Abstract
Endoplasmic reticulum-associated degradation (ERAD) employs membrane-bound ubiquitin ligases and the translocation-driving ATPase p97 to retrotranslocate misfolded proteins for proteasomal degradation. How retrotranslocated polypeptides bearing exposed hydrophobic motifs or transmembrane domains (TMD) avoid aggregation before reaching the proteasome is unclear. Here we identify a ubiquitin ligase-associated multiprotein complex comprising Bag6, Ubl4A, and Trc35, which chaperones retrotranslocated polypeptides en route to the proteasome to improve ERAD efficiency. In vitro, Bag6, the central component of the complex, contains a chaperone-like activity capable of maintaining an aggregation-prone substrate in an unfolded yet soluble state. The physiological importance of this holdase activity is underscored by observations that ERAD substrates accumulate in detergent insoluble aggregates in cells depleted of Bag6, or of Trc35, a cofactor that keeps Bag6 outside the nucleus for engagement in ERAD. Our results reveal a ubiquitin ligase-associated holdase that maintains polypeptide solubility to enhance protein quality control in mammalian cells.
Keywords: Bag6, Ubl4A-Trc35, ERAD/retrotranslocation, chaperone holdase, protein aggregation, proteasome, gp78, ubiquitin ligase
In eukaryotic cells, thousands of secretory and membrane proteins are translocated into the lumen of the endoplasmic reticulum (ER) or integrated into the ER membrane before being delivered to their final destinations. Despite the presence of a large number of ER chaperones that assist nascent polypeptides in folding and assembly, protein misfolding occurs frequently. To deal with this problem, eukaryotic cells have evolved a protein quality control mechanism, through which misfolded ER proteins are selectively retained and subsequently retrotranslocated into the cytosol. Retrotranslocated polypeptides are degraded by the ubiquitin proteasome system (Hirsch et al., 2009; Vembar and Brodsky, 2008). This garbage disposal process, named ER-associated degradation (ERAD) or retrotranslocation, alleviates ER stress to improve cell vitality (Ron and Walter, 2007). Interestingly, certain pathogens can co-opt the ERAD pathway to dispose of native proteins essential for immune defense. For example, the human cytomegalovirus (HCMV) genome encodes two proteins US2 and US11, each of which can downregulate newly synthesized major histocompatibility complex (MHC) class I heavy chain (HC) at the ER membrane (Lilley and Ploegh, 2005). These viral proteins can bind HC and target it to membrane dislocation complexes for retrotranslocation and degradation (Lilley and Ploegh, 2004; Ye et al., 2004).
ERAD employs a variety of chaperones and lectins such as BiP, Os9, EDEM, PDI, which selectively recognize terminally misfolded proteins carrying immature glycans, exposed hydrophobic regions, or unpaired cysteine residues (Buchberger et al., 2011). Misfolded polypeptides are then moved across the ER membrane to enter into the cytosol. The mechanisms underlying protein retrotranslocation are unclear, but existing evidence suggests that retrotranslocation of distinct cohorts of substrates are mediated by different membrane complexes. These complexes often contain one or more multi-spanning transmembrane ubiquitin ligases (E3) and the p97 ATPase. Although not yet proven, the dislocation complexes likely form a few protein conducting channels through which misfolded proteins exit the ER. Once reaching the cytosolic side of the membrane, substrates are polyubiquitinated by the E3s. In mammalian cells, several ERAD-dedicated E3s including Hrd1, gp78, TEB4, Kf-1, RMA1, and TRC8 have been identified (Hirsch et al., 2009). Ubiquitinated ERAD substrates are dislocated from the membrane by p97 and its associated cofactors (Bays et al., 2001; Jarosch et al., 2002; Rabinovich et al., 2002; Ye et al., 2001). p97 subsequently hands substrates off to the proteasome with the assistance of several shuttling factors.
Several lines of evidence indicate that the putative retrotranslocons cannot accommodate tightly folded polypeptides or protein aggregates possibly due to size constraints (Bhamidipati et al., 2005; Nishikawa et al., 2001; Okuda-Shimizu and Hendershot, 2007; Soetandyo et al., 2010). First, fusion of a tightly folded domain to the model ERAD substrate CPY* abolishes its retrotranslocation and degradation (Bhamidipati et al., 2005). Second, for unassembled TCRα chain, two intramembrane charged residues prevent inter-chain disulfide formation, which facilitates TCRα degradation by maintaining it in a retrotranslocation competent monomeric state (Soetandyo et al., 2010). Moreover, the retrotranslocation of non-secreted κ-LC requires the conversion of a partially oxidized precursor into a reduced monomer (Okuda-Shimizu and Hendershot, 2007). Lastly, many ER chaperones with unfolding activities have been identified to mediate retrotranslocation (Bernardi et al., 2008; Lee et al., 2010; Nishikawa et al., 2001; Ushioda et al., 2008). Together, these findings suggest that ERAD substrates are most likely retrotranslocated across the membrane in unfolded forms. Thus, dislocated polypeptides are expected to bear exposed hydrophobic motifs or TMDs prone to aggregation in the aqueous cytosolic environment. These hydrophobic motifs need to be properly shielded before substrates are safely targeted to the proteasome. Otherwise, substrate aggregation could prematurely terminate the degradation process. Here, we report a chaperone holdase complex that maintains the solubility of retrotranslocated polypeptides to promote ER protein quality control in mammalian cells.
Results
The Bag6 complex interacts with ERAD E3s
To identify factors that facilitate ERAD, we expressed in 293T cells the ER-associated ubiquitin ligase gp78, which is responsible for the degradation of many ERAD substrates (Fang et al., 2001; Shen et al., 2006; Song et al., 2005). We performed affinity purification to isolate gp78 and its associated proteins. The Coomassie-stained gel revealed several gp78-associated proteins (Fig. 1A). Peptide sequencing by mass spectrometry and immunoblotting identified many of them as known ERAD factors including p97, Npl4, Ube2g2, UbxD8, and Herp (Alexandru et al., 2008; Chen et al., 2006; Li et al., 2007; Schulze et al., 2005; Zhong et al., 2004). Interestingly, a ~150kD protein was identified as Bag6, a gp78 partner with no previously documented function in ERAD (Fig. S1A).
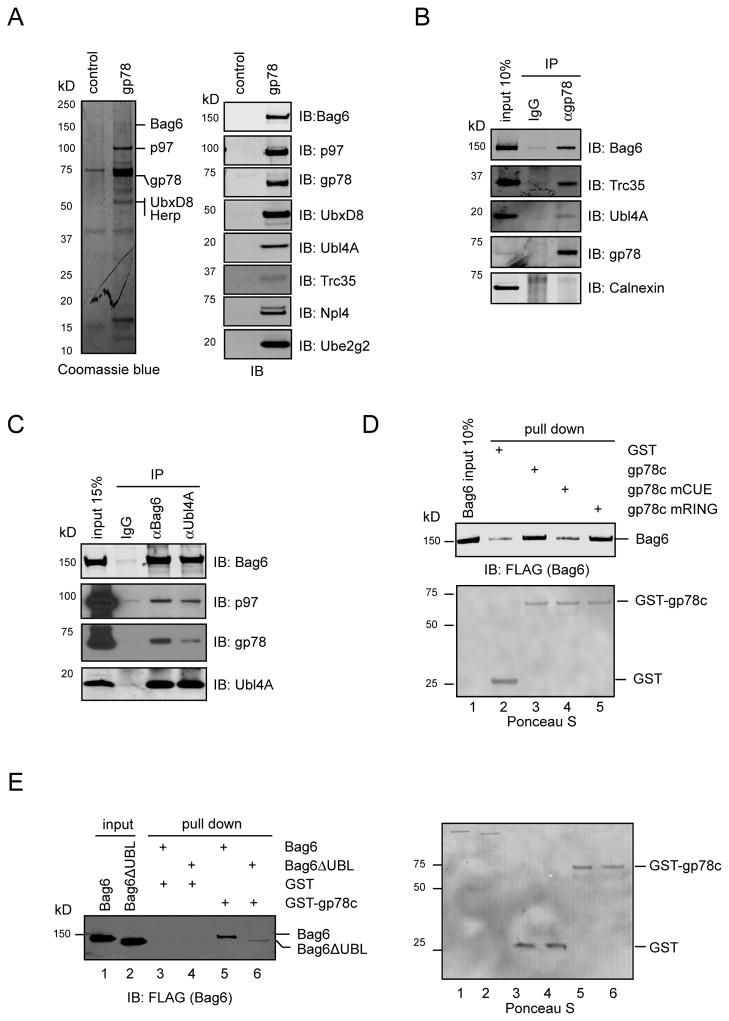
Figure 1. The Bag6 complex interacts with the gp78-p97 retrotranslocation complex.
