrix Homepage, NetAffx Analysis Center. Expressed sequence tag sequences without given gene identities were analyzed by PubMed database searches (standard nucleotide-nucleotide BLAST) (20). The microarray experiment was performed in duplicate with the threshold for differentially expressed up- and down-regulated genes set at >1.5-fold/1.5-fold (median value of both samples) and >2.0-fold/2.0-fold in at least one of the duplicate samples. The rat RG-U34A0 microarray contains ∼7,000 full-length sequences and ∼1,000 expressed sequence tags clusters. The data were subsequently imported into dCHIP software.
Real-time qPCR
Expression patterns of L-type pyruvate kinase, transmembrane protein27 (Tmem27), Bmp-3, Bmp-4, and insulin gene mRNA were examined using qPCR. INS-1 cells were harvested from culture at the appropriate time points. Total RNA was extracted using the RNeasy Mini kit (Qiagen). First-strand cDNA synthesis was performed using 1 μg of total RNA as template and Superscript II reverse transcriptase (Invitrogen) primed with 50-pmol random hexamers (New England Biolabs). Real-time qPCR was performed using the LightCycler 2.0 (Roche Diagnostics) and the QuantiTech SYBR Green PCR kit (Qiagen) according to the manufacturers' protocols and 25 pmol of gene-specific primer pair (Sigma-Genosys).
Specific PCR products (150–250 bp in length) for each gene analyzed were designed using Primer3 software (21). Primers were: GAGATCCCTGCGGAGAAGGT and ATTGGCCACATCGCTTGTCT for L-type prk; CATCTGTGCCCGTCTGGATT and CCAAGGGATCACAAGGGATG for Tmem27; TTACGGAAGGCCAAGCAAGA and GGGCTTCTCTGACGTGGCTA for Bmp-3; GCGTAGTCCCAAGCATCACC and ACAGTCCCCGTGGCAGTAGA for Bmp-4; AACAGCACCTTTGTGGTCCT and GTGCAGCACTGATCCACAAT for Ins1; and AGCCATCCAGGCTGTGTTGT and CAGCTGTGGTGGTGAAGCTG for β-actin.
Western Blotting
Cells were lysed in lysis buffer (62.5 mm Tris-Cl (pH 6.8), 2% SDS, 10% glycerin, and protease inhibitor mixture (Sigma)). Protein content was determined using the Pierce BCA Micro Protein Assay kit (Thermo Fisher Scientific). Samples were supplemented with 2-mercaptoethanol and denatured at 95 °C for 5 min. 20–50 μg of protein were separated by SDS-PAGE and blotted to nitrocellulose membranes (Protean BA 85; Schleicher & Schuell). The membranes were blocked with 1% BSA in Tris-buffered saline for 2 h at room temperature. Blots were probed with the following primary antibodies: goat polyclonal antibody to HNF1A (Santa Cruz Biotechnology, Santa Cruz, CA), diluted 1:1,000; mouse monoclonal anti-β-actin antibody (clone DM 1A; Sigma), diluted 1:5,000; polyclonal anti-BMP-3 antibody (H-73) (Santa Cruz Biotechnology), diluted 1:1,000; polyclonal anti-insulin antibody (H-86) (Santa Cruz Biotechnology), diluted 1:1,000; monoclonal anti-phospho-SMAD2/3 (Ser465/Ser467) (Cell Signaling Technology), diluted 1:1,000; rabbit polyclonal anti-phospho-SMAD1/5/8 (Ser463/Ser465//Ser426/Ser428), diluted 1:1,000; rabbit polyclonal anti-BMP-R2 antibody (H-300; Santa Cruz Biotechnology); and rabbit polyclonal anti-ALK-4 antibody (H-75) Santa Cruz Biotechnology), were diluted 1:1,000 in Tris-buffered saline containing 1% BSA. Antibodies were incubated overnight at 4 °C. The secondary antibodies were peroxidase-coupled goat anti-rabbit IgG (Sigma) or goat anti-mouse (Jackson Immunoresearch) diluted 1:10,000 in the same buffer. Antibody-conjugated peroxidase activity was visualized using the SuperSignal chemiluminescence reagent (Pierce) and imaged using a FujiFilm LAS-3000 imaging system (Fuji, Sheffield, UK).
Transfections and Luciferase Assays
A 2-kb fragment 5′ to exon 1 of the rat Bmp-3 gene was cloned using Kpn1 and HindIII restriction sites into the pGL3-basic vector (Promega) to generate the Bmp-3 promoter plasmid. For transfection analyzes, 105/cm2 INS-1 cells were seeded onto 6-well plates in RPMI 1640 medium 1 day prior to transfection. The cells were transfected with 4 μg of BMP-3 promoter plasmid using Lipofectamine 2000 (Invitrogen). Transfected cells were induced with doxycycline for 24 h and 48 h before harvesting. Reporter activity was measured using the Dual Luciferase® Reporter Assay System (Promega) and normalized to Renilla luciferase activity. Experiments were carried out in triplicate and repeated three times. Data are presented as n-fold expression of Bmp-3 promoter activity compared with non-induced controls.
