Abstract
Therapeutic strategies designed to treat HIV infection with combinations of antiviral drugs have proven to be the best approach for slowing the progression to AIDS. Despite this progress, there are problems with viral drug resistance and toxicity, necessitating new approaches to combating HIV-1 infection. We have therefore developed a different combination approach for the treatment of HIV infection in which an RNA aptamer, with high binding affinity to the HIV-1 envelope (gp120) protein and virus neutralization properties, is attached to and delivers a small interfering RNA (siRNA) that triggers sequence-specific degradation of HIV RNAs. We have tested the antiviral activities of these chimeric RNAs in a humanized Rag2−/−γc−/− (RAG-hu) mouse model with multilineage human hematopoiesis. In this animal model, HIV-1 replication and CD4+ T cell depletion mimic the situation seen in human HIV-infected patients. Our results show that treatment with either the anti-gp120 aptamer or the aptamer-siRNA chimera suppressed HIV-1 replication by several orders of magnitude and prevented the viral-induced helper CD4+ T cell decline. In comparison to the aptamer alone, the aptamer-siRNA combination provided more extensive inhibition, resulting in a significantly longer antiviral effect that extended several weeks beyond the last injected dose. The aptamer thus acts as a broad-spectrum HIV-neutralizing agent and an siRNA delivery vehicle. The combined aptamer-siRNA agent provides an attractive, nontoxic therapeutic approach for treatment of HIV infection.
INTRODUCTION
Therapeutic strategies designed to combat HIV/AIDS have primarily relied on small-molecule drugs. Current highly active antiretroviral therapy (HAART) treatment for HIV-1 has been therapeutically effective in most patients. However, drug resistance and toxicity remain a problem, with some individuals unresponsive to therapy (1). Alternative therapeutic strategies need to be developed to overcome these limitations. Nucleic acid aptamers, one new and potentially potent class of anti-HIV drugs, are nucleic acids that are selected from random sequence pools of RNAs under conditions that identify highly specific, tight binding to targeted molecules (2, 3). The low nanomolar binding affinities and exquisite binding specificity of aptamers to their targets have made them versatile tools for diagnostics, in vivo imaging, and therapeutics (4, 5). Another form of RNA-based therapeutics involving RNA interference (RNAi), can function as a gene-specific therapeutic option for controlling HIV-1 replication (6–9). In this approach, small interfering RNAs (siRNAs) targeted to HIV-1 transcripts or to host messenger RNAs (mRNAs) potently down-regulate viral gene expression and inhibit HIV-1 replication both in vitro and in vivo (6–13). Although siRNAs hold promise for treatment of HIV/AIDS, efficient, targeted, systemic delivery of siRNAs in vivo remains a principal challenge for clinical translation. Recently, cell type–specific aptamers have been combined with siRNAs to achieve cell-specific delivery of the siRNAs for selective target mRNA knockdown (14–19). We previously demonstrated that an anti–HIV-1 gp120 aptamer can neutralize HIV-1 infection and has the added bonus of delivering anti-HIV siRNAs into HIV-1–infected cells (17, 18). These encouraging cell culture results with aptamers and with aptamer-siRNA chimeras suggested the next step: to validate the anti-HIV efficacy of these agents in an in vivo model. In this regard, newly developed humanized mouse models such as the Rag2−/−γc−/− (RAG-hu) with a capacity for multilineage human hematopoietic cell engraftment are likely to be useful (6, 20–24). Previous studies have demonstrated that HIV-1–infected RAG-hu mice can sustain chronic viremia lasting for more than a year and display a continuous declining trend of CD4+ T cell levels, mirroring the main features of human HIV-1 infection (1, 25). These features are in contrast to those of the long-established SCID-hu and hu-PBL-SCID mouse models, which display only acute HIV infection (20). Thus, the RAG-hu mouse model of HIV-1 infection is an excellent model for in vivo pathogenesis studies and the evaluation of new anti-HIV therapeutic strategies. Here, we used the RAG-hu mouse model system to evaluate the efficacy of our anti-gp120 aptamer and aptamer-siRNA chimera (18).
RESULTS
Aptamer A-1 and aptamer-siRNA chimera (Ch A-1) inhibit HIV strains in vitro. We previously described the in vitro selection of aptamers directed against gp120 that bind the HIV-1 BaL envelope protein with low nanomolar dissociation constants (18). We fused the lead aptamer, designated as A-1, to an siRNA that targeted the HIV-1 tat/rev RNAs that encode early regulatory proteins required for replication. The resulting chimeric construct (Ch A-1) (Fig. 1) is designed to deliver the siRNA to HIV-1–infected cells, resulting in targeted, RNAi-mediated knockdown of tat/rev expression (18). In these studies, Ch A-1 inhibited tat/rev expression by more than 50% when compared with the A-1 aptamer alone (18). Both the A-1 aptamer and Ch A-1 inhibited the replication of CXCR4-tropic (NL4-3 and IIIB) and CCR5-tropic (BaL) strains of HIV-1 in cultured T lymphocytes (CEM cells) and in peripheral blood mononuclear cells (PBMCs) (18).
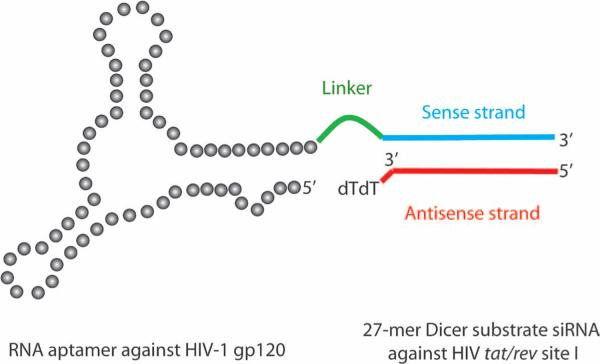
Fig. 1.
Schematic of anti-HIV-1 gp120 aptamer-siRNA chimera (Ch A-1). The aptamer and sense strand segment of the siRNAs contain nuclease-resistant 2′-fluoro uridine triphosphate and 2′-fluoro cytidine triphosphate. The aptamer portion of the chimera binds to gp120 protein, whereas the 27-mer Dicer substrate siRNA duplex is targeted to the HIV-1 tat/rev common exon. A linker (UU) between the aptamer and siRNA is indicated in green.
To better determine the range of envelope proteins that the A-1 aptamer and Ch A-1 could recognize, we examined their ability in vitro to inhibit HIV by using several CXCR4-tropic and CCR5-tropic laboratory and clinical HIV isolates. We carried out HIV-1 viral challenge assays in vitro in human PBMCs and CEM T cells. Cells were infected with the HIV-1 isolates, and then A-1 or Ch A-1 was added to the culture media 3 days later and virus levels were monitored. The A-1 aptamer and Ch A-1 inhibited different strains of HIV, as assessed by p24 concentrations, to varying degrees (fig. S1, A to F). The Ch A-1 chimera inhibited the two laboratory strains of HIV-1 better than did the A-1 aptamer alone (fig. S1, E and F). The Ch A-1 chimera, but not the aptamer alone, was able to suppress expression of the tat/rev gene, as would be expected from the tat/rev specificity of the siRNA component of the chimera (fig. S1, G and H). The siRNA alone did not suppress tat/rev expression, presumably because it was unable to enter the infected cells unless fused with the aptamer (fig. S1, G and H).
