Summary
The regulation of apoptosis is critical for controlling tissue homeostasis and preventing tumor formation and growth. Reactive Oxygen Species (ROS) generation plays a key role in such regulation. Here, we describe a HIF-1 target, ATIA (anti-TNFα-induced apoptosis), which protects cells against TNFα- and hypoxia-induced apoptosis. Through the generation of ATIA knockout mice, we show that ATIA protects cells from apoptosis through regulating the function of the mitochondrial antioxidant, thioredoxin-2, and ROS generation. ATIA is highly expressed in human glioblastoma and ATIA knockdown in glioblastoma cells renders them sensitive to hypoxia-induced apoptosis. Therefore, ATIA is not only a HIF-1 target that regulates mitochondrial redox pathways but a potentially diagnostic marker and therapeutic target in human glioblastoma.
Introduction
Apoptosis is a crucial physiological process for development and tissue homeostasis (Henson and Hume, 2006). Deregulation of apoptosis often leads to pathological conditions. For instance, lack of apoptosis can lead to autoimmune disorders or cancer, while excessive apoptosis can lead to immunodeficiency or neurodegenerative disease (Fadeel and Orrenius, 2005). Due to the importance of apoptosis in both physiological and pathological conditions, the underlying molecular mechanisms in the regulation of apoptosis have been intensively studied. For instance, apoptotic pathways mediated by the death receptors, including Fas and TNFR1, are some of the most studied of signal transduction events (Wajant, 2003). Many recent studies suggest that reactive oxygen species (ROS) play a key role in death receptor-induced apoptosis and necrosis (Lin et al., 2004; Ventura et al., 2004; Shen and Pervaiz, 2006; Morgan et al., 2008).
Reactive oxygen species (ROS) are often generated during normal signal transduction events, yet much of the ROS found within the cell, including superoxide anions, hydrogen peroxide and hydroxyl radicals (O2-, H2O2, and OH·) are often formed in the mitochondria during the electron transfer reactions of oxidative phosphorylation (Thannickal and Fanburg, 2000). Low cellular levels of ROS are usually maintained by systems of antioxidant enzymes and their substrates including the glutathione and thioredoxin systems. The thioredoxin proteins, 1 & 2, are critical regulators of reduction-oxidation (redox) balance in cells (Holmgren and Lu, 2010). Thioredexin 2 (TRX2), which is specifically expressed in the mitochondria, is required for cell viability and protects cells from apoptosis (Tanaka et al., 2002; Nonn et al., 2003). However, under pathological conditions, excessive ROS accumulation can induce apoptosis or necrosis.
Hypoxia is characterized by a decrease in oxygen within cells and plays a role in physiological processes, such as cell differentiation, proliferation, and viability (Hamanaka and Chandel, 2009). More commonly hypoxia is associated with pathophysiological conditions such as cancer, inflammation, and ischemia (Paul et al., 2004). Almost all solid tumors have regions of reduce oxygen concentration. Tumor cells adapt to low oxygen by inducing angiogenesis, increasing glucose consumption and switching to glycolysis. This response is regulated by two transcription factors, Hypoxia inducible factor 1 and 2 (HIF-1 and HIF-2) (Nakayama 2009; Semenza, 2010). HIF-1 is the main transcriptional factor activated as a result of decrease of oxygen in cells. Under normoxia, the alpha subunit (HIF-1α) of HIF-1 undergoes rapid proteasomal degradation. When oxygen is low HIF-1α is stabilized and translocated to the nucleus where it heterodimerizes with constitutively active HIF-1β to form an active transcription complex. HIF antagonizes apoptosis through activating prosurvival target genes and the HIF-mediated anti-apoptotic effect is critical for cells to survive under hypoxic conditions (Loor and Schumacker, 2008). Oxygenation and ROS levels are tightly linked. Evidence suggests that hypoxia triggers mitochondria to produce ROS and activates HIF-1 and in turn HIF-1 regulates the level of mitochondrial ROS through turning on its target genes (Fukuda et. al., 2007; Hamanaka and Chandel, 2010, Semenza, 2010).
For TNF receptor 1 (TNFR1) signaling, receptor-interacting protein 1(RIP1), TNF receptor (TNFR)-associated factor 2 (TRAF2), and Fas-associated death domain protein (FADD) are important effector molecules (Baud and Karin, 2001; Wajant et al., 2003). While FADD is essential for TNFα-induced apoptosis, RIP1 and TRAF2 are critical for TNFα-induced activation of the transcription factor NF-κB and mitogen-activated protein kinases (MAPKs) and loss of either RIP1 or TRAF2 leads to a dramatic increase of cell sensitivity to TNFα-induced apoptosis (Yeh et al., 1997; Kelliher et al., 1998). Our previous studies found that RIP1 and TRAF2 mediate ROS generation in TNFα-induced death (Lin et al., 2004; Kim et al., 2007). Therefore, to further understand the molecular mechanisms of TRAF2-mediated cell protection against TNFα-induced apoptosis and particularly, the regulation of ROS generation during the process, we attempted to identify additional proteins that protect cells against TNFα-induced apoptosis. Through screening a retroviral cDNA expression library for genes that protect TRAF2 null cells following TNFα treatment, we identified a novel anti-apoptotic gene, ATIA (anti-TNFα-induced apoptosis).
ATIA is highly expressed in some tissues including liver, lung, heart, and testis. The human ortholog of ATIA, vasorin, was reported to be a potential inhibitor of TGFβ signaling with a possible role in angiogenesis (Ikeda et al., 2004). We found that the expression of ATIA is dramatically reduced in TRAF2−/− cells and its expression under hypoxia is regulated by HIF-1. Using ATIA null mice and ATIA−/− MEFs, we show that ATIA protects cells against TNFα and hypoxia-induced apoptosis. Importantly, we found that the ATIA protein protects cells against TNFα-induced apoptosis through modulating the mitochondrial thioredoxin, TRX2. Finally, we show that ATIA protein is highly expressed in human glioblastoma. Knockdown of ATIA renders glioblastoma cells sensitive to hypoxia-induced apoptosis.
