Abstract
PIWI-interacting RNAs (piRNAs) are a class of small RNAs abundantly expressed in animal gonads. piRNAs that map to retrotransposons are generated by a “ping-pong” amplification loop to suppress the activity of retrotransposons. However, the biogenesis and function of other categories of piRNAs have yet to be investigated. In this study, we first profiled the expression of small RNAs in type A spermatogonia, pachytene spermatocytes, and round spermatids by deep sequencing. We then focused on the computational analysis of the potential piRNAs generated in the present study as well as other published sets. piRNAs mapping to retrotransposons, mRNAs, and intergenic regions had different length distributions and were differentially regulated in spermatogenesis. piRNA-generating mRNAs (PRMRs), whose expression positively correlated with their piRNA products, constituted one-third of the protein-coding genes and were evolutionarily conserved and enriched with splicing isoforms and antisense transcripts. PRMRs with piRNAs preferentially mapped to CDSs and 3′ UTRs partitioned into three clusters differentially expressed during spermatogenesis and enriched with unique sets of functional annotation terms related to housekeeping activities as well as spermatogenesis-specific processes. Intergenic piRNAs were divided into 2992 clusters probably representing novel transcriptional units that have not been reported. The transcripts of a large number of genes involved in spermatogenesis are the precursors of piRNAs, and these genes are intricately regulated by alternative splicing and antisense transcripts. piRNAs, whose regulatory role in gene expression awaits to be identified, are clearly products of a novel regulatory process that needs to be defined.
Keywords: piRNA, small RNA, microRNA, spermatogenesis, mitosis, meiosis, spermatogenesis, antisense transcript, alternative splicing
INTRODUCTION
Spermatogenesis in the mouse is a coordinated process that can be divided into three phases: the mitotic proliferation of spermatogonia, the meiotic division of spermatocytes, and the metamorphic change of spermatids into mature spermatozoa (Oakberg 1956). Several different types of spermatogonia that can be recognized morphologically include the undifferentiated Asingle, Apaired, and Aaligned spermatogonia, the differentiating A1-4 spermatogonia, the intermediate type I spermatogonia, and the type B spermatogonia that are committed to undergo meiosis (Oakberg 1956; de Rooij and Grootegoed 1998). The complex nature of spermatogenesis implies intricate gene regulation at both transcription and post-transcription levels. As an example, a large number of non-coding transcripts including antisense RNAs and microRNAs are highly expressed in mammalian testis (Amaral and Mattick 2008; Hayashi et al. 2008).
PIWI proteins are a subset of the Argonaute proteins and are expressed predominantly in the germline of a variety of organisms. They are essential for germ cell maintenance and spermatogenesis in Drosophila and mammals (Thomson and Lin 2009). MILI, MIWI, and MIWI2 are three mouse PIWI proteins that bind small RNAs of ∼24–31 nt termed PIWI-interacting RNAs (piRNAs) (Grivna et al. 2006; Lau et al. 2006; Aravin et al. 2007b, 2008; Lin 2007). piRNAs often initiate with a 5′ uracil and contain 2′-O-methyl groups at their 3′ ends (Saito et al. 2007) and are often found in clusters throughout the genome (Aravin et al. 2007b). PIWI proteins and piRNAs in Drosophila melanogaster are implicated in transposon silencing via the ‘‘ping-pong’’ mechanism of amplification (Aravin et al. 2007a). The murine transposon-derived piRNAs share features with the Drosophila repeat-associated piRNAs and are also proposed to control transposable elements (Klattenhoff and Theurkauf 2008). In addition to PIWI proteins, other proteins such as GASZ, MOV10L1, Sun1, Tdrd9, and Tdrd1 are also required for biogenesis and/or stability of mouse piRNAs (Chi et al. 2009; Ma et al. 2009; Reuter et al. 2009; Shoji et al. 2009; Zheng et al. 2010).
Mouse testes also accumulate abundant piRNAs that do not map to repeat sequences (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006), but much less is known about their biogenesis and function. In Drosophila ovaries, murine testes, and Xenopus eggs, 3′ UTRs of an extensive set of mRNAs are processed into piRNAs (Robine et al. 2009). To examine the expression profile of piRNAs at mitotic, meiotic, and post-meiotic stages of spermatogenesis and their sequence features, we isolated type A spermatogonia, pachytene spermatocytes, and round spermatids from mouse testes, collected small RNAs of 18–32 nt and identified their sequences by deep sequencing. We mapped the piRNAs to the genome and transcriptome and found that most of the piRNAs that map to a unique position in the genome are localized to the exons of protein-coding mRNAs. The sequence features of these piRNA-generating mRNAs (PRMRs) as well as their expression profile and potential functions during spermatogenesis were systematically analyzed. We propose that gene expression during spermatogenesis is regulated post-transcription by multiple processes including alternative splicing and antisense transcripts, both of which are positively correlated with the production of piRNAs.
RESULTS
Profiles of small RNAs in mouse spermatogenesis
To profile small RNA expression during spermatogenesis in postnatal mice, three types of germ cells were isolated using gravity density gradient sedimentation (Romrell et al. 1976; Bellve et al. 1977). Type A spermatogonia (A), pachytene spermatocytes (PS), and round spermatids (RS) were isolated from 8-d-old mice, 17-d-old mice, and adult mice, respectively. The three types of germ cells could be distinguished by their unique morphology under the phase contrast microscope. Their purities all exceeded 90%, which was further verified by measuring the expression of three marker genes that are uniquely expressed in the three types of germ cells (Sohlh1 for A, Scp3 for PS, and Prm2 for RS) using real-time RT-PCR (see Supplemental Fig. 1). Small RNAs of 18–36 nt were recovered from an agarose gel after total RNAs were separated by electrophoresis. The three small RNA libraries were prepared using the Illumina small RNA preparation kit and were deep-sequenced using Solexa technology.
