SUMMARY
Lipotoxicity is a metabolic stress response implicated in the pathogenesis of diabetes complications and has been shown to involve lipid-induced oxidative stress. To elucidate the molecular mechanisms of lipotoxicity, we used retroviral promoter trap mutagenesis to isolate a cell line that is resistant to lipotoxic and oxidative stress. We show that loss of three box C/D small nucleolar RNAs (snoRNAs) encoded in the ribosomal protein L13a (rpL13a) locus is sufficient to confer resistance to lipotoxic and oxidative stress in vitro and prevents the propagation of oxidative stress in vivo. Our results provide evidence for a previously unappreciated, non-canonical role for box C/D snoRNAs as regulators of metabolic stress response pathways in mammalian cells.
INTRODUCTION
Studies in mice and in humans have shown that lipid overload plays an important role in the pathogenesis of diabetic complications. In addition to hyperglycemia, free fatty acids and triglycerides are elevated in diabetes due to insulin resistance in both adipose depots and the liver. Hyperlipidemia leads to excessive delivery of fatty acid substrates to non-adipose tissues, resulting in lipid accumulation, which is associated with cellular dysfunction and cell death through the process of lipotoxicity (Unger, 1995). In diabetes and metabolic syndrome, lipotoxicity contributes to the pathogenesis of heart failure, renal dysfunction, steatohepatitis, and progressive β-cell insufficiency (Angulo, 2002; Jiang et al., 2005; Sharma et al., 2004; Shimabukuro et al., 1998).
To characterize the cellular mechanisms underlying lipotoxicity, many groups have employed cell culture models in which growth media is supplemented with excess fatty acids complexed to albumin to achieve free fatty acid levels in the pathophysiological range. Fibroblasts, myoblasts, pancreatic β-cells, hepatocytes, and endothelial cells demonstrate a time- and dose-dependent induction of cell death that is characterized by features of apoptosis (Cacicedo et al., 2005; de Vries et al., 1997; Listenberger et al., 2001; Maedler et al., 2003; Wei et al., 2006). This response is relatively specific for saturated free fatty acids and is augmented by the simultaneous exposure to elevated glucose concentrations (El-Assaad et al., 2003).
While increased triglyceride stores are the sine qua non of lipid overload in non-adipose cells, in vitro and in vivo studies suggest that sequestration of excess lipid in triglyceride droplets is cytoprotective (Listenberger et al., 2003; Liu et al., 2009). However, the relatively limited capacity for storage of excess fatty acids in triglyceride droplets in non-adipose cells is rapidly exceeded in hyperlipidemic states. This is followed by initiation of several cellular stress response pathways. Fatty acid-induced activation of NADPH oxidase and mitochondrial dysfunction due to remodeling of organelle membranes lead to oxidative stress in a variety of cell types (Inoguchi et al., 2000; Ostrander et al., 2001). The observation that anti-oxidants mitigate lipotoxic cell death supports a central role for oxidative stress in lipotoxicity (Borradaile et al., 2006b; Listenberger et al., 2001). Excess fatty acids also induce the endoplasmic reticulum (ER) stress response pathway, which may be precipitated by oxidative stress and/or by deleterious remodeling of ER membranes (Borradaile et al., 2006b; Cnop et al., 2007). Oxidative and ER stress responses to lipid overload have been demonstrated not only in cell culture models of lipotoxicity, but also in mouse models of diabetes (Ozcan et al., 2004). Nonetheless, the precise molecular mechanisms through which lipids induce these pathways remain to be elucidated.
To identify genes critical for the lipotoxic response, we performed a genetic screen in Chinese hamster ovary (CHO) cells using retroviral promoter trap mutagenesis to create single gene disruptions and positive selection for survival under lipotoxic growth conditions. Herein, we describe a mutant cell line in which the promoter trap has disrupted the locus for ribosomal protein L13a (rpL13a). Unexpectedly, our studies reveal that the portions of this gene essential for lipotoxicity are three highly conserved box C/D small nucleolar RNAs (snoRNAs) embedded within the rpL13a introns, rather than the protein-coding exonic sequences. Our findings suggest a previously unsuspected role for snoRNAs in the regulation of metabolic stress in mammalian cells.
RESULTS
Disruption of one rpL13a allele confers resistance to palmitate-induced apoptosis
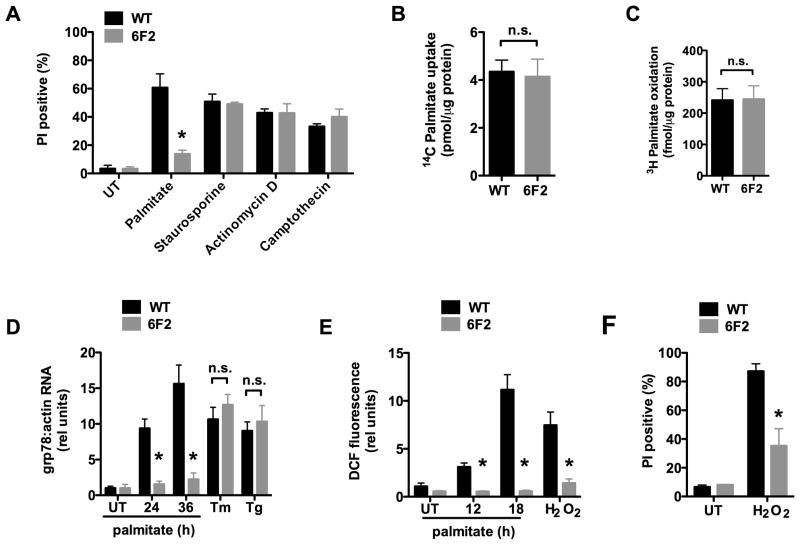
To identify genes critical for the cellular lipotoxic response, we performed a genetic screen in CHO cells, with mutagenesis by transduction with ROSAβgeo retrovirus at low multiplicity of infection to achieve, on average, one insertion per ten genomes. Although the integrated provirus contains a cDNA cassette for a β-galactosidase-neomycin phosphotransferase fusion protein, it lacks its own promoter, and thus its transcript is expressed only if the retrovirus inserts downstream of an active promoter and splice donor site. Mutagenized cells that survived a round of neomycin selection were then treated for 48 h in media supplemented with a lipotoxic concentration of palmitate (500 μM) to model pathophysiological states. Under these conditions, wild type (WT) cells were killed, but mutant line 6F2 survived.
Since palmitate-induced cell death occurs through activation of apoptosis, we first tested whether 6F2 cells retained the ability to activate these cell death pathways. Using flow cytometry, we quantified cell death by propidium iodide (PI) staining and apoptosis by TUNEL staining in parental WT and mutant 6F2 cells treated with palmitate or with three other inducers of apoptosis (Figure 1A & supplemental Figure 1A). Consistent with their isolation in a positive screen under lipotoxic conditions, 6F2 cells were significantly protected from palmitate-induced cell death and apoptosis compared to WT cells. However, 6F2 mutants were not significantly different from WT cells with respect to cell death or apoptosis induction following treatment with the other apoptosis inducers. These observations indicate that 6F2 mutant cells have intact cell death pathways, yet they are resistant to apoptosis induced by lipotoxic conditions.
Figure 1. 6F2 cells are resistant to palmitate-induced lipotoxicity.
(A) Wild type (WT) and mutant 6F2 CHO cells were untreated (UT) or supplemented with 500 μM palmitate for 48h or with 80 nM staurosporine, 2 μM actinomycin D, or 10 μM camptothecin for 24 h. Cell death was quantified by propidium iodide (PI) staining and flow cytometry on 104 cells/sample. (B) WT and 6F2 cells were incubated with 14C-palmitate for 1 min. Uptake of labeled palmitate was assessed by scintillation counting. (C) WT and 6F2 cells were incubated with 3H-palmitate for 2 h. 3H2O production as a measure of fatty acid oxidation was assessed by scintillation counting. (D) WT and 6F2 cells were treated with palmitate for indicated times, or with the ER stress inducers, tunicamycin (Tm, 2.5 μg/ml) or thapsigargin (Tg, 1 μM) for 6 h. grp78 expression was analyzed by quantitative real-time PCR (qRT-PCR) and normalized to β-actin expression. (E) WT and 6F2 cells were treated with palmitate for indicated times or with 500 μM H2O2 for 1 h. ROS generation was quantified by CM-H2DCFDA (DCF) labeling and flow cytometry on 104 cells/sample. (F) WT and 6F2 cells were UT or treated with 2.3 mM H2O2 for 20 h. Cell death was quantified by PI staining and flow cytometry on 104 cells/sample. All data expressed as mean ± SE for 3 independent experiments. * p < 0.01 for mutant 6F2 vs. WT; n.s., not significant. (See also supplemental Figure 1.)
