Abstract
The γ-hydroxy-1,N2-propano-2′-deoxyguanosine adduct (γ-OH-PdG) was introduced into 5′-d(GCTAGCXAGTCC)-3′·5′-d(GGACTCGCTAGC)-3′ (X = γ-OH-PdG). In the presence of excess peptide KWKK, 13C isotope-edited NMR revealed the formation of two spectroscopically distinct DNA–KWKK conjugates. These involved the reaction of the KWKK N-terminal amino group with the N2-dG propylaldehyde tautomer of the γ-OH-PdG lesion. The guanine N1 base imino resonance at the site of conjugation was observed in isotope-edited 15N NMR experiments, suggesting that the conjugated guanine was inserted into the duplex and that the guanine imino proton was protected from exchange with water. The conjugates could be reduced in the presence of NaCNBH3, suggesting that they existed, in part, as imine (Schiff base) linkages. However, 13C isotope-edited NMR failed to detect the imine linkages, suggesting that these KWKK conjugates existed predominantly as diastereomeric carbinolamines, in equilibrium with trace amounts of the imines. The structures of the diastereomeric DNA–KWKK conjugates were predicted from potential energy minimization of model structures derived from the refined structure of the fully reduced cross-link [Huang, H., Kozekov, I. D., Kozekova, A., Rizzo, C. J., McCullough, A., Lloyd, R. S., and Stone, M. P. (2010) Biochemistry, 24, 6155−6164]. Molecular dynamics calculations carried out in explicit solvent suggested that the conjugate bearing the S-carbinolamine linkage was the major species due to its potential for intramolecular hydrogen bonding. These carbinolamine DNA–KWKK conjugates thermally stabilized duplex DNA. However, the DNA–KWKK conjugates were chemically reversible and dissociated when the DNA was denatured. In this 5′-CpX-3′ sequence, the DNA–KWKK conjugates slowly converted to interstrand N2-dG:N2-dG DNA cross-links and ring-opened γ-OH-PdG derivatives over a period of weeks.
Introduction
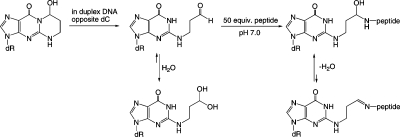
Acrolein (Scheme 1, 1), a potential mutagen and carcinogen,1,2 reacts with dG to form 3-(2-deoxy-β-d-erythro-pentofuranosyl)-5,6,7,8-tetrahydro-8-hydroxypyrimido[1,2a] purin-10(3H)-ones, γ-OH-PdG diastereomeric adduct 2.2,3 Adduct 2 has been detected in animal and human tissues2 and have been suggested to be involved in mutagenesis and carcinogenesis from exposure to acrolein.4 Methods for site-specific synthesis of 2 in oligodeoxynucleotides have been developed.5,6 When placed into DNA opposite dC at neutral pH, 2 opens spontaneously to aldehyde 3 and the corresponding aldehydrol 4.7 Aldehyde 3 potentially reacts with primary amines in proteins to yield DNA–protein conjugates (DPCs); DNA containing the structurally related 1,N2-dG adduct arising from crotonaldehyde exposures yields DNA–topoisomerase I conjugates.8 Indeed, DPCs have been detected in cultured human lymphoma cells exposed to acrolein.9 If not repaired, such DPCs are anticipated to interfere with both DNA replication and transcription.
Scheme 1. Acrolein-Derived γ-OH-PdG Adduct in the Presence of the Peptide KWKK When Placed Opposite 2′-Deoxycytosine in the 5′-CpX-3′ (X = N2-modified dG) Sequence Context.
The asterisks indicate the isotope-edited carbon or nitrogen.
The mechanisms by which cells repair DPCs are of considerable interest. One potential mechanism involves partial proteolysis, leading to the formation of DNA–peptide conjugates, as substrates for nucleotide excision repair (NER). This model is consistent with several observations. Inhibition of nuclear proteosomal protein degradation greatly reduces the repair of DPCs.10 The proteolytically active 20S core of the 26S proteasome localizes to the nucleus,11,12 and inhibition of proteasome function with lactacystin results in the inhibition of the repair of formaldehyde-induced DPCs, in normal, XP-A, and XP-F fibroblasts.10 When topoisomerase I is conjugated to DNA in the presence of camptothecin, proteolysis occurs in a ubiquitin-dependent fashion, giving rise to a ladder of DNA–polypeptide products.13
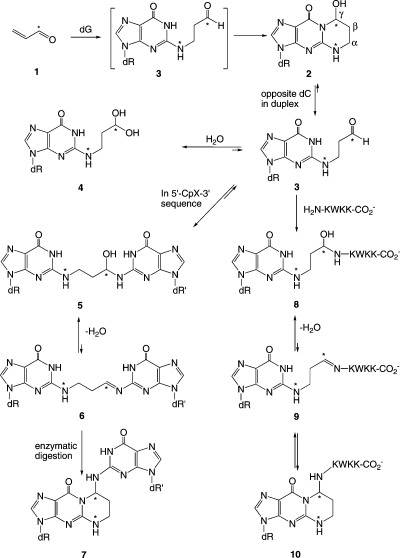
Kurtz and Lloyd14,15 trapped a DNA–KWKK conjugate involving a trimethylene linkage between the N-terminal amine of the peptide and N2-dG upon insertion of γ-OH-PdG adduct 2 into an oligodeoxynucleotide duplex, followed by exposure to a molar excess of the KWKK peptide, and NaCNBH3 treatment. Similar chemistry has been used to link peptides to γ-OH-1,N2-propano-2′-deoxyadenosine (γ-OH-PdA).16 The observation that the peptide conjugate is reducible implies the presence of an imine 9, in equilibrium with a conjugated carbinolamine 8. However, carbinolamine 8, imine 9, and potentially the pyrimidopurinone 10 conjugates exist in equilibrium (Scheme 1), and monitoring the composition of the mixture in situ was of considerable interest. All three conjugated species may influence the biological processing of DNA–KWKK conjugates.
The site-specific introduction of a 13C label at the γ carbon, and a 15N label at N2-dG enables the equilibrium chemistry of γ-OH-PdG adduct 2 to be monitored, in situ.17 Using this approach, we demonstrate that the native DNA–KWKK linkage exists predominantly as a pair of diastereomeric carbinolamines (Scheme 2), in equilibrium with trace amounts of the corresponding imine (Schiff base) linkages. The structures of the diastereomeric DNA–KWKK conjugates appear similar to the refined structure of the fully reduced cross-link.18 Molecular dynamics calculations carried out in explicit solvent suggested that the conjugate bearing the S-carbinolamine linkage is the major species due to its potential for intramolecular hydrogen bonding. The native carbinolamine DNA–KWKK conjugates stabilize the DNA to thermal denaturation. Upon dissociation in the 5′-CpX-3′ sequence, the native DNA–KWKK conjugates slowly convert to interstrand N2-dG:N2-dG DNA cross-links (5,6) or revert to the N2-dG aldehyde adducts (3, 4).
Scheme 2. Numbering Scheme of the Carbinolamine N2-dG:N-Lys DNA–KWKK Conjugates in Which the X7N2 or Cγ of the γ-Hydroxytrimethylene Tether was Isotope-Edited (In Red Color).
Materials and Methods
Materials
The oligodeoxynucleotide 5′-d(GGACTCGCTAGC)-3′ was synthesized and purified by the Midland Certified Reagent Co. (Midland, TX). It was purified by anion-exchange chromatography. Site-specific 13C- or 15N-labeled γ-OH-PdG adducts were synthesized and incorporated into 5′-d(GCTAGCXAGTCC)-3′ (X = γ-OH-PdG) using established procedures.17 The oligodeoxynucleotides were characterized by MALDI-TOF mass spectrometry; their purity was assessed by capillary gel electrophoresis and HPLC. Oligodeoxynucleotides were desalted on Sephadex G-25. The concentrations of the oligodeoxynucleotides were measured by UV absorption at 260 nm.19 The strands were annealed in 10 mM NaH2PO4, 100 mM NaCl, and 50 μM EDTA (pH 7.0). The solutions were heated to 95 °C for 10 min, then slowly cooled to room temperature. The duplex oligodeoxynucleotides were purified using DNA grade hydroxyapatite with a gradient from 10 to 200 mM NaH2PO4 (pH 7.0) in 100 mM NaCl, 50 μM EDTA, and desalted by elution from Sephadex G-25. The tetrapeptide KWKK was synthesized using standard solid phase peptide synthesis methods employing Fmoc protecting groups and purified by reverse-phase HPLC. It was characterized by MALDI-TOF MS and NMR spectroscopy. The concentration of the peptide was determined by UV absorption at 280 nm.