A, Identification of the Bag6 complex as an interaction partner of gp78. Cell extracts from 293T cells transfected with FLAG-tagged gp78 or a control plasmid were subject to immunoprecipitation (IP) with FLAG antibody. IB, immunoblotting B, C, Endogenous interactions between the Bag6 and the gp78-p97 complexes. 293T cell extract was subject to IP analyses using the indicated antibodies. D, Bag6 directly binds the cytosolic domain of gp78 via the CUE domain. The indicated GST-tagged proteins were immobilized on glutathione beads and incubated with purified Bag6. E, The UBL domain in Bag6 is required for gp78 interaction. Purified Bag6 or a Bag6 variant lacking UBL was incubated with glutathione beads containing GST-gp78c or GST. The right panel shows the Ponceau S-stained membrane used for immunoblotting (left panel). See also Figure S1
Bag6 was recently identified as the central component of a stable three-protein complex that also contains Ubl4A and Trc35, which promotes membrane targeting of ER tail-anchored (TA) proteins (Leznicki et al., 2010; Mariappan et al., 2010). Using Bag6, Ubl4A and Trc35 specific antibodies, we confirmed that these proteins all associated with gp78-FLAG (Fig. 1A). Moreover, each component of the Bag6 complex could be co-precipitated with endogenous gp78 by a gp78 antibody, whereas the ER lectin calnexin failed to bind gp78 (Fig. 1B). Reciprocal immunoprecipitation showed that both Bag6 and Ubl4A antibodies precipitated the Bag6 complex together with endogenous gp78 and p97 (Fig. 1C). These results demonstrate an interaction between the Bag6 complex and the gp78-p97 retrotranslocation complex in the cell.
To further define the interaction between the Bag6 complex and gp78, we performed a series of pulldown experiments using purified proteins. A direct interaction between gp78 and Bag6 could be established using full length Bag6 and a GST-tagged cytosolic domain of gp78 (gp78c) (Fig. 1D, lane 3 versus lane 2). A ligase-defective gp78c mutant (mRING) also interacted with Bag6 (lane 5). However, a gp78c CUE mutant defective in ubiquitin recognition (Chen et al., 2006) failed to bind Bag6 (lane 4). On the Bag6 side, deletion of its N-terminal ubiquitin-like (UBL) domain dramatically impaired the interaction with gp78, albeit not completely abolishing it (Fig. 1E, lane 6 versus lane 5; Fig. S1B). Thus, Bag6 appears to have two binding sites for gp78 with the one localized in the UBL domain being recognized by the gp78 CUE domain. Hrd1, another ERAD specific E3 also interacted with Bag6 (Fig. S1C). Hrd1 does not contain a CUE domain. Accordingly, the Hrd1-Bag6 interaction was independent of the Bag6 UBL domain (Fig. S1D).
Trc35 retains Bag6 in the cytosol
We next characterized the subcellular localization of the Bag6 complex. Subcellular fractionation and immunoblotting showed that the Bag6 complex was present in both cytosol and an ER-containing membrane fraction (Fig. S2). This observation was intriguing because Bag6 was predicted to be a nuclear protein with a nuclear localization signal (NLS) (Desmots et al., 2005; Minami et al., 2010; Nguyen et al., 2008). We postulated that a cytosolically localized interacting partner might maintain a cytosolic pool of Bag6, which could associate with ERAD specific ubiquitin ligases. Indeed, we found by a series of immunostaining experiments that Trc35, but not Ubl4A could mask Bag6 NLS to retain it in the cytoplasm. Our data showed that endogenous Bag6 was present in both nucleus and cytoplasm (Fig. 2A, panels 1–3), whereas overexpressed Bag6 was mostly localized to the nucleus (Fig. 2B, panel 1) likely because of insufficient Trc35. Consistent with this interpretation, overexpression of Trc35, which was a cytosolically localized protein by itself (Fig. 2B, panel 2), almost completely eliminated the nuclear localization of both endogenous and overexpressed Bag6, resulting in a cytosolic staining pattern for Bag6 (Fig. 2A, panels 4–6; Fig. 2B, panels 7–9). By contrast, Ubl4A itself was localized in both cytosol and nucleus (Fig. 2B, panel 3), and co-expression of Bag6 with Ubl4A enriched both proteins in the nucleus (Fig. 2B, panels 4–6). Bag6 apparently can carry Ubl4A into the nucleus when endogenous Trc35 is insufficient to keep overexpressed Bag6 in the cytosol.
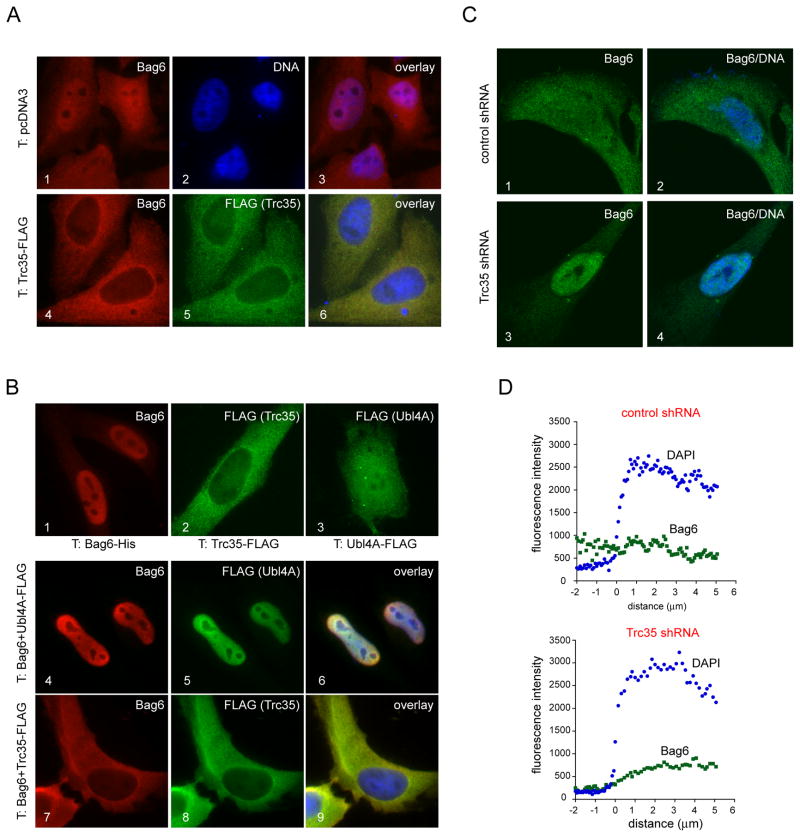
Figure 2. Trc35 maintains a cytosolic pool of Bag6.
A, Trc35 expression alters the subcellular localization of endogenous Bag6. HeLa cells transfected (T) with a control vector (panels 1–3) or a Trc35-FLAG expressing plasmid were stained with anti-Bag6 (red) and -FLAG (green) antibodies. Where indicated, DNA was stained with DAPI (blue). B, Trc35, but not Ubl4A, retains overexpressed Bag6 in the cytoplasm. HeLa cells transfected with the indicated plasmids either individually (panels 1–3) or in combination (panels 4–9) were stained to visualize Bag6 (red), FLAG-tagged proteins (green), and DNA (blue). C, D, Knockdown of Trc35 increases nuclear localization of Bag6. HeLa cells transfected with either a control or Trc35 shRNA construct were stained with a Bag6 antibody (red) and DAPI (blue). Shown are representative cells imaged by a confocal microscope. The graphs in D show the profiles of averaged Bag6 (green) and DAPI (blue) fluorescence intensity across the nuclear envelope boundary (indicated by position 0) measured by a confocal microscope (n=10 cells). See also Figure S2
We also examined the effect of Trc35 knockdown on Bag6 localization. Immunostaining with a Bag6 antibody showed that in control cells Bag6 was localized both in the cytosol and in the nucleus (Fig. 2C, panels 1, 2). Confocal microscopy analyses showed on average no significant change in fluorescence intensity across the nuclear envelop boundary (Fig. 2C, D). In contrast, ~60% of the cells treated with a Trc35 shRNA construct displayed an apparent enrichment of Bag6 in the nucleus (Fig. 2C, panels 3, 4; Fig. 2D). These data further support the notion that Trc35 is required for maintaining a cytosolic pool of Bag6.