Results
Identification of ATIA as a putative anti-apoptotic protein
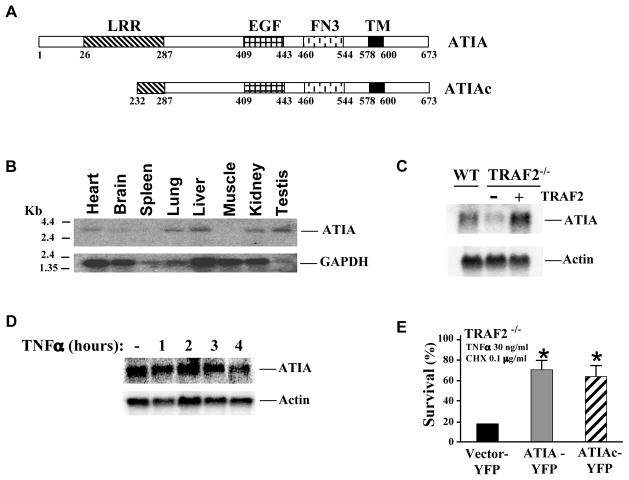
To screen for potential anti-apoptotic genes that protect TRAF2−/− cells from TNFα-induced apoptosis, we constructed a retroviral cDNA expression library with mRNA isolated from wild type (wt) MEFs and used this library to infect TRAF2−/− cells (Figure S1A). TRAF2−/− MEFs were challenged with TNFα plus a low concentration of cycloheximide (CHX). The resulting TNFα-resistant cells were pooled and the genomic DNA was isolated. The cDNAs were recovered by PCR with primers corresponding to the viral vector and identified by DNA sequencing (Figure S1A). Through such a screening approach, we identified an unknown gene at the time and named it as ATIA. The ATIA gene encodes a polypeptide with 673 amino acid residues, which has a predicted molecular weight of 72 kDa. A few years later the human ortholog of ATIA, vasorin, was reported (Ikeda et al., 2004) and the sequence of mouse vasorin was deposited in GenBank (Gene ID: 246154). Like vasorin, ATIA protein has a N-terminal leucine-rich repeat (LRR), an EGF signature, a fibronectin 3 domain (FN3) and a C-terminal transmembrane domain (Figure 1A). Interestingly, we found that a truncated ATIA (aa 232-673) was present in most of the cells that survived during our screening. Therefore, we designated this truncated ATIA as ATIAc (Figure 1A). As shown in Figure 1B, ATIA is highly expressed in tissues such as liver, lung, kidney, and testis. Importantly, ATIA expression is dramatically reduced in the TRAF2−/− cells while the ectopic expression of TRAF2 restores the level of ATIA (Figure 1C). However, ATIA expression is not inducible by TNFα in wt cells (Figure 1D). To assess the anti-apoptotic function of ATIA, we generated ATIA-YFP and ATIAc-YFP constructs and transfected these plasmids into TRAF2−/− cells. Then the cells were treated with TNFα plus CHX. As shown in Figure 1E, cell survival was significantly increased in ATIA or ATIAc transfected cells. A similar protective effect of ATIA was also observed in MCF7 cells, a mammary cancer cell line that is sensitive to TNFα-induced apoptosis (Figure S1B). Therefore, ATIA is a potential anti-apoptotic gene that protects cells against TNFα-induced apoptosis.
Figure 1. Schematic presentation and expression of ATIA.
(A) N-terminal leucine-rich repeat (LRR), an EGF signature, a fibronectin3 domain (FN3) and a C-terminal transmembrane domain are highlighted.
(B) Northern blot analysis of ATIA mRNA expression in different tissues.
(C) ATIA expression is down-regulated in TRAF2−/− cells. Total RNA was isolated from wild type, TRAF2−/− and TRAF2 reconstituted MEFs and analyzed by Northern blot.
(D) ATIA is not induced by TNFα treatment. WT MEF cells were treated with TNFα for the indicated times, and total RNA was isolated. ATIA mRNA was detected by Northern blot.
(E) Both ATIA full-length and ATIAc protect cells from TNFα-induced apoptosis. TRAF2−/− MEFs were transiently transfected with YFP-tagged ATIA full-length, ATIAc and vector alone and treated with TNFα and CHX for 5 hours. The percent YFP-positive and PI negative cells for each treatment group was determined by FACS. *p < 0.05 versus vector-YFP control. Error bars represent SEM.
Cellular localizations of ATIA
Vasorin was reported to localize to the cell plasma membrane (Ikeda et al., 2004). To confirm that ATIA has a similar cellular localization, the ATIA-YFP and ATIAc-YFP constructs were transfected into wt MEFs. We found that the expression of the full-length ATIA-YFP gives two products, a major one at about 130 kDa and a minor one at about 110 kDa (Figure 2A). Since the predicted molecular weight of ATIA-YFP is about 97 kDa without the signal peptide, the larger sizes of the products are apparently due to additional modifications of the proteins that we found are at least partially due to glycosylation (data not shown). The ATIAc-YFP construct gives a product at the predicted 75 kDa (Figure 2A). Confocal microscopy demonstrated that the majority of the cells transfected with the full-length ATIA had intense labeling of the plasma membrane while surprisingly, cells also showed a punctuate pattern in the cytoplasm (Figure 2B, left top panel). However, cells transfected with the truncated ATIAc demonstrated only punctuate expression and no membrane expression (Figure 2B, left bottom panel). Since this punctuate pattern resembled the mitochondrial localization, it raised the possibility that ATIA also localized to the mitochondria. To confirm this, the transfected cells were stained with the mitochondrial marker, MitoTracker. As shown in Figure 2B, the punctuate patterns of ATIA and ATIAc colocalized with the MitoTracker staining in transfected cells. These results suggest that ATIA may localize to both cell plasma membrane and mitochondria. Since the expression of ATIA-YFP plasmid yields two different sized products, these two products may represent the ATIA protein localized to the plasma membrane and mitochondria, respectively.
Figure 2. Subcellular localization and protective role of full-length and truncated ATIA.
(A) Western blot analysis of the expression of ATIA-YFP and ATIAc-YFP transiently transfected into wt MEFs. The cell lysates were immunoblotted and analyzed with the indicated antibodies.
(B) Confocal microscopy of live wt MEFs transiently transfected with ATIA-YFP and ATIAc-YFP constructs. Mitochondrial staining was performed by MitoTracker.
(C) Western blot analysis of pronase-treated ATIA-YFP or ATIAc-YFP transfected MEFs. The cells lysates were immunoblotted with the indicated antibodies.
(D) Western blot analysis of endogenous ATIA after pronase treatment of MEFs followed by cellular fractionation. The pronase treated and untreated cytosolic (C), mitochondrial (M) fractions and the total cell lysates (T) were blotted with the indicated antibodies. AIF is a mitochondrial specific control whereas HSP90 is cytosolic specific. TNFR1 is a control for membrane specific proteins. The (*) indicates a non-specific band.
To further verify the membrane and mitochondrial localizations of ATIA, we next tested whether the upper band is localized to the cell plasma membrane by treating cells with pronase, a proteolytic enzyme that removes exposed membrane proteins. Wt MEFs were transfected with ATIA-YFP or ATIAc-YFP followed by pronase or no treatment. As shown in Figure 2C, the upper band of the full-length ATIA almost disappeared upon pronase digestion, but the lower band of ATIA and ATIAc were unaffected. This suggests that the upper band represents the membrane-localized ATIA protein whereas the lower band and the truncated ATIAc are not expressed on the cell surface. We then performed cell fractionation experiments with both transfected and endogenous ATIA proteins. As shown in Figure 2D and Figure S2, ATIA proteins were detected in the mitochondrial fractions, but not in the cytosolic fractions. The anti-ATIA antibody detects the endogenous ATIA protein at two different sizes in total cell lysate: an upper band (100 kDa) and a lower band (80 kDa) (Figure S7A). The mitochondrial protein AIF and the cytosolic protein HSP90 or JNK were used as controls to show the relative specificity of the fractionation experiments. Taken together, these results support the idea that the upper band of ATIA is localized to the plasma membrane and the smaller form of ATIA to the mitochondria and they are designated as mem-ATIA and mito-ATIA, respectively (Figure 2D). In addition, since we found that ATIAc protects TRAF2−/− cells from TNFα plus CHX (Figure 1E), these results imply that mito-ATIA is accountable for its anti-apoptotic effect.