Preparation of DNA–KWKK Conjugates
The DNA duplex containing the site-specific γ-OH-PdG adduct was dissolved at 0.8 mM strand concentration in 280 μL of 10 mM NaH2PO4, 100 mM NaCl, and 50 μM EDTA (pH 7.0). Fifty equivalents of KWKK were added, and pH was adjusted to 7.0 by the addition of dilute HCl or NaOH. The reaction was incubated at 5 °C for 8 weeks and was monitored by either 13C or 15N HSQC NMR. Excess KWKK was removed by gel filtration with Sephadex G-25. The product was lyophilized immediately. The DNA–KWKK conjugates were characterized by MALDI-TOF mass spectrometry: observed at m/z of 4272.5 (calcd for carbinolamine 8 [M-H2O-1]: 4272.7). The complementary strand was observed at m/z of 3645.6 (calcd for M-1: 3645.2).
NMR
Experiments were performed on a Bruker Avance 500 spectrometer using a cryogenic probe. Samples were at 0.8 mM strand concentration and were dissolved in 280 μL of 10 mM NaH2PO4, 100 mM NaCl, and 50 μM EDTA (pH 7.0). Samples for observation of nonexchangeable protons were exchanged with D2O and suspended in 99.996% D2O. The pH was adjusted with dilute DCl or NaOD solutions. Chemical shifts of protons were referenced to water. Chemical shifts of carbon and nitrogen resonances were not precisely referenced to an internal standard. Data were processed using FELIX (Accelrys Inc., San Diego, CA) on Silicon Graphics workstations (Silicon Graphics Inc., Fremont, CA). For all experiments, a relaxation delay of 1.5 s was used. For assignment of exchangeable protons, NOESY experiments used the WATERGATE sequence.20 Spectra were recorded with 512 real data in the t1 dimension and 2048 real data in the t2 dimension, and were zero-filled during processing to create a matrix of 1024 × 512 real points. The mixing time was 250 ms. For assignment of nonexchangeable protons, NOESY experiments used TPPI quadrature detection and a mixing time of 250 ms. The spectra were recorded with 512 real data in the t1 dimension and 2048 real data in the t2 dimension, and were zero-filled during processing to create a matrix of 1024 × 1024 real points. 15N HSQC experiments21−23 were performed with WATERGATE water suppression.20,24 The spectra were recorded with 256 real data in the t1 dimension and 1024 real data in the t2 dimension and were zero filled to create a matrix of 512 × 256 real points. The 2D 15N NOESY-HSQC pulse program was modified from the corresponding 3D pulse program25,26 with 128 points centered at 81 ppm. The water signal was suppressed using the WATERGATE method. The mixing time was 250 ms. The spectrum was recorded with 512 real data in the t1 dimension and 2048 real data in the t2 dimension, and was zero-filled during processing to create a 1024 × 512 matrix. 13C HSQC experiments were carried out using standard 1H-detected pulse programs with States-TPPI phase cycling.21−23 The spectra were recorded with 256 real data in the t1 dimension and 1024 real data in the t2 dimension, and were zero filled to create a matrix of 512 × 256 real points. The lifetimes of the peptide conjugates were determined with the 13C-labeled sample using 13C HSQC NMR. The sample was incubated for one month at 37 °C in 10 mM NaH2PO4, 100 mM NaCl, and 50 μM EDTA (pH 7.0).
Thermal Melting Studies
Melting temperatures of the DNA–KWKK conjugates were measured in 10 mM NaH2PO4, 100 mM NaCl, and 50 μM EDTA (pH 7.0) by UV spectroscopy at 260 nm. The strand concentration was 10 μM. The thermal scan proceeded from 15 to 85 °C. The melting temperatures were calculated by differentiating the absorbance profiles.
Molecular Modeling
Starting structures were created from refined structures of the fully reduced trimethylene DNA–peptide conjugate,18 using the program INSIGHT II (Accelrys, Inc., San Diego, CA). The partial charges of the γ-hydroxytrimethylene tether were obtained from density function theory (DFT) calculations using a neutral total charge, utilizing the B3LYP/6-31G* basis set and the program Gaussian.27 Potential energy minimization calculations were conducted with the program AMBER28 and the parm99 force field. The pairwise generalized Born (GB) model29,30 was used to simulate implicit waters. The cutoff radius for nonbonding interactions was 18 Å. A 1000-iteration potential energy minimization was performed, using the conjugate gradient algorithm.
Molecular Dynamics Simulations
The simulations were performed in explicit water using the AMBER force field with the particle mesh Ewald (PME) method.31,32 The potential energy-minimized structures of the R- and S-carbinolamine DNA–KWKK conjugates were used as the starting structures. The conjugates were surrounded by a 10.0 Å TIP3P water box in each direction. A total of 20 Na+ ions were added to neutralize the duplex. Bond lengths involving hydrogen were fixed using the SHAKE algorithm.33 The cutoff radius for nonbonded interactions was 10 Å. The conjugate was first energy-minimized for 2,500 iterations. The MD simulation was then carried out with constant volume at 300 K for 10,000 iterations with an integrator time of 1 fs. The MD simulation at constant pressure was performed at 300 K for 5 ns with an integrator time of 2 fs. The PTRAJ program from the AMBER package28 [v. 10] was used to analyze the trajectories. The rmsd values of the trajectories were referenced to the starting structures. A distance of less than 3.5 Å and an angle of greater than 120° between the potential hydrogen donor and acceptor were used as criteria for hydrogen bond formation. Helicoidal analysis of the backbone was carried out using CURVES.34,35
Results
Synthesis of DNA–KWKK Conjugates
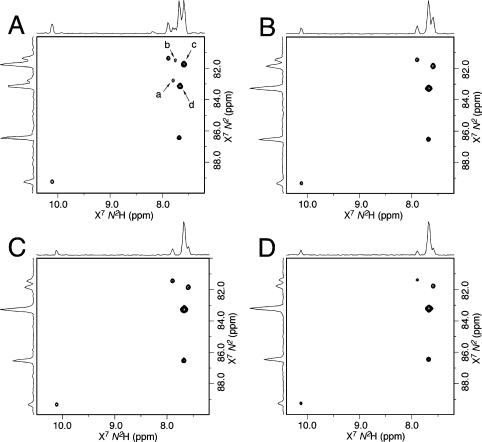
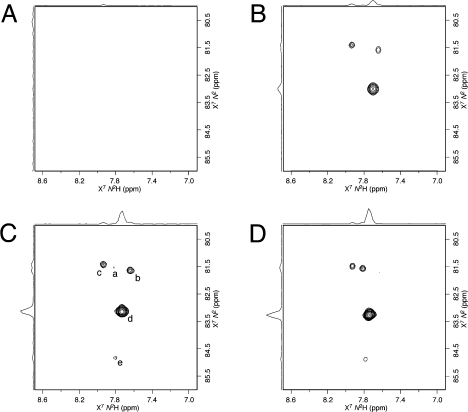
The intramolecular formation of N2-dG:N2-dG DNA interstrand cross-links (5,6) competes with the bimolecular association of the KWKK peptide with the DNA (Scheme 1). To minimize the formation of interstrand DNA cross-links, excess KWKK was used. To avoid producing DNA–KWKK conjugates resulting from the reaction with the amino groups of the lysine side chains, the reaction was carried out at pH 7.0 such that the lysine side chains were protonated.36 Using a 15N-labeled sample, the reaction was monitored by 15N HSQC NMR (Figure 1). Two X7N2 → N2H correlations arising from the DNA–KWKK conjugate 8 became stronger as time elapsed. Simultaneously, the X7N2 → N2H correlations of other γ-OH-PdG derivatives became weaker and disappeared. The intensities of the three cross-peaks arising from the natural abundance of the excess KWKK amides remained constant. At 5 °C, the reaction approached equilibrium after 6 weeks. After 8 weeks, the excess peptide was removed by gel filtration to facilitate the analyses of the DNA–KWKK conjugate. A total of less than 10% of nonconjugated oligodeoxynucleotides and DNA interstrand cross-links was also observed (Figure 2). These could not be separated from the DNA–KWKK conjugate product without causing the dissociation of the DNA–KWKK conjugates. The DNA–KWKK conjugates were characterized by MALDI-TOF mass spectrometry.
