Abstract
In the Chlorophyceae, the chloroplast genome is extraordinarily fluid in architecture and displays unique features relative to other groups of green algae. For the Chaetophorales, 1 of the 5 major lineages of the Chlorophyceae, it has been shown that the distinctive architecture of the 223,902-bp genome of Stigeoclonium helveticum is consistent with bidirectional DNA replication from a single origin. Here, we report the 182,759-bp chloroplast genome sequence of Schizomeris leibleinii, a member of the earliest diverging lineage of the Chaetophorales. Like its Stigeoclonium homolog, the Schizomeris genome lacks a large inverted repeat encoding the rRNA operon and displays a striking bias in coding regions that is associated with a bias in base composition along each strand. Our results support the notion that these two chaetophoralean genomes replicate bidirectionally from a putative origin located in the vicinity of the small subunit ribosomal RNA gene. Their shared structural characteristics were most probably inherited from the common ancestor of all chaetophoralean algae. Short dispersed repeats account for most of the 41-kb size variation between the Schizomeris and Stigeoclonium genomes, and there is no indication that homologous recombination between these repeated elements led to the observed gene rearrangements. A comparison of the extent of variation sustained by the Stigeoclonium and Schizomeris chloroplast DNAs (cpDNAs) with that observed for the cpDNAs of the chlamydomonadalean Chlamydomonas and Volvox suggests that gene rearrangements as well as changes in the abundance of intergenic and intron sequences occurred at a slower pace in the Chaetophorales than in the Chlamydomonadales.
Keywords: plastid genome evolution, chloroplast gene rearrangements, introns, replication origin, short dispersed repeats, Chlamydomonadales
Introduction
The chloroplast genome is evolving in a dynamic fashion in the chlorophyte class Chlorophyceae (Chlorophyta). Comparative analyses of the complete chloroplast genome sequences from Chlamydomonas reinhardtii (Chlamydomonadales), Scenedesmus obliquus (Sphaeropleales), Stigeoclonium helveticum (Chaetophorales), Oedogonium cardiacum (Oedogoniales), and Floydiella terrestris (Chaetopeltidales) revealed that each of the five orders recognized in this green algal class is characterized by a distinctive genome architecture (Maul et al. 2002; Bélanger et al. 2006; de Cambiaire et al. 2006; Brouard et al. 2008, 2010). In addition to uncovering the dynamic evolution of the chlorophycean chloroplast genome, these comparative studies resolved the branching order of the five chlorophycean lineages and allowed the reconstruction of a hypothetical scenario depicting the origins of some of the observed genomic changes (Brouard et al. 2010). Two major clades were identified, the CS clade (Chlamydomonadales and Sphaeropleales) and the OCC clade (Oedogoniales, Chaetopeltidales, and Chaetophorales), and the Oedogoniales was identified as an early diverging branch of the OCC clade (Turmel et al. 2008; Brouard et al. 2010). These phylogenetic results received strong independent support from structural characters at the levels of gene content, intron content, gene order, and gene structure. More recently, the collection of chlorophycean chloroplast genomes was further enriched with the nearly complete chloroplast DNA (cpDNA) sequence of Volvox carteri (Chlamydomonadales) (Smith and Lee 2009) and the complete cpDNA sequence of Dunaliella salina (Chlamydomonadales) (Smith et al. 2010).
Among all completely sequenced green algal genomes, those of chlorophycean green algae display the largest sizes (size ranges from 161 kb in Scenedesmus to 521 kb in Floydiella) and the lowest retention of ancestral genomic features (Maul et al. 2002; Bélanger et al. 2006; de Cambiaire et al. 2006; Brouard et al. 2008; Smith and Lee 2009; Brouard et al. 2010; Smith et al. 2010). The gene repertoire of chlorophycean genomes (94–100 genes) is consistently reduced relative to streptophyte (charophycean and land plant) and other chlorophyte (prasinophycean, trebouxiophycean, and ulvophycean) cpDNAs, with all chlorophycean cpDNAs lacking eight conserved genes (accD, chlI, minD, psaI, rpl19, ycf20, ycf62, and trnR(ccg)) relative to the two ulvophycean cpDNAs sequenced thus far (Pombert et al. 2005, 2006). Moreover, chlorophycean genomes have retained a very limited number of ancestral gene clusters from the bacterial progenitor of primary chloroplasts. Like most green plant cpDNAs, five of the completely sequenced chlorophycean genomes (those of the Chlamydomonadales, Sphaeropleales, and Oedogoniales) exhibit two copies of a large inverted repeat (IR) sequence separated by single-copy regions; however, gene contents of the single-copy regions differ remarkably between distinct chlorophycean orders and strongly deviate from the ancestral patterns observed for the prasinophyceans Nephroselmis and Pyramimonas, the trebouxiophyceans Pedinomonas, Parachlorella and Oocystis, and all streptophytes having an IR in their chloroplast genome (Turmel, Otis, and Lemieux 1999, 2009; de Cambiaire et al. 2006; Brouard et al. 2008; Turmel, Gagnon, et al. 2009). Several alterations at the level of gene structure, notably the fragmentation of the rpoB gene in two separate open reading frames (ORFs) and the expansion of the coding regions of clpP, rps3, and rps4 (Bélanger et al. 2006; de Cambiaire et al. 2006; Brouard et al. 2008, 2010) distinguish all chlorophycean cpDNAs from their homologs in the Chlorophyta. In contrast, genomic changes such as the breakup of four genes (petD, psaA, psaC, rbcL) by putatively trans-spliced group II introns and the fragmentation of rpoC1 and rps2 arose following the emergence of specific lineages (Bélanger et al. 2006; de Cambiaire et al. 2006; Brouard et al. 2008, 2010). Unlike most of their chlorophyte counterparts, several chlorophycean cpDNAs are intron rich, although the distribution and abundance of both group I and group II introns are highly variable. Similarly, the prevalence of short dispersed repeats in the chloroplast genomes of Chlamydomonas, Volvox, Stigeoclonium, and Floydiella contrasts sharply with the low frequency observed for such sequences in the Scenedesmus and Oedogonium chloroplasts.
The extremely high variability in chloroplast genome architecture observed among the chlorophyceans examined so far emphasizes the need for sampling additional taxa in order to document the extent of changes found within each order, validate the evolutionary scenario inferred by Brouard et al. (2010), and better understand the evolutionary forces shaping the genome. The focus of the present study is on the Chaetophorales and Chlamydomonadales. The chloroplast genome of Schizomeris leibleinii, a representative of the earliest diverging lineage (Schizomeridaceae) of the Chaetophorales (Buchheim et al. 2001; Caisova et al. 2011) was fully sequenced and compared with that of its distant chaetophoralean relative S. helveticum (which belongs to a clade containing branched taxa of the Chaetophoraceae) as well as with other previously sequenced chloroplast genomes from the Chlorophyceae, including those of the chlamydomonadalean algae Chlamydomonas and Volvox.
The IR-lacking chloroplast genome of Stigeoclonium is rich in introns and dispersed short repeats and is presumed to share a common loss of the IR with the cpDNA of Floydiella (Chaetopeltidales) (Brouard et al. 2010). It displays a remarkable pattern of gene distribution in which genes on one half of the genome are encoded by the same strand and those on the other half are encoded by the alternative strand (Bélanger et al. 2006). As found in prokaryotic genomes that replicate bidirectionally from a single origin (Grigoriev 1998; Tillier and Collins 2000a, 2000b; Guy and Roten 2004), this strand bias in coding regions is closely associated with a bias in GC composition along each strand. Analysis of the cumulative GC skew has proven useful to identify the origin and terminus of replication in prokaryotic genomes (Grigoriev 1998; Guy and Roten 2004), and the application of this method to the Stigeoclonium chloroplast genome disclosed a putative replication origin in the trnS(gga)-rrs intergenic region and a putative terminus in the psbD-tufA intergenic region (Bélanger et al. 2006).