After adapter sequences were trimmed, 6,917,849, 7,323,697, and 8,221,820 small RNA reads of 16–32 nt were obtained from A, PS, and RS, respectively. These reads were first mapped to the mouse genome using PASS (Campagna et al. 2009) and then to the transcriptome (Ensembl Release 59). piRNAs mapped to retrotransposons, mRNAs, and intergenic regions were regarded as retrotransposon-derived piRNAs (RTPRs), mRNA-derived piRNAs (MRPRs), and intergenic piRNAs (IGPRs). Accordingly, the piRNA-generating mRNAs were named PRMRs. A number of small RNAs were mapped to the borders of two spliced exons. For subsequent analysis, only sequences that had a perfect match to either the genome or the transcriptome were considered. Consequently, about 4.3 million, 5.8 million, and 6.3 million reads corresponding to 1.2 million, 0.9 million, and 0.9 million unique sequences were acquired for A, PS, and RS, respectively. Seventy-two percent of sequences in A, 82% in PS, and 83% RS were mapped to unique loci on the mouse genome (Table 1). These small RNA sequences were compared with known ncRNAs (miRNAs, rRNAs, tRNAs, snRNAs, snoRNAs, etc.). We identified 1.8 million reads in A (43% of the total reads) corresponding to 438 miRNAs (on average 4000 reads per miRNA), out of which 338 have at least 10 reads in A. In contrast, only about 0.4 million reads in PS (7% total reads) and 0.3 million in RS (5%) were identified for 348 and 364 miRNAs, respectively. The average abundances were about 1000 and 800 reads per miRNA for these two cell types, respectively. All miRNAs detected in the three cell types and their reads are shown in Supplemental Table 1.
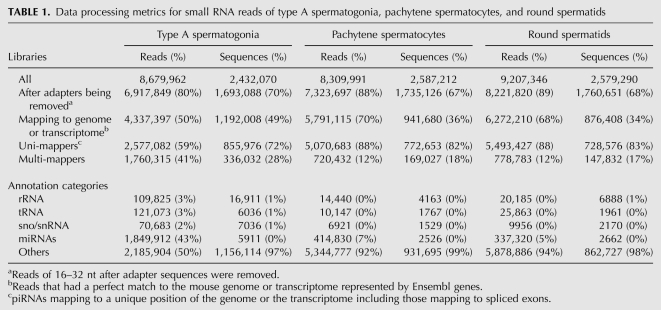
TABLE 1.
Data processing metrics for small RNA reads of type A spermatogonia, pachytene spermatocytes, and round spermatids
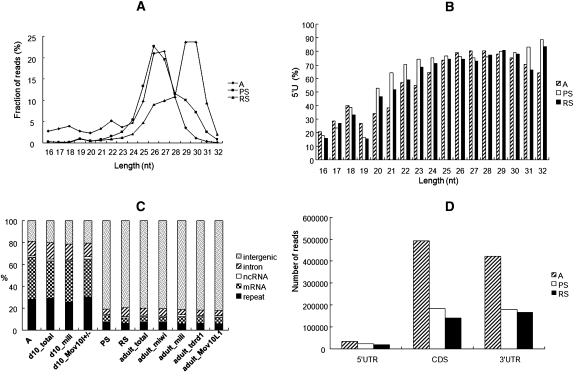
After the known ncRNA reads were filtered out, the remaining reads that were perfectly mapped to either the mouse genome or transcriptome were further analyzed for the three cell types. Based on their size distributions, right-skewed peaks starting from 24 nt and ending at 32 nt were observed (Fig. 1A). The peaks for A and PS were centered around 26 nt, while the one for RS was centered around 30 nt. When the percentages of small RNAs that contained a 5′U were plotted against their lengths, it was seen that the percentages exceeded 60% when the sizes were bigger than 24 nt for all three cell types (Fig. 1B). Therefore, we thought that these small RNAs bigger than 24 nt were not randomly degraded RNA fragments but were potential piRNAs given the characteristics of piRNAs reported by others (Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Aravin et al. 2007b). As a result, 837,777, 791,614, and 725,731 unique piRNA sequences were derived for A, PS, and RS, respectively. If a contig was defined as a set of overlapping piRNAs mapping to the same genomic locus, 352,678, 143,840, and 132,104 contigs were derived for the three cell types, respectively. The average reads per contig in A, PS, and RS were 2.7, 30.8, and 37.8, respectively.
FIGURE 1.
Small RNA profiles at different stages of spermatogenesis. (A) Size distribution of small RNAs in type A spermatogonia (A), pachytene spermatocytes (PS), and round spermatids (RS). (B) Distribution of percentages of piRNAs with a 5′U in length. (C) Distribution of piRNAs from 11 libraries in different categories. (D) Reads of three types of MRPRs in three cell types.
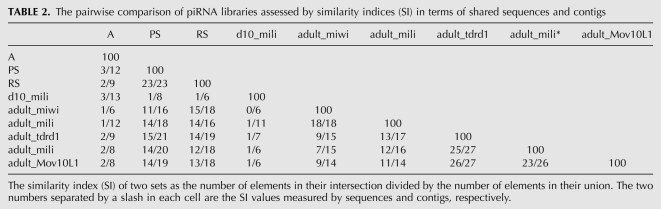
The three data sets generated in the present study were compared with those generated by others, which are either total small RNAs or small RNAs immunoprecipitated by antibodies against, for example, MILI, MIWI, Tdrd1, and MOV10L1 from mice 10 days postpartum (dpp) or adult mice (Supplemental Table 2). We defined the similarity index (SI) of two sets as the number of elements in their intersection divided by the number of elements in their union. For the sake of consistency, all data sets were reprocessed in parallel with the same procedure. The mouse adult testis MILI-IP piRNA data, GSM400968 (Reuter et al. 2009) and GSM475280 (Robine et al. 2009), were data sets generated independently by two groups. The SI of these two sets of piRNAs represented by sequences was 12%, and was only 16% even if piRNAs were represented by contigs, indicating that each of these two libraries covered only a small fraction of all contigs. However, the shared contigs of these two libraries covered >40% of all reads in each of the libraries, suggesting that each library was able to detect the highly productive contigs while it may miss less productive ones. The comparisons between more data sets in pairs were shown in Table 2. It can be seen that these libraries only represent limited samples of all possible piRNAs, and the coverages are far from completion.
TABLE 2.
The pairwise comparison of piRNA libraries assessed by similarity indices (SI) in terms of shared sequences and contigs
Genomic mapping of piRNAs showed that MRPRs and RTPRs were mainly expressed in A and 10-dpp testis, while IGPRs were mainly expressed in PS and RS and adult testis (Fig. 1C). Therefore, piRNAs generated at different stages in spermatogenesis are different in terms of their sequence origins.