To investigate the regulation of ATIA mitochondrial localization, we generated a set of serial deletion mutants of ATIA-YFP as shown in Figure 3A. These plasmids were transfected into wt MEFs and the localization of each mutant ATIA was analyzed by confocal microscopy. As shown in Figure 3B and data not shown, the ATIA (287-673) mutant localizes to mitochondrial, but the ATIA (409-673) mutant lost the specificity of mitochondrial localization, indicating the aa 287-409 region of ATIA is needed for targeting ATIA to the mitochondria. This portion of ATIA was sufficient to specifically target YFP to the mitochondria (Figure 3C, top panel). Further analysis of this region revealed that amino acids 287-316 of ATIA is the mitochondrial targeting sequence of ATIA (Figure 3C, middle and bottom panels). More importantly, the ATIA (287-673) mutant, which was localized to mitochondria, but not the ATIA (409-673) mutant, which was cytoplasmic, was adequate to protect TRAF2−/− cells from TNFα-induced apoptosis (Figure 3D). This further suggests that the mitochondrial localization of ATIA is critical for protecting cells.
Figure 3. Schematic presentation of truncated forms of ATIA and their localization.
(A) The various truncated forms of ATIA with loss of specific domains are shown.
(B & C) Confocal microscopy of live wt MEFs transiently transfected with various constructs. Mitochondrial staining was performed by MitoTracker.
(D) ATIA−/− MEFs were transiently transfected with YFP-tagged ATIA vector alone, full-length, 287–673 or 409–673. Comparable expression of these plasmids was determined by Western blot analysis with anti-GFP antibody (right panel). The transfected cells were treated with TNFα and CHX for 8 hours (left panel). The percent YFP-positive and PI negative cells for each treatment group was determined by FACS. *p<0.002 versus control with vector. Error bars represent SEM
ATIA is a target of HIF-1
Since ATIA expression requires TRAF2, but is not regulated by TNFα (Fig. 1), we then examined the promoter sequence of ATIA for potential regulator(s) of its expression. We found that there are several target sites for different transcription factors including Sp1 and HIF-1 in the ATIA promoter. To test whether ATIA is a HIF-1 target, we first examined the expression of ATIA under hypoxic conditions. We treated wt MEFs with either CoCl2 (a chemical inducer of HIF-1) or 0.1% or 1% hypoxia. Under these conditions, expression of HIF-1α is induced by 4-8 hours and is followed by a significant increase in ATIA expression (Figure 4A, B, and S3A) as well as ATIA mRNA (Figure 4C, D, and S3B). In addition, as shown in Figure 4E and Figure S3C, in wt MEFs and human A172 cells respectively, the HIF binding site in ATIA promoter was specifically pulled down by a HIF-1α antibody, and not the IgG control antibody. More importantly, when expression of HIF-1α was knocked down in wt MEFs, little increase in ATIA expression was observed following CoCl2 or hypoxia as compared to control cells transfected with non-targeting siRNA (Figure 4F, G, and S3D). This increase in hypoxia-induced expression of ATIA is seen in TRAF2−/− cells as well (Figure S3E). These results suggest that ATIA is a target of HIF-1. Since there is no detectable HIF-1α in wt and TRAF2−/− cells (Figure S3E), HIF-1 is not involved in the regulation of the basal levels of ATIA in these cells.
Figure 4. Hypoxia induces ATIA expression.
(A&B) Western blot analysis of wt MEFs treated with (A) CoCl2 or (B) physiological hypoxia for the indicated times. The cell lysates were immunoblotted with the indicated antibodies.
(C&D) Real time PCR of total RNA isolated from wt MEFs treated with (C) CoCl2 or
(D) 0.1% hypoxia for the indicated times. For (C) *p<0.04 versus untreated control and for (D) *p< 0.03 versus untreated control. Error bars represent SEM.
(E) ChIP Analysis of wt MEFs treated with 0.1% hypoxia for 8 hr. Chromatin immunoprecipitation was done with anti-HIF-1α antibody and goat IgG isotype control.
(F) Western blot analysis following CoCl2 treatment of wt MEFs transfected with 50 pMol of non-targeting (NT) or HIF-1α (pool#1) siRNA. The cells lysates were immunoblotted with the indicated antibodies.
(G) Western blot analysis following 0.1% hypoxia treatment of wt MEFs transfected with 50 pMol of non-targeting (NT) or HIF-1α (pool#2) siRNA. The cell lysates were immunoblotted with the indicated antibodies.
ATIA protects cells against TNFα and hypoxia-induced apoptosis
To further study the function of ATIA, we generated ATIA knockout (ko) mice in which 495bp (a. a. 287-452) of the ATIA gene was deleted (Figure S4A). The presence of the mutant ATIA allele in ES cells was confirmed by PCR and DNA sequencing (Figure S4B, data not shown). The offspring of ATIA ko mice were genotyped by PCR (Figure S3C) and the deletion of ATIA was also confirmed by RT-PCR (data not shown). After ATIA+/− heterozygotic mice were backcrossed to C57BL/6 mice for 8 generations, the resulting heterozygous mice were bred to each other and the homozygous wt and ko progeny were used for in vivo experiments. ATIA ko mice appear normal phenotypically, however, male ATIA ko mice have impaired fertility.