The Bag6 complex interacts with TCRα
To test the possible involvement of the Bag6 complex in ERAD, we assessed its association with TCRα, a model ERAD substrate whose degradation requires gp78 (Chen et al., 2006). Because substrate binding to ERAD machinery factors occurs in a dynamic fashion, we treated cells stably expressing YFP-tagged TCRα with the proteasome inhibitor MG132 to halt the constant flow of substrate to the proteasome. As expected, when TCRα was immunoprecipitated, several known ERAD factors including p97, gp78, and the proteasome could be detected in the precipitated TCRα complex in a MG132 dependent manner (Fig. S3A). The Bag6 complex also interacted with TCRα in a similar manner (Fig. 3A; Fig. S3A). The interaction of Bag6 with TCRα must occur in the cell because a similar co-immunoprecipitation experiment performed with MG132-treated cell extract containing exogenously added recombinant Bag6 failed to detect significant interactions between TCRα and the exogenous Bag6 (Fig. S3B). Interestingly, when p97 level was knocked down, interactions of TCRα with Bag6 and the proteasome were reduced (Fig. 3B), indicating that like the proteasome, Bag6 encountered TCRα following its retrotranslocation by p97. The Bag6-TCRα complex was stable in both NP40 and RIPA lysis buffer, although the latter contained the strong detergent sodium dodecyl sulfate (SDS), which disrupted protein interactions, as revealed by the significant loss of TCRα interaction with p97 (Fig. 3C). Together, these results indicate that Bag6 may directly interact with TCRα during retrotranslocation in the cell.
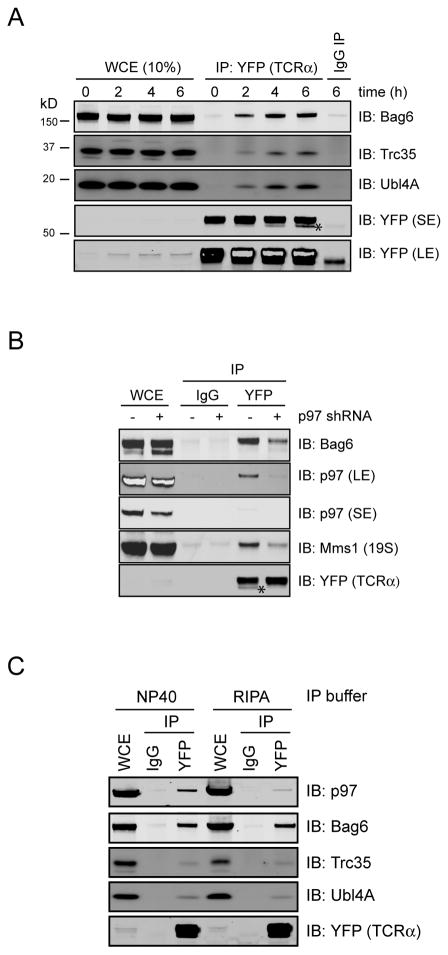
Figure 3. The Bag6 complex interacts with the retrotranslocation substrate TCRα.
A, 293T cells stably expressing YFP-tagged TCRα were treated with MG132 (20μM) for the indicated time periods. TCRα-YFP-containing complexes were immunoprecipitated from NP40 lysis buffer-generated cell extracts and analyzed by immunoblotting. Where indicated, a fraction of the whole cell extract (WCE) was analyzed directly by immunoblotting. Asterisk indicates a deglycosylated TCRα species. LE, long exposure; SE, short exposure. B, As in A, except that siRNA transfected cells were treated with MG132 for 6h. C, The Bag6-TCRα complex is stable in the RIPA buffer. 293T cells expressing YFP-tagged TCRα were treated with MG132 (20μM) for 4h. Cells were lysed in either a NP40-containing lysis buffer or the RIPA buffer. Cell extracts were subject to immunoprecipitation and immunoblotting by the indicated antibodies. See also Figure S3
The Bag6 complex facilitates ERAD
To determine whether the Bag6 complex functions in ERAD, we first tested whether the degradation of TCRα was impaired in Bag6 knockdown cells. When the level of TCRα was measured by flow cytometry and immunoblotting, it was indeed increased in Bag6 knockdown cell (Fig. 4A). Knockdown of Bag6 also increased the steady state level of another ERAD substrate CD4 in cells expressing the HIV protein Vpu (Fig. 4B) as well as a TCRα variant lacking TMD (see below). By contrast, the degradation of a cytosolic proteasome substrate (Ub-V-GFP) was largely unaffected by Bag6 knockdown (Fig. 4C).
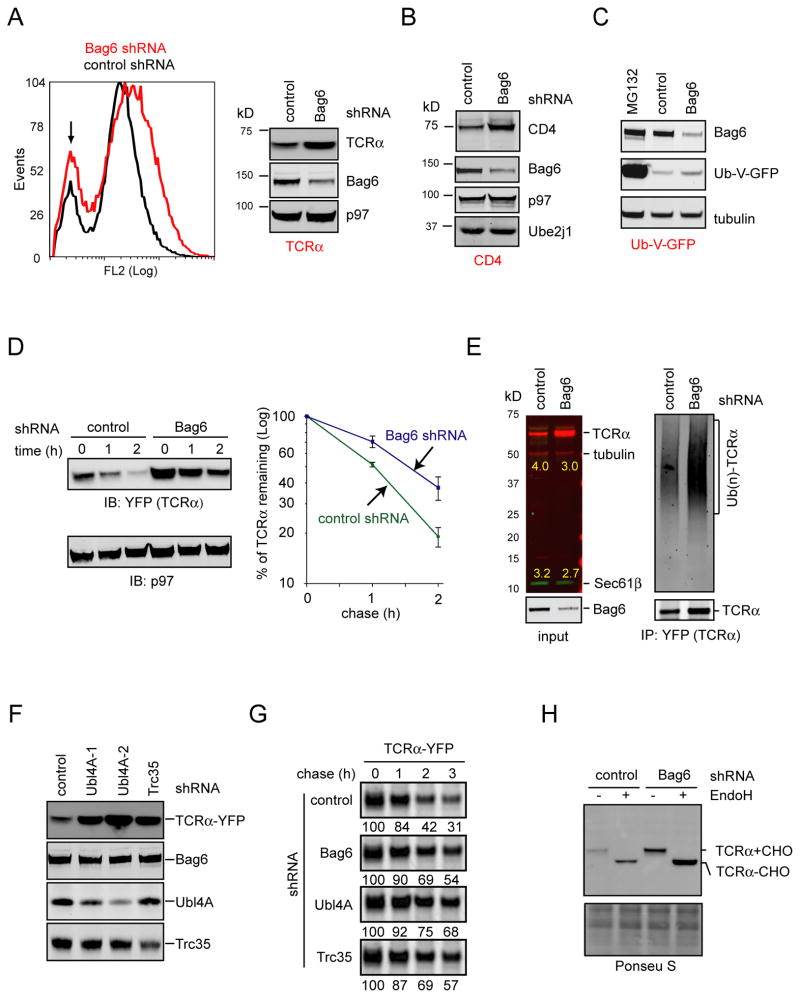
Figure 4. The Bag6 complex is required for efficient degradation of misfolded ER proteins.
