Abstract
Accurate control of tissue-specific gene expression plays a pivotal role in heart development, but few cardiac transcriptional enhancers have thus far been identified. Extreme non-coding sequence conservation successfully predicts enhancers active in many tissues, but fails to identify substantial numbers of heart enhancers. Here we used ChIP-seq with the enhancer-associated protein p300 from mouse embryonic day 11.5 heart tissue to identify over three thousand candidate heart enhancers genome-wide. Compared to other tissues studied at this time-point, most candidate heart enhancers are less deeply conserved in vertebrate evolution. Nevertheless, the testing of 130 candidate regions in a transgenic mouse assay revealed that most of them reproducibly function as enhancers active in the heart, irrespective of their degree of evolutionary constraint. These results provide evidence for a large population of poorly conserved heart enhancers and suggest that the evolutionary constraint of embryonic enhancers can vary depending on tissue type.
Heart disease is a leading cause of mortality in infants and adults1,2. Despite extensive screening of protein-coding regions, the genetic basis of many cardiac defects is unknown3–5. While variants of gene regulatory sequences have been suggested to play a role6, their contribution has been difficult to evaluate because the genomic locations and activity patterns of regulatory sequences active in the heart remain largely obscure. Among the different types of regulatory sequence, transcriptional enhancers are particularly challenging to identify as they can be located at large genomic distances from the genes they regulate7. While extreme evolutionary sequence conservation has proven a valuable tool for the identification of developmental enhancers in general8–14, relatively few heart enhancers have been identified by this approach. In the largest existing datasets of in vivo embryonic enhancers identified through extreme sequence conservation13,14, less than 2% of tested sequences were found to be heart enhancers compared to 16%, 14% and 5% for forebrain, midbrain and limb enhancers respectively. This raises the possibilities that at this time-point in embryonic development there are either fewer enhancers active in heart than in other tissues, or that the conservation properties of heart enhancers differs from those of other tissues, rendering them unidentifiable by comparative genomic approaches. To resolve this issue, we sought an alternative genomic approach for identifying heart enhancers that is independent of the requirement for evolutionary DNA constraint.
The transcriptional co-activator protein p300 is expressed nearly ubiquitously in mouse embryogenesis15 and can bind to a wide spectrum of active tissue-specific enhancers. Exploiting these properties, chromatin immuno-precipitation with p300 directly from animal tissues coupled with massively parallel sequencing (ChIP-seq) can accurately predict the genomic location and tissue specificity of active developmental enhancers16–19. To obtain an initial genome-wide set of candidate enhancer sequences active in the heart, we performed p300 ChIP-seq on heart tissue from approximately 270 embryonic day 11.5 (e11.5) mouse embryos. Enrichment analysis20 of this dataset identified 3,597 regions that do not overlap known promoters but were significantly enriched in p300 binding and were therefore considered candidate heart enhancers. For comparison across different embryonic tissues, we applied the same ChIP-seq analysis approach to e11.5 forebrain, midbrain, and limb and identified 2,759, 2,786 and 3,839 p300-enriched regions in these tissues, respectively (see Methods and Supplementary Tables 1–4). The vast majority (84%) of p300 peaks in the heart do not overlap p300 peaks found in any of the other three tissues examined. These results indicate that p300 binding in the developing heart identifies a subset of non-coding regions that are distinct from putative enhancers active in other embryonic structures.
