Summary
Lovastatin and other statins inhibit HMG-CoA reductase, which carries out an early step in the sterol biosynthesis pathway. Statins lower cholesterol and are widely prescribed to prevent heart disease, but like many drugs, they can interact with nutritionally acquired metabolites. To probe these interactions, we explored the effect of a diverse library of metabolites on statin effectiveness using a Saccharomyces cerevisiae model. In yeast, treatment with lovastatin results in reduced growth. We combined lovastatin with the library of metabolites, and found that copper and zinc ions impaired the ability of the statin to inhibit yeast growth. Using an integrated genomic and metabolomic approach, we found that lovastatin plus metal synergistically upregulated some sterol biosynthesis genes. This altered pattern of gene expression resulted in greater flux through the sterol biosynthesis pathway and an increase in ergosterol levels. Each sterol intermediate level was correlated with expression of the upstream gene. Thus, the ergosterol biosynthetic response induced by statin is enhanced by copper and zinc. In cultured mammalian cells, these metals also rescued statin growth inhibition. Because copper and zinc impair the ability of statin to reduce sterol biosynthesis, dietary intake of these metals could have clinical relevance for statin treatment in humans.
Introduction
The metabolic landscape of organisms is immensely complex, comprising a highly regulated network of thousands of metabolites and enzymes. The goal of many drugs is to perturb the metabolic landscape in ways that ameliorate disease. Despite the general success of this approach, drugs and metabolites can interact in unexpected and deleterious ways. Documented interactions of this type are numerous, with some having grave clinical consequences. For example, many drugs have their metabolism affected by grapefruit juice, which inhibits the cytochrome P450 CYP3A4.1 Warfarin is an anticoagulant whose action is compromised by the consumption of foods rich in vitamin K.2 Monoamine oxidase inhibitors have potentially fatal interactions with foods containing large amounts of tyramine.3 These well-known examples involve a metabolite that interacts in a direct biochemical manner with the drug in question (e.g. by competing for the same enzyme). However, other metabolites are likely to interact with drugs in more complex and unpredictable ways by altering gene expression and regulatory networks. Identifying and understanding such complex interactions could significantly enhance the efficacy of drug treatments and reduce side effects, but requires adopting a systems-level perspective.
Statin drugs inhibit HMG-CoA reductase, which is the rate-limiting enzyme in the synthesis of cholesterol. Thus, statins are used to treat high cholesterol and atherosclerosis. Statin therapy reduces the incidence of strokes and heart attacks, and the broad application of these drugs has revolutionized the prevention of heart disease.4 However, statins have poorly understood pleiotropic effects, despite being one of the first drugs developed with a specific molecular target in mind.5 Furthermore, they can have significant side effects, including musculoskeletal deterioration and rhabdomyolysis. Though these side effects are rare (1 case per 10,000 patients per year), they are significant because of the staggering number of patients taking statins (29.7 million in the U.S. in 2005).6, 7
We chose to search for statin-metabolite interactions because statins are among the most widely prescribed drugs in the world, so even small nutritional impacts on statin effectiveness could be of clinical benefit. Furthermore, nutritional metabolites such as vitamin D are known to modulate statin effectiveness in humans.8 Searching for drug-metabolite interactions is daunting because metabolites comprise a highly diverse class of molecules, including 714 compounds that are annotated in the Saccharomyces Genome Database as endogenous metabolites in the budding yeast Saccharomyces cerevisiae. In addition to being both substrates for and products of protein action, metabolites have profound regulatory effects, ranging from simple enzymatic product inhibition to allostery to participation in complex signaling cascades that regulate gene expression programs. Furthermore, exogenous metabolites, acquired for nutritive purposes or used as chemical defenses, greatly expand the diversity of metabolites a cell might encounter. We hypothesized that perturbation of metabolite levels in the context of a phenotype dependent on drug treatment could provide rich information about interactions between statins and metabolites.
In S. cerevisiae, statins inhibit the yeast homologs of HMG-CoA reductase and lower the level of the yeast cholesterol analog, ergosterol.9, 10 Statin treatment produces dose-dependent growth inhibition in yeast. By combining exposure of yeast to a library of 52 metabolites along with lovastatin treatment, we found that the divalent metal ions zinc, copper and manganese alleviated statin-mediated growth inhibition. Because no obvious mechanistic connection between ergosterol biosynthesis and divalent metal ions exists, we characterized these effects using an integrated metabolomic and genomic approach. At the metabolomic level, we measured ergosterol biosynthetic intermediates by spectroscopy and gas chromatography– mass spectrometry; at the genomic level, we examined gene expression. We found that divalent metal ions activate a complex transcriptional program that results in more efficient ergosterol biosynthesis. These metals also alleviated statin-mediated growth inhibition in cultured mammalian cells, revealing that interactions between divalent metals and statins are not species-specific. Because increased metal exposure enhances ergosterol levels, altered metal status could decrease the effectiveness of statin treatment in humans.
Results and Discussion
Zinc and copper rescue statin toxicity in a yeast-based screen
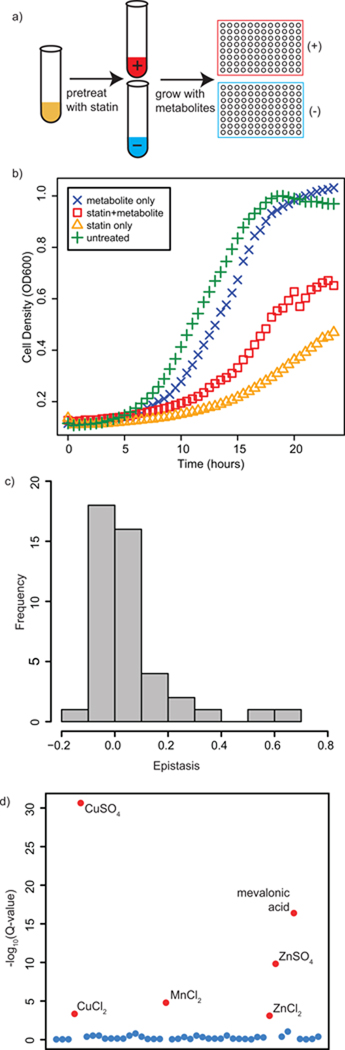
To maximize the effect that exogenously added metabolites would have on statin-treated yeast, we used yeast bearing a PDR5 deletion. PDR5 encodes an ABC cassette efflux pump responsible for the removal of a variety of small molecules, including xenobiotics and drugs.11 In the presence of lovastatin, the knockout strain reached cell densities approximately 40% that of the PDR+ strain (Supplementary Fig. 1). The assay was performed in synthetic complete media to minimize the availability of metabolites in the media and to avoid undefined components in autolyzed yeast extract.
Yeast cultures were pretreated overnight with lovastatin to exhaust the intracellular ergosterol reserve and then diluted into fresh media containing metabolites from the library. Growth was measured every 30 minutes for 24 hours (Fig. 1a, b). We chose a lovastatin concentration that inhibited yeast growth by approximately 50%, enabling the identification of metabolites that enhanced or rescued statin toxicity (Fig. 1b, CuSO4 is shown).
Figure 1.
A growth-based assay of drug-metabolite interactions reveals divalent metal ion-mediated rescue of statin growth inhibition. (a) Yeast cultures were pretreated with statin to deplete ergosterol reserves. After pretreatment, the yeast were diluted into fresh media containing the metabolite of interest. Growth was followed spectrophotometrically in a microplate format. (b) A typical set of growth curves from the assay that show metabolite rescue, CuSO4 in this case. Untreated cultures reach saturation after approximately 15 hours (green plusses) whereas statin treatment slows growth (gold triangles). The added metabolite was slightly toxic (blue Xs), but addition of the metabolite to statin-treated cultures resulted in increased growth (red squares). Growth curves generated for yeast grown in the presence of metabolites and/or statin were used to calculate fitness scores (W) for each condition relative to untreated cultures. Epistasis values were calculated from fitness scores (epistasis = Wstatin+metabolite − Wmetabolite * Wstatin); epistasis values less than zero indicate synthetic lethality whereas epistasis values greater than zero indicate rescue. (c) Shown is a histogram of epistasis values for all metabolites. The majority of metabolites failed to influence statin-mediated growth inhibition, with a mean epistasis value close to zero. (d) The results of significance testing performed on the distribution of epistasis values show that the divalent metal ions zinc, copper and manganese rescued statin growth inhibition regardless of counterion. Red points indicate metabolites with a false discovery rate adjusted P-value (Q-value) less than 0.05.