A, Knockdown of Bag6 stabilized TCRα. TCRα-YFP expressing cells were transfected with the indicated shRNA constructs. TCRα-YFP levels in cells were determined by flow cytometry and immunoblotting. A small population of cells does not express TCRα-YFP, as indicated by the arrow. Unless specified, cell extracts were prepared using a NP40-containing lysis buffer. B, Cells expressing CD4 and Vpu were treated with the indicated shRNA. Whole cell extracts were analyzed by immunoblotting with the indicated antibodies. C, As in B, except that cells expressing Ub-V-GFP were used. Where indicated, cells were treated with MG132 (20μM) for 12h. D, Cells expressing TCRα-YFP and the indicated shRNAs were subject to cycloheximide chase analyses to determine the half life of TCRα. The graph shows the quantification results from 3 independent experiments. Error bars, S.D. (n=3). E, TCRα accumulated in Bag6 knockdown cells in polyubiquitinated form. Where indicated, TCRα was immunoprecipitated under a denaturing condition and blotted for the indicated proteins. F, Knockdown of Ubl4A or Trc35 caused accumulation of TCRα in cells. Detergent extracts of TCRα-YFP expressing cells transfected with the indicated shRNA constructs were analyzed by immunoblotting. G, Pulse chase analysis of TCRα degradation in 293T cells expressing the indicated knockdown shRNAs together with TCRα. The numbers indicate the relative levels of TCRα. H, Whole cell extracts prepared from TCRα-YFP expressing cells transfected with the indicated shRNA constructs were treated with Endo H. See also Figure S4
When cycloheximide chase was used to compare the half life of TCRα in control and Bag6 knockdown cells, we repeatedly found a moderate increase in the half life of TCRα in Bag6 knockdown cells (Fig. 4D). Thus, the accumulation of TCRα in Bag6 knockdown cells can be at least in part attributed to increased protein stability. In agreement with this notion, polyubiquitinated TCRα was accumulated in Bag6 knockdown cells (Fig. 4E). Stabilization of TCRα was also seen when Ubl4A and Trc35 were individually knocked down (Fig. 4F, G; Fig. S4A), suggesting that Bag6 acts in conjunction with these two cofactors to increase ERAD efficiency.
The ERAD defects associated with depletion of the Bag6 complex cannot be simply attributed to the previously proposed chaperoning function of Bag6 in biosynthesis of ER TA proteins such as the translocon component Sec61β or the ER-associated ubiquitin E2 enzyme Ube2j1 for the reasons outlined below. First, although the Bag6 complex can increase the efficiency and fidelity of membrane targeting for some TA proteins in vitro, an essential role in TA protein biogenesis in intact cells for Bag6 has not been demonstrated (Leznicki et al., 2010; Mariappan et al., 2010). In fact, yeast strains lacking ether GET4 or GET5, the functional homologues of Trc35 and Ubl4A, respectively, are viable, whereas deletion of single ER TA proteins such as BOS1 can cause lethality. This strongly suggests that the Bag6 complex and its yeast functional ortholog are not absolutely essential for TA protein membrane targeting in the cell, as suggested previously (Mariappan et al., 2010). Consistent with this view, we found that TCRα accumulated in Bag6 knockdown cells co-migrated with TCRα from control cells on a SDS-PAGE gel. This type I membrane protein apparently still carried N-glycan, which was confirmed by Endo H treatment (Fig. 4H). Likewise, Endo H treatment also caused a shift in molecular weight of polyubiquitinated TCRα isolated from Bag6 knockdown cells (Fig. S4B). Thus, TCRα accumulated in Bag6 knockdown cells must be originated from the ER. This scenario would not occur if knockdown of Bag6 had impaired membrane insertion of Sec61β. Second, neither the protein levels nor the solubility of Sec61β and Ube2j1 was affected by depletion of Bag6 or Ubl4A, even though ERAD defect under the same condition was manifest (Fig. 4B, E; Fig. S4A). These TA proteins are apparently targeted to the ER membranes in Bag6 knockdown cells because otherwise they would be rapidly degraded by the proteasome due to the presence of the exposed TMD in the cytosol (Hessa et al., 2011). We therefore conclude that TA proteins biogenesis is largely unaffected in cells containing reduced levels of the Bag6 complex. Perhaps, a redundant mechanism(s) exists in the cell to ensure normal levels of TA protein biosynthesis in the absence of Bag6 (see below). Thus, the most plausible interpretation of our data is that the Bag6 complex has a direct function in ERAD independent of its role in TA protein biogenesis.
Bag6 maintains the solubility of ERAD substrates
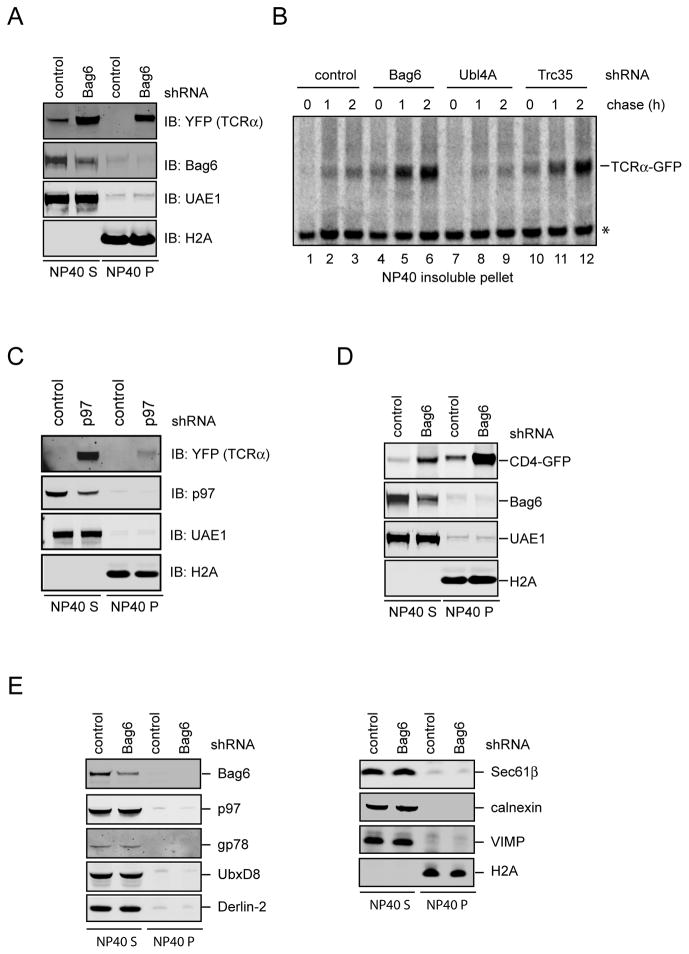
When Bag6 knockdown cells were examined by fluorescence microscopy, we noticed that the TCRα-YFP signal often displayed an uneven pattern with many bright speckles, indicative of protein aggregation (Fig. S5A). Indeed, sequential extraction experiments showed that the NP40 insoluble pellet derived from Bag6 knockdown cells, but not from control cells, contained a large amount of TCRα, which could be resolubilized by the Laemmli buffer (Fig. 5A). Pulse chase experiment confirmed a time dependent accumulation of TCRα in an NP40 insoluble form in Bag6 knockdown cells, which was not observed in control cells (Fig. 5B, lanes 4–6 versus lanes 1–3). The TCRα aggregation phenotype was specifically associated with Bag6 depletion because knockdown of the dislocation driving ATPase p97 caused TCRα to accumulate primarily in NP40 soluble fractions (Fig. 5C). In addition to TCRα, Bag6 depletion also caused other ERAD substrates including CD4 (Magadan et al., 2010) and a TCRα variant lacking TMD (Soetandyo et al., 2010) to accumulate in NP40-insoluble fractions (Fig. 5D; Fig. S5B). By contrast, under the same Bag6 depletion condition, neither the ERAD machinery proteins such as gp78, p97, UbxD8, Derlin-2, VIMP, nor the ER resident proteins Sec61β and calnexin, accumulated in NP40 insoluble fractions (Fig. 5E). Together, these results suggest that Bag6 contains a chaperone-like activity that acts on ERAD substrates to maintain their solubility, which improves their degradation efficiency.
Figure 5. The Bag6 complex maintains the solubility of ERAD substrates to facilitate their turnover.
