Abstract
The fastest-evolving regions in the human and chimpanzee genomes show a remarkable excess of weak (A,T) to strong (G,C) nucleotide substitutions since divergence from their common ancestor. We investigated the phylogenetic extent and possible causes of this weak to strong (W→S) bias in divergent sequences (BDS) using recently sequenced genomes and recombination maps from eight trios of eukaryotic species. To quantify evidence for BDS, we inferred substitution histories using an efficient maximum likelihood approach with a context-dependent evolutionary model. We then annotated all lineage-specific substitutions in terms of W→S bias and density on the chromosomes. Finally, we used the inferred substitutions to calculate a BDS score—a log odds ratio between substitution type and density—and assessed its statistical significance with Fisher's exact test. Applying this approach, we found significant BDS in the coding and noncoding sequence of human, mouse, dog, stickleback, fruit fly, and worm. We also observed a significant lack of W→S BDS in chicken and yeast. The BDS score varies between species and across the chromosomes within each species. It is most strongly correlated with different genomic features in different species, but a strong correlation with recombination rates is found in several species. Our results demonstrate that a W→S substitution bias in fast-evolving sequences is a widespread phenomenon. The patterns of BDS observed suggest that a recombination-associated process, such as GC-biased gene conversion, is involved in the production of the bias in many species, but the strength of the BDS likely depends on many factors, including genome stability, variability in recombination rate over time and across the genome, the frequency of meiosis, and the amount of outcrossing in each species.
Keywords: fast-evolving sequence, clustered substitution, fixation bias, genome analysis, biased gene conversion
Introduction
The recent sequencing of the genomes of many closely related species has created a powerful new paradigm for investigating the evolutionary processes that generate the diversity of life on Earth. Comparing the complete human genome sequence to that of a chimpanzee, our closest living relative, Dreszer et al. (2007) demonstrated that the most divergent regions of both genomes show a striking W→S substitution bias and that this association is correlated with recombination rates. This bias in divergent sequences (BDS) is not limited to neutrally evolving sequences and can significantly impact substitution patterns in conserved noncoding sequences (Pollard, Salama, King et al. 2006) and protein-coding exons (Berglund et al. 2009; Ratnakumar et al. 2010), suggesting the possibility of significant functional consequences. These observations have profound implications regarding the interpretation of adaptive evolution in fast-evolving sequences of the human genome and our understanding of the evolutionary forces driving divergence between closely related species in general.
In this paper, we explore two fundamental questions about BDS and what the phenomenon tells us about genome evolution and function. First, is BDS unique to the hominoids or a more widespread phenomenon? The recent sequencing of many closely related eukaryotic species enables us to investigate the phylogenetic extent of BDS. Second, what evolutionary processes produce BDS? Based on the patterns of BDS found in human, Dreszer et al. (2007) argued that GC-based gene conversion (gBGC) (Duret and Galtier 2009) is the cause of BDS. gBGC is a nonadaptive evolutionary process that favors the fixation of weak alleles near the double-strand breaks that initiate recombination events. Episodes of gBGC in a genomic region could produce an association between W→S substitutions and substitution density (BDS) by driving W→S mutations to fixation in recombination hotspots. We investigate the origin of BDS by examining correlations between BDS and genomic variables, including recombination rates, in different species.
To examine BDS in a broader phylogenetic context, we characterized recent substitutions in eight trios of eukaryotic organisms including: human (Homo sapiens), mouse (Mus musculus), dog (Canis familiaris), chicken (Gallus gallus), stickleback (Gasterosteus aculeatus), fruit fly (Drosophila melanogaster), worm (Caenorhabditis elegans), and yeast (Saccharomyces cerevisiae). We selected these species based on availability of sequenced genomes for a closely related comparison taxon and an outgroup, quality of the genome assemblies, and (if possible) availability of recombination maps.
To enable these genome-wide analyses, we inferred substitution histories using maximum likelihood and a context-dependent model of evolution with the PHAST package (Hubisz et al. 2011). This approach accounts directly for CpG hypermutability and other context effects that can lead to incorrect inference of substitution type (e.g., W→S vs. not W→S) in parsimony-based analyses. Next, we analyzed substitution patterns using a new efficient statistical test for the association of nucleotide substitution types with substitution rates and genome annotations. Most previous studies of substitution patterns in divergent regions have focused on discrete predefined elements across the genome; in contrast, our approach is more broad and allows a flexible continuous definition of “divergent” based on the density of substitution across the genome. Our work extends the approach of Dreszer et al. (2007) and provides a more rigorous statistical framework for measuring associations between substitution type and density.
Using these tools, we confirm the previously observed pattern of BDS in the human genome and its association with elevated recombination rates. Our analysis of non-primate clades shows that BDS is common outside of human, though not universal. When BDS is present, it exists in both coding and noncoding sequence and is often, but not always, correlated with high rates of recombination. This correlation, paired with the lack of W→S bias in within-species polymorphisms, argues that a recombination-driven fixation bias for strong nucleotides, such as that produced by gBGC, may be involved in the production of BDS, especially when there is variation in strength and location of gBGC over time. Overall, the strong evidence we find for BDS in many eukaryote genomes highlights the importance of understanding its causes and developing statistical models of DNA and protein evolution that incorporate these observations.
Materials and Methods
Data
Genome sequences and multiple sequence alignments of recent assemblies for all species (fig. 1) were downloaded from the University of California, Santa Cruz (UCSC) Genome Browser (Kent et al. 2002). The genome alignments were constructed from syntenic pairwise alignments which were then multiply aligned using the UCSC/MULTIZ alignment pipeline (Kent et al. 2003; Blanchette et al. 2004). When more than one precomputed alignment was available for a reference species, we chose the most phylogenetically restricted. For chicken, we did not consider the microchromosomes (International Chicken Genome Sequencing Consortium 2004) in our analysis. The following genome assemblies were used (UCSC identifiers): hg18, panTro2, rheMac2, mm9, rn4, cavPor2, canFam2, felCat3, bosTau4, galGal3, taeGut1, anoCar1, gasAcu1, oryLat1, fr2, dm3, droSim1, droYak2, ce6, caePb2, caeJap1, sacCer2, sacPar, sacMik. Conservation scores for each reference species were downloaded from the Genome Browser; phyloP (Pollard et al. 2010) scores were used when available, otherwise phastCons (Siepel et al. 2005; Hubisz et al. 2011) scores were used. Species trees and divergences were taken from the phastCons tree models estimated from 4-fold degenerate sites using phyloFit (Siepel et al. 2005; Hubisz et al. 2011).