Chromatin Immunoprecipitation Assay
Approximately 105/cm2 INS-1 cells were seeded onto 10-cm dishes in RPMI 1640 medium 1 day prior to treatment. The cells were induced with doxycycline for 48 h in 6 mm glucose to overexpress WT-HNF1A. Cells were harvested and resuspended in 10 ml of ice-cold PBS, and proteins and DNA were cross-linked with 1% formaldehyde for 10 min at 37 °C followed by quenching with 2 mm glycine. Cells were pelleted and resuspended in 500 μl of sonication buffer (1% Triton X-100, 0.1% deoxycholate, 50 mm Tris-Cl (pH 8.0), 150 mm NaCl, 0.1% AEBSF, 1% SDS). Cells were sonicated for 5 s on ice using a Branson Digital Sonifier 250 (Danbury, CT). Correct shearing of the DNA was confirmed on an agarose gel to ensure fragments were in the range of 200–500 bp. Samples were added to 1 ml with sonication buffer and precleared with a 50% slurry of protein A/G-agarose (Santa Cruz Biotechnology) containing 2 μg of fish sperm DNA and 1% FCS on a rotary wheel for 30 min. The beads were removed by centrifugation and the supernatant collected and incubated with 5 μg of either a rabbit polyclonal anti-HNF1α antibody (Santa Cruz Biotechnology) or an isotype control antibody overnight on a rotary wheel at 4 °C in the presence of 2 μg of fish sperm DNA, 1% FCS, and 45 μl of a 50% slurry of protein A/G-agarose. The beads were pelleted by centrifugation and washed sequentially with PBS, sonication buffer, TSE I buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.1, 150 mm NaCl), TSE II buffer (same as TSE I with addition of 500 mm NaCl), TSE III buffer (0.25 mm LiCl, 1% NP-40, 1% deoxycholate, 1 mm EDTA, 10 mm Tris-HCl, pH 8.1) and followed by a final wash in PBS. The DNA including control input was extracted with 300 μl of extraction buffer (1% SDS, 0.1% NaHCO3) and incubated at 65 °C for 4 h to reverse cross-links followed by purification using the Qiagen PCR purification kit according to the manufacturer's instructions. PCR was performed using the following primers which yielded a PCR product of 249 bp: sense, 5′-TAGGGCAACAAATTGTGCAG-3′; antisense, 5′-TGCTCTTTCTCGGTTGTGTG-3′.
Immunohistochemistry
Paraffin-embedded pancreatic sections from transgenic mice expressing a DN-HNF1A mutant under the control of rat insulin promoter DN-HNF1A and control wild-type C57BL/6JBomTac mice-were deparaffinized (22) and incubated overnight at 4 °C with the rabbit polyclonal anti-BMP-3 (Santa Cruz Biotechnology) diluted 1:20). Slides were incubated for 1 h at room temperature in Alexa Fluor 568-labeled anti-rabbit secondary antibody (Molecular Probes) (1:200). Slides were incubated again overnight at 4 °C with the second primary guinea pig anti-insulin antibody (DakoCytomation A0564; 1:20) followed by 1 h at room temperature in FITC labeled anti-goat secondary antibody (Jackson Immunoresearch; 1:200) and mounted in Vectashield with DAPI (Vector Shield, Vector Laboratories). Images were taken with a Zeiss LSM710 confocal microscope equipped with a 40 × 1.3 NA oil immersion objective (Carl Zeiss). FITC was excited at 488 nm with an argon laser, using a 488/543-nm multichroic beam splitter; the emission was collected at a 490- to 550-nm spectral band. Rhodamine was excited at 543 nm with a helium neon laser, using the same beam splitter, and the emission was collected at a 570–650-nm spectral band. DAPI was excited using the 405-nm DPSS laser and a 405-nm dichroic beam splitter. DAPI fluorescence was detected in the 420–480-nm spectral band. Images were processed using ImageJ software.
Digital pictures were taken of islets of Langerhans, recognized by their distinct morphology. Immunohistochemistry of mouse pancreatic slice cultures was conducted as described above. Insulin expression levels were assessed by measuring the fluorescence intensity of antibody staining in islets across the optical slices. Image z-stacks were taken using an LSM 5 live equipped with a 40 × 1.3 NA oil immersion objective using a 489-nm DPSS laser for excitation of FITC (Carl Zeiss). The optical slice thickness was set to 1.8 nm; fluorescence emission was detected at 495–555 nm. Hoechst staining was assessed as a reference for nuclei. Stacks of optical slices were taken at intervals of 1 μm. Resultant image stacks were analyzed using ImageJ. The mean fluorescence intensity of an islet and the intensity of the islet cells peripheral to the slice were determined. Intensity of insulin staining was calculated as the percent of average islet intensity. Data were averaged from at least three similarly treated slices, and the experiment was repeated with similar results.
Statistics
Results were expressed as means ± S.E. Statistical analysis was conducted using the SPSS version15.0 software package for Windows (SPSS Inc.). Differences between treatments were analyzed by Student's t test, the Mann-Whitney U test, or one-way analysis of variance (ANOVA) with post hoc Tukey-Kramer multiple comparison test. Differences were considered to be significant at p ≤ 0.05.
RESULTS
Microarray Analysis of the Transcriptional Response of Rat INS-1 Cells Inducibly Expressing the Pro291fsinsC-HNF1A Frameshift Mutation
The time course of hnf1a gene induction in Pro291fsinsC-HNF1A INS-1 cells is represented by absolute qPCR (Fig. 1A). Data are presented as cDNA copy number/μl of the hnf1a gene expression. Data are represented as means ± S.E. from n = 3 cultures. The experiment was repeated three times with similar results. * p < 0.05 indicates the difference from non-induced controls.
FIGURE 1.
Induction of Hnf1a gene expression in Pro291fsinsC-HNF1A INS-1 cells with subsequent mRNA expression analysis. A, Pro291fsinsC-HNF1A INS-1 cells were induced with doxycycline for 0–48 h, and Hnf1a gene expression was determined using absolute real-time qPCR. Data are presented as cDNA copy number/μl of the Hnf1a gene expression. Data are represented as means ± S.E. (error bars) from n = 3 cultures. The experiment was repeated three times with similar results. *, p < 0.05 indicates the difference from non-induced controls. B, cells were treated as described above, and whole cell lysates were analyzed by Western blotting on 12% SDS-PAGE. Membranes were probed with a polyclonal antibody recognizing HNF1A. β-Actin served as a loading control. C and D, Pro291fsinsC-HNF1A INS-1 cells were treated as in A, and L-type Prk gene mRNA expression (C) and Tmem27 mRNA expression (D) were determined using real-time qPCR. Experiments were carried out in triplicate, repeated three times, and normalized to β-actin. Data are represented as mean ± S.E., and statistical analysis was carried out as in A. *, p < 0.05 compared with non-induced controls.