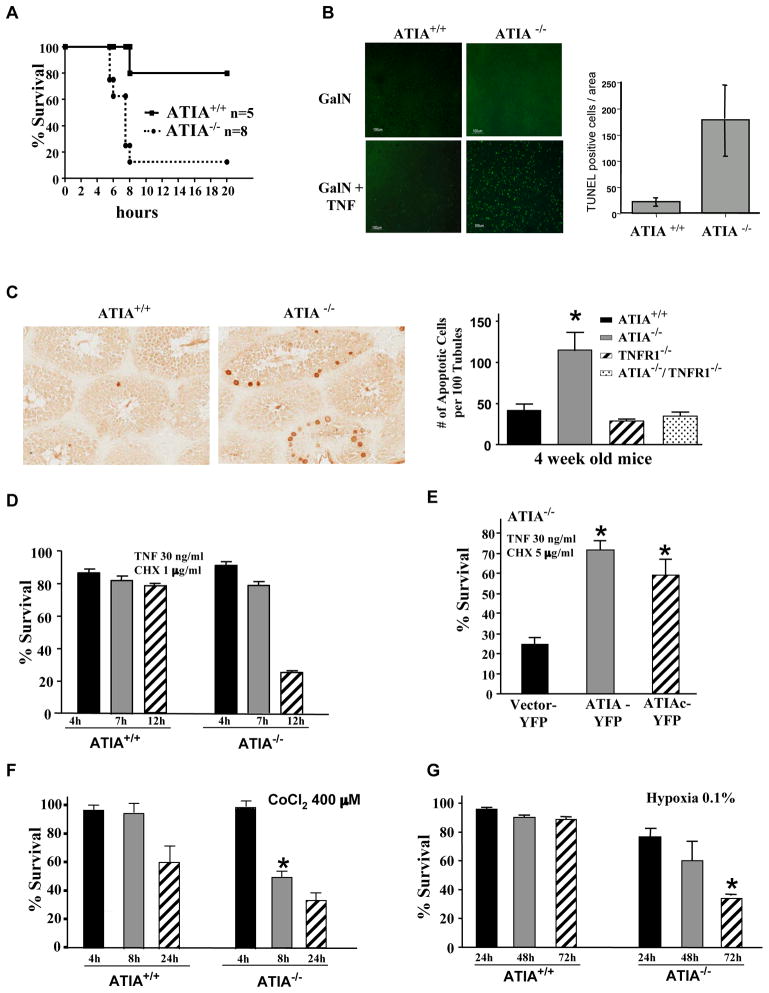
To determine whether ATIA is able to protect from TNFα-induced apoptosis in vivo, we used a model of TNFα-induced hepatitis (Tiegs et al., 1989). ATIA−/− and ATIA+/+ litter mates from heterozygote crosses were pretreated with a metabolic inhibitor of transcription, D-galactosamine (GalN) (Decker and Keppler, 1974) followed by intravenous injection of a sub-lethal dose of TNFα. By 8 hr post-treatment, 20% of ATIA+/+ mice as compared to more than 85% of ATIA−/− mice were moribund (Figure 5A). Histological analysis of H&E stained liver sections 4 hrs after TNFα administration demonstrated increased apoptosis in livers of ATIA−/− mice, while ATIA+/+ litter mates showed little apoptosis (Figure S4D). This observation was further confirmed by TUNEL staining as a significant number of TUNEL-positive cells were present in the ATIA−/− livers as compared to ATIA+/+ livers (Figure 5B). Therefore, ATIA−/− hepatocytes are more sensitive to TNFα-induced apoptosis. In addition, because ATIA is highly expressed in the testis (Figure 1B) and male ATIA−/− mice have impaired fertility, we also examined whether there is increased apoptosis in ATIA−/− testis. To do so, we collected testis samples from 4 week old ATIA+/+ and ATIA−/− littermates and performed TUNEL analysis to check the status of apoptosis. As shown in Figure 5C (enlarged in Figure S4E), there is a dramatic increase in apoptotic cells within the compartment containing Sertoli cells, spermatogonia and primary spermatocytes in ATIA−/− testis as compared to ATIA+/+ littermates. Importantly, the impaired fertility phenotype of ATIA−/− male mice was reversed when ATIA−/− mice were crossed with TNFR1−/− mice, resulting in apoptotic scores in the ATIA−/−/TNFR1−/− mice similar to that of wt mice (Figure 5C, right panel). These data indicated that the deletion of ATIA leads to the increase of apoptosis in the testis that is mediated by TNFR1. Taken together, the above studies strongly suggest that ATIA has a protective role against TNFα-induced apoptosis in vivo.
Figure 5. ATIA protects from TNFα-and hypoxia-induced apoptosis.
(A) ATIA+/+ and ATIA−/− mice were injected with GalN and TNFα. Survival curves of ATIA+/+ (n=5) and ATIA−/− (n=8) after treatment.
(B) TUNEL staining of liver tissue isolated from ATIA+/+ and ATIA−/− mice 4 hrs after administration of GalN alone or GalN and TNFα. Increased apoptosis is seen in GalN and TNFα treated ATIA−/− as compared to the ATIA+/+ liver tissue (left panel). The level of TUNEL staining is quantified to show increased apoptosis in ATIA−/− livers as compared to wild type when treated with TNFα and GalN for 4 hours (right panel).
(C) Representative TUNEL staining of testis tissue sections from 4 week old ATIA+/+ and ATIA−/− mice (left panels). Arrows indicated TUNEL positive cells. Apoptosis in the testis of different mice as indicated was quantified by the number of apoptotic cells per 100 tubules (right panel). Data shown is the average of 3 mice for each group. *p<0.03 as compared to wild type mice.
(D) ATIA−/− MEFs are more sensitive to TNFα-induced apoptosis. ATIA+/+ and ATIA−/− MEFs were treated with TNFα and CHX for the indicated times. Cell survival was measured by MTT assay and was calculated by comparing TC treatment group to CHX treatment group.
(E) ATIA−/− MEFs were reconstituted with Vector-YFP, ATIA-YFP or ATIAc-YFP and treated with TNFα and CHX for 12 hrs. Propidium iodide staining was used to determine cell viability. The percent YFP-positive cells for each treatment group was determined by FACS. *p<0.005 as compared to with vector-YFP.
(F) ATIA+/+ and ATIA−/− MEFs were treated with CoCl2 for the indicated times. Cell survival was measured by MTT assay. Survival ratio was calculated by comparing each treatment group to untreated cells. *p<0.0003 as compared to untreated control.
(G) ATIA+/+ and ATIA−/− MEFs were treated with physiological hypoxia. Cell survival was measured by FACS analysis of annexin-positive and PI negative cells for each treatment group. Survival ratio was calculated by comparing each treatment group to untreated cells. *p< 0.003 as compared to untreated control.
Error bars represent SEM.
We next examined TNFα-induced apoptosis in ATIA−/− MEFs, which were verified by genomic PCR analysis and western blotting with the anti-ATIA antibody (Figure S4F). To do so, a time course of TNFα-induced apoptosis and varying doses of CHX were performed in wt and ATIA−/− cells. As shown in Figure 5D and S4G, ATIA−/− cells were more sensitive to TNFα-induced apoptosis compared to wt cells, suggesting that the loss of ATIA increases the sensitivity of these cells to TNFα-induced apoptosis. But however, ATIA has little effect on TNF-induced necrosis (Figure S4H). Additionally, as shown in Figure S4I-M, ATIA shows a protective effect on H2O2 and UV-induced cell death, but has no effect on doxorubicin, etoposide and staurosporine-induced apoptosis. In addition, we found that the loss of ATIA has no effect on TNFα-induced activation of NF-κB and JNK pathways (Figure S4N, O). When ATIA was ectopically expressed in ATIA−/− MEFs, it restored the resistance of the cells to TNFα-induced apoptosis (Figure 5E). To verify our observation that mito-ATIA protects cells against TNFα-induced apoptosis (Figure 1E), we also ectopically expressed ATIAc in ATIA−/− MEFs. As shown in Figure 5E, ATIAc rescued cells as efficiently as did the full length ATIA. Therefore, these results further confirmed that the mito-ATIA is sufficient to protect cells against TNFα-induced apoptosis.
We then investigated the effect of loss of ATIA on hypoxia-induced death. We treated ATIA+/+ and ATIA−/− MEFs with CoCl2 or hypoxia. As shown in Figure 5F and G, ATIA−/− MEFs showed significantly greater sensitivity to cell death under hypoxic conditions than did wt cells. Therefore, the presence of ATIA also protects cells from hypoxia-induced apoptosis.