Retrotransposon-derived piRNAs
The piRNAs are required for suppressing retrotransposable elements in the germline of various species containing Drosophila, zebrafish, and mouse (Aravin et al. 2007b; Brennecke et al. 2007; Houwing et al. 2007). As indicated in Figure 1C, RTPRs at different stages only constitute of small portions of total piRNAs. Three major classes of retrotransposable elements exist in mammalian genomes: LTRs, LINEs, and SINEs. Representative elements of each class that produced the larger number of piRNAs were SINE/B1, LINE/L1_MM, and LTR/IAPEZI. As shown by Supplemental Figure 2, the abundance of both the sense and the antisense piRNAs in the three classes drop along with the differentiation of A into PS and RS.
piRNAs are processed from one-third of mouse mRNAs
MRPRs from A, 10-dpp testis, or MILI-IP piRNAs constitute the largest group among all categories (Fig. 1C). The majority of MRPRs mapped to exons of mRNAs, and some (∼1%) mapped across splice sites (Supplemental Fig. 3), while few mapped to introns, strongly suggesting the MRPRs are generated from spliced mRNAs. In the subsequent analysis, only MRPRs with at least 10 mapped piRNAs were considered. Consequently, 13,883, 6023, and 4742 PRMRs were identified in A, PS, and RS, respectively, corresponding to 6896, 3218, and 2595 genes. Collectively, the three PRMR sets contained 15,288 unique PRMRs from 7559 genes. According to Ensembl Genes (Release 59), 47,676 mouse mRNAs corresponding to 22,809 genes are annotated. Hence, around one-third of the mouse mRNAs were processed into piRNAs during spermatogenesis. Three thousand nine hundred and fifteen pseudogenes corresponding to 1452 parent genes were obtained from the pseudogene database (http://www.pseudogene.org/mouse/). Eighty percent of parent genes had at least one piRNA, and 63% had at least 10 piRNAs. Therefore, a parent gene of pseudogenes is more likely a PRMR and tends to produce more piRNAs.
piRNAs preferentially map to 3′ UTR and CDS regions of mRNAs
piRNAs mainly mapped to 3′ UTRs and CDSs but infrequently to 5′ UTRs of PRMRs (Fig. 1D). It was interesting to notice that piRNAs tended to map to CDSs when their abundance was low but tended to map to 3′ UTRs when their abundance was high, as indicated by the distribution of piRNAs in A (Fig. 2). A PRMR was defined as either a CDS-PRMR or a 3′UTR-PRMR if the percentage of either its CDS-piRNAs or its 3′UTR-piRNAs is at least 70%. Sixty-seven percent and 21% of PRMRs in A are CDS-PRMRs and 3′UTR-PRMRs, respectively. On average, 32 piRNAs corresponding to 15 contigs mapped to a CDS-PRMR, while 113 piRNAs corresponding to 33 contigs mapped to a 3′UTR-PRMR.
FIGURE 2.
Distribution of piRNAs in 5′ UTR, CDS, and 3′ UTR regions as a function of the abundance of piRNAs in PRMRs. PRMRs were sorted and grouped according to the number of piRNAs that they contain. The group interval was 20. The percentages of piRNAs in the three regions were plotted for each group. As can be seen, piRNAs tend to map to 3′ UTR as their abundance increases.
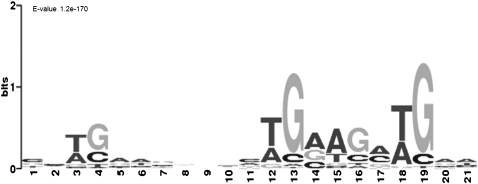
Considering the uneven distribution of piRNAs in PRMRs, we wondered whether piRNA processing from these precursors was guided by any RNA motif(s) recognized by the processing machinery. To this end, the piRNA-enriched regions of 100 nt were extracted from PRMRs and were scanned with the MEME software (Bailey and Elkan 1994). As shown in Figure 3, CDS-PRMRs in all three cell types were enriched with motifs that contain several core sequences mainly composed of TGAA (Fig. 3; Supplemental Fig. 4). In contrast, 3′UTR-PRMRs were not enriched with any motif significantly.
FIGURE 3.
The motif enriched in type A spermatogonia CDS-PRMRs identified by MEME software prediction. (E-value) The statistical significance of the motif.
PRMRs are more conserved and have more isoforms
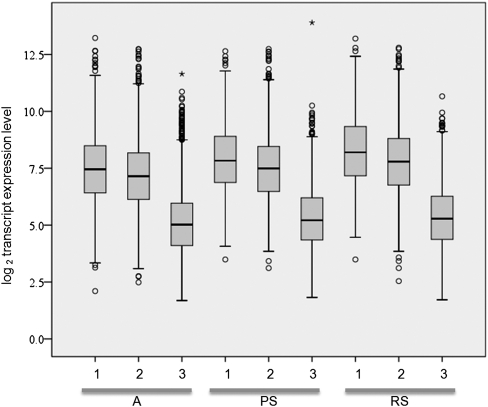
Based on the NCBI HomoloGene database (Release 64), 94% of the mouse PRMRs have homologs in humans, while 40% have homologs in both humans and fruit flies. In contrast, only 60% and 7% of non-PRMR mRNAs have homologs in humans and in both humans and fruit flies. PRMRs have significantly higher number of isoforms (on average 4.2 isoforms per PRMR) than non-PRMR mRNAs (2.2 isoforms per mRNA; P < 10−4, unpaired t-test). 68% of PRMRs have at least two isoforms, which produce different amount of piRNAs. For example, the mRNA of Cbx5 (chromobox homolog 5), which produces the greatest number of piRNAs in A, has four isoforms generating 1469, 364, 67, and 15 piRNAs (Supplemental Fig. 5). The testicular expression of different isoforms was compared using the exon microarray data set GSE15998 downloaded from the NCBI GEO database. The mRNA levels of the isoforms producing the highest amount of piRNAs were significantly higher than that of the isoforms producing the lowest amount of piRNAs when only isoform pairs with at least twofold difference in piRNA abundance are considered (P < 10−4). PRMRs without isoforms are as abundant as those PRMR isoforms generating the highest amount of piRNAs (Fig. 4).
FIGURE 4.
Box plot of the expression levels of splicing isoforms of PRMRs in three cell types: (1) PRMRs without isoforms; (2, 3) the two isoforms that contain the highest and lowest number of piRNAs. Splicing isoforms of 1, 2, and 3 were identified from the mouse testis exon expression data set GSE15998, and the expression levels were plotted using the SPSS software.
Expression of CDS-PRMRs and 3′UTR-PRMRs is developmentally regulated
The expression of CDS-PRMRs and 3′UTR-PRMRs in different germ cell types was analyzed using two microarray mRNA expression data sets—GSE4193 (Namekawa et al. 2006) and GSE2736 (Shima et al. 2004). Expression levels of CDS-PRMRs in three cell types were significantly higher than those of 3′UTR-PRMR (P < 10−4, unpaired t-test). The expression of CDS-PRMRs and 3′UTR-PRMRs of piRNAs immunoprecipitated by antibodies against MILI, TDRD, and MOV10L1 in 10-dpp or adult testis were also compared based on the GSE926 data set, consisting of mRNA expression profiles at different stages of mouse spermatogenesis (Shima et al. 2004). As can be seen from Supplemental Table 3, the levels of CDS-PRMRs were consistently higher than 3′UTR-PRMRs regardless of the sources of piRNAs or the stages of spermatogenesis.