In non-induced INS1 cells no increase in Pro291fsinsC-HNF1A gene expression in copies/μl was detected. Western blot analysis confirmed the induction of Pro291fsinsC-HNF1A (Fig. 1B). Concomitant with the induction of the Pro291fsinsC-HNF1A mutant was a significant reduction in mRNA levels of L-type pyruvate kinase gene (Fig. 1C) and Tmem27 (Fig. 1D), both known HNF1A target genes, confirming previous results in this cellular system (17, 23–25).
Using an unbiased microarray approach (Affymetrix GeneChip, Rat RG-U34A) to investigate transcriptome changes in INS-1 cells following induction of the mutant protein for 24 h we identified several genes with altered expression in response to Pro291fsinsC-HNF1A. Cutoff for up-regulated genes was set at 2.0-fold and 0.5-fold for down-regulated genes. Differentially expressed genes of interest were classified into distinct functional subgroups. These included metabolic regulation, signal transduction, transport activity, differentiation, inflammatory response, and regeneration of beta cells (Table 1). Expression levels of selected genes were confirmed by real-time qPCR techniques (supplemental Fig. 1).
TABLE 1.
Differentially expressed genes classified into distinct functional subgroups
| Accession | Gene |
|---|---|
| Down-regulated genes | |
| Signal transduction | |
| X13905cds | Ras-related protein 1B |
| rcAA859896 | Myristoylated alanine-rich protein kinase C substrate |
| D49785 | Mitogen-activated protein kinase 12 |
| X13905cds | Ras-related protein 1B |
| Metabolic regulation | |
| X05684 | L-type pyruvate kinase |
| NM 0538262 | Pyruvate dehydrogenase kinase |
| M73714 | Aldehyde dehydrogenase family 3 |
| Transcription factors | |
| AB017044 | Forkhead box A3 |
| Growth factor and differentiation | |
| S77492i and D63860 | Bone morphogenic protein 3 |
| M93257s | Interleukin 6 |
| Cytoskeletal proteins/transport activity | |
| rcAA866465s | Tropomyosin 3 |
| rcAI145444 | Neurabin 1 |
| U14398 | Synaptotagmin IV |
| Up-regulated genes | |
| Signal transduction | |
| AF072439 | Growth hormone-releasing hormone |
| M93257s | Catechol-O-methytransferase |
| Metabolic regulation | 3α-Hydroxysteroid dehydrogenase |
| D17310s | |
| Transcription factors | |
| X55955 | Forkhead box A1 |
| NM 012742 | Forkhead box A2 |
| Transport activity | |
| AB013112s | Aquaporin 9 |
| D13962 | Solute carrier family 22 |
| Growth factor and differentiation | |
| L20869 | Pancreatitis-associated protein III |
| U93306 | VEGF receptor |
| U69278 | Ephrin receptor |
| Inflammation | |
| L22190 | Serum amyloid A |
| D50558g | CD86 antigen |
| AF072411 | CD36 antigen |
Among the genes identified in this microarray study were several known target genes of HNF1A including Pklr, Foxa3, and Pdk-1, and all were found to be down-regulated in response to Pro291fsinsC-HNF1A (16, 17, 26, 27). Foxa2, which is subject to transcriptional repression by HNF1A (28), was found to be up-regulated. Interestingly, a number of potential novel target genes regulated by HNF1A were also identified on the Rat RG-U34A microarray. In particular, we observed a significant down-regulation of the TGF-β superfamily member BMP-3 as well as up-regulation of Cd36 and PAP III.
Suppression of the Bmp-3 Gene and Protein Expression in INS-1 Cells Inducibly Expressing the Pro291fsinsC-HNF-1A Frameshift Mutation
BMPs are members of the TGF-β superfamily, a large family of growth factors, including TGF-β and activins. They are critically involved in tissue differentiation and development and signal via type I and type II TGF-β/BMP receptors (9). To validate the down-regulation of Bmp-3 mRNA expression, we examined its expression by qPCR. Analysis of Bmp-3 mRNA expression revealed a significant time-dependent decrease in expression in Pro291fsinsC-HNF1A cells, evident from 8 h onward (Fig. 2A). In contrast, INS-1 cells inducibly expressing WT-HNF1A cells showed a time-dependent increase in Bmp-3 gene expression (Fig. 2B). Down-regulation of Bmp-3 mRNA expression was also observed in INS-1 cells expressing the artificial dominant negative HNF1A mutant (SM6) (Fig. 2C).
FIGURE 2.
BMP-3 and BMP-4 expression in INS-1 cells. Pro291fsinsC-HNF1A INS-1 cells, WT-HNF1A cells, and DN-HNF1A cells were treated with 500 ng/ml doxycycline for 8, 16, 24, and 48 h as indicated. Cells were harvested and total RNA isolated. Bmp-3 (A–C) or Bmp-4 (D) mRNA expression was determined using real-time qPCR following normalization to β-actin. Experiments were carried out in triplicate and analyzed using one-way ANOVA with post hoc Tukey-Kramer test. Data are represented as mean ± S.E. (error bars), and statistical analysis was carried out as in A. *, p < 0.05 compared with non-induced controls. E and F, Pro291fsinsC-HNF1A INS-1 cells were treated as above and harvested for protein. Whole cell lysates were analyzed by Western blotting on 15% SDS-PAGE. Membranes were probed with an anti-BMP-3 polyclonal antibody. Probing with β-actin served as a loading control.