ATIA modulates TRX2 oxidation/reduction
To understand the mechanism of ATIA-mediated protection against apoptotic stimuli, we sought to identify ATIA-interacting proteins. A mouse liver cDNA library was therefore screened by yeast-two hybrid analysis using full length ATIA as the bait (Figure S5A). After two rounds of screening, we isolated only one potential ATIA-interacting protein, thioredoxin 2 (TRX2). Interestingly, it has been shown that TRX2 is specifically expressed in the mitochondria and essential for cell survival (Tanaka et al., 2002; Nonn et al., 2003). Deletion of TRX2 causes massive apoptosis, accumulation of intracellular ROS and early embryonic lethality in homozygous mice (Nonn et al., 2003). Therefore, the identification of TRX2 as a potential ATIA-interacting protein raised the possibility that ATIA protects cells against apoptosis through regulating the activity of TRX2.
To confirm the interaction between ATIA and TRX2, we examined the ATIA and TRX2 interaction by GST pull-down assay. To do so, ATIA-YFP protein was expressed in wt MEFs and cell lysate was mixed with GST-TRX2 fusion protein or GST protein as a control. As shown in Figure 6A, GST-Trx2, but not GST, pulled down ATIA-YFP protein with a preference for the mito-ATIA-YFP, though the protein level of the mem-ATIA-YFP is much more abundant than the mito-ATIA-YFP. This result suggested that TRX2 interacts with mito-ATIA with a much higher affinity. Similar results were obtained with GST pull-down assay of endogenous ATIA protein (Figure S5B).
Figure 6. ATIA modulates TRX2 oxidization and ROS production.
(A) In vitro GST pull down assay. ATIA-YFP was transfected into wt MEFs. Cell lysates were applied to GST- and GST-TRX2 beads in lysis buffer. Bound ATIA protein was detected by western blot using anti-GFP.
(B–D) Different TRX2 redox states in ATIA+/+ and ATIA−/− MEF cells. ATIA+/+ and ATIA−/− MEF cells were treated with H2O2 (B) or TNFα and CHX (C) or CoCl2 (D). Separation of reduced and oxidized TRX2 bands is performed by AMS alkylation of thiols followed by SDS-PAGE.
(E–F) ATIA+/+ and ATIA−/− MEFs were treated with TNFα and CHX or CoCl2 for indicated times, stained with DCFDA and analyzed by flow cytometry. Grey histograms represent background level of ROS with no treatment.
(G) ATIA+/+ and ATIA−/− MEFs were treated with TNFα and CHX in the presence of BHA (100 mM) or NAC (2.5 mM) for 10 hrs. Cell survival was measured by MTT assay and was calculated by comparing TC treatment group to CHX and BHA or NAC treatment group. *p<0.003 as compared to cells treated with just TNFα and CHX.
(H) ATIA+/+ and ATIA−/− MEFs were treated with CoCl2 for 16 hrs. Cell survival was measured by MTT assay. Survival ratio was calculated by comparing each treatment group to untreated cells. *p<0.002 as compared to CoCl2 treated cells.
Error bars represent SEM.
TRX2 is a major player in scavenging ROS generated in the mitochondria (Holmgren and Lu, 2010). Particularly, it has been shown that TRX2 is specifically oxidized in response to extra cellular stimuli such as TNFα and H2O2, and the redox state of TRX2 reflects the regulation of TRX2 function (Hansen et al., 2006). We examined the redox state of TRX2 using redox Western analysis with 4-acetamido-4′-maleimidylstibene-2,2′-disulfonic acid in ATIA−/− cells following different treatments. First, we found that the deletion of ATIA does not affect the expression of TRX2 in ATIA−/− cells comparing to wild type MEFs (Figure S5C). However, as shown in Figure 6B and 6C, while TRX2 is mainly in the reduced form in wild type MEFs, the basal amount of the oxidized TRX2 protein is increased in ATIA−/− MEFs. Following treatments, the level of the oxidized form of TRX2 is only slightly increased (H2O2) or not changed at all (TNFα/CHX) in wild type MEFs. In contrast, in ATIA−/− MEFs, almost all of the TRX2 protein becomes oxidized in response to either treatment. Similarly, while the level of oxidized TRX2 is increased somewhat in wt cells, CoCl2 treatment caused all of TRX2 to become oxidized in ATIA−/− cells. These results suggest that ATIA is essential for maintaining the reduced state of TRX2.
Because oxidized TRX2 is the inactive form of the protein, we next examined if ROS levels are altered in ATIA−/− cells. To do so, we stained wt and ATIA−/− MEFs with CM-H2 DCFDA (DCFDA), a cell-permeable fluorescence dye that reacts to a broad spectrum of ROS, at different time points following TNFα or CoCl2. As shown in Figure 6E and 6F, both TNFα plus CHX and CoCl2 caused an increase of ROS accumulation in ATIA−/− cells, whereas in wt MEFs no to little increase was detected. To test the role of the increased ROS in TNFα-or CoCl2-induced apoptosis, we pretreated cells with the ROS scavengers NAC or BHA before TNFα plus CHX or CoCl2 treatment. As shown in Figure 6G and 6H, ROS scavengers prevented cell death in ATIA−/− cells. Additionally, the knockdown of TRX2 in wt MEFs greatly rendered the cells sensitive to TNFα or CoCl2-induced apoptosis while the decrease of TRX2 level in ATIA−/− cells did not potentiate the sensitivity of these cells to apoptosis (Figure S5D, E), suggesting that the increased sensitivity of ATIA−/− MEFs to TNFα or hypoxia-induced apoptosis is mainly due to the loss of TRX2 function. Therefore, these results indicated that the protective effects of ATIA are likely mediated by maintaining TRX2 in a reduced form that is capable of ameliorating ROS damage.
ATIA protein is highly expressed in human glioblastoma
Hypoxic conditions are often found in tumors and the hypoxic response mediated through HIF-1 contributes to tumor survival and progression. Since ATIA is a HIF-1 target that protects against apoptosis, it could be involved in aspects of tumorigenesis. To explore this possibility, we first examined the levels of ATIA (vasorin) protein in several types of cancers and normal tissues with Western blots purchased from ProSci Inc. As shown in Figure 7A, ATIA protein level is dramatically increased in brain tumors as compared to normal brain, using an anti-ATIA (vasorin) antibody. We then checked the ATIA protein levels in several human brain tumor cell lines including A-172 (glioblastoma) and CCF-STTG1 (astrocytoma) and found that both A-172 and CCF cells have high ATIA expression (Figure 7B). These results raised the possibility that ATIA may be highly expressed in some human brain tumors. We then performed an immuno-histological study of human glioblastoma tissue samples. As shown in Figure 7C, cells within tumors are highly positive for anti-ATIA staining while the normal tissue staining was negligible. We examined four different human glioblastoma samples and found three of the four to be highly positive for anti-ATIA staining. To further confirm this observation, we examined human tissue arrays consisting of different types of human brain tumors (Biomax, US). As summarized in Figure 7D, only glioblastoma and astrocytoma samples are positive for ATIA and no ATIA positive sample was detected in other types of brain tumors or normal tissues. Particularly, almost 70% of the 86 glioblastoma samples were ATIA positive. Therefore, ATIA is highly expressed in human glioblastoma.
Figure 7. ATIA is highly expressed in human glioblastomas.
(A) ATIA protein was detected in brain tumor by Western blotting.
(B) Western blotting of human brain tumor cell lines with the indicated antibodies.
(C) Sections of human glioblastoma samples stained with H&E and anti-vasorin antibody.
