Abstract
Hypoxia is a widely occurring condition experienced by diverse organisms under numerous physiological and disease conditions. To probe the molecular mechanisms underlying hypoxia responses and tolerance, we performed a genome-wide screen to identify mutants with enhanced hypoxia tolerance in the model eukaryote, the yeast Saccharomyces cerevisiae. Yeast provides an excellent model for genomic and proteomic studies of hypoxia. We identified five genes whose deletion significantly enhanced hypoxia tolerance. They are RAI1, NSR1, BUD21, RPL20A, and RSM22, all of which encode functions involved in ribosome biogenesis. Further analysis of the deletion mutants showed that they minimized hypoxia-induced changes in polyribosome profiles and protein synthesis. Strikingly, proteomic analysis by using the iTRAQ profiling technology showed that a substantially fewer number of proteins were changed in response to hypoxia in the deletion mutants, compared with the parent strain. Computational analysis of the iTRAQ data indicated that the activities of a group of regulators were regulated by hypoxia in the wild-type parent cells, but such regulation appeared to be diminished in the deletion strains. These results show that the deletion of one of the genes involved in ribosome biogenesis leads to the reversal of hypoxia-induced changes in gene expression and related regulators. They suggest that modifying ribosomal function is an effective mechanism to minimize hypoxia-induced specific protein changes and to confer hypoxia tolerance. These results may have broad implications in understanding hypoxia responses and tolerance in diverse eukaryotes ranging from yeast to humans.
Keywords: genome-wide screen, proteomic analysis, iTRAQ, regulatory network, stress response
living organisms ranging from aquatic organisms to plants and humans can experience episodes of low oxygen availability, namely, hypoxia. Hypoxia can occur in plant roots when a field is flooded, in yeast during fermentation, or in humans at high altitudes or as a result of a heart attack or stroke. Because oxygen is critical for the survival and development of many living organisms, particularly eukaryotes, living organisms ranging from yeast to humans have developed sophisticated mechanisms to respond and adapt to the changes in oxygen levels in the environment (12, 127). In the wilderness, several species, including fossorial mammals, diving animals, and birds living at high altitudes, have acquired hypoxia tolerance, which enables them to survive under conditions of limited oxygen supply (9, 84, 92). In humans, hypoxia is associated with an array of pathological conditions, such as cancer and stroke. Mechanisms of oxygen sensing and regulation have been implicated in a wide array of physiological and pathological processes (35, 99, 127). Thus, studying hypoxia responses and resistance/tolerance is of fundamental and broad biological significance (12). The discovery of genes that are not previously known to control hypoxic sensitivity or hypoxia tolerance may reveal novel mechanisms of oxygen sensing and hypoxia responses. This may lead to the development of novel therapies for diseases whose progression is modulated by hypoxic sensitivity (118).
Because performing genome-wide functional screens in higher eukaryotes is still difficult, we performed a screen to identify genes that affected hypoxic tolerance in the yeast Saccharomyces cerevisiae. We used the yeast knockout strain collection with >5,000 viable knockout strains (Open Biosystems) (117) to identify strains that grew better under hypoxia than in air, compared with the wild-type parent strain. After a series of screens and confirmations, we identified a small group of five genes whose deletion substantially enhanced hypoxia tolerance. These five genes are RAI1, NSR1, BUD21, RPL20A, and RSM22. They are all involved in ribosome biogenesis (32, 111, 121, 126). To elucidate the molecular mechanism underlying enhanced hypoxia tolerance of the deletion strains, we performed biochemical and proteomic experiments. Interestingly, the deletion strains minimized the effect of hypoxia on protein synthesis and ribosome profiles. Additional experiments showed that hypoxia did not induce, but somewhat suppressed, unfolded protein response (UPR) (78), in the wild-type parent and knockout strains in most cases. Genome-wide proteomic analysis using iTRAQ (isobaric tags for relative and absolute quantitation) profiling technology (94) showed that hypoxia altered the levels of >200 proteins in the wild-type parent strain, including 48 proteins involved in ribosomal functions. Strikingly, the number of hypoxia-altered proteins was substantially reduced in the knockout strains with enhanced hypoxia tolerance. Particularly, the targets of a group of regulators, including Cst6 and Rap1, were regulated by hypoxia in the wild-type parent strain, but their regulation was diminished in the deletion strains. These results provide novel insights into the molecular mechanisms underlying hypoxia responses and tolerance.
MATERIALS AND METHODS
Yeast strains and plasmids.
The yeast knockout/deletion and parent BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) strains were purchased from Open Biosystems. Yeast strains with GCN4 or IRE1 deleted were generated by transforming yeast strains with PCR products containing the LEU2 gene in the middle and 44 bps of sequences flanking the open reading frame (ORF) sequence of GCN4 or IRE1 on each end. Knockout strains were confirmed by using PCR analysis of the corresponding genomic DNA. Sequences for PCR primers are available upon request. Reporters driven by unfolded protein response elements (UPREs) were provided by Dr. Peter Walter (85).
Cell growth and β-galactosidase assays.
Yeast cells were grown in synthetic complete media, as described previously (45, 56). Hypoxic (∼10 ppb O2) growth condition was created by using an anaerobic chamber (Coy Laboratory) and by filling the chamber with a mixture of 5% H2 and 95% N2 in the presence of palladium catalyst (45, 56). The oxygen level in the chamber was monitored by using the model 10 gas analyzer (Coy Laboratory). The precise level of oxygen was also detected by using a CHEMetrics rhodazine oxygen detection kit (K-7511) with the minimum detection limit at 1 ppb and a range of 0–20 ppb (56). The hypoxic state was further confirmed by measuring oxygen-controlled promoter activities, including UAS1/CYC1, ANB1, and OLE1 (45, 50, 67). Sustained cell growth under hypoxic conditions was supplemented with Tween 80 and ergosterol, as described previously (13, 45).
For β-galactosidase assays, yeast cells bearing reporter plasmids were grown in synthetic complete media in air or in a hypoxia chamber. Cells were collected after they reached an absorbance at 600 nm of ∼1.0–1.5. Collected cells were then subjected to chloroform permeabilization β-galactosidase assays (in Miller units), as described previously (45). For tunicamycin treatments, 1 μg/ml was added to the cell cultures (85).
Identification of yeast strains with enhanced hypoxia tolerance and measurement of cell growth rate.
The yeast strain collection with 5,007 viable knockout strains, as well as the wild-type parent cells, was replicated onto 96-well plates containing synthetic complete medium by using a 96-well pin tool. Cells were grown in air and in a hypoxia chamber, to an extent that allowed visual detection of differences in the growth rate of different colonies. The growth of all colonies in air and in the hypoxia chamber was scored compared with the wild-type cells. The colonies that exhibited better growth in the hypoxia chamber than in air were identified. Note that this was done compared with the wild-type cells. For example, if a strain grew significantly worse than the parent strain in air but grew about the same in a hypoxia chamber, this strain would be identified as one with enhanced hypoxia tolerance. Colonies were viewed and scored by two different researchers, and scores were assigned based on their visual estimation of cell growth on plates. In this visual stage, 148 strains were identified to have varying degrees of enhanced hypoxia tolerance. Subsequently, 20 strains with the highest scores were subjected to further quantitative measurement. They were grown in liquid cultures in air or in a hypoxia chamber. We detected cell growth rates by measuring their absorbance at multiple time points. For each strain, measurements were taken from at least three independent cultures. The measurements were repeated twice. These quantitative measurements confirmed that six of the strains exhibited substantially improved cell growth rate under hypoxia compared with the wild-type parent strain. One of the strains, which has the deletion of YGR160W, should essentially be the same as Δnsr1, since YGR160W overlaps with NSR1 and is not likely to encode an expressed protein. Therefore, only five strains were further studied. Eventually, the growth of these five strains was monitored continuously until they reached stationary phase, in air and in a hypoxia chamber. Cell growth was supplemented with Tween 80 and ergosterol. The growth curves of strains under hypoxic and normoxic conditions were plotted. The doubling times of various strains in air or under hypoxia were calculated based on the slopes of the growth curves in the exponential growth phase.
Measurement of protein synthesis and polyribosome profile analysis.
To measure the levels of protein synthesis in the wild-type and knockout strains, cells were grown in synthetic complete medium in air or in a hypoxia chamber to an absorbance (600 nm) of ∼0.6. Then 2.8 μCi of l-[35S]-methionine was added and incubated for 90 min. Cells were collected and radiolabeled proteins were extracted and measured as described (10, 101). Briefly, cells were resuspended in 20% cold trichloroacetic acid (TCA) and then heated at 100°C. The precipitate was collected by filtration through Whatman GF/C glass microfiber filters and washed with 20% TCA. The radioactivity incorporation was measured using a scintillation counter, and the protein levels in strains were expressed as the radioactivity (cpm) divided by the absorbance of cells (A600).
Polyribosome preparation and analysis were performed based on previously described procedures (7, 27). Briefly, yeast cells were grown to an A600 of 0.6–0.8, and cycloheximide was added to a final concentration of 0.1 mg/ml. Cells were collected and resuspended in lysis buffer containing 10 mM Tris·HCl (pH 7.4), 0.1 M NaCl, 30 mM MgCl2, 50 μg/ml of cycloheximide, 200 μg/ml heparin, and 0.2% diethyl pyrocarbonate. Cells were then permeabilized by agitation with glass beads. Cell lysates were collected, and A260 was measured. Approximately 6 A260 units of lysates were layered onto a 7–50% sucrose gradient, in a buffer containing 7.50 mM Tris·HCl (pH 7.4), 70 mM NH4Cl, 12 mM MgCl2, 1 mM dithiothreitol, 50 μg/ml of cycloheximide, 200 μg/ml heparin, and 0.1% diethyl pyrocarbonate (16 ml). The gradients were centrifuged in a Beckman SW32.1 rotor at 30,000 rpm for 4 h at 4°C and fractionated, and the sucrose gradient profiles were recorded by measuring A260.
To get an estimate of the levels of ribosomes associated with a defined amount of synthesized proteins in different cells, cell lysates containing 800 μg of proteins were analyzed by using the 7–50% sucrose gradient as described above. Then, fractions containing monosomes and polysomes were collected and combined. The levels of RNA in the fractions were then detected by measuring A260. The levels of RNA should reflect the levels of ribosomes correlating with the loaded amount of proteins synthesized in the cells.
Extraction of proteins and iTRAQ analysis.
Yeast wild-type and knockout cells were grown in air or in a hypoxia chamber in synthetic complete media to an A600 of ∼0.8–1.0. The cells were then collected, and extracts were prepared as described previously (45, 94). Briefly, cells were harvested and resuspended in four packed cell volumes of lysis buffer (0.1 M triethylammonium bicarbonate, 0.1% vol/vol Triton X-100, 6 M Guanidine) containing the protease inhibitor cocktail (Roche, cat. no. 11836153001). Cells were then permeabilized by agitation with three packed cell volumes of glass beads. Extracts were collected and centrifuged at 4°C at 13,000 rpm for 15 min in an Eppendorf 5417R centrifuge. Protein concentrations were measured by using the bicinchoninic acid protein assay kit (Pierce, Rockford, IL). Proteins were reduced by 2 mM Tris-(2-carboxyethyl) phosphine and alkylated by 5 mM iodoacetamide. Proteins were then precipitated by the addition of six volumes of cold acetone, collected by centrifugation, and dried in air. Subsequently, proteins were digested by trypsin and labeled with reagents from the 8plex iTRAQ labeling kit (Applied Biosystems) by following the recommended protocol. The 8plex reagents contain eight reporter group ions with molecular masses ranging from 113 to 121, allowing the detection and comparison of eight samples. We mixed 100 μg of labeled proteins from each sample together and further analyzed them. The eight samples processed were from aerobic and hypoxic parent BY4741, Δnsr1, Δrsm22, and Δrpl20a cells.
The iTRAQ samples were analyzed by the Penn State Hershey Mass Spectrometry Core facility. Prior to mass spectrometric (MS) analysis, protein samples were fractionated and separated into 15 fractions by using two-dimensional liquid chromatography. Fractions were analyzed in a data-dependent manner on an ABI 4800 MALDI TOF-TOF. MS/MS spectra were taken by using up to 2,500 laser shots per spectrum at Laser Power 3000, with CID gas Air at 1.2 to 1.3 × 10−6 Torr. Spectra were identified with 95% confidence. The database used for searches was the yeast (cerevisiae) National Center for Biotechnology Information (NCBI) database sequences containing 36,621 protein sequences, plus 156 common lab contaminants. The total protein sequences searched in database plus contaminants plus concatenated reverse decoy database contained 73,406 sequences. The local false discovery rate for the lowest ranking protein ID was <5%.
MS analysis was repeated three times. Bionformatic analyses of the data were performed by using the algorithms in the ProteinPilot software. The details of the software can be found elsewhere (e.g., http://www3.appliedbiosystems.com/cms/groups/psm_marketing/documents/generaldocuments/cms_042786.pdf). The analyses were performed following the established ways developed by the Penn State Hershey Mass Spectrometry Core facility (http://www.hmc.psu.edu/core/proteins_MassSpec/Mass_Spec_Protein_Instrumentation.htm) (17). For detected proteins, the ProteinPilot software package was used to compute protein expression ratios between hypoxia and air for each replicate experiment. We restricted analysis to proteins whose ratios were significantly different from 1.0 as assessed by ProteinPilot. To further restrict to more pronounced changes in protein expression, we applied the following additional cut-offs: 1) The protein was found to be increased or decreased in all three hypoxic samples compared with aerobic samples; 2) The ratio of the protein from at least two of the three biological replicates was >1.2 (for upregulated) or <0.8 (for downregulated); 3) The ratio average from three biological replicates was >1.3 (for upregulated) or <0.7 (for downregulated). To confirm the statistical significance of the identified proteins, we performed permutation analysis to determine the P values for the number of upregulated or downregulated proteins between air and hypoxia. We found that the total number of changed proteins identified is highly statistically significant (P values <0.001).
