Abstract
BACKGROUND
In the southern region of the United States, such as in Louisiana and Texas, there are autochthonous cases of leprosy among native-born Americans with no history of foreign exposure. In the same region, as well as in Mexico, wild armadillos are infected with Mycobacterium leprae.
METHODS
Whole-genome resequencing of M. leprae from one wild armadillo and three U.S. patients with leprosy revealed that the infective strains were essentially identical. Comparative genomic analysis of these strains and M. leprae strains from Asia and Brazil identified 51 single-nucleotide polymorphisms and an 11-bp insertion–deletion. We genotyped these polymorphic sites, in combination with 10 variable-number tandem repeats, in M. leprae strains obtained from 33 wild armadillos from five southern states, 50 U.S. outpatients seen at a clinic in Louisiana, and 64 Venezuelan patients, as well as in four foreign reference strains.
RESULTS
The M. leprae genotype of patients with foreign exposure generally reflected their country of origin or travel history. However, a unique M. leprae genotype (3I-2-v1) was found in 28 of the 33 wild armadillos and 25 of the 39 U.S. patients who resided in areas where exposure to armadillo-borne M. leprae was possible. This genotype has not been reported elsewhere in the world.
CONCLUSIONS
Wild armadillos and many patients with leprosy in the southern United States are infected with the same strain of M. leprae. Armadillos are a large natural reservoir for M. leprae, and leprosy may be a zoonosis in the region. (Funded by the National Institute of Allergy and Infectious Diseases and others.)
Leprosy (Hansen’s Disease) is a chronic infectious disease caused by Mycobacterium leprae.1,2 Though often considered a disease of antiquity, it is found most commonly today in tropical and semitropical regions, and a total of 249,007 new cases were reported globally in 2008.3 Genomic polymorphisms have allowed us to trace the historical spread of leprosy around the world, as human populations migrated.4 The disease was not present in the New World before Columbus discovered the American continents but rather appears to have been introduced here from Europe and Africa during colonization. Early case reports suggest that leprosy was already well established among settlers in the vicinity of New Orleans by the 1750s,5 and autochthonous transmission of the infection continues in the region today.
Leprosy is rare in the United States, with only about 150 new cases reported each year. The majority of these affected people lived or worked abroad in leprosy-endemic areas and may have acquired their disease there. However, about a third of all patients in the United States report no foreign residence and appear to have acquired their disease from local sources — although most are unable to recall any known contact with a person who had leprosy. These cases arise most frequently in Texas and Louisiana,6 but the range of endemic involvement appears to be expanding to other states.7,8
M. leprae is an obligate intracellular pathogen that cannot be cultivated on artificial laboratory mediums. The only animal in which leprosy is reliably recapitulated is the nine-banded armadillo (Dasypus novemcinctus).1 Its unique susceptibility to experimental infection with M. leprae was first demonstrated in the 1970s, and armadillos have been the primary animal model for leprosy since.9 However, M. leprae infection also occurs naturally among some free-ranging armadillos.10
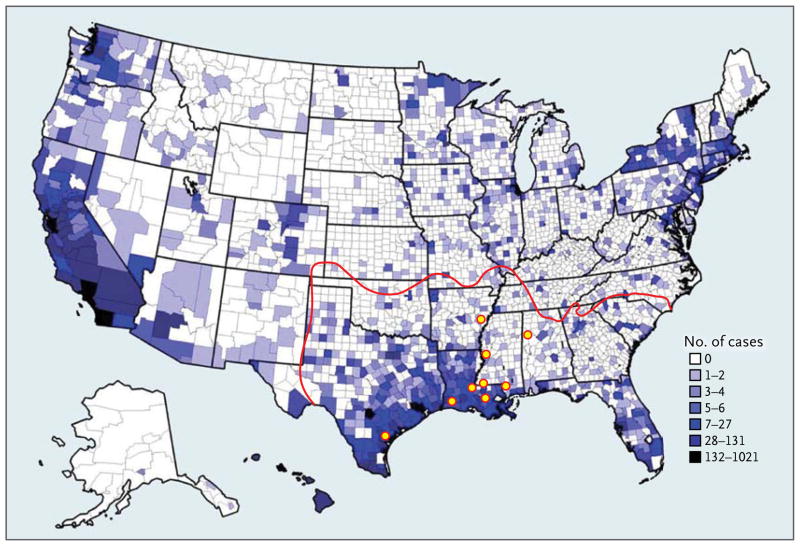
The origins of M. leprae infection among armadillos, the geographic range of the infected animals, and the potential risks infected armadillos present to people have been topics of concern. The infection originated among armadillos decades before they were ever used in leprosy research,11 and numerous surveys have confirmed that armadillos in the southern United States are a large natural reservoir for M. leprae; its prevalence exceeds 20% in some locales.10,12 Infected armadillos have been reported in Alabama, Arkansas, Louisiana, Mississippi, Texas (Fig. 1), and Mexico.13–15 Several case reports have suggested that armadillos may be a source of M. leprae for some U.S. patients,7,8,16,17 and contact with armadillos has been shown to be a significant risk factor for leprosy in three U.S. case–control studies.18–21
Figure 1. Distribution of Leprosy in the United States.