A, TCRα accumulates in a NP40-insoluble state in Bag6 knockdown cells. Cells expressing TCRα-YFP and the indicated shRNAs were first extracted by the NP40 lysis buffer (NP40 S). The NP40 insoluble pellet fractions were resolubilized in the Laemmli buffer (NP40 P). B, Knockdown of the individual component of the Bag6 complex differentially affects the solubility of TCRα. 293T cells expressing TCRα-YFP and the indicated shRNAs were pulse labeled with 35S-Met/Cys and then incubated in a medium containing excess unlabeled Met and Cys. The NP40 insoluble materials from the cells were resolubilized by a buffer containing 2% SDS and 5mM DTT followed by dilution into the NP40 buffer before immunoprecipitation analyses. C, As in A, except that cells transfected with a shRNA construct targeting p97 were analyzed. D, As in A, except that cells expressing CD4-GFP and Vpu were used. E, As in A, except that samples were immunoblotted with antibodies against the indicated proteins. See also Figure S5
We next tested whether the Bag6-interacting proteins Trc35 and Ubl4A were required for the chaperone-like activity of Bag6 in the cell. Pulse chase analyses demonstrated that TCRα-YFP also formed NP40-insoluble aggregates in Trc35 knockdown cells, and the kinetics of TCRα aggregation in Trc35 knockdown cells was comparable to that in Bag6-depleted cells (Fig. 5B). This result was anticipated given the established role of Trc35 in maintaining a cytosolic pool of Bag6 for engagement in ERAD (Fig. 2). Intriguingly, knockdown of Ubl4A did not cause TCRα to accumulate in NP40 insoluble fractions (Fig. 5B), despite that its depletion had a strong inhibitory effect on ERAD (Fig. 4G; Fig. S4A). Thus, Ubl4A is dispensable for the chaperone function of Bag6, although its activity is strictly required for ERAD.
Bag6 contains a chaperone holdase activity
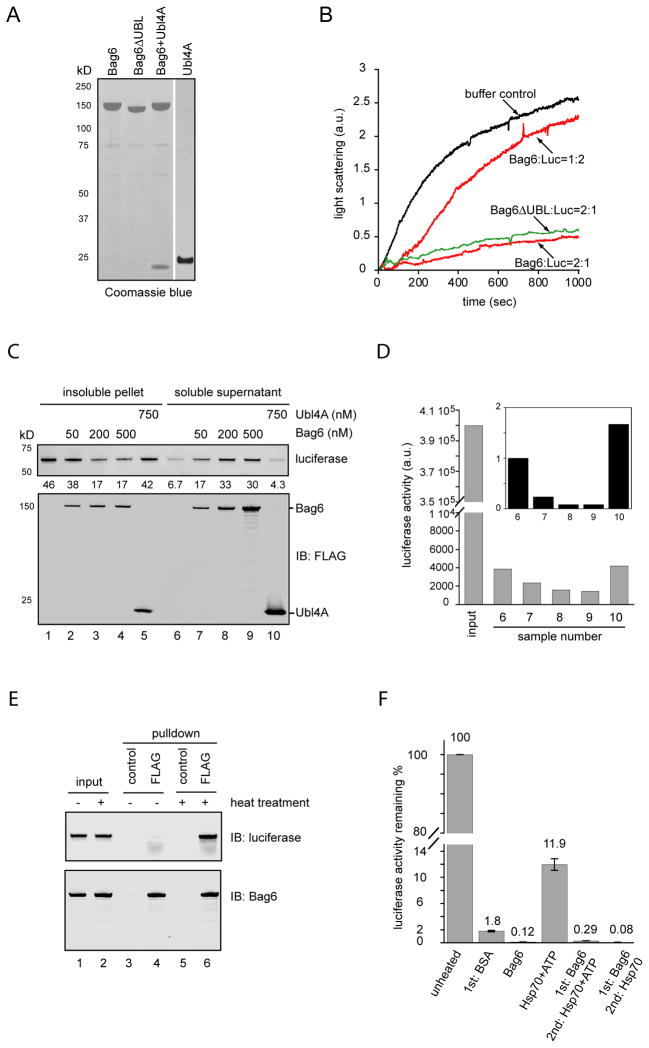
To understand the nature of the Bag6 chaperone activity, we tested whether purified recombinant Bag6 could prevent protein aggregation in vitro. We used luciferase as a model substrate because its thermodynamic instability caused it to aggregate at 42ºC. To eliminate potential confounding effects from factors co-purified with Bag6, particularly the previously reported Bag6-binding protein Hsp70 (Thress et al., 2001), we purified Bag6 from 293T cells to more than 90% homogeneity under a high salt condition (Fig. 6A). Immunoblotting showed no significant contamination of Hsp70 proteins in the purified Bag6 sample (Fig. S6A). Light scattering experiments demonstrated a concentration dependent suppression of luciferase aggregation by Bag6 (Fig. 6B). This activity did not require the Bag6 UBL domain because an UBL-deleted Bag6 mutant had a similar activity as wild type Bag6. Moreover, ATP was not required since the reactions did not contain any nucleotides. We also tested purified Ubl4A and found that it did not suppress luciferase aggregation. On the other hand, a purified Bag6-Ubl4A complex had a similar activity as Bag6 alone (Fig. S6B). We were unable to test Trc35 in this assay because we could not purify Trc35 due to poor expression and solubility. Interestingly, when bovine serum albumin (BSA) was used as a negative control, it caused a small amount of luciferase to remain in soluble fractions after heating when compared to the buffer control (Fig. S6C). The mechanism of this observation was unclear, but light scattering experiments showed that the effect BSA on luciferase aggregation was insignificant compared to that of Bag6 (Fig. S6D).
Figure 6. Bag6 has a chaperone holdase activity that maintains an aggregation-prone substrate in an unfolded soluble state.
A, A Coomassie blue-stained gel shows the purified proteins. B, Bag6 suppresses luciferase aggregation. Luciferase (150nM) was incubated with either buffer or the indicated amount of purified Bag6 or a UBL domain-deleted Bag6 mutant (ΔUBL) at 42 ºC. The scattered light at 330nm was measured by a spectrometer. C, Luciferase (150nM) was incubated with either buffer or the indicated amount of purified proteins at 42 ºC for 20min. Luciferase aggregates were sedimented by centrifugation. The pellet and supernatant fractions were analyzed by immunoblotting. The numbers show the relative luciferase level. D, The luciferase activity in the supernatant fractions from C was determined. Input shows the activity of an equal amount of native luciferase. The inset shows the relative luciferase activity normalized by protein levels determined in C. E, Bag6 binds denatured luciferase. Luciferase was incubated with Bag6 at a 1:2 molar ratio either on ice (−) or at 42 ºC (+) for 20min. Soluble fractions were subject to immunoprecipitation with FLAG antibody to pull down Bag6. F, Luciferase captured by Bag6 cannot be refolded by Hsp70. Luciferase (80nM) was heat inactivated in the presence of BSA (160nM), Bag6 (160nM) or a Hsp70 chaperone mixture containing Hsp70 (150nM), HOP (200nM), Hdj1 (300nM) and ATP 5mM. After denaturation, samples were incubated at 25 ºC for 30min. Where indicated, the same Hsp70 chaperon mixture was added with or without ATP during the second incubation. Error bars indicate the mean of two independent experiments. See also Figure S6
Bag6 may suppress luciferase aggregation by promoting luciferase refolding or by sequestering it in an unfolded soluble state. To discriminate between these possibilities, we monitored the folding status of luciferase by measuring the luciferase activity after heat denaturation. If Bag6 promoted luciferase refolding, one would expect to detect more luciferase activity in the soluble fractions when Bag6 was present. As expected, incubation of luciferase at 42ºC reduced its activity by 100-fold (Fig. 6D, sample 6 versus input). The remaining luciferase activity likely resulted from spontaneous refolding after denaturation. Remarkably, despite that more luciferase was present in the Bag6-containing supernatant fractions (Fig. S6C; Fig. 6C), lesser activity was detected (Fig. 6D, samples 7–9 versus samples 6, 10). We thus conclude that denatured luciferase in Bag6-containing samples remains unfolded and must be still bound by Bag6. Indeed, an interaction between heat-denatured luciferase and Bag6 could be readily detected by co-immunoprecipitation, whereas native luciferase failed to bind Bag6 (Fig. 6E). Because incubation of the luciferase-Bag6 complex with an Hsp70 folding system comprising purified Hdj1, HOP, and Hsp70 did not lead to significant luciferase refolding (Fig. 6F), luciferase captured by Bag6 apparently could not be spontaneously released. Intriguingly, when Bag6 and Hsp70 were present at the same time during luciferase denaturation, substoichiometric amount of Bag6 (the ratio of Bag6: Hsp70 at 1:8) could inhibit the refolding activity of Hsp70 by ~80% (Fig. S6E). This is most likely due to competition of Bag6 and Hsp70 for substrate binding. Taken together, our results suggest that Bag6 does not act as a conventional chaperone that assists proteins in folding. Instead, it functions as a molecular holdase that binds denatured polypeptides to sequester them in an unfolded yet soluble state.