Protein names and ORF names were assigned based on NCBI protein IDs by using the PIR databases (http://pir.georgetown.edu/cgi-bin/batch.pl). Gene Ontology (GO) analysis of hypoxia-altered proteins was performed by using Funspec (http://funspec.med.utoronto.ca/). The identification for targets of regulators was based on the data published previously (40, 47, 68). The interaction network was identified and plotted by using Pathway Studio (version 7.1, Ariadne Genomics). The P values for all shown GO and network groups are <0.001.
RESULTS
Genome-wide screening identifies five genes whose deletion enhances hypoxia tolerance.
To systematically identify genes that are crucial for mediating hypoxia responses and tolerance, we took advantage of the yeast knockout strain collection with >5,000 viable knockout strains (117). The initial screen was performed by visually screening cells grown on 96-well plates in a hypoxia chamber or in air. In this screen, we identified 148 strains that showed varying degrees of improved growth under hypoxia, using the wild-type parent strain as a reference standard. Of those, 20 strains showed substantial improvement in growth under hypoxia compared with the parent strain. We then monitored the growth of these 20 strains in liquid cultures by measuring their absorbance at 600 nm. These quantitative measurements allowed us to identify six strains that grew substantially better under hypoxia compared with the parent strain.
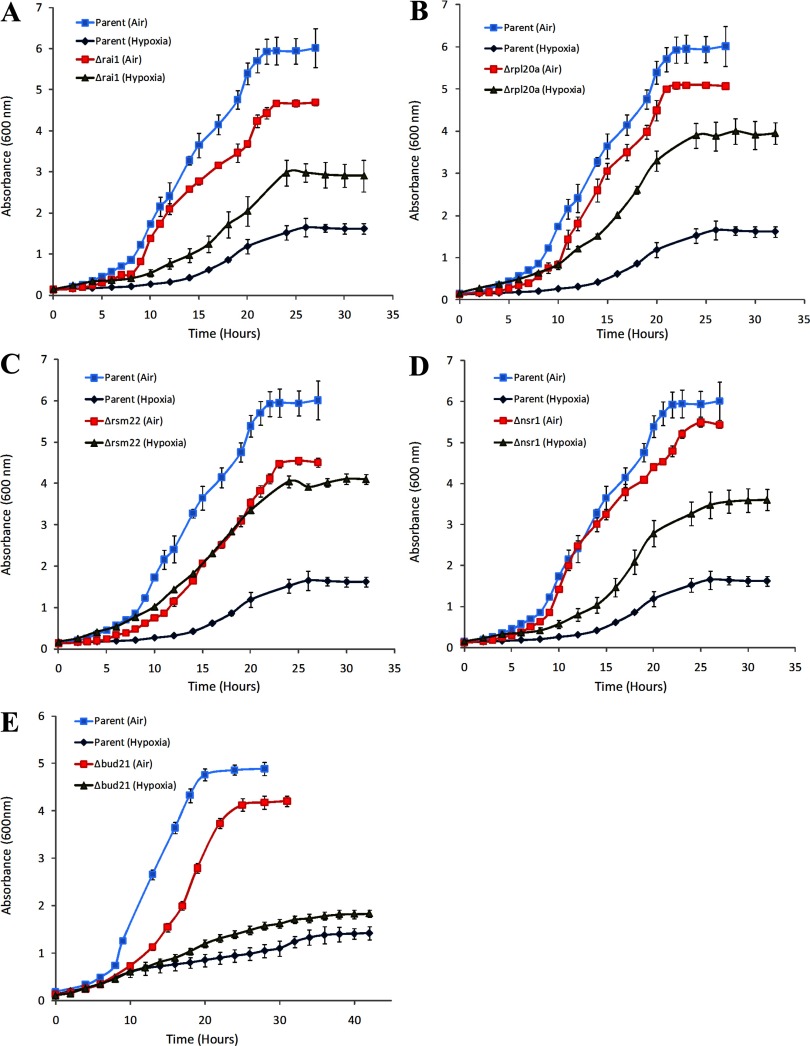
These six strains are knockout strains of RAI1, NSR1, BUD21, RPL20A, RSM22, and YGR160W. YGR160W overlaps with NSR1 on the opposite strand and is unlikely to be an expressed gene (SGD). Thus, it is highly likely that the phenotype of Δygr160w derives from the deletion of NSR1. Furthermore, the identification of such two overlapping deletions strongly supports the validity of this screen. For further analyses, we focused on RAI1, NSR1, BUD21, RPL20A, and RSM22. Figure 1 shows the growth curves of Δrai1, Δnsr1, Δrpl20a, Δbud21, and Δrsm22 compared with the parent. Clearly, these deletion strains grew substantially better under hypoxia compared to the parent, although they grew worse than the parent in air. Intriguingly, all five genes, RAI1, NSR1, BUD21, RPL20A, and RSM22, encode functions involved in ribosome biogenesis (32, 111, 121, 126): Rai1 and Nsr1 proteins are required for pre-rRNA processing and ribosome biogenesis; Bud21, Rpl20a, and Rsm22 proteins are components of the small ribosomal subunit (SSU) processosome, the large (60S) ribosomal subunit, and the mitochondrial small ribosomal subunit, respectively.
Fig. 1.
The comparison of cell growth in air and under hypoxia between the parent BY4741 and Δrai1 (A), Δrpl20a (B), Δrsm22 (C), Δnsr1 (D), or Δbud21 (E) knockout strains. Yeast cells were grown in air or under hypoxia in synthetic complete medium containing glucose. Cells were collected at the indicated time points, and absorbance at 600 nm was measured. The data plotted here are averaged from at least 3 independent cultures. Welch 2-sample t-tests were performed to determine the statistical significance of the difference between the growth of hypoxic and normoxic wild-type or mutant cells. The P values were calculated by using the R program. The P values for the data at time points greater than 15 h were <0.005.
Deletion of the genes relating to ribosome biogenesis minimizes the effects of hypoxia on protein synthesis and polyribosome profiles.
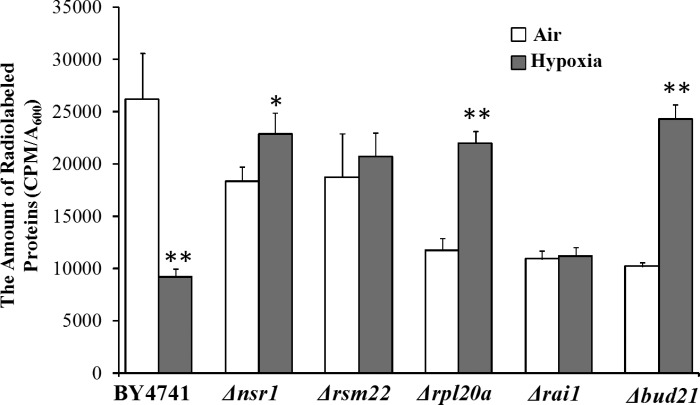
Tο gain insights into how the deletion of such ribosomal genes enhances hypoxia tolerance, we examined the effects of their deletion on global protein synthesis. We measured and compared the levels of global protein synthesis by the parent and knockout strains, grown in air and under hypoxia. As expected, in air, protein synthesis was less efficient in the knockout strains compared with the parent, likely due to reduced ribosome function (Fig. 2). Likewise, protein synthesis in the wild-type parent strain was reduced under hypoxia compared with that in air. Notably, in hypoxic Δnsr1, Δbud21, Δrsm22, and Δrpl20a cells, the levels of total protein synthesis were not significantly reduced (Fig. 2). The levels of total protein synthesis in hypoxic Δbud21 and Δrpl20a cells were somewhat increased compared with the normoxic cells (Fig. 2). The data suggest that the effect of hypoxia on protein synthesis is reversed or minimized in the deletion strains.
Fig. 2.
A comparison of the levels of total protein synthesis in the parent BY4741 and knockout strains. Yeast cells were grown in air or under hypoxia for 24 h, to an A600 of ∼0.6. Then, the cells were labeled with l-[35S]-methionine for 90 min. Cells were collected and lysed. Proteins were collected by TCA precipitation. Radiolabeled proteins were detected by using a scintillation counter. Data plotted here are averages from 5 independent replicates. Welch 2-sample t-tests were performed to compare hypoxic with normoxic wild-type or mutant cells. The P values were calculated by using the R program. *P values of < 0.005; **P values of < 0.001.
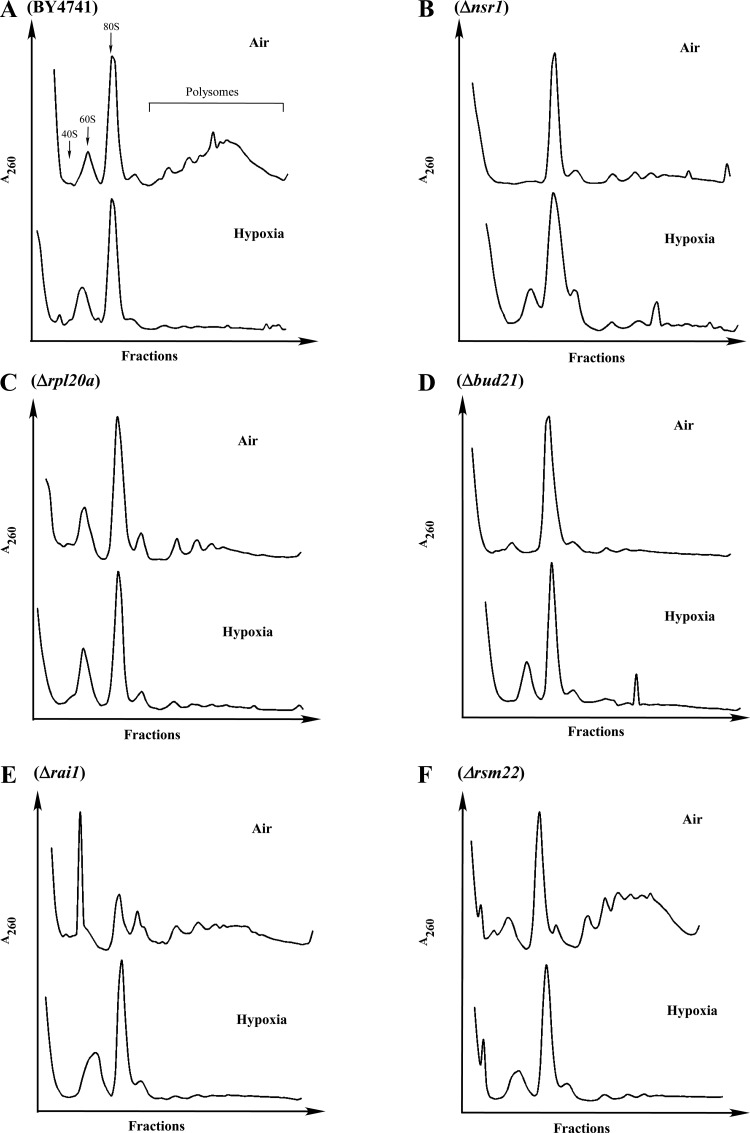
To probe the molecular basis for this reversal of hypoxia effect, we performed polyribosome profile analysis (Fig. 3). In the wild-type parent cells (Fig. 3A), under hypoxia, the level of polyribosomes was significantly diminished. This is consistent with a previous study examining the effect of hypoxia on polyribosome profiles (61). Except for RSM22 (Fig. 3F), which encodes for mitochondrial ribosomal protein of the small subunit, deletion of other genes diminished the levels of polyribosomes to a varying degree, even in normoxic cells (Fig. 3, B–E). Consequently, in these deletion strains, the effect of hypoxia on the levels of polyribosomes was greatly diminished (Fig. 3, B–E). These results suggest that the deletion of genes involved in cytosolic ribosome biogenesis diminishes the effect of hypoxia on protein synthesis by preadopting a hypoxic polyribosome profile.
Fig. 3.
The polyribosome profiles of the wild-type parent and deletion mutant cells. Polyribosome preparation was performed by using wild-type BY4741 (A), Δnsr1 (B), Δrpl20a (C), Δbud21 (D), Δrai1 (E), and Δrsm22 (F) cells, grown under normoxic and hypoxic conditions, respectively. Then polyribosome profiles were analyzed by using 7–50% sucrose gradients.
Hypoxia does not induce the activity of reporters driven by unfolded protein response element.