Counties in which leprosy cases have been reported are shown, with darker color indicating a greater total numbers of cases since 1894, according to the National Hansen’s Disease Registry. The currently estimated range of armadillos is outlined in red. Yellow circles indicate approximate locations of wild armadillos infected with Mycobacterium leprae — in Arkansas, Alabama, Louisiana, Mississippi, and Texas — suggesting that the central Gulf Coast is an area of endemic transmission to people. Leprosy cases in counties outside the armadillos’ range are due to familial contact, foreign exposure, or unknown sources.
Here, we describe our use of whole-genome sequencing, single-nucleotide polymorphism (SNP) typing, and variable-number tandem-repeat (VNTR) analysis to compare M. leprae obtained from wild armadillos and patients in the United States with leprosy and to understand better the role of armadillos in perpetuating leprosy in this country.
METHODS
STUDY DESIGN
We performed an ecologic cohort study to determine the type and frequency of M. leprae strains in U.S. patients, assess the degree to which they are genetically similar to strains from wild armadillos, and compare the geographic locations of the patients and armadillos. Statistical analysis was performed with the use of SAS software (version 9.2). The studies of the patients did not require board approval (on the basis of exemption category 4) and the requirement of written informed consent was waived by the institutional review board of Louisiana State University (Baton Rouge).
M. leprae Strains and DNA
Details of the M. leprae strains obtained from 50 patients with leprosy and 33 wild armadillos in the United States are given in Table S1 in the Supplementary Appendix (available with the full text of this article at NEJM.org). DNA from four genome-sequenced reference strains described previously22 and two foreign reference standards (LWM26 Philippines and 43926 Brazil) were also included.
Armadillo DNA
Armadillos were captured from the wild in five southern U.S. states (Fig. 1, and Table S1 in the Supplementary Appendix). DNA was isolated from 1-g specimens of liver, spleen, or lymph-node tissue by means of homogenization and was extracted with the use of a DNeasy kit (Qiagen).
Skin Biopsy in Patients
Skin-biopsy specimens are regularly obtained from patients who attend the National Hansen’s Disease Program outpatient clinic (Baton Rouge, LA) or are referred there for diagnosis. The specimens are stored in optimum-cutting-temperature compound (VWR) and frozen. We used frozen specimens from unrelated patients with multibacillary leprosy who presented between 1993 and 2007. Each sample was thawed, washed with phosphate-buffered saline, and processed by means of a DNeasy kit. Skin-biopsy specimens from 64 Venezuelan patients with leprosy living in 12 provinces were collected between 2007 and 2009, preserved in 70% ethanol, and processed for use in this study as described previously.4
GENOME RESEQUENCING AND ANALYSIS
Genome sequences of the human reference strains of M. leprae NHDP-98 (from a Mexican-born patient residing in Texas) and NHDP-55 (from another patient residing in Texas) (see Table S1 in the Supplementary Appendix) and the wild-armadillo–derived strain I-30 were obtained from DNA-fragment libraries, sequenced (36-Cycle Sequencing Kit v.1), and analyzed (Genome Analyzer II [Illumina]), as described previously.22 Sequence reads were mapped onto the consensus genome sequence of the M. leprae reference strain TN with the use of mapping quality scores.23 All sequence variations were confirmed by another means (BigDye Terminator v.3.1 cycle sequencing on an ABI3130XL DNA sequencer [Applied Biosystems]).