We report here that, although the Schizomeris genome is more compact and substantially rearranged relative to the Stigeoclonium cpDNA, it also lacks an IR and, as revealed by a cumulative GC skew analysis, displays a putative origin of replication in the same region. The two chaetophoralean genomes share very similar noncoding sequences in this region, supporting their putative role as an origin of replication and as anticipated, these sequences are also conserved at the corresponding cpDNA locus in the unbranched alga Uronema belkae (Chaetophoraceae). Moreover, our comparative analyses of the two available pairs of closely related chlorophycean cpDNAs suggests that the chloroplast genome evolved more conservatively in the Chaetophorales than in the Chlamydomonadales.
Materials and Methods
Strains and Culture Conditions
Schizomeris leibleinii (UTEX LB 1228) and U. belkae (UTEX 1179) were obtained from the Culture Collection of Algae at the University of Texas at Austin and were grown in C medium (Andersen et al. 2005) and the modified Volvox medium (McCracken et al. 1980), respectively. Both cultures were subjected to alternating 12-h light–dark periods.
Cloning and Sequencing of the Schizomeris Chloroplast Genome
An A + T-rich organellar DNA fraction was obtained by CsCl-bisbenzimide isopycnic centrifugation as described earlier (Turmel, Lemieux, et al. 1999). This DNA fraction was sheared by nebulization to produce 2,000- to 4,000-bp fragments that were subsequently cloned into the pSMART-HCKan plasmid (Lucigen Corporation). Positive clones were selected by hybridization of the plasmid library with the original DNA used for cloning. DNA templates were amplified using the Illustra TempliPhi Amplification Kit (GE Healthcare) and sequenced as described previously (Turmel et al. 2005). The DNA sequences were edited and assembled using SEQUENCHER 4.7 (Gene Codes Corporation). Genomic regions absent in the clones analyzed were directly sequenced from polymerase chain reaction (PCR)-amplified fragments using internal primers. Alternatively, PCR-amplified fragments were subcloned using the TOPO TA cloning kit (Invitrogen) before sequencing.
Amplification of the Putative Replication Origin of Uronema cpDNA
We wished to determine whether the conserved noncoding sequences we identified in the cpDNA regions containing the putative origins of replication of Schizomeris and Stigeoclonium are also found at the corresponding locus in the chaetophoralean Uronema. We thus amplified by PCR the spacer between the psbF and rrs genes of Uronema using total cellular DNA and the following primers: 5′-CGTTGAATAAATTGCATAGCAG-3′ and 5′-CAACTAGCTAATCAGACGCAAG-3′. The resulting fragment of 4,385 bp was sequenced using the latter primers as well as internal primers.
Sequence Analyses
Genes and ORFs were identified as described previously (Brouard et al. 2010). Boundaries of introns were located by modeling intron secondary structures (Michel et al. 1989; Michel and Westhof 1990) and by comparing the sequences of intron-containing genes with those of intronless homologs using diverse programs of the FASTA package (Pearson and Lipman 1988; Pearson et al. 1997). Regions of the genome sequence containing nonoverlapping repeated elements were mapped with REPEATMASKER (http://www.repeatmasker.org/) running under the WU-BLAST 2.0 search engine (http://blast.wustl.edu/), using the repeats ≥30 bp identified with REPFIND of the REPuter 2.74 program (Kurtz et al. 2001) as input sequences.
The sidedness index (Cs) was determined as described by Cui et al. (2006) using the formula Cs = (n − nSB)/(n − 1), where n is the total number of genes in the genome and nSB is the number of sided blocks, that is, the number of blocks including adjacent genes on the same strand. The strand bias in base composition was calculated for the whole genome. For the entire genome sequence, the GC skew, that is, the sum of values (G − C)/(G + C), where C and G represent the number of occurrences of these two nucleotides, was calculated for windows of length 5,000, starting with nucleotides 150,000–155,000 and continuing by shifting 500 nucleotides downstream along the strand for each new window.
Homology searches in intergenic regions were performed with the BlastN tool of the NCBI web server against the nucleotide collection (nr/nt) using the Schizomeris intergenic sequences as queries and the default parameters.
Gene Order Analyses
Gene pairs exhibiting identical gene polarities in Schizomeris and other chlorophycean cpDNAs were identified using a custom-built program. The GRIMM web server (Tesler 2002) was used as described previously (Bélanger et al. 2006) to infer the minimal number of gene permutations by inversions in all possible pairwise comparisons of chlorophycean cpDNAs. The data set used in the latter analyses consisted of 93 genes; note that pieces of the rpoB gene and all exons of the genes containing trans-spliced introns were coded as distinct fragments (for a total of 98 gene loci). Because GRIMM requires that the compared genomes have exactly the same gene content, duplicated genes, including those within 1 of the 2 copies of the IR, as well as genes lacking in some of the genomes analyzed were excluded from the data set and the genomes were considered as linear molecules. In the case of the duplicated trnS(gcu) gene in the Schizomeris genome, the copy being part of the gene pair shared with the Stigeoclonium genome (trnS(gcu)-ycf1) was retained for analysis. The data set used for inferring the scenario of gene rearrangements between the IR-lacking Schizomeris and Stigeoclonium cpDNAs was constructed in the same manner and comprised 99 genes, for a total of 104 gene loci. In this analysis, the two compared genomes were considered as circular molecules.
Phylogenetic Analyses of Sequence Data
The deduced amino acid sequences of individual chloroplast protein-coding genes shared by eight chlorophyceans (for names of taxa and accession numbers of chloroplast genome sequences, see footnote of table 1) and the ulvophycean Pseudendoclonium akinetum were aligned using MUSCLE 3.7 (Edgar 2004). A total of 61 genes met this criterion: atpA, B, E, F, H, I, ccsA, cemA, clpP, ftsH, petB, D, G, L, psaA, B, C, J, psbA, B, C, D, E, F, H, I, J, K, L, M, N, T, Z, rbcL, rpl2, 5, 14, 16, 20, 23, 36, rpoA, B, C1, C2, rps2, 3, 4, 7, 8, 9, 11, 12, 14, 18, 19, tufA, ycf1, 3, 4, 12. The amino acid alignments were then converted into alignments of codons. The poorly aligned and divergent regions in each codon alignment were removed using GBLOCKS 0.91b (Castresana 2000) and the option –t = c for exclusion of complete codons. After concatenation of the codon alignments, a maximum likelihood tree was inferred from the resulting data set using Treefinder (version of October 2008) and the GTR + Γ4 model of nucleotide substitutions. Evolutionary distances were estimated with CODEML in PAML (Yang 1997, 2007) using the same data set and the tree generated by Treefinder as a constraint. The analysis was run using all default parameters, except that the free-ratio model was selected to allow the ratio of synonymous and nonsynonymous substitution rates to vary along each branch in the tree.
Table 1.