Figure 1.
15N HSQC of the reaction of γ-OH-PdG containing oligodeoxynucleotide with the peptide KWKK (1:50, pH 7.0). (A) Immediately after KWKK was added. (B) At 5 °C for 2 weeks. (C) At 5 °C for 4 weeks. (D) At 5 °C for 8 weeks. X7N2 → N2H correlations were assigned as follows: a, γ-OH-PdG 2; b, N2-aldehyde 3; c, N2-aldehydrol 4; d, DNA–KWKK conjugates 8. Other cross-peaks arose from the natural abundances of excess KWKK amides.
Figure 2.
15N HSQC spectra of the 15N-labeled DNA–KWKK conjugates at different temperatures at pH 5.3: (A) at 10 °C, (B) at 20 °C, (C) at 30 °C, and (D) at 40 °C. X7N2 → N2H correlations were assigned to (a) N2-aldehyde 3, (b) N2-aldehydrol 4, (c) DNA interstrand cross-link 5, (d) DNA–KWKK conjugates 8, and (e) unassignable, might be arising from DNA–KWKK conjugates 9, 10, or a side-chain byproduct.
NMR Spectroscopy
Figure 2 shows the 15N HSQC spectra of the 15N-labeled DNA–KWKK conjugates at different temperatures at pH 5.3. The X7N2 → N2H correlations were broad at 10 °C, and they were sharper at higher temperatures. A similar phenomenon was observed for the Cγ → Hγ correlations in the 13C HSQC spectra. This was attributed to aggregation of the DNA–KWKK conjugates due to the electrostatic interaction of a positively charged peptide with negatively charged DNA. The spectra at neutral pH and the 1H resonances of the DNA–KWKK conjugates were compared with those in acidic and basic solutions. No pH dependence was observed. Satisfactory spectra were recorded at pH 5.3, pH 7.0, and pH 8.9 and 25 and 30 °C for the exchangeable protons and at 37 °C for the nonexchangeable protons. However, some of the exchangeable proton resonances remained broad under these conditions.
Spectroscopic Characterization of the DNA–KWKK Conjugate Linkage
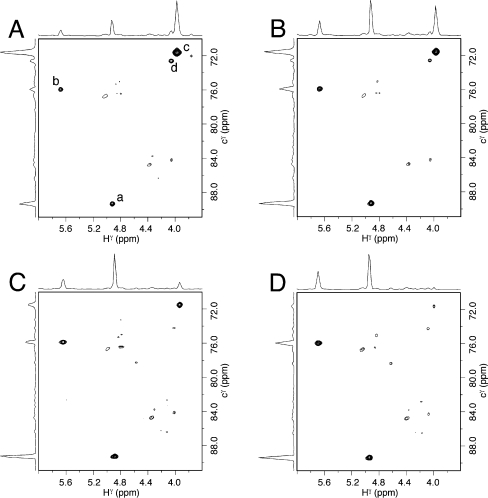
Figure 3A displays the isotope-edited 13C HSQC spectrum of the Cγ DNA–KWKK conjugates. Two Cγ → Hγ correlations were assigned to the conjugates. They were in equilibrium with N2-dG:N2-dG interstrand cross-links (5,6) and ring-opened species (3,4) derived from the γ-OH-PdG adduct (Scheme 1). Kurtz et al.14 reported that the γ-OH-PdG induced DNA–KWKK conjugates were reducible by NaCNBH3, suggesting that imine conjugate 9 was present. However, compared with the resonances of imine carbon and hydrogen,37 the resonances of Cγ and Hγ of both DNA–KWKK conjugates were located significantly upfield, suggesting that neither of the two NMR-detectable DNA–KWKK conjugates was imine 9.
Figure 3.
13C HSQC spectra of the 13C labeled DNA–KWKK conjugates at pH 7.0: (A) immediately after isolation, (B) at 37 °C for 1 week, (C) at 37 °C for 2 weeks, and (D) at 37 °C for 4 weeks. Cγ → Hγ correlations were assigned to (a) N2-aldehydrol 4, (b) DNA interstrand cross-link 5, (c) major DNA–KWKK conjugate 8, and (d) minor DNA–KWKK conjugate 8. Other cross-peaks were assigned to the C → H correlations of natural abundance.
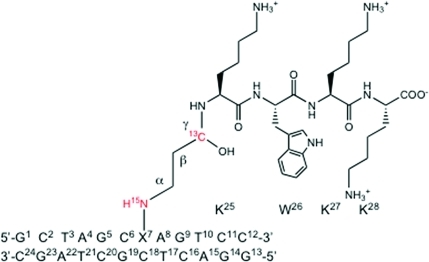
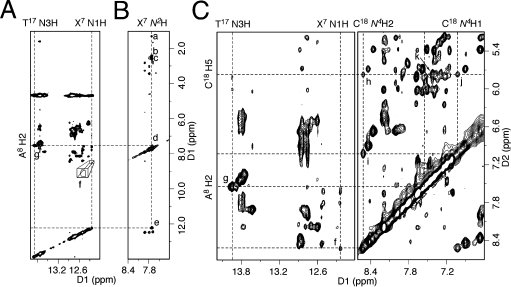
Isotope-edited 15N HSQC spectra of the X7N2 15N-labeled DNA–KWKK conjugates are displayed in Figure 2. The X7N2 → N2H correlations of two DNA–KWKK conjugates were not resolved, although the cross-peaks appeared to split at 40 °C. Corresponding to the X7N2 → N2H correlations, two sets of NOEs were observed in the 15N NOESY-HSQC spectrum (Figure 4B). The slightly downfield X7N2H resonance was assigned to the major DNA–KWKK conjugate and the upfield X7N2H resonance was assigned to the minor. Each conjugate exhibited a strong NOE between X7N2H and guanine (X7 or G19) N1H imino protons. The NOESY data suggested that all nucleotides of the major DNA–KWKK conjugate adopted the anti conformation about the glycosyl bond. In B-DNA, the anti conformation places the neighboring G19 N1H 5.0–6.0 Å from the X7N2H amino proton. In contrast, the X7 N1H imino proton in carbinolamine 8 is ∼2.0 Å from the X7N2H amino proton. Therefore, these strong NOEs were assigned to X7N2H → X7 N1H correlations. This was consistent with the X7 N1H → C18N4H2, X7N2H → Hα1, X7N2H → Hα2, X7N2H → Hβ, and X7N2H → A8 H2 NOEs (Figure 4). Pyrimidopurinone 10 lacks an imino proton. The observation of the X7 N1H resonance proved that these two DNA–KWKK conjugates were diastereomeric carbinolamines 8. This conclusion was also consistent with the chemical shifts of the Hγ protons. Pyrimidopurinone 10 has a chemical structure similar to that of γ-OH-PdG and the pyrimidopurinone dG-dG cross-link. Compared with their Hγ protons,17,38−40 the Hγ resonances of the two DNA–KWKK conjugates were shifted significantly upfield.
Figure 4.