SEQUENCE AND PHYLOGENETIC ANALYSES
Details of the primers used for genotyping are given in Table S2 in the Supplementary Appendix, and the genotyping scheme is shown in Figure 2. Polymerase-chain-reaction (PCR) assays were performed as described previously,22 and amplicons were treated with exonuclease I and shrimp alkaline phosphatase (USB) before sequencing (BigDye Terminator). Sequence data were compared as described previously,24,25 and minimum-spanning-tree analysis based on SNPs and VNTRs was performed (BioNumerics software, version 6.1 [Applied Maths]).
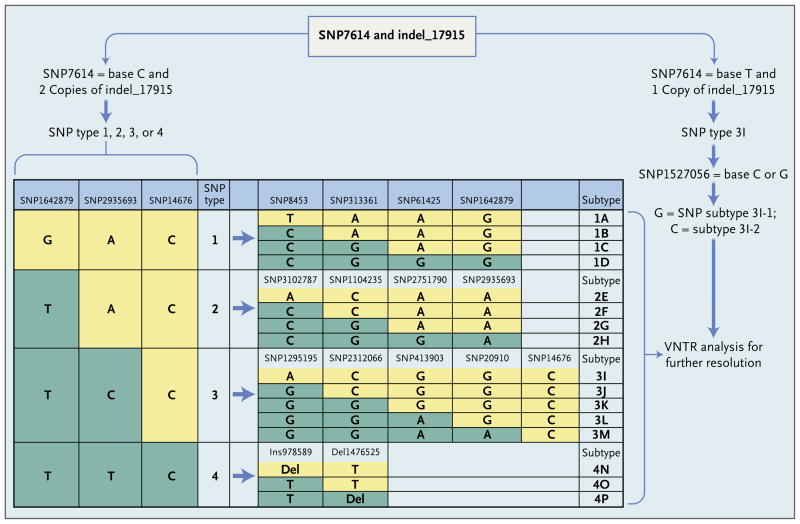
Figure 2. Genotyping of Mycobacterium leprae Strains.
SNP7614 and indel_17915 allowed for rapid and unambiguous identification of M. leprae strains containing SNP type 3I. Type 3I SNPs can be further subdivided into types 3I-1 and 3I-2 on the basis of SNP1527056 and four other SNPs (not shown). Samples with two copies of indel_17915 are classified into major SNP types 1, 2, 3, and 4 (as previously described4) and then further subtyped as a single letter from A though P, as shown, on the basis of the listed SNPs (which are representative of a panel of 84 SNPs22). Further high-resolution classification was then based on analysis of 10 variable-number tandem repeats (VNTRs).
VNTR TYPING
The primers used for VNTR typing at 10 loci26 are given in Table S3 in the Supplementary Appendix. Nei’s index of diversity27 (D = 1−Σ[allele frequency]2) based on 475 M. leprae strains28 was used to eliminate hypervariable loci (i.e., those with D>0.85) from consideration. PCR amplicons were sequenced (BigDye Terminator) to determine copy number.
RESULTS
GENOME SEQUENCING AND SNP IDENTIFICATION
Using Illumina technology, we obtained deep coverage of the genome sequences from M. leprae reference strains NHDP-55, NHDP-98, and the wild-armadillo–derived strain I-30 (Table 1, and Table S4 in the Supplementary Appendix). The resultant sequence reads were mapped onto the genome sequence of the TN reference strain (from India, SNP type 1A)29 and compared with the other sequenced M. leprae reference strains: Br4923 (from Brazil, SNP type 4P), NHDP-63 (from the United States, SNP type 3I), and Thai53 (from Thailand, SNP type 1A).22 This analysis confirmed the exceptionally high level of sequence conservation (99.995% identity), even among M. leprae strains of widely different geographic origins, and identified all four U.S.-derived genomes as SNP type 3I.
Table 1.
Single-Nucleotide Polymorphisms (SNPs) and Indels in 3I-Type Mycobacterium leprae Genomes Found in the United States.*
| M. leprae Strain | Average Coverage† | Markers Specific to 3I Type | Markers Differing within 3I Type | |||
|---|---|---|---|---|---|---|
| SNP | Indel | SNP | Indel | |||
| 1 bp | 11 bp | 1 bp | ||||
| NHDP-63 | 46 | 49 | 2 | 1 | 3 | 2 |
|
| ||||||
| NHDP-55 | 57 | 49 | 2 | 1 | 8 | 1 |
|
| ||||||
| NHDP-98 | 78 | 49 | 2 | 1 | 5 | 1 |
|
| ||||||
| I-30 | 22 | 49 | 2 | 1 | 1 | |
The SNPs were identified by means of comparative genomic methods based on M. leprae reference standards (strains TN, Br4923, and Thai53). NHDP denotes National Hansen’s Disease Program.