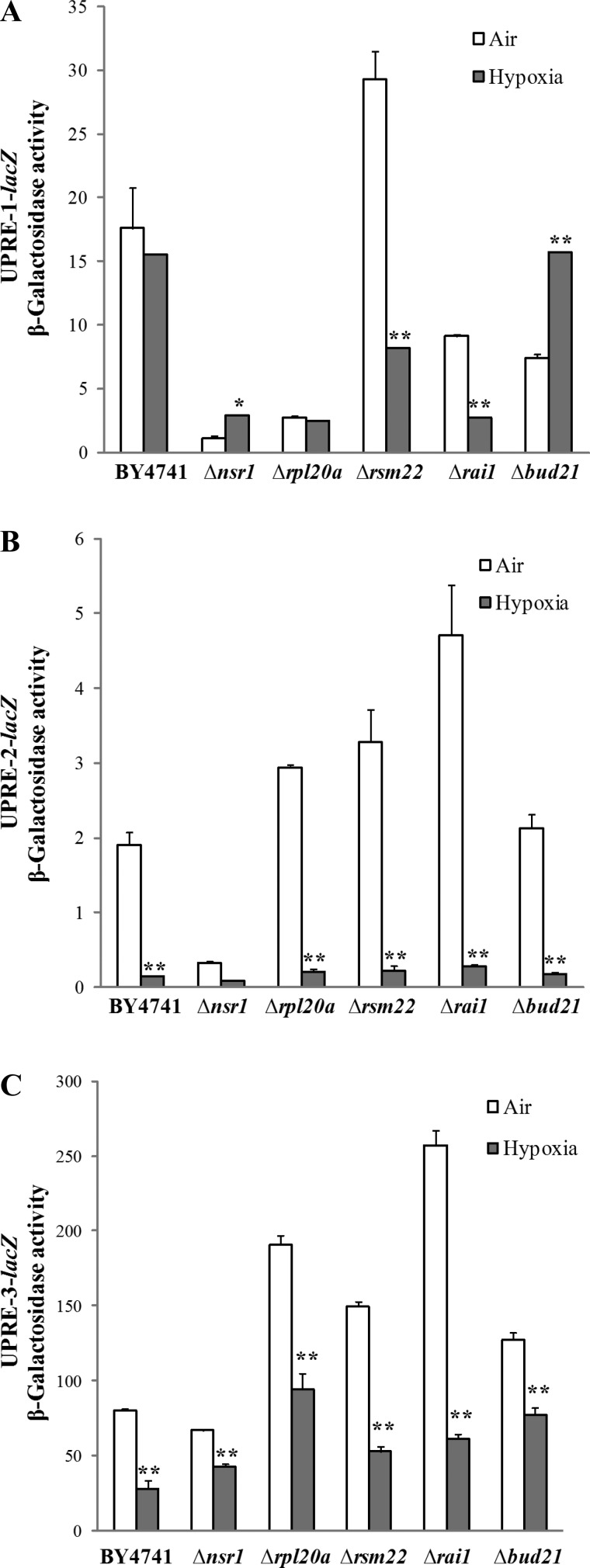
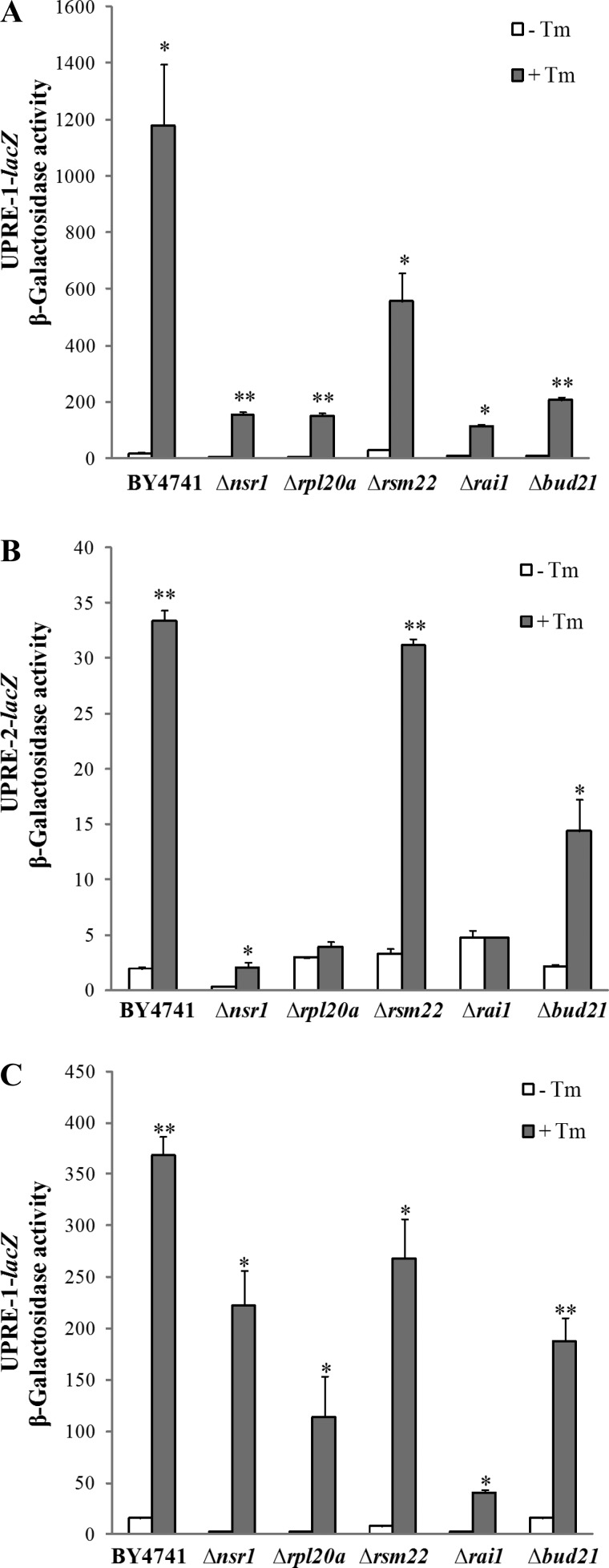
A previous study has suggested that hypoxia induces UPR and that reducing unfolded protein toxicity may contribute to hypoxia tolerance in Caenorhabditis elegans (2). We reasoned that UPR may be involved in hypoxia tolerance in yeast as well. We therefore examined the effect of hypoxia on UPR in the wild-type and knockout strains by using reporters driven by the UPRE identified previously (85). We measured the activities of three UPRE-driven reporters bearing three different UPREs (Fig. 4). These UPREs were identified from analysis of all UPR target genes and should be very representative of UPR (85, 113). We found that hypoxia did not induce, but suppressed, the activity of the UPRE-driven reporters in the wild-type parent strain and in knockout strains, in most cases (Fig. 4). Only in Δnsr1 and Δbud21 cells, the activity of UPRE-1-driven reporter was somewhat induced by hypoxia. As a control, we showed that the activity of all three reporters was highly induced by tunicamycin [a potent inducer of endoplasmic reticulum (ER) stress and UPR] in the wild-type and knockout strains, in most cases (Fig. 5, A and B). Even in hypoxic cells, tunicamycin strongly induced UPR (see Fig. 5C). This demonstrates that the activity of these reporters indeed reflects UPR (85). These results suggest that hypoxia does not induce UPR in the wild-type parent strain and the mutant strains, except for Δnsr1 and Δbud21.
Fig. 4.
The effect of hypoxia on the activity of unfolded protein response element (UPRE)-driven reporters in the parent BY4741 and knockout strains. A: the effect of hypoxia on the activity of the UPRE-1-driven reporter. B: the effect of hypoxia on the activity of the UPRE-2-driven reporter. C: the effect of hypoxia on the activity of the UPRE-3-driven reporter. The parent BY4741, Δrai1, Δrpl20a, Δrsm22, Δnsr1, and Δbud21 cells bearing a UPRE-driven reporter were grown in air or in a hypoxia chamber. Cells were collected, and β-galactosidase activities (in Miller units) were measured and plotted here. Data shown here are averages of data from at least three independent cultures. Welch 2-sample t-tests were performed to compare hypoxic with normoxic wild-type or mutant cells. The P values were calculated by using the R program. *P values of < 0.005; **P values of < 0.001.
Fig. 5.
The effect of tunicamycin (Tm) on the activity of UPRE-driven reporters in the parent BY4741 and knockout strains. A: the effect of Tm on the activity of the UPRE-1-driven reporter in normoxic cells. B: the effect of Tm on the activity of the UPRE-2-driven reporter in normoxic cells. C: the effect of Tm on the activity of the UPRE-1-driven reporter in hypoxic cells. The parent BY4741, Δrai1, Δrpl20a, Δrsm22, Δnsr1, and Δbud21 knockout cells bearing a UPRE-driven reporter were grown in the presence (+) or absence (−) of Tm. Cells were collected, and β-galactosidase activities (in Miller units) were measured and plotted here. Data shown here are averages of data from at least three independent cultures. Welch 2-sample t-tests were performed to compare Tm-treated with untreated wild-type or mutant cells. The P values were calculated by using the R program. *P values of < 0.005; **P values of < 0.001.
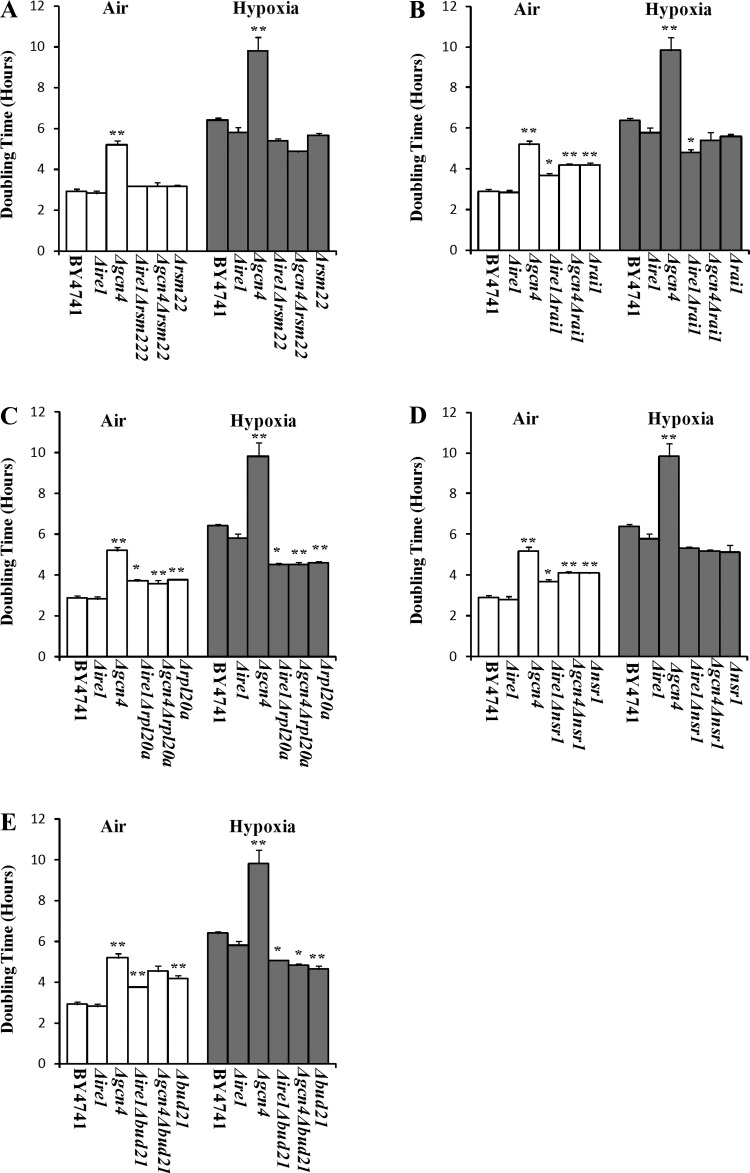
To further ascertain the role of UPR in hypoxia tolerance, we decided to examine the effect of knocking out key components of the UPR signaling pathway (16, 39, 78, 85) on the hypoxia tolerance of the mutants with enhanced hypoxia tolerance. Gcn4 was shown to be essential for UPR and is required for the induction of a majority of UPR target genes (85). Ire1 is a key component mediating UPR signaling (54, 78). We therefore examined the effect of deleting GCN4 and IRE1 on the hypoxia tolerance of the mutant strains, respectively. As shown in Fig. 6, deletion of IRE1 per se slightly reduced the doubling time of the wild-type cells (i.e., enhanced cell growth) both in air and under hypoxia. Deletion of GCN4 per se substantially reduced cell growth in both aerobic and hypoxic parent cells. Remarkably, deletion of GCN4 or IRE1 in Δrai1, Δrpl20a, and Δrsm22 did not increase, but somewhat reduced, their doubling times in the double knockout strains (see Fig. 6, A–C). In Δnsr1 and Δbud21 cells, deletion of GCN4 or IRE1 slightly increased the doubling times in the double knockout strains (Fig. 6, D and E). These results are consistent with the results from the measurement of reporter activities. They show that the deletion of UPR signaling components does not abolish hypoxia tolerance in the strains with ribosome biogenesis genes deleted. They suggest that hypoxia does not induce UPR and that UPR is not responsible for enhanced hypoxia tolerance in Δrai1, Δrpl20a, and Δrsm22 cells. UPR may be somewhat induced and partially contributes to enhanced hypoxia tolerance in Δnsr1 and Δbud21 cells. Together, these results suggest that multiple mechanisms may contribute to enhanced hypoxia tolerance.
Fig. 6.
The effects of deletion of GCN4 and IRE1 on the doubling times of the wild-type parent and knockout strains with enhanced hypoxia tolerance. A: a comparison of the effect of deletion of GCN4 and IRE1 on the doubling times of the wild-type parent and Δrsm22 strains. B: a comparison of the effect of deletion of GCN4 and IRE1 on the doubling times of the wild-type parent and Δrai1 strains. C: a comparison of the effect of deletion of GCN4 and IRE1 on the doubling times of the wild-type parent and Δrpl20a strains. D: a comparison of the effect of deletion of GCN4 and IRE1 on the doubling times of the wild-type parent and Δnsr1 strains. E: a comparison of the effect of deletion of GCN4 and IRE1 on the doubling times of the wild-type parent and Δbud21 strains. The data plotted here are averages from at least 3 independent cultures. Welch 2-sample t-tests were performed to compare hypoxic with normoxic wild-type or mutant cells. The P values were calculated by using the R program. *P values of < 0.005; **P values of < 0.001.
Genome-wide proteomic analysis shows that hypoxia-induced changes in the numbers of specific proteins are minimized in strains with enhanced hypoxia tolerance.