General Features of Schizomeris and Other Sequenced Chlorophycean cpDNAs
| OCC Clade |
CS Clade |
|||||||
| Oedogoniales | Chaetopeltidales | Chaetophorales |
Chlamydomonadales |
Sphaeropleales | ||||
| Feature | Oc | Ft | Sh | Sl | Cr | Vca | Ds | So |
| Size (bp) | ||||||||
| Total | 196,547 | 521,168 | 223,902 | 182,759 | 203,827 | 461,064 | 269,044 | 161,452 |
| IR | 35,492 | —b | —b | —b | 22,211 | 15,948 | 14,409 | 12,022 |
| SC1c | 80,363 | —b | —b | —b | 81,307 | 227,676 | 127,339 | 72,440 |
| SC2d | 45,200 | —b | —b | —b | 78,088 | 200,100 | 112,887 | 64,968 |
| A + T (%) | 70.5 | 65.5 | 71.1 | 72.8 | 65.5 | 57.0 | 67.9 | 73.1 |
| Sidedness index | 0.74 | 0.91 | 0.95 | 0.97 | 0.87 | 0.83 | 0.86 | 0.88 |
| Conserved genes (no.)e | 99 | 97 | 97 | 98 | 94 | 94 | 94 | 96 |
| Introns | ||||||||
| Fraction of genome (%) | 17.9 | 3.4 | 10.9 | 13.4 | 7.0 | 4.4 | 10.3 | 7.9 |
| Group I (no.) | 17 | 19 | 16 | 24 | 5 | 3 | 21 | 7 |
| Group II (no.) | 4 | 7 | 5 | 9 | 2 | 6 | 2 | 2 |
| Intergenic sequencesf | ||||||||
| Fraction of genome (%) | 22.6 | 77.8 | 44.4 | 31.5 | 49.2 | 75.3 | 54.1 | 34.3 |
| Average size (bp) | 370 | 3,824 | 975 | 538 | 937 | 3,405 | 1,347 | 517 |
| Short repeated sequencesg | ||||||||
| Fraction of genome (%) | 1.3 | 49.9 | 17.8 | 1.2 | 16.8 | 45.5 | 8.6 | 3.0 |
NOTE.—Abbreviations: Oc, Oedogonium cardiacum, NC_011031 (Brouard et al. 2008); Ft, Floydiella terrestris, NC_014346 (Brouard et al. 2010); Sh, Stigeoclonium helveticum, NC_008372 (Bélanger et al. 2006); Sl, Schizomeris leibleinii, HQ700713 (this study); Cr, Chlamydomonas reinhardtii, NC_005353 (Maul et al. 2002); Vc, Volvox carteri, GU084820 (Smith and Lee 2010); Ds, Dunaliella salina, GQ250046 (Smith et al. 2010); So, Scenedesmus obliquus, NC_008101 (de Cambiaire et al. 2006).
Values provided for the Volvox cpDNA should be considered as estimates because some intergenic regions could not be entirely sequenced.
Because the Floydiella, Stigeoclonium, and Schizomeris cpDNAs lack an IR, only the total sizes of these genomes are given.
Single-copy region with the larger size.
Single-copy region with the smaller size.
Conserved genes refer to free-standing coding sequences usually present in chloroplast genomes. Genes present in the IR as well as others duplicated were counted only once.
ORFs < 130 codons showing no sequence similarity with known genes were considered as intergenic sequences.
Nonoverlapping repeated elements ≥30 bp were identified as described in the Materials and Methods.
Data Deposition
The annotated sequences of the S. leibleinii chloroplast genome and of the U. belkae psbF-rrs spacer have been deposited in GenBank under the accession numbers HQ700713 and HQ700714, respectively.
Results and Discussion
The Schizomeris Chloroplast Genome Shares a Distinctive Architecture with the Stigeoclonium Genome
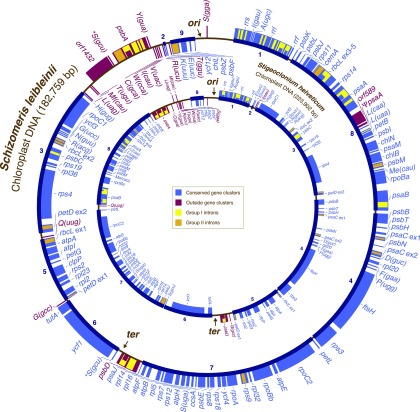
At 182,759 bp, the circular-mapping chloroplast genome of Schizomeris is 41-kb smaller than the Stigeoclonium genome (fig. 1). Table 1 summarizes the general features of the Schizomeris genome and compares them with those of other chlorophycean cpDNAs. Like its homologs in Stigeoclonium and the more distantly related Floydiella, the Schizomeris chloroplast genome lacks an IR, thus strengthening the hypothesis that the IR was lost before the divergence of the Chaetophorales and the Chaetopeltidales (Brouard et al. 2010). The repertoire of conserved chloroplast genes in Schizomeris only differs from that of Stigeoclonium by the presence of trnT(ggu) (supplementary table S1, Supplementary Material online); other distinguishing features of the Schizomeris genome include the complete duplication of the trnS(gcu) and partial duplication of psaA. In the supplementary section S1 of the Supplementary Material online, we discuss about the possible origin of trnT(ggu) and report other cases of gene duplications in green plant chloroplast genomes. The Schizomeris genome contains 12 extra introns compared with its Stigeoclonium counterpart (table 1). Nine of the 33 introns it displays (for the characteristics of all introns, see supplementary table S2 and section S2, Supplementary Material online) occur at positions that have not been reported so far in green plants. An updated compilation of known insertion sites for group I introns in completely sequenced chlorophyte genomes is presented in supplementary figure S1 (Supplementary Material online). In contrast to the Stigeoclonium and Floydiella genomes (table 1), the intergenic regions of the Schizomeris cpDNA display a small proportion of short dispersed repeats (see supplementary section S3 and fig. S2, Supplementary Material online).
FIG. 1.—
Gene maps of the Schizomeris and Stigeoclonium chloroplast genomes. Coding sequences on the outside of the map are transcribed in a clockwise direction. Genes included in the nine blocks of colinear sequences (numbered 1 to 9) shared between Schizomeris and Stigeoclonium are shown in blue, whereas rearranged or unique genes are shown in red. Group I and group II introns are shown in yellow and orange, respectively; intron ORFs are distinguished from exons by narrower boxes. Arrows mark the putative origin (ori) and terminus (ter) of replication. The duplicated Schizomeris trnS(gcu) is indicated by an asterisk. The rpoB gene consists of two separate ORFs (rpoBa and rpoBb) that are not associated with sequences typical of group I or group II introns. Two free-standing ORFs containing more than 130 codons (orf589 and orf1432) were identified. These ORFs, which are found upstream of the partially duplicated psaA gene and the duplicated trnS(gcu) copy, show no homology with any known DNA sequences. tRNA genes are indicated by the one-letter amino acid code followed by the anticodon in parentheses (Me, elongator methionine; Mf, initiator methionine).
The Schizomeris genome resembles its Stigeoclonium counterpart in displaying a striking bias in the distribution of genes between the two DNA strands. In the Schizomeris cpDNA, all the conserved genes in the 120-kb segment extending from trnS(gga) to trnG(gcc) are located on the same strand, whereas most of the remaining genes are encoded on the opposite strand (fig. 1). Two large blocks of genes encoded by distinct strands are also observed in the Stigeoclonium cpDNA; however, each block occupies approximately one half of the genome (fig. 1). The sidedness index (Cs) is a helpful parameter to estimate the propensity of adjacent genes to be located on the same DNA strand in a given genome (Cui et al. 2006). The calculated values for the Schizomeris and Stigeoclonium cpDNAs (Cs = 0.97 and Cs = 0.95, respectively) are substantially higher than those reported for other chlorophycean cpDNAs (table 1).