(D) Brain tumor tissue arrays with normal brain tissue controls were immunostained with anti-vasorin antibody. The data from three different brain tumor arrays was compiled and the percentage of ATIA positive cases was determined.
(E–F) A-172 cells were transfected with 50 pMol of non-targeting (NT) or ATIA siRNA and 24 hrs later treated with CoCl2 or hypoxia for the indicated times. Cell survival was measured by MTT assay (E) or by FACS of annexin-positive and PI negative cells for each treatment group (F). Survival ratio was calculated by comparing each treatment group to untreated cells. *p< 0.003 (E) or *p< 0.01 (F) as compared to untreated control. Error bars represent SEM.
Since hypoxia is critical for brain tumor development and ATIA is highly expressed in glioblastomas, we then tested whether ATIA protects glioblastoma cells from apoptosis. As shown in Figure 7E and F, the knockdown of ATIA with siRNA under either CoCl2 or hypoxic conditions increases cell death in A-172 cells when compared to a control siRNA. These cells were also sensitized to cell death induced by TNFα (Figure S6) when ATIA was knocked down. These results suggest that high levels of ATIA in glioblastoma cells are important for its protective effect and may protect them from hypoxia-induced apoptosis, thus contributing to tumor progression.
Discussion
Apoptosis is a biological process that plays a critical role in controlling tissue homeostasis. The molecular mechanisms of apoptosis have been intensively studied, however many aspects of the regulation of apoptosis remain vague. For instance, ROS is a key component of redox control and regulation of the machinery of apoptosis, but the various ways in which such regulation is achieved are largely uncharacterized (Circu and Aw, 2010). In this study, using TNFα-induced apoptosis in TRAF2−/− MEFs as the model, we have identified a HIF-1 target, ATIA, which protects cells against TNFα- and hypoxia- induced apoptosis. While ATIA level is not increased following TNFα treatment, its basal expression requires TRAF2 and its expression is induced under hypoxic conditions by HIF-1. Through the generation of ATIA ko mice, we have demonstrated the physiological role of ATIA as a survival factor and showed that ATIA protects cells from apoptosis through regulating oxidation/reduction of the mitochondrial antioxidant enzyme, thioredoxin 2 and the generation of ROS. Our findings demonstrate that ATIA is a key regulatory factor of the mitochondrial redox pathway. Furthermore, we found that ATIA is highly expressed in human glioblastoma and knockdown of ATIA expression in glioblastoma cells renders them sensitive to TNFα- and hypoxia-induced apoptosis. Therefore, ATIA is not only a HIF-1 target that regulates the mitochondrial redox pathway but also a potential diagnostic marker and therapeutic target of human glioblastoma.
Protein cellular localization is critical for its biological function and the functions of many proteins are regulated through precise control of their cellular localizations. Therefore, the same protein may have distinct physiological functions when localized to different cellular compartments. Our microscopy and cell fractionation experiments indicated that the two forms of ATIA protein localize to the plasma membrane and the mitochondria respectively. Based on our N-terminal sequencing results of the two ATIA proteins, we found that both products are the full length proteins coded by the ATIA gene (data not shown). Consequently, it is most likely that the different sizes and the different cellular localizations of the two ATIA products are due to the different modifications of the protein. The human ortholog of ATIA, vasorin, was reported as a potential inhibitor of TGFβ signaling and to localize to the cell plasma membrane (Ikeda et al., 2004). Since the truncated ATIAs, ATIAc and ATIA (287-673), do not localize to the plasma membrane, but still protect ATIA−/− MEFs from TNFα-induced apoptosis, it is unlikely that the interaction of ATIA with TGFβ plays any significant role in the ATIA anti-apoptotic effect. It is plausible that the plasma membrane localization of ATIA allows it to regulate TGFβ signaling while its mitochondrial localization of the protein ensures protection against apoptosis.
Our study suggests that ATIA protects cells from TNFα- and hypoxia-induced apoptosis. However, ATIA expression is regulated by hypoxia, but not TNFα. Our findings that ATIA protein level is low in TRAF2−/− cells and is restored when TRAF2 is ectopically expressed suggest that ATIA basal expression requires TRAF2. Studies with TRAF2 deletion have suggested that TRAF2 protects cells against TNFα-induced apoptosis independently of NF-κB, and the Sp1 transcription factor, LKLF, is a downstream effector of TRAF2 against TNFα-induced apoptosis (Lin et al., 2003; Yeh et al., 1997). It is possible that TRAF2 regulates ATIA basal expression through Sp1 since there are several Sp1 sites in ATIA promoter. Our current study provides evidence that the NF-κB-independent anti-apoptotic pathway employed distinct machinery from what we know about the NF-κB-mediated protection and reveals an addtional regulation of TNFα-induced apoptosis. Although it has been shown previously that TRX2 protects cells against apoptosis, our study connects the NF-κB-independent anti-apoptotic pathways to the mitochondrial redox pathway through the ATIA link. Since ATIA level is quite high in p65−/− cells (data not shown), which are highly sensitive to TNFα-induced apoptosis, it is unlikely that ATIA plays a role in NF-κB-dependent anti-apoptotic pathway.
Thioredoxins play a key role in the balance of cellular redox homeostasis. TRX1 is the major redox protein in the cytoplasm and is localized to the cytoplasm/nucleus (Holmgren and Lu, 2010). TRX2 is specific to the mitochondria and functions independently of TRX1 (Hansen et al., 2006). Our data suggests that the lower molecular weight band of ATIA is localized in the mitochondria and modulates the function of TRX2. It is possible that ATIA functions as a scaffold protein to facilitate the reduction process of TRX2 by its reductase, TRXR2. In addition, the ROS level is elevated in ATIA−/− cells, suggesting that ATIA may protect against TNFα-and hypoxia-induced apoptosis by regulating ROS generation. Alternatively, since the anti-apoptotic effect of TRX2 is not fully understood yet, it is possible that some target proteins of TRX2, whose redox state is regulated by ATIA, may be key regulators of TNFα- and hypoxia-induced apoptosis.
ATIA protein protects from both TNFα and hypoxia-induced apoptosis. While the basal ATIA expression is needed for protecting cells from TNFα-induced apoptosis, the elevated level of ATIA by HIF-1α is likely pivotal for preventing hypoxia-induced cell death. TNFα plays a critical role in diverse cellular events. Opposing effects of TNFα on cancer have been described: a high dose of TNFα (acute inflammation) has anti-neoplastic effects, such as direct cytotoxicity on certain types of cancer, while endogenous low-doses of TNFα (chronic inflammation) promote cancer development. HIF-1 antagonizes apoptosis through activating its target genes and these HIF-1-mediated genes are critical for tumor cells to survive under hypoxic conditions. Therefore, ATIA may be critical for the tumorigenesis of some tumors, such as human glioblastoma, by blocking TNFα and hypoxia-induced apoptosis and allowing TNFα to fulfill its tumor promoting function. Consequently, it is important to study the potential involvement of ATIA in the development of human glioblastoma and to explore the therapeutic value of ATIA in cancer treatment in the future. Our discovery that ATIA is highly expressed in human glioblastoma, but not in normal brain tissues, suggests that ATIA could also be a valuable diagnostic marker for this poorly studied, deadly disease.