Bag6 is involved in US11-induced ERAD
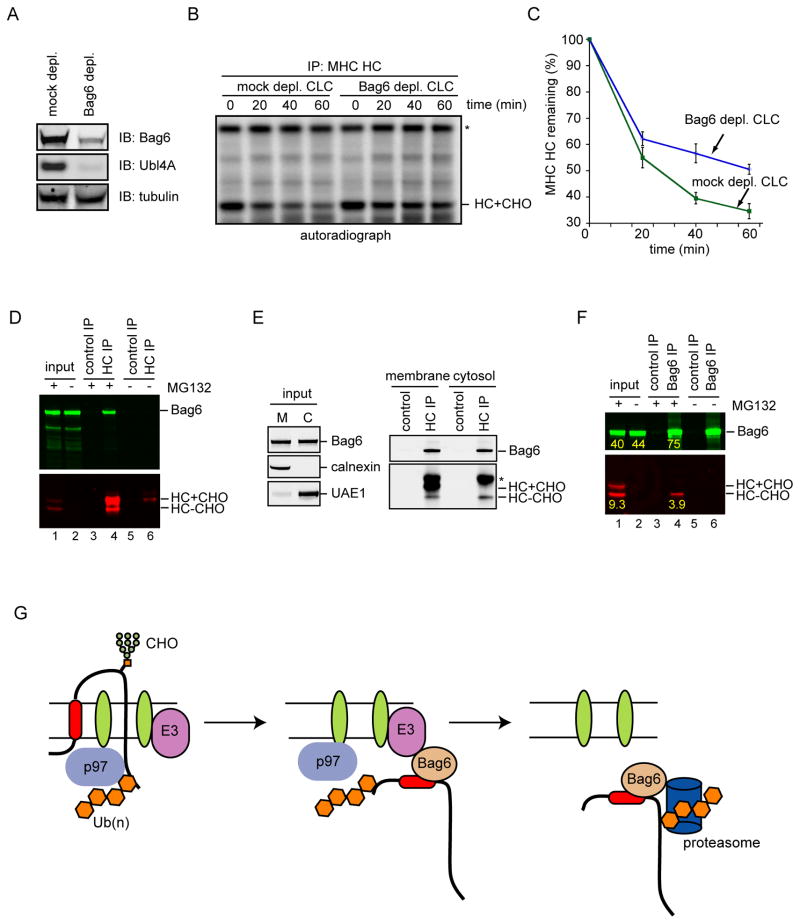
To further demonstrate that the Bag6 complex is directly involved in ERAD, we tested whether the Bag6 complex was required for the degradation of MHC class I heavy chain (HC) in permeabilized A9 cells. A9 cells stably expressed the HCMV protein US11 and a hemagglutinin (HA)-tagged HC (HLA-A2). Under the influence of US11, newly synthesized HC is constantly degraded by the ERAD pathway (Shamu et al., 1999). We and others have previously established an in vitro permeabilized cell assay that recapitulates HC degradation in vivo (Shamu et al., 1999; Soetandyo and Ye, 2010; Ye et al., 2001). The assay used a low concentration of the detergent digitonin to permeabilize the plasma membrane of the cells that contain radiolabeled nascent HC. This allows efficient removal of the cytosol by centrifugation. The degradation of HC can be reconstituted by incubating the membrane fraction with cytosol of a defined source in the presence of ATP and ubiquitin (Fig. S7A). To test the involvement of the Bag6 complex in this process, we treated permeabilized A9 cells with high salt, which removed most membrane bound Bag6 complex (data not shown). We then incubated the membrane with either mock depleted cow liver cytosol (CLC) or CLC depleted of the Bag6 complex using an Ubl4A antibody (Fig. 7A). HC was rapidly degraded when A9 cell-derived membranes were incubated with control CLC. In contrast, HC degradation was significantly attenuated when Bag6-depleted CLC was used (Fig. 7B, C).
Figure 7. Bag6 is involved in US11-induced ERAD of MHC class I HC.
A, Depletion of the Bag6 complex from CLC was confirmed by immunoblotting with antibodies against Bag6 and Ubl4A. B, Depletion of the Bag6 complex attenuates the degradation of HC in permeabilized A9 cells. Experiments were performed as outlined in Fig. S7A. Asterisk indicates a non-specific band. C, Quantification of HC turnover from 3 independent experiments. Error bars, S.D. (n=3). D, Bag6 interacts with HC in US11 cells. HC+CHO and HC-CHO indicate glycosylated and deglycosylated HC, respectively. E, Bag6 binds HC both at the ER membrane and in the cytosol. MG132-treated A9 cells were permeabilized by the detergent digitonin. Cells were then fractionated into a membrane (M) and a cytosol (C) fraction before immunoprecipitation and immunoblotting analysis. Asterisk indicates IgG. F, Bag6 specifically interacts with deglycosylated HC. As in D, except that immunoprecipitation was performed with anti-Bag6 antibody. The number shows the fluorescence intensity of the indicated bands. G, A model for the role of the Bag6 complex in ERAD. Association of Bag6 with ER-bound E3s positions it in proximity to substrates undergoing retrotranslocation, which allows Bag6 to efficiently capture dislocated substrates bearing aggregation prone hydrophobic motifs or TMD. Bag6 may escort substrates to the proteasome to enhance degradation efficiency. See also Figure S7
Because we could not purify Trc35 and perform an add-back experiment to rectify the ERAD defect associated with depletion of the Bag6 complex, we further examined the role of Bag6 in US11-induced ERAD by analyzing its interaction with HC. Similar to TCRα, a significant fraction of Bag6 could be co-precipitated with HC in proteasome-inhibited cells (Fig. 7D). Importantly, proteasome inhibition caused the accumulation of a deglycosylated HC species (Fig. 7D) that comprised a membrane-bound retrotranslocation intermediate and a fully dislocated species in the cytosol (Fig. 7E). Fractionation experiments showed that Bag6 interacted with HC both in the membrane and in the cytosol (Fig. 7E). Because only deglycosylated HC specifically co-precipitated with Bag6 (Fig. 7F, Fig. S7B), Bag6 must bind a HC dislocation intermediate at the ER membrane and accompany it into the cytosol for proteasome degradation. Quantification of the binding experiment showed that ~25% of the dislocated HC was captured by Bag6 (Fig. 7F). This was unlikely caused by dissociation of HC from Bag6 during immunoprecipitation given the strong interaction of Bag6 with substrates (Fig. 3C; Fig. 6F). Thus, another cytosolic holdase(s) may act in parallel with Bag6 to capture dislocated HC. Collectively, the ERAD defect associated with Bag6 depletion and the physical interaction of retrotranslocated HC with Bag6 strongly suggest that Bag6 is directly involved in US11-induced retrotranslocation.
Discussion
A chaperoning role for Bag6 in ERAD
In this study, we establish the Bag6-Ubl4A-Trc35 complex as a key mediator in a previously unknown chaperoning step in ERAD. We show that the ERAD substrate TCRα accumulates in polyubiquitinated form in Bag6 depleted cells. Moreover, since the association of Bag6 with TCRα is reduced upon p97 depletion, Bag6 likely acts downstream of p97-mediated dislocation. Our characterization of Bag6 interaction with the retrotranslocation substrate HC further supports this conclusion. As a type I membrane protein carrying a single N-glycan in its ER luminal domain, HC is retrotranslocated in glycosylated form. Once emerged from a putative retrotranslocon, deglycosylation occurs rapidly by a cytosolic N-glycanase. The deglycosylated HC is then released into the cytosol for degradation by the proteasome (Blom et al., 2004; Shamu et al., 1999). Unlike many other ERAD substrates, retrotranslocation and degradation of HC can be uncoupled (Wiertz et al., 1996). When the proteasome function is impaired, HC accumulates in the cytosol as a fully dislocated degradation intermediate lacking N-glycan. We show that Bag6 preferentially interacts with deglycosylated HC both on the membrane and in the cytosol. Therefore, the initial substrate binding by Bag6 likely occurs on the membrane while HC is undergoing retrotranslocation and deglycosylation. Bag6 may even be required for deglycosylation because both TCRα and HC are stabilized entirely in the sugar-containing form when Bag6 was knocked down. Since the translocation-driving ATPase p97 does not form stable interactions with the proteasome, ERAD substrates are thought to be transferred from p97 to the proteasome via some shuttling factors (Hirsch et al., 2009; Vembar and Brodsky, 2008). Our data suggest that Bag6 may be an integral part of a shuttling system, which recognizes a retrotranslocation intermediate on the ER membrane and accompany it to the proteasome (Fig. 7G). In this regard, it is conceivable that ERAD substrates with slow transport kinetics or those carrying strong hydrophobic motifs may be particularly prone to aggregation in the cytosol, and therefore be more dependent on a holdase to maintain solubility while being targeted to the proteasome.