Average coverage is defined as the average number of consensus sequence reads obtained from the strain; there were no gaps except for the dispersed repeats that could not be distinguished owing to the short read length from Illumina sequencing.
On detailed comparison of these seven genome sequences, 52 markers were found only in the SNP type 3I strains. These 3I strains differed among themselves at 21 positions (Table 1, and Table S4 in the Supplementary Appendix). One 11-bp indel (indel_17915) was particularly important, since the 3I strains have only one copy of the sequence (TTGGTGGTGTA, in pseudogene ML0014), whereas all other M. leprae strains have two copies.
GENOTYPING OF M. LEPRAE STRAINS
SNP Analysis and Classification
We classified M. leprae obtained from 33 wild armadillos, 50 biopsy specimens from U.S. patients, and 4 foreign reference strains, using the algorithm shown in Figure 2. Armadillos were sampled in the five states known to harbor the sylvan infection (Fig. 1).14 Among the 50 U.S. patients examined (see Table S1 in the Supplementary Appendix), 39 reported a residence history in areas of the United States or Mexico where endemic exposure to armadilloborne M. leprae was possible, and 29 of these 39 had no history of foreign residence.
SNP analysis revealed seven types of M. leprae strains in the United States, including four found in patients with no history of foreign residence (Fig. 3). Some exotic strains may have become endemic over time among patients with no history of foreign residence or may now occur in the United States as a result of unreported exposure (e.g., SNP type 1A, which is more commonly associated with the Philippines, in Patient H-02).22 Among patients with possible exposure by means of foreign residence only, the SNP type was typical for strains previously reported from the foreign location. SNP type 3I, generally associated with European–American populations, was most abundant in our samples, found in those from all 33 armadillos and 26 of the 29 patients with no history of foreign residence.
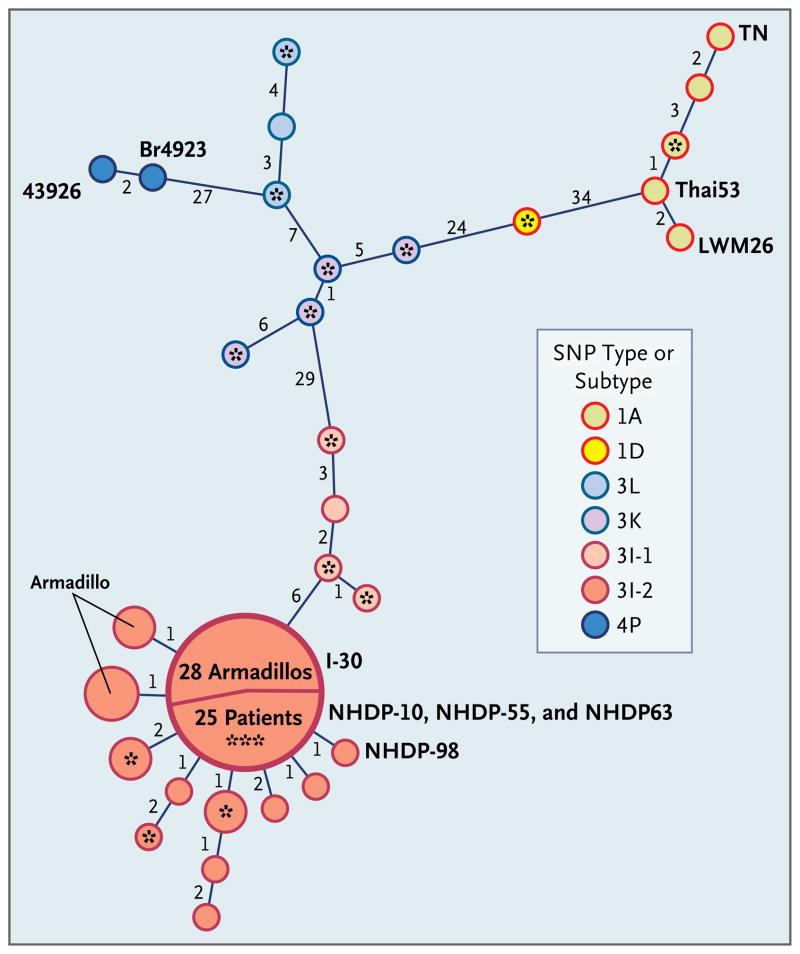
Figure 3. Minimum-Spanning Phylogenetic Tree of Mycobacterium leprae Genotypes Based on Analysis of Single-Nucleotide Polymorphisms (SNPs) and Variable-Number Tandem Repeats (VNTRs).