We next investigated the specificity of Bmp-3 down-regulation compared with other members of the TGF-β superfamily. Several members of the TGF-β superfamily, including receptors and downstream signal transducers, were present on the chip and remained unchanged from the Affymetrix Gene array data (Bmp-2-like, Bmp-4, Bmp-6, Bmp-7, and Bmp-type IA receptor, type III Tgf-β receptor gene, LTBP-3, LTBP-2, the Smads1–3, and Smads7 and 8 (Table 2). However, because BMP-4 has previously been identified to be involved in pancreatic function (29), it was therefore further analyzed by qPCR. Bmp-4 mRNA expression was not altered during Pro291fsinsC-HNF1A induction (Fig. 2D). Furthermore, qPCR analysis was also performed on several members of the TGF-β superfamily members (Tgf-β1, Tgf-β2, Tgf-β3, and activin β-A and activin β-B) and demonstrated that there was no prominent down-regulation of other genes of the TGF-β superfamily at either 8 h or 16 h (supplemental Fig. 2).
TABLE 2.
Members of the TGF-β superfamily present on the Affymetrix GeneArray
| Accession | Gene | Change |
|---|---|---|
| D63860 and S77492 | BMP-3 | Decreased |
| U66298 | BMP-6 | No change |
| Z22607 | BMP-4 | No change |
| D29769 | BMP-7 | No change |
| S75359 | Bone morphogenetic protein type IA receptor | No change |
| L20678 | BMP-2-like | No change |
| M80784 | Type III TGF-β receptor | No change |
| M96643 | TGF-β2 | No change |
| X52498 | TGF-β1 | No change |
| AF016900 | Latent TGF3-β-binding protein 3 | No change |
| AF016901 | Latent TGF3β-binding protein 2 | No change |
| AF067727 and U66478 | SMAD1 | No change |
| AB017912 | SMAD2 | No change |
| AF042499 | SMAD7 | No change |
| AF012347 | SMAD8 | No change |
| U66479 | SMAD3 | No change |
We also investigated BMP-3 protein levels by Western blotting in INS-1 cells inducibly expressing either Pro291fsinsC-HNF1A or WT-HNF1A. BMP-3 protein expression was significantly reduced upon induction of Pro291fsinsC-HNF1A mutant at 8, 16, and 24 h compared with non-induced controls with expression virtually absent at 48 h (Fig. 2E). Expression of WT-HNF1A resulted in increased expression of BMP-3 protein (Fig. 2F). These data suggested that BMP-3 expression was controlled by HNF1A.
HNF1A Represses Bmp-3 Promoter Activity in INS-1 Cells Inducibily Expressing the Pro291fsinsC-HNF1A Mutation
We next investigated whether the HNF1A mutant directly reduced Bmp-3 promoter activity. A 2-kb fragment of Bmp-3 sequence 5′ to the transcription initiation site was identified by the ENSEMBL database and cloned into the pGL3 basic vector. To assess the effects of HNF1A expression on the activity of the Bmp-3 promoter we transfected WT-HNF1A and Pro291fsinsC-HNF1 INS-1 cells with the promoter construct and monitored the relative luciferase expression levels following induction for 24 and 48 h. Luciferase activity assays demonstrated that WT-HNF1A significantly increased the trans-activation of the Bmp-3 promoter (Fig. 3A), whereas the induction of the Pro291fsinsC-HNF1A mutation decreased the trans-activation of the Bmp-3 promoter compared with non-induced controls (Fig. 3B). To investigate whether the effects of HNF1A on the Bmp-3 promoter were mediated via direct binding to the promoter we first examined the putative promoter sequence for HNF1A binding sites. Using the ALGGEN Promo database, we identified a potential HNF1A binding site in the Bmp-3 promoter fragment (Fig. 3C). To assess whether HNF1A could bind within this region we performed ChIP experiments using an HNF1A antibody or an IgG isotype control. As demonstrated in Fig. 3C, amplification of a region encompassing the identified HNF1A binding site was evident in WT-HNF1A overexpressing cells but not in non-induced or IgG controls (Fig. 3D). Taken together, these data suggest that HNF1A can bind the Bmp3 promoter directly.
FIGURE 3.
Repression of BMP-3 promoter activity in Pro291fsinsC-HNF1A INS-1 cells. A and B, WT-HNF1A (A) and Pro291fsinsC-HNF1A (B) INS-1 cells were transfected with a vector containing the firefly luciferase gene under control of the BMP-3 promoter alongside a Renilla luciferase reporter plasmid (pRL-TK) as an internal transfection control. INS-1 cells were treated with doxycycline (Dox; 500 ng/ml) for 24 or 48 h. Luminescence activity was normalized to the activity of the co-transfected RL-TK-luc and expressed as -fold of increase with respect to controls. Experiments were repeated three times. Statistical significance was determined using one-way ANOVA with post hoc Tukey-Kramer multiple comparison test. *, p < 0.05 compared with non-induced controls. C, schematic of BMP-3 promoter and the identified potential HNF1A binding site is shown. The HNF1A binding site is identified in block capitals, and the base pair numbers indicated are relative to the start of the first exon. D, WT-HNF1A INS-1 cells were treated with 500 ng/ml doxycycline for 48 h. Non-induced INS-1 cells served as a control. Cells were harvested and fixed, and ChIP was performed using an HNF1A antibody or an IgG isotype control. The enrichment of the HNF1A target site was assessed by PCR with a specific primer set that spanned the identified HNF1A binding site. Samples were run on 2% agarose gel. Experiments were repeated twice with similar results. Error bars, S.E.