Prior studies suggest that lipotoxic pathways can be mitigated by activation of pathways through which palmitate is metabolized (Borradaile et al., 2006b; Listenberger et al., 2003). Therefore, it was possible that the palmitate resistance of 6F2 mutant cells might have resulted from a defect in fatty acid uptake or increased ability to metabolize exogenous palmitate. We quantified cellular uptake of 14C-palmitate in WT and 6F2 cells under lipotoxic conditions (Figure 1B). The absence of a significant difference in lipid uptake between 6F2 and WT cells suggests that the defect in 6F2 cells is downstream of the cellular lipid transport machinery. Moreover, β-oxidation of exogenous palmitate was not affected by the mutation in 6F2 cells that conferred resistance to lipotoxicity (Figure 1C). Thus, resistance to lipotoxicity in the mutant line did not simply result from increased efficiency of palmitate metabolism.
Previous studies in cultured cells and in mouse models have implicated ER stress and oxidative stress pathways in the pathogenesis of lipotoxicity (Ozcan et al., 2004). We hypothesized that one or both of these stress pathways might be affected in 6F2 cells. We first compared the induction of ER stress in 6F2 and WT cells following treatment with palmitate. Palmitate treatment in WT CHO cells resulted in a time-dependent increase in expression of the protein-folding chaperone grp78 (Figure 1D). This induction was significantly reduced in mutant 6F2 cells suggesting resistance to ER stress induction. Consistent with this observation, 6F2 cells also showed delayed and diminished splicing of XBP-1 pre-mRNA and induction of CHOP expression (supplemental Figure 1B & D). However, 6F2 and WT cells were indistinguishable in these measures following treatment with the chemical ER stress inducers, tunicamycin and thapsigargin (Figure 1D and supplemental Figure 1 C & E). Together, these results suggest that 6F2 cells are specifically resistant to palmitate-induced ER stress, while the general ER stress response machinery remains intact.
ER stress in lipotoxicity may be induced, in part, downstream of lipid-induced oxidative stress. To assess palmitate-induced oxidative stress we measured levels of intracellular reactive oxygen species (ROS) using the dihydrofluorescene derivative, CM-H2DCFDA (6-carboxy-2′,7′-dicholorodihydrofluorescein diacetate, di(acetoxymethyl ester) or DCF). Palmitate treatment resulted in a time-dependent increase in DCF fluorescence in WT CHO cells, indicative of an increase in ROS (Figure 1E). This response was abrogated in 6F2 cells. When ROS production was more directly induced by treatment with hydrogen peroxide (H2O2), 6F2 cells showed ROS levels that were markedly blunted compared to WT cells. Furthermore, 6F2 cells were protected from H2O2-induced cell death when compared to WT cells (Figure 1F). Together, these data suggest that 6F2 cells are resistant not only to palmitate-induced oxidative stress, but also to more generalized oxidative stress induction.
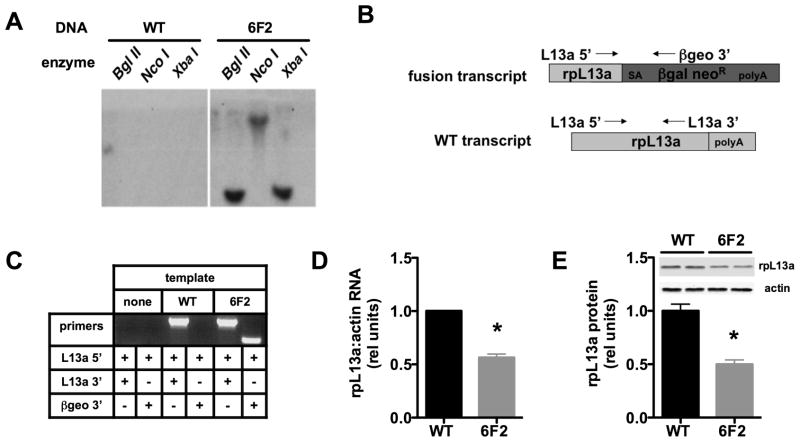
The promoter trap mutagenesis approach we employed facilitates identification of the disrupted gene, because following a single proviral insertion, a unique fusion transcript is generated that includes the 5′ end of the disrupted gene’s mRNA followed by the ROSAβgeo sequence. To confirm the existence of a single genomic integration, we performed Southern blot analysis of genomic DNA from 6F2 cells (Figure 2A). Labeled probe complementary to proviral sequences hybridized to only one band in each of three digests of 6F2 DNA, indicating that there is likely a single proviral integration in the mutant line. 5′RACE (rapid amplification of cDNA ends) revealed the disrupted gene to be ribosomal protein L13a (rpL13a) (Figure 2B). This integration was confirmed by PCR, which revealed (Figure 2C) a fusion transcript in 6F2, but not WT cells, and a WT transcript in both cell types. This suggests that in addition to the trapped allele, 6F2 cells retain an intact rpL13a allele and thus represent a model of haploinsufficiency. Consistent with this model, 6F2 cells have a ~50% reduction in rpL13a mRNA (Figure 2D) and protein (Figure 2E) expression compared to WT cells. Of note, although this protein is a constituent of the large 60S ribosomal subunit, haploinsufficient 6F2 cells showed no decrease in overall rates of protein synthesis compared to WT cells under either normal growth or lipotoxic conditions (supplemental Figure 2).
Figure 2. 6F2 cells are haploinsufficient for rpL13a.
(A) Genomic DNA isolated from WT and 6F2 mutant cells was digested with Bgl II, Nco I, and Xba I and analyzed by Southern blotting with 32P-labeled probe derived from ROSAβgeo retroviral sequences. (B) Upon transduction with the ROSAβgeo retrovirus, the provirus containing the splice acceptor (SA), the promoterless β-galactosidase-neomycin resistance cassette (βgal neoR), and polyadenylation (polyA) sequences integrated into the rpL13a gene in 6F2 cells. Diagram shows resulting ROSAβgeo fusion transcript containing 5′ end of rpL13a mRNA and βgal neoR sequences and the endogenous WT rpL13a mRNA. Forward (L13a 5′) and reverse (L13a 3′) primers for rpL13 and reverse proviral primer (βgeo 3′) are shown with arrows. (C) PCR was performed with the indicated primers on cDNA from WT and 6F2 cells to detect endogenous rpL13a and fusion transcript expression. (D) Basal rpL13a RNA expression in WT and 6F2 mutant cells was determined by qRT-PCR and normalized to β-actin expression. (E) Basal rpL13a protein expression in WT and 6F2 mutant cells was determined by western blotting and quantified by densitometry. Representative blot shown for rpL13a and actin loading control. All data expressed as mean ± SE for 3 independent experiments. * p < 0.01 for mutant 6F2 vs. WT. (See also supplemental Figure 2.)
Palmitate treatment induces rpL13a snoRNA accumulation in CHO cells
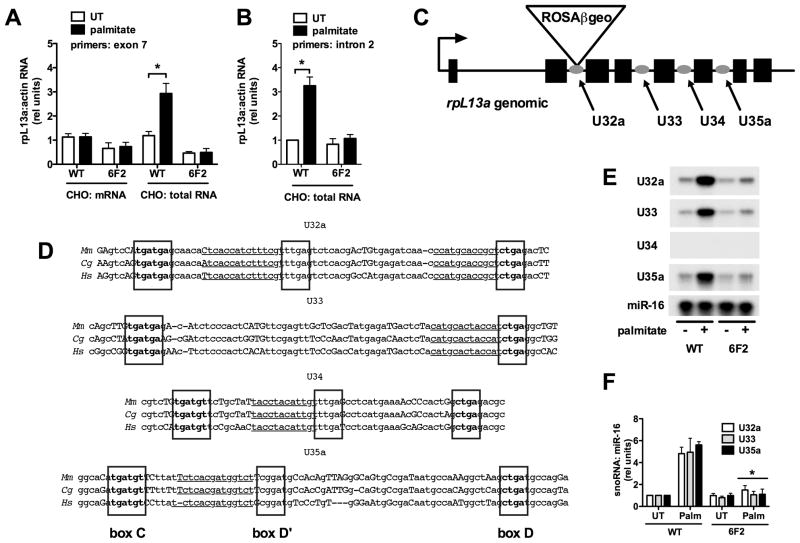
We hypothesized that rpL13a might play a role in palmitate-induced cell death as a lipotoxicity-inducible gene, as has been the case for disrupted genes in two previously identified mutants from our laboratory (Borradaile et al., 2006a; Brookheart et al., 2009). To test for rpL13a induction, we quantified rpL13a RNA by quantitative real-time PCR (qRT-PCR). mRNA levels (oligo-dT-primed cDNA) did not increase in WT or 6F2 cells under lipotoxic conditions (Figure 3A). On the other hand, using total RNA (random hexamer-primed cDNA) as template, we observed a 3-fold increase in rpL13a RNA in WT, but not in 6F2 cells under lipotoxic conditions (Figures 3A & B). These findings suggested that in the setting of cellular mechanisms for tight regulation of rpL13a mRNA levels, there is specific induction of rpL13a pre-mRNA under lipotoxic conditions. This prompted us to examine the non-coding regions of the rpL13a locus.