Expansions of NOESY spectra at pH 5.3: (A) NOESY at 30 °C, (B) 15N NOESY-HSQC at 30 °C, and (C) NOESY at 25 °C. NOEs of the major DNA–peptide conjugate were assigned to (a) X7N2H → Hβ, (b) X7N2H → Hα1, (c) X7N2H → Hα2, (d) X7N2H → A8 H2, (e) X7N2H → X7 N1H, (f) X7 N1H → C18N4H2, (g) T17 N3H → A8 H2, (h) C18N4H2 → C18 H5, (i) C18N4H2 → C18N4H1, (j) C18N4H1 → C18 H5, and (k) C18 H6 → C18 H5.
Thermal Melting Studies
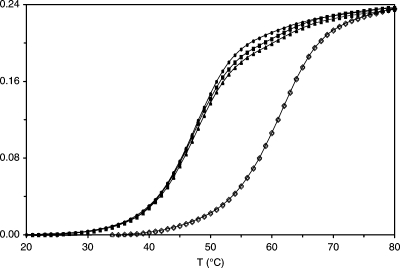
The thermal melting of the DNA–KWKK conjugates was monitored using UV spectroscopy at pH 7 in 100 mM NaCl (Figure 5). The DNA–KWKK conjugates exhibited a melting temperature (Tm) of 62 ± 1 °C. Compared with the unmodified DNA (Tm: 60 °C) and the DNA containing the γ-OH-PdG adduct (Tm: 48 °C), the formation of these DNA–KWKK conjugates stabilized the DNA.
Figure 5.
Melting studies of the DNA–KWKK conjugates by UV–vis (254 nm) at pH 7.0 in 100 mM NaCl: —◊— first experiment; —▲— second experiment; —◼— third experiment; —●— fourth experiment.
When the denatured DNA duplex containing the DNA–KWKK conjugates was reannealed, and the melting experiment repeated, a melting transition corresponding to DNA containing the γ-OH-PdG adduct was observed together with a weaker melting transition corresponding to the DNA–KWKK conjugates. Further thermal cycling showed that the melting transition of the DNA–KWKK conjugates became weaker in the third melting cycle and disappeared in the fourth. Thus, the DNA–KWKK conjugates reversed when the DNA was denatured. NMR experiments agreed with this conclusion. When the 13C-labeled DNA–KWKK conjugates were denatured at 85 °C for 30 min and reannealed, the Cγ → Hγ correlations of stereoisomeric carbinolamine conjugates 8 disappeared in the 13C HSQC spectrum, and simultaneously, the Cγ → Hγ correlations of N2-propylaldehyde 3 and N2-propanoaldehydrol 4 appeared.
The lifetimes of the carbinolamine DNA–KWKK conjugates 8 in DNA were evaluated by 13C HSQC NMR using a 13C-labeled sample (Figure 3). Immediately after isolation, two strong Cγ → Hγ correlations were observed. They became weaker as time elapsed. The minor carbinolamine diastereomer was unobservable after 2 weeks and the major after 4 weeks. They slowly converted to the unconjugated N2-propylaldehyde 3 (its Cγ → Hγ correlation not shown in Figure 3), N2-propanoaldehydrol 4, and DNA interstrand cross-links 5. Thus, the stereoisomeric carbinolamine DNA–KWKK conjugates have longer lifetimes than do the DPCs induced by formaldehyde whose half-life times are less than 1 day in vitro,10 and their lifetimes are comparable to the lifetime of DPC induced by malondialdehyde.41
NMR Resonance Assignments
Exchangeable protons of the major DNA–KWKK conjugate were assigned by comparing the 15N HSQC filtered NOESY spectrum with the nonisotope-edited NOESY spectrum (Figure 4). The X7 N1H imino resonance of the major DNA–KWKK conjugate broadened at 30 °C. Notably, it still exhibited a weak X7 N1H → C18N4H2 NOE in acidic solution (Figure 4A). This assignment was supported by NOEs including C18N4H2 → C18N4H1, C18N4H2 → C18 H5, C18N4H1 → C18 H5, and C18 H5 → C18 H6 (Figure 4C). The X7 N1H → C18N4H2 NOE suggested that the X7·C18 base pair maintained Watson–Crick hydrogen bonding. No evidence was observed for protonated G·C Hoogsteen base pairing often observed for 1,N2-dG adducts in acidic solution.42−44 Neither the C18N4H1 or C18N4H2 resonances shifted downfield compared to other cytosine amino protons.
The nonexchangeable protons were assigned on the basis of the NOE sequential connectivity of the base H6/H8 protons with 2′-deoxyribose H1′ protons.45,46 Complete sequential NOESY connectivities were observed for both modified and complementary strands of the major DNA–peptide conjugate. The X7 H8 → C6 H1′ NOE was weaker than other NOEs of the aromatic protons with their 5′-neighboring H1′ protons, indicating the X7·C18 and C6·G19 base pairs underwent small perturbations. The chemical shifts of aromatic H6/H8 and sugar H1′ protons were compared with those in the corresponding unmodified oligodeoxynucleotide. Except for C6, X7, and C18 H1′ protons, very small chemical shift perturbations were observed, indicating that the major DNA–peptide conjugate maintained B-DNA geometry. C6, X7, and C18 H1′ protons shifted downfield by 0.26, 0.25, and 0.34 ppm, respectively. These remarkable chemical shift perturbations suggested structural perturbations at base pairs X7·C18 and C6·G19. The deoxyribose H3′ and H2′(′′) protons were also assigned. Significant downfield shifts were not observed for the H2′(′′) protons, suggesting the nucleotides did not adopt the syn conformation.47−50
Molecular Modeling
The diastereomeric DNA–KWKK conjugates were modeled using the refined structure of the fully reduced trimethylene conjugate18 as a starting structure. The carbinolamine conjugates 8 of either R- or S-configurations were built by adding a hydroxyl group to the Cγ of the tether. Potential energy minimization was carried out to provide model structures. Figure 6 displays the modified region of structures viewed from the minor groove. Consistent with NMR data, the structures maintained Watson–Crick hydrogen bonding at the site of conjugation. The KWKK peptide was located in the minor groove.
Figure 6.
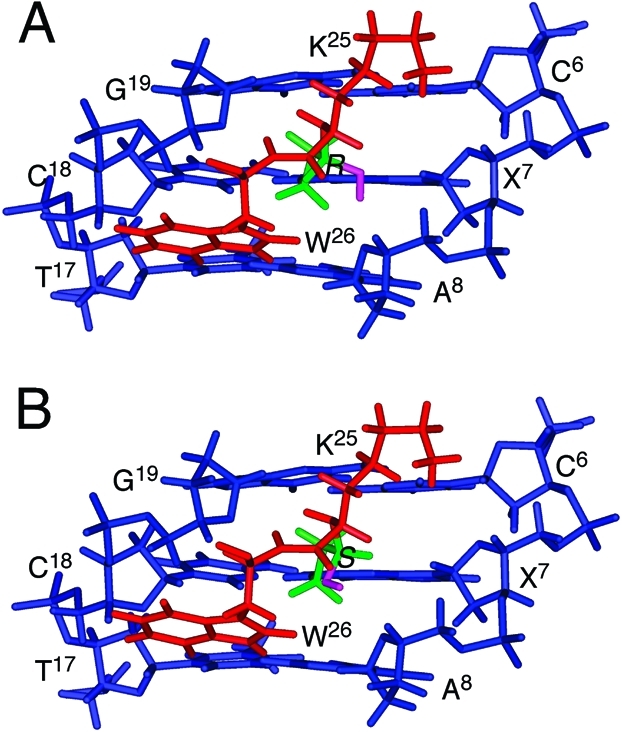
Expanded model structures of the diastereomeric carbinolamine N2-dG:N-Lys DNA–KWKK conjugates viewed from the minor groove: (A) (R)-configuration, (B) (S)-configuration. Blue, red, green, and pink sticks represent oligonucleotide, peptide, trimethylene, and the hydroxyl group, respectively.