2′-Fluoro–modified Ch A-1 is partially resistant to nuclease degradation in mouse serum
We have previously detected the 2′-fluoro–modified, aptamer-delivered siRNA after 7 days of incubation in HIV-1–infected cell culture (17). To determine the nuclease resistance of 2′-fluoro–modified Ch A-1 in serum in the absence of HIV-1 cells, we incubated Ch A-1 in 50 or 5% mouse serum (normal mouse serum) for different lengths of time at 37°C. After 24 hours of incubation, ~30% of the full-length 2′-fluoro–modified Ch A-1 was detectable in 50% serum and more than 50% was detectable in 5% serum (fig. S2, A and B). Together, these results suggested that enough Ch A-1 should be present to be internalized and functionally active in HIV-1–infected cells in vivo.
A-1 and Ch A-1 suppress HIV-1 viral loads in viremic RAG-hu mice
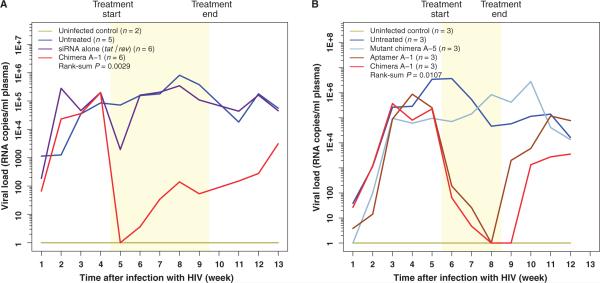
To evaluate the potential efficacy of Ch A-1 in vivo, we tested its antiviral efficacy in the humanized Rag2−/−γc−/− (RAG-hu) mouse model of HIV-1 infection (20, 24). RAG-hu mice infected with HIV-1 NL4-3 became viremic by 3 weeks after infection, with viral loads averaging 105 per milliliter, indicating an established infection. We first compared the ability of the Ch A-1 chimera and the tat/rev siRNA alone (as a control) to inhibit HIV. Six animals were given five weekly injections of 0.25 nmol (0.38 mg/kg) of Ch A-1 or 0.25 nmol (0.15 mg/kg) of tat/rev siRNA alone. Plasma viral loads were monitored to determine treatment efficacy (Fig. 2A, fig. S3A, and table S1A). A general pattern of decreased viral loads was seen after treatment in most chimera Ch A-1–treated mice, unlike the HIV-1–infected, untreated, and siRNA-treated controls, and this difference reached statistical significance with a rank sum P = 0.0029 (Fig. 2A, fig. S3A, and table S3A). The viral loads were suppressed to below detectable values in all the Ch A-1–treated mice within a week of treatment (at week 5, as shown), and this suppression persisted throughout the treatment period in most mice. Four of six mice had undetectable viral loads even up to 3 weeks after treatment, indicating the sustained efficacy of the Ch A-1 chimera (table S1A). In two of the Ch A-1–treated mice (J276, J247), the first 3 weeks of suppression were followed by viral rebound, suggestive of the emergence of Ch A-1 resistance. Even though these animals had high viral loads at weeks 9 and 7, respectively, their CD4-CD3 T cell ratios (CD4+ T cells) did not decline (table S2A). The tat/rev siRNA alone treatment did not provide viral suppression for more than 1 week. To determine the relative contribution of the siRNA component of the Ch A-1 chimera, in a separate experiment, we treated additional groups of three viremic animals with 0.25 nmol (0.22 mg/kg) of A-1, Ch A-1 (0.38 mg/kg), and a mutant aptamer-tat/rev siRNA chimera Ch A-5 (0.38 mg/kg) (see details in Materials and Methods). The A-5 aptamer portion of Ch A-5 has previously been shown to have poor affinity for gp120, and so this construct would be expected to be ineffective (18). We monitored plasma viral loads to ascertain the efficacy of each treatment (Fig. 2B). Viral loads were strongly suppressed in A-1– and Ch A-1–treated mice compared to the untreated group, reaching statistical significance (P = 0.0107). As expected, there was no notable viral suppression in the mice treated with the control, mutant aptamer-siRNA Ch A-5 (Fig. 2B, fig. S3B, and table S1B). The viral levels remained suppressed in all of the A-1– and Ch A-1–treated mice throughout the 3-week treatment period (weeks 6, 7, and 8). In the A-1–treated animals, this reduction during treatment was followed by moderate suppression continuing for an additional 2 weeks. In contrast, all of the Ch A-1–treated animals had undetectable viral loads at week 9, 1 week after the last injection (Fig. 2B, fig. S3B, and table S1B), and reduced viral loads persisted through week 12 in the Ch A-1–treated animals. In contrast, viral loads returned to pretreatment levels by week 11 in the animals treated with the A-1 aptamer alone. The enhanced duration of HIV-1 suppression afforded by the inclusion of the siRNA in Ch A-1 over the A-1 aptamer alone is statistically significant (P = 0.04) (tables S1A and S3A).
Fig. 2.
Aptamer A-1 and chimera Ch A-1, but not a mutant chimera, suppress viral loads in HIV-1–infected RAG-hu mice. Mice were infected with HIV-1 at week 0, and 5 weeks later, weekly treatments were started. The treatment period is indicated by the yellow region. (A) Viral loads in uninfected mice (n = 2), untreated mice (n = 5), tat/rev siRNA only–treated mice (n = 6), and ChA-1–treated mice (n = 6). (B) In a separate experiment, viral loads in uninfected mice (n = 3), untreated mice (n = 3), A-1 aptamer–treated mice (n = 3), Ch A-1 chimera–treated mice (n = 3), andmutantA-5 chimera–treated mice (n = 3). The viral RNA was detected through qRT-PCR as described in Materials and Methods. Thus, if there was no detectable viral RNA, we established this as a value of 1 (100) to allow for the use of logarithmic values on the y axis. P values for the effects of A-1 and Ch A-1 are indicated and were calculated as described in Materials and Methods. The data for individual mice are presented in fig. S3 and table S1.
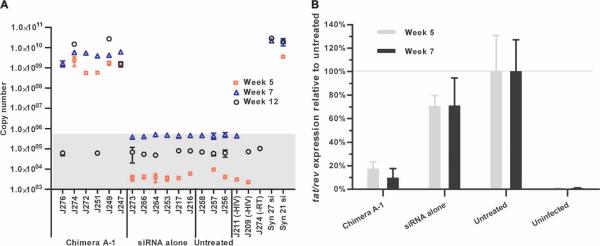
To validate that the Ch A-1 delivered the anti-tat/rev siRNA to infected T lymphocytes, we collected PBMCs after the first and third treatments (weeks 5 and 7), and 3 weeks after the last treatment (week 12), in the first experimental set of animals (Fig. 2A). We analyzed small RNAs for the presence of the tat/rev siRNA by real-time TaqMan quantitative reverse transcription polymerase chain reaction (qRT-PCR) assays. The tat/rev siRNA was detectable in PBMCs from all of the Ch A-1–treated mice at weeks 5 and 7, and in three of the animals 3 weeks after the last injection (week 12) (Fig. 3A). In contrast, no siRNAs were detected in the PBMCs of mice treated with tat/rev siRNA only. In table S1, we report the death of a single Ch A-1–treated animal. We believe that it is unlikely that this death resulted from toxicity of the Ch A-1, because no other deaths were observed in any of the other treated animals. Death of this animal was more likely the result of the infrequent, but well-documented, spontaneous morbidity that occurs among severely immunocompromised mice (26).