Figure 3. rpL13a-encoded box C/D snoRNAs are induced in palmitate treated CHO cells.
(A, B, E, F) WT and 6F2 cells were untreated (UT) or supplemented with palmitate for 48 h. (A, B) cDNA synthesis was primed with either oligo dT (mRNA) or random hexamers (total RNA). (A) rpL13a expression was analyzed by qRT-PCR, using primers specific to exon 7, and normalized to β-actin expression. (B) rpL13a expression was analyzed by qRT-PCR, using primers specific to intron 2, and normalized to β-actin expression. (C) rpL13a gene organization showing location of ROSAβgeo promoter trap insertion and locations of intronic U32a, U33, U34 and U35a snoRNAs. (D) Comparison of rpL13a snoRNA sequences from mouse (Mm), hamster (Cg) and human (Hs) species. C, D and D′ box sequences are indicated in boxed, bolded text. Regions with rRNA antisense homology are underlined. Conserved and non-conserved nucleotides are displayed in lower and upper case letters, respectively. (E) Small RNA was harvested and used in RNase protection assay with 32P- labeled hamster rpL13a snoRNA probes or miR-16 probe as control. (F) Autoradiograms from RNase protection experiments as in (E) were quantified by densitometry. All data expressed as mean ± SE for 3 independent experiments. * p < 0.01 for palmitate treated vs. untreated. (See also supplemental Figure 3.)
The genomic structure of rpL13a is highly conserved across diverse species with respect to intron-exon architecture. Furthermore, in addition to expected conservation of protein-coding sequences, all mammalian loci for rpL13a contain four highly conserved intronic box C/D small nucleolar RNAs (snoRNAs) that are predicted to be processed during splicing of the rpL13a pre-mRNA transcript (Figure 3C) (Nicoloso et al., 1996). These snoRNAs, U32a, U33, U34, and U35a, are located within introns 2, 4, 5 and 6, respectively, and range in size from 61–82 nucleotides. We cloned the rpL13a genomic sequence from CHO cells and found that U32a, U33, U34, and U35a are conserved 94, 87, 91 and 81%, respectively, between hamster and humans (Figure 3D). Our RACE results indicated that the proviral insertion in 6F2 mutant cells is downstream of rpL13a exon 2, which is predicted not only to disrupt expression of the processed rpL13a mRNA, but also expression of the full length pre-mRNA and the intronic snoRNAs.
Since lipotoxic conditions induced the expression of total RNA from the rpL13a locus, we hypothesized that rpL13a snoRNAs would also accumulate under stress conditions. We assessed the expression of rpL13a-encoded snoRNAs under lipotoxic conditions by RNase protection assays with 32P-labeled probes specific for the snoRNA sequences (Figure 3E & F). Levels of the microRNA miR-16 did not change under these conditions and served as a loading control. In WT CHO cells, U32a, U33 and U35a snoRNAs are expressed at low levels under basal conditions and increased following palmitate treatment, whereas U34 was not detected under basal or lipotoxic conditions. In mutant 6F2 cells, induction of U32a, U33 and U35a was markedly attenuated, consistent with the lack of induction of total rpL13a RNA (Figure 3A & B). We also observed palmitate induction of U32a, U33 and U35a snoRNAs in WT C2C12 murine myoblasts, which demonstrate similar sensitivity to lipotoxic conditions (supplemental Figure 3A–G). Moreover, these snoRNAs were induced by saturated fatty acids known to cause lipotoxicity (myristic, palmitic and stearic acids), but not by unsaturated palmitoleic acid and oleic acid, which are well tolerated by cells (supplemental Figure 3H). Together these findings suggest a previously undescribed, conserved role for snoRNAs as mediators of lipotoxicity.
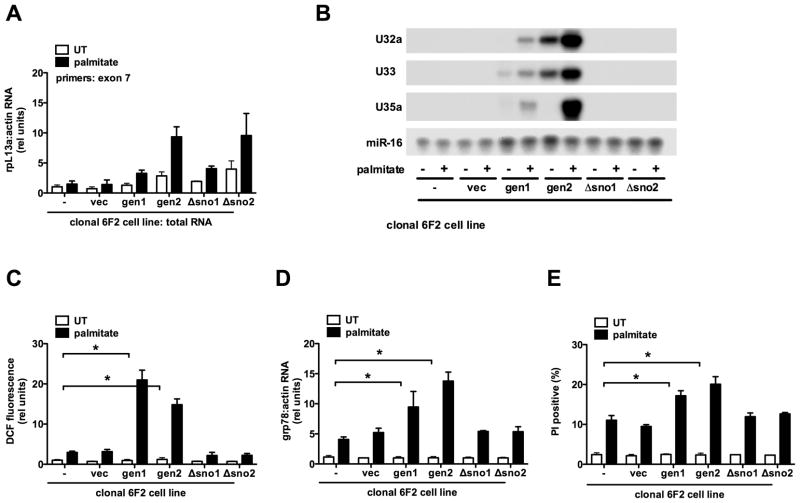
Re-expression of rpL13a genomic sequence complements 6F2 cells
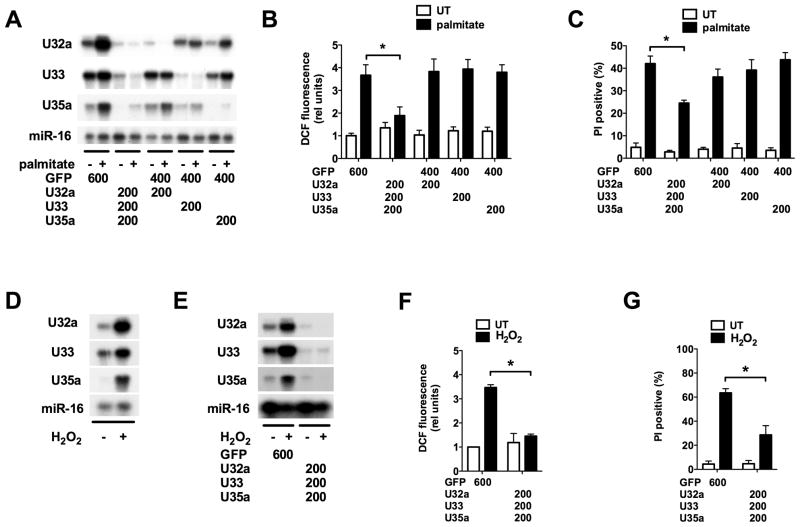
To determine if the rpL13a genomic locus is sufficient to restore palmitate-sensitivity, we generated stable cell lines in the mutant 6F2 background. Mutant cells were transfected with a plasmid containing 4.3 kb from the murine rpL13a genomic locus, including all eight exons and intervening introns, expression of which was driven by 1 kb of the endogenous rpL13a promoter (gen). As a control, mutant cells were also transfected with a similar construct in which all four snoRNAs were removed, but promoter and exon-intron structure was otherwise intact (Δsno). Stable clonal gen and Δsno lines with comparable low basal and palmitate-inducible expression of total rpL13a RNA were isolated and compared with 6F2 cells transfected with vector sequences alone (vec) and parental 6F2 cells (−) (Figure 4A). Complementation of 6F2 cells with murine sequences enabled us to distinguish between expression of hamster rpL13a sequences from the remaining single allele in 6F2 cells and expression from exogenously supplied murine sequences, because the RNase protection assays are sensitive to the small differences between hamster and murine sequences. We detected palmitate-inducible expression of murine specific rpL13a snoRNAs U32a, U33 and U35a in cell lines gen1 and gen2 (Figure 4B). As expected, no expression of the murine snoRNAs was detected in the Δsno1, Δsno2 or vec lines. Concomitant with palmitate-inducible expression of the snoRNAs in gen1 and gen2, we observed significant increases in palmitate-mediated ROS generation, grp78 mRNA induction, and cell death (Figure 4C–E). Palmitate-induced ROS generation, grp78 induction and cell death were indistinguishable in Δsno1, Δsno2, vec and untransfected 6F2 cells. These data demonstrate that genomic rpL13a sequences complement the phenotype in 6F2 cells and strongly support a specific role for rpL13a-encoded snoRNAs as mediators of lipotoxicity. Additionally, while transient transfection of 6F2 cells with CMV promoter-driven rpL13a cDNA and genomic constructs supported higher levels of expression of the rpL13a mRNA, only the genomic construct restored palmitate-induced oxidative stress (supplemental Figure 4). This further supports a model in which rpL13a intronic sequences are critical for lipotoxicity.