Experimental Procedures
Reagents
TNFα was purchased from R&D Inc. Antibodies were purchased: anti-pJNK and anti-V5 from Invitrogen; anti-GST from Cell Signaling; anti-IκBα and anti-AIF from Santa Cruz; anti-JNK from Pharmingen; anti-actin and anti GFP, from SIGMA; anti-TNFR1, anti-vasorin and anti-HIF-1α from R&D; anti-TRX2 from ABCAM. Anti-ATIA was made by Rockland.
Treatment conditions
In all experiments, TNFα was used at 30ng/ml, CHX at 100ng/ml for TRAF2−/− and 1 μg/ml for other cells or as indicated.
Cell Culture and Transfection
ATIA−/− MEFs were generated from ATIA ko mice. Cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% calf serum, 2 mM glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were transfected with plasmids using Lipofectamine and Plus reagent (Invitrogen). Brain tumor cell lines A172, IMR-32 and CCF-STTG1 were purchased from ATCC.
Cytotoxicity assay
The cell death was determined by 3-(4,5-dimethylthiazol-2yl)-2,5-diphenyltetrazolium bromide (MTT) assay. MTT absorbance was read at 570 nm. To determine cell survival by FACS analysis, we transfected TRAF2−/− or ATIA−/− cells with different plasmids. 24 hours later, the cells were treated with CHX (1μg/ml) or/and TNFα for 18 or 8 hours. Propidium iodide staining (PI) was used to determine cell viability. The percent YFP-positive cells for each treatment group was determined by FACS. Survival ratio was calculated by comparing TNFα plus CHX treatment group to CHX treatment group for each plasmid.
Northern blot
ATIA cDNA probes were prepared with a random primer kit (Stratagene) (Fig 1B) or with a STRIP-EZ kit (Ambion) (Fig 1C, D). For Figure 1B, a mouse tissue RNA blot was from Clonetech. For Figure 1C and D, each sample had 15 ug of total RNA. For loading control, the blots were stripped and reprobed with a GADPH cDNA probe or actin cDNA probe.
Immunoblot analysis
Cells were collected and lysed in M2 buffer (20 mM Tris, pH 7, 0.5% NP40, 250 mM NaCl, 3 mM EDTA, 3 mM EGTA, 2 mM DTT, 0.5 mM PMSF, 20 mM β-glycerol phosphate, 1 mM sodium vanadate, 1 μg/ml Leupeptin). Cell lysates were separated by SDS-PAGE and analyzed by immunoblot. The proteins were visualized by enhanced chemiluminescence (ECL), according to the manufacturer’s (Pierce) instruction.
Hypoxia
Cells were incubated in normaxia (20%O2) or hypoxia (0.1% or 1% O2) for various times. Hypoxia was achieved by incubating the cells in a sealed chamber in which there was 0.1% O2 or 1%O2, 5% CO2 and the remainder N2. This chamber was then placed in an incubator at 37oC. Hypoxia was chemically induced by CoCl2 (400 μM or as indicated) added to the media.
Confocal Imaging
Confocal microscopy was performed with cells transiently transfected with YFP-tagged ATIA constructs. In order to determine mitochondrial localization, the cells were co-stained with MitoTracker Red (Molecular Probes) according to manufactures protocol.
Pronase treatment
Monolayers of cells were washed with HBSS and incubated in 1 ml of HBSS containing 0.01% of pronase (Calbiochem) at 37°C for 10 min. Pronase activity was blocked with ice cold complete media.
Mitochondrial fractionation
Cytosolic and mitochondrial fractions of cell lysates were obtained using the Mitochondrial Fractionation Kit (Active Motif). Fractionation was performed according to the protocol provided.
Animal Treatment
The experiments were conducted in accordance with ACUC guidelines. ATIA ko and wt siblings at 8–10 weeks were sensitized with intraperitoneal administration of 700 mg/Kg of Galactosamine (GalN). After 30 minutes, a sub-lethal dose of recombinant mouse TNFα in pyrogen-free saline (2 ug/Kg) was administered intravenously. 4 hours later livers were harvested and the liver sections were analyzed by H&E and TUNEL (R&D).
GST-, GST-TRX2 Pull-Down Assay
GST and GST-TRX2 fusion proteins were affinity purified on glutathione-Sepharose beads (Pharmacia). Cell lysates (400 μg) expressing GFP-tagged ATIA were incubated with 30 μg of GST or GST-TRX2 bound to glutathione-Sepharose in the lysis buffer overnight at 4°C. The beads were washed 4–6 times with the lysis buffer and the bound ATIA proteins were removed by boiling in SDS buffer and resolved on SDS-PAGE for Western blot analysis.
Detection of ROS Accumulation
Cells were cultured in phenol red-free medium and treated with TNFα and CHX for the indicated time periods. The 5(6)-chloromethyl-2-7-dichlorodihydrofluorescence diacetate (DCFDA, 1μM) from Molecular Probes was added 30 min before collecting cells. The stained cells were analyzed with a flow cytometer (FACSCalibur, BD Biosciences) and data were processed with the FlowJo program (BD Biosciences).
Measuring Red-Ox
Cells were treated with H2O2 (1 mM) for 10 minutes or CHX (1ug/ml) and TNFα for the indicated times. The redox state of TRX2 was determined by performing western analysis with 4-acetamido-4′-maleimidylstilbene-2, 2′-disulfonic acid (AMS) as described by (Halvey et al., 2005) based on the original method of (Damdimopoulos et al., 2002).
Brain tumor Arrays
ATIA (vasorin) protein detected in several types of cancers and normal tissues by Western blots from ProSci Inc. The brain tumor array (GL803), glioblastoma tissue array (BS17018) and the multiple brain cancer and normal adjacent tissue array (GL1001) were purchased from BioMax. The slides were stained with anti-vasorin antibody using immunohistochemistry procedures as described in Supplemental methods.
Statistical Analysis
Error bars represent standard error of mean. The p value was determined using Student’s t test and p<0.05 versus respective control is significant.