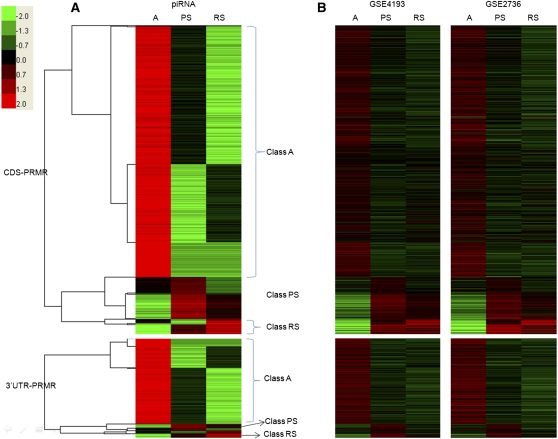
CDS-PRMRs and 3′UTR-PRMRs were clustered according to piRNA abundance in three cell types, and three classes were identified in each category based on which cell type had the highest level of piRNAs (Fig. 5A). For example, the piRNA level of Class A PRMRs was the highest in A among all three cell types. CDS-PRMRs and 3′UTR-PRMRs were also clustered according to their mRNA levels in three cell types using data sets GSE4193 and GSE2736, and the same three classes were again identified (Fig. 5B). In other words, abundances of piRNAs and mRNAs were positively correlated.
FIGURE 5.
Heat maps of piRNA and mRNA expression profiles of CDS-PRMRs and 3′UTR-PRMRs. (A) Heat map of piRNA expression profiles of PRMRs. Expression intensities are displayed from green (low expression) to red (high expression). Expression profiles are clustered using hierarchical clustering. Data were visualized with Treeview. According to the clustering results, the CDS-PRMRs and 3′UTR-PRMRs were divided into three groups (Class A, PS, and RS), respectively. (B) Heat map of mRNA expression profiles of PRMRs according to GEO data sets GSE4193 and GSE2736. The order of rows is the same as in A.
The mRNA levels of each cluster of the two categories were checked in 15 mouse tissues using the microarray data sets GSE15998 and GSE1133 (Su et al. 2004). The levels of PRMRs in Class A and Class PS are significantly higher than the mean of all genes in most of the tissues. The levels of PRMRs in Class PS and Class RS in testis are significantly higher than in other tissues (Supplemental Fig. 6).
PRMRs are enriched with unique sets of functional annotation terms
The enrichment of functional annotation terms (FATs) in these classes was analyzed using the Functional Annotation Tool of DAVID (Dennis et al. 2003; Huang et al. 2009). Genes expressed at stages of A, PS, and RS were used as reference sets for Class A, PS, and RS PRMRs, respectively. FATs with enrichment score ≥3 were regarded as enriched (Supplemental Table 4). Class A CDS-PRMRs were enriched with FATs such as translation, RNA processing, nucleotide binding, proteolysis, chromosome organization, ribosome biogenesis, ATPase activity, RNA localization, mitosis, and DNA repair, suggesting that the proteins of this cluster were mainly involved in regulation of gene expression. This is consistent with the high-level expression of these genes in different tissues. Cluster RS CDS-PRMRs were enriched with FATs related to spermatogenesis, and this was consistent with their higher expression in testis. Class A 3′UTR-PRMRs were enriched with FATs related to zinc ion binding and transcription. It was noted that the enriched FAT nucleotide binding contained the highest number of genes in Class A CDS-PRMRs, and most of these genes are not well studied. However, some of them such as ERCC3, INO80, CHD3, PMS2, BLM, MSH2, and LIG4 are reported to participate in chromatin remodeling, DNA repair, and genome maintenance (Kim et al. 1999; Mills et al. 2004; Chu et al. 2010; Jiang et al. 2010; Larocque and Jasin 2010; Rybanska et al. 2010; van Oers et al. 2010). For example, mice with a mutation in the ATP-binding domain of MLH1 were sterile due to its inability to interact with meiotic chromosomes of pachynema (Avdievich et al. 2008).
PRMRs were enriched with antisense transcripts
It has been well known that siRNAs and miRNAs both target to complementary mRNAs to regulate the stability and translation of the latter. Moreover, retrotransposon-derived piRNAs also target retrotransposon mRNAs for their degradation through the ping-pong amplification loop. We noticed that majority of piRNAs only mapped to the sense strand of PRMRs, indicating that the mechanism for the generation of MRPRs might be different from that for RTPRs. However, it is still possible that the sense–antisense complement is established but ping-pong amplification does not ensue for MRPR processing. This hypothesis implies the enrichment of antisense transcripts in PRMRs. Indeed, when PRMRs were compared with the ESTs and mRNAs sequences in the UCSC database using the BLAT software with a stringent condition (length of reverse complement match ≥100 nt and percentage of identity ≥99%) (Kent 2002), a majority of PRMRs had antisense transcripts. For instance, 64% PRMRs in A had antisense transcripts, while only 20% non-PRMR mRNAs had antisense transcripts (P < 10−4). According to the gbCdnaInfo table (the information table about ESTs and mRNAs) of the UCSC database, most of these antisense transcripts were from unknown genes, and no information about their expression pattern was available. In addition to EST data, SAGE (serial analysis of gene expression) tags were also used to search for antisense transcripts (Velculescu et al. 1995). PRMRs were compared with the LongSAGE data set GSE4726 (Siddiqui et al. 2005), and antisense transcripts from testis tissue were identified for 52% PRMRs in A. In contrast, only 5% of non-PRMR transcripts had antisense transcripts identified from this data set. Again, this difference is statistically significant (P < 10−4). The enrichment of antisense transcripts for PRMRs was also confirmed in other sets of PRMRs. Some antisense transcripts were derived from the antisense strand of a pseudogene of a PRMR. Indeed, Chan et al. (2006) have reported that pseudogenes are another source of antisense transcripts.
Intergenic piRNA clusters
Most piRNAs in PS and RS mapped to intergenic regions. Their locations on the chromosomes might provide clues about their production at transcriptional and post-transcriptional levels. It is known that piRNAs tend to form clusters along the chromosomes. IGPRs from 11 small RNAs libraries (A, PS, RS, d10_total, d10_mili, adult_total, adult_mili, adult_miwi, adult_tdrd1, adult_mili*, adult_Mov10L1) were partitioned into 2992 clusters when the clusters were presumed to be 10,000 nt long and to contain at least 50 piRNA sequences. The average cluster length was 32,000 nt with the largest one being 356,000 nt and the shortest one being 75 nt. Ninety-seven percent of the clusters are within 100,000 nt. The average piRNA contigs in each cluster were 3.2 per 1000 nt (Supplemental Fig. 7).