Figure 4. rpL13a genomic sequence containing box C/D snoRNAs complements mutant 6F2 cells.
(A–E) Mutant 6F2 cells were stably transfected with a construct containing genomic rpL13a murine sequence, genomic rpL13a murine sequence with deletion of U32a, U33, U34, and U35a, or the empty vector. Clonal cell lines were isolated and analyzed untreated (UT) or supplemented with palmitate for 48 h. Graphs show untransfected 6F2 cells (−), a stable 6F2 line harboring vector sequences only (vec), and lines expressing genomic (gen1, gen2) or snoRNA-deleted (Δsno1, Δsno2) rpL13a sequences. (A) cDNA synthesis was primed with random hexamers and rpL13a expression analyzed by qRT-PCR and normalized to β-actin. Primers were chosen from exon 7 sequence that is completely conserved between mouse and hamster. (B) Small RNA was used in RNase protection assay with murine-specific rpL13a snoRNA probes or mir-16 probe as control. (C) ROS generation was quantified by DCF labeling and flow cytometry. (D) cDNA synthesis was primed with oligo dT and grp78 expression analyzed by qRT-PCR and normalized to β-actin. (E) Cell death was assessed by PI staining and flow cytometry on 104 cells/sample. All data expressed as mean ± SE for 3 independent experiments. * p < 0.01. (See also supplemental Figure 4.)
Directed knockdown of rpL13a snoRNAs protects against lipotoxicity
To extend these findings to another cell type and to test directly whether snoRNA expression is necessary for induction of lipotoxicity, we nucleofected WT murine C2C12 myoblasts with phosphorothioate-modified anti-sense oligos (ASO) complementary to the 20-nucleotide sequence upstream of the box D motifs in U32a, U33 and U35a (Ideue et al., 2009). As these sequences are not represented in the murine rpL13a mRNA sequence and do not have significant homologies to other known sequences in the NCBI databases, this approach provided a means for examining the specific contributions of each of the snoRNAs. ASO were nucleofected alone and in combination into cells and compared to a control ASO directed against green fluorescent protein (GFP) sequence. Nucleofected cells were assayed under basal and lipotoxic (palmitate-supplemented) conditions. Using ASO directed against each snoRNA, we achieved efficient knockdown of U32a, U33 and U35a snoRNAs individually without affecting the expression of the other rpL13a-encoded snoRNAs, indicating that each ASO preferentially targeted its specific snoRNA species and not the pre-mRNA (Figure 5A). ASO directed against U35a did not affect expression of the highly related snoRNA U35b, further supporting the specificity of this approach (supplemental Figure 5A). A cocktail of all three ASO effectively and simultaneously knocked down palmitate-induced expression of all three snoRNAs (Figure 5A). This was associated with a 50% decrease in palmitate-induced ROS and 40% decrease in palmitate-induced cell death compared to control cells (Figures 5B & C). Knockdown of any one of the snoRNAs alone was not sufficient to significantly decrease palmitate-induced ROS generation or PI positivity. As a control, knockdown of three unrelated box C/D snoRNAs did not protect against palmitate-induced oxidative stress or death (supplemental Figure 5B–D). These findings are consistent with the palmitate-resistant phenotype we observe in rpL13a-deficient, palmitate-resistant 6F2 mutant cells. Furthermore, they directly implicate induction of the three rpL13a-encoded snoRNAs in palmitate-induced ROS and cell death. Our data support a model in which the three snoRNAs function in concert to promote palmitate-induced oxidative stress, since loss of a single snoRNA alone does not abrogate this response.
Figure 5. Antisense oligo (ASO)-mediated knockdown of rpL13a-encoded snoRNAs protects C2C12 murine myoblasts from palmitate-induced and generalized oxidative stress and cell death.
(A–C) C2C12 myoblasts were nucleofected with the indicated ASOs, designed to specifically target U32a, U33 or U35a snoRNAs or green fluorescent protein (GFP) as a control. Nucleofected cells were then untreated (UT) or supplemented with palmitate for 24 h. (A) Small RNA was used in RNase protection assay with rpL13a snoRNA probes or miR-16 probe as control. (B) ROS generation was quantified by DCF labeling and flow cytometry. (C) Cell death was quantified by PI staining and flow cytometry. (D) Following treatment of C2C12 myoblasts with 750 μM H2O2 for 20 h, small RNA was harvested and used in RNase protection assay as in (A). (E–G) C2C12 myoblasts were nucleofected with ASOs to target U32a, U33 or U35a snoRNAs or GFP and were then UT or treated with H2O2. (E) Small RNA was analyzed by RNase protection. (F) ROS generation was quantified by DCF labeling. (G) Cell death was quantified by PI staining. All data expressed as mean ± SE for 3 independent experiments. * p < 0.01. (See also supplemental Figure 5.)
rpL13a snoRNAs are mediators of general oxidative stress
In 6F2 cells, genetic disruption of rpL13a not only protects against lipotoxicity, but also leads to decreased ROS-generation and cell death in response to H2O2 treatment (Figure 1E and F), suggesting that the rpL13a locus serves as a mediator of generalized oxidative stress. To test whether the rpL13a snoRNAs play a role in generalized oxidative stress, we treated C2C12 cells with H2O2 and assessed snoRNA induction. Similar to lipotoxic conditions, treatment with H2O2 increased U32a, U33 and U35a snoRNA expression (Figure 5D). ASO-mediated knockdown of the snoRNAs was effective in blocking H2O2-induced expression of U32a, U33, and U35a (Figure 5E) and resulted in a significant decrease in H2O2–induced ROS generation (Figure 5F) compared to GFP control. Furthermore, snoRNA knockdown resulted in a 55% reduction in H2O2-induced cell death in C2C12 cells (Figure 5G). Together, these data implicate rpL13a snoRNAs in the more general cellular response to oxidative stress.
rpL13a snoRNAs function as non-canonical box C/D snoRNAs
Canonical box C/D snoRNAs participate in ribonucleoproteins that localize to nucleoli. In S. cerevisiae and in X. laevis, box C/D snoRNAs serve as guides that target 2′-O-methylation of rRNAs with which they share short stretches of antisense homology (Kiss-Laszlo et al., 1996). Although they lack some sequence features of canonical 2′-O-methylation guide snoRNAs (internal box C′ sequence not well-conserved, U33 lacks box D′, rRNA complementarity not upstream of box D in U35a), U32a, U33 and U35a each contain 10–12 nucleotide stretches of complementarity to rRNA sites of 2′-O-methylation (Figure 3D), suggesting a potential role as guide RNAs for 2′-O-methylation of G1328 in 18S and A1511 in 28S (U32a), U1326 in 18S (U33), and C4506 in 28S (U35a) rRNAs (Nicoloso et al., 1996). We reasoned that if the mechanism of action of snoRNAs U32a, U33 and U35a in lipotoxic and oxidative stress involved 2′-O-methylation of these rRNAs, modifications of these rRNA sites should be diminished in 6F2 compared to WT cells under metabolic stress conditions when the snoRNAs are induced in WT cells. However, primer extension studies showed no differences in the extent of modification of these rRNA sites between WT and 6F2 cells under basal or palmitate-treated conditions (supplemental Figure 6). These data indicate that under basal and lipotoxic conditions, either residual expression of U32a, U33 and U35a in 6F2 cells is sufficient to support these modifications of rRNAs, or this function is subserved by other molecules in eukaryotic cells. Furthermore, at a point in the lipotoxic response at which absence of snoRNA induction is readily apparent and functionally correlates with resistance to lipotoxicity in 6F2 cells, there is no corresponding change in 2′-O-methylation of rRNAs.