Statin treatment in yeast at high concentrations and for sustained periods can cause degradation of mitochondrial DNA and the consequent appearance of the petite phenotype.9, 12, 13 We established that at the concentration of statin and for the period of treatment we employed, petite formation did not occur (Supplementary Fig. 1).
We screened a small library designed to cover a spectrum of metabolite classes, including enzyme cofactors and metals as well as nutritional and nutriceutical compounds (Table 1). Metabolites were added at high concentration (typically mM) unless toxicity was observed, in which case lower concentrations were chosen (Supplementary Table 1). We generated growth curves from triplicate cultures that were averaged and transformed into a fitness measurement. We examined the fitness impact of each metabolite on the background of statin treatment by calculating chemical epistasis values (Epistasis = Fitnessstatin+metabolite − Fitnessstatin*Fitnessmetabolite, Supplementary Table 1).14 The epistasis values comprised a distribution with a mean score = 0.073 +/− 0.022 (Fig. 1c). Statistical analysis of the distribution of epistasis scores revealed that the divalent metal ions zinc, copper and manganese rescued statin toxicity (Fig. 1d). We included in the screen two positive controls: mevalonic acid, the product of HMG-CoA reductase; and ergosterol, the endpoint of the pathway. Mevalonic acid produced a strong rescue effect, in accord with previous studies.9, 10 Ergosterol did not rescue, likely because it is sparingly soluble in water and therefore not bioavailable to yeast after dilution into the culture. Indeed, reports of the effectiveness of ergosterol in the rescue of yeast treated with sterol synthesis inhibitors are conflicting, likely for this reason.9
Table 1.
Library of metabolites
| Coenzymes & Cofactors | Nutriceuticals | Nutrients | amino acids | metals |
|---|---|---|---|---|
| coenzyme A | resveratrol | myoinositol | L-arginine | lithium chloride |
| coenzyme Q10 | curcumin | taurine | L-glutamate | zinc chloride |
| vitamin B12 | caffeine | 5-hydroxytryptophan | L-tyrosine | zinc sulfate |
| methylcobalamin | genistein | L-ascorbic acid | iron sulfate | |
| NAD | daidzein | propionic acid | iron chloride | |
| FAD | carotene | linolenic acid | manganese chloride | |
| pyridoxal 5’ phosphate | docosahexaenoic acid | mevalonic acid | copper chloride | |
| thiamine pyrophosphate | eicosapentaenoic acid | cholesterol | copper sulfate | |
| biotin | astaxanthin | sodium selenite | ||
| nicotinic acid | betaine | calcium chloride | ||
| riboflavin | citrate | chromium chloride | ||
| tetrahydrobiopterin | L-glutathione (reduced) | nickel sulfate | ||
| thiamine pyrophosphate | pyruvic acid | |||
| folinic acid | ||||
| folic acid | ||||
| D-pantothenic acid |
Given that the three hits from the screen were metals, we tested a variety of additional metals in our assay. Other divalent metals, including iron, nickel and calcium, did not rescue statin growth inhibition. Additionally, neither chromium nor lithium rescued statin growth inhibition. By testing both chloride and sulfate salts of copper and zinc, we established that the rescue is independent of the counterion present. Furthermore, the copper and zinc rescue effects were dose-dependent, occurring maximally over a narrow range of concentrations (1–2 mM) just below the point at which toxicity dominated (Supplementary Fig. 1). From these data, we conclude that the rescue effects observed for copper, zinc and manganese are due to the metals themselves, and cannot be generalized to other metals.
We performed experiments to rule out possible confounding features of our assay. Testing in mating type α yeast revealed that the rescue effect was not specific to the mating type a strain used in the screen (Supplementary Fig. 1). Copper is highly oxidatively active, and zinc has been reported to induce oxidative stress responses in yeast at high concentrations.15, 16 Therefore, we examined whether hydrogen peroxide, a strong oxidant, could rescue statin growth inhibition and found that it did not. Thus, the rescue effects are not strain-or mating type-specific nor do they arise from the induction of a general oxidative stress response.
Lovastatin inhibits ergosterol biosynthesis by preventing the synthesis of mevalonic acid, an early intermediate in the pathway. Mevalonic acid is also used by the cell to generate heme and coenzyme Q10 as well as to prenylate proteins. Statin-mediated toxicity in yeast arises from a lack of protein prenylation9 as well as ergosterol starvation. To investigate whether these non-ergosterol products of mevalonic acid play a role in the metal rescue effect, we tested ketoconazole, an antifungal agent that inhibits Erg11, an enzyme that acts after the pathway is committed to ergosterol.17 We asked whether copper and zinc rescue ketoconazole-mediated growth inhibition in yeast. Both metals produced a rescue effect similar to that seen when they are combined with lovastatin (Supplementary Fig. 1). These data show that the metals act by restoring ergosterol biosynthesis rather than by modulating other products of mevalonic acid.
Zinc and copper increase the level of ergosterol and its precursors in lovastatin-treated yeast
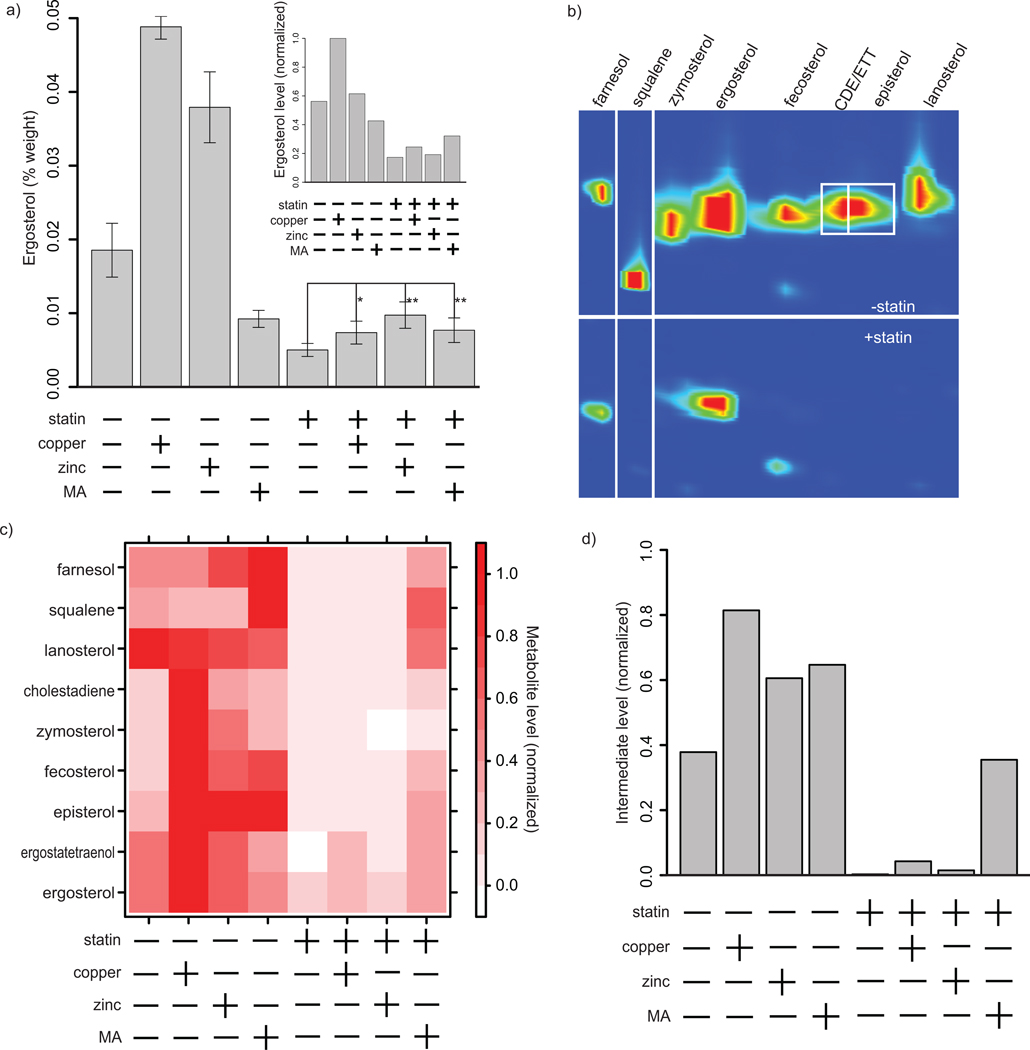
We reasoned that metal rescue of lovastatin growth inhibition was likely mediated by an increase in ergosterol levels. We tested this hypothesis by measuring the ergosterol content of yeast cultures by spectroscopic analysis. We found that lovastatin treatment reduced ergosterol to one quarter the level found in untreated cells (Fig. 2a). Treatment with zinc or copper alone resulted in an approximately twofold increase in ergosterol level relative to untreated cells. In the statin-treated cultures, the addition of zinc or copper also led to a twofold increase in ergosterol relative to statin treatment alone. The addition of mevalonic acid alone suppressed ergosterol biosynthesis, likely the result of the very high mevalonic acid concentrations used. The mechanism by which mevalonic acid supplementation produces this result is unclear; however, the sterol biosynthesis pathway is subject to feedback inhibition.18 In the lovastatin-treated culture, mevalonic acid produced an increase in ergosterol levels equivalent to that observed with lovastatin plus zinc or copper.