BMP-3 Is Expressed in Beta Cells in Mouse Pancreas and Stimulates Insulin Expression in INS-1 Cells and Beta Cells
Because a physiological role for BMP-3 in the pancreas has not yet been described, we next investigated whether BMP-3 and insulin were co-expressed in beta cells in vivo. Therefore, we performed immunohistochemistry on pancreatic tissue from rat insulin promoter DN-HNF1A and control wild-type C57BL/6JBomTac mice (22). The populations of beta cells immunopositive for insulin also showed prominent reactivity against BMP-3 and demonstrated a strong co-localization of BMP-3 and insulin in the islets from wild-type mice (Fig. 4A). In contrast, DN-HNF1A islets showed significantly reduced expression of BMP-3 in islet cells (Fig. 4). Moreover, reduced expression of BMP-3 correlated with reduced levels of insulin expression.
FIGURE 4.
BMP-3 and Insulin co-localization in pancreatic islets in vivo. A, paraffin-embedded pancreatic sections from 3-month-old wild-type C57BL/6JBomTac control mice and rat insulin promoter DN-HNF1A transgenic mice were double stained using antibodies against BMP-3 and insulin. Primary antibodies were recognized by secondary antibodies coupled to Alexa Fluor 568 (red) for BMP-3 and to FITC (green) for insulin, respectively. Cell nuclei were stained with DAPI. Confocal images were taken using 1-μm-thick optical section. Scale bar, 50 μm. In islets derived from WT animals the average pixel intensity of the BMP-3 fluorescence was significantly higher than in transgenic mice (TG). Data are represented as means ± S.E. (error bars); * p < = 0.05, Mann-Whitney U test; n = 39 (WT) and 35 (TG) pooled from three separate animals.
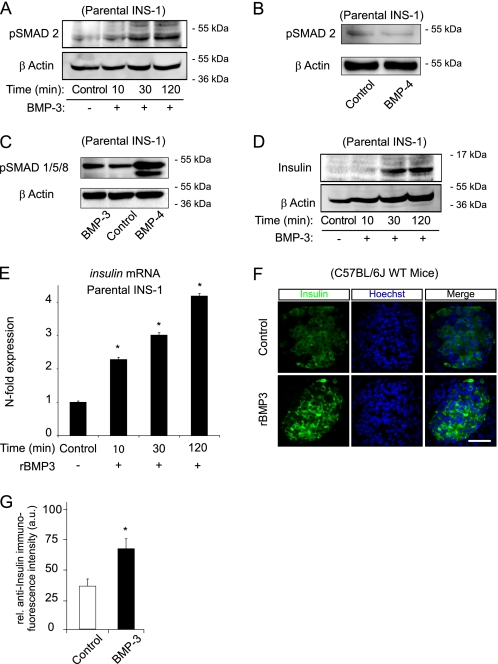
Control experiments demonstrated the expression of the canonical BMP receptors, BMP-R2 and ALK-4, in INS-1 cells (see Fig. 6, B and C). Next we explored the possibility that BMP-3 was involved in the regulation of insulin expression. The TGF-β type I/activin-receptor like serine/threonine kinase (ALK) signaling pathways initiate the ligand-induced phosphorylation of the receptor-SMAD proteins in their C termini (10). The canonical activation pathway of SMADs 1, 5, and 8 involves phosphorylation by BMP receptors, whereas SMAD2 and 3 are activated by TGF-βs, activins and BMP-3, respectively (30, 31). Indeed, treatment of parental INS-1 cells with 10 ng/ml of recombinant BMP-3 showed an increase in the C-terminal phosphorylation of SMAD2/3 (Fig. 5A). In contrast, treatment of parental INS-1 cells with 10 ng/ml recombinant human BMP-4 did not show any increased phosphorylation of SMAD2/3 (Fig. 5B), but instead increased the phosphorylation of SMAD1/5/8 (Fig. 5C), demonstrating the specificity of the BMP-3/SMAD2 response. Treatment of parental INS-1 cells with rBMP-3 potently increased insulin protein (Fig. 5D) and gene expression (Fig. 5E). Furthermore, murine organotypic pancreatic slice cultures treated with rBMP-3 for 24 h had significantly increased insulin immunofluorescence compared with control treated slices (Fig. 5, F and G).
FIGURE 6.
Treatment of INS1 cells with recombinant BMP-3 protein reverses the insulin-deficiency phenotype in HNF1A-MODY cells. A–C, INS-1 cells were incubated with doxycycline for 8, 16, 24, and 48 h to induce the expression of the Pro291fsinsC-HNF1A mutant protein. Cells were harvested for protein, and whole cell lysates were analyzed by Western blotting. Membranes were probed with polyclonal antibodies directed against phosphorylated-SMAD2 (A), BMP-receptor 2 (B), or activin-like-kinase-receptor-4 (C). β-Actin was used as a loading control. D and E, INS-1 cells were cultured in 2% serum overnight. Cells were induced with doxycycline (Dox) to overexpress the Pro291fsinsC-HNF1A mutant (D) or DN-HNF1A mutant (E) and treated with 10 ng/ml rBMP-3 in 0.05% serum at 6 mm glucose for 24 h. Cells were harvested for RNA, and insulin gene expression was analyzed by real-time qPCR and normalized to β-actin. Experiments were carried out in triplicate. Data are represented as mean ± S.E. (error bars). *, p < 0.05 different from non-induced controls. F, Pro291fsinsC-HNF1A INS-1 cells were induced with doxycycline for 24 h. Cells were then treated with 10 ng/ml of rBMP-3 for 20, 30, 60, 120, and 240 min. Whole cell lysates were analyzed by Western blotting. Membranes were probed with a polyclonal antibody raised against phosphorylated-SMAD2, and β-actin served as a loading control.
FIGURE 5.