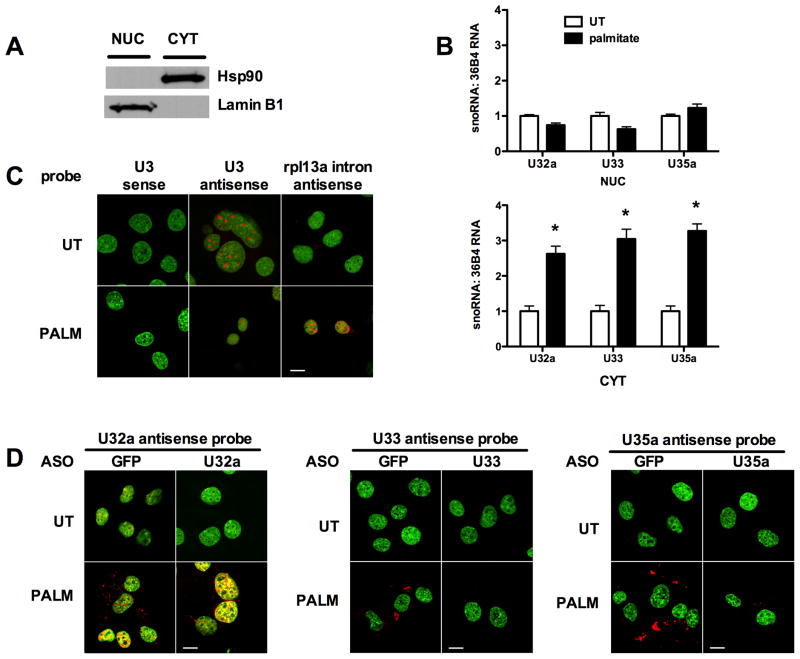
We hypothesized that if U32a, U33 and U35a were involved in functions other than modification of ribosomal RNAs, then under lipotoxic stress conditions, they may have a subcellular distribution distinct from canonical box C/D snoRNAs, which are nucleolar. Following palmitate treatment of C2C12 cells, we isolated nuclear and cytosolic RNAs and quantified U32a, U33 and U35a by qRT-PCR. With palmitate treatment U32a, U33 and U35a increase in the cytoplasm, whereas levels of these snoRNAs remain unchanged in the nucleus (Figure 6A and B). Accumulation of rpL13a snoRNAs in the cytosol under lipotoxic conditions was confirmed by fluorescence in situ hybridization. As expected, anti-sense probe for snoRNA U3 demonstrated strong nucleolar localization, and this was unaffected by lipotoxic stress (Figure 6C). Staining for the rpL13a snoRNAs was perfomed in cells nucleofected with control ASO (GFP) or with ASO targeting each of the rpL13a snoRNAs. Consistent with data from RNase protection and qPCR assays, in control nucleofected cells expression of U32a, U33 and U35a was low under normal growth conditions and increased under lipotoxic conditions (Figure 6D, GFP-nucleofected panels). Prominent staining for each of these snoRNAs was observed in the cytoplasm, but not nucleoli. The probe for U32a also stains non-nucleolar regions of the nucleus. Cytoplasmic staining for the rpL13a snoRNAs under lipotoxic conditions was markedly diminished when the snoRNAs were depleted by specific ASOs that target each snoRNA (Figure 6D, U32a, U33 and U35a ASO-nucleofected panels). Cytoplasmic staining for U32a, U33 and U35a was also distinct from the nuclear pattern observed under lipotoxic conditions using a probe specific for intron 1 (Figure 6C), indicating that the cytosolic distributions of the rpL13a snoRNAs do not simply reflect localization of the pre-mRNA. The nuclear staining for U32a resembled the staining for intron 1 and was not diminished with ASOs that target U32a, suggesting that this represents detection of the pre-mRNA, which is not targeted by U32a ASOs. Together our biochemical and in situ hybridization data support a model in which U32a, U33 and U35a snoRNAs act in non-canonical roles in the cytoplasm during lipotoxic stress.
Figure 6. U32a, U33 and U35a accumulate in the cytoplasm under lipotoxic conditions.
(A, B) C2C12 cells were untreated (UT) or treated with palmitate for 24 h. Cells were separated into cytosolic (CYT) and nuclear (NUC) fractions by sequential detergent solubilization. (A) Fractions were analyzed by western blotting for a cytosolic marker, hsp90, and for a nuclear marker, lamin B1. (B) Total RNA was prepared from the fractions and analyzed for rpL13a snoRNA abundance relative to 36B4 by qRT-PCR. Graphs show mean ± SE from a representative experiment (n = 3). * p < 0.05 for palmitate treated vs. UT. (C, D) C2C12 cells were analyzed by in situ hybridization under basal conditions (UT) and following 24 h treatment with palmitate (PALM) using specific snoRNA or control probes (red). Nuclei were stained with SYTOX Green. (C) Cells were probed with U3 antisense probe for known nucleolar snoRNA, control U3 sense probe, and control rpL13a intron 1 antisense probe. (D) Control GFP ASO-nucleofected and specific snoRNA ASO-nucleofected cells were examined by in situ hybridization with antisense probes for U32a, U33 and U35a. Bars, 10 μm. (See also supplemental Figure 6.)
In vivo expression of rpL13a snoRNAs
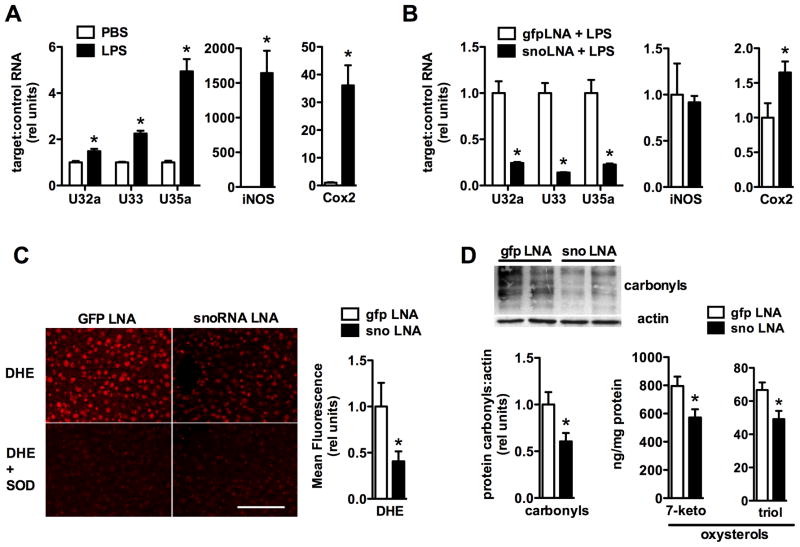
Our studies of the rpL13a snoRNAs show that these molecules function in the cellular response to lipotoxic and oxidative stress. To extend these findings to an in vivo model of oxidative stress, we examined the expression of rpL13a snoRNAs in a well-established model of lipopolysaccharide (LPS)-mediated liver injury that is characterized by inflammation, steatosis and oxidative stress (Bautista and Spitzer, 1990). Compared to saline-injected control mice, liver tissue from LPS-treated mice showed significant up-regulation of U32a, U33 and U35a snoRNAs (Figure 7A), demonstrating that the rpL13a snoRNAs are induced in vivo in response to metabolic stress.
Figure 7. rpL13a snoRNAs are required for oxidative stress in vivo.
(A) Mice were injected intraperitoneally with lipopolysaccharide (LPS), or equivalent volume of phosphate-buffered saline (PBS) as control, and liver tissue was harvested 12 h later. RNA isolated from livers was used for quantification of rpL13a snoRNAs (relative to 36B4) or iNOS and Cox2 (relative to actin). Graph shows mean ± SE from a representative experiment with N = 3 (PBS) and N = 4 (LPS) animals per group. * p < 0.05 for LPS vs. PBS. (B, C, D) Mice were pretreated with three serial doses of ASOs targeting rpL13a snoRNAs or GFP as control prior to LPS injection and analysis of liver tissue. (B) snoRNA, iNOS, and Cox2 expression in the liver was quantified as in (A). (C) Representative images show frozen sections of liver tissue stained with DHE and parallel sections in which staining was performed in the presence of pegylated superoxide dismutase (SOD) as control. Scale bar, 100 μm. Graph shows quantification of fluorescence intensity. (D) Quantification of protein carbonylation by western blotting and tissue oxysterols (7-ketocholesterol, 7-keto; 3β,5α,6β-cholestantriol, triol) 24 h following LPS. n = 4 to 6 per group. * p < 0.05 for GFP vs. snoRNA knockdown.
Based on these findings, we analyzed the effects of loss-of-function of the rpL13a snoRNAs in the LPS-mediated liver injury model. To achieve specific knockdown of the snoRNAs in vivo, mice were treated with three serial intraperitoneal injections of antisense locked nucleic acid oligonucleotides directed against each of the three snoRNAs or directed against GFP as a control prior to LPS injection. Antisense oligonucleotides directed against the snoRNAs achieved 72, 84, and 74% knockdown of U32a, U33 and U35a, respectively, in liver tissue following LPS injection without diminishing LPS-induced inflammation (Figure 7B). Knockdown of the snoRNAs mitigated LPS-induced oxidative stress as demonstrated by dihydroethidium staining for superoxide and oxidative damage to liver tissue proteins and lipids (Figures 7C and D). These findings indicate that rpL13a snoRNAs are required in vivo for propagation of oxidative stress.