Figure 2.
Copper and zinc increase levels of ergosterol and ergosterol biosynthetic intermediates. (a) Ergosterol was measured by spectrophotometric quantitation after extraction with n-heptane and by GCxGC-MS (inset). Comparison of ergosterol levels in untreated (left-hand four bars) and statin-treated (right-hand four bars) yeast cultures revealed a reduction upon statin treatment. In either untreated or statin-treated cultures, addition of copper (*, P < 0.05) or zinc (**, p < 0.01) resulted in an approximately twofold increase in ergosterol levels. Mevalonic acid (MA) decreased ergosterol levels in untreated cultures and produced an increase in ergosterol in statin-treated cultures (**, p < 0.01) that was equivalent to that observed with zinc or copper. (b) GCxGC-MS was used to analyze intermediate levels in the ergosterol biosynthesis pathway (CDE = cholestadiene, ETT = ergostatetraenol). A two-dimensional gas chromatogram of statin-treated versus untreated cultures is shown; red indicates high intermediate levels. (c) Pathway intermediate levels are quantified for eight ergosterol precursors as well as ergosterol. Intensities are normalized within each compound to enable comparison between conditions. (d) An average of the levels of all eight intermediates, normalized within each intermediate, are shown.
We explored ergosterol biosynthesis in more detail by profiling levels of intermediates in this pathway using two-dimensional gas chromatography and mass spectrometry (GCxGC-MS). A heptane-based extraction isolated these highly hydrophobic intermediates. Using standards and chemical library information,19 we identified eight ergosterol pathway intermediates as well as ergosterol itself (Fig. 2b, c). We quantified each of these intermediates after treatment with combinations of lovastatin, zinc, copper and mevalonic acid. Copper, zinc or mevalonic acid alone induced increases in intermediate levels (PCu = 4e-11, PZn = 0.0001, Pmevalonic acid = 3e-5, t-test) (Fig. 2c, d). In contrast, statin treatment alone led to a striking depletion of intermediates, with most being reduced to immeasurably low concentrations (P = 1.4e-12, t-test). Treatment with copper, zinc or mevalonic acid along with lovastatin increased the levels of these intermediates relative to their low levels in the presence of the statin alone (PCu = 0.003, PZn = 0.02, Pmevalonic acid = 5e-16, t-test).
As a complement to our spectroscopic approach, we used GCxGC-MS to examine ergosterol levels in yeast treated with combinations of copper, zinc and lovastatin (Fig. 2a, inset). Copper alone produced a large and significant increase in the level of ergosterol (P = 2e-6, t-test). Zinc alone produced a small increase that is not significant. Mevalonic acid alone reduced ergosterol levels (P = 0.001, t-test). Treatment with lovastatin alone reduced ergosterol by 70%. In the presence of statin, both copper and zinc increased the diminished ergosterol level due to the statin, although this increase was small and statistically significant only for copper (P = 0.046, t-test). As expected, mevalonic acid treatment resulted in an increase in ergosterol level. These observations are in general agreement with the ergosterol levels determined by spectrophotometric measurement (Fig. 2a).
These metabolite profiling data reveal a detailed picture of the effect of copper, zinc and lovastatin on yeast ergosterol biosynthesis. Lovastatin treatment eliminated ergosterol precursors and reduced ergosterol levels by approximately 70%. Copper and zinc stimulated ergosterol production, increasing levels of ergosterol precursors as well as ergosterol itself. This metal-mediated enhancement of ergosterol biosynthesis occurred in the context of lovastatin-treated yeast. Thus, metal enhancement of ergosterol biosynthesis likely mediates the observed rescue of lovastatin-mediated growth inhibition.
Gene expression analysis of metal-mediated rescue of lovastatin-treated yeast
To understand the basis for metal modulation of ergosterol biosynthesis, we analyzed gene expression. Expression data were collected for yeast treated with lovastatin, with copper or zinc, or with both the statin and a metal. We searched for genes whose expression was significantly affected by both lovastatin and metal treatment using a linear model-based analysis. We found 321 such genes at a false discovery rate of 5%. Clustering of these genes identified six major groups; of these, four were primarily responsive to statin and two were primarily responsive to metal (Supplementary Fig. 2). Gene Ontology (GO) analysis of these groups revealed that statin treatment in general modulated the expression of sterol, lipid and cell wall biosynthetic genes, whereas metal treatment modulated the expression of mitochondrial and metal binding/transport genes (Supplementary Table 2).
The expression level of many genes in yeast is dependent on growth rate.20 Given that statin treatment resulted in decreased growth rate (doubling time for untreated = 5.5 hours, statin = 10.75 hours), we checked whether the transcriptional changes observed were due to such growth rate-dependent expression effects. Of the 321 genes found to be significantly up- or downregulated, only 17 had growth rate-dependent slopes of similar sign in the Brauer et al. data set.20 Thus, the transcriptional changes do not originate primarily from changes in growth rate.
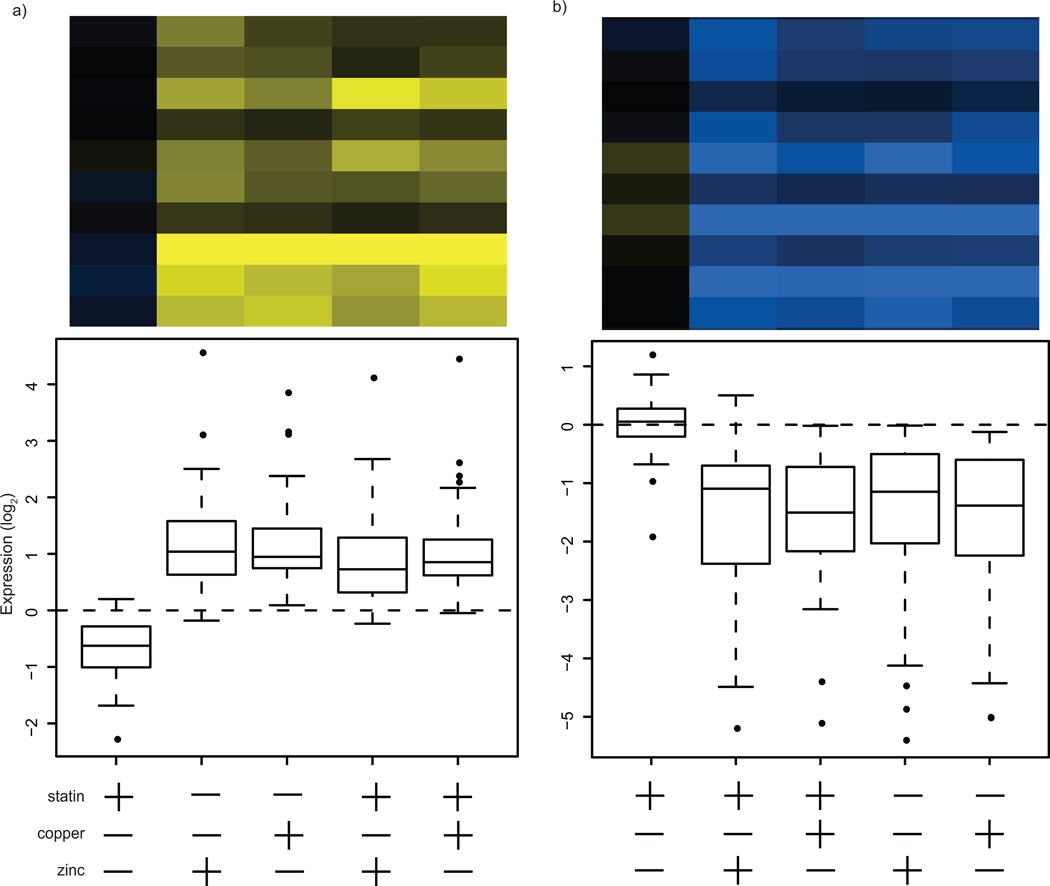
The yeast transcriptional response to zinc and, in particular, to copper has been extensively studied;15, 16, 21–24 our results generally agree with previous observations. The two clusters that responded primarily to metal (metal responsive, MR) contain many genes known to respond to changes in metal availability, including metalloproteins and metal transporters (Supplementary Table 2). MR cluster #1 contains genes whose expression was upregulated in response to zinc or copper (Fig. 3). MR cluster #1 genes are representative of metal-stress responses and include the metallothioneins CUP1 and CUP2. A large number of mitochondria-associated genes are also found in this cluster; mitochondrial metabolism is sensitive to metal and oxidoreductive perturbations.25
Figure 3.