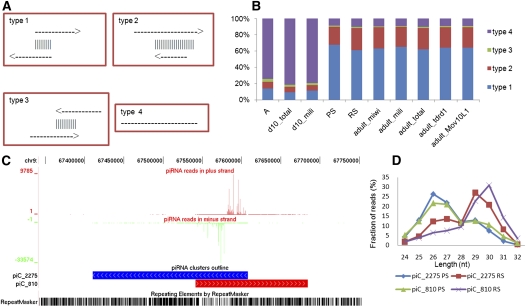
Four types of clusters were discovered based on whether their piRNAs mapped to both sense and antisense strands and how the mapped regions overlapped (Fig. 6A): type 1, 2, and 3 clusters were piRNAs mapped to both strands with different types of overlaps, while type 4 were piRNAs mapped to only the sense strand. piRNAs from A and 10-dpp testis were mainly from the type 4 clusters, while those from PS and later-stage testis were from type 1 and type 2 clusters (Fig. 6B). Eighty-six percent of piRNA reads of clusters mapped to the overlapping regions. Similar to MRPRs, IGPRs also preferentially mapped one strand rather than the other. This is illustrated by two clusters shown in Figure 6C, which contained many piRNAs belonging to type 1. Moreover, both sense and antisense strand-mapped piRNAs had the 5′ ends biased to U, but the 10A preference was not observed. It seemed that the mechanism of intergenic piRNA generation was different from the ping-pong generation mechanism of transposon-derived piRNAs, where piRNAs from the one strand have the 5′U bias, while those from the complementary strand have the 10A bias. It was observed that piRNAs from the same cluster showed different length distributions at different stages of spermatogenesis (Fig. 6D).
FIGURE 6.
Types of intergenic piRNA clusters. (A) Four types of clusters of IGPRs are illustrated according to the locations of piRNAs in sense and antisense strands. Arrows indicate the transcriptional orientations of the piRNA clusters. The vertical lines indicate the overlap region of two clusters. Types 1–3 show pairs of clusters with different overlapping patterns. Type 4 is a single cluster that does not overlap with other clusters. (B) Distribution of IGPRs from different libraries in these four types. (C) Annotation of a two type 1 clusters. piRNA clusters on the plus strands (red boxes) and minus strands (blue boxes). The vertical coordinate indicates the number of piRNA reads. (Green bars) Negative values show piRNAs that are on the minus strand of the chromosome. (Red bars) Positive values indicate piRNAs that are on the plus strand. Repeat elements were marked at the bottom. (D) The length distribution of the sense and antisense piRNAs of two type 1 clusters in pachytene spermatocytes (PS) and round spermatid (RS) piRNAs. It is noted that the lengths of piRNAs from the same cluster at different stages have different distributions.
DISCUSSION
It was striking to see that 43% of the 4.3 million small RNA reads were from the 438 miRNAs in A, while this percentage dropped to 7% and 5% in PS and RS despite the total reads increasing slightly to 5.8 million and 6.3 million, respectively. Conversely, the percentages of piRNA reads increased from 50% in A to 92% in PS to 94% in RS. Moreover, the major types of piRNAs switched from MRPR and RTPRs in A to IGPRs in PS and RS. Even for IGPRs, their average lengths changed from 26 nt in PS to 30 nt in RS. Therefore, it is apparent that different types of small RNAs vary during spermatogenesis in a well-coordinated manner. A similar change in major small RNA classes also occurs during pre-implantation development (Ohnishi et al. 2010).
It is puzzling that piRNAs are processed from mRNAs of one-third of the protein-coding genes. The observations that piRNAs mainly mapped to within exons and some to the borders of two spliced exons indicate that piRNAs are processed from spliced mRNAs. Our observation that piRNAs preferentially mapped to 3′ UTRs and then to CDSs but only infrequently to 5′ UTRs suggested that their generation is well regulated. PRMR genes not only are more conserved but also have more splicing isoforms than non-PRMR genes. The expression of CDS-PRMRs are about twofold higher than 3′UTR-PRMRs. PRMRs are either involved in the basic activities of cells such as chromosome remodeling, transcription, RNA processing, and proteolysis and are more abundantly expressed than the average of all mRNAs or are involved in spermatogenesis-specific activities and are more abundantly expressed in testis than in other tissues. All these features indicated that only certain groups of mRNAs are selected to be PRMRs. However, the selection could be made only when multiple standards are met by a candidate. For example, as also observed by Robine et al. (2009), some highly expressed transcripts in testes are not PRMRs. While one such standard could be the presence of a motif containing the TGAA core sequence as identified in most CDS-PRMRs, others could be higher-order structures that could not be easily discovered.
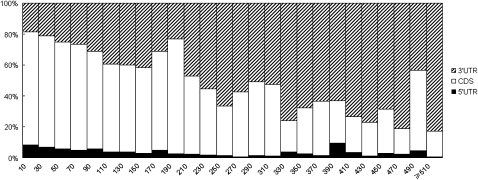
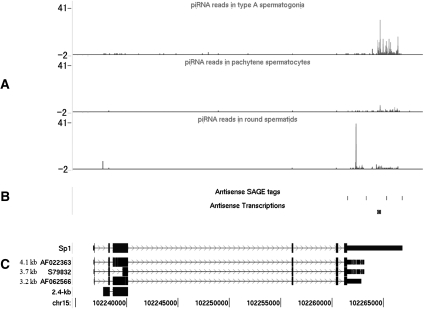
Since PRMRs have more splicing isoforms, we examined the distribution of truncated regions in relation to piRNAs in these isoforms. Longer isoforms produced more piRNA reads than shorter ones. Eighty percent of 3′UTR-PRMRs had isoforms that are truncated at the 3′ UTRs, resulting in the reduction of piRNA reads to ≤20% on the short isoforms compared with the longer ones. For example, the 3′ UTR of the transcription factor Sp1 has many mapped piRNAs (Fig. 7). Persengiev et al. (1995) have reported that the normal 8.2-kb transcript started to disappear in the pachytene stage of meiosis, and a germ-cell-specific 2.4-kb isoform with a truncated 3′ UTR appeared in late-meiotic and early-haploid cells. Thomas et al. (2005) further reported that a few 3–4-kb Sp1 transcripts, all with a truncated 3′ UTR, were detected in a pachytene cDNA library. More examples similar to Sp1 include Rnf4, Nr6a1, Snon2, Pdpk1, and Tsn (Gu et al. 1998; Dong et al. 2002; Pero et al. 2003; Yang et al. 2003; Itman et al. 2009). For CDS-PRMRs, the truncated exons did not locate in the CDSs preferentially. Therefore, it seems that PRMRs are regulated both by alternative splicing and a process related to the production of piRNA generation, and these two processes could be coupled in the case of 3′UTR-PRMRs. Consistent with our results, Robine et al. (2009) reported lately that the 3′ UTRs of mRNAs could be processed into piRNAs in Drosophila ovaries, murine testes, and Xenopus eggs; in addition, IP experiments were performed to show that MILI and MIWI, proteins bind directly to 3′ UTRs. In addition, PRMRs were also more conserved evolutionarily and probably involved in gene regulation at all levels from chromosome remodeling to protein degradation as well as spermatogenesis-specific activities. All these features suggested that the expression of both housekeeping and cell-type-specific genes was intricately regulated at multiple levels in the complex process of spermatogenesis.