DISCUSSION
In this study, we combined promoter trap mutagenesis in CHO cells with a positive selection for survival in the setting of metabolic stress in order to characterize molecular mediators of lipotoxicity. While the isolation of palmitate-resistant 6F2 cells identified the rpL13a gene as a mediator of lipotoxicity, the results of complementation experiments in 6F2 cells and directed knockdown of snoRNAs in WT murine myoblasts demonstrate that snoRNAs within this locus, rather than the encoded protein, are critical mediators of lipotoxic cell death in both the hamster and murine species. Consistent with a primary role for these snoRNAs, lipotoxic stress strongly induces expression of U32a, U33 and U35a, but has no effect on the steady state levels of the mRNA encoding rpL13a protein. Our observations that these snoRNAs are similarly induced following exposure to H2O2, that 6F2 cells are resistant to H2O2-induced ROS and cell death, and that knockdown of the snoRNAs protects against H2O2 cytotoxicity together implicate these snoRNAs as mediators of the general response to oxidative stress. We have extended these findings in vivo by showing that the rpL13a snoRNAs are induced in response to LPS-mediated liver injury and are required for the propagation of oxidative stress in LPS-treated livers.
The rpL13a locus is highly conserved across species. As a constituent of the 60S ribosomal subunit, the encoded rpL13a protein is likely to play an important role in peptide synthesis. Like many ribosomal proteins, it is predicted to reside as a globular domain on the surface of the large ribosomal subunit, and may contribute more to regulation of peptide production than to catalysis of peptide synthesis (Moore and Steitz, 2003). Thus, it is not entirely surprising that the 6F2 mutant, which is haploinsufficient for rpL13a protein, has rates of overall protein synthesis that are indistinguishable from WT cells, a finding consistent with failure of directed knockdown of rpL13a protein to change global translational activity or alter translational fidelity (Chaudhuri et al., 2007). Extra-ribosomal roles for large and small ribosomal subunit proteins have also been described, including a role for rpL13a in translational silencing (Mazumder et al., 2003). However, the function of the rpL13a locus in lipotoxicity is unlikely to involve this mechanism, since polysome association of rpL13a and expression from a reporter construct containing a GAIT element in the 3′ untranslated region were unaffected by lipotoxicity (not shown).
A key observation that facilitated our understanding of the contributions of the rpL13a locus to lipotoxicity was the extent of sequence conservation of this gene beyond the protein-coding exons. Four box C/D snoRNAs, U32a, U33, U34 and U35a located in rpL13a introns 2, 4, 5 and 6, respectively, are highly conserved across mammalian species both in terms of their primary sequence and their position within the locus. While box C/D snoRNAs function as guides for 2′-O-methylation in yeast (Lowe and Eddy, 1999), the sequences of the U32a, U33, U34 and U35a snoRNAs and their genomic organization diverge substantially from yeast to mammals. In mammals, a role in the modification of ribosomal RNAs has not been demonstrated for these snoRNAs. We show here in a stable mutant cell line that decreased basal expression of U32a, U33 and U35a snoRNAs and loss of palmitate induction of these snoRNAs is not associated with changes in 2′-O-methylation of predicted rRNA targets, yet is associated with resistance to lipotoxicity. While it is possible that these snoRNAs contain more than one functional guide sequence to target multiple substrates including rRNAs (Cavaille et al., 1996; Tycowski et al., 1998), their accumulation in the cytosol during lipotoxicity suggests a non-nucleolar function for these snoRNAs. An independently transcribed box C/D snoRNA, U8, has been shown to be exported from the nucleus (Watkins et al., 2007). The present study provides evidence for intronic snoRNAs in the cytosol. Our data are most consistent with a model in which the rpL13a snoRNAs function in lipotoxicity and oxidative stress response pathways as non-canonical box C/D snoRNAs. Our results demonstrate that U32a, U33 and U35a function coordinately in lipotoxic and oxidative cellular stress responses in vitro and in propagation of oxidative stress in vivo. Thus, our study provides evidence for snoRNA regulation of metabolic stress response pathways.
Increases in rpL13a total RNA are concomitant with induction of U32a, U33 and U35a, suggesting that induction of these snoRNAs is regulated, either at a transcriptional level or at the level of pre-mRNA stabilization. Under basal conditions, mutant 6F2 cells have one half the level of expression of rpL13a mRNA and protein and diminished U32a, U33 and U35a expression compared to WT cells, consistent with a model of haploinsufficiency. However, 6F2 cells fail to induce expression of total rpL13a RNA and the U32a, U33 and U35a snoRNAs upon lipotoxic stress, despite retaining one intact allele for rpL13a. Although CHO cells are diploid by cytogenetic and chemical criteria, a large body of evidence indicates that they are haploid at many loci by gene expression and functional criteria (Siminovitch, 1976). As a consequence, there are many CHO cell mutants in which mutation of only one of two alleles produces a null or near-null phenotype (e.g., (Cao et al., 1996)). Furthermore, evidence is emerging that snoRNA loci, particularly in the brain, are sites of developmentally programmed epigenetic changes that affect gene expression (Leung et al., 2009). Thus, the remaining allele in mutant 6F2 cells may no longer support transcriptional induction under cellular stress conditions due to changes in chromatin structure of the rpL13a locus. While lipotoxicity and oxidative stress induce rpL13a in total RNA pools and production of U32a, U33, and U35a, these stresses do not affect rpL13a mRNA levels. This implies that cells have an ability to regulate levels of rpL13a mRNA tightly, possibly by selective elimination of the mRNA. Although U34 is not similarly induced by lipotoxic or oxidative stress in fibroblasts or myoblasts, coordinate induction of all four rpL13a snoRNAs in murine embryonic stem cells under lipotoxic stress (our unpublished observations) suggests that there may be cell type-specific or developmentally-regulated mechanisms for regulation of snoRNA processing or stability.
Our observations in the LPS-induced model of hepatic tissue injury confirm that the rpL13a snoRNAs are induced in the setting of oxidative stress in vivo. Moreover, our in vivo knockdown data show that these snoRNAs are required for elements of the oxidative stress response, including the production of superoxide, protein carbonyls, and oxysterols. Because LPS has pleiotropic effects in vivo and because ASO knockdown in vivo is limited by liver toxicity, it is not surprising that in vivo knockdown of these snoRNAs provided only a partial reduction in the oxidative stress response and did not blunt the release of serum transaminases (not shown). In future studies, genetic approaches to loss of function and/or blunting of additional effector pathways (e.g., inflammatory signaling) may be required to block liver injury entirely. Nonetheless, our in vivo knockdown studies show that the rpL13a snoRNAs are required for the full induction of tissue oxidative stress in the murine LPS model, thus providing in vivo evidence for a role of snoRNAs in metabolic stress.
There are a number of mechanisms through which rpL13a snoRNAs may function in metabolic stress. Although some snoRNAs serve as substrates for Dicer-mediated cleavage and function as precursors for miRNA biogenesis (Ender et al., 2008), our RNase protection analyses of the rpL13a snoRNAs provide no evidence that U32a, U33 or U35a are processed to produce smaller miRNAs. Alternatively, non-canonical snoRNAs may serve as guides in ribonucleoproteins that catalyze chemical modifications of non-ribosomal RNA species through RNA editing, alternative splicing, methylation or acetylation. In mammalian cells, some snoRNAs have been shown to target small nuclear RNAs and RNA pol II transcripts (Cavaille et al., 1996; Tycowski et al., 1998). Two box C/D snoRNAs, HBII-52 and HBII-85, have been implicated in adenosine-to-inosine editing and/or alternative splicing of the serotonin receptor 5-HT2cR, through a mechanism involving complementarity of the snoRNA antisense elements to exonic sequence of the serotonin receptor pre-mRNA proximal to a splice junction (Kishore and Stamm, 2006). Localization of rpL13a snoRNAs in the cytosol suggests that they could function in regulation of mRNA translation, possibly through targeted modification or sequestration of specific RNAs. Given the manifold changes in gene expression during the lipotoxic and oxidative stress responses (Biden et al., 2004; Han et al., 2008; Shenton et al., 2006), the coordinate function of the rpL13a snoRNAs, and the possibility that snoRNA-directed modifications of targets may not be reflected by changes in abundance of those RNAs, identification of specific targets for each of these snoRNAs will be a complex task and the subject of future investigations.
Our genetic screen demonstrates that a mutation in a single gene can render cells resistant not only to lipotoxicity, but also to generalized oxidative stress. Previous biochemical studies in diverse cell types have correlated lipotoxic stress with the generation of ROS (Borradaile et al., 2006a; Inoguchi et al., 2000). Our study provides evidence of a functional link between the progress of lipotoxic cell death and the deleterious cellular response to oxidative stress. While transcriptional, post-translational and signaling mechanisms are known to contribute to the cellular response to oxidative stress (Han et al., 2008; Houstis et al., 2006), our findings are the first to demonstrate a role for snoRNAs in the cellular response to this environmental perturbation. The rpL13a snoRNAs may not only play a critical role in the metabolic stress of lipid overload, but also in the many pathophysiological processes that involve oxidative stress-induced cell damage and death.