Gene expression clusters primarily responsive to metal treatment. Measurement of gene expression following treatment of yeast cultures with statin, zinc/copper or both resulted in clusters of genes with particular expression patterns. Each panel represents a cluster and contains a representative clustergram as well as a plot of the mean expression level for treatment conditions relative to untreated cultures. (a) Metal responsive (MR) cluster #1 contains genes that are primarily upregulated in response to metal. (b) Metal responsive (MR) cluster #2 contains genes that are primarily upregulated in response to metal. The scale shown is log2.
MR cluster #2 contains genes that were downregulated in response to metals (Fig. 3). Included in this cluster are genes that characterize the primary yeast response to high zinc concentrations, which is mediated by Zap1 repression of genes carrying zinc responsive elements.23, 24, 26 Genes from this regulon that were downregulated by zinc include ZPS1, ZRT1, ZRT2 and ZRT3. Other genes that were downregulated include FET5, FET4, CTR1 and FRE1, all of which play a role in the uptake or transport of copper. Many of the genes found in both MR clusters relate to iron metabolism, which is also disrupted by high copper and zinc concentrations.16, 21
Treatment with statins or other antifungal agents, along with the study of a variety of genetic modifications that interfere with ergosterol metabolism, has revealed that yeast respond to ergosterol deprivation primarily through the transcription factors Ecm22 and Upc2, which act on Sterol Responsive Elements (SREs) to upregulate many ergosterol biosynthetic genes as well as the DAN/TIR cell wall biosynthesis gene families.10, 17, 27, 28 UPC2 expression is expected to increase during ergosterol deprivation.29 We observed a sevenfold increase in UPC2 expression upon lovastatin treatment. However, this expression change was not considered significant in our model, which was designed to identify genes whose expression changed in response to both metal and statin treatment. Since UPC2 expression did not respond to metal treatment, the metal-mediated rescue of statin growth inhibition is likely not effected by Upc2. ECM22 expression is reported to decrease during ergosterol deprivation,29 but we did not observe a significant change upon statin treatment.
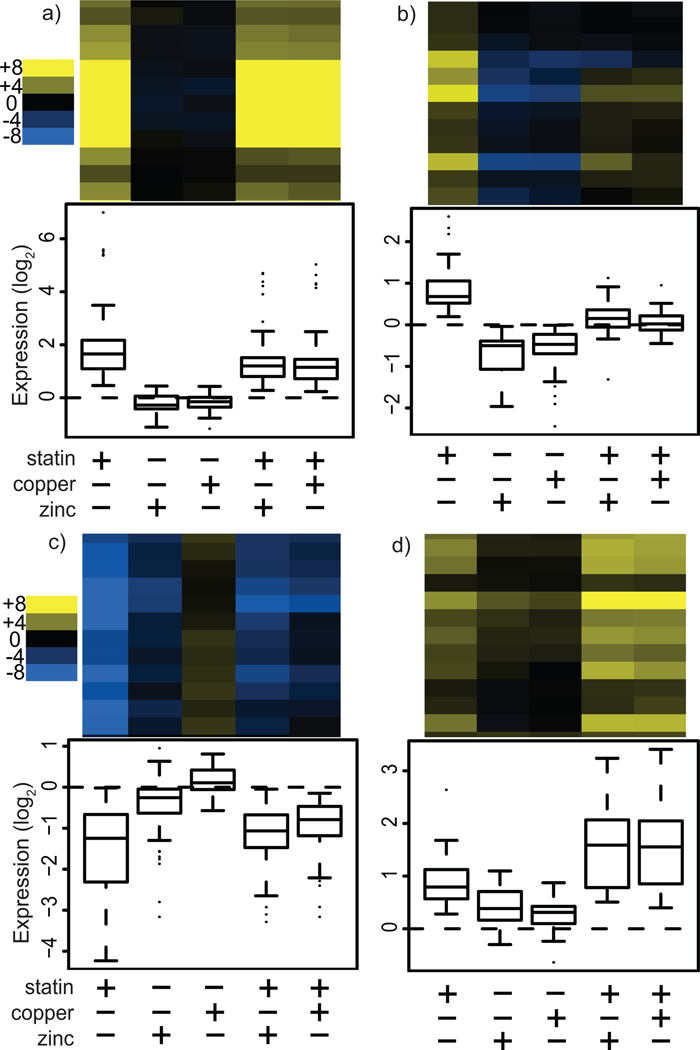
Four clusters of primarily statin-responsive (SR) genes showed an expression pattern that was also modulated by metal treatment. SR cluster #1 contains 72 genes whose expression was increased greatly by statin alone, was unaffected by metal alone, and was diminished by statin plus metal relative to statin alone (Fig. 4a, see also Supplementary Fig. 2). Based on GO analysis, this cluster is enriched for sterol and lipid biosynthetic as well as cell wall genes (Supplementary Table 2). These genes characterize the stereotypic SRE-driven response which upregulates ergosterol and cell wall biosynthesis.
Figure 4.
Gene expression clusters primarily responsive to statin treatment. Measurement of gene expression following treatment of yeast cultures with statin, metal (copper or zinc) or both resulted in clusters of genes with particular expression patterns. Each panel represents a cluster and contains a representative clustergram as well as a box plot of the expression level of all genes in the cluster for each treatment condition relative to untreated cultures. (a) Statin-responsive (SR) cluster #1 contains genes that are primarily upregulated in response to statin. (b) SR cluster #2 genes are upregulated by statin and downregulated by metal treatment. (c) SR cluster #3 contains genes that are primarily downregulated in response to statin. (d) SR cluster #4 genes are upregulated both by statin and metals, and show a positive synergistic effect. The scale shown is log2.
SR cluster #2 contains 29 genes that were upregulated in response to statin alone, downregulated in response to metal alone, and upregulated more weakly or not at all in response to statin plus metal relative to statin alone (Fig. 4b). No GO terms are enriched in this cluster, which contains a diverse array of seemingly unrelated genes. SR cluster #3 contains 94 genes that were downregulated in response to statin alone, unaffected by metal alone, and downregulated more weakly in response to statin plus metal relative to statin alone (Fig. 4c). GO analysis reveals that this large cluster is enriched for amino acid and nucleotide biosynthetic genes.
SR cluster #4 contains 27 genes whose expression was upregulated weakly by statin alone, was upregulated weakly by metal alone, and was enhanced by statin plus metal relative to statin alone (Fig. 4d). Thus, in this cluster, metal and statin treatments synergize to increase gene expression levels, which suggests that these genes could account for the observed metal-mediated rescue of statin growth inhibition. This hypothesis is strengthened by GO analysis, which revealed that this cluster is enriched for sterol biosynthetic genes (Bonferroni-corrected P = 2.66e-7). In fact, SR cluster #2 contains a large fraction (5 of 14) of the sterol biosynthesis genes identified by our model as having a significant response to the combination of lovastatin and metal treatment.