Fig. 3.
The detection and function of tat/rev siRNA in PBMCs from treated RAG-hu mice. (A) Detection of the siRNA sequences in PBMCs during (weeks 5 and 7 after infection) and after (week 12) treatment with Ch A-1 and siRNA only, and in untreated mice and uninfected mice (animals from Fig. 2A). One minus RT control, J274 (−RT), also was set up. The synthetic 27- and 21-mer tat/rev siRNAs were used as positive controls. The background copy number of siRNA is <106 (gray area). Error bars indicate SD (n = 4 measurements per sample). Some of the error bars are not discernible in the graph because their relative percentage was less than 5%. (B) Expression of tat/rev gene transcripts at the first and third weeks of treatment (weeks 5 and 7 after infection) with Ch A-1 and siRNA only and in untreated and uninfected animals (animals from Fig. 2A). Gene expression was normalized to that of a representative HIV-1–infected untreated mouse.
We also measured tat/rev gene expression in PBMCs of these mice with qRT-PCR. Our results showed a 75 to 90% reduction in the levels of tat/rev transcript in chimera Ch A-1–treated mice at both the first and third weeks of treatment (weeks 5 and 7) and a 30% reduction in mice treated with siRNA only compared to untreated mice (Fig. 3B).
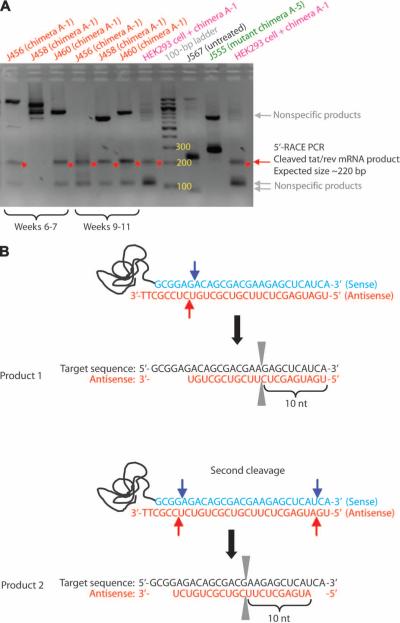
To confirm that the tat/rev siRNA was indeed functioning through an RNAi mechanism in vivo, we performed modified 5′-RACE [rapid amplification of complementary DNA (cDNA) ends] PCR on tat/rev RNAs from PBMCs of the second group of treated animals (represented in Fig. 2B). The intracellular enzyme Ago2 cleaves between bases 10 and 11 relative to the 5′ end of the siRNA guide strand (27, 28). Thus, the RACE PCR product sequence analyses should reveal the linker at the base 10 nucleotides downstream from the 5′ end of the siRNA guide strand. PCR bands of the predicted lengths were detected in the PBMC samples from mice treated with the chimera Ch A-1 after three nested PCRs (Fig. 4A). No appropriately sized products were generated from total RNA of PBMCs of untreated animals or from those treated with the mutant chimera A-5. The individual clones were characterized by DNA sequencing to verify that they contained the expected PCR products. Two different cleavage sites were found in the samples from the animals treated with Ch A-1 (Fig. 4B). One cleavage takes place between positions 10 and 11 from the 5′ end of the siRNA antisense strand (Fig. 4B). The second RACE product was displaced by two nucleotides along the mRNA, suggesting that a different Dicer product of the tat/rev siRNA generated a guide strand shifted by two bases relative to the first cleavage product (Fig. 4B). These data show that the siRNAs delivered to the cells by Ch A-1 are processed intracellularly and incorporated into the RNA-induced silencing complex (RISC), thus triggering sequence-specific degradation of the HIV tat/rev target RNA.
Fig. 4.
5′-RACE PCR analysis of in vivo tat/rev siRNA delivered by Ch A-1. (A) Nested PCR products were resolved in an agarose gel; specific siRNA-mediated RACE PCR cleavage mRNA products are marked by a red arrow. (B) DNA sequence analyses of cloned RACE PCR products. The positions of the two different siRNA-directed cleavage sites in the tat/rev target RNA are indicated. nt, nucleotides.
A-1 and Ch A-1 protect against HIV-1–mediated CD4+ T cell depletion
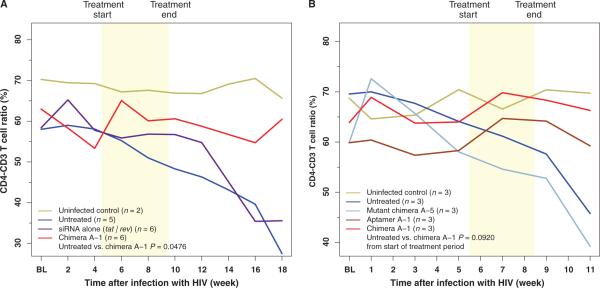
A major characteristic of HIV-1 infection is helper CD4+ T cell loss during the acute stage of infection with a return to a set point for several months or years, followed by a gradual depletion leading to AIDS (1, 29). Therefore, prevention of CD4+ helper T cell loss would contribute to immune reconstitution and restoration of immune function. To determine whether treatment of HIV-1–infected RAG-hu mice with the Ch A-1 chimera could protect against depletion of CD4+ T cells, we measured CD4+ T cells (expressed as the CD4-CD3 T cell ratio) in peripheral blood collected at weekly intervals during and after treatment (Fig. 5, fig. S4, and table S2). In the first set of animals tested (as described in Fig. 2A), Ch A-1 significantly protected against T cell depletion compared to the untreated group (P = 0.0476) (Fig. 5A, fig. S4A, and table S2A). In control, uninfected mice (HIV negative), the levels of CD4+ T cells remained relatively stable within a 5% variation range, whereas in untreated HIV-1–infected mice, the CD4+ T cell levels began to decline beginning at 4 weeks after infection to below 50% of the starting values at 18 weeks after infection. In contrast, the CD4+ T cells in Ch A-1–treated mice remained at or near the levels found in un-infected mice, and this value remained stable well beyond the last treatment, indicating that chimera A-1 provided protection against CD4+ T cell depletion. Although the Ch A-1 and A-1 treatment seemed to protect against T cell depletion in the second group of animals, this change did not reach statistical significance (P = 0.0920), possibly as a result of the shorter duration of the A-1 and Ch A-1 injections (3 weeks versus 5 weeks for the first group of animals) (Fig. 5B, fig. S4B, and table S2B).
Fig. 5.