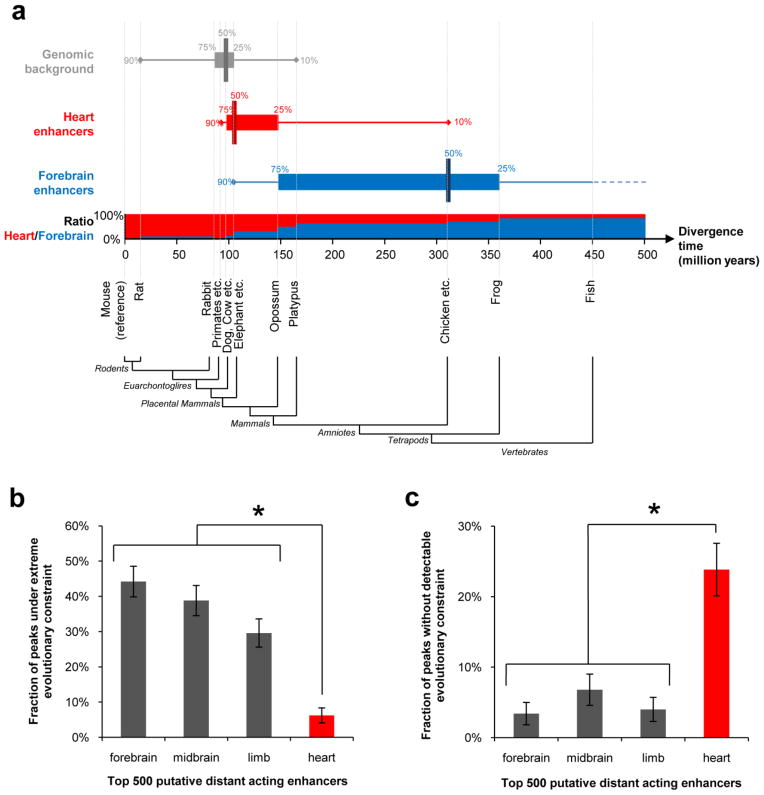
To evaluate potential differences in conservation properties of enhancers between tissues, we compared the evolutionary conservation depth of candidate heart and forebrain enhancers (the two tissues for which conservation-based predictions were least and most successful, respectively; see Methods). Most (65%) predicted heart enhancers are detectably conserved only among placental mammals, whereas the majority (56%) of predicted forebrain enhancers are conserved between mammals and birds (Fig. 1a). Using the median divergence time of species with detectable conservation as an approximate measure of evolutionary conservation depth, predicted forebrain enhancers are almost three times as deeply conserved as predicted heart enhancers (310 million years and 105 million years, respectively, Fig. 1a). The difference between the two tissues is particularly pronounced at the extremes of the conservation spectrum. Heart enhancers are nine-fold more abundant than forebrain enhancers among sequences conserved only within rodents, whereas predicted forebrain enhancers are seven times more frequent than heart enhancers among sequences conserved between mammals and fish (Fig. 1a). Predicted limb and midbrain enhancers exhibit an intermediate degree of evolutionary conservation compared with heart and forebrain enhancers (Supplementary Fig. 1), consistent with the frequency of enhancers in these tissues in comparative genomic datasets13,14. Notably, there is substantial overlap between the conservation profiles of heart enhancers and matched random genomic regions (Fig. 1a and Supplementary Fig. 2), suggesting that in contrast to forebrain enhancers a sizable proportion of heart enhancers active at this time-point cannot be confidently distinguished from surrounding genomic sequence by evolutionary conservation alone.
Figure 1. Weak evolutionary conservation of candidate heart enhancers identified by p300-binding in embryos at e11.5.
In order to compare equally sized high-confidence samples, analyses were performed on the 500 most significantly p300-bound regions from each tissue (see Methods). a) Comparison of the evolutionary conservation properties of heart and forebrain enhancers identified through p300 binding in e11.5 tissues. Conservation depth of enhancers was defined as the estimated divergence time from mouse of the most distantly related species with aligned genomic sequence22,36 (see Methods). Median (vertical bar), 25 to 75% percentile (horizontal bar) and 10 to 90% percentile (horizontal line) intervals of conservation depth are shown for forebrain enhancers (blue), heart enhancers (red) and the genomic background (10,000 randomly selected regions from the mouse genome with size, sequence mappability and repeat composition matched to candidate enhancers). The horizontal axis represents the mouse evolutionary lineage, with vertical dashed lines indicating the estimated divergence times37–39 of species or groups of species with sequenced genomes included in the analysis. For each interval on the mouse lineage, the bar chart shows the ratio of forebrain enhancers to heart enhancers among enhancers that are maximally conserved to that interval. b,c) 1 kb regions flanking p300 peaks from each tissue were assigned the score of the most highly constrained overlapping vertebrate phastCons element in the mouse genome21. b) Fraction of candidate enhancers that are under strong evolutionary constraint (score > 600). c) Fraction of candidate enhancers that are under no detectable constraint (no overlapping vertebrate constrained element). Error bars represent 95% binomial proportion confidence interval. *, P < 10−14, Fisher’s Exact Test, one-tailed.