We performed a search for transcription factors that could be responsible for the observed expression changes. A search of YEASTRACT30 reveals consensus binding sites for the transcription factors Sfp1, Hap1 and Yap1 in the promoter regions of at least four of these five genes. Given that Sfp1 is a stress-sensitive regulator of ribosomal protein expression,31 that Yap1 activates a large transcriptional program in response to oxidative stress32 and that Hap1 responds to changes in oxygen availability,33 any of these transcription factors could be responsible for the metal effect. However, few of the other known targets of these factors were differentially expressed (7% for Yap1, 6% for Sfp1 and 13% for Hap1), making any of these factors unlikely to be responsible.
Intriguingly, the five ergosterol biosynthetic genes in SR cluster #4 (DAP1, ERG11, ERG5 , CYB5 and ERG1) are all involved in oxidoreductive reactions. Metal-induced oxidative stress might account for the differential regulation of these genes. However, we did not observe rescue of lovastatin-mediated growth inhibition upon treatment with hydrogen peroxide, a powerful inducer of oxidative stress. Nevertheless, perturbation of metal ion homeostasis induced by excess zinc and copper treatment could cause the observed upregulation of these genes after metal treatment.
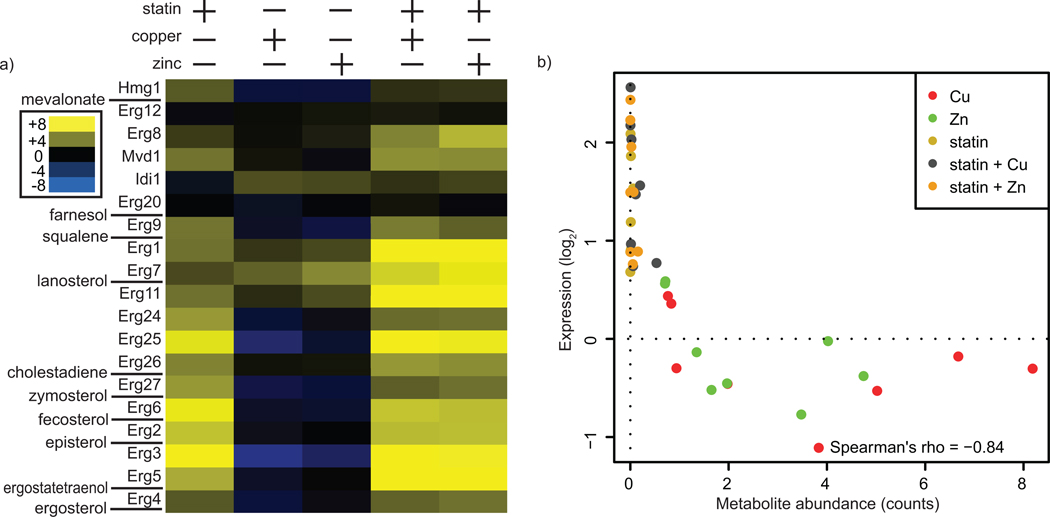
Enhanced Expression of Ergosterol Biosynthetic Genes Correlates with Metabolite Levels
Treatment of yeast with lovastatin, copper or zinc resulted in profound changes in both gene expression and metabolite levels within the ergosterol biosynthesis pathway. We combined our metabolomic and expression datasets to better understand the interplay between metabolite levels and gene expression. As shown, treatment with lovastatin or metal increased the expression of ergosterol biosynthetic genes (Fig. 5a). Examination of these data in the context of ergosterol pathway intermediates reveals that the level of an intermediate inversely correlated with the expression of the gene immediately downstream in the biosynthesis pathway (which encodes the protein for which the intermediate in question is a substrate) (Fig. 5b). Lovastatin-mediated induction of ergosterol biosynthetic genes by as little as twofold relative to untreated cultures was sufficient to reduce the level of the substrate metabolite to immeasurably small levels.
Figure 5.
Ergosterol metabolites and biosynthetic gene expression levels are correlated. (a) Expression data for each gene in the ergosterol biosynthetic pathway is shown, in descending order. Ergosterol intermediates measured in this study (see Figure 2) are indicated. The scale shown is log2. (b) Intermediate levels are plotted against the expression level of the gene immediately downstream of each intermediate (i.e. the protein for which the intermediate is a substrate). For each condition, pathway intermediate and expression data were normalized to the untreated condition.
Zinc and copper rescue growth inhibition in cultured HeLa cells
Many facets of sterol synthesis and regulation are conserved between yeast and humans, including most of the biosynthetic machinery.18 However, mammalian sterol homeostasis is achieved mostly by the Sterol Response Element Binding Proteins (SREBPs) whereas S. cerevisiae regulates sterol homeostasis primarily through the Upc2 and Ecm22 transcription factors, which are not homologous to SREBPs. Given these differences, we asked whether zinc and copper could rescue statin growth inhibition in a mammalian cell culture system.
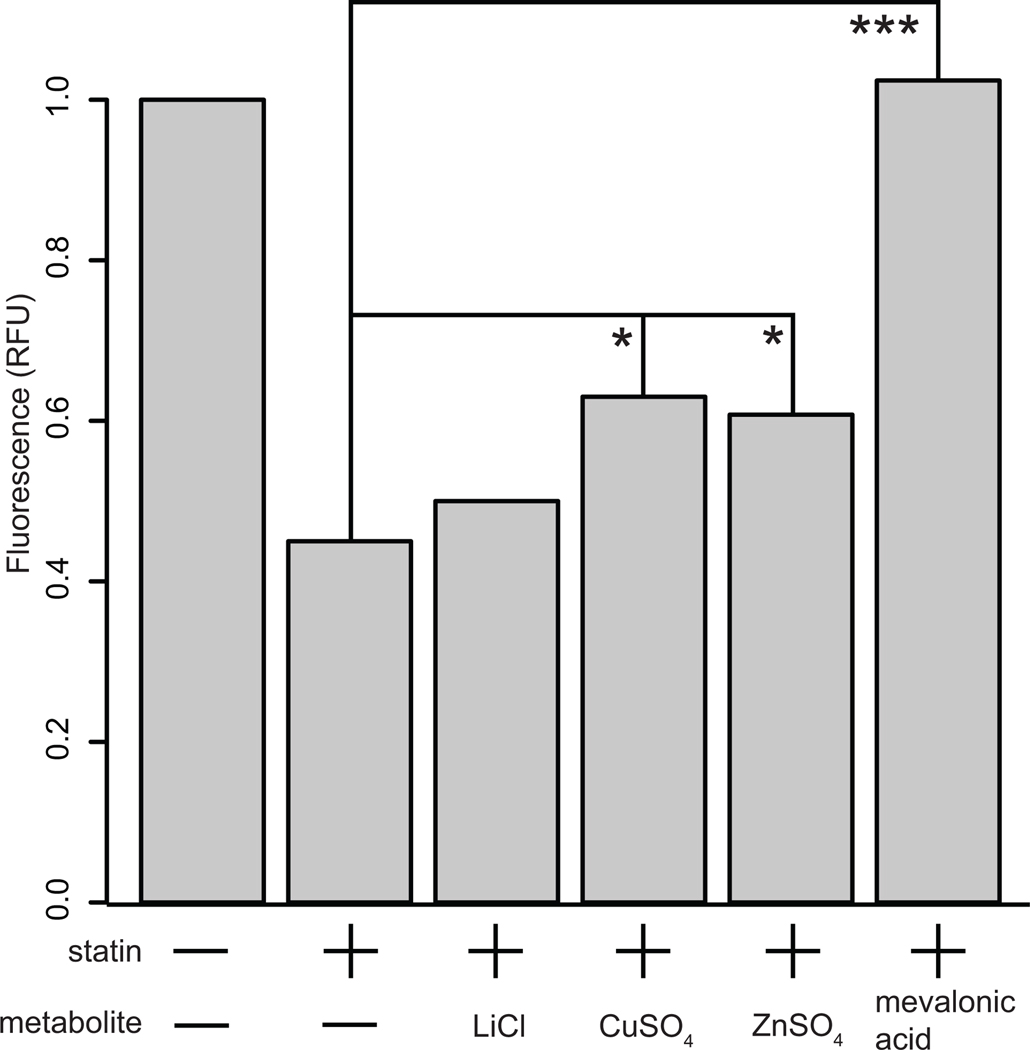
We treated HeLa cells with lovastatin to obtain an approximately 50% reduction in cell density. Treatment with zinc or copper produced a significant rescue of this statin growth inhibition (copper 40.4% increase, zinc 35% increase) (Fig. 6). Lithium, which failed to rescue statin growth inhibition in the yeast assay, did not result in a significant increase in HeLa cell growth. Mevalonic acid, the positive control in the yeast assay, produced a complete rescue of statin growth inhibition in HeLa cells. These results, showing consistent patterns in the activity of the two metals in lovastatin-treated yeast and mammalian cells, suggest that copper and zinc intake may be of relevance to human patients taking statins.