A Bidirectional Mode of cpDNA Replication from a Single Origin Appears to Have Been Maintained in the Chaetophorales
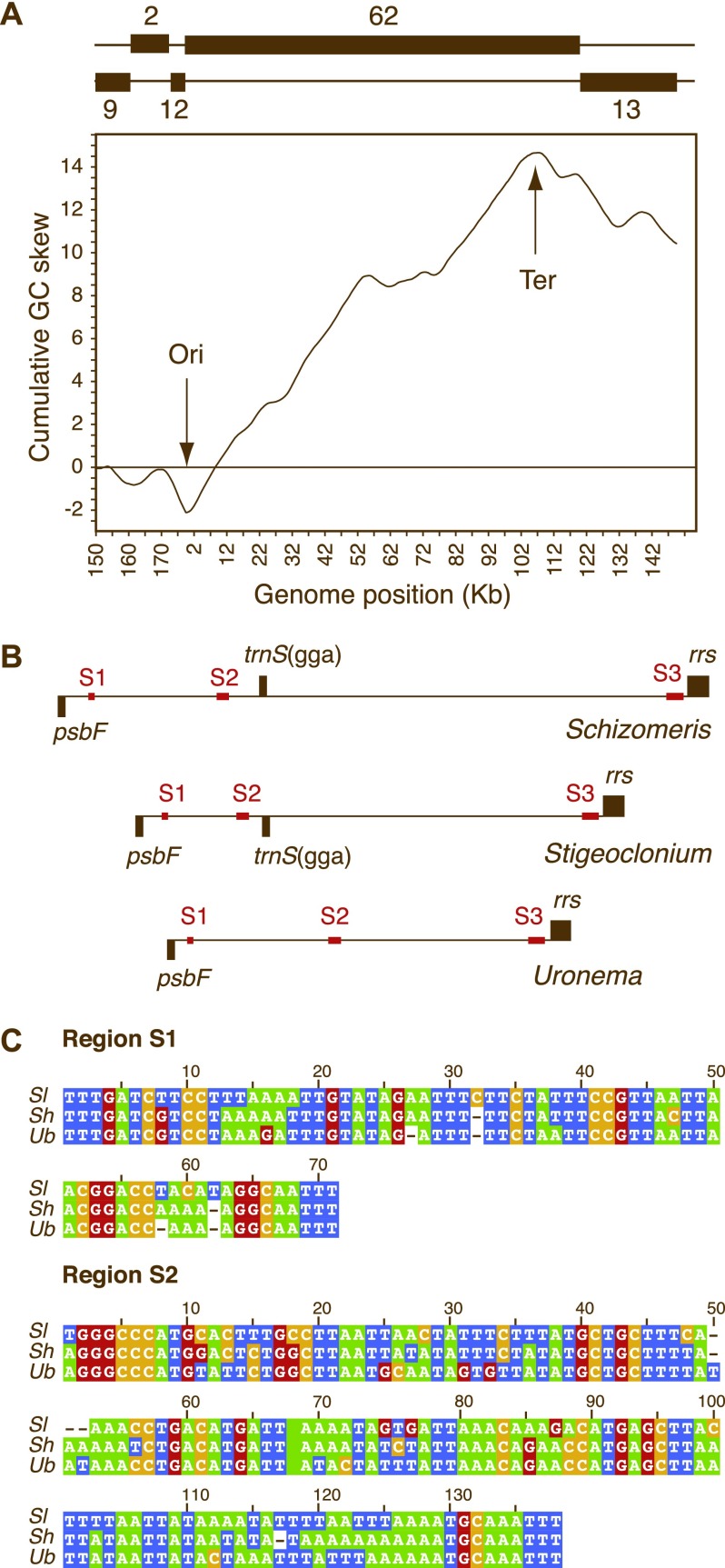
We investigated whether the Schizomeris genome resembles its Stigeoclonium counterpart in exhibiting a bias in GC composition along each strand. As shown in figure 2A, the plot of the cumulative GC skew in the Schizomeris genome has a V-shape, with the minimum and maximum lying on opposite ends of the genome. This profile is similar to those reported for the Stigeoclonium cpDNA and prokaryotic genomes that replicate bidirectionally from a single origin. The putative origin of replication in the Schizomeris cpDNA (corresponding to the minimum) was localized in the psbF-trnS(gga) spacer, a site that coincides with a switch in coding strand and actually marks one of the boundaries of the block containing the 62 consecutive genes on the same strand (figs. 1 and 2A). According to our cumulative GC skew analysis, the putative terminus of replication (corresponding to the maximum) resides in the psbD-trnS(gcu) spacer located in the 62-gene block (fig. 2A). This site is 12.5 kb away from the switch in coding strand observed between trnG(gcc) and the first exon of petD (fig. 1).
FIG. 2.—
Identification of the putative origin and terminus of replication in Schizomeris cpDNA. (A) Plot of cumulative GC skew over the entire genome sequence. The cumulative GC skew was calculated as indicated in the Materials and Methods; base at position 150,000 was arbitrarily selected to represent the starting point. Above the plot is a representation of the coding strand, either the strand whose sequence has been deposited in GenBank (the top strand) or the alternate strand (bottom strand). The numbers of conserved genes encoded in the filled boxes are indicated. (B) Positions of three highly conserved noncoding sequences (S1, S2, and S3) in the Schizomeris, Stigeoclonium, and Uronema cpDNA regions containing a putative origin of replication. (C) Alignments of the Schizomeris (Sl), Stigeoclonium (Sh), and Uronema (Ub) S1 and S2 sequences.
The putative origin and terminus of replication in the Schizomeris genome map at almost the same positions as those identified in the Stigeoclonium genome (Bélanger et al. 2006). The putative origin in the Stigeoclonium cpDNA was located in the trnS(gga)-rrs spacer, that is, in the spacer immediately adjacent to that carrying the putative origin in the Schizomeris cpDNA. In this context, it should be pointed out that the putative origins in the Schizomeris and Stigeoclonium genomes lie in a region of gene synteny, with the exception that the trnS(gga) gene differs in polarity (fig. 1). Given the relatively high degree of synteny reported here for the two chaetophoralean genomes (see below), the inversion responsible for the change in polarity of trnS(gga) might account for our observation that the Schizomeris and Stigeoclonium origins map to distinct spacers. Like its Schizomeris counterpart, the terminus of replication in the Stigeoclonium cpDNA was mapped to the spacer downstream of psbD. Again here, it is obvious that this region was implicated in genomic rearrangements: in Schizomeris, trnG(gcc), and psbD lie on opposite borders of the syntenic block encoding trnS(gcu), ycf1, and tufA (block 6 in fig. 1), whereas in Stigeoclonium, these two genes are next to each other just beside the latter syntenic block. These rearrangements, which apparently included the inversion of the region containing the block 6 and trnG(gcc), might explain why the switch in coding strand associated with the replication terminus corresponds to the second highest peak in the cumulative GC skew diagram of the Schizomeris genome (fig. 2A). In support of this hypothesis, disruptions of linearity appearing at local minima and maxima in GC skew analysis of prokaryotic genomes have been proposed to represent recent genome rearrangements (Grigoriev 1998).
In an attempt to identify conserved motifs that may be involved in the replication of the Schizomeris and the Stigeoclonium cpDNAs, we performed BlastN searches against the NCBI database using Schizomeris intergenic sequences as queries. Only 6 of the 105 Schizomeris intergenic spacers (psbB-psbT, rps8-psbE, rpoBex2-rpl32, rps2-clpP, psbF-trnS(gga), and trnS(gga)-rrs) displayed regions of significant homology (E values ranging from 7 × 10−5 to 8 × 10−17) with known sequences. As expected, all significant hits matched with the Stigeoclonium genome. More importantly, our searches revealed that three short Schizomeris noncoding sequences (S1, S2, and S3) in the region extending from psbF to rrs have counterparts in the Stigeoclonium cpDNA (fig. 2B). The 71-bp S1 is situated 247-bp upstream from psbF, whereas the 138-bp S2, also found within the psbF-trnS(gga) spacer, is 1,321-bp away from S1 and 327-bp upstream from trnS(gga). These two sequence elements represent the best candidates for a replication origin. Considering that the 187-bp S3 is located 40-bp upstream from rrs in both genomes, it would seem more compatible with a role in the transcriptional regulation of the ribosomal RNA operon. In contrast, no putative termination site could be detected in the intergenic spacers corresponding to the maximum or second highest peaks in our cumulative GC skew analysis. All other intergenic sequences that were found to be conserved in our BlastN analyses are possibly involved in transcriptional or translational regulation.