EXPERIMENTAL PROCEDURES
Additional details of methods are available online in the Extended Experimental Procedures
Cell culture
CHO-K1 and C2C12 cells were cultured, supplemented with 500 μM palmitate (fatty acid to bovine serum albumin (BSA) molar ratio of 2:1), and assayed for cell death and apoptosis as described (Brookheart et al., 2009; Listenberger et al., 2001). For reactive oxygen species detection, cells were loaded with 5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester (CM-H2DCFDA, Invitrogen) and analyzed by flow cytometry. Mutants were generated by transducing CHO cells with the ROSAβgeo retroviral promoter trap as described (Borradaile et al., 2006a; Friedrich and Soriano, 1991). The endogenous gene disrupted by retroviral insertion was identified by 5′ rapid amplification of cDNA ends (SMART RACE cDNA amplification kit, Clontech). Cells were assayed for uptake of 14C-palmitate as described (Brookheart et al., 2009). Palmitate oxidation rates were determined using [9,10-3H]palmitic acid as described (Djouadi et al., 2003). To assess rates of protein synthesis, cells were pulse-labeled for with 35S-Cys/Met TCA-precipitatable proteins were quantified.
Quantitative real Time PCR (qRT-PCR)
TRIzol reagent (Invitrogen) was used to isolate total RNA, and mirVana miRNA isolation kit (Ambion) was used to isolate small RNA. cDNA synthesis used SuperScript First-strand Synthesis System (Invitrogen). For snoRNA analyses stem-loop oligos were used as previously described (Feng et al., 2009). qRT-PCR was performed using SYBR Green PCR master mixture (Applied Biosystems) and an ABI Prism 7500 Fast Real-Time PCR System. Relative quantification of gene expression was performed using the comparative threshold method.
snoRNA probe synthesis and RNase digestion
Probes were generated using Megashortscript kit (Ambion). RNA was isolated from cells and hybridized to 32P-labeled RNA probes (mirVana miRNA Detection kit), followed by RNase digestion and ethanol precipitation. RNA was separated by 10 or 15% polyacrylamide electrophoresis and visualized by autoradiography.
Expression of rpL13a genomic constructs
Hamster genomic, murine mRNA, and murine genomic rpL13a sequences were generated from CHO DNA, C2C12 cDNA, and a murine chromosome 7 BAC clone (RP24-235B15, CHORI), respectively, using Platinum Taq Hi-Fidelity polymerase (Invitrogen). QuikChange II Site-directed Mutagenesis Kit (Stratagene) was used to create mutant constructs. Constructs were transfected into 6F2 mutant cells using Lipofectamine 2000 (Invitrogen), followed by antibiotic selection for stable cell lines.
snoRNA knockdown in vitro
2-O-methyl-modified anti-sense oligos (ASOs) were designed to specifically target murine U32a, U33, U35a, U50, U57, and U60 snoRNA sequences according to Ideue et al (Ideue et al., 2009) and nucleofected into C2C12 myoblasts (Amaxa). An ASO targeting sequence from GFP was used as a control.
Immunoblot analyses
Total cell lysates or subcellular fractions isolated by sequential detergent solubilization (Holden and Horton, 2009) were immunoblotted using a polyclonal rabbit α-hamster rpL13a antibody (1:2000), monoclonal α-actin (Sigma), monoclonal α-CHOP-10 (Santa Cruz), α-hsp90 (Stressgen), and α-lamin B1 (Abcam) antibodies. Visualization used horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) and chemiluminescence (PerkinElmer Life Sciences).
Mapping of 2′-O-methyl modification by primer extension
Determination of 2′-O-methyl modifications was as described (Lowe and Eddy, 1999).
In situ hybridization of snoRNA probes
Synthesis, labeling, and hybridization of RNA probes were as described (Darzacq et al., 2002).
Mouse model of LPS-mediated oxidative stress and in vivo snoRNA knockdown
Mice were injected intraperitoneally (IP) with LPS (8 mg/kg) or equivalent volume of PBS and euthanized 12–24 h later for analysis of liver tissue. For knockdown experiments, locked nucleic acid (LNA)-modified ASOs (Exiqon) to specifically target snoRNAs or to target GFP as a control were injected IP every other day for three doses (2.5 mg/kg of total LNA per injection) prior to LPS administration. Detection of superoxide was performed on frozen sections using dihydroethidium (DHE, Invitrogen) and PEG-SOD (Sigma) (Miller et al., 2008). Quantification of liver protein carbonyls was performed using OxyBlot Protein Oxidation Detection Kit (Chemicon) and quantification of liver oxysterols were as described (Porter et al., 2010).
Supplementary Material
HIGHLIGHTS.
Lipotoxic and oxidative stress induce rpL13a snoRNAs, U32a, U33, and U35a
rpL13a snoRNAs are critical mediators of oxidative stress and lipotoxicity
Metabolic stress induces rpL13a snoRNAs in the cytosol
rpL13a snoRNAs are induced and required for propagation of oxidative stress in vivo
Acknowledgments
This work was supported by grants to JES from NIH (DK064989) and the Burroughs Wellcome Foundation (1005935), to CIM from NIH (DK077577), to RTB from NIH (DK077583), and to DSO from NIH (HL103001). Support from the Washington University Diabetes Research Training Center (DK020579), the Washington University Diabetic Cardiovascular Disease Metabolomics Facility, NIH T32 HL07081 and NIH T32 HL07275 is also acknowledged. MAB is employed by Integrated DNA Technologies (IDT), which offers oligonucleotides for sale similar to some of the compounds described in the manuscript. IDT is, however, not a publicly traded company, and MAB does not own any shares/equity in IDT.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Angulo P. Nonalcoholic fatty liver disease. N Engl J Med. 2002;346:1221–1231. doi: 10.1056/NEJMra011775. [DOI] [PubMed] [Google Scholar]
- Bautista AP, Spitzer JJ. Superoxide anion generation by in situ perfused rat liver: effect of in vivo endotoxin. Am J Physiol. 1990;259:G907–912. doi: 10.1152/ajpgi.1990.259.6.G907. [DOI] [PubMed] [Google Scholar]
- Biden TJ, Robinson D, Cordery D, Hughes WE, Busch AK. Chronic effects of fatty acids on pancreatic beta-cell function: new insights from functional genomics. Diabetes. 2004;53(Suppl 1):S159–165. doi: 10.2337/diabetes.53.2007.s159. [DOI] [PubMed] [Google Scholar]
- Borradaile NM, Buhman KK, Listenberger LL, Magee CJ, Morimoto ET, Ory DS, Schaffer JE. A critical role for eukaryotic elongation factor 1A-1 in lipotoxic cell death. Mol Biol Cell. 2006a;17:770–778. doi: 10.1091/mbc.E05-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borradaile NM, Han X, Harp JD, Gale SE, Ory DS, Schaffer JE. Disruption of endoplasmic reticulum structure and integrity in lipotoxic cell death. J Lipid Res. 2006b;47:2726–2737. doi: 10.1194/jlr.M600299-JLR200. [DOI] [PubMed] [Google Scholar]
- Brookheart RT, Michel CI, Listenberger LL, Ory DS, Schaffer JE. The non-coding RNA gadd7 is a regulator of lipid-induced oxidative and endoplasmic reticulum stress. J Biol Chem. 2009;284:7446–7454. doi: 10.1074/jbc.M806209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacicedo JM, Benjachareowong S, Chou E, Ruderman NB, Ido Y. Palmitate-induced apoptosis in cultured bovine retinal pericytes: roles of NAD(P)H oxidase, oxidant stress, and ceramide. Diabetes. 2005;54:1838–1845. doi: 10.2337/diabetes.54.6.1838. [DOI] [PubMed] [Google Scholar]
- Cao G, Goldstein JL, Brown MS. Complementation of mutation in acyl-CoA:cholesterol acyltransferase (ACAT) fails to restore sterol regulation in ACAT-defective sterol-resistant hamster cells. J Biol Chem. 1996;271:14642–14648. doi: 10.1074/jbc.271.24.14642. [DOI] [PubMed] [Google Scholar]
- Cavaille J, Nicoloso M, Bachellerie JP. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- Chaudhuri S, Vyas K, Kapasi P, Komar AA, Dinman JD, Barik S, Mazumder B. Human ribosomal protein L13a is dispensable for canonical ribosome function but indispensable for efficient rRNA methylation. RNA. 2007;13:2224–2237. doi: 10.1261/rna.694007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnop M, Ladriere L, Hekerman P, Ortis F, Cardozo AK, Dogusan Z, Flamez D, Boyce M, Yuan J, Eizirik DL. Selective inhibition of eukaryotic translation initiation factor 2 alpha dephosphorylation potentiates fatty acid-induced endoplasmic reticulum stress and causes pancreatic beta-cell dysfunction and apoptosis. J Biol Chem. 2007;282:3989–3997. doi: 10.1074/jbc.M607627200. [DOI] [PubMed] [Google Scholar]