FIGURE 7.
Annotation of a PRMR gene, Sp1, with piRNAs from the cell types and antisense transcripts identified by SAGE tags and ESTs, as well as splicing isoforms. (A) The distribution of piRNA reads on the sp1 locus in three cell types. piRNAs are enriched in the 3′ UTR. (B) (Solid bars) Antisense SAGE tags identified from the testis and antisense transcripts. (C) The structure of five sp1 isoforms with exons mapped to the plus strand of chromosome 15.
The fact that piRNAs were mainly generated from the sense strands of PRMRs even though antisense transcripts were transcribed indicated that the mechanism for MRPRs is different from that for RTPRs. The presence of antisense transcripts has been recognized as a pervasive feature of mammalian genomes (Faghihi and Wahlestedt 2009), and high-level expression of antisense transcripts in testis has been reported in several species (Werner et al. 2009). However, their enrichment in PRMRs deserves more investigation. Although it is possible that these antisense transcripts could directly regulate the transcription of PRMRs during or after their transcription either through a transcription collision mechanism or through their direct interaction with promoters of sense transcripts, it is also likely that they regulate their sense strands through forming complementary duplexes. The consequences of the sense–antisense duplexes, if they exist, could be complex because each of the diverse outcomes such as changes in mRNA stability and translation, blocking the accessibility of miRNAs, and formation of endogenous siRNAs are all possible (Faghihi and Wahlestedt 2009).
MILI-KO resulted in reduction in piRNA expression (Aravin et al. 2008). Robine et al. (2009) reported that the abundance of 3′UTR-PRMRs did not change in MILI−/− testes compared with MILI+/− at 10 dpp, and Unhavaithaya et al. (2009) reported that levels of MILI-associated mRNAs also did not change in MILI-KO 24-dpp testis. Therefore, MILI probably does not have an essential role in the processing of piRNAs but may protect piRNAs from degradation. It has been shown that MILI proteins were associated with translational machinery and positively regulate translation (Grivna et al. 2006; Unhavaithaya et al. 2009).
Based on these results acquired in this study and previous reports, we propose a hypothetical model for gene regulation at the post-transcriptional level centered on the production of piRNAs as follows: In A that undergo mitosis actively, the expression of a set of genes that is involved in gene regulation is required. The translation of these PRMRs is up-regulated when they are targeted by antisense transcripts, and MILI protein could be recruited as a component of this regulatory machinery. When spermatogonia change their division from mitosis to meiosis, some genes need to be down-regulated, and the degradation of their PRMRs and the concurrent production of piRNAs are induced. For a subset of PRMRs whose proteins are needed at later stages, their mRNAs could be transcribed at an earlier time point and are stabilized by the formation of a sense–antisense duplex, and this phenomenon is more popular at post-meiotic stages than early ones. Whether MRPRs themselves have regulatory function during spermatogenesis remains as an open question, although some indication has been reported (Saito et al. 2009).
The majority of piRNAs generated in PS and RS are IGPRs, which have been reported by previous studies but whose production and function have not been well addressed. According to the distribution of inter contig distances of 11 piRNA libraries, we identified 2992 clusters that could be novel transcription units. IGPRs are more like PRMRs in terms of the presence of antisense transcripts as well as their preferential mapping to one strand. Four types of clusters can be identified, and three of them have antisense transcripts identified. Although piRNAs map to both strands, their abundance levels are very different even in the overlapping regions. The IGPRs do not have the 10A bias, although the 5′U preference is kept. Therefore, the production of IGPRs may not be mediated by the “ping-pong” amplification loop. Marcon et al. (2008) have observed that some piRNAs are located in the meiotic nucleus of the male mouse. Given the large amount of IGPRs generated at meiosis and post-meiotic stages during which chromosomes are dramatically remodeled, these piRNAs could be heavily involved in chromosome remodeling.
In summary, we have identified a large amount of piRNAs that can be divided into three major classes (RTPRs, MRPRs, and IGPRs) based on their mapping to either retrotransposons, mRNAs, or intergenic regions. While it has been reported that the production of RTPRs and their anti-retrotransposon function are the two sides of one coin, we propose that MRPRs are products of gene regulation at the post-transcriptional level, although they may also serve other regulatory functions, and that IGPRs could be involved in chromosome remodeling, a prominent feature of spermatogenesis. We also propose that antisense transcripts may play an important role for the production of piRNAs, although only the sense strands are processed to generate piRNAs. It is apparent that these hypotheses are based on correlated observations derived from computational analysis rather than on experimental results. However, they are valuable guides for the design of future experiments by which they can either be justified or rejected.
MATERIALS AND METHODS
Isolation of germ cells from postnatal mice
Because the first wave of spermatogenesis in postnatal mice occurs in a sequential and synchronized manner, different types of germ cells appear in the testis on particular days postpartum (dpp) and can be isolated using mice of different ages. Type A spermatogonia (A), pachytene spermatocytes (PS), and round spermatids (RS) were isolated from 8-dpp, 17-dpp, and adult mice using the unit gravity sedimentation procedure as described by others (Romrell et al. 1976; Bellve et al. 1977). Briefly, 25, 15, and five mice of corresponding ages were used for isolating A, PS, and RS. Total testis cells were prepared by digestion of decapsulated testes with collagenase (1 mg/mL, 2 min, 37°C) and trypsin (0.25%, 5 min, 37°C) after albuginea was removed. About 108 dispersed cells were suspended in MEM medium and bottom-loaded into a glass cylinder 12.5 cm in diameter followed by 600 mL of BSA solution of 2%–4% gradient in DMEM (Hyclone). After 3 h of sedimentation at unit gravity, the cell fractions (10 mL/fraction) were collected from the bottom of the cylinder at a rate of 10 mL/min. The purity of the three cell types all exceeded 90% based on morphological evaluation and was confirmed by quantitative real-time RT-PCR evaluation of cell-type-specific marker genes (Supplemental Fig. 1).