To further probe the molecular mechanisms underlying hypoxia tolerance, we decided to perform proteomic analyses. We detected and compared the changes in protein levels induced by hypoxia in the wild-type parent strain and in three deletion strains with enhanced hypoxia tolerance: Δnsr1, Δrsm22, and Δrpl20a. These three deletion strains were selected because they exhibited strong hypoxia tolerance (Fig. 1) and because they represent different functional groups. Nsr1 represents proteins involved in ribosome biogenesis; Rpl20A represents cytosolic ribosome subunits; and Rsm22 represents mitochondrial ribosomal subunits. We used iTRAQ profiling technology (1, 94). This technique allowed us to detect and compare the levels of all cellular proteins in samples prepared from multiple yeast cultures grown under various conditions, by using MS. We used 8plex iTRAQ analysis to simultaneously detect and compare protein levels in eight different samples. They are protein samples prepared from aerobic and hypoxic wild-type parent, Δnsr1, Δrsm22, and Δrpl20a cells. We repeated the iTRAQ analysis three times successfully. Over 700 proteins were identified and quantified every time. We then used all data from three independent iTRAQ experiments to identify upregulated and downregulated proteins by hypoxia in the wild-type, Δnsr1, Δrsm22, and Δrpl20a cells. A combined list of the three sets of original data is shown in Supplemental Table S1.1 We performed a series of statistical analyses and filtering steps to identify hypoxia-altered proteins; see materials and methods.
We identified and compared hypoxia-altered proteins in the wild-type parent, Δnsr1, Δrsm22, and Δrpl20a cells (Table 1). We also performed GO analysis of the hypoxia-altered proteins to identify the distinct functional categories of proteins. The result of GO analysis is summarized in Table 1. A complete list of different functional categories of hypoxia-upregulated proteins in the wild-type cells is shown in Table 2, while a complete list of hypoxia-downregulated proteins is shown in Table 3. For a complete list of hypoxia-altered proteins in knockout cells, please see Supplemental Table S1. These results suggest several important points or conclusions: First, the data clearly show that the number of hypoxia-altered proteins was dramatically reduced in the knockout strains with enhanced hypoxia tolerance (Table 1). Only a fraction of hypoxia-altered proteins in the wild-type parent cells were altered in the knockout strains (Tables 2 and 3). Second, strikingly, the number of hypoxia-altered ribosomal and related proteins was even more dramatically reduced in knockout strains (Tables 1–3): fewer than half of those altered in the wild-type strain were altered in the knockout strains. The changes in many upregulated or downregulated proteins in the wild-type cells did not occur in the knockout strains (Tables 2 and 3). These proteins mainly included components of ribosome subunits, as well as some involved in regulating ribosome activity. Another class of proteins whose changes were reduced was metal ion binding proteins (Tables 1–3). These results show that at the molecular and genome-wide levels, a major difference between the wild-type and knockout strains with enhanced hypoxia tolerance is that fewer changes in the levels of specific proteins were induced by hypoxia in the strains with enhanced hypoxia tolerance.
Table 1.
Summary of proteins whose levels are altered by hypoxia in the wild-type and mutant strains
| Protein Category | BY4741 | Δrsm22 | Δnsr1 | Δrpl20a |
|---|---|---|---|---|
| Total | 233:122+111− | 132:84+48− | 135:70+65− | 80:46+34− |
| Ribosome and related proteins | 48 | 17 | 20 | 10 |
| Protein folding and stabilization | 12 | 12 | 10 | 10 |
| Ergosterol synthesis | 9 | 5 | 8 | 2 |
| Protein transport | 6 | 2 | 2 | 4 |
| Metal ion binding | 30 | 10 | 17 | 10 |
| Oxidation reduction | 14 | 13 | 14 | 10 |
+Upregulated proteins; −downregulated proteins.
Table 2.
List of upregulated proteins in the wild-type BY4741 strain
| Protein ID | ORF Name | Common Name | Δrsm22 | Δnsr1 | Δrpl20a |
|---|---|---|---|---|---|
| Ribosome and related proteins | |||||
| gi 6321315 | YGL123W | RPS2 | |||
| gi 730687 | YHL015W | RPS20 | |||
| gi 85695429 | YJL190C | RPS22A | |||
| gi 9755341 | YMR143W | RPS16A | down | ||
| gi 6320949 | YER102W | RPS8B | up | ||
| gi 642297 | YDR025W | RPS11A | |||
| gi 6323196 | YLR167W | RPS31 | |||
| gi 730453 | YJL191W | RPS14B | |||
| gi 6681849 | YNL162W | ||||
| gi 747904 | YLR075W | RPL10 | |||
| gi 914973 | YPR102C | RPL11A | |||
| gi 730454 | YHL001W | RPL14B; RPL14A | |||
| gi 730534 | YLR029C | RPL15A | |||
| gi 6322284 | YJL177W | RPL17B | |||
| gi 6324452 | YOL120C | RPL18A | |||
| gi 940843 | YOR312C | RPL20A; RPL20B | |||
| gi 6321587 | YGR148C | RPL24B | up | ||
| gi 6324445 | YOL127W | RPL25 | |||
| gi 6324637 | YOR063W | RPL3 | |||
| gi 6435679 | YGL030W | RPL30 | |||
| gi 914877 | YML073C | RPL6A | |||
| gi 717063 | YLR448W | RPL6B | down | ||
| gi 6324351 | YNR024W | up | up | up | |
| gi 6322507 | YJR047C | ANB1 | up | up | up |
| gi 6320863 | YER025W | GCD11 | |||
| gi 6322769 | YKL081W | TEF4 | down | ||
| gi 6321724 | YGR285C | ZUO1 | up | ||
| gi 6325018 | YPL237W | SUI3 | |||
| gi 6320788 | YEL047C | up | |||
| gi 6319724 | YBR247C | ENP1 | up | ||
| gi 887610 | YMR229C | RRP5 | |||
| Protein folding and stabilization | |||||
| gi 730923 | YDR188W | CCT6 | |||
| gi 417149 | YAL005C | SSA1 | up | ||
| gi 717089 | YJR045C | SSC1 | |||
| gi 6323002 | YLL026W | HSP104 | up | ||
| gi 99031946 | YPL240C | HSP82 | |||
| gi 6320420 | YDR214W | AHA1 | |||
| gi 729768 | YNL209W | SSB2 | up | ||
| Ergosterol synthesis | |||||
| gi 730943 | YPL028W | ERG10 | up | up | |
| gi 927536 | YML126C | ERG13 | up | ||
| gi 854482 | YML008C | ERG6 | up | up | |
| gi 6324811 | YOR237W | HES1 | up | up | |
| gi 6324371 | YNR043W | MVD1 | up | ||
| gi 6321795 | YHR007C | ERG11 | up | up | |
| gi 6321614 | YGR175C | ERG1 | up | up | |
| gi 6323129 | YLR100W | ERG27 | up | ||
| gi 730126 | YHR042W | NCP1 | up | up | |
| Metal ion binding | |||||
| gi 6321203 | YGL234W | ADE5,7 | |||
| gi 728694 | YGL156W | AMS1 | up | ||
| gi 6319279 | YAL038W | CDC19 | |||
| gi 870734 | YNL118C | DCP2 | |||
| RRRRRgi 731716 | YHR132C | ECM14 | |||
| gi 817866 | YMR108W | ILV2 | |||
| gi 854590 | YJR016C | ILV3 | |||
| gi 6322746 | YKL103C | LAP4 | up | ||
| gi 6321429 | YGL009C | LEU1 | |||
| gi 6319380 | YBL091C | MAP2 | |||
| gi 6321679 | YGR240C | PFK1 | |||
| gi 6320016 | YDL185W | TFP1 | |||
| gi 809445 | YPR074C | TKL1 | down | ||
| RRRRRgi 6324515 | YOL057W | ||||
| gi 809587 | YPR022C | ||||
| gi 49065653 | YCL030C | HIS4 | up | ||
| gi 6322434 | YJL026W | RNR2 | up | up | |
| Oxidation reduction | |||||
| gi 790495 | YGR204W | ADE3 | |||
| gi 6324950 | YOR374W | ALD4 | |||
| gi 6322666 | YKL182W | FAS1 | up | up | |
| gi 854531 | YPL231W | FAS2 | up | up | |
| gi 6319986 | YDL215C | GDH2 | up | ||
| gi 854428 | YDR044W | HEM13 | up | up | up |
| gi 7245389 | YJR139C | HOM6 | |||
| gi 849175 | YDR353W | TRR1 | |||
| gi 927720 | YDR453C | TSA2 | up | up | |
| gi 6321399 | YGL039W | up | up | ||
| gi 683675 | YDL022W | GPD1 | up | ||
| Others | |||||
| gi 6321290 | YGL148W | ARO2 | |||
| gi 642806 | YDR001C | NTH1 | |||
| gi 99031872 | |||||
| RRRRRgi 886910 | YGR218W | CRM1 | |||
| gi 6323367 | YLR335W | NUP2 | |||
| gi 817876 | YMR116C | ASC1 | |||
| gi 6322174 | YIL015W | BAR1 | |||
| gi 731024 | YHR208W | BAT1 | |||
| gi 6321721 | YGR282C | BGL2 | |||
| gi 6325209 | YPL048W | CAM1 | up | ||
| gi 84029497 | YLR270W | DCS1 | |||
| gi 854478 | YML012W | ERV25 | |||
| gi 732946 | YMR246W | FAA4 | up | ||
| gi 88192429 | YER136W | GDI1 | |||
| gi 6319809 | YCL040W | GLK1 | up | ||
| gi 728687 | YDR232W | HEM1 | |||
| gi 6320896 | YER055C | HIS1 | |||
| gi 3793 | YGL253W | HXK2 | down | ||
| gi 731512 | YER127W | LCP5 | down | ||
| gi 6321540 | YGR103W | NOP7 | |||
| gi 172140 | YMR205C | PFK2 | |||
| gi 6319673 | YBR196C | PGI1 | |||
| gi 728653 | YMR006C | PLB2 | up | ||
| gi 994830 | YNL055C | POR2; POR1 | up | up | |
| gi 6320235 | YDR032C | PST2 | up | ||
| gi 4255 | YBR218C | PYC2 | down | ||
| gi 929855 | YNL098C | RAS2 | |||
| gi 6320066 | YDL135C | RDI1 | up | ||
| gi 172372 | YBR049C | REB1 | down | ||
| gi 74645057 | YDL204W | RTN2 | |||
| gi 730701 | YER043C | SAH1 | |||
| gi 6319691 | YBR214W | SDS24 | up | ||
| gi 807961 | YMR079W | SEC14 | |||
| gi 6322594 | YJR134C | SGM1 | |||
| gi 6321225 | YGL213C | SKI8 | |||
| gi 6323207 | YLR178C | TFS1 | up | up | |
| gi 6319638 | YBR162C | TOS1 | |||
| gi 6325053 | YPL203W | TPK2 | |||
| gi 836765 | YFR010W | UBP6 | |||
| gi 7546471 | YEL021W | URA3 | down | ||
| gi 6321531 | YGR094W | VAS1 | |||
| gi 6324804 | YOR230W | WTM1 | up | up | |
| gi 913916 | YKL196C | YKT6 | |||
| gi 6322723 | YKL126W | YPK1 | |||
| gi 74676373 | YLR301W | up | |||
| gi 6319530 | YBR056W | ||||
| gi 854503 | YNL134C | ||||
Those proteins that were upregulated or downregulated in the mutant strains are indicated. ORF, open reading frame.
Table 3.