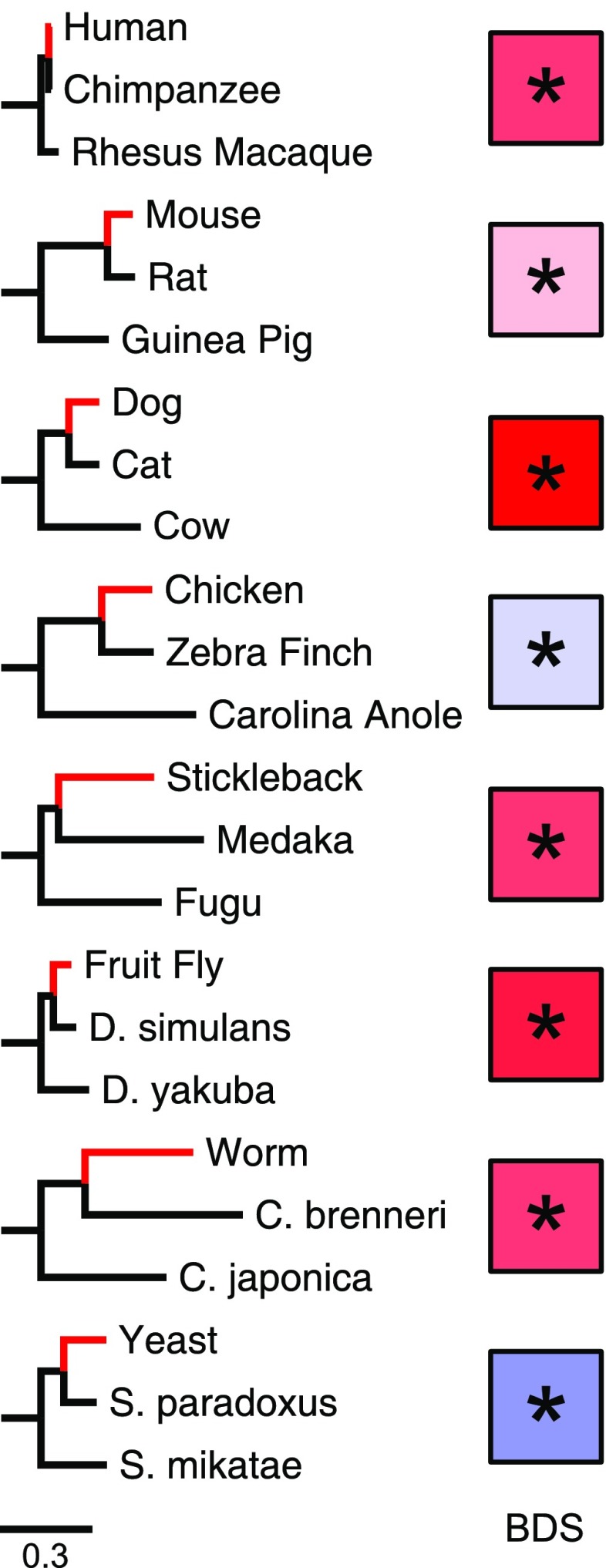
FIG. 1.—
The patterns of BDS across eight eukaryotic lineages. Each trio of species contains a reference (red branch), comparison, and outgroup species. Substitutions occurring on the branch leading to the reference species from the last common ancestor with the comparison species were considered. The color of each box reflects the strength of the BDS in the associated species. Warm colors (reds) indicate W→S BDS, and cool colors (blues) indicate a preference against W→S substitutions in fast-evolving regions. Asterisks indicate a significant deviation from expected substitution patterns. BDS statistics for each species are given in table 1.
The raw single nucleotide polymorphism (SNP) data for human and mouse come from dbSNP (Sherry et al. 2001) release 130 and 128, respectively. SNPs for chicken were identified by the Beijing Genomics Institute and downloaded from the UCSC Genome Browser bgiSNP track.
Recombination rate data were obtained from a variety of sources. For human, we used the combined recombination map from the HapMap project (The International Hapmap Consortium 2007), as well as the deCODE genetics male and female maps which are based on 15,257 Icelandic parent–offspring pairs (Kong et al. 2010). Mouse recombination data were downloaded from the Mouse Map Converter (Jackson Laboratories 2009), which is based on SNP analysis across 46 families (Shifman et al. 2006; Cox et al. 2009). The chicken recombination map from Groenen et al. (2009) is based on SNPs across three mapping populations. For fruit fly, the Drosophila Recombination Rate Calculator (Fiston-Lavier et al. 2010), which compares genetic and physical maps of the genome to infer recombination rates, was used.
Analysis
Our BDS analysis pipeline consists of several steps. For each clade of interest, we started with an alignment of three species: reference, comparison (sister taxon), and outgroup. Our analysis workflow is as follows (details below). First, the three-species alignments were filtered for alignment quality. Then substitution histories were computed using maximum likelihood and a context-dependent evolutionary model. Given the set of expected recent substitutions, the association between patterns of substitution and substitution density (BDS) was quantified. Finally, the strength of the BDS across the genome was correlated with several other genomic features.
Data processing and analysis were performed using custom programs written in R (R Development Core Team 2009) and Python with SciPy (Jones et al. 2001) and matplotlib (Hunter 2007).
Alignment Filtering.
In order to study patterns of substitution between species, it is crucial that the data are not polluted by false substitutions introduced by alignment errors. We filtered all alignments in a consistent manner across each set of species using several criteria that could be applied in each clade considered. First, repetitive sequences as identified by the UCSC Genome Browser alignment pipeline were not considered. These regions were identified using the Tandem Repeats Finder and RepeatMasker (Smit and Hubley 2008–2010) and are indicated by lowercase letters in the alignments. (See the Genome Browser documentation for more details.) Next, the quality of the alignment around each position was considered. If there were any insertions or deletions between the reference and comparison species within five base pairs (bp) of a position, then it was not considered. Finally, all positions in regions of the genome that had significant homology to another region of the genome were filtered out. These duplicated or repetitive regions are often difficult to align to other species due to their similarity. These regions were removed using the Genome Browser's chainSelf track of significant alignments of a genome with itself.
Identification of Recent Substitutions.
We are interested in substitutions that occurred in the reference species since its divergence from the last common ancestor with the comparison species—for example, on the human lineage after its last common ancestor with chimpanzee. These branches of interest are indicated in red in figure 1. After filtering the alignments as described above, we fit a context-dependent dinucleotide phylogenetic model to the alignments for each chromosome using maximum likelihood. We used the general unrestricted dinucleotide model with strand symmetry (U2S) (Siepel and Haussler 2004). This phylogenetic model was fit to the alignments with phyloFit from the PHAST software package (Hubisz et al. 2011). Using the model, we computed (also using phyloFit) the posterior expected number of substitutions of each type on each branch of the tree for each site in the alignment.
Calculation of BDS.
Given the inferred probabilities of each type of substitution on the branch of interest at each site across each genome, we quantified the BDS with a log odds ratio that relates the density and pattern of substitutions across a genomic region. The odds ratio is based on a two-by-two contingency table in which each possible substitution was classified as 1) W→S or not W→S and 2) in a divergent sequence or not. Any substitution from an A or T in the ancestor to G or C in the reference species was W→S, and all others were not. Each position was classified as divergent/not divergent based on the substitution density in a genomic window of a given size around it. The expected number of substitutions of each type on the reference branch at this position was then added to the relevant cell of the contingency table.