To further evaluate differences in conservation properties, we compared the evolutionary constraint of enhancers from all four tissues using pre-computed evolutionary constraint scores (phastCons21 scores) generated from multi-vertebrate genome alignments22. Overall, only 6% of candidate heart enhancers overlap genome regions that are under extremely high constraint (score >600), compared to 44%, 39% and 30% of candidate forebrain, midbrain and limb enhancers respectively (P < 10−22, Fisher’s Exact Test; Fig. 1b). Conversely, the fraction of candidate heart enhancers that do not overlap detectably constrained sequences (24%) is four- to seven-fold greater than for candidate enhancers from other studied tissues (P < 10−14, Fisher’s Exact Test; Fig. 1c, Supplementary Table 5 and Supplementary Fig. 3). Importantly, these observations are robustly maintained when sequence constraint is determined from subsets of vertebrate species covering shorter evolutionary distances (Supplementary Fig. 4a–d) and using relaxed stringency thresholds for defining p300 peaks (Supplementary Fig. 4e–h). Taken together, these results show pronounced differences in the degree of evolutionary sequence constraint of p300-binding regions across tissues, with candidate heart enhancers under weaker constraint compared to other tissues. This observation provides a plausible explanation for the poor performance of extreme sequence conservation in identification of heart enhancers at this time-point, and suggests that p300 binding might identify a sizable population of weakly conserved heart enhancers that are likely not identifiable by existing comparative genomic approaches.
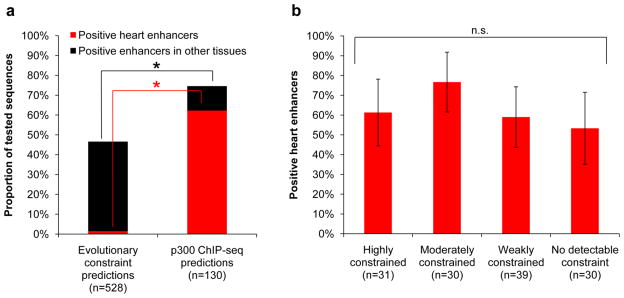
To assess the in vivo activity of p300-based predictions of heart enhancers, we tested 130 candidate heart enhancers in a transgenic mouse enhancer assay13,23 (Supplementary Fig. 5). In total, 97 sequences (75%) were found to be reproducible tissue-specific enhancers in e11.5 embryos (the p300 ChIP-seq time-point), of which the vast majority (81/97, 84%) were active in the developing heart (Supplementary Table 6). This represents a greater than 29-fold increase in specificity for heart enhancers over previous approaches based on extreme evolutionary constraint, in which only 8 out of 282 (3%) sequences confirmed to be enhancers in the same transgenic assay were active in the heart (P < 10−55, Fisher’s Exact Test; Fig. 2a). Importantly, the accuracy of heart enhancer predictions was found to be independent of the sequence constraint of the tested sequences, with no significant difference in the frequency of positive heart enhancers among the highly conserved sequences (19/31, 61%), compared with sequences overlapping no conservation (16/30, 53%; P > 0.1, Fisher’s Exact Test). These results suggest that p300 is an accurate predictor of enhancer activity independent of sequence conservation, and confirm the in vivo activity of weakly and apparently non-constrained heart enhancers (Fig. 2b).
Figure 2. In vivo testing of p300 heart enhancer predictions.
a) Comparison of the frequency of positive heart enhancers among previously tested sequences predicted on the basis of extreme evolutionary constraint13,14, and sequences predicted by heart p300 ChIP-seq. *, P < 10−55, Fisher’s Exact Test, one-tailed. b) Frequency of positive heart enhancers among tested sequences exhibiting different degrees of evolutionary sequence constraint (highly constrained, score > 450; moderately constrained, score 350–450; weakly constrained score <350; No detectable constraint, no overlapping constrained element). Error bars represent 95% binomial proportion confidence interval. n.s., not significant (P > 0.05, all pair-wise comparisons, Fisher’s Exact Test, two-tailed).