Increased insulin expression following treatment with rBMP-3. A–D, parental INS-1 cells were precultured in reduced serum overnight and treated with 10 ng/ml rBMP-3 for 10, 30, and 120 min or 10 ng/ml rBMP-4 for 1 h in 0.05% serum medium with 6 mm glucose as indicated. Whole cell lysates were analyzed by Western blotting. Membranes were probed with polyclonal antibodies raised against phosphorylated-SMAD2 (A and B), phosphorylated SMAD1/5/8 (C), or insulin (D). Anti-β-actin immunoreactivity served as loading control. E, parental INS-1 cells were precultured in reduced serum overnight and treated with 10 ng/ml rBMP-3 for 10, 30, and 120 min in 0.05% serum medium with 6 mm glucose. After treatment, mRNA expression of insulin gene was analyzed by real-time qPCR. Expression levels were normalized to controls. Data are represented as means ± S.E. (error bars) from n = 3 cultures.*, p < 0.05 different from controls. F and G, mouse organotypic pancreatic slice cultures were incubated for 24 h with 50 ng/ml rBMP-3 or vehicle and stained with anti-insulin antibody (green); nuclei were counterstained with Hoechst (blue). Quantification of insulin immunofluorescence staining intensity was evaluated from confocal stacks of 20 islets per treatment in three independent experiments. Islets treated with rBMP-3 show significant higher levels of peripheral insulin staining. Data are represented as mean ± S.E., and statistical analysis was carried out using the Mann-Whitney U test. *, p < 0.05 compared non-treated controls. Scale bar, 50 μm.
Treatment with Recombinant BMP-3 Reverses the Pro291fsinsC-HNF1A-induced Decrease in Insulin Expression
We next determined whether exogenous BMP-3 could reverse the Pro291fsinsC-HNF1A-induced decrease in insulin gene expression previously observed in INS-1 cells (32). Western blotting experiments revealed a prominent down-regulation in the levels of phosphorylated SMAD2/3 upon induction of the mutant (Fig. 6A), demonstrating that indeed signaling through SMAD2 was significantly disturbed after the induction of Pro291fsinsC-HNF1A. In contrast, we found no evidence for a down-regulation of BMP-R2 (Fig. 6B) or ALK-4 protein levels (Fig. 6C). Next, Pro291fsinsC INS-1 cells were simultaneously treated with doxycycline and 10 ng/ml rBMP-3 for 24 h. Treatment of INS-1 cells inducibly expressing the Pro291fsinsC-HNF1A or DN-HNF1A mutant with 10 ng/ml rBMP-3 was sufficient to prevent the decrease in insulin gene expression (Fig. 6, D and E). In agreement with these data, the C-terminal phosphorylation of SMAD2/3 was significantly increased in Pro291fsinsC-HNF1A-expressing cells after treatment with 10 ng/ml rBMP-3 (Fig. 6F).
DISCUSSION
The present study demonstrates several important findings contributing to the understanding of mutant HNF1A-induced defects in the expression of genes involved in tissue differentiation and homeostasis. These include (i) the inducible expression of the common, frameshift mutation, Pro291fsinsC-HNF1A leads to the potent down-regulation of Bmp-3 at the mRNA and protein level in insulin-secreting INS-1 cells; (ii) Dual Luciferase Reporter assays reveal that the HNF1A mutant reduces Bmp-3 promoter activity, which results in the decrease in the transcriptional activity of the Bmp-3 gene and corresponding protein expression; (iii) mouse islet cells express BMP-3 in beta cells; (iv) treatment of INS-1 cells and organotypic pancreatic slice cultures with exogenous recombinant BMP-3 increases protein levels of insulin; and (v) treatment of INS-1 cells with BMP-3 completely rescues the decrease in insulin expression induced by the HNF1A frameshift mutation. Our study therefore suggests a new role for the TGF-β superfamily member BMP-3 in the regulation of insulin production and indicates that loss of BMP-3 function may be involved in the pathophysiology of HNF1A-MODY.
Given the importance of beta cell dysfunction in HNF1A-MODY, we utilized high density oligonucleotide microarrays and real-time qPCR techniques to carry out an unbiased analysis of HNF1A-induced defects in the expression of novel genes involved in beta cell function, differentiation, and regeneration. Interestingly, this Genechip array, in combination with dCHIP software, identified several known and multiple putative novel HNF1A target genes down-regulated in INS-1 cells inducibly expressing the Pro291fsinsC-HNF1A mutant. We also found several genes to be significantly up-regulated in response to Pro291fsinsC-HNF1A induction, which could be a result of compensatory or complex gene regulation. Among the genes identified, we found a significant effect of the HNF1A mutant on the expression of Bmp-3. Subsequent qPCR analyses suggested Bmp-3 to be a novel HNF1A target gene. To understand the regulation of expression of Bmp-3, we cloned and sequenced a 2.0-kb promoter fragment of the Bmp-3 gene 5′ to the transcription initiation site which was identified using the ENSEMBL database. Notably, Dual Luciferase Reporter assay experiments showed that induction of the Pro291fsinsC-HNF1A frameshift mutation decreased the transcriptional activity of the Bmp-3 promoter significantly compared with non-induced controls, suggesting that the BMP-3 cytokine is a direct transcriptional target of physiological HNF1A activity. Indeed, we could demonstrate enriched binding of HNF1A to a region within the Bmp-3 promoter with a potential HNF1A binding site.