Figure 6.
Zinc and copper rescue statin growth inhibition in cultured mammalian cells. HeLa cells were treated with statin in combination with lithium (negative control), copper, zinc and mevalonic acid (positive control). Cell growth was measured by calcein dye uptake. Statin treatment produced a significant growth inhibition. Mevalonic acid effected complete rescue (***, p < 0.001), whereas copper (*, p <0.05) and zinc (*, p < 0.05) led to a partial rescue. Lithium, which did not produce a rescue effect in yeast, failed to ameliorate statin growth inhibition in HeLa cells.
Conclusion
As no obvious mechanistic link explains how metals influence ergosterol synthesis, we employed an integrated approach that analyzed both gene expression and metabolite profiles. Gene expression measurements of yeast treated with statin and copper or zinc revealed that the drug and the metals co-regulate the expression of many genes. Thus, unlike many other drug-metabolite interactions in which the drug and metabolite act upon the same enzyme, the interaction between lovastatin and these metals occurs on the level of the entire ergosterol biosynthetic pathway.
Metabolomic analysis showed that under ergosterol starvation stress, the ergosterol biosynthetic pathway becomes highly efficient in terms of intermediate utilization. Metabolic flux through the pathway is increased to enable the production of as much ergosterol as possible, at a cost of maintaining an elevated expression of the ergosterol biosynthetic machinery. Indeed, others have observed enhanced conversion of ergosterol precursors into ergosterol during ergosterol starvation.34 However, the metals were not capable of restoring ergosterol to the level in cells not treated with statin, supporting the idea that ergosterol is growth limiting even in statin-treated cultures that also contained copper or zinc. Growth rates increased as much as possible, given the lower rate of ergosterol biosynthesis.
Interactions between nutritive metabolites and drugs are important in humans. We showed that zinc and copper were capable of rescuing statin-induced growth inhibition in cultured mammalian cells as well as yeast. Our results suggest that by upregulating sterol biosynthesis, excess copper and zinc could reduce the effectiveness of statin treatment in humans. Intriguingly, large amounts of dietary copper cause significant increases in cholesterol levels in rats,35 consistent with our findings. Furthermore, statin treatment in humans alters plasma copper and zinc levels.36–38 These data highlight the importance of metabolite-statin interactions, and suggest that further investigation into the clinical relevance of copper and zinc in the context of hypercholesterolemia and statin treatment is warranted. Copper and zinc intake can be regulated, and levels of these metals are easy to assay in human plasma. Thus, management of the levels of these metals could constitute a new way to increase the effectiveness of statin treatment.
Experimental
General methods
All chemicals and reagents were obtained from Sigma (St. Louis, MO) unless otherwise indicated. BY4741a, BY4742α and PDR5 knockout strains thereof were obtained from Open Biosystems (Huntsville, AL) and used without further modification. Cell culture media was obtained from GIBCO/Invitrogen (Carlsbad, CA). Computational and statistical analyses were carried out using the R statistical computing program. Lovastatin was reconstituted at 10 mg/mL with 0.25% (w/v) NaOH in 85% DMSO/H2O. The reconstituted lovastatin was then hydrolyzed at 60 °C for 1 hour in a shaker incubator.
Data availability
Microarray data will be deposited in the GEO database at the time of publication.
Bioscreen growth assay
Yeast (BY4741a or BY4742α) carrying the pdr5Δ mutation was grown to saturation in SC media + 2% glucose for 20–24 hr at 30 °C with constant agitation. The culture was back diluted to 0.01 OD600 (-statin) or 0.09 OD600 (+statin, 5 µg/mL) and these pretreatment cultures were grown for an additional 18–20 hr, at which point an OD600 of approximately 0.4–0.6 was reached. Cultures were spun down and resuspended in fresh media (SC, 2% glucose, 0.25 mM EDTA) either with or without statin (5 µg/mL) at an OD600 of 0.1 for the assay. Metabolites were added as indicated and the vehicle (DMSO) concentration was adjusted to 0.1% (v/v) in all cases. Triplicate 100 µl samples were loaded into 100 well Bioscreen plates (Growth Curves USA, Piscataway, NJ). Plates were incubated at 30 °C with maximum shaking and OD600 readings were taken every 30 min for 24 hr. Of the 52 metabolites we tested, we were able to obtain growth data for 41. Ketoconazole growth curves were obtained in an analogous manner for 0.03, 0.3, 1.5, 3 and 30 µg/mL ketoconazole.
Bioscreen data analysis
Triplicate data from each run of the Bioscreen growth assay were averaged and normalized to the maximum OD600 value of the untreated control. The time at which the untreated and statin-treated cultures reached 50% of their maximum OD600 value was identified (T50untreated and T50treated). The OD600 value for each metabolite-treated culture at the appropriate time (T50untreated or T50treated) was used to compute the fitness score, W = [OD600 metabolite / OD600 untreated]. Epistasis was calculated using the product method where epistasis = Wstatin/metabolite − Wstatin * Wmetabolite.14 P-values were calculated with R using a linear model to quantify the effect of each metabolite treatment on the observed epistasis values. These were corrected for multiple testing using false discovery rate control, at a 5% false discovery rate.39, 40
RNA isolation, labeling and hybridization
The yeast strain BY4741a pdr5Δ was grown as for the Bioscreen assay, and harvested at approximately 50% of the eventual maximum OD600 (~0.4 for +statin, ~0.8 for –statin). Cultures were collected using a 0.45 µm vacuum filter with nylon filter membranes and the cell cake was rapidly frozen in liquid nitrogen and stored at −80 °C. RNA isolation was initiated by adding 4 mL lysis buffer (0.1 M EDTA, 0.5% SDS, 0.01 M Tris pH 7.5) to the frozen filter membrane and vortexing thoroughly. Then, 3 mL acid phenol was added and the tube was vortexed and placed in a 65 °C water bath for 1 hr with additional vortexing every 20 min. The filter paper was removed with tweezers and the liquid poured into a phase lock gel tube (5Prime, Gaithersburg, GA), incubated at 4 °C for 10 min and centrifuged for 10 min at 1880 RCF. 3 mL of chloroform was added, and the tube was inverted 3 times to mix and then centrifuged at 1880 RCF for 10 min. The chloroform extraction was repeated and the aqueous layer was poured into a new 15 mL conical tube. The RNA was precipitated by adding 1/10 volume 3 M sodium acetate and 2 volumes of 100% ethanol followed by mixing and incubation overnight at −20 °C. The precipitated RNA was pelleted and washed twice with 70% ethanol, then reconstituted in 250 µL deionized, RNAse-free H2O. For dye labeling, 5 µg of RNA were purified using Qiagen RNeasy kit (Qiagen, Valencia, CA), and a reference RNA sample was created by pooling all 6 conditions. RNA was labeled and hybridized to Agilent S. cerevisiae v2 (8 × 15k) microarrays following the Agilent two-color gene expression protocol v5.7 (Agilent Technologies, Santa Clara, CA), although only 25% of the recommended amounts of Cy3 and Cy5 dyes were used in each reaction. Slides were washed according to the Agilent protocol and then scanned using an Agilent High Resolution C scanner. There were two randomly chosen conditions that were replicated per slide; these technical replicates were used to assess RNA labeling and hybridization quality. Three independent biological replicates were performed according to the procedure outlined above.