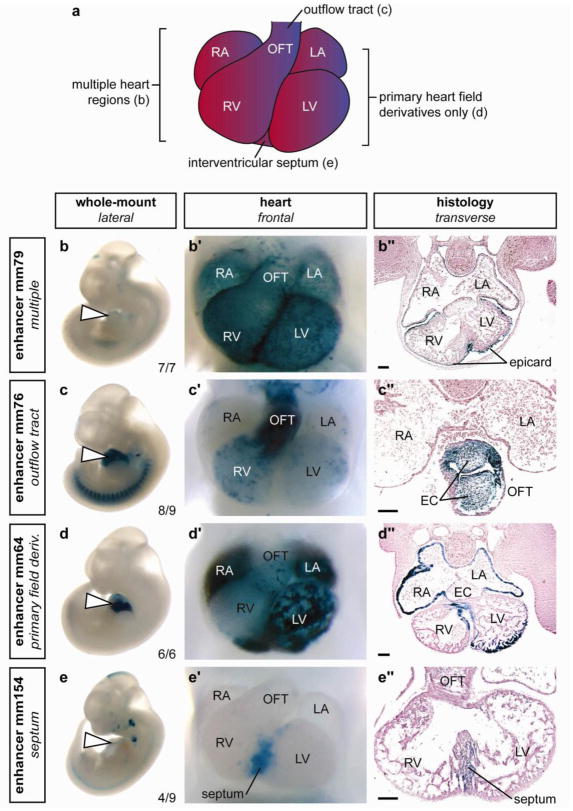
The heart encompasses several anatomical subregions and cell types at e11.5, which include the precursor structures of the definitive functional compartments (atria, ventricles), as well as transient structures and cell populations that have critical functions in heart development and disease24. To examine the spatial diversity of the identified heart enhancers, we annotated reproducible reporter staining patterns within whole-mount stained embryos (Fig. 3b–e, Supplementary Table 7). Essentially all anatomical sub-regions are reproducibly targeted by at least one of the identified in vivo enhancers (Fig. 3a,b′–e′, Supplementary Table 7). To characterize expression patterns in more detail, we examined transverse sections of hearts from representative embryos, which revealed examples of enhancers with activity in each of the major tissue types and discrete lineages within the developing heart (Fig. 3b″–e″, Supplementary Table 7). The observed patterns included regions of the developing heart such as the interventricular septum, which is a common site of structural defects in cases of congenital heart disease2 (Fig. 3d), suggesting the potential for this enhancer identification approach to uncover regulatory regions with relevance to human disease. Both strongly and weakly constrained enhancer sequences exhibited highly reproducible staining in the heart, with no significant correlation between the reproducibility of expression patterns and their respective sequence constraint (Supplementary Fig. 6). Notably, the enhancers identified in these studies exhibited highly restricted expression patterns, with 51/81 (63%) of enhancers driving reporter gene expression exclusively in the developing heart (Supplementary Tables 6 and 7). These data indicate that p300 binding identifies enhancers with activity throughout the developing heart, and with no detectable regional or tissue-specific bias.
Figure 3. Examples of the diverse structural and cell type specificities of p300 ChIP-seq identified cardiac enhancers.
a) Schematic of mouse embryonic day 11.5 heart. b–e) Side views of whole embryos, b′–e′) magnified ventral views of hearts and b″– e″) transverse sections through the heart region are shown for each of four representative X-gal-stained embryos with in vivo enhancer activity at e11.5. Element ID and reproducibility of expression patterns are indicated alongside whole embryo images. b) Enhancer with activity exclusively in epicardium in all anatomical regions of the heart. c) Enhancer with activity primarily in outflow tract endocardium and in all endocardial cushion (EC) mesenchyme. d) Enhancer primarily active in derivatives of the primary heart field (atrial and ventricular myocardium and a small region of the interventricular septum). e) Enhancer with activity predominantly in the muscular portion of the interventricular septum. RA, right atrium; LA, left atrium; RV, right ventricle; LV, left ventricle; OFT, outflow tract; epicard, epicardium. The bars in panels showing transverse sections are equal to 100 μm. The complete in vivo expression dataset is available online40.
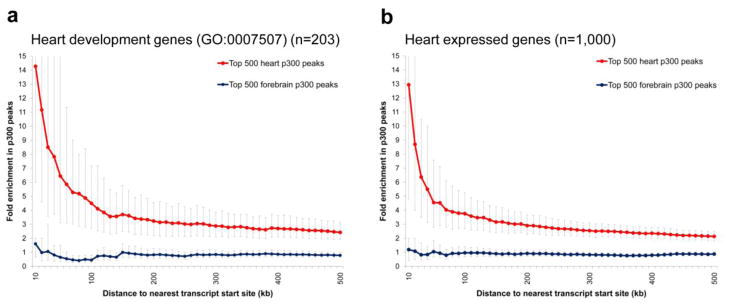
Several of the enhancers validated through our in vivo studies are located near genes with well-described roles in heart development or function (Supplementary Fig. 7), and exhibit activity patterns consistent with the expression of those genes (Supplementary Note). To assess a possible global enrichment of predicted heart enhancers near genes implicated in heart development, we determined the frequency of heart p300 peaks near genes annotated with the gene ontology25 (GO) term ‘heart development’, and near genes that are expressed during heart development26. There is a more than three-fold enrichment in heart p300 peaks within 100 kb of the transcript start sites (excluding the promoter region) of known ‘heart-development’ genes (P < 10−5, Fisher’s Exact Test), increasing to over fourteen-fold enrichment for peaks within 10 kb (P < 10−4, Fisher’s Exact Test, Fig. 4a). Similarly, there is a more than 13-fold enrichment in heart p300 peaks within 10 kb of the transcript start sites of the 1,000 genes most highly expressed in embryonic heart (P < 10−4, Fisher’s Exact Test, Fig. 4b), with enrichment also observed near genes that are specifically over-expressed in the heart compared with the whole embryo (Supplementary Fig. 8). As negative controls, we observed no pronounced enrichment of forebrain p300-binding sites near heart genes (P > 0.1, Fisher’s Exact Test, Fig. 4a,b) and no enrichment of p300-binding sites in other tissues near heart genes (Supplementary Figs. 8–10). In addition to these findings, candidate heart enhancers are enriched in binding sites for transcription factors with known roles in heart gene regulatory networks (Supplementary Fig. 11). Together, these results support a global role for p300-associated candidate heart enhancers in tissue-specific transcriptional activation of neighboring genes during heart development and provide further evidence that a substantial proportion of the heart-specific p300-binding sites identified in this study are bona fide heart enhancers.