Molecular Dynamics Simulations
To assess the potential hydrogen bonding interactions of the carbinol hydroxyl group in the DNA–KWKK conjugates, a series of MD simulations were carried out in explicit water. Table 1 lists hydrogen bonding occupations of the hydroxyl group by potential hydrogen bond acceptors. For both isomers, the carbinol hydroxyl showed minimal intermolecular hydrogen bonding with water. The occupancy of an intramolecular hydrogen bond between the hydroxyl group and G19 O4′ was greatest for the S-configuration of the carbinolamine link 8. The S-isomer also exhibited a low occupancy with regard to the formation of a hydrogen bond with K25 C=O. The hydroxyl group in the R-configuration exhibited minimal occupancy of intramolecular hydrogen bonds with A8 O4′ or X7 N3.
Table 1. Hydrogen Bond Occupancies of the Carbinol Hydroxyl Group by the Acceptors during MD Simulations.
| hydrogen bond occupation (%)a |
||
|---|---|---|
| receptor | R-configuration | S-configuration |
| X7 N3 | 0.2 | 0.0 |
| A8 O4′ | 5.2 | 0.0 |
| G19 O4′ | 0.0 | 29.6 |
| K25 C=O | 0.0 | 4.2 |
| water | 8.8 | 2.2 |
Criteria for the hydrogen bond formation: distance <3.5 Å and angle >120°.
Discussion
DNA–protein conjugates (DPCs) arise from cellular exposures to ionizing radiation, ultraviolet light, and a variety of chemicals and metals.51 If not repaired, DPCs are anticipated to interfere with both DNA replication and transcription; consequently, the mechanisms by which cells recognize and repair DPCs are of considerable interest. A possible mechanism for DPC repair involves proteolysis, leading to the formation of DNA–peptide conjugates, as substrates for nucleotide excision repair (NER).10−13 Two spectroscopically detectable DNA–KWKK conjugates are produced by reaction of the acrolein-derived γ-OH-PdG lesion with a molar excess of the peptide KWKK. These are formed by the N-terminal amine of lysine;14 the lysine side-chain amines are protonated at pH 7.36,52 The existence of imine 9 is surmised because the conjugate can be reduced with NaCNBH3.14 However, these conjugates potentially exist as an equilibrium mixture of carbinolamine 8, imine 9, and pyrimidopurinone 10 (Scheme 1). Carbinolamines are intermediates of imine formation and dissociation;53,54 those arising from primary- or secondary-amines often dehydrate to imines.55−57 Indeed, the N2-dG-lysine conjugate formed by the malondialdehyde-induced M1dG adduct favors the imine structure.58
Formation of Diasteromeric Carbinolamine Linkages
The present data reveal that at equilibrium, these acrolein-derived DNA–KWKK conjugates exist predominantly as diastereomeric carbinolamines 8, in equilibrium with trace amounts of the imine 9. NMR resonances attributed to the Hγ and Cγ nuclei of imine 9 are not observed in 13C HSQC experiments. Pyrimidopurinone cross-link 7 has been isolated following enzymatic digestion of DNA interstrand cross-links formed by the γ-OH-PdG lesion.40,59 However, pyrimidopurinone 10 is excluded for the acrolein-derived DNA–KWKK conjugates as it lacks an imino proton, whereas the two observable conjugates exhibit imino resonances. The upfield shifts observed for the Cγ and Hγ resonances and the observation of Watson–Crick hydrogen bonding of base pair X7·C18 in acidic solution therefore lead to the conclusion that these DNA–KWKK conjugates exist predominately as diastereomeric carbinolamines 8 (Scheme 2).
At equilibrium the diastereomeric carbinolamine DNA–KWKK conjugates are not present in equal amounts. Their absolute stereochemical configurations cannot be established from the present NMR data. We anticipate that hydrogen bonding interactions are important in stabilizing the formation of these carbinolamine DNA–KWKK conjugates and in determining the stereochemistry. Molecular dynamics calculations carried out in explicit solvent suggest that intramolecular hydrogen bonds might form between the carbinol and the proximate X7 N3, A8 O4′, G19 O4′, and K25 carboxyl oxygen. Molecular dynamics simulations suggest that the S-carbinol is more likely to form an intramolecular hydrogen bond than the R-carbinol (Table 1). Thus, it is anticipated that the S-carbinolamine is the major DNA–KWKK conjugate and that the R-carbinolamine is the minor one.
In DNA, these KWKK conjugates exist for weeks (Figure 3). However, upon denaturation of the duplex the DNA–KWKK conjugates reverse rapidly (Figure 5). This may also explain the MALDI-TOF mass spectrum of the N2-dG:N-Lys conjugates, indicating the presence of the imine 9 and/or pyrimidopurinone 10 linkages. This is attributed to denaturation of the DNA during the MS experiments, allowing carbinolamine DNA–peptide conjugates 8 to convert to imine 9 and/or pyrimidopurinone 10.
The N2-dG:N2-dG DNA interstrand cross-links arising from the γ-OH-PdG adduct in the 5′-CpX-3′ sequence also exist predominantly as diastereomers of carbinolamine 5.17,44,60 This is attributed to the observation that carbinolamine 5 is accommodated within the minor groove, with minimum perturbation of B-DNA structure; in contrast, imine 6 and pyrimidopurinone 7 interstrand linkages perturb DNA base pairing and base stacking.17,44,61 Additionally, the potential for hydrogen bonding between the carbinol and the DNA probably stabilizes the linkages and may account for the reaction stereoselectivity.62 Similarly, carbinolamine linkages have been trapped in the active sites of type I aldolases; their presence as compared to imines has been attributed to their stabilization by hydrogen bonding interactions.63−65
Structures of Carbinolamine DNA–KWKK Conjugates
Recently, we reported the structure of the reduced conjugate formed between the KWKK peptide and the γ-OH-PdG adduct.18 Watson–Crick hydrogen bonding was conserved at the site of conjugation, the peptide was oriented in the minor groove, and the tryptophan indolyl group did not intercalate into the DNA. At the site of conjugation, the guanine N2 amine nitrogen and the amine nitrogen of the N-terminal lysine were in the trans-configuration with respect to the Cα or Cγ of the trimethylene tether, respectively. The reduced DNA–KWKK conjugate thermally stabilized the DNA.
The refined structure of the reduced cross-link provided a basis for modeling of the native diastereomeric carbinolamine conjugates. The minimal chemical shift changes noted for the DNA protons and their similar values as compared to those of the reduced conjugate suggest that the diastereomeric carbinolamine DNA–KWKK conjugates have structures similar to that of the reduced DNA–KWKK conjugate. Molecular modeling suggests that the major stereoisomer of the carbinolamine DNA–KWKK conjugate places the peptide in the minor groove, similar to that observed for the reduced analogue (Figure 6). Base pair X7·C18 conserves Watson–Crick hydrogen bonding. However, the major carbinolamine conjugate exhibits a weak X7 H8 → C6 H1′ NOE correlation, while the C6·G19 and X7·C18 base pairs exhibit small perturbations.
1,N2-propano-2′-deoxyguanosine (PdG) has been used to model the 1,N2-dG adducts, and the PdG·C base pair has been well-studied.43,66 In acidic solution, PdG adopts the syn conformation about the glycosyl bond to enable protonated Hoogsteen hydrogen bonding with the complementary cytosine. The evident Watson–Crick hydrogen bonding of X7·C18 of the major DNA–KWKK conjugate was consistent with carbinolamine 7 being the predominant species.
Nucleotide Excision Repair of DNA–KWKK Conjugates
These lesions are anticipated to be substrates for the NER pathway. The kinetics of incision of UvrABC have been measured using either KWKK- or KFHEKHHSHRGY-peptide DNA conjugates or the T4-pdg DPC, conjugated to duplex DNA via γ-OH-PdG chemistry.14,67 In all instances, all UvrABC proteins were necessary to catalyze the incision reactions. Incision of DNA containing the KFHEKHHSHRGY-peptide conjugate at either the abasic or γ-OH-PdG site is significantly greater than the incision of DNA containing a fluorescein-conjugated dT.68 The KFHEKHHSHRGY-peptide attached via an abasic site is incised 3-times more efficiently than that when conjugated via γ-OH-PdG.