Given the resulting two-by-two contingency table for a genomic region of interest, we calculated the log odds ratio and associated statistics in the standard manner after rounding the expected number of substitutions in each cell to the nearest whole number. This log odds ratio quantifies the strength of association between W→S bias and sequence divergence. If substitutions in divergent windows exhibit an excess of W→S changes, then the log odds ratio is greater than zero. It is less than zero if these divergent regions contain fewer W→S substitutions than expected. We refer to this log odds ratio as the BDS score and use the terms “bias,” “BDS,” and “W→S BDS” to refer to increased W→S substitution in regions of high divergence. The genomic regions over which the BDS score was calculated may be small sequence blocks (as used in the correlation analysis), a set of regions across the genome (as in the coding sequence analysis), or the entire genome. The significance of the BDS score was assessed with Fisher's exact test (FET). All reported P values are from the FET unless otherwise indicated.
We explored a range of window sizes and density cutoffs; figure 2 demonstrates the robustness of our results to any specific cutoff. When a single cutoff was required, we used window size of 1,000 bp and a substitution density such that as near to 5% of all substitutions as possible were assigned to the divergent group. Because the reference–comparison species pairs we examined have different levels of sequence divergence, this threshold varies in absolute value across species (i.e., it is lower for species that are less diverged and higher for species that are more diverged).
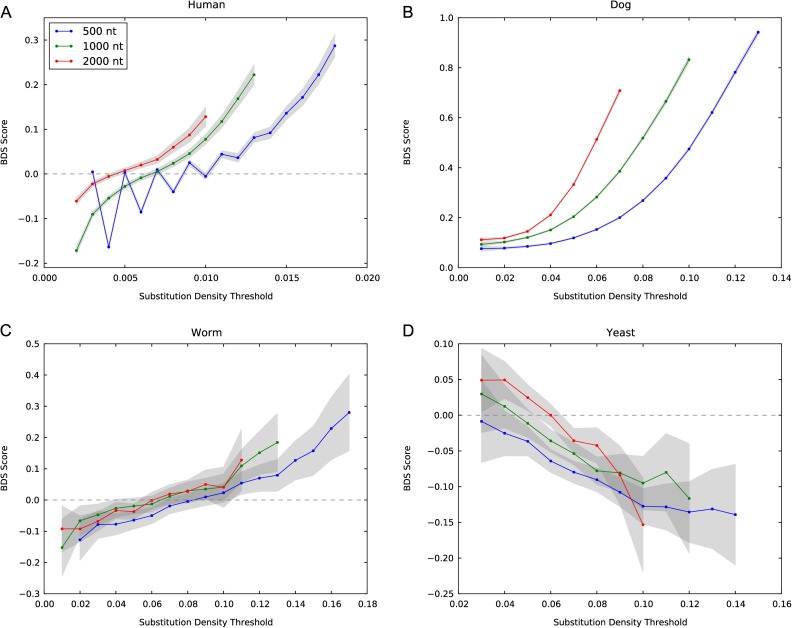
FIG. 2.—
Divergent sequences are significantly W→S biased in many, but not all, eukaryotes. Genomic regions with a high divergence show a significant enrichment for W→S substitutions (BDS) compared with regions with fewer substitutions. BDS is found in human (A), mouse, dog (B), stickleback, fly, and worm (C). In contrast, chicken and yeast (D) do not exhibit significant W→S BDS. These patterns are not sensitive to the size of the window (500–2000 bp) or the substitution density threshold. The gray bars indicate 95% confidence intervals for the BDS score. This figure includes substitution densities for which between 1% and 99% of substitutions are considered divergent. Table 1 gives BDS statistics for all species considered, and plots are provided for the other four species in supplementary figure S1 (Supplementary Material online).
Correlation of Genomic Features across the Genome.
To explore the correlation of data that vary across the genome, we selected an appropriate block size on which the features could be quantified. (This was often limited by the scale of the recombination maps available or the number of expected substitutions in a region.) Nonoverlapping blocks of 1 Mb were used for all species except for fly, worm, and yeast, where we used blocks of 10 and 100 kb due to their smaller genomes. We created a vector for each feature being compared, for example, BDS and recombination rate, across each corresponding block of the genome and calculated the Spearman rank correlation across all blocks. We also calculated and plotted the Spearman correlation of smoothed versions of the data. The data were smoothed by convolution with a seven-block Hanning window. To evaluate the significance of the difference in the correlation of a genomic feature with two other features, for example, in the comparison of BDS with male and female recombination rate, empirical P values were obtained by bootstrapping with 10,000 comparisons to random features.
Results
BDS Occurs in Vertebrates and Invertebrates
We calculated BDS scores and P values for eight diverse eukaryotic lineages (fig. 1). Figure 2A gives the BDS computed over substitutions on the human lineage since divergence from its last common ancestor with chimp for a range of window sizes and substitution density thresholds. For each window size, there is significant BDS; that is, the fastest-evolving regions show a significant enrichment for W→S nucleotide substitutions (increasing curves; all P ≈ 0). This result is in agreement with that reported in Dreszer et al. (2007) using parsimony to infer substitutions and a different statistical test for association between substitution type and density.
Extending the analysis to other lineages, we find significant BDS in mouse (P = 5.2 × 10−5), dog (P ≈ 0), stickleback (P ≈ 0), fruit fly (P ≈ 0), and worm (P = 5.1 × 10−5) (fig. 1 and table 1). As in the human genome, the patterns of BDS in these species are not sensitive to the particular window size and density thresholds used (fig. 2 and supplementary fig S1, Supplementary Material online). However, we do observe interspecies differences in the magnitude of the BDS score. Dog exhibits the strongest bias, whereas mouse has the weakest statistically significant W→S BDS.
Table 1.
BDS Statistics in Eight Eukaryotic Species.
| Divergent Regions |
Nondivergent Regions |
Branch | |||||
| Species | W→S | Not W→S | W→S | Not W→S | Length | BDS | P value |
| Human | 0.445 | 0.555 | 0.425 | 0.575 | 0.01 | 0.12 | ≈0 |
| Mouse | 0.419 | 0.581 | 0.415 | 0.585 | 0.17 | 0.02 | 5.2 × 10−5 |
| Dog | 0.512 | 0.488 | 0.423 | 0.577 | 0.20 | 0.52 | ≈0 |
| Chicken | 0.392 | 0.608 | 0.401 | 0.599 | 0.34 | −0.05 | 5.3 × 10−6 |
| Stickleback | 0.439 | 0.561 | 0.418 | 0.582 | 0.43 | 0.13 | ≈0 |
| Fruit fly | 0.297 | 0.703 | 0.270 | 0.730 | 0.13 | 0.19 | ≈0 |
| Worm | 0.311 | 0.689 | 0.295 | 0.705 | 0.81 | 0.11 | 5.1 × 10−5 |
| Yeast | 0.426 | 0.574 | 0.442 | 0.558 | 0.25 | −0.10 | 5.8 × 10−7 |
NOTE.—BDS is a log odds ratio quantifying the association between W→S substitution and the density of substitution. P values are computed using FET. Branch lengths are given in expected substitutions per site. All statistics are based on a cluster size of 1,000 bp and a density threshold that results in approximately 5% of substitutions being placed in the divergent class.