Minimum-spanning-tree analysis was performed with the use of combined VNTR and SNP data from human and armadillo M. leprae strains. Each circle represents a genotype (human unless marked as armadillo) based on the combined data, with the circle size directly proportional to the number of strains with the corresponding genotype. Numbers along the links between circles indicate the number of loci that differ between the genotypes on either side of the link. Three fully sequenced reference M. leprae strains (TN, Thai53, and Br492322,29) are labeled, as are two other reference strains (LWM26 and 43926) of foreign origin. Samples from patients with a history of foreign residence are indicated with an asterisk (with three asterisks indicating three patients). The 114 polymorphisms investigated include 84 SNPs described previously22 and 30 identified during our study; 10 VNTRs were also analyzed. The large circle illustrates the predominance of the 3I-2-v1 M. leprae genotype in our study, with 25 patients and 28 armadillos having this identical genotype.
To improve the resolution of our data for M. leprae 3I strains, we also surveyed for 30 of the 52 newly discovered markers. In addition to the 11-bp indel, 24 of 30 markers were restricted to SNPs of type 3I, irrespective of the source of the strain. However, in four 3I strains identified in patients, five SNPs contained ancestral bases and may represent intermediate sequences arising during the evolutionary divergence of 3I strains from their common ancestor. The strains with ancestral bases were classified as having SNPs of the subtype 3I-1 to differentiate them from the more divergent strains classified as 3I-2 strains found in all armadillos and most indigenous U.S. patients (Fig. 3, and Table S5 in the Supplementary Appendix).
To gain more insight into the distribution of 3I-1 strains in the Americas, we examined biopsy specimens from 64 Venezuelan patients infected with M. leprae strains containing SNP type 3I. Of the 64 specimens, 48 (75%) belong to the strain subtype 3I-1 and 16 (25%) belong to strain subtype 3I-2. In addition, we found 3I-1 subtypes in patients from Brazil, Puerto Rico, and the Dominican Republic. Therefore, the prevalence of 3I-1 and 3I-2 strains in North America is significantly different (P<0.001) from their prevalence in the Caribbean and South America (see Table S6 in the Supplementary Appendix).
VNTR AND MINIMUM-SPANNING-TREE ANALYSES
Owing to the remarkable conservation of the M. leprae genome, SNP analysis is of limited power. Accordingly, we used 10 polymorphic VNTR markers to enhance discrimination (see Table S4 in the Supplementary Appendix) of strain subtypes.26 Minimum-spanning-tree analysis of the combined SNP and VNTR profiles was performed to examine relationships among strains (Fig. 3). The resulting SNP–VNTR genotypes (see Table S1 in the Supplementary Appendix) confirm a high degree of homogeneity between M. leprae from armadillos and most indigenous U.S. cases of leprosy (Fig. 3). Among wild armadillos, 28 of the 33 strains showed complete genetic identity with respect to the SNP–VNTR genotype (3I-2-v1); the remaining 5 strains comprised two genotypes, 3I-2-v14 (2 specimens) and 3I-2-v13 (3 specimens) that varied at one VNTR locus (see Table S5 in the Supplementary Appendix). Similarly, 25 of the 39 patients (64%) with a history of residence in areas in which exposure to M. leprae from armadillos was possible — including 22 of the 29 patients with no history of foreign residence — also carried the 3I-2-v1 strain. The 3I-2-v1 genotype appears to be unique and highly distinctive. This combination of VNTR alleles was not found within a database of VNTR genotypes identified around the world,28 and allele frequencies in that database suggest a probability of random reassortment of the VNTR v1 genotype of only 1 in 10,000.28
In our study, 3I-2-v1 was the only genotype found in more than two patients. The combination of SNP and VNTR genotyping is highly discriminatory and confirms a significant association between the M. leprae strain infecting armadillos and many U.S. patients. The 3I-2-v1 strain was significantly associated with a history of residence in areas where M. leprae–infected armadillos have been found (P<0.001). People with leprosy who live in areas with infected armadillos and who have no history of foreign residence have a significantly increased risk of presenting with infection with the 3I-2-v1 strain, as compared with any other strain (odds ratio, 16.5; 95% confidence interval [CI], 4.2 to 64.7; P<0.001) (see Table S6 in the Supplementary Appendix).