Genotoxicity of DNA–KWKK Conjugates
Modified oligodeoxynucleotides generated by conjugating KWKK to γ-OH-PdG have been inserted into a single-stranded pMS2 shuttle vector replicated in COS-7 cells.16 Replication bypass of the γ-OH-PdG-mediated DNA–KWKK conjugate results in mutations at the site of modification at an overall frequency of ∼8.4%. The γ-OH-PdG-mediated DNA–KWKK conjugate yields predominantly single base substitutions, with a prevalence of G to T transversions.
The ability of the DNA–peptide conjugates generated by linking KWKK to γ-OH-PdG to be bypassed by both human and E. coli polymerases in vitro have also been investigated.69 Human DNA polymerase κ catalyzes efficient, error-free bypass of KWKK conjugated via γ-OH-PdG. These lesions block E. coli polymerases II, III, and V, but not polymerase IV. Cells deficient in polymerase IV are inefficient in replicating these lesions. It will now be of interest to determine the crystal structures of the complexes between the DNA–KWKK conjugate and Y-family DNA polymerases that bypass bulky minor groove lesions.
Summary
Treatment of an oligodeoxynucleotide containing a site-specific γ-OH-PdG lesion with excess peptide KWKK at neutral pH produced two stereoisomeric carbinolamine N2-dG:N-Lys DNA–KWKK conjugates. In DNA, these conjugates exhibited lifetimes of weeks at neutral pH. Hydrolysis of the conjugates was accelerated by denaturation of the DNA. Upon hydrolysis of the peptides in the 5′-CpX-3′ sequence, N2-dG:N2-dG DNA interstrand cross-links and ring-opened γ-OH-PdG derivatives appeared. Potential energy minimization, followed by MD simulations carried out in explicit solvent predict that the S-isomer is the major conjugate, probably due to the formation of an intramolecular hydrogen bond. Molecular modeling suggests that the carbinolamine DNA–KWKK conjugates exist with the peptide located in the minor groove.
This work was supported by NIH grant P01 ES-05355 (to C.J.R., R.S.L., and M.P.S.). Funding for the NMR spectrometers was supplied by Vanderbilt University, by NIH grant RR-05805, and the Vanderbilt Center in Molecular Toxicology, P30 ES-00267.
Supporting Information Available
Figures S1, gel filtration of the reaction mixture by Sephadex G25; S2, MALDF-TOF MS of the DNA–KWKK conjugates; S3, NOE connectivity of base aromatic H6/H8 protons with deoxyribose H1′ protons; S4, chemical shift perturbations; S5, force constants used for the carbinolamine linkage; S6, molecular dynamics simulations of the R- and S-carbinolamine DNA–KWKK. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Chung F. L.; Zhang L.; Ocando J. E.; Nath R. G. (1999) Role of 1,N2-propanodeoxyguanosine adducts as endogenous DNA lesions in rodents and humans. IARC Sci. Publ. 150, 45–54. [PubMed] [Google Scholar]
- Chung F. L.; Nath R. G.; Nagao M.; Nishikawa A.; Zhou G. D.; Randerath K. (1999) Endogenous formation and significance of 1,N2-propanodeoxyguanosine adducts. Mutat. Res. 424, 71–81. [DOI] [PubMed] [Google Scholar]
- Nath R. G.; Ocando J. E.; Chung F. L. (1996) Detection of 1,N2-propanodeoxyguanosine adducts as potential endogenous DNA lesions in rodent and human tissues. Cancer Res. 56, 452–456. [PubMed] [Google Scholar]
- Nath R. G.; Chung F.-L. (1994) Detection of exocyclic 1,N2-propanodeoxyguanosine adducts as common DNA lesions in rodents and humans. Proc. Natl. Acad. Sci. U.S.A. 91, 7491–7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khullar S.; Varaprasad C. V.; Johnson F. (1999) Postsynthetic generation of a major acrolein adduct of 2′-deoxyguanosine in oligomeric DNA. J. Med. Chem. 42, 947–950. [DOI] [PubMed] [Google Scholar]
- Nechev L. V.; Kozekov I. D.; Brock A. K.; Rizzo C. J.; Harris T. M. (2002) DNA adducts of acrolein: Site-specific synthesis of an oligodeoxynucleotide containing 6-hydroxy-5,6,7,8-tetrahydropyrimido[1,2-a]purin-10(3H)-one, an acrolein adduct of guanine. Chem. Res. Toxicol. 15, 607–613. [DOI] [PubMed] [Google Scholar]
- de los Santos C.; Zaliznyak T.; Johnson F. (2001) NMR characterization of a DNA duplex containing the major acrolein-derived deoxyguanosine adduct γ-OH-1,-N2-propano-2′-deoxyguanosine. J. Biol. Chem. 276, 9077–9082. [DOI] [PubMed] [Google Scholar]
- Dexheimer T. S.; Kozekova A.; Rizzo C. J.; Stone M. P.; Pommier Y. (2008) The modulation of topoisomerase I-mediated DNA cleavage and the induction of DNA-topoisomerase I crosslinks by crotonaldehyde-derived DNA adducts. Nucleic Acids Res. 36, 4128–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M.; Zhitkovich A.; Harris M.; Paustenbach D.; Gargas M. (1997) DNA-protein cross-links produced by various chemicals in cultured human lymphoma cells. J. Toxicol. Environ. Health 50, 433–449. [DOI] [PubMed] [Google Scholar]
- Quievryn G.; Zhitkovich A. (2000) Loss of DNA-protein crosslinks from formaldehyde-exposed cells occurs through spontaneous hydrolysis and an active repair process linked to proteosome function. Carcinogenesis 21, 1573–1580. [PubMed] [Google Scholar]
- Brooks P.; Fuertes G.; Murray R. Z.; Bose S.; Knecht E.; Rechsteiner M. C.; Hendil K. B.; Tanaka K.; Dyson J.; Rivett J. (2000) Subcellular localization of proteasomes and their regulatory complexes in mammalian cells. Biochem. J. 346, 155–161. [PMC free article] [PubMed] [Google Scholar]
- Lafarga M.; Berciano M. T.; Pena E.; Mayo I.; Castano J. G.; Bohmann D.; Rodrigues J. P.; Tavanez J. P.; Carmo-Fonseca M. (2002) Clastosome: A subtype of nuclear body enriched in 19S and 20S proteasomes, ubiquitin, and protein substrates of proteasome. Mol. Biol. Cell 13, 2771–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai S. D.; Liu L. F.; Vazquez-Abad D.; D’Arpa P. (1997) Ubiquitin-dependent destruction of topoisomerase I is stimulated by the antitumor drug camptothecin. J. Biol. Chem. 272, 24159–24164. [DOI] [PubMed] [Google Scholar]
- Kurtz A. J.; Lloyd R. S. (2003) 1,N2-deoxyguanosine adducts of acrolein, crotonaldehyde, and trans-4-hydroxynonenal cross-link to peptides via Schiff base linkage. J. Biol. Chem. 278, 5970–5976. [DOI] [PubMed] [Google Scholar]