To confirm that the three abovementioned noncoding sequence elements shared by Schizomeris and Stigeoclonium are also conserved in other chaetophoraleans, we sequenced the region spanning psbF and rrs in U. belkae, a member of the clade comprising the unbranched taxa of the Chaetophoraceae (Caisova et al. 2011). As anticipated, all three noncoding elements were retrieved at the same relative positions in Uronema; however, the trnS(gga) gene was found to be missing from the region analyzed. The conservation of S1, S2, and S3 in the Uronema psbF-rrs spacer further supports the functional importance of these sequences and strengthens the above conclusions regarding the potential role of S1 and S2 in the initiation of DNA replication.
Our finding that the Stigeoclonium and Schizomeris cpDNAs share a putative replication origin in the vicinity of the ribosomal RNA operon mirrors the situation observed for the two other IR-lacking chloroplast genomes that were proposed to replicate bidirectionally from a single origin: the genome of the euglenoid Euglena gracilis (Morton 1999) and the genome of the parasitic green alga Helicosporidium sp. (Trebouxiophyceae) (de Koning and Keeling 2006). In the case of Euglena, it was shown that the replication origin inferred by the GC skew analysis (Morton 1999) matched the single locus previously identified by electron microscopic analysis of replication intermediates (Koller and Delius 1982; Ravel-Chapuis et al. 1982). In contrast, all other chloroplast genomes whose replication origin was studied by diverse biochemical methods were found to contain multiple origins either in the large IR, close to the ribosomal RNA operon, as in Pisum sativum (Meeker et al. 1988), Nicotiana tabacum (Kunnimalaiyaan and Nielsen 1997a, 1997b), and Oenothera hookeri (Chiu and Sears 1992; Sears et al. 1996) or in the single-copy regions as in Chlamydomonas where oriA and oriB lie in the vicinity of rpl16 and chlL, respectively (Waddell et al. 1984; Chang and Wu 2000).
According to the standard model of cpDNA replication, which is based on electron microscopic studies of pea and maize cpDNA molecules, replication starts at two displacement loop initiation sites located about 7-kb apart on opposite strands (Kolodner and Tewari 1975). The displacement loops expand unidirectionally toward each other to form a Cairns (theta)-type structure, which expands bidirectionally until completion of two daughter molecules. Replication may then continue by a rolling circle mechanism (Heinhorst and Cannon 1993; Day and Madesis 2007). Krishnan and Rao (2009) recently tested the standard model of replication by investigating the accumulation of the adenine to guanine deaminations on the displaced single strands. In accordance with this model, local deamination gradients in virtually all the analyzed land plant cpDNAs were shown to increase bidirectionally from the center of each single-copy region toward the pairs of replications origins localized within the IR. In this study, the authors also reported on the basis of Blast searches that most completely sequenced chloroplast genomes bear at least one or more homologs of the replication origins found in the tobacco chloroplast genome. In our opinion, the Blast signal detected in chlorophyte genomes could simply reflect the conservation of the trnI(gau) and orf35/ycf1 sequences that overlap the replication origins in the N. tabacum cpDNA.
The Mechanisms Underlying the Rearrangements between the Schizomeris and Stigeoclonium cpDNAs Remain Unclear
Nine blocks of colinear sequences, encoding a total of 84 conserved genes, are conserved between the Schizomeris and Stigeoclonium genomes (fig. 1). Most of the conserved genes residing outside these syntenic blocks (genes shown in red in fig. 1) encode tRNAs and, intriguingly, they tend to be located near the putative origin of replication.
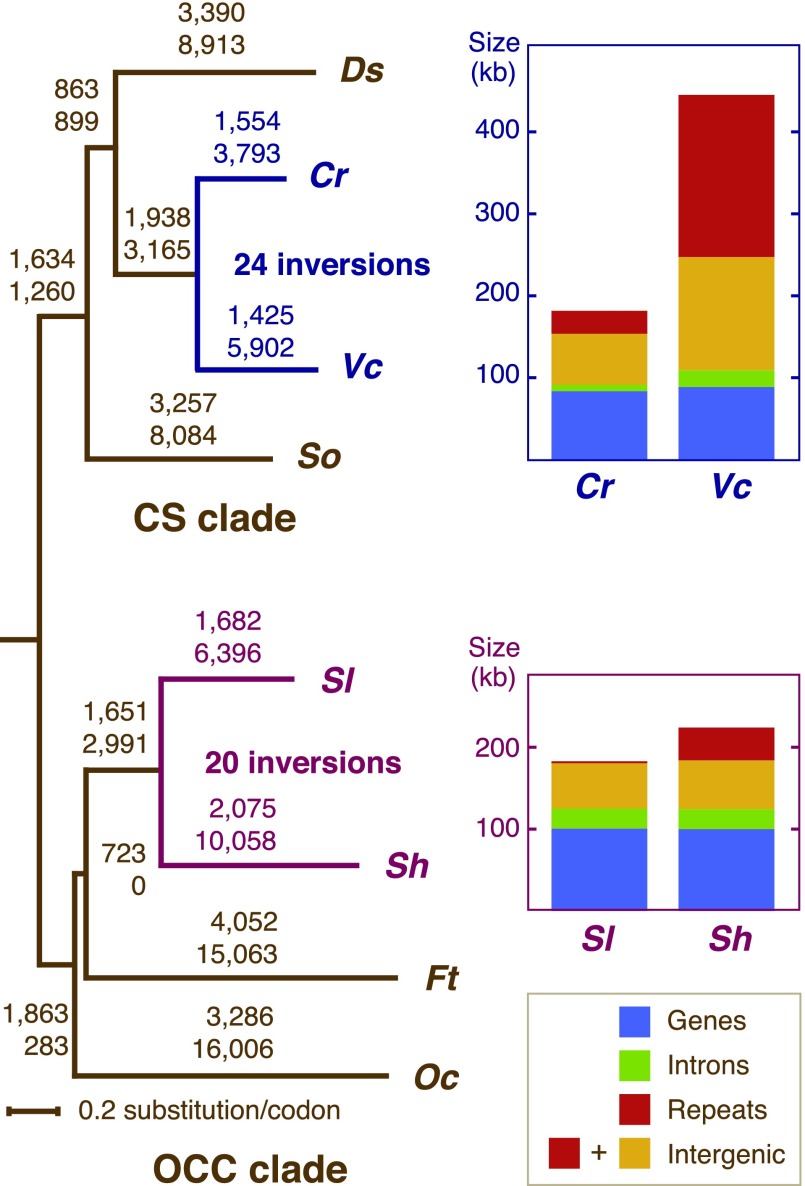
To assess the level of conservation that the Schizomeris and Stigeoclonium genomes exhibit compared with their chlorophycean counterparts, we carried out pairwise comparisons using GRIMM and a data set of 98 gene loci derived from all eight sequenced chlorophycean genomes (table 2). In these analyses, we assumed that all gene rearrangements occurred by inversions. Although inversions are thought to be the predominant mode of gene rearrangement in chloroplast genomes (Palmer 1991; Boudreau and Turmel 1995, 1996), we cannot exclude the possibility that other evolutionary mechanisms, such as transpositions or duplicative transpositions are responsible for some of the observed changes in gene order between the Stigeoclonium and Schizomeris cpDNAs. GRIMM computes inversion distances, that is, the minimal number of inversions that would be required to convert the gene order of a given genome to those of other genomes. Although more than 80 inversions are generally computed when genomes of chlorophyceans belonging to different orders are compared, analyses involving the chaetophoraleans Stigeoclonium and Schizomeris and the chlamydomonadaleans Chlamydomonas and Volvox yielded 20 and 24 inversions, respectively (table 2).