Protection of RAG-hu mice from CD4+ T cell loss by the Ch A-1 chimera. CD4+ T cell levels were assessed by FACS at each indicated week before and after siRNA treatment. BL, baseline measurement. Treatment duration is indicated by the yellow region. (A) Mice from the first experiment (shown in Fig. 2A) group. Uninfected mice (n = 2), untreated mice (n = 5), tat/rev siRNA–treated mice (n = 6), and chimera Ch A-1–treated mice (n = 6). (B) Mice from second experiment (shown in Fig. 2B). CD4-CD3 T cell ratios were assessed by FACS at each indicated week before and after treatment. P values for both experiments were determined as described in Materials and Methods. For baseline measurements, each individual mouse was bled twice before HIV-1 infection and the CD4-CD3 values were averaged within treatment groups to establish a baseline CD4-CD3 level. The data for individual mice are presented in fig. S4 and table S2.
Ch A-1 chimera does not trigger type I interferon response in vivo
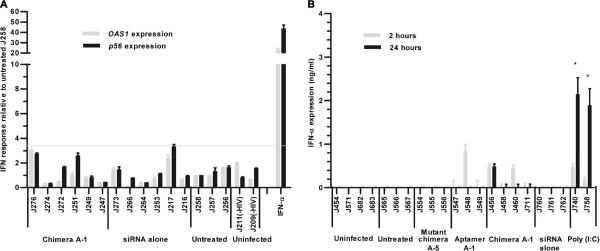
Systemic administration of synthetic siRNA duplexes can activate innate immune responses, inducing cytokines such as tumor necrosis factor-α (TNF-α), interleukin-6 (IL-6), and interferons (IFNs), particularly IFN-α (30, 31). Such nonspecific immune activation by siRNA-based drugs could contribute to the antiviral effects but can also create undesirable toxicities. We therefore assessed the potential induction of type I IFN-a-regulated gene expression using qRT-PCR expression assays on PBMC-derived RNAs isolated from Ch A-1–treated and control mice (Fig. 6A). We used IFN-α–treated PBMCs as a positive control for up-regulation of p56 and OAS1 gene expression. We did not observe type I IFN responses in PBMCs from any of the animals at any time after treatment (Fig. 6A). Using an enzyme-linked immunosorbent assay (ELISA), we also directly monitored IFN-α in HIV-1–uninfected and HIV-1–infected animals with or without treatment 2 and 24 hours after the injection of the experimental RNAs (or saline for the controls) (Fig.6B). IFN-– (<1 ng/ml) was detected in blood of one of the A-1–treated and two of the Ch A-1–treated animals, but these concentrations are several times lower than those induced by poly I:C (polyinosinepolycytosine) (Fig. 6B), a double-stranded RNA activator of innate IFN responses. Because we did not see an IFN response in infected or uninfected, untreated animals, we concluded that the virus itself was not triggering an antiviral IFN response.
Fig. 6.
Effect of in vivo administration of Ch A-1 on type I IFN. (A) The effect of aptamer A-1 and chimera A-1 on expression of type I IFN response genes (p56 and OAS1) at week 7 after infection. IFN-α-treated, infected human PBMCs were used as a positive control. Gene expression was normalized to the gapdh mRNA. Error bars indicate SD (n =4). (B) Expression of IFN-α 2 or 24 hours after RNA injection as measured by an ELISA. Poly I:C-treated, HIV-infected RAG-hu mice were used as a positive control. Error bars indicate SD (n = 3).
Deep sequence analyses of viral RNAs reveal mutations in envelope and tat/rev target mutations
Development of viral resistance to drugs, a result of the generation of viral escape mutants, is a common setback with HIV-1 therapies. To assess the possible emergence of viral mutants in the gp120 HIV gene env and tat/rev sequences (the targets of the aptamer and siRNA, respectively), we carried out deep sequencing analyses of the two targeted regions within these genes. Our goal was to determine whether a specific class of mutations predominated in the samples from treated and untreated, HIV–infected animals. To have enough material for these analyses, we used pooled virus from serum collected from Ch A-1–treated animals at weeks 7 to 9 of the first set of experiments. We compared this sample with virus collected from animals not receiving antiviral treatment and determined the mutations unique to the Ch A-1–treated animals (fig. S5). Illumina deep sequence alignments of the targeted env domain from Ch A-1 chimera–treated animals with that from the non-treated animals revealed an average frequency of unique point mutants of less than 3% (frequency is defined as the number of reads with mutations as a percentage of total reads). Figure S5 lists all the point mutations in the HIV-1 env-targeted domain unique to the Ch A-1–treated animals. More mutations were detected in the gp120 domain (32 mutations) than in the gp41 domain (4 mutations). There were only seven point mutations in the targeted tat/rev sequence, with an average of 0.23% frequency increase above the samples analyzed from untreated animals (fig. S5). These results suggested that Ch A-1 exerted some selective pressure on the HIV-1 gp120 env gene. Nevertheless, even for this gene, the mutation frequency was less than 5% above that in untreated animals and, most important, no dominant mutations were found.
DISCUSSION
Here, we have tested an RNA-based combinatorial approach for treatment of HIV-1 infection in a humanized mouse model that supports HIV-1 replication and CD4+ T cell decline as a consequence of HIV-1 infection. We fused an anti-HIV tat/rev siRNA to an aptamer directed to the gp120 protein found on the surface of HIV-infected cells to achieve cell type–specific delivery of the siRNA. Before our studies, single-chain antibody-siRNA chimeras targeting either gp120-expressing cells or the T cell–specific CD7 receptor had been shown to functionally deliver anti–HIV-1 siRNAs to cells in vivo (6, 32). In these studies, only the siRNA component has been shown to have antiviral activity. In our study, we observed that the aptamer itself exerted potent antiviral activity, which was enhanced by aptamer-mediated delivery of an anti–HIV-1–delivered siRNA.
The humanized mouse model in which we tested the aptamer-siRNA chimera Ch A-1 provided a stringent and relevant assessment for its anti–HIV-1 activity in human cells. An advantage of this mouse model over nonhuman primates is that the humanized mice can be infected with any strain of HIV-1 that is capable of infecting humans, whereas the nonhuman primates require the use of SIV (simian immunodeficiency virus) or SIV-HIV hybrids. Thus, therapeutic approaches shown to be effective in the humanized mouse model do not have to be redesigned for human applications.
In our humanized mouse model, HIV-1 infection resulted in high levels of viral replication before the injection of the A-1 aptamer and Ch A-1 chimeric construct. Both the A-1 aptamer and the Ch A-1 chimera decreased these high viral loads by several logs in all the treated animals within a week of intravenous administration. The suppression of viral load averaged three orders of magnitude or greater, relative to controls, and persisted throughout and beyond the treatment period in seven of the nine animals tested in this study. In addition, using real-time RT-PCR and RACE PCR assays, we demonstrated that the aptamer-delivered siRNA is functionally active in vivo. Our results provide an in vivo validation of the anti–HIV-1 efficacy of the anti-gp120 aptamersiRNA Ch A-1 chimera.
The inhibitory functions of both the A-1 aptamer and siRNA moieties in the chimera make this approach potentially superior to the other siRNA delivery approaches reported previously (6, 32). In these other studies, only the siRNA acted as an anti-HIV agent. We used the aptamer both as an HIV-neutralizing agent and as a vehicle for delivering an siRNA for triggering RNAi-mediated down-regulation of the HIV-1 tat/rev transcripts.