List of downregulated proteins in the wild-type BY4741 strain
| Protein ID | ORF Name | Common Name | Δrsm22 | Δnsr1 | Δrpl20a |
|---|---|---|---|---|---|
| Ribosome and related proteins | |||||
| gi 6322681 | YKL167C | MRP49 | down | ||
| gi 6324451 | YOL121C | RPS19A | |||
| gi 895891 | YJR123W | RPS5 | |||
| gi 6321653 | YGR214W | RPS0A | |||
| gi 927770 | YDR500C | RPL37B | down | ||
| gi 927724 | YDR447C | RPS17B; RPS17A | |||
| gi 6325006 | YPL249C-A | RPL36B | |||
| gi 6322272 | YJL189W | RPL39 | down | ||
| gi 6320122 | YDL081C | RPP1A | |||
| gi 6320073 | YDL130W | RPP1B | |||
| gi 6324534 | YOL039W | RPP2A | up | ||
| gi 927315 | YDR382W | RPP2B | |||
| gi 6324433 | YOL139C | CDC33 | |||
| gi 992578 | YAL003W | EFB1 | down | ||
| gi 6320801 | YEL034W | HYP2 | down | down | |
| gi 6322794 | YKL056C | ||||
| gi 6323953 | YMR295C | ||||
| Protein folding and stabilization | |||||
| gi 6323562 | YML078W | CPR3 | down | ||
| gi 854502 | YNL135C | FPR1 | up | ||
| gi 6324806 | YOR232W | MGE1 | |||
| gi 940372 | YOR265W | RBL2 | up | ||
| gi 731762 | YHR193C | EGD2 | |||
| Protein transport | |||||
| gi 6324344 | YNR017W | MAS6 | |||
| gi 731508 | YER120W | SCS2 | |||
| gi 836710 | YFL045C | SEC53 | |||
| gi 730698 | YLR250W | SSP120 | |||
| gi 6321519 | YGR082W | TOM20 | down | ||
| RRRRRgi 46562125 | YOR069W | VPS5 | |||
| Metal ion binding | |||||
| gi 6323326 | YLR295C | ATP14 | down | ||
| gi 6319426 | YBL045C | COR1 | down | ||
| gi 6321846 | YHR053C | CUP1-1; CUP1-2 | down | ||
| RRRRRgi 6322803 | YKL048C | ELM1 | |||
| gi 805042 | YPR062W | FCY1 | down | ||
| gi 6729856 | YBR011C | IPP1 | |||
| gi 729984 | YER003C | PMI40 | |||
| gi 6321251 | YGL187C | COX4 | down | ||
| gi 6321842 | YHR051W | COX6 | up | ||
| gi 6322531 | YJR070C | LIA1 | |||
| gi 6980688 | YJR104C | SOD1 | down | down | down |
| gi 88984161 | YGR234W | YHB1 | down | ||
| gi 984175 | YDR143C | SAN1 | down | ||
| Oxidation reduction | |||||
| gi 731458 | YER042W | MXR1 | |||
| gi 6319407 | YBL064C | PRX1 | up | up | |
| gi 836788 | YFR033C | QCR6 | down | down | |
| gi 6322633 | YKL216W | URA1 | up | ||
| gi 731693 | YHR104W | GRE3 | |||
| Others | |||||
| gi 6320240 | YDR035W | ARO3 | |||
| gi 73921293 | YOL109W | ZEO1 | down | down | |
| gi 6324298 | YNL030W | – | down | ||
| gi 9864778 | YOR240W | – | |||
| gi 728682 | YDR226W | ADK1 | |||
| gi 18266826 | YCL050C | APA1 | |||
| gi 731820 | YIL062C | ARC15 | |||
| gi 728968 | YER177W | BMH1 | |||
| gi 74676483 | YPL178W | CBC2 | down | ||
| gi 83578104 | YOR074C | CDC21 | |||
| gi 6324328 | YNR001C | CIT1 | |||
| gi 6319585 | YBR109C | CMD1 | up | ||
| gi 664873 | YLR429W | CRN1 | |||
| gi 798919 | YDR068W | DOS2 | |||
| gi 974204 | YBR078W | ECM33 | |||
| gi 6319655 | YBR177C | EHT1 | down | down | |
| gi 6323112 | YLR083C | EMP70 | |||
| gi 6319814 | YCL035C | GRX1 | up | up | |
| gi 927781 | YDR513W | GRX2 | down | ||
| gi 731801 | YIL041W | GVP36 | down | ||
| gi 74655018 | YDL223C | HBT1 | |||
| gi 6319439 | YBL032W | HEK2 | down | ||
| gi 6324297 | YNL031C | HHT2 | |||
| gi 731467 | YER057C | HMF1 | |||
| gi 728681 | YDR225W | HTA1 | down | ||
| gi 6320020 | YDL181W | INH1 | down | down | |
| gi 731523 | YER146W | LSM5 | |||
| gi 965089 | YPL004C | LSP1 | down | down | down |
| gi 6320071 | YDL131W | LYS21 | down | ||
| gi 6321332 | YGL106W | MLC1 | down | ||
| gi 791112 | YNL074C | MLF3 | down | ||
| gi 731437 | YER009W | NTF2 | up | down | |
| gi 730269 | YIR006C | PAN1 | |||
| gi 755798 | YGR193C | PDX1 | |||
| gi 6319441 | YBL030C | PET9 | down | down | |
| gi 6321523 | YGR086C | PIL1 | down | ||
| gi 6320798 | YEL037C | RAD23 | |||
| gi 6322288 | YJL173C | RFA3 | |||
| gi 83754253 | YOL005C | RPB11 | |||
| gi 6320106 | YDL097C | RPN6 | down | ||
| gi 83754248 | YPR187W | RPO26 | down | ||
| gi 6319854 | YCR009C | RVS161 | down | ||
| gi 6325038 | YPL218W | SAR1 | up | ||
| gi 6320049 | YDL153C | SAS10 | down | ||
| gi 74676529 | YOR007C | SGT2 | |||
| gi 886922 | YGR229C | SMI1 | |||
| gi 731411 | YEL026W | SNU13 | down | ||
| gi 119389383 | YML010W YGR063C | SPT5 | up | ||
| gi 3417405 | YDL185W | TFP1 | |||
| gi 6323072 | YLR043C | TRX1 | up | up | |
| gi 914876 | YDR092W | UBC13 | |||
| gi 6322828 | YKL024C | URA6 | |||
| gi 731777 | YIL008W | URM1 | down | ||
| gi 6324907 | YOR332W | VMA4 | |||
| gi 836758 | YFR003C | YPI1 | up | up | |
| gi 6320589 | YDR381W | YRA1 | |||
| gi 6324221 | YNL108C | up | |||
| gi 33438776 | YCR075W-A | ||||
| gi 74676469 | YOR051C | ||||
| gi 6325072 | YPL184C | ||||
| gi 790501 | YGR210C | ||||
| gi 786320 | YPR172W | ||||
| gi 33438816 | YIL002W-A | ||||
| gi 74676371 | YLR287C | ||||
| gi 798921 | YDR070C | ||||
Those proteins that were upregulated or downregulated in the mutant strains are indicated.
To determine which regulators may be involved in mediating hypoxia-induced changes in protein levels and in minimizing the changes in the deletion strains, we identified regulators whose targets were preferentially enriched among the hypoxia-altered proteins identified by iTRAQ. We used the previous databases that identified the targets for various regulators (40, 47, 68). As shown in Table 4, the targets of 21 regulators, including Aft1, Cst6, Mga2, and Rap1, were found to be regulated by hypoxia. This suggests that these 21 regulators were affected by hypoxia and that they played significant roles in mediating hypoxia responses. The identification of Mga2, a previously known hypoxia-responsive transcription factor (46, 51), supports the validity of our proteomic data. Interestingly, in the deletion strains, the targets of many regulators were no longer affected by hypoxia. Particularly, targets of only one and five regulators of the 21 regulators (Table 4) were found be regulated in Δrpl20a and Δnsr1 cells, respectively. In Δrsm22 cells, targets of 12 of the 21 regulators were affected. In addition, targets of five other regulators were also affected. These results strongly suggest that in Δrpl20a and Δnsr1 cells, fewer regulators were affected by hypoxia, thereby reducing the number of hypoxia-altered targets.
Table 4.
List of regulators whose targets are regulated by hypoxia
| Regulator | BY4741 | Δnsr1 | Δrpl20a | Δrsm22 |
|---|---|---|---|---|
| Aft1 | 16 | 16 | ||
| Cst6 | 88 | 36 | 41 | |
| Fhl1 | 16 | |||
| Gcr1 | 96 | 48 | 27 | 50 |
| Gln3 | 10 | |||
| Hap4 | 7 | 8 | ||
| Hfi1 | 32 | 20 | ||
| Hsf1 | 63 | 29 | 33 | |
| Mga2 | 17 | 20 | ||
| Rap1 | 78 | 37 | ||
| Ric1 | 9 | 7 | ||
| Rim101 | 9 | |||
| Sin3 | 18 | 13 | ||
| Sin4 | 35 | |||
| Sfp1 | 57 | 29 | ||
| Snf2 | 43 | |||
| Snf6 | 50 | |||
| Spt10 | 53 | 25 | ||
| Spt20 | 55 | 27 | 22 | |
| Swi3 | 33 | |||
| Tup1 | 26 | |||
| Hap2 | 8 | |||
| Cin5 | 13 | |||
| Met4 | 3 | |||
| Msn2 | 7 | |||
| Msn4 | 9 | |||
| Taf14 | 8 |
DISCUSSION
In this report, we describe our effort to identify genes controlling hypoxia tolerance or sensitivity in an unbiased, genome-wide manner and to probe the molecular mechanisms underlying hypoxia tolerance. Our data reveal several important findings: First, we found that all five identified genes whose deletion led to significant improvement in hypoxia tolerance are involved in cytosolic or mitochondrial ribosome biogenesis. Second, we found that the levels of total protein synthesis in most knockout strains with enhanced hypoxia tolerance, unlike that in the wild-type strain, were not diminished by hypoxia. Additionally, polyribosome profile analysis shows that under hypoxia, the effect of deleting ribosome biogenesis genes was minimized. Third, our genome-wide proteomic analysis showed that hypoxia caused substantially fewer numbers of changes in specific proteins in the knockout strains with enhanced hypoxia tolerance. Fourth, target analysis suggests that in Δrpl20a and Δnsr1 cells, fewer regulators were affected by hypoxia, thereby reducing the number of targets affected by hypoxia. Finally, we found that hypoxia did not induce UPR in the wild-type parent strain, and the disruption of the UPR signaling pathway did not abolish the enhanced hypoxia tolerance in the knockout strains. Although some chaperones involved in protein folding, such as Ssa1, were induced by hypoxia (See Table 2), the genes and proteins characteristics of UPR (113) were not induced. These findings provide novel insights into the molecular events underlying hypoxia responses and tolerance. Below we discuss the implications of our findings in the broader context of understanding hypoxia response and tolerance in eukaryotes.
Altered ribosome biogenesis and protein synthesis can have a significant impact on hypoxia response and tolerance.
Due to the variations in oxygen availability in the wilderness, many vertebrate animal species, including diving mammals and birds, reptiles, amphibians, and fishes, have developed the ability to survive hypoxia (9, 84, 92). One key strategy in hypoxia adaptation and tolerance is to conserve cellular energy via profound metabolic suppression (9, 84, 92). This strategy is also used by hypoxia-tolerant Drosophila melanogaster strains isolated in the laboratory (6, 129). Generally speaking, organisms can conserve energy by downregulating energy turnover and by upregulating energy efficiency. One mechanism to conserve energy is the suppression of protein synthesis (43). This has been found in diverse organisms ranging from flies to mammals (28, 42, 76, 89, 90, 103, 109, 124). Likewise, in yeast, protein synthesis is suppressed in response to hypoxia, as shown by the reduced total protein synthesis in the wild-type parent cells under hypoxia (Fig. 2). However, in the selected deletion strains with enhanced hypoxia tolerance, the cells continue to proliferate well, despite hypoxia (Fig. 1). In this case, protein synthesis needs to be maintained to support cell proliferation, as shown in Fig. 2. This is different from hypoxia tolerance exhibited by hypoxia-tolerant animals in the wilderness (9, 84, 92), which generally involves the suppression of all activities and no increase in cell proliferation. In our selected yeast strains with increased cell proliferation (compared to the parent), it appears that the cells acquired hypoxia tolerance by optimizing the efficiency of protein synthesis by altering ribosome biogenesis.
In this report, the data show that the deletion of RAI1, NSR1, BUD21, RPL20A, or RSM22 significantly increased yeast cell growth rate under hypoxia, leading to enhanced hypoxia tolerance. All five genes encode functions required for ribosome biogenesis (32, 111, 121, 126). Except for Bud21, the other four proteins, Rai1, Nsr1, Rpl20A, and Rsm22 have homologs in mammalian cells. Rai1 and Nsr1 proteins are required for pre-rRNA processing and ribosome biogenesis (64, 65, 111). Rai1 binds to and stabilizes the exoribonuclease Rat1 (111). It has decapping endonuclease activity and is required for pre-rRNA processing (52). The mammalian homolog of Rai1 is DOM3Z, whose function is not yet characterized (121). Nsr1 is a nucleolar protein that binds nuclear localization sequences (64). It is required for pre-rRNA processing and ribosome biogenesis (65). Its mammalian homolog is nucleolin. Interestingly, nucleolin has been shown to mediate translational control of collagen prolyl 4-hydroxylase in response to hypoxia (23). Hence, it is conceivable that this protein can affect protein synthesis in many eukaryotes, although its precise mechanisms of action may differ in different organisms. Bud21 is a component of small ribosomal subunit (SSU) processosome that contains U3 snoRNA (36, 83). It is involved in rRNA processing and 40S ribosome subunit assembly (37). Rpl20A and Rsm22 proteins are components of the large (60S) ribosomal subunit and the mitochondrial small ribosomal subunit, respectively (79, 96, 104). The mammalian homolog of Rpl20A is the ribosome protein L18a. It has been suggested that L18a may interact with RNA and promote translation (20). Rsm22 also has a mammalian homolog, suggesting that it is important for the function of the mitochondrial ribosome (96).