Table 2.
Minimal Numbers of Inversions Accounting for Gene Rearrangements between Chlorophycean cpDNAs
| Number of Gene Inversionsa |
|||||||
| cpDNA | Ft | Sh | Sl | Cr | Vc | Ds | So |
| Oc | 73 | 80 | 78 | 81 | 79 | 80 | 82 |
| Ft | — | 72 | 70 | 84 | 83 | 82 | 85 |
| Sh | — | 20 | 89 | 90 | 88 | 89 | |
| Sl | — | 90 | 90 | 90 | 87 | ||
| Cr | — | 24 | 51 | 58 | |||
| Vc | — | 51 | 62 | ||||
| Ds | — | 58 | |||||
NOTE.—Abbreviations: Oc, Oedogonium cardiacum; Ft, Floydiella terrestris; Sh, Stigeoclonium helveticum; Sl, Schizomeris leibleinii; Cr, Chlamydomonas reinhardtii; Vc, Volvox carteri; Ds, Dunaliella salina; So, Scenedesmus obliquus.
Inversions were computed using GRIMM (Tesler 2002).
In an attempt to produce a more parsimonious scenario of rearrangements, a separate GRIMM analysis of the Schizomeris and Stigeoclonium genomes was carried out using a data set of 104 gene loci. Here, the two IR-lacking chloroplast genomes were treated as circular molecules instead of linear molecules. The reversal scenario that was generated consists of 18 steps (supplementary fig. S3, Supplementary Material online), thus representing an economy of two steps. Interestingly, all the intermediate genome configurations in this scenario display a strong coding strand bias that may be compatible with a bidirectional mode of replication from a single origin. The only chloroplast genome rearrangements that were tolerated during the evolution of the Chaetophorales were most probably those that maintained an architecture compatible with bidirectional replication from a single origin.
Genes encoding tRNAs are located in the vicinity of most of the inversion endpoints, and some tRNA genes are implicated in more than one inversion (supplementary fig. S3, Supplementary Material online). For example, trnT(ugu) borders five inversion endpoints. The presence of tRNA genes near inversion endpoints has been previously observed in comparative studies of chloroplast genomes (Howe et al. 1988; Hiratsuka et al. 1989; Shimada and Sugiura 1989; Knox et al. 1993; Hoot and Palmer 1994; Haberle et al. 2008); however, the precise role of these genes in the promotion of gene rearrangements is unclear. It has been hypothesized that tRNA genes directly participate in intermolecular recombination via pairing with similar sequences (Hiratsuka et al. 1989).
Considering that genome rearrangements have been correlated with the presence of short repeats in land plant chloroplasts (Milligan et al. 1989; Shimada and Sugiura 1989; Tsai and Strauss 1989; Palmer 1991; Cosner et al. 1997; Chumley et al. 2006; Cai et al. 2008; Haberle et al. 2008; Guisinger et al. 2010), we compared the locations of the Schizomeris repeats relative to the inversion endpoints that distinguish the Schizomeris and Stigeoclonium genomes (supplementary figs. S2 and S3, Supplementary Material online). No strong correlation, however, was observed between these two features. Most repeats are randomly dispersed in the Schizomeris genome (supplementary fig. S2, Supplementary Material online). Analysis of the characterized repeats in the Stigeoclonium genome also failed to establish a link between inversion endpoints and dispersed repeats. As reported here for Schizomeris, dispersed repeats in previously examined chlorophycean genomes rarely exceed 60 bp (supplementary section S3, Supplementary Material online). These sequences may be too short and/or too scarce to act as substrates for homologous recombination events. Lengths of repeats have been shown to have a significant impact on recombination frequency (Day and Madesis 2007). For example, plastid transformation experiments using Chlamydomonas revealed that homologous recombination events require repeats of 150–200 bp and that deletions of 18- to 37-bp repeats near a recombination hot spot had no influence on recombination frequency (Newman et al. 1992). Alternatively, it is possible that the short dispersed repeats present in chaetophoralean genomes promote recombination and that we have failed to recognize this role because the Schizomeris and Stigeoclonium cpDNAs show extraordinary differences in their repeated sequences. Examination of chloroplast genomes from very closely related chlorophyceans will be needed to better understand how they evolve and whether they play a role in gene rearrangements.
Mode and Tempo of Chloroplast Genome Evolution in the Chaetophorales and Chlamydomonadales
The availability of cpDNA sequences from two pairs of closely related chlorophycean algae (Chlamydomonas and Volvox representing the Chlamydomonadales and Schizomeris and Stigeoclonium representing the Chaetophorales) offers the opportunity to test whether the chloroplast genome evolves under similar constraints in the OCC and CS lineages. The histograms shown in figure 3 compare the lengths of coding and noncoding (introns, repeats, and intergenic spacers) sequences in the four chlorophycean genomes. Clearly, the major change in size that the chloroplast genome experienced in the Chaetophorales is accounted for by dispersed repeats. In contrast, the chlamydomonadalean genomes sustained changes not only in the abundance of repeats but also in the abundance of intron and intergenic sequences. The higher variability observed for the Chlamydomonadales at the level of noncoding sequences is somewhat intriguing considering that the evolutionary distance between Chlamydomonas and Volvox (12,701 substitutions), as estimated by the numbers of accumulated synonymous and nonsynonymous substitutions in 61 protein-coding genes, is 1.6-fold smaller than that separating Schizomeris and Stigeoclonium (20,211 substitutions) (fig. 3). Interestingly, the higher variability in the abundance of noncoding sequences is also accompanied by a higher rate of genome rearrangements in the Chlamydomonadales. Using evolutionary distances as a means to normalize the number of genome reversals in the two pairs of related algae, we estimated that the Chlamydomonas/Volvox pair experienced almost twice as many inversions per distance unit (1.9 inversions/1,000 nucleotide substitutions) compared with the Schizomeris/Stigeoclonium pair (1.0 inversion/1,000 nucleotide substitutions) (fig. 3).
FIG. 3.—
Sequence divergence, gene rearrangements, and variation in the abundance of diverse categories of sequences in the Schizomeris/Stigeoclonium and Chlamydomonas/Volvox chloroplast genomes. The tree on the left shows the evolutionary distances as inferred from 61 concatenated protein-coding genes as well as the number of inversions for each pair of compared genomes. The numbers of nonsynonymous (upper numbers) and synonymous (lower numbers) nucleotide substitutions estimated by CODEML are shown along each branch. The histograms on the right illustrate the lengths of coding sequences, introns, repeats, and intergenic spacers excluding the repeats. Note that the sequences within the IR were counted only once. Abbreviations: Oc, Oedogonium cardiacum; Ft, Floydiella terrestris; Sh, Stigeoclonium helveticum; Sl, Schizomeris leibleinii; Cr, Chlamydomonas reinhardtii; Vc, Volvox carteri; Ds, Dunaliella salina; So, Scenedesmus obliquus.
At this point, we can offer no compelling explanation as to why the chloroplast genome evolves more conservatively at the levels of gene order and genome size in the Chaetophorales than in the Chlamydomonadales. The presence/absence of a stabilizing IR structure cannot be invoked because the more highly conserved genomes found in the chaetophoralean lineage both lack an IR. Considering that short dispersed repeats can be gained or lost rapidly over evolutionary time, perhaps such repeats were more abundant in ancestral genomes of the Chlamydomonadales compared with those of the Chaetophorales, thus favoring a higher level of recombination in the former lineage.