Supplementary Material
Highlights
ATIA protects against apoptosis by regulating TRX2 function and ROS generation
ATIA localizes to the membrane and the mitochondria
ATIA expression is regulated by HIF-1α under hypoxia
Loss of ATIA in glioblastoma renders cells sensitive to hypoxia-induced apoptosis
Acknowledgments
We would like to thank Drs. W. C. Yeh for TRAF2−/− MEFs and Dr. W. Min for the GST-TRX2 plasmid. We also thank Dr. K. D. Aldape for providing us with human glioblastoma samples. We thank Dr. Alfredo A. Molinolo for his expertise in histopathology. This research was supported by the Intramural Research Program of Center for Cancer Research, NCI, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baud V, Karin M. Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol. 2001;11:372–377. doi: 10.1016/s0962-8924(01)02064-5. [DOI] [PubMed] [Google Scholar]
- Circu ML, Aw TY. Reactive oxygen species, cellular redox systems, and apoptosis. Free Radic Biol Med. 2010;48:749–762. doi: 10.1016/j.freeradbiomed.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damdimopoulos AE, Miranda-Vizuete A, Pelto-Huikko M, Gustafsson JA, Spyrou G. Human mitochondrial thioredoxin. Involvement in mitochondrial membrane potential and cell death. J Biol Chem. 2002;277:33249–33257. doi: 10.1074/jbc.M203036200. [DOI] [PubMed] [Google Scholar]
- Decker K, Keppler D. Galactosamine hepatitis: key role of the nucleotide deficiency period in the pathogenesis of cell injury and cell death. Rev Physiol Biochem Pharmacol. 1974:77–106. doi: 10.1007/BFb0027661. [DOI] [PubMed] [Google Scholar]
- Fadeel B, Orrenius S. Apoptosis: a basic biological phenomenon with wide-ranging implications in human disease. J Intern Med. 2005;258:479–517. doi: 10.1111/j.1365-2796.2005.01570.x. [DOI] [PubMed] [Google Scholar]
- Fukuda R, Zhang H, Kim J-w, Shimoda L, Dang CV, Semenza GL. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129:111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- Halvey PJ, Watson WH, Hansen JM, Go YM, Samali A, Jones DP. Compartmental oxidation of thiol-disulphide redox couples during epidermal growth factor signalling. Biochem J. 2005;386:215–219. doi: 10.1042/BJ20041829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanaka RB, Chandel NS. Mitochondrial reactive oxygen species regulate hypoxic signaling. Curr Opin in Cell Biol. 2009;21:894–899. doi: 10.1016/j.ceb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JM, Zhang H, Jones DP. Mitochondrial thioredoxin-2 has a key role in determining tumor necrosis factor-alpha-induced reactive oxygen species generation, NF-kappaB activation, and apoptosis. Toxicol Sci. 2006;91:643–650. doi: 10.1093/toxsci/kfj175. [DOI] [PubMed] [Google Scholar]
- Henson PM, Hume DA. Apoptotic cell removal in development and tissue homeostasis. Trends Immunol. 2006;27:244–250. doi: 10.1016/j.it.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Holmgren A, Lu J. Thioredoxin and thioredoxin reductase: current research with special reference to human disease. Biochem Biophys Res Commun. 2010;396:120–124. doi: 10.1016/j.bbrc.2010.03.083. [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Imai Y, Kumagai H, Nosaka T, Morikawa Y, Hisaoka T, Manabe I, Maemura K, Nakaoka T, Imamura T, et al. Vasorin, a transforming growth factor beta-binding protein expressed in vascular smooth muscle cells, modulates the arterial response to injury in vivo. Proc Natl Acad Sci U S A. 2004;101:10732–10737. doi: 10.1073/pnas.0404117101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher MA, Grimm S, Ishida Y, Kuo F, Stanger BZ, Leder P. The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity. 1998;8:297–303. doi: 10.1016/s1074-7613(00)80535-x. [DOI] [PubMed] [Google Scholar]
- Kim YS, Morgan MJ, Choksi S, Liu ZG. TNF-induced activation of the Nox1 NADPH oxidase and its role in the induction of necrotic cell death. Mol Cell. 2007;26:675–687. doi: 10.1016/j.molcel.2007.04.021. [DOI] [PubMed] [Google Scholar]
- Lin Y, Choksi S, Shen HM, Yang QF, Hur GM, Kim YS, Tran JH, Nedospasov SA, Liu ZG. Tumor necrosis factor-induced nonapoptotic cell death requires receptor-interacting protein-mediated cellular reactive oxygen species accumulation. J Biol Chem. 2004;279:10822–10828. doi: 10.1074/jbc.M313141200. [DOI] [PubMed] [Google Scholar]
- Lin Y, Ryan J, Lewis J, Wani MA, Lingrel JB, Liu ZG. TRAF2 exerts its antiapoptotic effect by regulating the expression of Kruppel-like factor LKLF. Mol Cell Biol. 2003;23:5849–5856. doi: 10.1128/MCB.23.16.5849-5856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loor G, Schumacker PT. Role of hypoxia-inducible factor in cell survival during myocardial ischemia-reperfusion. Cell Death Differ. 2008;15:686–690. doi: 10.1038/cdd.2008.13. [DOI] [PubMed] [Google Scholar]
- Morgan MJ, Kim YS, Liu ZG. TNFalpha and reactive oxygen species in necrotic cell death. Cell Res. 2008;18:343–349. doi: 10.1038/cr.2008.31. [DOI] [PubMed] [Google Scholar]
- Nakayama K. Cellular signal transduction of the hypoxia response. J Biochem. 2009;146:757–765. doi: 10.1093/jb/mvp167. [DOI] [PubMed] [Google Scholar]
- Nonn L, Williams RR, Erickson RP, Powis G. The absence of mitochondrial thioredoxin 2 causes massive apoptosis, exencephaly, and early embryonic lethality in homozygous mice. Mol Cell Biol. 2003;23:916–922. doi: 10.1128/MCB.23.3.916-922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul SA, Simons JW, Mabjeesh NJ. HIF at the crossroads between ischemia and carcinogenesis. J Cell Physiol. 2004;200:20–30. doi: 10.1002/jcp.10479. [DOI] [PubMed] [Google Scholar]
- Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20:51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HM, Pervaiz S. TNF receptor superfamily-induced cell death: redox-dependent execution. Faseb J. 2006;20:1589–1598. doi: 10.1096/fj.05-5603rev. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Hosoi F, Yamaguchi-Iwai Y, Nakamura H, Masutani H, Ueda S, Nishiyama A, Takeda S, Wada H, Spyrou G, et al. Thioredoxin-2 (TRX-2) is an essential gene regulating mitochondria-dependent apoptosis. The EMBO journal. 2002;21:1695–1703. doi: 10.1093/emboj/21.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thannickal VJ, Fanburg BL. Reactive oxygen species in cell signaling. Am J Physiol Lung Cell Mol Physiol. 2000;279:L1005–1028. doi: 10.1152/ajplung.2000.279.6.L1005. [DOI] [PubMed] [Google Scholar]
- Tiegs G, Wolter M, Wendel A. Tumor necrosis factor is a terminal mediator in galactosamine/endotoxin-induced hepatitis in mice. Biochem Pharmacol. 1989;38:627–631. doi: 10.1016/0006-2952(89)90208-6. [DOI] [PubMed] [Google Scholar]
- Ventura JJ, Cogswell P, Flavell RA, Baldwin AS, Jr, Davis RJ. JNK potentiates TNF-stimulated necrosis by increasing the production of cytotoxic reactive oxygen species. Genes Dev. 2004;18:2905–2915. doi: 10.1101/gad.1223004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajant H. Death receptors. Essays in biochemistry. 2003;39:53–71. doi: 10.1042/bse0390053. [DOI] [PubMed] [Google Scholar]
- Wajant H, Pfizenmaier K, Scheurich P. Tumor necrosis factor signaling. Cell Death Differ. 2003;10:45–65. doi: 10.1038/sj.cdd.4401189. [DOI] [PubMed] [Google Scholar]
- Yeh WC, Shahinian A, Speiser D, Kraunus J, Billia F, Wakeham A, de la Pompa JL, Ferrick D, Hum B, Iscove N, et al. Early lethality, functional NF-kappaB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity. 1997;7:715–725. doi: 10.1016/s1074-7613(00)80391-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.