Reversing the roles of the reference and comparison species, we also find BDS in chimpanzee and several additional genomes (data not shown). However, inference of substitution histories is more difficult in comparison species, whose genome assemblies tend to be lower quality and often lack the resources for proper alignment quality filtering.
We also calculated a variation of the BDS score that considers only W→S and S→W substitutions. This resulted in slightly elevated scores compared with considering all substitution types. For example, in human, the bias increased from 0.12 to 0.14.
Chicken and Yeast Show a Significant Lack of W→S BDS
We do not observe W→S BDS in either chicken or yeast (table 1). In contrast to the other vertebrates analyzed, chicken shows a small but significant strong-to-weak (S→W) bias in fast-evolving regions (P = 5.3 × 10−6). Similarly, yeast shows a significant excess of S→W nucleotide changes in divergent regions (P = 5.8 ×10− 7, fig. 2D). This pattern in fixed substitutions is consistent with the S→W mutation bias observed in yeast (Lynch et al. 2008). In the next sections, we investigate several aspects of BDS that help us to interpret these observations. We consider possible explanations in the Discussion.
Variation in BDS between Species Is Not Due to Their Divergence
The species trios analyzed exhibit a range of evolutionary distances between the reference and comparison species (fig. 1; table 1). It is possible that the length of the branch considered might affect 1) our power to detect BDS and 2) the estimated strength of the BDS between the two species compared. Several observations argue against such biases, however. First, significant BDS patterns were observed in all comparisons. Hence, even though branch length likely influences the test's power, we are still able to detect a signal in each lineage. Additionally, the strength of the BDS observed between two species is not well correlated with their divergence (table 1). We also calculated BDS in substitutions between human and a series of increasingly divergent species that mirror the divergences of the other eukaryotic species sets considered here. In three of the four comparisons (human with tarsier, tree shrew, and opossum), significant bias was still identifiable between human and the more distant comparison species (supplementary table S1, Supplementary Material online). However, the fact that W→S BDS was not found in one of the comparisons (human with marmoset) suggests that the sources of BDS may not be constant over time or that our ability to detect BDS depends upon the quality of the comparison genome. We also tested the use of increasingly divergent outgroup species and found no major influence on patterns of BDS. Thus, our method is able to detect bias within the range of divergence found in the species sets we analyzed, including those used to quantify BDS in chicken and yeast. We therefore conclude that factors other than evolutionary distance appear to drive the variation in BDS between lineages.
BDS Occurs in Both Coding and Noncoding Regions
Coding and noncoding sequences are often under very different patterns of evolutionary constraint. The protein-coding fractions of the genomes we considered vary from around 2% in human to 73% in yeast (table 2). Thus, if substitution bias is different in coding and noncoding regions, this could influence our conclusions about the phylogenetic distribution of BDS. In particular, if BDS was absent or very weak in coding regions, we would have less power to detect genome-wide BDS in species with a high fraction of coding DNA, such as worm and yeast.
Table 2.
BDS Is Present in Coding Regions.
| Species | Percent Coding | Coding BDS |
| Human | 2.4 | 0.51 |
| Mouse | 2.3 | 0.11 |
| Dog | 1.5 | 0.83 |
| Chicken | 3.0 | −0.07 |
| Stickleback | 8.2 | 0.14 |
| Fruit fly | 18.5 | 0.51 |
| Worm | 27.9 | 0.36 |
| Yeast | 72.9 | −0.11 |
NOTE.—Bold indicates significant BDS.
To investigate this issue, we calculated BDS scores separately for coding regions in each species. Table 2 demonstrates that in species with significant genome-wide BDS, significant bias is present in coding sequence considered alone. Interestingly, coding regions generally exhibit stronger BDS than noncoding sequence. Similarly, coding sequence in yeast and chicken show S→W BDS, in agreement with the genome-wide evidence of S→W BDS in these species. The S→W bias in chicken coding regions is of similar magnitude to the genome-wide amount, but is not significant. This is not surprising given the weak signal in chicken genome wide. These results argue that patterns of bias in coding and noncoding sequences are usually in agreement and that the different fraction of coding sequence in different genomes is unlikely to be the source of their different bias patterns.
Intraspecies BDS Varies within and between Chromosomes
In the previous sections, a single BDS score was computed for each species to quantify genome-wide patterns of bias. To profile variation in BDS within genomes, we computed BDS scores across the chromosomes of each species in windows ranging in length from 10 kb to 1 Mb.
The strength of BDS varies across the chromosomes of each species (fig. 3). Some sections of the chromosome have significant bias, whereas others do not exhibit any BDS. This variation was observed previously in human (Dreszer et al. 2007), where a significant increase in BDS was found near the telomeres of most human and chimp chromosomes. Although variance in the BDS score is universal, increased BDS near telomeres is not a general phenomenon in all species we considered.
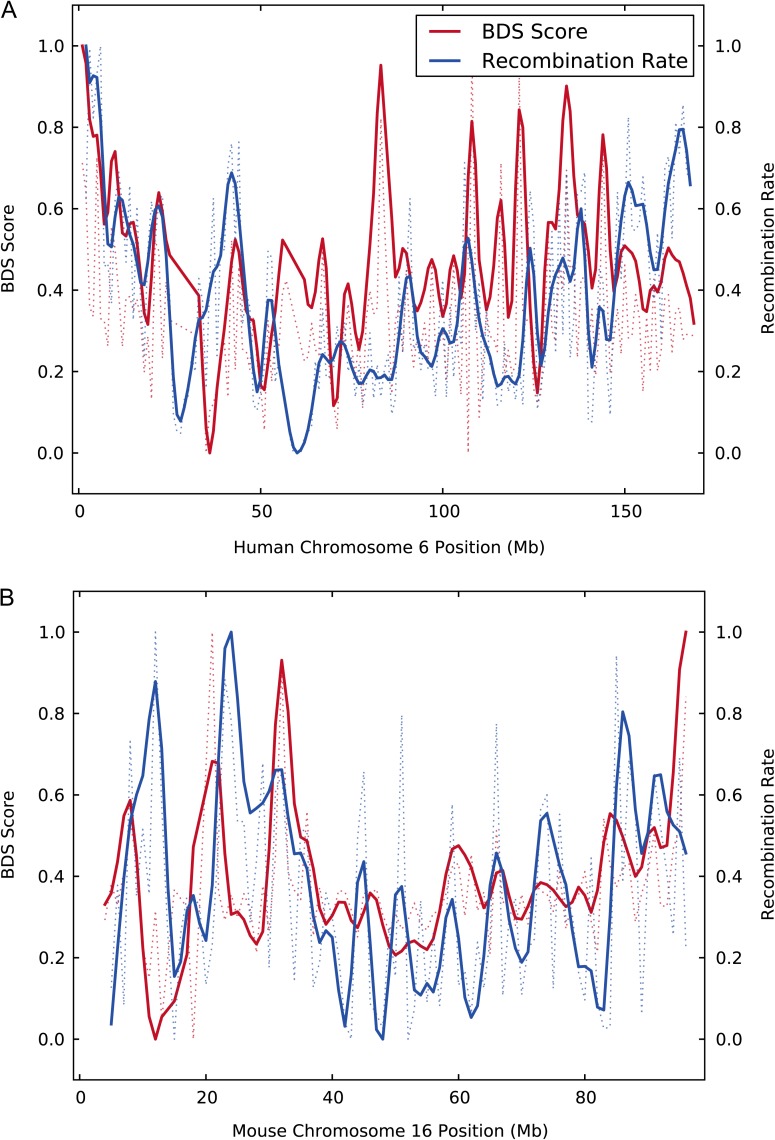
FIG. 3.—
BDS is often correlated with recombination rate. The BDS varies in strength along chromosomes and is significantly correlated with sex-averaged recombination rate in (A) human chromosome 6 (ρ = 0.36 smoothed, P = 9 × 10−7) and (B) mouse chromosome 16 (ρ = 0.36 smoothed, P = 2 × 10−4). The smoothed data are plotted with solid lines, and the raw values are indicated with dashed lines. See table 3 for genome-wide correlation statistics for these and other species on the raw data. For ease of visualization and comparison in this figure, the data have been scaled so that the minimum value is 0 and the maximum is 1.