Conclusions
By analyzing the chloroplast genome from a representative of the earliest diverging lineage of the Chaetophorales, we have gained insights into the ancestral structure and evolutionary dynamics of cpDNA in this major group of the Chlorophyceae. The Schizomeris genome was found to resemble its Stigeoclonium homolog with respect to the absence of the IR and the presence of a strong bias in coding strand and nucleotide composition along each DNA strand. This distinctive architecture probably represents the ancestral condition of the chaetophoralean chloroplast genome and is most likely the result of evolutionary pressures to retain a bidirectional mode of DNA replication from a single origin. The conserved motifs we uncovered in the vicinity of the ribosomal RNA operon may play a role in the initiation of DNA replication. Our study also revealed that short dispersed repeats evolve at a rapid pace in the genomic landscape of the Chlorophyceae; however, the chaetophoralean genomes appear to be more conserved in gene order and display less variation in the abundance of intergenic and intron sequences than their chlamydomonadalean homologs. Clearly, chloroplast genome sequences from additional representatives of the OCC and CS lineages are needed to identify the factors underlying the tremendous plasticity of the chloroplast genome in the Chlorophyceae.
Supplementary Material
Supplementary figures S1–S3, tables S1 and S2, and sections S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This study was supported by a grant from the Natural Sciences and Engineering Research Council of Canada (to M.T. and C.L.).
References
- Andersen RA, Berges JA, Harrison PJ, Watanabe MM. Appendix A—recipes for freshwater and seawater media. In: Andersen RA, editor. Algal culturing techniques. Burlington (MA): Elsevier Academic Press; 2005. pp. 429–538. [Google Scholar]
- Bélanger A-S, et al. Distinctive architecture of the chloroplast genome in the chlorophycean green alga Stigeoclonium helveticum. Mol Gen Genomics. 2006;276:464–477. doi: 10.1007/s00438-006-0156-2. [DOI] [PubMed] [Google Scholar]
- Boudreau E, Turmel M. Gene rearrangements in Chlamydomonas chloroplast DNAs are accounted for by inversions and by the expansion/contraction of the inverted repeat. Plant Mol Biol. 1995;27:351–364. doi: 10.1007/BF00020189. [DOI] [PubMed] [Google Scholar]
- Boudreau E, Turmel M. Extensive gene rearrangements in the chloroplast DNAs of Chlamydomonas species featuring multiple dispersed repeats. Mol Biol Evol. 1996;13:233–243. doi: 10.1093/oxfordjournals.molbev.a025560. [DOI] [PubMed] [Google Scholar]
- Brouard J-S, Otis C, Lemieux C, Turmel M. Chloroplast DNA sequence of the green alga Oedogonium cardiacum (Chlorophyceae): unique genome architecture, derived characters shared with the Chaetophorales and novel genes acquired through horizontal transfer. BMC Genomics. 2008;9:290. doi: 10.1186/1471-2164-9-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouard JS, Otis C, Lemieux C, Turmel M. The exceptionally large chloroplast genome of the green alga Floydiella terrestris illuminates the evolutionary history of the Chlorophyceae. Genome Biol Evol. 2010;2:240–256. doi: 10.1093/gbe/evq014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchheim MA, Michalopulos EA, Buchheim JA. Phylogeny of the Chlorophyceae with special reference to the Sphaeropleales: a study of 18S and 26S rDNA data. J Phycol. 2001;37:819–835. [Google Scholar]
- Cai Z, et al. Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J Mol Evol. 2008;67:696–704. doi: 10.1007/s00239-008-9180-7. [DOI] [PubMed] [Google Scholar]
- Caisova L, Marin B, Sausen N, Proschold T, Melkonian M. Polyphyly of Chaetophora and Stigeoclonium within the Chaetophorales (Chlorophyceae), revealed by sequence comparisons of nuclear-encoded SSU rRNA genes. J Phycol. 2011;47:164–177. doi: 10.1111/j.1529-8817.2010.00949.x. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Chang CH, Wu M. The effects of transcription and RNA processing on the initiation of chloroplast DNA replication in Chlamydomonas reinhardtii. Mol Gen Genet. 2000;263:320–327. doi: 10.1007/s004380051174. [DOI] [PubMed] [Google Scholar]
- Chiu WL, Sears BB. Electron microscopic localization of replication origins in Oenothera chloroplast DNA. Mol Gen Genet. 1992;232:33–39. doi: 10.1007/BF00299134. [DOI] [PubMed] [Google Scholar]
- Chumley T, et al. The complete chloroplast genome sequence of Pelargonium x hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol. 2006;23:2175. doi: 10.1093/molbev/msl089. [DOI] [PubMed] [Google Scholar]
- Cosner ME, Jansen RK, Palmer JD, Downie SR. The highly rearranged chloroplast genome of Trachelium caeruleum (Campanulaceae): multiple inversions, inverted repeat expansion and contraction, transposition, insertions/deletions, and several repeat families. Curr Genet. 1997;31:419–429. doi: 10.1007/s002940050225. [DOI] [PubMed] [Google Scholar]
- Cui L, et al. Adaptive evolution of chloroplast genome structure inferred using a parametric bootstrap approach. BMC Evol Biol. 2006;6:13. doi: 10.1186/1471-2148-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day A, Madesis P. DNA replication, recombination, and repair in plastids. In: Bock R, editor. Cell and molecular biology of plastids. Berlin (Germany): Springer; 2007. pp. 65–119. [Google Scholar]
- de Cambiaire J-C, Otis C, Lemieux C, Turmel M. The complete chloroplast genome sequence of the chlorophycean green alga Scenedesmus obliquus reveals a compact gene organization and a biased distribution of genes on the two DNA strands. BMC Evol Biol. 2006;6:37. doi: 10.1186/1471-2148-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning AP, Keeling PJ. The complete plastid genome sequence of the parasitic green alga Helicosporidium sp. is highly reduced and structured. BMC Biol. 2006;4:12. doi: 10.1186/1741-7007-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev A. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 1998;26:2286–2290. doi: 10.1093/nar/26.10.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol Biol Evol. 2011;28:583–600. doi: 10.1093/molbev/msq229. [DOI] [PubMed] [Google Scholar]
- Guy L, Roten CA. Genometric analyses of the organization of circular chromosomes: a universal pressure determines the direction of ribosomal RNA genes transcription relative to chromosome replication. Gene. 2004;340:45–52. doi: 10.1016/j.gene.2004.06.056. [DOI] [PubMed] [Google Scholar]
- Haberle RC, Fourcade HM, Boore JL, Jansen RK. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J Mol Evol. 2008;66:350–361. doi: 10.1007/s00239-008-9086-4. [DOI] [PubMed] [Google Scholar]
- Heinhorst S, Cannon GC. DNA replication in chloroplasts. J Cell Sci. 1993;104:1–9. [Google Scholar]
- Hiratsuka J, et al. The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet. 1989;217:185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- Hoot SB, Palmer JD. Structural rearrangements, including parallel inversions, within the chloroplast genome of Anemone and related genera. J Mol Evol. 1994;38:274–281. doi: 10.1007/BF00176089. [DOI] [PubMed] [Google Scholar]
- Howe CJ, Barker RF, Bowman CM, Dyer TA. Common features of three inversions in wheat chloroplast DNA. Curr Genet. 1988;13:343–350. doi: 10.1007/BF00424430. [DOI] [PubMed] [Google Scholar]
- Knox EB, Downie SR, Palmer JD. Chloroplast genome rearrangements and the evolution of giant lobelias from herbaceous ancestors. Mol Biol Evol. 1993;10:414–430. [Google Scholar]