Microarray data analysis
The Agilent Feature Extractor software was used, with default settings, to generate log ratios for each spot. Probes that were not significantly above background were filtered out. Probes corresponding to individual genes were averaged, as were technical replicates. To identify genes that were differentially regulated under different treatment conditions, we employed a two-stage, linear mixed model approach.41, 42 Linear mixed models were fit using the lme4 package.43 First, a normalization model was fit:
where Ygij is the log ratio from each gene g, treatment i, and biological replicate j. μ represents the mean value, T the treatment, R the biological replicate, TR the interaction between treatments and replicates and ε the error. TR is treated as a random effect. Residuals from this model, rgij, represent normalized log ratios. The treatment effect for each gene was then modeled individually:
F statistics were computed for each gene and significance was assessed using an empirical null distribution (generated for each gene by randomizing treatment factors (n = 1000)). P-values were computed from the empirical null distribution; genes whose p-values were equal to 0 were tested against an expanded null distribution (n = 6000). Genes whose p-values were still equal to 0 were then considered to have a p-value equal to 1.67e-4 (1/6000). The list of p-values was corrected for multiple testing using false discovery rate control.39, 40 The resulting genes were clustered using a centroid-linked hierarchical clustering algorithm in the Cluster 3.0 software44 and visualized using HIDRA.45 Gene Ontology (GO) analysis was performed using HIDRA with a p-value cutoff of 0.05 and Bonferroni correction. To eliminate overlapping and redundancy, representative ontologies were selected and included in Table 1.
HeLa growth assay
HeLa cells were grown to ~80% confluence in D-MEM media with 10% fetal bovine serum and 1% penicillin/streptomycin. Cells were washed with PBS and trypsinized for 1–2 min with 0.5 mL trypsin EDTA, and then the trypsin was neutralized with 4.5 mL media. Cells were counted in a hemocytometer and diluted to a concentration of 20,000 cells/mL. Lovastatin was used at a concentration of 20 µM. For the assay, cells were incubated with metabolites in 250 µl of media in 96 well plates for 48 hours at 37 °C, 5% CO2. Media was removed by aspiration and 100 µl of 1 µM calcein AM (#80013, Biotium Inc, Hayward, CA) was added to the wells and incubated at 37° C for 30 min; fluorescence was measured on a Victor V3 plate reader (Perkin Elmer, Waltham, MA) (ex: 485 nm, em: 535 nm).
Ergosterol extraction and quantitation assay
Ergosterol was quantified following the method of Arthington-Skaggs.46 Yeast were grown and treated as in the Bioscreen assay except that the final cultures incubated at 30 °C with constant agitation for 48 hours. Cultures were spun down in pre-weighed 15 mL conicals at 1500 × g for 5 min and washed with ddH2O. Pellets were dried for 10 min under vacuum to remove excess water and weighed. 3 mL alcoholic KOH (25 g KOH, 35 mL ddH2O and filled to 100 mL with 100% EtOH) was added to the yeast pellets. This suspension was vortexed for 1 min, transferred to an 8 mL borosilicate glass tube and heated in an 85 °C water bath for 1 hr. Tubes were cooled to room temperature, 3 mL n-heptane was added to each tube and then the tubes were vortexed for 3 min. 1 mL of the n-heptane layer was removed to a 1.7 mL Eppendorf tube; the solvent was removed under vacuum for 45 min with no heat. The resulting solid was resuspended in 200 µL fresh n-heptane and then 800µL 100% EtOH was added. Samples were read every 2 nm between 230 and 300 nm on a 1420 Victor V3 plate reader (PerkinElmer, Waltham, MA) spectrophotometer. Ergosterol content was calculated as a percentage of the wet weight of the cell pellet by the following equation: {[(A282/290)*F]-[(A230/518)*F]}/pellet weight, where F is the factor for dilution in ethanol and 290 and 518 are the E values (in percentages per centimeter) determined for crystalline ergosterol and 24(28)DHE, respectively.
Sterol metabolite profiling by GCxGC-MS
Heptane extractions were performed as described above; solvent was removed from 500 µL extract using a SpeedVac on medium heat. Excess water was removed by adding 100 µL of methylene chloride and drying again. Trimethylsilylation derivatization was done in glass similarly to the previously described methods (Mohler et al 2008; Humston et al. 2008). Briefly, to each sample, we added 30 µL of a 20 mg/mL solution of methoxyamine in pyridine to protect carbonyl groups. We heated the samples at 30 °C for 90 min. Then, we added 70 µL of MSTFA (N-methyl-N-trifluoroacetamide) with 1% TCMS (trichloromethylsilane) (catalog #TS-48915, Thermo Fisher Scientific, Waltham, MA) and incubated at 60 °C for 60 min. The samples were assayed immediately after derivatization on a Leco 4D GCxGC-TOFMS system (Leco, St. Joseph, MI). The primary column is a 20 m × 250 µm i.d. × 0.5 µm RTX-5MS film (Restek, Bellefonte, PA) and the secondary column is a 2 m × 180 µm i.d. × 0.2 µm RTX-200MS film (Restek). Injections of 1 µL were made in split mode with a split ratio of 1:5. The inlet was set to 280 °C, the transfer line was set to 305 °C. Flow rate for the carrier gas, helium, was 1 mL/min. Initial oven temperatures were 60 °C for the primary oven, 75 °C for the secondary oven. Modulator temperature was maintained at 30 °C above the primary oven temperature. Oven temperatures were increased at a rate of 7 °C/min to final temperatures of 325 °C and 340 °C, respectively. The total run time was approximately 50 min. The modulation time for the second dimension was 5 seconds with 0.4 s hot, 2.1 s cold. The ion source was set at 250 °C and data were collected at a rate of 100 spectra per second after a 7 min solvent delay.
GCxGC-MS data analysis and metabolite identification
Data were processed using the Chromatof 4.22 software for deconvolution and peak calling. Metabolites were quantified using the m/z 73 ion as a reference, the mass to charge ratio of the fragment released from the derivatized molecules. Using unique mass as defined by default in the software does not significantly change the results. For squalene, we quantified based on m/z 81 because derivatization of squalene is not efficient. Each compound was identified by searching acquired spectra against the NIST library, the commercially available Fiehn library19 and sterol standards we assayed (ergosterol, zymosterol, lanosterol and squalene). We confirmed identities of additional peaks by making heptane extracts (described above) from yeast strains deleted for genes of the ergosterol biosynthetic pathway, which accumulate upstream intermediates. These strains were lacking ERG2, ERG3 , ERG4, ERG5, ERG6, or ERG28. The combination of these techniques allowed us to identify most of the sterol intermediates we observed among the yeast extracts.
Supplementary Material
Acknowledgements
We thank Josh Akey and Maitreya Dunham for help with microarray methods and data analysis. Ted Young and Maitreya Dunham gave helpful comments on the manuscript. Adam Quintero provided assistance during the early stages of this effort. This work was supported by the NIH P41RR11823 to S.F.; D.M.F. (F32GM084699) and S.J.C. (F32DK080608) are recipients of Ruth L. Kirchstein NRSA awards. S.F. is an investigator of the Howard Hughes Medical Institute.