BDS strength also varies between chromosomes. To compare the bias between chromosomes, we computed the BDS score for each chromosome in each species. In human, all chromosomes show peaks of significant BDS, but there is significant variation in the strength of the bias overall on different chromosomes (P < 2.2 × 10−16., Woolf test). The overall GC content of a chromosome is strongly correlated with recombination rate and negatively correlated with chromosome length in many species, including human (Fullerton et al. 2001) and chicken (International Chicken Genome Sequencing Consortium 2004). To frame BDS patterns in the context of these previous findings, we correlated its strength on each human chromosome with these features. Chromosomal BDS was not significantly correlated with the chromosome's GC content (Spearman ρ = 0.04), recombination rate (ρ = − 0.01), or length (ρ = 0.04). A similar lack of correlation of chromosomal BDS with GC content, recombination rate, and length was observed in all other species with significant BDS.
Local BDS Is Often Correlated with Recombination Rate
To investigate patterns of BDS at a finer scale, we calculated Spearman rank correlations of BDS in 1 Mb blocks with several genome features that vary across chromosomes: sex-averaged recombination rate, evolutionary conservation, GC content, and substitution rate (table 3). Stickleback, worm, and yeast were not included in the recombination correlation analysis due to lack of appropriate recombination maps. Our findings support the previous result in human, obtained using different methods, that recombination rate and BDS are correlated (Dreszer et al. 2007). This pattern occurs in other species as well; three of the four species (human, mouse, and fly) that have significant genome-wide BDS and recombination rate data show a significant correlation between these variables. The one exception is dog, which has experienced a recent pseudogenization of the PRDM9 gene (Oliver et al. 2009). This loss may explain the strong BDS in dog and the lack of correlation between BDS and recombination (see Discussion). BDS is not consistently correlated with GC content, substitution density, or conservation, although specific species do show modest significant correlations with some of these variables. Most notably, three out of the six species with genome-wide BDS have a positive correlation between BDS and evolutionary sequence conservation.
Table 3.
Correlation of BDS with Other Genome Features.
| Recombination | GC | Substitution | ||
| Species | Rate | Percent | Density | Conservation |
| Human | 0.16 | 0.05 | 0.08 | −0.01 |
| Mouse | 0.08 | 0.10 | −0.02 | 0.09 |
| Dog | 0.02 | −0.09 | −0.04 | 0.11 |
| Chicken | −0.09 | −0.12 | −0.03 | 0.06 |
| Stickleback | N/A | 0.02 | 0.08 | 0.02 |
| Fruit Fly | 0.10 | 0.15 | −0.05 | 0.14 |
| Worm | N/A | 0.03 | −0.02 | −0.11 |
| Yeast | N/A | 0.07 | −0.06 | 0.03 |
NOTE.—Each value is the Spearman correlation of BDS with the specified feature across blocks of the genome. Bold indicates a significant correlation at the 0.01 level. Blocks of 1 Mb were used for all species, except for fly (100 kb), worm (10 kb), and yeast (10 kb). The raw values for each feature were used; the correlation coefficients generally increase if the data are smoothed before calculation (e.g., see fig. 3).
Overall, our analyses suggest that BDS and recombination rate are often but not always correlated. It also appears that fluctuations in BDS are not directly associated with GC percent, divergence, or conservation; however, factors that indirectly influence these features may be relevant to the production of BDS patterns in some species.
BDS and Recombination Are More Strongly Correlated in Males
In the species for which we have sex-specific recombination data, we also considered the correlation of these rates with BDS separately for the two sexes. Previous studies of human Alu repeats (Webster et al. 2005) and substitution hotspots (Dreszer et al. 2007) indicate that biases associated with recombination may have a sex-specific impact. We find that BDS in human and mouse is more strongly associated with male recombination rate than with female (ρ = 0.152 vs. 0.119 in human, P ≈ 0.042; ρ = 0.093 vs. 0.014 in mouse, P ≈ 0.001). In dog, BDS does not show a significant correlation with sex-averaged recombination rate. However, when looking at male and female rates separately, the correlation with the male rate is significantly greater than with female (ρ = 0.058 vs. − 0.039, P ≈ 0.001), and though it is small in magnitude, the correlation between male recombination rate and BDS is significant (P = 0.018).
In contrast, when looking sex-specific rates in chicken, a species that does not show significant BDS overall, neither male or female recombination rates are significantly associated with BDS across the genome (ρ = − 0.079 and − 0.074, respectively), and the difference between the sex-specific correlations is not significant. These results suggest that when significant genome-wide BDS is present, the spatial correlation of BDS and recombination along a species' chromosomes is consistently higher in males. Sex-specific recombination data from additional species would help to test and further explore this hypothesis.
BDS Is Not Present in SNPs
Our test can also be applied to SNP data to study bias in population-level sequence variation. Dreszer et al. (2007) found no W→S bias in regions of the human with a high density of SNPs, suggesting that a fixation bias rather than a mutation bias was likely responsible for the BDS. To explore the phylogenetic extent of this pattern, we considered SNPs in human, mouse, and chicken. SNPs for each species were downloaded from the UCSC Table Browser (Karolchik et al. 2004). The ancestral variant was identified by parsimony using the alignment with the comparison species. Then, BDS scores were computed for SNPs using the same methods as used for fixed differences. SNPs for which no comparison species was present or with indeterminate ancestry were not considered in the analysis.
SNPs in human, mouse, and chicken do not exhibit W→S BDS (supplementary fig. S4, Supplementary Material online). This contrasts with the patterns in recent fixed substitutions in human and mouse. In fact, there is a significant lack of W→S changes in regions with the highest SNP density; the BDS scores are − 0.095 for human SNPs (P ≈ 0), − 0.018 for mouse (P = 3.3 × 10−5), and − 0.177 for chicken (P = 8.5 × 10−6). Thus, just as the BDS in recent fixed substitutions is present in species other than human, the previously observed lack of BDS in human SNPs also appears to be a general pattern. This result suggests that BDS is unlikely to be driven by local variation in mutation rates and patterns.