The high potency and enhanced duration of viral suppression of the Ch A-1 chimera may be attributed to the dual anti-HIV activities of the aptamer and the aptamer-delivered siRNA. In contrast to antibody-mediated siRNA delivery, which requires biological production of the antibodies and creation of chimeric antibody–cationic peptide conjugates, chimeric aptamer-siRNAs can be synthesized as a single unit or as two separate but combinable units with either biochemical or chemical methods (18). Because these RNA-based molecules are not antigenic, they can be administered repeatedly without concerns about generating the potentially undesirable immunological responses expected with antibody-peptide conjugates. The aptamer-siRNA technology can be potentially adapted to avert viral resistance with a cocktail of gp120 aptamers of different specificities combined with siRNAs directed against different mRNA targets (both viral and host).
Before aptamer and aptamer-siRNA chimeras can be used to treat HIV-1 infection in humans, several additional tests will need to be performed in animal models, including biodistribution, dose-response analyses, and the effects of this treatment on latent viral infection. Once the parameters of pharmacokinetics and dynamics are determined in animal models, we envision that the first dual-function aptamer-siRNA chimeras could be tested in human clinical trials, perhaps first in patients who harbor virus resistant to anti-retroviral drugs.
MATERIALS AND METHODS
Reagents
Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich, all restriction enzymes were obtained from New England BioLabs, and all cell culture products were purchased from Gibco. Sources for the other reagents were as follows: DuraScribe T7 transcription kit (Epicentre Biotechnologies), Silencer siRNA Labeling kit (Ambion), Hoechst 33342 (nuclear dye for live cells) (Molecular Probes, Invitrogen), random primers (Invitrogen), SuperScript III RT kit (Invitrogen), and Bio-Spin 30 Columns (Bio-Rad). CHO-Env transfectants (CHO-WT and CHO-EE), the HIV-1 NL4-3 and HIV-1 BaL viruses, and clinical isolates (X4-strain: HIV-1 92UG021; R5-strain: HIV-1 RU570 and HIV-1 98CN009) were obtained from the AIDS Research and Reference Reagent Program. Normal mouse serum was obtained from Jackson ImmunoResearch Laboratories.
Generation of aptamer-siRNA chimera RNA (Ch A-1) by in vitro transcription
The design, synthesis, and in vitro efficacies of the aptamer A-1 and aptamer-siRNA conjugates have been described in detail (18). siRNA Dicer substrate sense, antisense strand RNAs, and DNA oligonucleo-tides were purchased from Integrated DNA Technologies: tat/rev Site I 27-mersiRNA, 5′-GCGGAGACAGCGACGAAGAGCUCAUCA-3′ (sense) and 5′-UGAUGAGCUCUUCGUCGCUGUCUCCGCdTdT-3′ (antisense); aptamer A-1, 5′-GGGAGGACGAUGCGGAAUUGAGGGACCACGCGCUGCUUGUUGUGAUAAGCAGUUUGUCGUGAUGGCAGACGACUCGCCCGA-3′; chimera A-1, 5′-GGGAGGACGAUGCGGAAUUGAGGGACCACGCGCUGCUUGUUGUGAUAAGCAGUUUGUCGUGAUGGCAGACGACUCGCCCGAUUGCGGAGACAGCGACGAAGAGCUCAUCA-3′ (sense); chimera A-5, 5′-GGGAGGACGAUGCGGGAAACUAGUUUGAAUAAUGGUGUAGAGGAGGGUCAAUAGUUUCGUUGGUGCAGACGACUCGCCCGAUUGCGGAGACAGCGACGAAGAGCUCAUCA-3′ (sense) and 5′-UGAUGAGCUCUUCGUCGCUGUCUCCGCdTdT-3′ (antisense). The sense strands of the chimeras are underlined. The italic UU is the linker between the aptamer and siRNA portions. The assembly of these chimeric constructs was described previously (17, 18), and a schematic is presented in Fig. 1.
Serum stability assay
Five micrograms of refolded Ch A-1 was incubated at 37°C in 100 μl of RPMI 1640 medium (Mediatech) containing 50 or 5% (v/v) mouse serum (normal mouse serum; Jackson ImmunoResearch Laboratories), resulting in an RNA concentration of 50 ng/μl. At 0 min to 72 hours, 10-μl aliquots of the reaction were withdrawn and 10 μl of 2× loading buffer containing 8 M urea, 120 EDTA, and 0.1% SDS was added to each sample. The mixtures were heated at 95°C for 5 min and then stored at −80°C until all incubations were completed. Each mixture (20 μl) was analyzed by electrophoresis in an 8% denaturing polyacrylamide gel, and the RNAs were visualized after ethidium bromide staining with an ultraviolet transilluminator.
Generation and HIV-1 infection of humanized Rag2−/−γc−/− mice (RAG-hu mice)
Humanized BALB/c-Rag2−/−γc−/− mice were prepared as described (20, 24) with human fetal liver–derived CD34+ cells. Briefly, neonatal mice were conditioned by irradiating at 3.5 Gy and then injected intrahepatically with 0.5 × 106 to 1 × 106 human CD34+ cells. About 12 weeks after reconstitution, mice were screened for human cell engraftment. Blood was collected by tail bleeds, and red blood cells were lysed with the Whole Blood Erythrocyte Lysing Kit (R&D Systems). The white blood cell fraction was stained with antibodies against the human panleukocyte marker CD45 (Caltag) and FACS (fluorescence-activated cell sorting)–analyzed as described (20). To infect human cell–reconstituted RAG-hu mice, we injected HIV-1 NL4-3 (1.2 × 105 IU) in a 100-μl volume intraperitoneally at least 12 weeks after cell engraftment. Viral loads were examined weekly, and viremia was established in all the mice by 3 weeks. Treatment was done by intravenous injection on the last day of week 4 with 0.25 nmol of experimental RNAs [tat/rev siRNA (0.15 mg/kg) or Ch A-1 (0.38 mg/kg)] in a 40-μl volume, followed by another the next day. Later, the injections were continued on a weekly basis for 4 weeks. In the second in vivo treatment experiment, the same amounts as described above of mutant chimera Ch A-5, aptamer A-1, or chimera Ch A-1 in a 40-μl volume were administered at 5 weeks after infection as above and continued for only three weekly injections.
Measurement of viral load in plasma
To quantify cell-free HIV-1 by qRT-PCR, we extracted RNA from 25 to 50 μl of EDTA-treated plasma with the QIAamp Viral RNA kit (Qiagen). cDNAs were produced with SuperScript III reverse transcriptase (Invitrogen) with a primer set specific for the HIV-1 long terminal repeat (LTR) sequence, and qPCR was performed with the same primer set and an LTR-specific probe with Supermix UDG (Invitrogen) as described (20). If there was no detectable viral RNA, we established this as a value of 1 (100) to allow for the use of logarithmic values on the y axis.