- Minko I. G.; Kozekov I. D.; Harris T. M.; Rizzo C. J.; Lloyd R. S.; Stone M. P. (2009) Chemistry and biology of DNA containing 1,N2-deoxyguanosine adducts of the α,β-unsaturated aldehydes acrolein, crotonaldehyde, and 4-hydroxynonenal. Chem. Res. Toxicol. 22, 759–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minko I. G.; Kozekov I. D.; Kozekova A.; Harris T. M.; Rizzo C. J.; Lloyd R. S. (2008) Mutagenic potential of DNA-peptide crosslinks mediated by acrolein-derived DNA adducts. Mutat. Res. 637, 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y. J.; Kim H. Y.; Huang H.; Slutsky A.; Minko I. G.; Wang H.; Nechev L. V.; Kozekov I. D.; Kozekova A.; Tamura P.; Jacob J.; Voehler M.; Harris T. M.; Lloyd R. S.; Rizzo C. J.; Stone M. P. (2005) Spectroscopic characterization of interstrand carbinolamine cross-links formed in the 5′-CpG-3′ sequence by the acrolein-derived γ-OH-1,N2-propano-2′-deoxyguanosine DNA adduct. J. Am. Chem. Soc. 127, 17686–17696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H.; Kozekov I. D.; Kozekova A.; Rizzo C. J.; McCullough A. K.; Lloyd R. S.; Stone M. P. (2010) Minor groove orientation of the KWKK peptide tethered via the N-terminal amine to the acrolein-derived 1,N2-γ-hydroxypropanodeoxyguanosine lesion with a trimethylene linkage. Biochemistry 49, 6155–6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaluzzi M. J.; Borer P. N. (2004) Revised UV extinction coefficients for nucleoside-5′-monophosphates and unpaired DNA and RNA. Nucleic Acids Res. 32, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotto M.; Saudek V.; Sklenar V. (1992) Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 2, 661–665. [DOI] [PubMed] [Google Scholar]
- Palmer A. G. III; Rance M.; Wright P. E. (1991) Intramolecular motions of a zinc finger DNA-binding domain from Xfin characterized by proton-detected natural abundance 13C heteronuclear NMR spectroscopy. J. Am. Chem. Soc. 113, 4371–4380. [Google Scholar]
- Kay L. E.; Keifer P. A.; Saarinen T. (1992) Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 114, 10663–10665. [Google Scholar]
- Schleucher J.; Schwendinger M.; Sattler M.; Schmidt P.; Schedletzky O.; Glaser S. J.; Sorensen O. W.; Griesinger C. (1994) A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed field gradients. J. Biomol. NMR 4, 301–306. [DOI] [PubMed] [Google Scholar]
- Sklenar V.; Piotto M.; Leppik R. a. S., V. (1993) Gradient-Tailored water suppression for 1H-15N HSQC Experiments optimized to retain full sensitivity. J. Magn. Reson. 102, 241–245. [Google Scholar]
- Mori S.; Abeygunawardana C.; Johnson M.; vanZul P. C. M. (1995) Improved sensitivity of HSQC spectra of exchanging protons at short iterscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J. Magn. Reson. 108, 94–98. [DOI] [PubMed] [Google Scholar]
- Talluri S.; Wagner G. (1996) An Optimized 3D NOESY-HSQC. J. Magn. Reson. 112, 200–205. [DOI] [PubMed] [Google Scholar]
- Frisch M. J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Montgomery J. A., Vreven T., Kudin K. N., Burant J. C., Millam J. M., Iyengar S. S., Tomasi J., Barone V., Mennucci B., Cossi M., Scalmani G., Rega N., Petersson G. A., Nakatsuji H., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Klene M., Li X., Knox J. E., Hratchian H. P., Cross J. B., Adamo C., Jaramillo J., Gomperts R., Stratmann R. E., Yazyev O., Austin A. J., Cammi R., Pomelli C., Pomelli J., Ochterski W., Ayala P. Y., Morokuma K., Voth G. A., Salvador P., Dannenberg J. J., Zakrzewska V. G., Daniels A. D., Farkas O., Rabuck A. D., Raghavachari K., and Ortiz J. V. (2004) Gaussian 03, Gaussian, Inc., Wallingford, CT.
- Case D. A.; Cheatham T. E. 3rd; Darden T.; Gohlke H.; Luo R.; Merz K. M. Jr.; Onufriev A.; Simmerling C.; Wang B.; Woods R. J. (2005) The AMBER biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins G. D.; Cramer C. J.; Truhlar D. G. (1995) Pairwise solute descreening of solute charges from a dielectric medium. Chem. Phys. Lett. 246, 122–129. [Google Scholar]
- Hawkins G. D.; Cramer C. J.; Truhlar D. G. (1996) Parametrized models of aqueous free energies of solvation based on pairwise descreening of solute atomic charges from a dielectric medium. J. Phys. Chem. 100, 19824–19839. [Google Scholar]
- Essmann U.; Perera L.; Berkowitz M. L.; Darden T.; Lee H.; Pedersen L. G. (1995) A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593. [Google Scholar]
- Konerding D. E.; Cheatham T. E. r.; Kollman P. A.; James T. L. (1999) Restrained molecular dynamics of solvated duplex DNA using the particle mesh Ewald method. J. Biomol. NMR 13, 119–131. [DOI] [PubMed] [Google Scholar]
- Ryckaert J. P.; Ciccotti G.; Berendsen H. J. C. (1977) Numerical integration of cartesian equations of motion of a system with constraints: Molecular dynamics of N-alkanes. J. Comput. Phys. 23, 327–341. [Google Scholar]
- Stofer E.; Lavery R. (1994) Measuring the geometry of DNA grooves. Biopolymers 34, 337–346. [DOI] [PubMed] [Google Scholar]
- Lavery R.; Sklenar H. (1988) The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic-acids. J. Biomol. Struct. Dyn. 6, 63–91. [DOI] [PubMed] [Google Scholar]
- Poon D. K.; Schubert M.; Au J.; Okon M.; Withers S. G.; McIntosh L. P. (2006) Unambiguous determination of the ionization state of a glycoside hydrolase active site lysine by 1H-15N heteronuclear correlation spectroscopy. J. Am. Chem. Soc. 128, 15388–15389. [DOI] [PubMed] [Google Scholar]
- Breitmaier E., and Voelter W. (1987) Carbon-13 NMR Spectroscopy: High-Resolution Methods and Applications in Organic Chemistry and Biochemistry, 3rd completely revised ed., VCH Publishers, New York. [Google Scholar]
- Nechev L. V.; Harris C. M.; Harris T. M. (2000) Synthesis of nucleosides and oligonucleotides containing adducts of acrolein and vinyl chloride. Chem. Res. Toxicol. 13, 421–429. [DOI] [PubMed] [Google Scholar]
- Kozekov I. D.; Nechev L. V.; Moseley M. S.; Harris C. M.; Rizzo C. J.; Stone M. P.; Harris T. M. (2003) DNA interchain cross-links formed by acrolein and crotonaldehyde. J. Am. Chem. Soc. 125, 50–61. [DOI] [PubMed] [Google Scholar]
- Wang H.; Kozekov I. D.; Harris T. M.; Rizzo C. J. (2003) Site-specific synthesis and reactivity of oligonucleotides containing stereochemically defined 1,N2-deoxyguanosine adducts of the lipid peroxidation product trans-4-hydroxynonenal. J. Am. Chem. Soc. 125, 5687–5700. [DOI] [PubMed] [Google Scholar]
- Voitkun V.; Zhitkovich A. (1999) Analysis of DNA-protein crosslinking activity of malondialdehyde in vitro. Mutat. Res. 424, 97–106. [DOI] [PubMed] [Google Scholar]
- Singh U. S.; Moe J. G.; Reddy G. R.; Weisenseel J. P.; Marnett L. J.; Stone M. P. (1993) 1H NMR of an oligodeoxynucleotide containing a propanodeoxyguanosine adduct positioned in a (CG)3 frameshift hotspot of Salmonella typhimurium hisD3052: Hoogsteen base-pairing at pH 5.8. Chem. Res. Toxicol. 6, 825–836. [DOI] [PubMed] [Google Scholar]
- Weisenseel J. P.; Reddy G. R.; Marnett L. J.; Stone M. P. (2002) Structure of an oligodeoxynucleotide containing a 1,N2-propanodeoxyguanosine adduct positioned in a palindrome derived from the Salmonella typhimurium hisD3052 gene: Hoogsteen pairing at pH 5.2. Chem. Res. Toxicol. 15, 127–139. [DOI] [PubMed] [Google Scholar]
- Kim H. Y.; Voehler M.; Harris T. M.; Stone M. P. (2002) Detection of an interchain carbinolamine cross-link formed in a CpG sequence by the acrolein DNA adduct γ-OH-1,N2-propano-2′-deoxyguanosine. J. Am. Chem. Soc. 124, 9324–9325. [DOI] [PubMed] [Google Scholar]