BMP-3 and other members of the TGF-β superfamily mediate gene expression in diverse cell types and are involved in a wide range of biological functions in the pancreas, including cell proliferation, differentiation, neogenesis, and apoptosis (9, 10, 33). BMP-3 was originally thought to be a trophic factor required for bone growth because of its ability to induce bone and cartilage development (34). However, subsequent gene targeting studies revealed that BMP-3 is indeed antiosteogenic, but plays an instructive role during early pancreatic development (35, 36). Activated TGF-β superfamily members signal via dual type II and type I transmembrane/activin-like kinase serine/threonine kinase receptors and effector SMAD transcription factors (9, 10). Ligand binding and subsequent receptor activation lead to the phosphorylation and activation of SMADs which translocate to the nucleus and regulate the transcription of target genes. The ligand-specific SMADs, SMAD1, SMAD2, SMAD3, and SMAD5, are phosphorylated by activated type I TGF-β receptors. SMAD1 and SMAD5 are specific for all members of the BMP family of proteins except BMP-3. BMP-3 signals through phosphorylation of the SMADs2/3, similar to TGF-βs and activins (30, 31). It has been reported previously that TGF-β1 promotes the development of endocrine cells such as insulin-containing beta cells (37) and that activin receptor inactivation in pancreatic beta cells leads to the development of diabetes (12, 38). We here demonstrate a significant disturbance of BMP-3/SMAD2 signaling axis in response to the expression of Pro291fsinsC-HNF1A, which was reversed upon reintroduction of BMP-3. It is therefore conceivable that signaling through SMAD2 and SMAD3 is of vital importance for differentiation and maintenance of beta cell function. In support of these data, previous work has demonstrated that the conditional, pancreas-specific expression of the inhibitory SMAD7 protein, which inhibits TGF-β, activin, and BMP signaling, induces a reversible diabetic phenotype in mice (39, 40).
In contrast to BMP-3, we found little evidence for a dysregulation of other TGF-β superfamily members after induction of the Pro291fsinsC-HNF1A mutation, including BMP-4. An important role for BMP-4 and its receptor BMPR1a in beta cell function has been demonstrated previously (29). Using gene targeting approaches, mice with diminished BMP-4/BMPR1A signaling in beta cells showed decreased expression of genes involved in insulin production and release, glucose sensing, and secretion-stimulus coupling, as well as incretin signaling. These mice eventually developed diabetes due to impaired insulin secretion. Our data suggested that in the context of HNF1A-MODY and defective HNF1A signaling, BMP-3 signaling rather than BMP-4-induced pathways appears to be primarily disturbed.
Of note, further experiments demonstrated that BMP-3 was sufficient to activate insulin expression in INS-1 cells as well as in pancreatic islets. Treatment with BMP-3 also reversed the decrease in insulin expression after Pro291fsinsC-HNF1A induction, suggesting that repression of the Bmp-3 promoter led to the subsequent disturbance of insulin mRNA and protein expression. However, it should also be stated that the insulin gene promoter has direct HNF1A binding sites (17) which contribute to the decrease in insulin gene expression in response to Pro291fsinsC-HNF1A induction. Indeed, the insulin gene promoter is subject to a complex transcriptional regulation (41), and further work is necessary to determine the possible interaction between promoter elements and BMP-3/SMAD2 signaling in the regulation of insulin gene transcription.
In conclusion, our studies suggest a critical link between HNF1A-MODY-induced alterations in gene expression and growth factors of the TGF-β superfamily that control tissue differentiation and gene expression. We further propose that the BMP-3/SMAD2 pathway may play a role in regulating beta cell function and may hence present a new therapeutic target for the treatment of HNF1A-MODY or type 2 diabetes.
Supplementary Material
This work was supported by Health Research Board Grant RP/2008/14 and Science Foundation Ireland Grant 08/IN1/1949 (to J. H. M. P.) and Health Research Board Grants RP/2004/220 and RP/2007/316 (to M. M. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- MODY
- maturity-onset diabetes of the young
- AEBSF
- 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride
- BMP
- bone morphogenetic protein
- DN
- dominant negative
- HNF
- hepatocyte nuclear factor
- qPCR
- quantitative PCR
- rBMP-3
- recombinant BMP-3.
REFERENCES
- 1. Fajans S. S. (1990) Diabetes Care 13, 49–64 [DOI] [PubMed] [Google Scholar]
- 2. Bell G. I., Polonsky K. S. (2001) Nature 414, 788–791 [DOI] [PubMed] [Google Scholar]
- 3. Yamagata K., Furuta H., Oda N., Kaisaki P. J., Menzel S., Cox N. J., Fajans S. S., Signorini S., Stoffel M., Bell G. I. (1996) Nature 384, 458–460 [DOI] [PubMed] [Google Scholar]
- 4. Frayling T. M., Evans J. C., Bulman M. P., Pearson E., Allen L., Owen K., Bingham C., Hannemann M., Shepherd M., Ellard S., Hattersley A. T. (2001) Diabetes 50, S94–100 [DOI] [PubMed] [Google Scholar]
- 5. Byrne M. M., Sturis J., Menzel S., Yamagata K., Fajans S. S., Dronsfield M. J., Bain S. C., Hattersley A. T., Velho G., Froguel P., Bell G. I., Polonsky K. S. (1996) Diabetes 45, 1503–1510 [DOI] [PubMed] [Google Scholar]