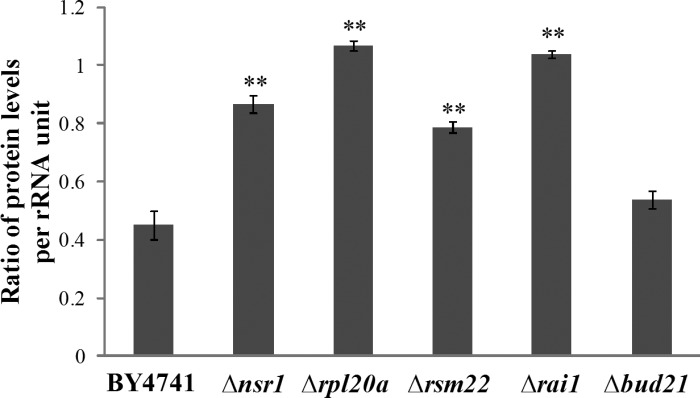
Ribosome biosynthesis consumes a vast amount of resources in rapidly growing cells; it accounts for 60% of total transcription, 50% of RNA polymerase II transcription, and 90% of mRNA splicing (25, 30, 115, 116). It is therefore a key control point for cell growth and division in both yeast and mammalian cells (8, 19). Thus, it would be advantageous for cells to minimize the production of ribosomes, particularly the wasteful, uncoordinated production of different components, and optimize their efficiency. Here, data from proteomic analyses show that the expression of a high number of ribosomal proteins is altered by hypoxia in wild-type cells (Table 1). Specifically, under hypoxia, some ribosomal proteins are upregulated, whereas some others are downregulated (Tables 1–3). This would likely make ribosome biosynthesis and ribosomal function less efficiently. It appears that the deletion of certain genes involved in ribosome biogenesis, such as nsr1, rsm22, and rpl20a, would rectify, at least partially, such an unnecessary consumption of cellular energy. As such, cells would become more tolerant to hypoxia. Polyribosome profile analysis (Fig. 3) showed that under hypoxia, the levels of polyribosomes are greatly reduced, and that the deletion of the genes involved in ribosome biogenesis preadopts a hypoxia-like polyribosome profile and minimizes the effect of hypoxia. Under hypoxia, in deletion mutants, ribosomes may act more efficiently, thereby minimizing the negative effects of hypoxia. Indeed, when we estimated the efficiency of ribosomes in the cells, we found that ribosomes were much less efficient in protein synthesis in hypoxic parent cells compared with the normoxic parent cells (ratio <0.5, Fig. 7). Notably, in Δrpl20a, Δrai1, and Δnsr1 cells, ribosome efficiency in hypoxic cells was significantly increased compared with the wild-type cells, although the efficiency in hypoxic Δrsm22 and Δnsr1 cells was still less than in the corresponding normoxic cells (ratio <1.0). This increase in ribosome efficiency correlates with enhanced growth in hypoxic deletion mutant cells (Fig. 1). Recent evidence suggests that the efficiency of ribosome and protein synthesis can result from enhanced ribosome recycling and/or reduced ribosome congestion (29, 58). Deletion of one of the five genes may significantly reverse the adverse effect of hypoxia on ribosome efficiency by affecting both ribosome recycling and congestion.
Fig. 7.
The ratio of total protein levels per rRNA unit in hypoxic cells to those in normoxic cells. Cell extracts containing proteins and ribosomes were prepared from the indicated hypoxic and normoxic cells. Extracts containing 800 μg of proteins were fractionated by using 7–50% sucrose gradient as described in materials and methods. Fractions containing ribosomes were pooled. The rRNA levels were detected by measuring A260. The rRNA levels in the fractions should reflect the amounts of ribosomes. Ribosome efficiency in hypoxic cells compared with normoxic cells was estimated by dividing the total protein levels per rRNA unit in hypoxic parent or mutant cells with those in the corresponding normoxic cells. Welch 2-sample t-tests were performed to compare hypoxic with normoxic wild-type or mutant cells. The P values were calculated by using the R program. **P values of < 0.001.
Interestingly, recent experiments (107) showed that a specific reduction of 60S ribosome subunit levels, due to gene deletion or chemical inhibition, slows aging in yeast. Similarly, a recent screen in C. elegans (2) showed that a reduction-of-function mutant of an arginyl-transfer RNA (t-RNA) synthetase, an enzyme essential for protein synthesis, causes hypoxia resistance. These and our results all point to one thing: Altering the production and function of the protein synthesis machinery to avoid unnecessary consumption of cellular energy can be an effective mechanism in cellular adaptation to the environment and in survival under stress.
It is worth noting that the expression of the five genes, RAI1, NSR1, BUD21, RPL20A, and RSM22, is generally suppressed by hypoxia and by an array of other stress conditions in yeast. Their expression has been found to be downregulated in hypoxic cells (56, 60), in aging cells (125), in cells experiencing heat shock (21, 34), in cells treated with hydrogen peroxide or arsenite (100, 112), in cells treated with rapamycin (21, 41, 48, 93), and in cells under nitrogen starvation (11). These results suggest that the suppression of their function may be generally beneficial for cells to survive and tolerate stress. This is consistent with our data showing that their deletion causes enhanced hypoxia tolerance.
Suppression of hypoxia responsiveness of a network of regulators may underlie minimized protein changes and hypoxia tolerance in the deletion strains.
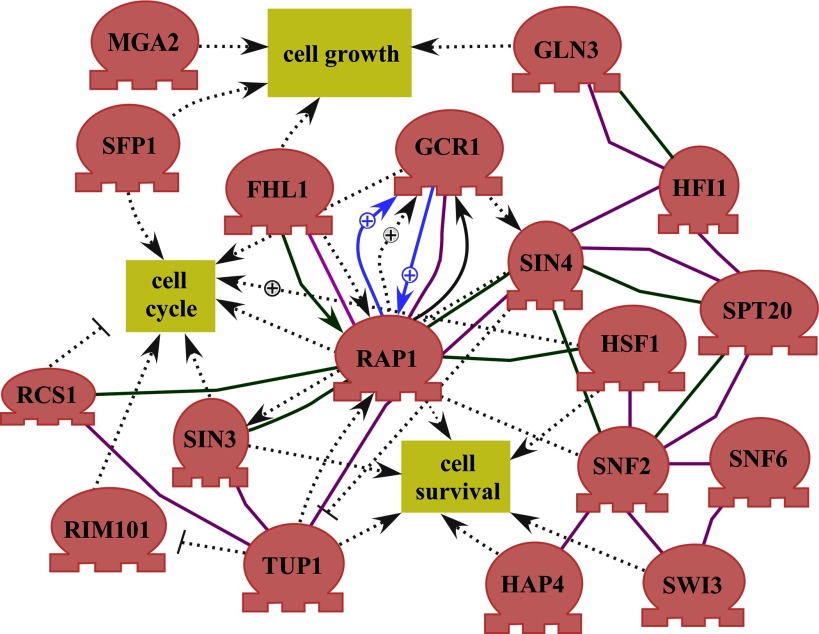
Computational analysis of proteomic data showed that many targets of a group of regulators were selectively altered by hypoxia in the wild-type parent cells (Table 4). This suggests that the activities of these regulators may be regulated by hypoxia, and they may mediate the changes in their targets induced by hypoxia. Network analysis of these regulators showed that many regulators are highly connected and interact with each other genetically and biochemically (Fig. 8) (5, 18, 44, 59, 72, 80, 81, 88, 91, 95, 98, 106, 120, 123, 128). Many of the regulators are components of global regulatory complexes. They include the components of the SWI/SNF complex (Snf2, Snf6, and Swi3) (14, 62, 87, 102), the SAGA complex (Hfi1 and Spt20) (38, 108, 119), and the Sin3-Rpd3 histone deacetylase complex (Sin3) (53, 63). The other regulators, Aft1, Cst6, Gcr1, Hsf1, Mga2, Rap1, Sfp1, and Spt10, are known DNA-binding transcriptional regulators (SGD) (15, 22, 33, 49, 50, 55, 66, 71, 122). Notably, many of the regulators are involved in the control of cell growth, cell cycle, and cell survival (Fig. 8). This provides an explanation for reduced cell growth under hypoxia (Fig. 1). Further characterization of the regulators shown in Fig. 8 should reveal how they contribute to the decreased protein changes in hypoxia-tolerant yeast strains and may uncover novel mechanisms underlying oxygen sensing.
Fig. 8.
The cell growth and regulatory network formed by regulators whose targets were altered by hypoxia. The network was constructed by using Pathway Studio (version 7.1, Ariadne Genomics). Shown here are the regulators that exhibit interactions and connections with other regulators or cell growth. Each line or arrow indicates a previously identified biochemical or genetic interaction. “+” indicates a positive effect, while a stop line indicates a negative effect.
These results suggest that a network of regulators are involved in mediating hypoxia response and tolerance in yeast. They may provide insights into the mechanisms underlying hypoxia tolerance in higher eukaryotes. In mammalian cells, one important mechanism underlying hypoxia response involves the hypoxia-inducible factors (HIFs) and their regulators prolyl hydroxylases (PHDs) and an asparaginyl hydroxylase named FIH (3, 24, 69, 77). In air, PHDs and FIH modify HIFα subunits and cause their degradation. PHDs and FIH sense oxygen because they use oxygen as a substrate. Although there is no homolog of PHDs and FIH found in yeast, analogous mechanisms operate to mediate oxygen sensing and hypoxia responses. For example, oxygen regulation of the Mga2 regulator activity is mediated by proteasome-dependent processing of the protein (46, 50, 51). Additionally, yeast can sense oxygen via heme synthesis, because oxygen is a required substrate for heme synthesis (45).
Our data show that in the deletion mutants, particularly in Δrpl20a and Δnsr1 cells, the number of regulators whose activity is affected by hypoxia is greatly reduced (Table 4). This suggests that in Δrpl20a and Δnsr1 cells, the changes in the activities of these regulators by hypoxia are suppressed. As a result, the changes in their targets are also suppressed, leading to increased hypoxia tolerance. Our analysis suggests that one molecular mechanism underlying hypoxia tolerance is the suppression of hypoxia-responsive regulators, thereby minimizing changes in protein synthesis and cellular energy consumption. Notably, this idea is also consistent with results from studies of hypoxia tolerance in other eukaryotes. For example, deficiency or inhibition of oxygen sensor PHD1 induces hypoxia tolerance in mice (4). Perhaps the suppression of hypoxia responsiveness of oxygen regulators is a common way to gain hypoxia tolerance, although the underlying molecular mechanisms of energy conservation may vary in different cases.
In Δrsm22 cells, many targets of 12 of the 21 regulators were still altered by hypoxia (Table 4). This suggests that the suppression of the regulators may not be as important to hypoxia tolerance in Δrsm22 cells compared with Δrpl20a and Δnsr1 cells. Interestingly, in Δrsm22 cells, targets of additional five regulators were altered by hypoxia (Table 4), suggesting that the induced hypoxia responsiveness of these five regulators may contribute to hypoxia tolerance. Four of these regulators, Cin5, Met4, Msn2, and Msn4, are transcription factors induced by stress. Cin5 (Yap4) is a bZIP (basic leucine zipper) transcription factor of the yeast AP1 family (26). It is induced by several stress conditions, such as oxidative and hyperosmotic stress (82, 86, 105). It also mediates pleiotropic drug resistance and salt tolerance (31, 74). Met4 is also a bZIP transcription factor (57). It may also be involved in stress response (114). Msn2 and Msn4 mediate the general stress response in yeast (73, 97). Evidently, in Δrsm22 cells, these stress regulators are selectively induced by hypoxia. Very likely, both the induction of stress regulators and partial suppression of the hypoxia responsiveness of other regulators (Table 4 and Fig. 8), contribute to the induced expression of their targets and enhanced hypoxia tolerance.
Hypoxia does not induce UPR in yeast.
Hypoxia has been shown to induce UPR in mammalian cancer cells (118). However, our data show that hypoxia does not induce UPR generally in yeast, although the levels of several chaperones, such as Ssa1 and Hsp104, were induced by hypoxia (Table 2). The activity of reporters driven by UPREs was not induced by hypoxia in the wild-type cells. Furthermore, very few (<10) of the previously identified >300 UPR target genes were induced by hypoxia, based on previous microarray data (56, 113) and the proteomic data shown here (Tables 2 and 3). Only in Δnsr1 and Δbud21 cells, hypoxia somewhat increased the activity of the UPRE-1-driven reporter (Fig. 4A). Likewise, in these two strains, deletion of the UPR signaling component, GCN4 or IRE1, did not further decrease the doubling time of Δnsr1 and Δbud21 cells under hypoxia (Fig. 6, D and E). In other strains with enhanced hypoxia tolerance, the deletion of GCN4 or IRE1 caused further decrease of doubling time under hypoxia (Fig. 6, A–C). These results show that UPR is not generally induced by hypoxia and that lack of UPR signaling does not abolish enhanced hypoxia tolerance. Only in Δnsr1 and Δbud21 cells, UPR may play a minor role in hypoxia responses and enhanced hypoxia tolerance. Notably, deletion of GCN4 greatly increased the doubling times of wild-type cells in both air and under hypoxia (Fig. 6, A–E), likely due to its important role in controlling amino acid metabolism. Strikingly, under hypoxic conditions, its deletion did not, however, cause a significant increase in the doubling times of the knockout strains with enhanced hypoxia tolerance (Fig. 6, A–E). This further supports the idea that UPR is not critical for enhanced hypoxia tolerance in these selected strains with enhanced hypoxia tolerance. UPR is a cry for help in response to ER stress. However, many organisms, particularly the yeast S. cerevisiae, likely constantly experience variations in oxygen availability in their natural environment. They have likely adapted to hypoxia, so that no ER stress is induced by hypoxia. Hence, UPR is not necessary to cope with hypoxia.
In summary, our studies here have identified a network of regulators (Table 4 and Fig. 8) that are likely to play important roles in mediating hypoxia responses and tolerance. Further investigation of these potential regulators can provide an in-depth understanding of the molecular mechanisms underlying hypoxia responses and tolerance. Our results suggest that an efficient mechanism to enhance hypoxia tolerance is to minimize the molecular changes induced by hypoxia. At the gene expression level, hypoxia is known to induce changes in the expression of a large number of genes in diverse organisms ranging from yeast to humans (56, 60, 70, 75, 110). Likewise, our data here show that the levels of many proteins are also changed by hypoxia in wild-type yeast cells (Tables 1–3). However, it appears that most hypoxia-induced changes are not necessary or even beneficial to cell survival under hypoxia, because mutant cells with enhanced hypoxia tolerance exhibited fewer changes. Very likely, with fewer changes in specific proteins and regulators in the deletion strains, cells can use cellular energy in a more efficient manner, thereby leading to enhanced hypoxia tolerance. Perhaps lack of certain proteins involved in ribosome biogenesis is an efficient mechanism to minimize such hypoxia-induced changes.