Figure 4. Enrichment of heart p300 ChIP-seq peaks near genes implicated in heart development.
Enrichment of heart and forebrain p300 peaks in the proximity of transcript start sites of a) heart development genes (GO:0007507), and b) the 1000 most highly expressed genes in e11.5 mouse heart 26. Fold-enrichment was determined by comparing the observed frequency of peaks up to 500 kb away from transcript start sites to an equal number of randomized positions genome-wide. For each tissue, only the 500 most significant p300 peaks were considered. Similarly specific enrichment of heart peaks near heart genes is observed compared with limb and midbrain p300 data, whereas no enrichment of heart peaks is observed near control gene sets with no known role in heart development or with no expression in e11.5 hearts (see Supplementary Figs. 8–10). Bold lines represent average fold enrichment; error bars indicate confidence intervals (5th- and 95th-percentile, see Methods).
In summary, we have identified and functionally validated a large population of enhancers active in the e11.5 heart, and found them to be under substantially weaker evolutionary constraint than enhancers active in other anatomical regions at the same developmental stage, despite their association with the same transcriptional co-activator protein, p300. While the biological significance of this finding remains to be elucidated, this observation may be considered surprising given the evolutionary antiquity of the heart and the genetic pathways that control its development27.
The question of whether sequence constraint is a general hallmark of gene regulatory elements has important implications for locating their position within the genome, as well as for understanding their function and evolutionary origin28. Cell culture-derived transcription factor binding data, along with comparative genomic analyses of isolated in vivo characterized enhancers have provided first indications that not all functional non-coding sequences are detectably constrained29–33. However, the genomic scale at which such putatively functional non-constrained elements have specific and reproducible in vivo activities has remained elusive because experimental data were available only for selected loci34. Our results suggest that, at least for the time-point studied, existing conservation-based measures underestimate the proportion of the genome that has regulatory functions in vivo. This notion is further supported by a recent study combining evolutionary constraint with sequence motif analysis for genome-wide prediction of heart enhancers, which failed to identify many of the heart enhancers validated in vivo in the present study35 (Supplementary Table 8 and Supplementary Note). These results emphasize the importance of experimental approaches for the unbiased annotation of functional elements in the genome. Based on our transgenic assays, a considerable proportion of the thousands of genomic regions predicted by this study are likely to be true heart enhancers, providing a comprehensive genome-wide set of candidate sequences that will facilitate the exploration of regulatory elements in cardiac development and disease.
Methods
ChIP sequencing from mouse embryonic tissues
Embryonic heart and midbrain tissues were isolated from approximately 270 CD-1 strain embryos at e11.5 respectively by microdissection in cold PBS. Tissue samples were processed for ChIP and DNA sequencing as described previously19. Briefly, tissues were cross-linked in formaldehyde and cells dissociated in a glass douncer. Chromatin isolation, sonication, and immunoprecipitation using an anti-p300 antibody (rabbit polyclonal anti-p300; SC-585, Santa Cruz Biotechnology) were performed as previously described41,42. Approximately 0.1 ng of each ChIP DNA sample was sheared by sonication, end-repaired, ligated to sequencing adapters and amplified by emulsion PCR for 40 cycles43. Amplified ChIP DNA was sequenced for 36 cycles on the Illumina Genome Analyzer II as described previously19. P300 ChIP-seq data from e11.5 mouse forebrain and limb was previously published19, but reanalyzed for this study using the approach outlined below.