Sequencing of small RNA libraries
Total RNA was isolated from A, PS, and RS using Trizol reagent following a standard protocol. Ten micrograms of total RNA of each sample was separated by 15% denaturing PAGE gel and visualized by SYBR-gold staining. Small RNAs of 18–36 nt were gel-purified, and cDNAs were prepared using the Illumina small RNA preparation kit (Illumina) and were sequenced using the Illumina Genome Analyzer system.
Annotation of the small RNAs
Small RNA sequences from the raw sequencing data were processed sequentially as indicated by the flowchart in Supplemental Figure 8. Basically, adapter sequences were first removed, and reads with any ambiguous calls represented by Ns were discarded. Small RNA data sets generated by others from mouse testis and other non-testicle tissues were downloaded in the format of FASTQ data files from SRA (http://www.ncbi.nlm.nih.gov/sra) and are described in Supplemental Table 2. All reads were mapped to the mouse genome (mm9/Build 37) using PASS (Campagna et al. 2009). Reads that cannot be mapped perfectly to the genome were mapped to the transcriptome represented by Ensembl Genes (http://www.ensembl.org/index.html). Reads that mapped neither to the genome nor to the transcriptome perfectly were discarded.
Reads from known small RNAs such as miRNAs, rRNAs, tRNAs, snRNAs, and snoRNAs were identified by comparing with sequences in the miRBase database (v13, http://www.mirbase.org) and fRNAdb database (http://www.ncrna.org/frnadb/) with PASS. Sequences of 24–32 nt that were not from known small RNAs and mapped perfectly to the genome or transcriptome were classified as candidate piRNAs.
piRNAs were compared with sequences of repeats, exons, ncRNAs, and introns in separate files downloaded from the UCSC Genome Browser (http://genome.ucsc.edu/index.html) and were classified as a particular type if exact matches were found. piRNAs that have matches in more than one file were classified exclusively in the order of repeat > exons > ncRNA > introns. piRNAs that did not have matches in any of the files were classified as the intergenic type. piRNAs that map to repeats were further classified as three retrotransposon types (LRT, SINE, and LINE) according to the Repbase database (Release 14.10) downloaded from http://www.girinst.org. When comparison were made, at most three mismatches were allowed.
piRNA cluster analysis
IGPRs that map to unique loci of the genome from 11 libraries were used for cluster analysis. Because piRNAs are processed from long transcripts, we assumed that distances of piRNAs within clusters were much smaller than the distances between clusters. As a result, we expected that the number of clusters dropped quickly as the presumed cluster length increased in the beginning and reached a plateau when the cluster length increased to a certain value. Indeed, we observed that the number of clusters reached the plateau when the cluster length increased to 1000–10,000 nt regardless of how the minimum number of piRNAs in each cluster was set (Supplemental Fig. 9). Therefore, the average length of a cluster, or the average maximum distance between two clusters, was defined to be 10,000 nt.
Motif search
A 100-nt sliding window was used to identify the region of a PRMR that has the highest number of piRNA sequences. These regions of CDS-PRMRs and 3′UTR-PRMRs were used to search for any motif(s) using the MEME program (http://meme.sdsc.edu/meme/meme.html). The parameters of MEME (Bailey and Elkan 1994) are set so that on average one motif of 8–50 nt was identified in one sequence with the E-value not exceeding 0.01.
Heatmap construction
The CDS-PRMR and 3′UTR-PRMR were clustered according to their piRNAs expression in A, PS, and RS using Cluster (http://bonsai.hgc.jp/∼mdehoon/software/cluster/software.htm#ctv). We first log-transformed the read counts of each PRMR in the three cell types and centered values by the mean. Hierarchical clustering approach with a Spearman rank correlation similarity metric was used to objectively classify the various types of expression profiles. Data were visualized with the TreeView program (Saldanha 2004). According to the clustering results, the CDS-PRMRs and 3′UTR-PRMRs were divided into three classes, respectively. In Class A, the piRNAs were highly expressed in the type A spermatogonia. In Class PS, the piRNAs were highly expressed in the pachytene spermatocytes. And in Class RS, the piRNAs were highly expressed in the round spermatids.
Enrichment analysis of GO functions
The DAVID functional annotation tool (Dennis et al. 2003; Huang et al. 2009) was used to perform Gene Ontology classification of various classes of PRMRs. Genes expressed at different stages of spermatogenesis were used as reference sets for the corresponding classes of PRMRs. For example, the gene set expressed in A (labeled as P in GSE4193) was used as the reference set for Class A PRMRs. FATs from the ontologies of “biological processes” and “molecular function” were recorded. The enrichment score cutoff was set to 3.0.
Identification of antisense transcripts
In order to identify antisense transcripts, we downloaded the ESTs and mRNAs sequences from the UCSC Genome Browser. Because the orientation of many ESTs has been known to be mis-annotated, we combined multiple evidence to infer the correct orientation for mRNAs and ESTs, including sequence type (mRNA or EST), poly(A) signal/tail, and consensus splicing junctions according to the strategy by Zhang et al. (2007). That information on mRNAs and ESTs was imported from the GoldenPath tables estOrientInfo, est_direction, and mrnaOrientInfo (http://hgdownload.cse.ucsc.edu/goldenPath/mm9/database/). The Ensembl mRNA sequences were aligned with the sequences of mRNAs and ESTs that were reliably oriented by the BLAT tool (Kent 2002). Only BLAT alignments with nucleotide identity ≥99% and overlap length ≥100 nt were used.
Identification of antisense SAGE tags
The LongSAGE tags analysis was conducted according to the strategy of Ge et al. (2006). The LongSAGE tags of testis (GSM106609, GSM106634, GSM106645, GSM106666, GSM106674) samples from GSE4726 (Siddiqui et al. 2005) were used for the study (http://www.ncbi.nlm.nih.gov/geo/). All tags were matched through PASS to the mouse genome sequences (mm9).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article. The sequence data from this study have been submitted to the NCBI Gene Expression Omnibus under accession number GSE24822. A reviewer link to these data is at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=jdelfismoswmkra&acc=GSE24822. The information on piRNA clusters and PRMRs can be queried and obtained from the piRNA db (http://kbrb.ioz.ac.cn/piRNA/).
ACKNOWLEDGMENTS
This study was supported by the National Basic Research Program of China (2006CB944004) and the National Natural Science Foundation of China (30871406).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2648411.