GRANTS
This work was supported by National Institute of General Medical Sciences Grant GM-62246 (L. Zhang).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Bruce and Anne Stanley at the Mass Spectrometry Core Research Facility at Penn State for performing iTRAQ analysis of our samples. We also thank Dr. Peter Walter for providing UPR reporter plasmids. We are grateful to Dr. Xiaotu Ma for assistance in the analysis of iTRAQ proteomic data and to Steve Lianoglou and Dr. Christina Leslie for identifying targets of regulators.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1. Aggarwal K, Choe LH, Lee KH. Shotgun proteomics using the iTRAQ isobaric tags. Brief Funct Genomic Proteomic 5: 112–120, 2006. [DOI] [PubMed] [Google Scholar]
- 2. Anderson LL, Mao X, Scott BA, Crowder CM. Survival from hypoxia in C. elegans by inactivation of aminoacyl-tRNA synthetases. Science 323: 630–633, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aragones J, Fraisl P, Baes M, Carmeliet P. Oxygen sensors at the crossroad of metabolism. Cell Metab 9: 11–22, 2009. [DOI] [PubMed] [Google Scholar]
- 4. Aragones J, Schneider M, Van Geyte K, Fraisl P, Dresselaers T, Mazzone M, Dirkx R, Zacchigna S, Lemieux H, Jeoung NH, Lambrechts D, Bishop T, Lafuste P, Diez-Juan A, Harten SK, Van Noten P, De Bock K, Willam C, Tjwa M, Grosfeld A, Navet R, Moons L, Van dendriessche T, Deroose C, Wijeyekoon B, Nuyts J, Jordan B, Silasi-Mansat R, Lupu F, Dewerchin M, Pugh C, Salmon P, Mortelmans L, Gallez B, Gorus F, Buyse J, Sluse F, Harris RA, Gnaiger E, Hespel P, Van Hecke P, Schuit F, VanVeldhoven P, Ratcliffe P, Baes M, Maxwell P, Carmeliet P. Deficiency or inhibition of oxygen sensor Phd1 induces hypoxia tolerance by reprogramming basal metabolism. Nat Genet 40: 170–180, 2008. [DOI] [PubMed] [Google Scholar]
- 5. Arechiga-Carvajal ET, Ruiz-Herrera J. The RIM101/pacC homologue from the basidiomycete Ustilago maydis is functional in multiple pH-sensitive phenomena. Eukaryot Cell 4: 999–1008, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Azad P, Zhou D, Russo E, Haddad GG. Distinct mechanisms underlying tolerance to intermittent and constant hypoxia in Drosophila melanogaster. PLoS One 4: e5371, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baim SB, Pietras DF, Eustice DC, Sherman F. A mutation allowing an mRNA secondary structure diminishes translation of Saccharomyces cerevisiae iso-1-cytochrome c. Mol Cell Biol 5: 1839–1846, 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bernstein KA, Bleichert F, Bean JM, Cross FR, Baserga SJ. Ribosome biogenesis is sensed at the Start cell cycle checkpoint. Mol Biol Cell 18: 953–964, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bickler PE, Buck LT. Hypoxia tolerance in reptiles, amphibians, and fishes: life with variable oxygen availability. Annu Rev Physiol 69: 145–170, 2007. [DOI] [PubMed] [Google Scholar]
- 10. Blomberg A. Global changes in protein synthesis during adaptation of the yeast Saccharomyces cerevisiae to 0.7 M NaCl. J Bacteriol 177: 3563–3572, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bradley PH, Brauer MJ, Rabinowitz JD, Troyanskaya OG. Coordinated concentration changes of transcripts and metabolites in Saccharomyces cerevisiae. PLoS Comp Biol 5: e1000270, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bunn HF, Poyton RO. Oxygen sensing and molecular adaptation to hypoxia. Physiol Rev 76: 839–885, 1996. [DOI] [PubMed] [Google Scholar]
- 13. Burke PV, Kwast KE, Everts F, Poyton RO. A fermentor system for regulating oxygen at low concentrations in cultures of Saccharomyces cerevisiae. Appl Environ Microbiol 64: 1040–1044, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cairns BR, Kim YJ, Sayre MH, Laurent BC, Kornberg RD. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc Natl Acad Sci USA 91: 1950–1954, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chellappa R, Kandasamy P, Oh CS, Jiang Y, Vemula M, Martin CE. The membrane proteins, Spt23p and Mga2p, play distinct roles in the activation of Saccharomyces cerevisiae OLE1 gene expression. Fatty acid-mediated regulation of Mga2p activity is independent of its proteolytic processing into a soluble transcription activator. J Biol Chem 276: 43548–43556, 2001. [DOI] [PubMed] [Google Scholar]
- 16. Chen Y, Feldman DE, Deng C, Brown JA, De Giacomo AF, Gaw AF, Shi G, Le QT, Brown JM, Koong AC. Identification of mitogen-activated protein kinase signaling pathways that confer resistance to endoplasmic reticulum stress in Saccharomyces cerevisiae. Mol Cancer Res 3: 669–677, 2005. [DOI] [PubMed] [Google Scholar]
- 17. Culnan DM, Cooney RN, Stanley B, Lynch CJ. Apolipoprotein A-IV, a putative satiety/antiatherogenic factor, rises after gastric bypass. Obesity (Silver Spring) 17: 46–52, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Davie JK, Kane CM. Genetic interactions between TFIIS and the Swi-Snf chromatin-remodeling complex. Mol Cell Biol 20: 5960–5973, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dez C, Tollervey D. Ribosome synthesis meets the cell cycle. Curr Opin Microbiol 7: 631–637, 2004. [DOI] [PubMed] [Google Scholar]
- 20. Dhar D, Mapa K, Pudi R, Srinivasan P, Bodhinathan K, Das S. Human ribosomal protein L18a interacts with hepatitis C virus internal ribosome entry site. Arch Virol 151: 509–524, 2006. [DOI] [PubMed] [Google Scholar]
- 21. Duvel K, Santhanam A, Garrett S, Schneper L, Broach JR. Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol Cell 11: 1467–1478, 2003. [DOI] [PubMed] [Google Scholar]
- 22. Eriksson PR, Mendiratta G, McLaughlin NB, Wolfsberg TG, Marino-Ramirez L, Pompa TA, Jainerin M, Landsman D, Shen CH, Clark DJ. Global regulation by the yeast Spt10 protein is mediated through chromatin structure and the histone upstream activating sequence elements. Mol Cell Biol 25: 9127–9137, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fahling M, Mrowka R, Steege A, Nebrich G, Perlewitz A, Persson PB, Thiele BJ. Translational control of collagen prolyl 4-hydroxylase-alpha(I) gene expression under hypoxia. J Biol Chem 281: 26089–26101, 2006. [DOI] [PubMed] [Google Scholar]
- 24. Fandrey J, Gorr TA, Gassmann M. Regulating cellular oxygen sensing by hydroxylation. Cardiovasc Res 71: 642–651, 2006. [DOI] [PubMed] [Google Scholar]
- 25. Fatica A, Tollervey D. Making ribosomes. Curr Opin Cell Biol 14: 313–318, 2002. [DOI] [PubMed] [Google Scholar]
- 26. Fernandes L, Rodrigues-Pousada C, Struhl K. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol Cell Biol 17: 6982–6993, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fingerman I, Nagaraj V, Norris D, Vershon AK. Sfp1 plays a key role in yeast ribosome biogenesis. Eukaryot Cell 2: 1061–1068, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fraser KP, Houlihan DF, Lutz PL, Leone-Kabler S, Manuel L, Brechin JG. Complete suppression of protein synthesis during anoxia with no post-anoxia protein synthesis debt in the red-eared slider turtle Trachemys scripta elegans. J Exp Biol 204: 4353–4360, 2001. [DOI] [PubMed] [Google Scholar]
- 29. Fredrick K, Ibba M. How the sequence of a gene can tune its translation. Cell 141: 227–229, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene 313: 17–42, 2003. [DOI] [PubMed] [Google Scholar]
- 31. Furuchi T, Ishikawa H, Miura N, Ishizuka M, Kajiya K, Kuge S, Naganuma A. Two nuclear proteins, Cin5 and Ydr259c, confer resistance to cisplatin in Saccharomyces cerevisiae. Mol Pharmacol 59: 470–474, 2001. [DOI] [PubMed] [Google Scholar]
- 32. Gallagher JE, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, Baserga SJ. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev 18: 2506–2517, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Garcia-Gimeno MA, Struhl K. Aca1 and Aca2, ATF/CREB activators in Saccharomyces cerevisiae, are important for carbon source utilization but not the response to stress. Mol Cell Biol 20: 4340–4349, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Giaccia AJ, Simon MC, Johnson R. The biology of hypoxia: the role of oxygen sensing in development, normal function, and disease. Genes Dev 18: 2183–2194, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schafer T, Kuster B, Tschochner H, Tollervey D, Gavin AC, Hurt E. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol Cell 10: 105–115, 2002. [DOI] [PubMed] [Google Scholar]
- 37. Granneman S, Baserga SJ. Ribosome biogenesis: of knobs and RNA processing. Exp Cell Res 296: 43–50, 2004. [DOI] [PubMed] [Google Scholar]
- 38. Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev 11: 1640–1650, 1997. [DOI] [PubMed] [Google Scholar]
- 39. Griffith DA, Delipala C, Leadsham J, Jarvis SM, Oesterhelt D. A novel yeast expression system for the overproduction of quality-controlled membrane proteins. FEBS Lett 553: 45–50, 2003. [DOI] [PubMed] [Google Scholar]
- 40. Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA. Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hardwick JS, Kuruvilla FG, Tong JK, Shamji AF, Schreiber SL. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc Natl Acad Sci USA 96: 14866–14870, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Harrison JF, Haddad GG. Effects of oxygen on growth and size: synthesis of molecular, organismal, and evolutionary studies with Drosophila melanogaster. Annu Rev Physiol 73: 95–113, 2011. [DOI] [PubMed] [Google Scholar]
- 43. Hochachka PW, Buck LT, Doll CJ, Land SC. Unifying theory of hypoxia tolerance: molecular/metabolic defense and rescue mechanisms for surviving oxygen lack. Proc Natl Acad Sci USA 93: 9493–9498, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hogan DA, Kolter R. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 296: 2229–2232, 2002. [DOI] [PubMed] [Google Scholar]
- 45. Hon T, Dodd A, Dirmeier R, Gorman N, Sinclair PR, Zhang L, Poyton RO. A Mechanism of oxygen sensing in yeast: multiple oxygen-responsive steps in the heme biosynthetic pathway affect Hap1 activity. J Biol Chem 278: 50771–50780, 2003. [DOI] [PubMed] [Google Scholar]
- 46. Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell 102: 577–586, 2000. [DOI] [PubMed] [Google Scholar]
- 47. Hu Z, Killion PJ, Iyer VR. Genetic reconstruction of a functional transcriptional regulatory network. Nat Genet 39: 683–687, 2007. [DOI] [PubMed] [Google Scholar]
- 48. Huang J, Zhu H, Haggarty SJ, Spring DR, Hwang H, Jin F, Snyder M, Schreiber SL. Finding new components of the target of rapamycin (TOR) signaling network through chemical genetics and proteome chips. Proc Natl Acad Sci USA 101: 16594–16599, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huie MA, Baker HV. DNA-binding properties of the yeast transcriptional activator, Gcr1p. Yeast 12: 307–317, 1996. [DOI] [PubMed] [Google Scholar]
- 50. Jiang Y, Vasconcelles MJ, Wretzel S, Light A, Gilooly L, McDaid K, Oh CS, Martin CE, Goldberg MA. Mga2p processing by hypoxia and unsaturated fatty acids in Saccharomyces cerevisiae: impact on LORE-dependent gene expression. Eukaryot Cell 1: 481–490, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jiang Y, Vasconcelles MJ, Wretzel S, Light A, Martin CE, Goldberg MA. MGA2 is involved in the low-oxygen response element-dependent hypoxic induction of genes in Saccharomyces cerevisiae. Mol Cell Biol 21: 6161–6169, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jiao X, Xiang S, Oh C, Martin CE, Tong L, Kiledjian M. Identification of a quality-control mechanism for mRNA 5′-end capping. Nature 467: 608–611, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kasten MM, Dorland S, Stillman DJ. A large protein complex containing the yeast Sin3p and Rpd3p transcriptional regulators. Mol Cell Biol 17: 4852–4858, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kimata Y, Ishiwata-Kimata Y, Yamada S, Kohno K. Yeast unfolded protein response pathway regulates expression of genes for anti-oxidative stress and for cell surface proteins. Genes Cells 11: 59–69, 2006. [DOI] [PubMed] [Google Scholar]
- 55. Kroeger PE, Morimoto RI. Selection of new HSF1 and HSF2 DNA-binding sites reveals difference in trimer cooperativity. Mol Cell Biol 14: 7592–7603, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kundaje A, Xin X, Lan C, Lianoglou S, Zhou M, Zhang L, Leslie C. A predictive model of the oxygen and heme regulatory network in yeast. PLoS Comput Biol 4: e1000224, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kuras L, Thomas D. Functional analysis of Met4, a yeast transcriptional activator responsive to S-adenosylmethionine. Mol Cell Biol 15: 208–216, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kurata S, Nielsen KH, Mitchell SF, Lorsch JR, Kaji A, Kaji H. Ribosome recycling step in yeast cytoplasmic protein synthesis is catalyzed by eEF3 and ATP. Proc Natl Acad Sci USA 107: 10854–10859, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kuruvilla FG, Shamji AF, Schreiber SL. Carbon- and nitrogen-quality signaling to translation are mediated by distinct GATA-type transcription factors. Proc Natl Acad Sci USA 98: 7283–7288, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lai LC, Kosorukoff AL, Burke PV, Kwast KE. Dynamical remodeling of the transcriptome during short-term anaerobiosis in Saccharomyces cerevisiae: differential response and role of Msn2 and/or Msn4 and other factors in galactose and glucose media. Mol Cell Biol 25: 4075–4091, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lang KJ, Kappel A, Goodall GJ. Hypoxia-inducible factor-1alpha mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Mol Biol Cell 13: 1792–1801, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Laurent BC, Treitel MA, Carlson M. Functional interdependence of the yeast SNF2, SNF5, and SNF6 proteins in transcriptional activation. Proc Natl Acad Sci USA 88: 2687–2691, 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lechner T, Carrozza MJ, Yu Y, Grant PA, Eberharter A, Vannier D, Brosch G, Stillman DJ, Shore D, Workman JL. Sds3 (Suppressor of Defective Silencing 3) is an integral component of the yeast sin3â·rpd3 histone deacetylase complex and is required for histone deacetylase activity. J Biol Chem 275: 40961–40966, 2000. [DOI] [PubMed] [Google Scholar]