Flow cytometry
Whole blood was collected and red blood cells were lysed as reported (20, 24). Peripheral blood cells were stained by hCD3-PE and hCD4-PECy5 (Caltag) antibodies and analyzed with a Coulter EPICS XL-MCL FACS analyzer (Beckman Coulter). CD4+ T cell levels were calculated as a ratio of the entire CD3 population (CD4+CD3+-CD4−CD3+). To establish baseline CD4+ T cell ratios, we analyzed all mice before infection. Each individual mouse was bled two times before HIV-1 infection and averaged within treatment groups to establish a baseline CD4-CD3 level.
Detection of tat/rev siRNA
At 5, 7, and 12 weeks after infection (1, 3, and 9 weeks after treatment), blood samples were collected and small RNAs were isolated with MirVana miRNA isolation Kit (Applied Biosystems) according to the manufacturer's instruction. The siRNA quantification was performed with TaqMan MicroRNA Assay according to the manufacturer's recommended protocol (Applied Biosystems). Small RNA (10 ng), 0.2 μM stem-loop RT primer, RT buffer, 0.25 mM deoxynucleotide triphosphates, MultiScribe reverse transcriptase (3.33 U/ml), and ribonuclease inhibitor (0.25 U/ml) were used in 15-μl RT reactions for 30 min at 16°C, 30 min at 42°C, and 5 min at 85°C with the TaqMan MicroRNA RT kit (Applied Biosystems). For real-time PCR, 1.33 μl of cDNA, 0.2 mM TaqMan Probe, 1.5 mM forward primer, 0.7 mM reverse primer, and TaqMan Universal PCR Master Mix were added in 20-μl reactions for 10 min at 95°C and 40 cycles of 15 s at 95°C and 1 min at 60°C. All real-time PCR experiments were done with an iCycler iQ system (Bio-Rad). Primers were as follows: site I looped RT primer, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACAGCG-3′; site I forward primer, 5′-GCTGATGAGCTCTTCGTCG-3′; site I reverse primer, 5′-GTGCAGGGTCCGAGGT-3′; site I probe primer, 5′-6-FAM-TCGCACTGGATACGACACAGCGACGA-BHQ1-3′. In this case, a synthetic 27-mer duplex RNA was used as positive control.
Cell culture conditions
Human embryonic kidney (HEK) 293 and CEM cells were purchased from the American Type Culture Collection and cultured in Dulbecco's modified Eagle's medium (DMEM) and RPMI 1640 supplemented with 10% fetal bovine serum (FBS). CHO-WT and CHO-EE cells were obtained through the AIDS Research and Reference Reagent Program and grown in modified Glasgow minimal essential medium (GMEM-S) supplement with 400 μM methionine sulfoximine (MSX). Cells were cultured in a humidified 5% CO2 incubator at 37°C. PBMCs were obtained from healthy donors from the City of Hope National Medical Center. They were isolated from whole blood by centrifugation through a Ficoll-Hypaque solution (Histopaque-1077, Sigma). CD8 cells (T cytotoxic/suppressor cells) were depleted from the PBMCs by CD8 Dynabeads (Invitrogen) according to the manufacturer's instructions. CD8+ T cell–depleted PBMCs were washed twice in phosphate-buffered saline (PBS) and resuspended in culture media [RPMI 1640 with 10% FBS, 1× penicillin-streptomycin, and IL-2 (100 U/ml)]. Cells were cultured in a humidified 5% CO2 incubator at 37°C.
HIV-1 challenge and p24 antigen assays
CEM cells or human PBMCs were infected with HIV strains IIIB, NL4-3, or BaL for 5 days [multiplicity of infection (MOI), 0.001 or 0.005]. For clinical isolates, human PBMC-CD4+ cells were infected for 3 days (MOI, 0.001 or 0.0005). Before RNA treatments, the infected cells were gently washed with PBS three times to remove free virus.Next, 2 × 104 infected cells and 3 × 104 uninfected cells were incubated with refolded RNAs at 400 nM final concentration in 96-well plates at 37°C. The culture supernatants were collected at different times after treatment (3, 5, 7, and 9 days or 11 days). Here, the p24 antigen analyses were performed with a Coulter HIV-1 p24 Antigen Assay (Beckman Coulter) according to the manufacturer's instructions.
Determination of HIV-1 tat/rev expression
Human PBMCs were obtained from treated mice at 5 and 7 weeks after infection (1 and 3 weeks after treatment began), and total RNAs were isolated with STAT16 60 (Tel-Test “B”) according to the manufacturer's instructions. Residual DNA was digested with a DNA-free kit per the manufacturer's instructions (Ambion). cDNA was made with 2 μg of total RNA. RT was carried out with Moloney murine leukemia virus reverse transcriptase and random primers in a 15-μl reaction according to the manufacturer's instructions (Invitrogen). Expression of the tat/rev coding RNAs was analyzed by qRT-PCR with 2× iQ SYBRGreen MasterMix (Bio-Rad) and specific primer sets at a final concentration of 400 nM. gapdh expression was used for normalization of the qPCR data. Primers were as follows: NL4-3 or IIIB tat/rev, 5′-GGCGTTACTCGACAGAGGAG-3′ (forward) and 5′-TGCTTTGATAGAGAAGCTTGATG-3′ (reverse); BaL tat/rev, 5′-GAAGCATCCAGGAAGTCAGC-3′ (forward) and 5′-TGCTTTGATAGAGAAACTTGATGA-3′ (reverse); gapdh, 5′-CATTGACCTCAACTACATG-3′ (forward) and 5′-TCTCCATGGTGGTGAAGAC-3′ (reverse).
5′-RACE PCR assay to detect in vivo RNAi-mediated target mRNA cleavage
Total RNA was isolated from PBMCs of treated mice with STAT-60 as described above. Residual DNA was digested with the DNA-free kit per the manufacturer's instructions (Ambion). Subsequently, total RNAs (5 mg) were ligated to a GeneRacer adaptor (Invitrogen) without previous treatment. Ligated RNA was reverse-transcribed with a gene-specific primer 1 (GSP-Rev 1: 5′-CCACTTGCCACCCATCTTATAGCA-3′) and SuperScript III reverse transcriptase according to the manufacturer's instructions (Invitrogen). To detect cleavage products, we performed PCR and nested PCRs with primers complementary to the RNA adaptor (5′-cDNA primer: 5′-GGACACTGACATGGACTGAAGGAGTA-3′) and gene-specific primers (GSP-Rev 2: 5′-CCCAGAAGTTCCACAATCCTCGTT-3′; GSP-Rev 3: 5′-TGGTAGCTGAAGAGGCACAGGCTC-3′; GSP-Rev4: 5′-CGCAGATCGTCCCAGATAAGTGCTAA-3′). Amplification products were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining. The specific PCR products were recovered with a QIAquick Gel purification kit and then cloned into 17 TOPO TA cloning vector pCR 2.1-TOPO vector (Invitrogen). Individual clones were identified by DNA sequencing.