- 6. Lehto M., Tuomi T., Mahtani M. M., Widén E., Forsblom C., Sarelin L., Gullström M., Isomaa B., Lehtovirta M., Hyrkkö A., Kanninen T., Orho M., Manley S., Turner R. C., Brettin T., Kirby A., Thomas J., Duyk G., Lander E., Taskinen M. R., Groop L. (1997) J. Clin. Invest. 99, 582–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weir G. C., Bonner-Weir S. (2004) Diabetes 53, S16–21 [DOI] [PubMed] [Google Scholar]
- 8. Bouwens L., Rooman I. (2005) Physiol. Rev. 85, 1255–1270 [DOI] [PubMed] [Google Scholar]
- 9. Attisano L., Wrana J. L. (2002) Science 296, 1646–1647 [DOI] [PubMed] [Google Scholar]
- 10. Feng X. H., Derynck R. (2005) Annu. Rev. Cell Dev. Biol. 21, 659–693 [DOI] [PubMed] [Google Scholar]
- 11. Massagué J., Blain S. W., Lo R. S. (2000) Cell 103, 295–309 [DOI] [PubMed] [Google Scholar]
- 12. Rane S. G., Lee J. H., Lin H. M. (2006) Cytokine Growth Factor Rev. 17, 107–119 [DOI] [PubMed] [Google Scholar]
- 13. Ball E. M., Risbridger G. P. (2001) Dev. Biol. 238, 1–12 [DOI] [PubMed] [Google Scholar]
- 14. Jiang F. X., Stanley E. G., Gonez L. J., Harrison L. C. (2002) J. Cell Sci. 115, 753–760 [DOI] [PubMed] [Google Scholar]
- 15. Wobser H., Düssmann H., Kögel D., Wang H., Reimertz C., Wollheim C. B., Byrne M. M., Prehn J. H. (2002) J. Biol. Chem. 277, 6413–6421 [DOI] [PubMed] [Google Scholar]
- 16. Wang H., Maechler P., Hagenfeldt K. A., Wollheim C. B. (1998) EMBO J. 17, 6701–6713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang H., Antinozzi P. A., Hagenfeldt K. A., Maechler P., Wollheim C. B. (2000) EMBO J. 19, 4257–4264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Asfari M., Janjic D., Meda P., Li G., Halban P. A., Wollheim C. B. (1992) Endocrinology 130, 167–178 [DOI] [PubMed] [Google Scholar]
- 19. Bonner C., Bacon S., Concannon C. G., Rizvi S. R., Baquié M., Farrelly A. M., Kilbride S. M., Dussmann H., Ward M. W., Boulanger C. M., Wollheim C. B., Graf R., Byrne M. M., Prehn J. H. (2010) Diabetes 59, 2799–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 21. Rozen S., Skaletsky H. (2000) Methods Mol. Biol. 132, 365–386 [DOI] [PubMed] [Google Scholar]
- 22. Hagenfeldt-Johansson K. A., Herrera P. L., Wang H., Gjinovci A., Ishihara H., Wollheim C. B. (2001) Endocrinology 142, 5311–5320 [DOI] [PubMed] [Google Scholar]
- 23. Akpinar P., Kuwajima S., Krützfeldt J., Stoffel M. (2005) Cell Metab. 2, 385–397 [DOI] [PubMed] [Google Scholar]
- 24. Fukui K., Yang Q., Cao Y., Takahashi N., Hatakeyama H., Wang H., Wada J., Zhang Y., Marselli L., Nammo T., Yoneda K., Onishi M., Higashiyama S., Matsuzawa Y., Gonzalez F. J., Weir G. C., Kasai H., Shimomura I., Miyagawa J., Wollheim C. B., Yamagata K. (2005) Cell Metab. 2, 373–384 [DOI] [PubMed] [Google Scholar]
- 25. Miquerol L., Lopez S., Cartier N., Tulliez M., Raymondjean M., Kahn A. (1994) J. Biol. Chem. 269, 8944–8951 [PubMed] [Google Scholar]
- 26. Servitja J. M., Pignatelli M., Maestro M. A., Cardalda C., Boj S. F., Lozano J., Blanco E., Lafuente A., McCarthy M. I., Sumoy L., Guigó R., Ferrer J. (2009) Mol. Cell. Biol. 29, 2945–2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Farrelly A. M., Wobser H., Bonner C., Anguissola S., Rehm M., Concannon C. G., Prehn J. H., Byrne M. M. (2009) Diabetologia 52, 136–144 [DOI] [PubMed] [Google Scholar]
- 28. Wang H., Gauthier B. R., Hagenfeldt-Johansson K. A., Iezzi M., Wollheim C. B. (2002) J. Biol. Chem. 277, 17564–17570 [DOI] [PubMed] [Google Scholar]
- 29. Goulley J., Dahl U., Baeza N., Mishina Y., Edlund H. (2007) Cell Metab. 5, 207–219 [DOI] [PubMed] [Google Scholar]
- 30. Verrecchia F., Chu M. L., Mauviel A. (2001) J. Biol. Chem. 276, 17058–17062 [DOI] [PubMed] [Google Scholar]
- 31. Kawabata M., Imamura T., Miyazono K. (1998) Cytokine Growth Factor Rev. 9, 49–61 [DOI] [PubMed] [Google Scholar]
- 32. Wobser H., Bonner C., Nolan J. J., Byrne M. M., Prehn J. H. (2006) Diabetologia 49, 519–526 [DOI] [PubMed] [Google Scholar]
- 33. Kim S. K., MacDonald R. J. (2002) Curr. Opin. Genet. Dev. 12, 540–547 [DOI] [PubMed] [Google Scholar]
- 34. Sampath T. K., Muthukumaran N., Reddi A. H. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 7109–7113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bahamonde M. E., Lyons K. M. (2001) J. Bone Joint Surg. Am. 83, S56–62 [PubMed] [Google Scholar]
- 36. Kim S. K., Hebrok M. (2001) Genes Dev. 15, 111–127 [DOI] [PubMed] [Google Scholar]
- 37. Sanvito F., Herrera P. L., Huarte J., Nichols A., Montesano R., Orci L., Vassalli J. D. (1994) Development 120, 3451–3462 [DOI] [PubMed] [Google Scholar]
- 38. Yamaoka T., Idehara C., Yano M., Matsushita T., Yamada T., Ii S., Moritani M., Hata J., Sugino H., Noji S., Itakura M. (1998) J. Clin. Invest. 102, 294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kuang C., Xiao Y., Liu X., Stringfield T. M., Zhang S., Wang Z., Chen Y. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1858–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Smart N. G., Apelqvist A. A., Gu X., Harmon E. B., Topper J. N., MacDonald R. J., Kim S. K. (2006) PLoS Biol. 4, e39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hay C. W., Docherty K. (2006) Diabetes 55, 3201–3213 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.