- 64. Lee WC, Xue ZX, Melese T. The NSR1 gene encodes a protein that specifically binds nuclear localization sequences and has two RNA recognition motifs. J Cell Biol 113: 1–12, 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lee WC, Zabetakis D, Melese T. NSR1 is required for pre-rRNA processing and for the proper maintenance of steady-state levels of ribosomal subunits. Mol Cell Biol 12: 3865–3871, 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet 28: 327–334, 2001. [DOI] [PubMed] [Google Scholar]
- 67. Lowry CV, Zitomer RS. ROX1 encodes a heme-induced repression factor regulating ANB1 and CYC7 of Saccharomyces cerevisiae. Mol Cell Biol 8: 4651–4658, 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics 7: 113, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell 40: 294–309, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Manalo DJ, Rowan A, Lavoie T, Natarajan L, Kelly BD, Ye SQ, Garcia JG, Semenza GL. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood 105: 659–669, 2005. [DOI] [PubMed] [Google Scholar]
- 71. Marion RM, Regev A, Segal E, Barash Y, Koller D, Friedman N, O'Shea EK. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc Natl Acad Sci USA 101: 14315–14322, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Marquez JA, Pascual-Ahuir A, Proft M, Serrano R. The Ssn6-Tup1 repressor complex of Saccharomyces cerevisiae is involved in the osmotic induction of HOG-dependent and -independent genes. EMBO J 17: 2543–2553, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Martínez-Pastor MT, Marchler G, Schüller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J 15: 2227–2235, 1996. [PMC free article] [PubMed] [Google Scholar]
- 74. Mendizabal I, Rios G, Mulet JM, Serrano R, de Larrinoa IF. Yeast putative transcription factors involved in salt tolerance. FEBS Lett 425: 323–328, 1998. [DOI] [PubMed] [Google Scholar]
- 75. Mense SM, Sengupta A, Zhou M, Lan C, Bentsman G, Volsky DJ, Zhang L. Gene expression profiling reveals the profound upregulation of hypoxia-responsive genes in primary human astrocytes. Physiol Genomics 25: 435–449, 2006. [DOI] [PubMed] [Google Scholar]
- 76. Metter EJ, Yanagihara T. Protein synthesis in rat brain in hypoxia, anoxia and hypoglycemia. Brain Res 161: 481–492, 1979. [DOI] [PubMed] [Google Scholar]
- 77. Metzen E, Ratcliffe PJ. HIF hydroxylation and cellular oxygen sensing. Biol Chem 385: 223–230, 2004. [DOI] [PubMed] [Google Scholar]
- 78. Mori K. Signalling pathways in the unfolded protein response: development from yeast to mammals. J Biochem (Tokyo) 146: 743–750, 2009. [DOI] [PubMed] [Google Scholar]
- 79. Moritz M, Paulovich AG, Tsay YF, Woolford JL., Jr Depletion of yeast ribosomal proteins L16 or rp59 disrupts ribosome assembly. J Cell Biol 111: 2261–2274, 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Nagamine N, Kawada Y, Sakakibara Y. Identifying cooperative transcriptional regulations using protein-protein interactions. Nucleic Acids Res 33: 4828–4837, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Nakamura TM, Cooper JP, Cech TR. Two modes of survival of fission yeast without telomerase. Science 282: 493–496, 1998. [DOI] [PubMed] [Google Scholar]
- 82. Nevitt T, Pereira J, Azevedo D, Guerreiro P, Rodrigues-Pousada C. Expression of YAP4 in Saccharomyces cerevisiae under osmotic stress. Biochem J 379: 367–374, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ni L, Snyder M. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol Biol Cell 12: 2147–2170, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Nilsson GE, Renshaw GM. Hypoxic survival strategies in two fishes: extreme anoxia tolerance in the North European crucian carp and natural hypoxic preconditioning in a coral-reef shark. J Exp Biol 207: 3131–3139, 2004. [DOI] [PubMed] [Google Scholar]
- 85. Patil CK, Li H, Walter P. Gcn4p and novel upstream activating sequences regulate targets of the unfolded protein response. PLoS Biol 2: E246, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Pereira J, Pimentel C, Amaral C, Menezes RA, Rodrigues-Pousada C. Yap4 PKA- and GSK3-dependent phosphorylation affects its stability but not its nuclear localization. Yeast 26: 641–653, 2009. [DOI] [PubMed] [Google Scholar]
- 87. Peterson CL, Dingwall A, Scott MP. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc Natl Acad Sci USA 91: 2905–2908, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Piper PW, Harris NL, MacLean M. Preadaptation to efficient respiratory maintenance is essential both for maximal longevity and the retention of replicative potential in chronologically ageing yeast. Mech Ageing Dev 127: 733–740, 2006. [DOI] [PubMed] [Google Scholar]
- 89. Podrabsky JE, Lopez JP, Fan TW, Higashi R, Somero GN. Extreme anoxia tolerance in embryos of the annual killifish Austrofundulus limnaeus: insights from a metabolomics analysis. J Exp Biol 210: 2253–2266, 2007. [DOI] [PubMed] [Google Scholar]
- 90. Preedy VR, Smith DM, Sugden PH. The effects of 6 hours of hypoxia on protein synthesis in rat tissues in vivo and in vitro. Biochem J 228: 179–185, 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Pujol-Carrion N, Belli G, Herrero E, Nogues A, de la Torre-Ruiz MA. Glutaredoxins Grx3 and Grx4 regulate nuclear localisation of Aft1 and the oxidative stress response in Saccharomyces cerevisiae. J Cell Sci 119: 4554–4564, 2006. [DOI] [PubMed] [Google Scholar]
- 92. Ramirez JM, Folkow LP, Blix AS. Hypoxia tolerance in mammals and birds: from the wilderness to the clinic. Annu Rev Physiol 69: 113–143, 2007. [DOI] [PubMed] [Google Scholar]
- 93. Reinke A, Chen JC, Aronova S, Powers T. Caffeine targets TOR complex I and provides evidence for a regulatory link between the FRB and kinase domains of Tor1p. J Biol Chem 281: 31616–31626, 2006. [DOI] [PubMed] [Google Scholar]
- 94. Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 3: 1154–1169, 2004. [DOI] [PubMed] [Google Scholar]
- 95. Rudra D, Zhao Y, Warner JR. Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J 24: 533–542, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Saveanu C, Fromont-Racine M, Harington A, Ricard F, Namane A, Jacquier A. Identification of 12 new yeast mitochondrial ribosomal proteins including 6 that have no prokaryotic homologues. J Biol Chem 276: 15861–15867, 2001. [DOI] [PubMed] [Google Scholar]
- 97. Schmitt AP, McEntee K. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 93: 5777–5782, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Scott KL, Plon SE. Loss of Sin3/Rpd3 histone deacetylase restores the DNA damage response in checkpoint-deficient strains of Saccharomyces cerevisiae. Mol Cell Biol 23: 4522–4531, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Semenza GL. Hypoxia-inducible factor 1 and cancer pathogenesis. IUBMB Life 60: 591–597, 2008. [DOI] [PubMed] [Google Scholar]
- 100. Shapira M, Segal E, Botstein D. Disruption of yeast forkhead-associated cell cycle transcription by oxidative stress. Mol Biol Cell 15: 5659–5669, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Shenton D, Grant CM. Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem J 374: 513–519, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Smith CL, Horowitz-Scherer R, Flanagan JF, Woodcock CL, Peterson CL. Structural analysis of the yeast SWI/SNF chromatin remodeling complex. Nat Struct Biol 10: 141–145, 2003. [DOI] [PubMed] [Google Scholar]
- 103. Smith RW, Houlihan DF, Nilsson GE, Brechin JG. Tissue-specific changes in protein synthesis rates in vivo during anoxia in crucian carp. Am J Physiol Regul Integr Comp Physiol 271: R897–R904, 1996. [DOI] [PubMed] [Google Scholar]
- 104. Smits P, Smeitink JA, van den Heuvel LP, Huynen MA, Ettema TJ. Reconstructing the evolution of the mitochondrial ribosomal proteome. Nucleic Acids Res 35: 4686–4703, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Sollner S, Schober M, Wagner A, Prem A, Lorkova L, Palfey BA, Groll M, Macheroux P. Quinone reductase acts as a redox switch of the 20S yeast proteasome. EMBO Rep 10: 65–70, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sommariva E, Pellny TK, Karahan N, Kumar S, Huberman JA, Dalgaard JZ. Schizosaccharomyces pombe Swi1, Swi3, and Hsk1 are components of a novel S-phase response pathway to alkylation damage. Mol Cell Biol 25: 2770–2784, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, Pak DN, Fields S, Kennedy BK, Kaeberlein M. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell 133: 292–302, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev 64: 435–459, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Storey KB. Molecular mechanisms of anoxia tolerance. Int Congr Ser 1275: 47–54, 2004. [Google Scholar]
- 110. Swiderek H, Logan A, Al-Rubeai M. Cellular and transcriptomic analysis of NS0 cell response during exposure to hypoxia. J Biotechnol 134: 103–111, 2008. [DOI] [PubMed] [Google Scholar]
- 111. Sydorskyy Y, Dilworth DJ, Yi EC, Goodlett DR, Wozniak RW, Aitchison JD. Intersection of the Kap123p-mediated nuclear import and ribosome export pathways. Mol Cell Biol 23: 2042–2054, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Thorsen M, Lagniel G, Kristiansson E, Junot C, Nerman O, Labarre J, Tamas MJ. Quantitative transcriptome, proteome, and sulfur metabolite profiling of the Saccharomyces cerevisiae response to arsenite. Physiol Genomics 30: 35–43, 2007. [DOI] [PubMed] [Google Scholar]
- 113. Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101: 249–258, 2000. [DOI] [PubMed] [Google Scholar]
- 114. Tyrrell A, Flick K, Kleiger G, Zhang H, Deshaies RJ, Kaiser P. Physiologically relevant and portable tandem ubiquitin-binding domain stabilizes polyubiquitylated proteins. Proc Natl Acad Sci USA 107: 19796–19801, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet 33: 261–311, 1999. [DOI] [PubMed] [Google Scholar]
- 116. Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440, 1999. [DOI] [PubMed] [Google Scholar]
- 117. Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M'Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906, 1999. [DOI] [PubMed] [Google Scholar]
- 118. Wouters BG, Koritzinsky M. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nat Rev Cancer 8: 851–864, 2008. [DOI] [PubMed] [Google Scholar]
- 119. Wu PYJ, Ruhlmann C, Winston F, Schultz P. Molecular architecture of the S. cerevisiae SAGA complex. Mol Cell 15: 199–208, 2004. [DOI] [PubMed] [Google Scholar]
- 120. Xu Z, Norris D. The SFP1 gene product of Saccharomyces cerevisiae regulates G2/M transitions during the mitotic cell cycle and DNA-damage response. Genetics 150: 1419–1428, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Xue Y, Bai X, Lee I, Kallstrom G, Ho J, Brown J, Stevens A, Johnson AW. Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol Cell Biol 20: 4006–4015, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Yamaguchi-Iwai Y, Stearman R, Dancis A, Klausner RD. Iron-regulated DNA binding by the AFT1 protein controls the iron regulon in yeast. EMBO J 15: 3377–3384, 1996. [PMC free article] [PubMed] [Google Scholar]
- 123. Yamamoto N, Maeda Y, Ikeda A, Sakurai H. Regulation of thermotolerance by stress-induced transcription factors in Saccharomyces cerevisiae. Eukaryot Cell 7: 783–790, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Yanagihara T. Cerebral anoxia: effect on neuron-glia fractions and polysomal protein synthesis. J Neurochem 27: 539–543, 1976. [DOI] [PubMed] [Google Scholar]
- 125. Yiu G, McCord A, Wise A, Jindal R, Hardee J, Kuo A, Shimogawa MY, Cahoon L, Wu M, Kloke J, Hardin J, Mays Hoopes LL. Pathways change in expression during replicative aging in Saccharomyces cerevisiae. J Gerontol A Biol Sci Med Sci 63: 21–34, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Zabetakis D. Inheritance of suppressors of the drug sensitivity of a NSR1 deleted yeast strain. Yeast 16: 1147–1159, 2000. [DOI] [PubMed] [Google Scholar]
- 127. Zhang H, Semenza GL. The expanding universe of hypoxia. J Mol Med 86: 739–746, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Zhang S, Burkett TJ, Yamashita I, Garfinkel DJ. Genetic redundancy between SPT23 and MGA2: regulators of Ty-induced mutations and Ty1 transcription in Saccharomyces cerevisiae. Mol Cell Biol 17: 4718–4729, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Zhou D, Xue J, Lai JC, Schork NJ, White KP, Haddad GG. Mechanisms underlying hypoxia tolerance in Drosophila melanogaster: hairy as a metabolic switch. PLoS Genet 4: e1000221, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.