Processing of ChIP-sequence data
Unfiltered 36 bp sequence reads were aligned to the mouse reference genome (NCBI build 37, mm9) using BLAT as described previously19. P300-enriched regions were identified using QuEST (version 2.3)20 with parameters bandwidth = 60 bp, region_size = 600 bp, ChIP enrichment = 10, ChIP extension enrichment = 3, and no QuEST_align_RX_noIP file. Peaks with a regional q-value of less than 2 (equivalent to a Poisson-estimated false discovery rate of less than or equal to 1%) were removed. Peaks mapping to unassembled chromosomal contigs, centromeric regions, telomeric regions, segmental duplications, peaks consisting of >70% repeat sequence, peaks coinciding with enriched regions from an e11.5 forebrain control sample (input DNA), and peaks where >20% contributing reads originate in a self–chain alignment were removed as likely artifacts. To exclude likely promoter sequences, we removed all p300-bound regions for which the distance from the peak maximum to the nearest transcript start site (UCSC known genes22) was less than 1 kb. The remaining peaks represented candidate distant-acting enhancers with activity in specific tissues. For consistent methodology and to allow relative ranking of peaks within datasets, we reanalyzed previously generated p300 ChIP-seq data from forebrain and limb19 according to the scheme described above. Peak calling using QuEST was highly consistent with the previously used approach. At least 87% of forebrain or limb p300 peaks identified previously, including all previously in vivo tested sequences overlap a QuEST peak in the new analysis. Conversely, at least 95% of the top 1000 forebrain or limb p300 peaks identified using QuEST were identified in the previous analyses (data not shown).
Computational analyses of candidate distant-acting enhancers
To most accurately identify and compare properties of candidate distant-acting enhancer datasets, computational analyses were initially performed on the top 500 high confidence candidate distant-acting enhancers from each tissue. An equal number of peaks were selected from each tissue to enable statistically straightforward comparisons. For analyses of constraint and conservation, the top 500 candidate distant-acting enhancers were selected from p300 peaks more than 5 kb from the nearest transcript start sites in order to ensure maximum filtering of promoter-proximal regions which are likely to contain conserved functional sequences other than enhancers (e.g. PolII binding sites, unannotated exons). For conservation depth analyses, multiple sequence alignments for the 100 bp region centered on the peak maximum of selected regions were extracted from pre-computed 30-way alignments to the mouse genome22,36. Maximum conservation depth was evaluated from the most distantly related species with at least 50% bases aligned to the mouse reference across this region, and timescales were obtained from current estimates of divergence times among vertebrates37–39. To evaluate the conservation properties of the genome background, we used 100 bp regions centered on the midpoint of 10,000 random regions in the genome with matched size distribution and sequence mappability and subject to the same filtering procedure applied to p300 peaks. For sequence constraint analyses, selected regions were assigned the score of the highest-scoring phastCons element21 overlapping the 1 kb genomic interval centered on the peak maximum. The same approach was used for analyses using either the top 500 scoring or all candidate distant-acting enhancers (including elements up to 1 kb from the nearest transcript start site), and analyses in which phastCons conservation scores were derived from placental mammal or euarchontoglire multiple sequence alignments. Regions with no overlapping phastCons elements are referred to as ‘not detectably constrained’ (Supplementary Fig. 2).
Enrichment of candidate enhancers near genes involved in heart development was determined using the top 500 scoring p300 ChIP-seq peaks greater than 1 kb from the nearest transcript start site from each tissue. For each dataset, peak randomizations were generated by moving each peak to a random location on the same chromosome, excluding regions that are less than 50% mappable (determined by BLAT), or that fail the peak filtering procedure described above. Enrichment of p300 peaks near 203 ‘heart development’ genes (GO:0007507) was calculated as the average, from 1,000 peak randomizations, of the ratio of p300 peaks to randomized peaks at defined distances from the nearest heart gene transcript start site. To determine the specificity of enrichment in the vicinity of heart genes, 1000 control datasets of ‘non-heart genes’ were assembled by randomly selecting 203 genes (UCSC known genes) that are not annotated with the ‘heart development’ GO term. Enrichment near non-heart genes was calculated as the average, from 1000 total randomizations of peaks and control genes, of the ratio of p300 peaks to randomized peaks at defined distances from the nearest non-heart gene transcript start site. For analysis of candidate enhancers near heart-expressed genes, the 1,000 most highly expressed and 1,000 least expressed genes were identified from publicly available e11.5 mouse heart expression data (GEO Series GSE1479, samples GSM25153-GSM2555)26. For analysis of candidate enhancers near heart-specific genes (Supplementary figure 8), heart over- and under-expressed genes were identified by comparison with e11.5 whole embryo expression data19. Heart over-expressed genes are those with an expression level of >100 in e11.5 heart and at least 5-fold higher expression than in whole embryo. Conversely, heart under-expressed genes are those with an expression level of >100 in e11.5 whole embryo and at least 5-fold higher expression in the embryo compared with the heart. Enrichment of p300 peaks near heart-expressed and not expressed gene sets was calculated as the average, from 1,000 peak randomizations, of the ratio of p300 peaks to randomized peaks at defined distances from the nearest transcript start site. The same approach was used for analyses of all p300 peaks from each tissue, and gave similar results (Supplementary Figs. 8c/d and 9c/d).