BDS Patterns Are Robust to the Methodology Used to Infer Substitution Histories
The use of parsimony to infer substitution types can potentially introduce biases into analyses of substitution patterns (Eyre-Walker 1998; Hernandez et al. 2007). CpG hypermutability, which increases the probability of multiple substitutions at a site and depends on the dinucleotide context, is a particular concern. To avoid these possible biases, our results are based on genome-wide substitution histories reconstructed in a maximum likelihood framework using a context-dependent evolutionary model. For comparison, we also performed the analysis using 1) maximum likelihood with a noncontext-dependent strand-specific reversible model (SSREV) and 2) parsimony.
Using maximum likelihood with the SSREV model produces very similar conclusions to those obtained with the U2S context–dependent model (supplementary fig. S2, Supplementary Material online). This suggests that context-dependent effects, such as CpG hypermutability, do influence the results slightly, but not significantly. The use of parsimony also leads to qualitatively similar conclusions; however, worm and chicken change their BDS classification (supplementary fig. S3, Supplementary Material online). When comparing all three methods, the two maximum likelihood approaches agree most closely, suggesting that using parsimony may indeed have an impact on inferred substitution histories. This is likely the result of fairly long branches in several of the clades considered; these are more likely to have experienced multiple substitutions, which would be missed by parsimony. Within the maximum likelihood context, our results are robust to the use of a context-dependent model or not, indicating that CpG effects are not the driving force behind BDS.
Discussion
We have demonstrated that divergent regions of several metazoan genomes from human to worm are associated with elevated rates of W→S substitution relative to the rest of the genome. In contrast, chicken and yeast do not exhibit significant BDS.
Episodic Biased Gene Conversion Is a Likely Cause of BDS
In human and several other species with significant BDS, we found consistent BDS in coding as well as noncoding sequences, correlations between BDS scores and recombination rates (especially in the male sex), and no significant BDS in SNPs. All of these observations are consistent with a recombination-associated, nonadaptive fixation bias as a cause of BDS. gBGC is a likely candidate. This bias occurs when there is a weak–strong polymorphism in a recombination heteroduplex, and the DNA mismatch is preferentially repaired to the strong base pair. These GC-biased conversion blocks are thought to range between 200 bp and 2 kb in length (Duret and Galtier 2009). gBGC has been directly observed in yeast (Mancera et al. 2008), but obtaining experimental evidence for the action of gBGC is extremely challenging in other species. Nonetheless, gBGC has received considerable attention as a possible explanation for many dominant and unexplained genomic attributes, such as the large-scale variation in GC content (the so-called isochore structure) of mammalian genomes (Eyre-Walker and Hurst 2001; Galtier et al. 2001; Meunier and Duret 2004; Romiguier et al. 2010). By driving strong alleles to higher frequencies and ultimately to fixation around recombination hotspots, bursts of gBGC could result in an increase in substitution rates and a W→S–biased substitution pattern (Berglund et al. 2009). These evolutionary events could produce the BDS pattern.
Other evolutionary mechanisms, such as variation in mutation rates across the genome or natural selection for GC alleles (Eyre-Walker and Hurst 2001), could also produce BDS. However, the action of a biased mutation rate is not consistent with our observations. Specifically, the lack of W→S BDS in SNPs argues against mutation bias as a source of the BDS pattern. The relationship between BDS and selection is less clear. If natural selection on GC content drives BDS, we would expect consistent differences in bias between regions of high and low conservation, such as coding and noncoding sequence. In most of the taxa we examined, significant BDS is present in both coding and noncoding sequence, and higher in coding regions. In addition, three species (mouse, dog, and fly) show a significant correlation between genome-wide patterns of BDS and evolutionary conservation. But the three other species with significant BDS (human, stickleback, and worm) show no such correlation. If selection has a role in BDS, we might additionally expect BDS to be stronger in species with large effective population sizes due to the increased efficiency of selection; however, this pattern is not observed. Thus, selective forces may be involved in the creation of BDS in some lineages, perhaps in concert with gBGC, but they are unlikely to be the sole cause of BDS. Together, these results suggest that there may multiple causes and paths to the creation of BDS in genomes.
Why Is BDS Stronger in Some Species than Others?
A recent study of the evolution of GC content across the mammalian phylogeny suggests that its dynamics are not constant across the tree and are influenced by many factors related to life history and genome organization (Romiguier et al. 2010). The variation we observe in BDS strength across the taxa considered here suggests a similarly dynamic picture for BDS with many possible factors influencing its strength. For example, the phylogenetic extent of gBGC, a likely source of BDS, across eukaryotes and its effect on genome evolution are currently unknown. There is strong sequence-based evidence for gBGC in mammals (Duret and Galtier 2009), and a recent comprehensive analysis of meiosis products in yeast provided direct experimental evidence of gBGC (Mancera et al. 2008). There is also indirect evidence of gBGC in additional eukaryotic taxa, based on correlations between GC content and recombination rate or chromosome size found in birds (International Chicken Genome Sequencing Consortium 2004), turtles (Kuraku et al. 2006), flies (Marais et al. 2003), worms (Marais et al. 2001), and several other species (Glémin 2010).
If gBGC is a cause of BDS, our identification of BDS in several eukaryotic species adds to the mounting evidence for its importance in genome evolution. However, it also suggests that population characteristics and mating patterns can influence the strength of BDS. Generation time, effective population size, frequency of outcrossing, recombination pattern, and conversion bias will all influence the effectiveness of gBGC (Duret and Arndt 2008), and many of these traits vary between species or within species over evolutionary time. These factors complicate cross-species comparison.
For example, in light of the experimental evidence for gBGC in S. cerevisiae (Birdsell 2002; Mancera et al. 2008), our finding of a significant lack of W→S BDS in yeast might seem to argue against gBGC as a cause of the bias. However, yeast differs from the other species we analyzed in a number of relevant genetic and physiological dimensions. Several aspects of sensu stricto yeast biology could act to limit gBGC's mutagenic impact. Most wild yeasts studied to date have very low frequencies of sex and outcrossing; recent work estimates that S. paradoxus undergoes meiosis only once in every 1,000 generations and only 1% of matings are outcrossed (Tsai et al. 2008). Since gBGC requires both meiosis and heterozygosity, its effect may be reduced in yeast by a factor of approximately 105 compared with obligately outcrossed species (Marais et al. 2004; Glémin et al. 2006; Tsai et al. 2010; Harrison and Charlesworth 2011). The resulting reduced mutagenic impact of gBGC has been proposed as an explanation for several differences in genomic patterns between yeast and other eukaryotes, such as the conservation of recombination hotspots (Tsai et al. 2010). Thus, the lack of detectable BDS in yeast is not inconsistent with the theory that gBGC is involved in creating BDS in other species.