IFN assays
Total RNA was isolated from PBMCs of treated mice with STAT-60. Expression of mRNAs encoding p56 (CDKL2) and OAS1 were analyzed by qRT-PCR with 2× iQ SYBRGreen MasterMix (Bio-Rad) as described above and specific primer sets for these genes at final concentrations of 400 nM. Primers were as follows: p56 (CDKL2), 5′-TCAAGTATGGCAAGGCTGTG-3′ (forward) and 5′-GAGGCTCTGCTTCTGCATCT-3′ (reverse); OAS1, 5′-ACCGTCTTGGAACTGGTCAC-3′ (forward) and 5′-ATGTTCCTTGTTGGGTCAGC-3′ (reverse). gapdh expression was used for normalization of the qPCR data. In addition, 25 to 50 μl of EDTA-treated plasma was collected 2 and 24 hours after treatment. As a positive control for IFN-α induction, mice were injected with 5 μg of poly I:C (Sigma) intravenously in a 50-μl volume. IFN-α levels were evaluated by Human IFN-α1 ELISA Ready-SET-Go! (eBioscience).
Mutation assay
PBMCs were obtained from treated mice 3, 4, and 5 weeks after Ch A-1 injection, and total RNAs were isolated with STAT-60 (Tel-Test) according to the manufacturer's instructions. Residual DNA was removed with the DNA-free kit per the manufacturer's instructions (Ambion). cDNA was synthesized with 1 to 5 μg of total RNA. RT was carried out with SuperScript III reverse transcriptase and an oligo(dT)20 primer in a 20-μl reaction according to the manufacturer's instructions (Invitrogen). To detect viral RNAs, we performed PCR and nested PCRs using gene-specific primers (NL4-3 primer S1-F: 5′-TACAATGAATGGACACTAGAG-3′; NL4-3 primer S1-R: 5′-TTCTAGGTCTCGAGATACTG-3′; NL4-3 primer S2-F: 5′-AGGCGTTACTCGACAGAGGA-3′; NL4-3 primer S2-R: 5′-TGGCGAATAGCTCTATAAGC-3′; NL4-3 S3-F: 5′-ATGAGAGTGAAGGAGAAGTAT-3′; NL4-3 S3-R: 5′-AAGAGTAAGTCTCTCAAGCG-3′; NL4-3 S4-F: 5′-TCAGCACTTGTGGAGATGGG-3′; NL4-3 S4-R: 5′-TGGTGAATATCCCTGCCTAA-3′). Amplification products were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining. The specific PCR products were recovered with a QIAquick Gel purification kit. Illumina deep sequencing was carried out by the City of Hope DNA sequencing core, and data analyses were performed by the City of Hope Bioinformatics Core facility. As an alternative approach, gel-purified PCR products were also cloned into the TOPO TA cloning vector pCR 2.1-TOPO vector and individual clones were identified by DNA sequencing.
Illumina deep sequencing and data analysis
Treatment and control libraries of the target region of HIV-1 NL4-3 (from 5830 to 8785) were pooled into a single lane of a flow cell with an Illumina multiplexing sample preparation oligonucleotide kit. Each sample was labeled with a different six-base oligo as a bar code. Deep sequencing on the mixed sample was carried out with an Illumina Genome Analyzer (GA) II by running 43 cycles (36 cycles for sample read, 6 cycles for bar code, and 1 extra cycle for bar code offset estimation). Raw data from Illumina GA II were processed with the Solexa software pipeline v. 1.5.1 into 36–base pair shotgun reads. Sequences were aligned back to the HIV-1 NL4-3 genome [National Center for Biotechnology Information (NCBI): AF324493] with CLC Genomics Workbench, and sequences that mapped to more than one position with similar alignment scores were removed. Variations were detected in both treatment and control samples with Neighborhood Quality Standard (NQS) algorithm (33). Because of the experiment design, the mutation ratio of the variation caused by treatment could be very low. To distinguish these low-frequency mutations from the background noise (such as sequencing error, alignment bias, and system error), we used two thresholds. First, 1% variation rate or 100-fold variation coverage (coverage refers to the number of overlapping sequences used to build a region of the assembly) was used as quality control threshold for variation detection. This threshold makes it possible to detect all possible variations at each position and maintains reasonable quality of the variation. Second, a variation-to-noise ratio was used if there were multiple variations at the same location. We defined the variation-to-noise ratio as the ratio between the majority variation ratio and the sum of rest variations observed at this location. Here, the variation-to-noise ratio threshold was 10, and this threshold filtered out those variations that were caused by experimental background noise. Variations above the threshold were compared with the variations observed in controls to identify the mutations caused by treatment.
Statistical methods
The mouse viral loads and CD4-CD3 T cell ratios were plotted with a LOWESS (locally weighted scatterplot smoothing) smoother across values. For larger sample sizes, some type of linear model such as linear regression, multivariate analysis of variance (MANOVA), or generalized estimating equations are used. However, each of these requires a larger number of observations per group for stable variance estimation. Our approach transformed the data into a test of mean areas under the curve (AUCs), which may be compared using a t test, despite the small samples. Viral loads were first log-transformed before smoothing and then anti-transformed for plotting. Missing values were imputed with a last observation carried forward scheme. To measure the differences between mouse treatment groups, we considered a primary endpoint evaluating longitudinal behavior over time. Our method was to calculate a cumulative AUC for each mouse, and then compare aggregate mean AUCs between mouse groups. For any sequential time points (xi, xj), and their corresponding endpoints (yi, yj), the AUC was calculated with the area of a trapezoid: 0.5 × (xj – xi) × (yi + yj). The cumulative AUC of a single observation for the duration of the experiment was the cumulative sum of trapezoids. The Kruskal-Wallis rank sum ANOVA analog was used for the groupwise cumulative AUC comparisons. We compared pairwise group mean AUCs using t tests and exact permutation tests under the Hothorn and Hornik exactRankTests package for the R language (34–37). Details for deriving the permutation P values in general are discussed by Streitberg and Röhmel (38).
Supplementary Material
Acknowledgments
We thank NIH AIDS Research and Reference Reagents Program for HIV-1–related reagents used in this work. We thank E. Gilboa and J. Termini for critical reading of the manuscript and their constructive suggestions. The authors would like to thank J. Burnett for critical reading of this manuscript, City of Hope DNA sequencing core (H. Gao and J. Wang) for Solexa Deep Sequencing, and City of Hope Bioinformatics Core facility (X. Wu and H. Li) for prompt data analyses.
Funding: Supported by NIH RO1 grants AI057066 and AI073255 to R.A. and AI29329 and HL07470 to J.J.R. This work has also been facilitated by the infrastructure and resources provided by the Colorado Center for AIDS Research Grant P30 AI054907.