Transcription factor binding site analysis of the top 500 p300 peaks from heart and forebrain was performed by first dividing each dataset into equally sized bins containing the 250 most conserved and 250 least conserved candidate enhancers respectively. For each dataset a 200 bp region centered on the position of maximum p300 coverage was searched against motifs in the JASPAR database44 of non-redundant vertebrate transcription factor binding using MAST45. Only binding sites that were present in at least 15 (>5%) sequences from at least one dataset were retained for further analysis. Transcription factor binding site counts were then converted to fold enrichment in comparisons between forebrain and heart datasets with the same conservation score.
Transgenic mouse enhancer assay
Regions for in vivo testing were primarily selected to be representative of the sequence constraint properties (vertebrate phastCons scores) of all 3,597 candidate distant-acting heart enhancers identified through p300 binding. Peaks were otherwise selected based on rank peak score, and are moderately biased towards sequences greater than 5 kb from the nearest transcript start site (Supplementary Fig. 3). Enhancer candidate regions consisting of ~2 kb of mouse genomic DNA flanking the p300 peak were amplified by PCR from mouse genomic DNA (Clontech) and cloned into the Hsp68-promoter-LacZ reporter vector as previously described 9,23. Genomic coordinates of amplified regions are reported in Supplementary Table 5. Transgenic mouse embryos were generated by pronuclear injection and F0 embryos were collected at e11.5 and stained for β-galactosidase activity with 5-bromo-4-chloro-3-indolyl -D-galactopyranoside (X-Gal) as previously described9. Only patterns that were observed in at least three different embryos resulting from independent transgenic integration events of the same construct were considered reproducible. For all confirmed reproducible heart enhancers, close-up images of the heart were taken of at least one representative embryo, and expression patterns were classified according to X-Gal staining in broadly defined anatomical regions. Selected elements were subject to sectioning (see Supplementary Table 7). For detailed section analyses, embryos were collected at e11.5, fixed in 4% paraformaldehyde and stained with X-Gal overnight. X-Gal-stained embryos were then embedded in paraffin using standard methods. Transverse sections were cut at a thickness of 8 μm, and sections were counterstained with neutral fast red for visualization of embryonic structures by light microscopy and photographed.
Animal work
All animal work was performed in accordance with protocols reviewed and approved by the Lawrence Berkeley National Laboratory Animal Welfare and Research Committee.
Supplementary Material
Acknowledgments
The authors wish to thank Roya Hosseini and Sengthavy Phouanenavong for technical support; L.A.P. and E.M.R. were supported by grant HL066681, Berkeley-PGA, under the Programs for Genomic Applications, funded by National Heart, Lung, & Blood Institute, and L.A.P. by grant HG003988 funded by National Human Genome Research Institute. B.L.B. was supported by grants HL64658 and HL89707 from the National Heart, Lung, and Blood Institute. B.R. is supported by funding from National Human Genome Research Institute and the Ludwig Institute for Cancer Research. Research was conducted at the E.O. Lawrence Berkeley National Laboratory and performed under Department of Energy Contract DE-AC02-05CH11231, University of California.
Footnotes
Author Contributions
M.J.B, A.V. and L.A.P. wrote the manuscript. All authors contributed to data collection and analysis and provided comments on the manuscript.
The authors declare no competing financial interests.
Data availability
All raw sequences and processed data from p300 ChIP-seq experiments are publicly available from NCBI under accession numbers GSE22549 (heart and midbrain) and GSE13845 (forebrain and limb). Complete in vivo data sets are available from the Vista Enhancer Browser (http://enhancer.lbl.gov/).