- Koller B, Delius H. Origin of replication in chloroplast DNA of Euglena gracilis located close to the region of variable size. EMBO J. 1982;1:995–998. doi: 10.1002/j.1460-2075.1982.tb01283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner RD, Tewari KK. Chloroplast DNA from higher plants replicates by both the Cairns and the rolling circle mechanism. Nature. 1975;256:708–711. doi: 10.1038/256708a0. [DOI] [PubMed] [Google Scholar]
- Krishnan NM, Rao BJ. A comparative approach to elucidate chloroplast genome replication. BMC Genomics. 2009;10:237. doi: 10.1186/1471-2164-10-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunnimalaiyaan M, Nielsen BL. Chloroplast DNA replication: mechanism, enzymes and replication origins. J Plant Biochem Biotechnol. 1997a;6:1–7. [Google Scholar]
- Kunnimalaiyaan M, Nielsen BL. Fine mapping of replication origins (oriA and oriB) in Nicotiana tabacum chloroplast DNA. Nucleic Acids Res. 1997b;25:3681–3686. doi: 10.1093/nar/25.18.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, et al. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul JE, et al. The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell. 2002;14:2659–2679. doi: 10.1105/tpc.006155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken DA, Nadakavukaren MJ, Cain JR. A biochemical and ultrastructural evaluation of the taxonomic position of Glaucosphaera vacuolata Korsch. New Phytol. 1980;86:39–44. [Google Scholar]
- Meeker R, Nielsen B, Tewari KK. Localization of replication origins in pea chloroplast DNA. Mol Cell Biol. 1988;8:1216–1223. doi: 10.1128/mcb.8.3.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F, Umesono K, Ozeki H. Comparative and functional anatomy of group II catalytic introns—a review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Milligan BG, Hampton JN, Palmer JD. Dispersed repeats and structural reorganization in subclover chloroplast DNA. Mol Biol Evol. 1989;6:355–368. doi: 10.1093/oxfordjournals.molbev.a040558. [DOI] [PubMed] [Google Scholar]
- Morton BR. Strand asymmetry and codon usage bias in the chloroplast genome of Euglena gracilis. Proc Natl Acad Sci U S A. 1999;96:5123–5128. doi: 10.1073/pnas.96.9.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman SM, Harris EH, Johnson AM, Boynton JE, Gillham NW. Nonrandom distribution of chloroplast recombination events in Chlamydomonas reinhardtii: evidence for a hotspot and an adjacent cold region. Genetics. 1992;132:413–429. doi: 10.1093/genetics/132.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD. Plastid chromosomes: structure and evolution. In: Bogorad L, Vasil I, editors. The molecular biology of plastids. Cell culture and somatic cell genetics of plants. San Diego (CA): Academic Press; 1991. pp. 5–53. [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Wood T, Zhang Z, Miller W. Comparison of DNA sequences with protein sequences. Genomics. 1997;46:24–36. doi: 10.1006/geno.1997.4995. [DOI] [PubMed] [Google Scholar]
- Pombert JF, Lemieux C, Turmel M. The complete chloroplast DNA sequence of the green alga Oltmannsiellopsis viridis reveals a distinctive quadripartite architecture in the chloroplast genome of early diverging ulvophytes. BMC Biol. 2006;4:3. doi: 10.1186/1741-7007-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombert JF, Otis C, Lemieux C, Turmel M. The chloroplast genome sequence of the green alga Pseudendoclonium akinetum (Ulvophyceae) reveals unusual structural features and new insights into the branching order of chlorophyte lineages. Mol Biol Evol. 2005;22:1903–1918. doi: 10.1093/molbev/msi182. [DOI] [PubMed] [Google Scholar]
- Ravel-Chapuis P, Heizmann P, Nigon V. Electron microscopic localization of the replication origin of Euglena gracilis chloroplast DNA. Nature. 1982;300:78–81. [Google Scholar]
- Sears BB, Stoike LL, Chiu WL. Proliferation of direct repeats near the Oenothera chloroplast DNA origin of replication. Mol Biol Evol. 1996;13:850–863. doi: 10.1093/oxfordjournals.molbev.a025645. [DOI] [PubMed] [Google Scholar]
- Shimada H, Sugiura M. Pseudogenes and short repeated sequences in the rice chloroplast genome. Curr Genet. 1989;16:293–301. doi: 10.1007/BF00422116. [DOI] [PubMed] [Google Scholar]
- Smith DR, Lee RW. The mitochondrial and plastid genomes of Volvox carteri: bloated molecules rich in repetitive DNA. BMC Genomics. 2009;10:132. doi: 10.1186/1471-2164-10-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Lee RW. Low nucleotide diversity for the expanded organelle and nuclear genomes of Volvox carteri supports the mutational-hazard hypothesis. Mol Biol Evol. 2010;27:2244–2256. doi: 10.1093/molbev/msq110. [DOI] [PubMed] [Google Scholar]
- Smith DR, et al. The Dunaliella salina organelle genomes: large sequences, inflated with intronic and intergenic DNA. BMC Plant Biol. 2010;10:83. doi: 10.1186/1471-2229-10-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesler G. GRIMM: genome rearrangements web server. Bioinformatics. 2002;18:492–493. doi: 10.1093/bioinformatics/18.3.492. [DOI] [PubMed] [Google Scholar]
- Tillier ER, Collins RA. The contributions of replication orientation, gene direction, and signal sequences to base-composition asymmetries in bacterial genomes. J Mol Evol. 2000a;50:249–257. doi: 10.1007/s002399910029. [DOI] [PubMed] [Google Scholar]
- Tillier ER, Collins RA. Genome rearrangement by replication-directed translocation. Nat Genet. 2000b;26:195–197. doi: 10.1038/79918. [DOI] [PubMed] [Google Scholar]
- Tsai CH, Strauss SH. Dispersed repetitive sequences in the chloroplast genome of douglas-fir. Curr Genet. 1989;16:211–218. doi: 10.1007/BF00391479. [DOI] [PubMed] [Google Scholar]
- Turmel M, Brouard JS, Gagnon C, Otis C, Lemieux C. Deep division in the Chlorophyceae (Chlorophyta) revealed by chloroplast phylogenomic analyses. J Phycol. 2008;44:739–750. doi: 10.1111/j.1529-8817.2008.00510.x. [DOI] [PubMed] [Google Scholar]
- Turmel M, Gagnon MC, O'Kelly CJ, Otis C, Lemieux C. The chloroplast genomes of the green algae Pyramimonas, Monomastix, and Pycnococcus shed new light on the evolutionary history of prasinophytes and the origin of the secondary chloroplasts of euglenids. Mol Biol Evol. 2009;26:631–648. doi: 10.1093/molbev/msn285. [DOI] [PubMed] [Google Scholar]
- Turmel M, et al. The complete mitochondrial DNA sequences of Nephroselmis olivacea and Pedinomonas minor. Two radically different evolutionary patterns within green algae. Plant Cell. 1999;11:1717–1729. doi: 10.1105/tpc.11.9.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C. The complete chloroplast DNA sequence of the green alga Nephroselmis olivacea: insights into the architecture of ancestral chloroplast genomes. Proc Natl Acad Sci U S A. 1999;96:10248–10253. doi: 10.1073/pnas.96.18.10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C. The complete chloroplast DNA sequences of the charophycean green algae Staurastrum and Zygnema reveal that the chloroplast genome underwent extensive changes during the evolution of the Zygnematales. BMC Biol. 2005;3:22. doi: 10.1186/1741-7007-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C. The chloroplast genomes of the green algae Pedinomonas minor, Parachlorella kessleri, and Oocystis solitaria reveal a shared ancestry between the Pedinomonadales and Chlorellales. Mol Biol Evol. 2009;26:2317–2331. doi: 10.1093/molbev/msp138. [DOI] [PubMed] [Google Scholar]
- Waddell J, Wang XM, Wu M. Electron microscopic localization of the chloroplast DNA replicative origins in Chlamydomonas reinhardii. Nucleic Acids Res. 1984;12:3843–3856. doi: 10.1093/nar/12.9.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]