A history regarding contact with armadillos was available for 15 patients. Although 7 recalled no contact, 8 recalled having contact, including 1 who reported frequently hunting, cooking, and eating armadillos. Nine of the 15 patients were infected with the 3I-2-v1 strain. These data confirm interaction with armadillos by some of our patients, and suggest an increased likelihood of infection with the 3I-2-v1 strain as a result (odds ratio vs. having no contact, 4.0; 95% CI, 0.5 to 35.8; P = 0.314).
DISCUSSION
Combining the high discriminatory power of VNTR analysis26 with the robust common-roots approach of SNP typing22,30 has proved effective in molecular epidemiologic studies of tuberculosis,31 and was used in our study of leprosy to similar benefit. We show that a high percentage of unrelated leprosy cases in the southern United States involve infection with the same unique strain of M. leprae that occurs naturally among wild armadillos in the region. These armadillos are a large natural reservoir for M. leprae.13,14 The genome sequences of the predominant armadillo and human strains in this region are essentially identical, and M. leprae of this genotype has not been reported previously elsewhere in the world.28 Though it is difficult to establish specific causality, when these data are taken together, they strongly implicate armadillos as a source of infection. Therefore, leprosy appears to be a zoonosis in the southern United States.
Autochthonous leprosy is exceptionally rare in the United States, but exposure to armadillos in southern states is quite common. Three case–control studies have shown contact with armadillos to be a significant risk factor for leprosy in the United States,18,19,21 and our work shows that a single predominant strain is involved in most human and armadillo infections. Transmission of leprosy can be by direct or indirect means involving fomites but is thought to occur most frequently through long-term direct contact with an infected host.1 M. leprae is an obligate intracellular pathogen with limited capacity for survival in the environment.29 Frequent direct contact with armadillos and cooking and consumption of armadillo meat should be discouraged.
Armadillos must have acquired M. leprae from humans sometime after colonization of the New World, but the lack of diversity of strain types infecting the animals suggests that interspecies transfer of M. leprae is uncommon and inefficient. However, inter-armadillo transfer appears to be highly efficient, since the 3I-2-v1 strain is now found across five states. The near-uniformity of M. leprae strains recovered from armadillos in this area is consistent with recent acquisition or rapid emergence of the infection as armadillos expanded their range into the United States and high population densities developed.32 High prevalence rates among armadillos have been observed in parts of the southern United States only; armadillos in the eastern United States are not known to be infected with M. leprae. The eastern population originated from a separate introduction of armadillos into Florida, which expanded and has only relatively recently merged with the main U.S. population.13,14 Monitoring the eastern United States for possible spread of M. leprae will be informative.
Armadillos are found in the western hemisphere only. Their potential effect on leprosy control in the Americas depends on the rate at which the infection can spread to contiguous armadillo populations and its likelihood to emerge among other high-density armadillo populations. Biomarkers of M. leprae have been reported in armadillos in Argentina, Brazil, and Colombia,18,19,21 and contact with armadillos has been reported to be a risk factor for leprosy in Brazil.33 Application of advanced genotyping techniques in such areas may help elucidate leprosy transmission in other human populations and increase our understanding of the specific risk factors perpetuating this infection globally.
Susceptibility to leprosy is modulated by a number of complex genetic traits,34 and the majority of people appear to be naturally immune to M. leprae infection. There are currently no recognized pathological variant M. leprae strains, and the outcome of infection is influenced primarily by the individual host response. Early diagnosis and prompt drug therapy remain the most effective means to avoid the undesirable complications of leprosy.1 Physicians caring for patients with potential exposure to M. leprae by means of armadillos should consider leprosy in their differential diagnosis of chronic cutaneous lesions, especially those not responsive to common treatments.
Leprosy remains one of the most socially stigmatizing diseases; diagnosis can evoke profound anxiety in the patient, especially when there is no identifiable index case and the source of infection is unknown. Armadillos are the only known non-human reservoir of M. leprae. Recognition of this zoonotic link may provide relief to some patients by identifying a biologically plausible source of their disease.