References
- 1.Hoffman JI, Kaplan S, Liberthson RR. Prevalence of congenital heart disease. Am Heart J. 2004;147:425–39. doi: 10.1016/j.ahj.2003.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Lloyd-Jones D, et al. Heart disease and stroke statistics--2009 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2009;119:480–6. doi: 10.1161/CIRCULATIONAHA.108.191259. [DOI] [PubMed] [Google Scholar]
- 3.Pierpont ME, et al. Genetic basis for congenital heart defects: current knowledge: a scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation. 2007;115:3015–38. doi: 10.1161/CIRCULATIONAHA.106.183056. [DOI] [PubMed] [Google Scholar]
- 4.Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943–8. doi: 10.1038/nature06801. [DOI] [PubMed] [Google Scholar]
- 5.Bentham J, Bhattacharya S. Genetic mechanisms controlling cardiovascular development. Ann N Y Acad Sci. 2008;1123:10–9. doi: 10.1196/annals.1420.003. [DOI] [PubMed] [Google Scholar]
- 6.Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aparicio S, et al. Detecting conserved regulatory elements with the model genome of the Japanese puffer fish, Fugu rubripes. Proc Natl Acad Sci U S A. 1995;92:1684–8. doi: 10.1073/pnas.92.5.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 10.de la Calle-Mustienes E, et al. A functional survey of the enhancer activity of conserved non-coding sequences from vertebrate Iroquois cluster gene deserts. Genome Res. 2005;15:1061–72. doi: 10.1101/gr.4004805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woolfe A, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prabhakar S, et al. Close sequence comparisons are sufficient to identify human cis-regulatory elements. Genome Res. 2006;16:855–63. doi: 10.1101/gr.4717506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pennacchio LA, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 14.Visel A, et al. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet. 2008;40:158–60. doi: 10.1038/ng.2007.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao TP, et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–72. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 16.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 17.Xi H, et al. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007;3:e136. doi: 10.1371/journal.pgen.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–17. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 19.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–8. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valouev A, et al. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5:829–34. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–50. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn RM, et al. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009;37:D755–61. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kothary R, et al. A transgene containing lacZ inserted into the dystonia locus is expressed in neural tube. Nature. 1988;335:435–7. doi: 10.1038/335435a0. [DOI] [PubMed] [Google Scholar]
- 24.Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–48. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 25.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schinke M. NHLBI Program for Genomic Applications. Harvard Medical School; Nov, 2009. Genomics of Cardiovascular Development, Adaptation, and Remodeling. URL: http://www.cardiogenomics.org. [Google Scholar]
- 27.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–7. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper GM, Brown CD. Qualifying the relationship between sequence conservation and molecular function. Genome Res. 2008;18:201–5. doi: 10.1101/gr.7205808. [DOI] [PubMed] [Google Scholar]
- 29.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Margulies EH, et al. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007;17:760–74. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng Y, et al. Transcriptional enhancement by GATA1-occupied DNA segments is strongly associated with evolutionary constraint on the binding site motif. Genome Res. 2008;18:1896–905. doi: 10.1101/gr.083089.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–12. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King DC, et al. Finding cis-regulatory elements using comparative genomics: some lessons from ENCODE data. Genome Res. 2007;17:775–86. doi: 10.1101/gr.5592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fisher S, Grice EA, Vinton RM, Bessling SL, McCallion AS. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science. 2006;312:276–9. doi: 10.1126/science.1124070. [DOI] [PubMed] [Google Scholar]
- 35.Narlikar L, et al. Genome-wide discovery of human heart enhancers. Genome Res. doi: 10.1101/gr.098657.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blanchette M, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–15. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–21. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bininda-Emonds OR, et al. The delayed rise of present-day mammals. Nature. 2007;446:507–12. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- 39.Hedges SB. The origin and evolution of model organisms. Nat Rev Genet. 2002;3:838–49. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- 40.Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barrera LO, et al. Genome-wide mapping and analysis of active promoters in mouse embryonic stem cells and adult organs. Genome Res. 2008;18:46–59. doi: 10.1101/gr.6654808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Z, et al. A global transcriptional regulatory role for c-Myc in Burkitt’s lymphoma cells. Proc Natl Acad Sci U S A. 2003;100:8164–9. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blow MJ, et al. Identification of ancient remains through genomic sequencing. Genome Res. 2008;18:1347–53. doi: 10.1101/gr.076091.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bryne JC, et al. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–6. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.