Efficient degradation of misfolded ER proteins also requires the two Bag6 cofactors, Trc35 and Ubl4A. Our results suggest that one function of Trc35 is to maintain a cytosolic pool of Bag6 that can participate in ERAD, but our data do not exclude the possibility that Trc35 may have other functions essential for the holdase activity of Bag6 in the cell. The function of Ubl4A in ERAD is unclear. In budding yeast, GET5 and GET4 appears to regulate substrate handoff from Sgt2, a functional counterpart of Bag6, to the downstream chaperone GET3 (Jonikas et al., 2009; Wang et al., 2010). Ubl4A may have a similar role in substrate handoff to the proteasome, which would explain the lack of protein aggregation in Ubl4A knockdown cells.
Bag6 is a multi-functional chaperone holdase
We demonstrate that Bag6, the central component of the Bag6 complex, can maintain the solubility of ERAD substrates to improve degradation efficiency. This function is consistent with the holdase activity of Bag6, which is demonstrated using an in vitro assay. The holdase activity allows Bag6 to maintain substrates in an unfolded yet soluble state, which makes them preferred substrates of the proteasome because the proteasome cannot efficiently digest protein aggregates (Venkatraman et al., 2004).
Our finding that Bag6 acts as a holdase unifies the various facets of Bag6 functions around a common theme, and explains how Bag6 can operate in an array of seemingly unrelated biological processes. In essence, the holdase activity of Bag6 can shield hydrophobic patches or TMDs on diverse substrates, allowing them to be safely delivered to different cellular destinations without forming aggregation. In ERAD, the association of Bag6 with an ER-bound E3 renders it a preferred chaperone for capturing retrotranslocated polypeptides containing exposed aggregation-prone motifs. The substrates might remain bound to Bag6 until the chaperone-substrate complex encounters the proteasome or its associated ubiquitin receptors. This activity can be similarly employed to degrade misfolded cytosolic proteins (Minami et al., 2010) or cytosol-derived mislocalized membrane proteins including defective TA proteins carrying hydrophobic TMD (Hessa et al., 2011). The holdase concept also explains the proposed role of Bag6 and of its yeast counterpart Sgt2 in TA protein biogenesis (Mariappan et al., 2010; Wang et al., 2010). In this process, Bag6 holds ribosome-released nascent chains bearing aggregation-prone tail anchor to facilitate their membrane insertion.
Substrate recognition by Bag6
Bag6 was recently shown to interact with polyubiquitinated proteins in the cell (Minami et al., 2010). However, Bag6 does not contain any ubiquitin binding activity by itself (Fig. S6F). Thus, the interaction of Bag6 with retrotranslocation substrates must be mediated by a non-modified segment in substrates.
During TA protein biogenesis, Bag6 acts as a TMD selective chaperone that preferentially binds exposed TMD in substrates (Mariappan et al., 2010). Our results show that in the context of ERAD, TMD is not a prerequisite for Bag6 engagement because the degradation of a TMD-deleted TCRα variant also involves Bag6. We propose that substrate recognition by Bag6 may involve a long stretch of hydrophobic residues, but not necessarily a TMD. This type of motif is present in many dislocated soluble ERAD substrates that are unfolded during retrotranslocation. For example, the TMD-deleted TCRα contains several long stretches of hydrophobic residues, which may be the binding site(s) for Bag6. Our model is supported by several lines of evidence. First, the soluble protein luciferase, once unfolded, can be directly recognized by Bag6. Additionally, the degradation of a model proteasome substrate carrying a hydrophobic degron (not a TMD) fused to GFP is dependent on Bag6 (Minami et al., 2010), whereas the degradation of a folded substrate Ub-V-GFP does not require Bag6 presumably because it does not contains any exposed hydrophobic motifs. Lastly, Bag6 is known to have functions in the nucleus where no membrane protein is present (Nguyen et al., 2008).
If Bag6 recognizes hydrophobic segments in substrates, it needs to compete with a large number of cytosolic chaperones, which bind substrates via a similar manner (Bukau et al., 2006). Several mechanisms may act together to ensure efficient capture of substrates by Bag6. First, the difference in hydrophobicity may offer a gauge for chaperones to distinguish between different substrates. For example, Hsp70 is known to recognize substrates carrying short hydrophobic segments. A long hydrophobic segment in a substrate may increase its affinity to Bag6, making it a preferred chaperone. In agreement with this idea, luciferase contains at least two long stretches of hydrophobic residues, which explains why Bag6 can effectively compete with Hsp70 to capture denatured luciferase even at a concentration substantially lower than Hsp70. Second, the association of Bag6 with distinct cellular partners may further govern substrate recognition. It is possible that the interaction of Bag6 with the ribosome allows Bag6 to efficiently recognize nascent TA proteins (Mariappan et al., 2010), whereas its association with ER-anchored E3s could give Bag6 edge over other competing chaperones even for binding to soluble substrates carrying a lesser hydrophobic motif.
The redundancy of the holdase systems
Our results suggest that different ERAD substrates may have different levels of dependence on Bag6 function for degradation. For example, knockdown of Bag6 significantly stabilizes TCRα, but depletion of the Bag6 only moderately attenuates HC degradation in US11 cells. It is possible that another chaperone holdase may substitute Bag6 function to support HC degradation. Consistent with this interpretation, we found that only a fraction of the dislocated HC is bound to Bag6, although the presence of a TMD in this substrate predicts that all dislocated HC molecules should be accompanied by a holdase in the cytosol. Our results also show that knockdown of Bag6 does not have an apparent effect on the biogenesis of ER TA proteins, which further suggests the existence of multiple holdase systems with overlapping substrate specificity in the cell. This would also explain why yeast deletion mutants lacking GET4 and GET5 are viable, whereas deletion of many TA proteins singularly can often result in lethality.
Experimental procedures
Gene knockdown and various ERAD assays
To knock down Bag6 and its associated cofactors, 293T cells were transfected with a gene specific shRNA construct using Lipofectamine 2000 on Day 1 and Day 2. 72h post the first transfection, cells were harvested for various assays. For pulse chase experiments, 3.0×106 cells were harvested and incubated in 8ml starvation DMEM medium free of Met/Cys for 60 min, and then incubated in 0.3ml starvation medium containing 75μCi 35S-Met/Cys for 30 min. Cells were then incubated in a chase medium containing unlabeled Met (2.5mM) and Cys (1mM). 0.5×106 cells were removed at the indicated time points. Cell extracts were prepared in the NP40 lysis buffer. TCRα-YFP was immunoprecipitated using a GFP antibody. For cycloheximide chase experiments, cells treated with a DMEM medium containing 50μg/ml cycloheximide were harvested at different time points. Cell extracts were prepared in the NP40 lysis buffer for analyses by immunoblotting. To analyze the in vivo aggregation of ERAD substrates, cells transfected with plasmids that expressed various ERAD substrates together with shRNA knockdown constructs were first solubilized in the NP40 lysis buffer. The detergent insoluble fractions were further solubilized by the Laemmli buffer. To perform in vitro retrotranslocation assay, radiolabeled A9 cells were treated with a low concentration of the detergent digitonin to remove cytosol. Membranes were then washed and incubated with either mock depleted or Bag6 depleted cytosol to initiate HC retrotranslocation.
Miscellaneous biochemical assays
Luciferase (150nM) was incubated with Bag6 or other proteins at concentrations specified in the figure legends in a buffer containing 20mM HEPES 7.4, 5mM MgOAC, 50mM KCl, 1mM DTT. The samples were heated at 42 ºC and the scattered light was measured at 330nm using an AB2 luminescence Spectrometer (Thermo Scientific). Alternatively, the samples were subject to centrifugation at 20,000g for 10min. The pellet and supernatant fractions were analyzed by immunoblotting. Luciferase activity was measured using a luciferase reporter gene assay kit from Roche. Depletion of the Bag6 complex was performed as the following. Protein A agarose was first incubated with 1ml anti-Ubl4A serum or control IgG and then with 1ml cow liver cytosol (~40mg/ml). This procedure is repeated one more time to deplete more than 90% of the Bag6 complex. Endo H digestion experiments were carried out by incubating cell extracts containing 0.5% SDS with 2μl Endo H (New England BioLab) in the presence of 1X reaction buffer (50 mM sodium citrate pH 5.5) at 37ºC for 2h.