Several factors may contribute to the lower BDS observed in mouse than in human. For example, recombination rate is thought to be approximately two times higher on average in human than in mouse (Coop and Przeworski 2006), and it is also less variable across mouse chromosomes. Since BDS likely results from episodic gBGC (either in time or across the chromosome), this relative lack of variation would produce less difference between divergent and nondivergent sequences in mouse. In agreement with this interpretation, a very recent study found that substitution patterns are under different influences in primates and rodents with a weaker effect of gBGC in rodents (Clément and Arndt 2011).
In dog, in contrast to other species with BDS, BDS and recombination rate do not show a significant correlation across the genome. The PRDM9 gene, which is thought to determine the location of about 40% of human recombination hotspots, has been pseudogenized in dog (Oliver et al. 2009). This event likely dramatically influenced the recombination landscape of dog and thus may explain why current recombination patterns do not correlate well with historical patterns of bias over the entire branch to the ancestor of dog and cat. Further investigations are needed to determine exactly why BDS is so strong in the dog genome.
The chicken genome lacks significant W→S BDS despite an estimated recombination rate, on both macro- and microchromosomes, considerably higher than in human (International Chicken Genome Sequencing Consortium 2004). However, the chicken karyotype is thought to have been far more stable over time than that of most mammals and may resemble that of the ancestral amniote (Webster et al. 2006). Lack of chromosomal rearrangements removes a common source of variation in recombination rate across the genome over time. In addition, the cellular machinery for determining recombination hotspots may be different in birds because sauropsids lack PRDM9 as well (Oliver et al. 2009).
Finally, the varying quality and availability of genome sequence data complicate cross-species comparisons of BDS. Low-coverage genome sequences are more likely to create noise from false substitutions inferred from sequencing errors. Our sequence and alignment quality filters help correct for these differences. But in the end, some subsets of each genome may still be influenced by error.
The phylogenetic patterns of BDS identified here point to the need for future work integrating all relevant variables in a consistent model for BDS. Unfortunately, this approach awaits further data generation as many of the important variables are not yet well-characterized across multiple taxa.
gBGC and Selection may Jointly Shape the Evolution of Functional Sequences
The dramatic enrichment for W→S substitutions in and around human accelerated regions (HARs) (Pollard, Salama, King, et al. 2006; Katzman et al. 2010) and the presence of possibly deleterious BDS in coding sequence (Berglund et al. 2009; Ratnakumar et al. 2010) suggest a complex interaction between BDS, selection, and the evolution of functional DNA elements. If the substitutions driving BDS in coding regions are caused by gBGC, they may increase the susceptibility of a gene to malfunction, as would be expected from the accumulation of mildly deleterious alleles (Charlesworth B and Charlesworth D 1998).
The presence of BDS in many HARs prompted the suggestion that gBGC, rather than positive selection, may have generated the acceleration (Galtier and Duret 2007). Thus, we might expect that HARs showing strong evidence of gBGC would be less likely to have obtained new functions in human. HAR1 and HAR2 (HACNS1), the two fastest evolving HARs, have strikingly biased substitution patterns. However, there is strong experimental evidence of function maintenance in HAR1 (Pollard, Salama, Lambert, et al. 2006) and gain in HAR2 (Prabhakar et al. 2008)—a surprising result if the human-specific changes in these sequences were created by a purely neutral mutational process. Therefore, we hypothesize that in some evolutionary scenarios, gBGC substitutions may themselves lead to novel functions or may set the stage for later adaptive changes, perhaps due to compensatory substitutions driven by selection.
Conclusions
In this study, we used efficient statistical methods to highlight phylogenetic patterns of a substitution bias. Our analysis of BDS in many eukaryotes suggests that it is common outside of human. Episodic gBGC driven by recombination is likely to play a major role in the production of BDS, but a number of evolutionary and organismal factors are likely to influence its occurrence. These conclusions underscore the importance of developing models of sequence evolution that incorporate the action of gBGC and other processes that interact with selection (Hurst 2009). Several promising preliminary steps have been made in the modeling of gBGC and selection (Duret and Arndt 2008; Berglund et al. 2009; Ratnakumar et al. 2010; Glémin 2010). As more genomes are assembled and richer recombination and polymorphism data become available for multiple species, we will be able to develop a deeper understanding of the causes and effects of BDS across the tree of life.
Supplementary Material
Supplementary figures S1–S4 and table S1 are available at Genome Biology and Evolution online http://www.gbe.oxfordjournals.org/.
Acknowledgments
We thank Dennis Kostka for many helpful discussions and suggestions. This work was supported by the National Institute of General Medical Sciences at the National Institutes of Health [GM82901 to K.S.P.]; and the Alfred P. Sloan Foundation.
References
- Berglund J, Pollard KS, Webster MT. Hotspots of biased nucleotide substitutions in human genes. PLoS Biol. 2009;7(1):e1000026. doi: 10.1371/journal.pbio.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdsell JA. Integrating genomics, bioinformatics, and classical genetics to study the effects of recombination on genome evolution. Mol Biol Evol. 2002;19(7):1181–1197. doi: 10.1093/oxfordjournals.molbev.a004176. [DOI] [PubMed] [Google Scholar]
- Blanchette M, et al. Aligning multiple genomic sequences with the threaded blockset sligner. Genome Res. 2004;14(4):708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. Some evolutionary consequences of deleterious mutations. Genetica. 1998;102–103(1–6):3–19. [PubMed] [Google Scholar]
- Clément Y, Arndt PF. Substitution patterns are under different influences in primates and rodents. Genome Biol Evol. 2011;3:236–245. doi: 10.1093/gbe/evr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Przeworski M. An evolutionary view of human recombination. Nat Rev Genet. 2006;8(1):23–34. doi: 10.1038/nrg1947. [DOI] [PubMed] [Google Scholar]
- Cox A, et al. A new standard genetic map for the laboratory mouse. Genetics. 2009;182(4):1335–1344. doi: 10.1534/genetics.109.105486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreszer TR, Wall GD, Haussler D, Pollard KS. Biased clustered substitutions in the human genome: the footprints of male-driven biased gene conversion. Genome Res. 2007;17(10):1420–1430. doi: 10.1101/gr.6395807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Arndt PF. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008;4(5):e1000071. doi: 10.1371/journal.pgen.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu Rev Genomics Hum Genet. 2009;10(1):285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A. Problems with parsimony in sequences of biased base composition. J Mol Evol. 1998;47:686–690. doi: 10.1007/pl00006427. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Hurst LD. The evolution of isochores. Nat Rev Genet. 2001;2(7):549–555. doi: 10.1038/35080577. [DOI] [PubMed] [Google Scholar]