References Cited
- 1.Bailey DG, Spence JD, Munoz C, Arnold JM. Lancet. 1991;337:268–269. doi: 10.1016/0140-6736(91)90872-m. [DOI] [PubMed] [Google Scholar]
- 2.Kamali F, Wynne H. Annu. Rev. Med. 2010;61:63–75. doi: 10.1146/annurev.med.070808.170037. [DOI] [PubMed] [Google Scholar]
- 3.Rapaport MH. J. Clin. Psychiatry. 2007;68 Suppl 8:42–46. [PubMed] [Google Scholar]
- 4.Hebert PR, Gaziano JM, Chan KS, Hennekens CH. JAMA. 1997;278:313–321. [PubMed] [Google Scholar]
- 5.Endo A, Kuroda M, Tanzawa K. FEBS Lett. 1976;72:323–326. doi: 10.1016/0014-5793(76)80996-9. [DOI] [PubMed] [Google Scholar]
- 6.Law M, Rudnicka AR. Am. J. Cardiol. 2006;97:52C–60C. doi: 10.1016/j.amjcard.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 7.Stagnitti MN. Trends in Statins Utilization and Expenditures for the U.S. Civilian Noninstitutionalized Population, 2000 and 2005. Medical Expenditure Panel Survey; 2008. [Google Scholar]
- 8.Schwartz JB. Clin. Pharmacol. Ther. 2009;85:198–203. doi: 10.1038/clpt.2008.165. [DOI] [PubMed] [Google Scholar]
- 9.Callegari S, McKinnon RA, Andrews S, de Barros Lopes MA. FEMS Yeast Res. 2010;10:188–198. doi: 10.1111/j.1567-1364.2009.00593.x. [DOI] [PubMed] [Google Scholar]
- 10.Dimster-Denk D, Thorsness MK, Rine J. Mol. Biol. Cell. 1994;5:655–665. doi: 10.1091/mbc.5.6.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogers B, Decottignies A, Kolaczkowski M, Carvajal E, Balzi E, Goffeau A. J. Mol. Microbiol. Biotechnol. 2001;3:207–214. [PubMed] [Google Scholar]
- 12.Westermeyer C, Macreadie IG. FEMS Yeast Res. 2007;7:436–441. doi: 10.1111/j.1567-1364.2006.00194.x. [DOI] [PubMed] [Google Scholar]
- 13.Wikhe K, Westermeyer C, Macreadie IG. Biochem. Soc. Trans. 2007;35:1529–1532. doi: 10.1042/BST0351529. [DOI] [PubMed] [Google Scholar]
- 14.Mani R, St Onge RP, Hartman JL, Giaever G, Roth FP. Proc. Natl. Acad. Sci. USA. 2008;105:3461–3466. doi: 10.1073/pnas.0712255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yasokawa D, Murata S, Kitagawa E, Iwahashi Y, Nakagawa R, Hashido T, Iwahashi H. Environ. Toxicol. 2008;23:599–606. doi: 10.1002/tox.20406. [DOI] [PubMed] [Google Scholar]
- 16.Pagani MA, Casamayor A, Serrano R, Atrian S, Arino J. Mol. Microbiol. 2007;65:521–537. doi: 10.1111/j.1365-2958.2007.05807.x. [DOI] [PubMed] [Google Scholar]
- 17.Agarwal AK, Rogers PD, Baerson SR, Jacob MR, Barker KS, Cleary JD, Walker LA, Nagle DG, Clark AM. J. Biol. Chem. 2003;278:34998–35015. doi: 10.1074/jbc.M306291200. [DOI] [PubMed] [Google Scholar]
- 18.Espenshade PJ, Hughes AL. Annu. Rev. Genet. 2007;41:401–427. doi: 10.1146/annurev.genet.41.110306.130315. [DOI] [PubMed] [Google Scholar]
- 19.Kind T, Wohlgemuth G, Lee do Y, Lu Y, Palazoglu M, Shahbaz S, Fiehn O. Anal. Chem. 2009;81:10038–10048. doi: 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, Botstein D. Mol. Biol. Cell. 2008;19:352–367. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Bakel H, Strengman E, Wijmenga C, Holstege FC. Physiol. Genomics. 2005;22:356–367. doi: 10.1152/physiolgenomics.00055.2005. [DOI] [PubMed] [Google Scholar]
- 22.Rustici G, van Bakel H, Lackner DH, Holstege FC, Wijmenga C, Bahler J, Brazma A. Genome Biol. 2007;8:R73. doi: 10.1186/gb-2007-8-5-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu CY, Bird AJ, Chung LM, Newton MA, Winge DR, Eide DJ. BMC Genomics. 2008;9:370. doi: 10.1186/1471-2164-9-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyons TJ, Gasch AP, Gaither LA, Botstein D, Brown PO, Eide DJ. Proc. Natl. Acad. Sci. USA. 2000;97:7957–7962. doi: 10.1073/pnas.97.14.7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrero E, Ros J, Belli G, Cabiscol E. Biochim. Biophys. Acta. 2008;1780:1217–1235. doi: 10.1016/j.bbagen.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 26.Lyons TJ, Villa NY, Regalla LM, Kupchak BR, Vagstad A, Eide DJ. Proc. Natl. Acad. Sci. USA. 2004;101:5506–5511. doi: 10.1073/pnas.0306324101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuranda K, Francois J, Palamarczyk G. FEMS Yeast Res. 2009;10:14–27. doi: 10.1111/j.1567-1364.2009.00560.x. [DOI] [PubMed] [Google Scholar]
- 28.Bammert GF, Fostel JM. Antimicrob. Agents Chemother. 2000;44:1255–1265. doi: 10.1128/aac.44.5.1255-1265.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies BS, Rine J. Genetics. 2006;174:191–201. doi: 10.1534/genetics.106.059964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monteiro PT, Mendes ND, Teixeira MC, d'Orey S, Tenreiro S, Mira NP, Pais H, Francisco AP, Carvalho AM, Lourenco AB, Sa-Correia I, Oliveira AL, Freitas AT. Nucleic Acids Res. 2008;36:D132–D136. doi: 10.1093/nar/gkm976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marion RM, Regev A, Segal E, Barash Y, Koller D, Friedman N, O'Shea EK. Proc. Natl. Acad. Sci. USA. 2004;101:14315–14322. doi: 10.1073/pnas.0405353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J, Godon C, Lagniel G, Spector D, Garin J, Labarre J, Toledano MB. J. Biol. Chem. 1999;274:16040–16046. doi: 10.1074/jbc.274.23.16040. [DOI] [PubMed] [Google Scholar]
- 33.Ter Linde JJ, Steensma HY. Yeast. 2002;19:825–840. doi: 10.1002/yea.879. [DOI] [PubMed] [Google Scholar]
- 34.Leszczynska A, Burzynska B, Plochocka D, Kaminska J, Zimnicka M, Kania M, Kiliszek M, Wysocka-Kapcinska M, Danikiewicz W, Szkopinska A. PLoS One. 2009;4:e8499. doi: 10.1371/journal.pone.0008499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galhardi CM, Diniz YS, Faine LA, Rodrigues HG, Burneiko RC, Ribas BO, Novelli EL. Food Chem. Toxicol. 2004;42:2053–2060. doi: 10.1016/j.fct.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 36.Ghayour-Mobarhan M, Lamb DJ, Taylor A, Vaidya N, Livingstone C, Wang T, Ferns GA. J. Trace Elem. Med. Biol. 2005;19:61–67. doi: 10.1016/j.jtemb.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 37.Ghayour-Mobarhan M, Taylor A, Kazemi-Bajestani SM, Lanham-New S, Lamb DJ, Vaidya N, Livingstone C, Wang T, Ferns GA. Clin. Lab. 2008;54:321–329. [PubMed] [Google Scholar]
- 38.Leonhardt W, Kurktschiev T, Meissner D, Lattke P, Abletshauser C, Weidinger G, Jaross W, Hanefeld M. Eur J Clin Pharmacol. 1997;53:65–69. doi: 10.1007/s002280050338. [DOI] [PubMed] [Google Scholar]
- 39.Storey JD, Tibshirani R. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benjamini Y, Hochberg Y. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- 41.Cui X, Churchill GA. Genome Biol. 2003;4:210. doi: 10.1186/gb-2003-4-4-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolfinger RD, Gibson G, Wolfinger ED, Bennett L, Hamadeh H, Bushel P, Afshari C, Paules RS. J. Comput. Biol. 2001;8:625–637. doi: 10.1089/106652701753307520. [DOI] [PubMed] [Google Scholar]
- 43.Maechler M, Bates D. lme4: Linear mixed-effects models using S4 classes. http://lme4.r-forge.r-project.org/.
- 44.de Hoon MJ, Imoto S, Nolan J, Miyano S. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 45.Hibbs M, Wallace G, Dunham M, Li K, Troyanskaya O. Viewing the Larger Context of Genomic Data through Horizontal Integration. 2007 [Google Scholar]
- 46.Arthington-Skaggs BA, Jradi H, Desai T, Morrison CJ. J. Clin. Microbiol. 1999;37:3332–3337. doi: 10.1128/jcm.37.10.3332-3337.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data will be deposited in the GEO database at the time of publication.