Supplementary Material
Highlights.
The Bag6-Ubl4A-Trc35 complex associates with ERAD specific ubiquitin ligases
Bag6 interacts with dislocated ERAD substrates to facilitate their degradation
Bag6 contains a holdase activity that can suppress protein aggregation in vitro
Bag6 depletion causes dislocated ERAD substrates to aggregate in the cell
Acknowledgments
We thank S. Fang (University of Maryland, Baltimore, MD) for reagents, Michael Krause and Martin Gellert (NIDDK), and Tom Rapoport (Harvard Medical School) for critical reading of the manuscript. The research is supported by the NIH Intramural AIDS Targeted Antiviral Program (IATAP) and by the Intramural Research Program of the NIDDK.
Footnotes
Additional methods are available as online supporting information.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexandru G, Graumann J, Smith GT, Kolawa NJ, Fang R, Deshaies RJ. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell. 2008;134:804–816. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bays NW, Wilhovsky SK, Goradia A, Hodgkiss-Harlow K, Hampton RY. HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol Biol Cell. 2001;12:4114–4128. doi: 10.1091/mbc.12.12.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi KM, Forster ML, Lencer WI, Tsai B. Derlin-1 facilitates the retro-translocation of cholera toxin. Mol Biol Cell. 2008;19:877–884. doi: 10.1091/mbc.E07-08-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhamidipati A, Denic V, Quan EM, Weissman JS. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol Cell. 2005;19:741–751. doi: 10.1016/j.molcel.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Blom D, Hirsch C, Stern P, Tortorella D, Ploegh HL. A glycosylated type I membrane protein becomes cytosolic when peptide: N-glycanase is compromised. Embo J. 2004;23:650–658. doi: 10.1038/sj.emboj.7600090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A, Bukau B, Sommer T. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol Cell. 2011;40:238–252. doi: 10.1016/j.molcel.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Chen B, Mariano J, Tsai YC, Chan AH, Cohen M, Weissman AM. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc Natl Acad Sci U S A. 2006;103:341–346. doi: 10.1073/pnas.0506618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmots F, Russell HR, Lee Y, Boyd K, McKinnon PJ. The reaper-binding protein scythe modulates apoptosis and proliferation during mammalian development. Mol Cell Biol. 2005;25:10329–10337. doi: 10.1128/MCB.25.23.10329-10337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Ferrone M, Yang C, Jensen JP, Tiwari S, Weissman AM. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2001;98:14422–14427. doi: 10.1073/pnas.251401598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessa T, Sharma A, Mariappan M, Eshleman HD, Gutierrez E, Hegde RS. Protein targeting and degradation pathways are coupled for elimination of misfolded proteins. Nature. 2011 doi: 10.1038/nature10181. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458:453–460. doi: 10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- Jarosch E, Taxis C, Volkwein C, Bordallo J, Finley D, Wolf DH, Sommer T. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat Cell Biol. 2002;4:134–139. doi: 10.1038/ncb746. [DOI] [PubMed] [Google Scholar]
- Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, Schuldiner M. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SO, Cho K, Cho S, Kim I, Oh C, Ahn K. Protein disulphide isomerase is required for signal peptide peptidase-mediated protein degradation. Embo J. 2010;29:363–375. doi: 10.1038/emboj.2009.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leznicki P, Clancy A, Schwappach B, High S. Bat3 promotes the membrane integration of tail-anchored proteins. J Cell Sci. 2010;123:2170–2178. doi: 10.1242/jcs.066738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Tu D, Brunger AT, Ye Y. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature. 2007;446:333–337. doi: 10.1038/nature05542. [DOI] [PubMed] [Google Scholar]
- Lilley BN, Ploegh HL. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429:834–840. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- Lilley BN, Ploegh HL. Viral modulation of antigen presentation: manipulation of cellular targets in the ER and beyond. Immunol Rev. 2005;207:126–144. doi: 10.1111/j.0105-2896.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- Magadan JG, Perez-Victoria FJ, Sougrat R, Ye Y, Strebel K, Bonifacino JS. Multilayered mechanism of CD4 downregulation by HIV-1 Vpu involving distinct ER retention and ERAD targeting steps. PLoS Pathog. 2010;6:e1000869. doi: 10.1371/journal.ppat.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariappan M, Li X, Stefanovic S, Sharma A, Mateja A, Keenan RJ, Hegde RS. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature. 2010;466:1120–1124. doi: 10.1038/nature09296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami R, Hayakawa A, Kagawa H, Yanagi Y, Yokosawa H, Kawahara H. BAG-6 is essential for selective elimination of defective proteasomal substrates. J Cell Biol. 2010;190:637–650. doi: 10.1083/jcb.200908092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen P, Bar-Sela G, Sun L, Bisht KS, Cui H, Kohn E, Feinberg AP, Gius D. BAT3 and SET1A form a complex with CTCFL/BORIS to modulate H3K4 histone dimethylation and gene expression. Mol Cell Biol. 2008;28:6720–6729. doi: 10.1128/MCB.00568-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa SI, Fewell SW, Kato Y, Brodsky JL, Endo T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J Cell Biol. 2001;153:1061–1070. doi: 10.1083/jcb.153.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda-Shimizu Y, Hendershot LM. Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol Cell. 2007;28:544–554. doi: 10.1016/j.molcel.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich E, Kerem A, Frohlich KU, Diamant N, Bar-Nun S. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol Cell Biol. 2002;22:626–634. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Schulze A, Standera S, Buerger E, Kikkert M, van Voorden S, Wiertz E, Koning F, Kloetzel PM, Seeger M. The ubiquitin-domain protein HERP forms a complex with components of the endoplasmic reticulum associated degradation pathway. J Mol Biol. 2005;354:1021–1027. doi: 10.1016/j.jmb.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Shamu CE, Story CM, Rapoport TA, Ploegh HL. The pathway of US11-dependent degradation of MHC class I heavy chains involves a ubiquitin-conjugated intermediate. J Cell Biol. 1999;147:45–58. doi: 10.1083/jcb.147.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Ballar P, Fang S. Ubiquitin ligase gp78 increases solubility and facilitates degradation of the Z variant of alpha-1-antitrypsin. Biochem Biophys Res Commun. 2006;349:1285–1293. doi: 10.1016/j.bbrc.2006.08.173. [DOI] [PubMed] [Google Scholar]
- Soetandyo N, Wang Q, Ye Y, Li L. Role of intramembrane charged residues in the quality control of unassembled T-cell receptor alpha-chains at the endoplasmic reticulum. J Cell Sci. 2010;123:1031–1038. doi: 10.1242/jcs.059758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soetandyo N, Ye Y. The p97 ATPase dislocates MHC class I heavy chain in US2 expressing cells via an Ufd1-Npl4 independent mechanism. J Biol Chem. 2010 doi: 10.1074/jbc.M110.131649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song BL, Sever N, DeBose-Boyd RA. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol Cell. 2005;19:829–840. doi: 10.1016/j.molcel.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Thress K, Song J, Morimoto RI, Kornbluth S. Reversible inhibition of Hsp70 chaperone function by Scythe and Reaper. Embo J. 2001;20:1033–1041. doi: 10.1093/emboj/20.5.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321:569–572. doi: 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatraman P, Wetzel R, Tanaka M, Nukina N, Goldberg AL. Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol Cell. 2004;14:95–104. doi: 10.1016/s1097-2765(04)00151-0. [DOI] [PubMed] [Google Scholar]
- Wang F, Brown EC, Mak G, Zhuang J, Denic V. A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol Cell. 2010;40:159–171. doi: 10.1016/j.molcel.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiertz EJ, Jones TR, Sun L, Bogyo M, Geuze HJ, Ploegh HL. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell. 1996;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- Zhong X, Shen Y, Ballar P, Apostolou A, Agami R, Fang S. AAA ATPase p97/valosin-containing protein interacts with gp78, a ubiquitin ligase for endoplasmic reticulum-associated degradation. J Biol Chem. 2004;279:45676–45684. doi: 10.1074/jbc.M409034200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.