- Fiston-Lavier AS, Singh ND, Lipatov M, Petrov DA. Drosophila melanogaster recombination rate calculator. Gene. 2010;463(1–2):18–20. doi: 10.1016/j.gene.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Fullerton SM, Bernardo Carvalho A, Clark AG. Local rates of recombination are positively correlated with GC content in the human genome. Mol Biol Evol. 2001;18(6):1139–1142. doi: 10.1093/oxfordjournals.molbev.a003886. [DOI] [PubMed] [Google Scholar]
- Galtier N, Duret L. Adaptation or biased gene conversion? Extending the null hypothesis of molecular evolution. Trends Genet. 2007;23(6):273–277. doi: 10.1016/j.tig.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Galtier N, Piganeau G, Mouchiroud D, Duret L. GC-content evolution in mammalian genomes: the biased gene conversion hypothesis. Genetics. 2001;159(2):907–911. doi: 10.1093/genetics/159.2.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glémin S. Surprising fitness consequences of GC-biased gene conversion: I. Mutation load and inbreeding depression. Genetics. 2010;185(3):939–959. doi: 10.1534/genetics.110.116368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glémin S, Bazin E, Charlesworth D. Impact of mating systems on patterns of sequence polymorphism in flowering plants. Proc R Soc B Biol Sci. 2006;273(1604):3011–3019. doi: 10.1098/rspb.2006.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen MA, et al. A high-density SNP-based linkage map of the chicken genome reveals sequence features correlated with recombination rate. Genome Res. 2009;19(3):510–519. doi: 10.1101/gr.086538.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison RJ, Charlesworth B. Biased gene conversion affects patterns of codon usage and amino acid usage in the saccharomyces sensu stricto group of yeasts. Mol Biol Evol. 2011;28(1):117–129. doi: 10.1093/molbev/msq191. [DOI] [PubMed] [Google Scholar]
- Hernandez RD, Williamson SH, Zhu L, Bustamante CD. Context-dependent mutation rates may cause spurious signatures of a fixation bias favoring higher GC-content in humans. Mol Biol Evol. 2007;24(10):2196–2202. doi: 10.1093/molbev/msm149. [DOI] [PubMed] [Google Scholar]
- Hubisz MJ, Pollard KS, Siepel A. PHAST and RPHAST: phylogenetic analysis with space/time models. Brief Bioinform. 2011;12(1):41–51. doi: 10.1093/bib/bbq072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter JD. Matplotlib: a 2D graphics environment. Comput Sci Eng. 2007;9(3):90–95. [Google Scholar]
- Hurst LD. Genetics and the understanding of selection. Nat Rev Genet. 2009;10(2):83–93. doi: 10.1038/nrg2506. [DOI] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432(7018):695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Jackson Laboratories. Mouse map converter. 2009 [cited 2010 Mar 22]. Available from: http://cgd.jax.org/mousemapconverter/. [Google Scholar]
- Jones E, Oliphant T, Peterson P. SciPy: Open source scientific tools for Python. 2001. [cited 2010 Oct 12]. Available from: http://www.scipy.org/. [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32(Suppl 1):D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman S, Kern AD, Pollard KS, Salama SR, Haussler D. GC-biased evolution near human accelerated regions. PLoS Genet. 2010;6(5):e1000960. doi: 10.1371/journal.pgen.1000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, et al. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc Natl Acad Sci U S A. 2003;100(20):11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, et al. The Human Genome Browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467(7319):1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- Kuraku S, et al. cDNA-based gene mapping and GC3 profiling in the soft-shelled turtle suggest a chromosomal size-dependent GC bias shared by sauropsids. Chromosome Res. 2006;14(2):187–202. doi: 10.1007/s10577-006-1035-8. [DOI] [PubMed] [Google Scholar]
- Lynch M, et al. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc Natl Acad Sci U S A. 2008;105(27):9272–9277. doi: 10.1073/pnas.0803466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E, et al. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454(7203):479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais G, Charlesworth B, Wright S. Recombination and base composition: the case of the highly self-fertilizing plant arabidopsis thaliana. Genome Biol. 2004;5(7):R45. doi: 10.1186/gb-2004-5-7-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais G, Mouchiroud D, Duret L. Does recombination improve selection on codon usage? Lessons from nematode and fly complete genomes. Proc Natl Acad Sci U S A. 2001;98(10):5688–5692. doi: 10.1073/pnas.091427698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais G, Mouchiroud D, Duret L. Neutral effect of recombination on base composition in Drosophila. Genet Res. 2003;81(2):79–87. doi: 10.1017/s0016672302006079. [DOI] [PubMed] [Google Scholar]
- Meunier J, Duret L. Recombination drives the evolution of GC-content in the human genome. Mol Biol Evol. 2004;21(6):984–990. doi: 10.1093/molbev/msh070. [DOI] [PubMed] [Google Scholar]
- Oliver PL, et al. Accelerated evolution of the prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 2009;5(12):e1000753. doi: 10.1371/journal.pgen.1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20(1):110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, King B, et al. Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006;2(10):e168. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443(7108):167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Prabhakar S, et al. Human-specific gain of function in a developmental enhancer. Science. 2008;321(5894):1346–1350. doi: 10.1126/science.1159974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. Vienna (Austria): R Foundation for Statistical Computing; 2009. R: a language and environment for statistical computing. ISBN 3-900051-07-0. [Google Scholar]
- Ratnakumar A, et al. Detecting positive selection within genomes: the problem of biased gene conversion. Philos Trans R Soc B Biol Sci. 2010;365(1552):2571–2580. doi: 10.1098/rstb.2010.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romiguier J, Ranwez V, Douzery EJP, Galtier N. Contrasting GC-content dynamics across 33 mammalian genomes: relationship with life-history traits and chromosome sizes. Genome Res. 2010;20(8):1001–1009. doi: 10.1101/gr.104372.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifman S, et al. A high-resolution single nucleotide polymorphism genetic map of the mouse genome. PLoS Biol. 2006;4(12):e395. doi: 10.1371/journal.pbio.0040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15(8):1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Haussler D. Combining phylogenetic and hidden markov models in biosequence analysis. J Comput Biol. 2004;11(2–3):413–428. doi: 10.1089/1066527041410472. [DOI] [PubMed] [Google Scholar]
- Smit A, Hubley R. RepeatModeler Open-1.0. 2008–2010. [cited 2011 Jun 16]. Available from: http://www.repeatmasker.org. [Google Scholar]
- The International Hapmap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai IJ, Bensasson D, Burt A, Koufopanou V. Population genomics of the wild yeast Saccharomyces paradoxus: quantifying the life cycle. Proc Natl Acad Sci U S A. 2008;105(12):4957–4962. doi: 10.1073/pnas.0707314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai IJ, Burt A, Koufopanou V. Conservation of recombination hotspots in yeast. Proc Natl Acad Sci U S A.. 2010;107(17):7847–7852. doi: 10.1073/pnas.0908774107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MT, Axelsson E, Ellegren H. Strong regional biases in nucleotide substitution in the chicken genome. Mol Biol Evol. 2006;23(6):1203–1216. doi: 10.1093/molbev/msk008. [DOI] [PubMed] [Google Scholar]
- Webster MT, et al. Male-driven biased gene conversion governs the evolution of base composition in human Alu repeats. Mol Biol Evol. 2005;22(6):1468–1474. doi: 10.1093/molbev/msi136. [DOI] [PubMed] [Google Scholar]