Supplementary Material
Acknowledgments
Supported by Fondation Raoul Follereau, the American Leprosy Missions, the Society of Saint Lazarus, the IDEAL (Initiative for Diagnostic and Epidemiological Assays for Leprosy) Consortium, the Health Resources and Services Administration, and the National Institutes of Health, National Institute of Allergy and Infectious Diseases (grants RO1-AI47197-01A1 and NIAID IAA-2646).
We thank all the participants in this study, particularly J. Adams, K. Andrews, A. Carter, P. Brennan, B. Hall, K. Harshman, M. Kearney, S. Keas, J. Krahenbuhl, G. McCormick, N. Robbins, J. Spencer, R. Stevenson, B. Stryjewska, S. Uplekar, and H. Zhang; F. Knight, J. Kumaresan, J. Loughry, and C. McDonough for their assistance in armadillo surveys; and the Vital-IT Center of the Swiss Institute of Bioinformatics for computational support.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Scollard DM, Adams LB, Gillis TP, Krahenbuhl JL, Truman RW, Williams DL. The continuing challenges of leprosy. Clin Microbiol Rev. 2006;19:338–81. doi: 10.1128/CMR.19.2.338-381.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Britton WJ, Lockwood DN. Leprosy. Lancet. 2004;363:1209–19. doi: 10.1016/S0140-6736(04)15952-7. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization. Global leprosy situation, 2009. Wkly Epidemiol Rec. 2009;84:333–40. [PubMed] [Google Scholar]
- 4.Monot M, Honoré N, Garnier T, et al. On the origin of leprosy. Science. 2005;308:1040–2. doi: 10.1126/science/1109759. [DOI] [PubMed] [Google Scholar]
- 5.Badger LF. Leprosy in the United States. Public Health Rep. 1955;70:525–35. [PMC free article] [PubMed] [Google Scholar]
- 6.A summary of Hansen’s disease in the United States — 2008. Washington, DC: Department of Health and Human Services, Health Resources and Services Administration, National Hansen’s Disease Program; 2009. ( http://www.hrsa.gov/hansens/2008RegistryReport.pdf) [Google Scholar]
- 7.Abide JM, Webb RM, Jones HL, Young L. Three indigenous cases of leprosy in the Mississippi delta. South Med J. 2008;101:635–8. doi: 10.1097/SMJ.0b013e31816f8610. [DOI] [PubMed] [Google Scholar]
- 8.Lane JE, Walsh DS, Meyers WM, Klassen-Fischer MK, Kent DE, Cohen DJ. Borderline tuberculoid leprosy in a woman from the state of Georgia with armadillo exposure. J Am Acad Dermatol. 2006;55:714–6. doi: 10.1016/j.jaad.2006.02.070. [DOI] [PubMed] [Google Scholar]
- 9.Kirchheimer WF, Storrs EE. Attempts to establish the armadillo (Dasypus novemcinctus Linn.) as a model for the study of leprosy. I. Report of lepromatoid leprosy in an experimentally infected armadillo. Int J Lepr Other Mycobact Dis. 1971;39:693–702. [PubMed] [Google Scholar]
- 10.Walsh GP, Meyers WM, Binford CH. Naturally acquired leprosy in the nine-banded armadillo: a decade of experience 1975–1985. J Leukoc Biol. 1986;40:645–56. doi: 10.1002/jlb.40.5.645. [DOI] [PubMed] [Google Scholar]
- 11.Truman RW, Shannon EJ, Hagstad HV, Hugh-Jones ME, Wolff A, Hastings RC. Evaluation of the origin of Mycobacterium leprae infections in the wild armadillo, Dasypus novemcinctus. Am J Trop Med Hyg. 1986;35:588–93. doi: 10.4269/ajtmh.1986.35.588. [DOI] [PubMed] [Google Scholar]
- 12.Truman RW, Kumaresan JA, Mc-Donough CM, Job CK, Hastings RC. Seasonal and spatial trends in the detectability of leprosy in wild armadillos. Epidemiol Infect. 1991;106:549–60. doi: 10.1017/s0950268800067613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loughry WJ, Truman RW, Mc-Donough CM, Tilak MK, Garnier S, Delsuc F. Is leprosy spreading among nine-banded armadillos in the southeastern United States? J Wildl Dis. 2009;45:144–52. doi: 10.7589/0090-3558-45.1.144. [DOI] [PubMed] [Google Scholar]
- 14.Truman R. Leprosy in wild armadillos. Lepr Rev. 2005;76:198–208. [PubMed] [Google Scholar]