Footnotes
SUPPLEMENTARY MATERIAL www.sciencetranslationalmedicine.org/cgi/content/full/3/66/66ra6/DC1
Competing interests: J.J.R. is a paid consultant for Kylin Therapeutics, which uses bacteriophage-derived pRNAs as nanoparticles for delivery of aptamers and siRNAs, and is a cofounder and chairman of the Scientific Advisory Board of Dicerna Pharmaceuticals, which specializes in Dicer substrate siRNAs as therapeutic agents. J.J.R. and J. Zhou have a patent pending on cell type-specific aptamer-siRNA delivery system for HIV-1 therapy, and J.J.R., P.S., and J. Zhou have filed a patent disclosure for the in vitro evolution of the anti–HIV-1 gp120 aptamers. The other authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. The challenge of finding a cure for HIV infection. Science. 2009;323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 2.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 3.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 4.Chu TC, Marks JW, III, Lavery LA, Faulkner S, Rosenblum MG, Ellington AD, Levy M. Aptamer:toxin conjugates that specifically target prostate tumor cells. Cancer Res. 2006;66:5989–5992. doi: 10.1158/0008-5472.CAN-05-4583. [DOI] [PubMed] [Google Scholar]
- 5.Mayer G. The chemical biology of aptamers. Angew. Chem. Int. Ed. Engl. 2009;48:2672–2689. doi: 10.1002/anie.200804643. [DOI] [PubMed] [Google Scholar]
- 6.Kumar P, Ban HS, Kim SS, Wu H, Pearson T, Greiner DL, Laouar A, Yao J, Haridas V, Habiro K, Yang YG, Jeong JH, Lee KY, Kim YH, Kim SW, Peipp M, Fey GH, Manjunath N, Shultz LD, Lee SK, Shankar P. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell. 2008;134:577–586. doi: 10.1016/j.cell.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palliser D, Chowdhury D, Wang QY, Lee SJ, Bronson RT, Knipe DM, Lieberman J. An siRNA-based microbicide protects mice from lethal herpes simplex virus 2 infection. Nature. 2006;439:89–94. doi: 10.1038/nature04263. [DOI] [PubMed] [Google Scholar]
- 8.Rossi JJ. RNAi as a treatment for HIV-1 infection. Biotechniques. 2006:25–29. doi: 10.2144/000112167. [DOI] [PubMed] [Google Scholar]
- 9.Novina CD, Murray MF, Dykxhoorn DM, Beresford PJ, Riess J, Lee SK, Collman RG, Lieberman J, Shankar P, Sharp PA. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- 10.Rossi JJ, June CH, Kohn DB. Genetic therapies against HIV. Nat. Biotechnol. 2007;25:1444–1454. doi: 10.1038/nbt1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boden D, Pusch O, Ramratnam B. HIV-1-specific RNA interference. Curr. Opin. Mol. Ther. 2004;6:373–380. [PubMed] [Google Scholar]
- 12.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scherer L, Rossi JJ, Weinberg MS. Progress and prospects: RNA-based therapies for treatment of HIV infection. Gene Ther. 2007;14:1057–1064. doi: 10.1038/sj.gt.3302977. [DOI] [PubMed] [Google Scholar]
- 14.Chu T, Ebright J, Ellington AD. Using aptamers to identify and enter cells. Curr. Opin. Mol. Ther. 2007;9:137–144. [PubMed] [Google Scholar]
- 15.Chu TC, Twu KY, Ellington AD, Levy M. Aptamer mediated siRNA delivery. Nucleic Acids Res. 2006;34:e73. doi: 10.1093/nar/gkl388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dassie JP, Liu XY, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR, Meyerholz DK, McCaffrey AP, McNamara JO, II, Giangrande PH. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat. Biotechnol. 2009;27:839–849. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou J, Li H, Li S, Zaia J, Rossi JJ. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol. Ther. 2008;16:1481–1489. doi: 10.1038/mt.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou J, Swiderski P, Li H, Zhang J, Neff CP, Akkina R, Rossi JJ. Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells. Nucleic Acids Res. 2009;37:3094–3109. doi: 10.1093/nar/gkp185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNamara JO, II, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 20.Berges BK, Akkina SR, Folkvord JM, Connick E, Akkina R. Mucosal transmission of R5 and X4 tropic HIV-1 via vaginal and rectal routes in humanized Rag2−/−γc−/− (RAG-hu) mice. Virology. 2008;373:342–351. doi: 10.1016/j.virol.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denton PW, Garcia JV. Novel humanized murine models for HIV research. Curr. HIV/AIDS Rep. 2009;6:13–19. doi: 10.1007/s11904-009-0003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Legrand N, Ploss A, Balling R, Becker PD, Borsotti C, Brezillon N, Debarry J, de Jong Y, Deng H, Di Santo JP, Eisenbarth S, Eynon E, Flavell RA, Guzman CA, Huntington ND, Kremsdorf D, Manns MP, Manz MG, Mention JJ, Ott M, Rathinam C, Rice CM, Rongvaux A, Stevens S, Spits H, Strick-Marchand H, Takizawa H, van Lent AU, Wang C, Weijer K, Willinger T, Ziegler P. Humanized mice for modeling human infectious disease: Challenges, progress, and outlook. Cell Host Microbe. 2009;6:5–9. doi: 10.1016/j.chom.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Duyne R, Pedati C, Guendel I, Carpio L, Kehn-Hall K, Saifuddin M, Kashanchi F. The utilization of humanized mouse models for the study of human retroviral infections. Retrovirology. 2009;6:76. doi: 10.1186/1742-4690-6-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berges BK, Wheat WH, Palmer BE, Connick E, Akkina R. HIV-1 infection and CD4 T cell depletion in the humanized Rag2−/−γc−/− (RAG-hu) mouse model. Retrovirology. 2006;3:76. doi: 10.1186/1742-4690-3-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berges BK, Akkina SR, Remling L, Akkina R. Humanized Rag2−/−γc−/− (RAG-hu) mice can sustain long-term chronic HIV-1 infection lasting more than a year. Virology. 2010;397:100–103. doi: 10.1016/j.virol.2009.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foreman O, Kavirayani AM, Griffey SM, Reader R, Shultz LD. Opportunistic bacterial infections in breeding colonies of the NSG mouse strain. Vet. Pathol. doi: 10.1177/0300985810378282. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 28.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Moir S, Fauci AS. B cells in HIV infection and disease. Nat. Rev. Immunol. 2009;9:235–245. doi: 10.1038/nri2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, Manoharan M, Akira S, de Fougerolles A, Endres S, Hartmann G. Sequence-specific potent induction of IFN-a by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 31.Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat. Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 32.Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM, Feng Y, Palliser D, Weiner DB, Shankar P, Marasco WA, Lieberman J. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat. Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 33.Brockman W, Alvarez P, Young S, Garber M, Giannoukos G, Lee WL, Russ C, Lander ES, Nusbaum C, Jaffe DB. Quality scores and SNP detection in sequencing-by-synthesis systems. Genome Res. 2008;18:763–770. doi: 10.1101/gr.070227.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.David HA. Robust estimation in the presence of outliers. In: Launer RL, Wilkinson GN, editors. Robustness in Statistics. Academic Press; New York: 1979. pp. 61–74. [Google Scholar]
- 35.Guttman I, Smith DE. Investigation of rules for dealing with outliers in small samples from the normal distribution. I: Estimation of the mean. Technometrics. 1969;11:527–550. [Google Scholar]
- 36.R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2010. [Google Scholar]
- 37.Hothorn T, Hornik K. exactRankTests package for R Publisher. http://cran.r-project.org.
- 38.Streitberg B, Röhmel J. Exact distributions for permutation and rank tests: An introduction to some recently published algorithms. Stat. Softw. Newslett. 1986;12:10–17. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.