- Patel D. J.; Shapiro L.; Hare D. (1987) DNA and RNA: NMR studies of conformations and dynamics in solution. Q. Rev. Biophys. 20, 35–112. [DOI] [PubMed] [Google Scholar]
- Reid B. R. (1987) Sequence-specific assignments and their use in NMR studies of DNA structure. Q. Rev. Biophys. 20, 2–28. [DOI] [PubMed] [Google Scholar]
- Norman D.; Abuaf P.; Hingerty B. E.; Live D.; Grunberger D.; Broyde S.; Patel D. J. (1989) NMR and computational characterization of the N-(deoxyguanosin-8-yl)aminofluorene adduct [(AF)G] opposite adenosine in DNA: (AF)G[syn]:A[anti]pair formation and its pH dependence. Biochemistry 28, 7462–7476. [DOI] [PubMed] [Google Scholar]
- Mao B.; Hingerty B. E.; Broyde S.; Patel D. J. (1995) Solution conformation of [AF]dG opposite a −2 deletion site in a DNA duplex: intercalation of the covalently attached aminofluorene ring into the helix with base displacement of the C8-modified syn guanine and adjacent unpaired 3′-adenine into the major groove. Biochemistry 34, 16641–16653. [DOI] [PubMed] [Google Scholar]
- Cosman M.; Hingerty B.; Geacintov N. E.; Broyde S.; Patel D. J. (1995) Structural alignments of (+)- and (-)-trans-anti-benzo[a]pyrene-dG adducts positioned at a DNA template/primer junction. Biochemistry 34, 15334–15350. [DOI] [PubMed] [Google Scholar]
- Mao B.; Gorin A.; Gu Z.; Hingerty B. E.; Broyde S.; Patel D. J. (1997) Solution structure of the aminofluorene-intercalated conformer of the syn [AF]-C8-dG adduct opposite a −2 deletion site in the NarI hot spot sequence context. Biochemistry 36, 14479–14490. [DOI] [PubMed] [Google Scholar]
- Barker S.; Weinfeld M.; Murray D. (2005) DNA-protein crosslinks: Their induction, repair, and biological consequences. Mutat. Res. 589, 111–135. [DOI] [PubMed] [Google Scholar]
- Kurtz A. J.; Dodson M. L.; Lloyd R. S. (2002) Evidence for multiple imino intermediates and identification of reactive nucleophiles in peptide-catalyzed β-elimination at abasic sites. Biochemistry 41, 7054–7064. [DOI] [PubMed] [Google Scholar]
- Jencks W. P. (1969) Catalysis in Chemistry and Enzymology, McGraw-Hill, New York. [Google Scholar]
- Bruckner R. (2002) Advanced Organic Chemistry: Reaction Mechanisms, Harcourt/Academic Press, San Diego, CA. [Google Scholar]
- Jo B. H.; Nair V.; Davis L. (1977) Carbon-13 nuclear magnetic resonance studies of vitamin B6 Schiff base and carbinolamine formation in aqueous solution. 1. The adduct of pyridoxal 5′-phosphate and DL-alanine. J. Am. Chem. Soc. 99, 4467–4471. [DOI] [PubMed] [Google Scholar]
- Upthagrove A. L.; Nelson W. L. (2001) Carbinolamines, imines, and oxazolidines from fluorinated propranolol analogs. 19F NMR and mass spectral characterization and evidence for formation as intermediates in cytochrome P450-catalyzed N-dealkylation. Drug Metab. Dispos. 29, 1114–1122. [PubMed] [Google Scholar]
- Huizing G.; Segura J.; Beckett A. H. (1980) On the mechanism of metabolic N-dealkylation. Isolation of a relatively stable carbinolamine. J. Pharm. Pharmacol. 32, 650–651. [DOI] [PubMed] [Google Scholar]
- Szekely J.; Rizzo C. J.; Marnett L. J. (2008) Chemical properties of oxopropenyl adducts of purine and pyrimidine nucleosides and their reactivity toward amino acid cross-link formation. J. Am. Chem. Soc. 130, 2195–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao Y.; Hecht S. S. (2005) Synthesis and properties of an acetaldehyde-derived oligonucleotide interstrand cross-link. Chem. Res. Toxicol. 18, 711–721. [DOI] [PubMed] [Google Scholar]
- Stone M. P.; Cho Y.-J.; Huang H.; Kim H.-Y.; Kozekov I. D.; Kozekova A.; Wang H.; Lloyd R. S.; Harris T. M.; Rizzo C. J. (2008) Interstrand DNA cross-links induced by α,β-unsaturated aldehydes derived from lipid peroxidation and environmental sources. Acc. Chem. Res. 41, 793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y. J.; Wang H.; Kozekov I. D.; Kozekova A.; Kurtz A. J.; Jacob J.; Voehler M.; Smith J.; Harris T. M.; Rizzo C. J.; Lloyd R. S.; Stone M. P. (2006) Orientation of the crotonaldehyde-derived N2-[3-Oxo-1(S)-methyl-propyl]-dG DNA adduct hinders interstrand cross-link formation in the 5′-CpG-3′ sequence. Chem. Res. Toxicol. 19, 1019–1029. [DOI] [PubMed] [Google Scholar]
- Huang H.; Kim H. Y.; Kozekov I. D.; Cho Y. J.; Wang H.; Kozekova A.; Harris T. M.; Rizzo C. J.; Stone M. P. (2009) Stereospecific formation of the (R)-γ-hydroxytrimethylene interstrand N2-dG:N2-dG cross-link arising from the γ-OH-1,N2-propano-2′-deoxyguanosine adduct in the 5′-CpG-3′ DNA sequence. J. Am. Chem. Soc. 131, 8416–8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard J.; Grochulski P.; Sygusch J. (2001) Covalent intermediate trapped in 2-keto-3-deoxy-6- phosphogluconate (KDPG) aldolase structure at 1.95-A resolution. Proc. Natl. Acad. Sci. U.S.A. 98, 3679–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erskine P. T.; Newbold R.; Brindley A. A.; Wood S. P.; Shoolingin-Jordan P. M.; Warren M. J.; Cooper J. B. (2001) The x-ray structure of yeast 5-aminolaevulinic acid dehydratase complexed with substrate and three inhibitors. J. Mol. Biol. 312, 133–141. [DOI] [PubMed] [Google Scholar]
- Thorell S.; Schurmann M.; Sprenger G. A.; Schneider G. (2002) Crystal structure of decameric fructose-6-phosphate aldolase from Escherichia coli reveals inter-subunit helix swapping as a structural basis for assembly differences in the transaldolase family. J. Mol. Biol. 319, 161–171. [DOI] [PubMed] [Google Scholar]
- Huang P.; Eisenberg M. (1992) The three-dimensional structure in solution (pH 5.8) of a DNA 9-mer duplex containing 1,N2-propanodeoxyguanosine opposite deoxyadenosine. Restrained molecular dynamics and NOE-based refinement calculations. Biochemistry 31, 6518–6532. [DOI] [PubMed] [Google Scholar]
- Minko I. G.; Kurtz A. J.; Croteau D. L.; Van Houten B.; Harris T. M.; Lloyd R. S. (2005) Initiation of repair of DNA-polypeptide cross-links by the UvrABC nuclease. Biochemistry 44, 3000–3009. [DOI] [PubMed] [Google Scholar]
- Truglio J. J.; Croteau D. L.; Skorvaga M.; DellaVecchia M. J.; Theis K.; Mandavilli B. S.; Van Houten B.; Kisker C. (2004) Interactions between UvrA and UvrB: The role of UvrB’s domain 2 in nucleotide excision repair. EMBO J. 23, 2498–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minko I. G.; Yamanaka K.; Kozekov I. D.; Kozekova A.; Indiani C.; O’Donnell M. E.; Jiang Q.; Goodman M. F.; Rizzo C. J.; Lloyd R. S. (2008) Replication bypass of the acrolein-mediated deoxyguanine DNA-peptide cross-links by DNA polymerases of the DinB family. Chem. Res. Toxicol. 21, 1983–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.