- 15.Amezcua ME, Escobar-Gutiérrez A, Storrs EE, Dhople AM, Burchfield HP. Wild Mexican armadillo with leprosy-like infection. Int J Lepr Other Mycobact Dis. 1984;52:254–5. [PubMed] [Google Scholar]
- 16.Lumpkin LR, III, Cox GF, Wolf JE., Jr Leprosy in five armadillo handlers. J Am Acad Dermatol. 1983;9:899–903. doi: 10.1016/s0190-9622(83)70206-9. [DOI] [PubMed] [Google Scholar]
- 17.Hamilton HK, Levis WR, Martiniuk F, Cabrera A, Wolf J. The role of the armadillo and sooty mangabey monkey in human leprosy. Int J Dermatol. 2008;47:545–50. doi: 10.1111/j.1365-4632.2008.03722.x. [DOI] [PubMed] [Google Scholar]
- 18.Bruce S, Schroeder TL, Ellner K, Rubin H, Williams T, Wolf JE., Jr Armadillo exposure and Hansen’s disease: an epidemiologic survey in southern Texas. J Am Acad Dermatol. 2000;43:223–8. doi: 10.1067/mjd.2000.106368. [DOI] [PubMed] [Google Scholar]
- 19.Clark BM, Murray CK, Horvath LL, Deye GA, Rasnake MS, Longfield RN. Case-control study of armadillo contact and Hansen’s disease. Am J Trop Med Hyg. 2008;78:962–7. [PubMed] [Google Scholar]
- 20.Filice GA, Greenberg RN, Fraser DW. Lack of observed association between armadillo contact and leprosy in humans. Am J Trop Med Hyg. 1977;26:137–9. doi: 10.4269/ajtmh.1977.26.137. [DOI] [PubMed] [Google Scholar]
- 21.Thomas DA, Mines JS, Mack TM, Thomas DC, Rea TH. Armadillo exposure among Mexican-born patients with lepromatous leprosy. J Infect Dis. 1987;156:990–2. doi: 10.1093/infdis/156.6.990. [DOI] [PubMed] [Google Scholar]
- 22.Monot M, Honore N, Garnier T, et al. Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat Genet. 2009;41:1282–9. doi: 10.1038/ng.477. [DOI] [PubMed] [Google Scholar]
- 23.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–8. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonfield JK, Smith K, Staden R. A new DNA sequence assembly program. Nucleic Acids Res. 1995;24:4992–9. doi: 10.1093/nar/23.24.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 26.Gillis T, Vissa V, Matsuoka M, et al. Characterisation of short tandem repeats for genotyping Mycobacterium leprae. Lepr Rev. 2009;80:250–60. [PubMed] [Google Scholar]
- 27.Weir BS. Genetic data analysis II: methods for discrete population genetic data. 2. Sunderland, MA: Sinauer; 1996. [Google Scholar]
- 28.Hall BG, Salipante SJ. Molecular epidemiology of Mycobacterium leprae by structure-neighbor clustering. J Clin Microbiol. 2010;48:1997–2008. doi: 10.1128/JCM.00149-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cole ST, Eiglmeier K, Parkhill J, et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–11. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- 30.Achtman M. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu Rev Microbiol. 2008;62:53–70. doi: 10.1146/annurev.micro.62.081307.162832. [DOI] [PubMed] [Google Scholar]
- 31.Comas I, Homolka S, Niemann S, Gagneux S. Genotyping of genetically monomorphic bacteria: DNA sequencing in Mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS OEN. 2009;4(11):e7815. doi: 10.1371/journal.pone.0007815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taulman JF, Rosen LW. Recent range expansion and distributional limits of the nine-banded armadillo (Dasypus novemcinctus) in the United States. J Biogeogr. 1996;23:635–48. [Google Scholar]
- 33.Deps PD, Alves BL, Gripp CG, et al. Contact with armadillos increases the risk of leprosy in Brazil: a case control study. Indian J Dermatol Venereol Leprol. 2008;74:338–42. doi: 10.4103/0378-6323.42897. [DOI] [PubMed] [Google Scholar]
- 34.Zhang FR, Huang W, Chen SM, et al. Genomewide association study of leprosy. N Engl J Med. 2009;361:2609–18. doi: 10.1056/NEJMoa0903753. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.