- Darzacq X, Jady BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. Embo J. 2002;21:2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries JE, Vork MM, Roemen TH, de Jong YF, Cleutjens JP, van der Vusse GJ, van Bilsen M. Saturated but not mono-unsaturated fatty acids induce apoptotic cell death in neonatal rat ventricular myocytes. J Lipid Res. 1997;38:1384–1394. [PubMed] [Google Scholar]
- Djouadi F, Bonnefont JP, Munnich A, Bastin J. Characterization of fatty acid oxidation in human muscle mitochondria and myoblasts. Mol Genet Metab. 2003;78:112–118. doi: 10.1016/s1096-7192(03)00017-9. [DOI] [PubMed] [Google Scholar]
- El-Assaad W, Buteau J, Peyot ML, Nolan C, Roduit R, Hardy S, Joly E, Dbaibo G, Rosenberg L, Prentki M. Saturated fatty acids synergize with elevated glucose to cause pancreatic beta-cell death. Endocrinology. 2003;144:4154–4163. doi: 10.1210/en.2003-0410. [DOI] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Feng J, Wang K, Liu X, Chen S, Chen J. The quantification of tomato microRNAs response to viral infection by stem-loop real-time RT-PCR. Gene. 2009;437:14–21. doi: 10.1016/j.gene.2009.01.017. [DOI] [PubMed] [Google Scholar]
- Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- Han ES, Muller FL, Perez VI, Qi W, Liang H, Xi L, Fu C, Doyle E, Hickey M, Cornell J, et al. The in vivo gene expression signature of oxidative stress. Physiol Genomics. 2008;34:112–126. doi: 10.1152/physiolgenomics.00239.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden P, Horton WA. Crude subcellular fractionation of cultured mammalian cell lines. BMC Res Notes. 2009;2:243. doi: 10.1186/1756-0500-2-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houstis N, Rosen ED, Lander ES. Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature. 2006;440:944–948. doi: 10.1038/nature04634. [DOI] [PubMed] [Google Scholar]
- Ideue T, Hino K, Kitao S, Yokoi T, Hirose T. Efficient oligonucleotide-mediated degradation of nuclear noncoding RNAs in mammalian cultured cells. RNA. 2009;15:1578–1587. doi: 10.1261/rna.1657609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoguchi T, Li P, Umeda F, Yu HY, Kakimoto M, Imamura M, Aoki T, Etoh T, Hashimoto T, Naruse M, et al. High glucose level and free fatty acid stimulate reactive oxygen species production through protein kinase C--dependent activation of NAD(P)H oxidase in cultured vascular cells. Diabetes. 2000;49:1939–1945. doi: 10.2337/diabetes.49.11.1939. [DOI] [PubMed] [Google Scholar]
- Jiang T, Wang Z, Proctor G, Moskowitz S, Liebman SE, Rogers T, Lucia MS, Li J, Levi M. Diet-induced obesity in C57BL/6J mice causes increased renal lipid accumulation and glomerulosclerosis via a sterol regulatory element-binding protein-1c-dependent pathway. J Biol Chem. 2005;280:32317–32325. doi: 10.1074/jbc.M500801200. [DOI] [PubMed] [Google Scholar]
- Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- Kiss-Laszlo Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- Leung KN, Vallero RO, DuBose AJ, Resnick JL, LaSalle JM. Imprinting regulates mammalian snoRNA-encoding chromatin decondensation and neuronal nucleolar size. Hum Mol Genet. 2009;18:4227–4238. doi: 10.1093/hmg/ddp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listenberger LL, Han X, Lewis SE, Cases S, Farese RV, Jr, Ory DS, Schaffer JE. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc Natl Acad Sci U S A. 2003;100:3077–3082. doi: 10.1073/pnas.0630588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listenberger LL, Ory DS, Schaffer JE. Palmitate-induced apoptosis can occur through a ceramide-independent pathway. J Biol Chem. 2001;276:14890–14895. doi: 10.1074/jbc.M010286200. [DOI] [PubMed] [Google Scholar]
- Liu L, Shi X, Bharadwaj KG, Ikeda S, Yamashita H, Yagyu H, Schaffer JE, Yu YH, Goldberg IJ. DGAT1 expression increases heart triglyceride content but ameliorates lipotoxicity. J Biol Chem. 2009;284:36312–36323. doi: 10.1074/jbc.M109.049817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. A computational screen for methylation guide snoRNAs in yeast. Science. 1999;283:1168–1171. doi: 10.1126/science.283.5405.1168. [DOI] [PubMed] [Google Scholar]
- Maedler K, Oberholzer J, Bucher P, Spinas GA, Donath MY. Monounsaturated fatty acids prevent the deleterious effects of palmitate and high glucose on human pancreatic beta-cell turnover and function. Diabetes. 2003;52:726–733. doi: 10.2337/diabetes.52.3.726. [DOI] [PubMed] [Google Scholar]
- Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto PE, Fox PL. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell. 2003;115:187–198. doi: 10.1016/s0092-8674(03)00773-6. [DOI] [PubMed] [Google Scholar]
- Miller JD, Chu Y, Brooks RM, Richenbacher WE, Pena-Silva R, Heistad DD. Dysregulation of antioxidant mechanisms contributes to increased oxidative stress in calcific aortic valvular stenosis in humans. J Am Coll Cardiol. 2008;52:843–850. doi: 10.1016/j.jacc.2008.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PB, Steitz TA. The structural basis of large ribosomal subunit function. Annu Rev Biochem. 2003;72:813–850. doi: 10.1146/annurev.biochem.72.110601.135450. [DOI] [PubMed] [Google Scholar]
- Nicoloso M, Qu LH, Michot B, Bachellerie JP. Intron-encoded, antisense small nucleolar RNAs: the characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation of rRNAs. J Mol Biol. 1996;260:178–195. doi: 10.1006/jmbi.1996.0391. [DOI] [PubMed] [Google Scholar]
- Ostrander DB, Sparagna GC, Amoscato AA, McMillin JB, Dowhan W. Decreased cardiolipin synthesis corresponds with cytochrome c release in palmitate-induced cardiomyocyte apoptosis. J Biol Chem. 2001;276:38061–38067. doi: 10.1074/jbc.M107067200. [DOI] [PubMed] [Google Scholar]
- Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Gorgun C, Glimcher LH, Hotamisligil GS. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- Porter FD, Scherrer DE, Lanier MH, Langmade SJ, Molugu V, Gale SE, Olzeski D, Sidhu R, Dietzen DJ, Fu R, et al. Cholesterol oxidation products are sensitive and specific blood-based biomarkers for Niemann-Pick C1 disease. Sci Transl Med. 2010;2:56ra81. doi: 10.1126/scitranslmed.3001417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Adrogue JV, Golfman L, Uray I, Lemm J, Youker K, Noon GP, Frazier OH, Taegtmeyer H. Intramyocardial lipid accumulation in the failing human heart resembles the lipotoxic rat heart. Faseb J. 2004;18:1692–1700. doi: 10.1096/fj.04-2263com. [DOI] [PubMed] [Google Scholar]
- Shenton D, Smirnova JB, Selley JN, Carroll K, Hubbard SJ, Pavitt GD, Ashe MP, Grant CM. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J Biol Chem. 2006;281:29011–29021. doi: 10.1074/jbc.M601545200. [DOI] [PubMed] [Google Scholar]
- Shimabukuro M, Zhou YT, Levi M, Unger RH. Fatty acid-induced beta cell apoptosis: a link between obesity and diabetes. Proc Natl Acad Sci U S A. 1998;95:2498–2502. doi: 10.1073/pnas.95.5.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siminovitch L. On the nature of hereditable variation in cultured somatic cells. Cell. 1976;7:1–11. doi: 10.1016/0092-8674(76)90249-x. [DOI] [PubMed] [Google Scholar]
- Tycowski KT, You ZH, Graham PJ, Steitz JA. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol Cell. 1998;2:629–638. doi: 10.1016/s1097-2765(00)80161-6. [DOI] [PubMed] [Google Scholar]
- Unger RH. Lipotoxicity in the pathogenesis of obesity-dependent NIDDM. Genetic and clinical Implications. Diabetes. 1995;44:863–870. doi: 10.2337/diab.44.8.863. [DOI] [PubMed] [Google Scholar]
- Watkins NJ, Lemm I, Luhrmann R. Involvement of nuclear import and export factors in U8 box C/D snoRNP biogenesis. Mol Cell Biol. 2007;27:7018–7027. doi: 10.1128/MCB.00516-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Wang D, Topczewski F, Pagliassotti MJ. Saturated fatty acids induce endoplasmic reticulum stress and apoptosis independently of ceramide in liver cells. Am J Physiol Endocrinol Metab. 2006;291:E275–281. doi: 10.1152/ajpendo.00644.2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.