REFERENCES
- Amaral PP, Mattick JS 2008. Noncoding RNA in development. Mamm Genome 19: 454–492 [DOI] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J 2007a. The Piwi–piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ 2007b. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avdievich E, Reiss C, Scherer SJ, Zhang Y, Maier SM, Jin B, Hou H Jr, Rosenwald A, Riedmiller H, Kucherlapati R, et al. 2008. Distinct effects of the recurrent Mlh1G67R mutation on MMR functions, cancer, and meiosis. Proc Natl Acad Sci 105: 4247–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Bellve AR, Millette CF, Bhatnagar YM, O'Brien DA 1977. Dissociation of the mouse testis and characterization of isolated spermatogenic cells. J Histochem Cytochem 25: 480–494 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Campagna D, Albiero A, Bilardi A, Caniato E, Forcato C, Manavski S, Vitulo N, Valle G 2009. PASS: a program to align short sequences. Bioinformatics 25: 967–968 [DOI] [PubMed] [Google Scholar]
- Chan WY, Wu SM, Ruszczyk L, Law E, Lee TL, Baxendale V, Lap-Yin Pang A, Rennert OM 2006. The complexity of antisense transcription revealed by the study of developing male germ cells. Genomics 87: 681–692 [DOI] [PubMed] [Google Scholar]
- Chi YH, Cheng LI, Myers T, Ward JM, Williams E, Su Q, Faucette L, Wang JY, Jeang KT 2009. Requirement for Sun1 in the expression of meiotic reproductive genes and piRNA. Development 136: 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu WK, Hanada K, Kanaar R, Hickson ID 2010. BLM has early and late functions in homologous recombination repair in mouse embryonic stem cells. Oncogene 29: 4705–4714 [DOI] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: 3 doi: 10.1186/gb-2003-4-5-p3 [PubMed] [Google Scholar]
- de Rooij DG, Grootegoed JA 1998. Spermatogonial stem cells. Curr Opin Cell Biol 10: 694–701 [DOI] [PubMed] [Google Scholar]
- Dong LQ, Ramos FJ, Wick MJ, Lim MA, Guo Z, Strong R, Richardson A, Liu F 2002. Cloning and characterization of a testis and brain-specific isoform of mouse 3′-phosphoinositide-dependent protein kinase-1, mPDK-1 beta. Biochem Biophys Res Commun 294: 136–144 [DOI] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestedt C 2009. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol 10: 637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Wu Q, Jung YC, Chen J, Wang SM 2006. A large quantity of novel human antisense transcripts detected by LongSAGE. Bioinformatics 22: 2475–2479 [DOI] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202 [DOI] [PubMed] [Google Scholar]
- Grivna ST, Pyhtila B, Lin H 2006. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci 103: 13415–13420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Wu XQ, Meng XH, Morales C, el-Alfy M, Hecht NB 1998. The RNA- and DNA-binding protein TB-RBP is spatially and developmentally regulated during spermatogenesis. Mol Reprod Dev 49: 219–228 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Chuva de Sousa Lopes SM, Kaneda M, Tang F, Hajkova P, Lao K, O'Carroll D, Das PP, Tarakhovsky A, Miska EA et al. 2008. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS One 3: e1738 doi: 10.1371/journal.pone.0001738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. 2007. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129: 69–82 [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Itman C, Whiley PA, Zhou W, Meistrich M, Sahin Z, Loveland KL 2009. Regulated production of SnoN2 is a feature of testicular differentiation. Microsc Res Tech 72: 833–844 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Wang X, Bao S, Guo R, Johnson DG, Shen X, Li L 2010. INO80 chromatin remodeling complex promotes the removal of UV lesions by the nucleotide excision repair pathway. Proc Natl Acad Sci 107: 17274–17279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ 2002. BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. 1999. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity 10: 345–355 [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Theurkauf W 2008. Biogenesis and germline functions of piRNAs. Development 135: 3–9 [DOI] [PubMed] [Google Scholar]
- Larocque JR, Jasin M 2010. Mechanisms of recombination between diverged sequences in wild-type and BLM-deficient mouse and human cells. Mol Cell Biol 30: 1887–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE 2006. Characterization of the piRNA complex from rat testes. Science 313: 363–367 [DOI] [PubMed] [Google Scholar]
- Lin H 2007. piRNAs in the germ line. Science 316: 397. [DOI] [PubMed] [Google Scholar]
- Ma L, Buchold GM, Greenbaum MP, Roy A, Burns KH, Zhu H, Han DY, Harris RA, Coarfa C, Gunaratne PH, et al. 2009. GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLoS Genet 5: e1000635 doi: 10.1371/journal.pgen.1000635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcon E, Babak T, Chua G, Hughes T, Moens PB 2008. miRNA and piRNA localization in the male mammalian meiotic nucleus. Chromosome Res 16: 243–260 [DOI] [PubMed] [Google Scholar]
- Mills KD, Ferguson DO, Essers J, Eckersdorff M, Kanaar R, Alt FW 2004. Rad54 and DNA Ligase IV cooperate to maintain mammalian chromatid stability. Genes Dev 18: 1283–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namekawa SH, Park PJ, Zhang LF, Shima JE, McCarrey JR, Griswold MD, Lee JT 2006. Postmeiotic sex chromatin in the male germline of mice. Curr Biol 16: 660–667 [DOI] [PubMed] [Google Scholar]
- Oakberg EF 1956. A description of spermiogenesis in the mouse and its use in analysis of the cycle of the seminiferous epithelium and germ cell renewal. Am J Anat 99: 391–413 [DOI] [PubMed] [Google Scholar]
- Ohnishi Y, Totoki Y, Toyoda A, Watanabe T, Yamamoto Y, Tokunaga K, Sakaki Y, Sasaki H, Hohjoh H 2010. Small RNA class transition from siRNA/piRNA to miRNA during pre-implantation mouse development. Nucleic Acids Res 38: 5141–5151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pero R, Lembo F, Chieffi P, Del Pozzo G, Fedele M, Fusco A, Bruni CB, Chiariotti L 2003. Translational regulation of a novel testis-specific RNF4 transcript. Mol Reprod Dev 66: 1–7 [DOI] [PubMed] [Google Scholar]
- Persengiev SP, Saffer JD, Kilpatrick DL 1995. An alternatively spliced form of the transcription factor Sp1 containing only a single glutamine-rich transactivation domain. Proc Natl Acad Sci 92: 9107–9111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS 2009. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol 16: 639–646 [DOI] [PubMed] [Google Scholar]
- Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, Blower MD, Lai EC 2009. A broadly conserved pathway generates 3′UTR-directed primary piRNAs. Curr Biol 19: 2066–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romrell LJ, Bellve AR, Fawcett DW 1976. Separation of mouse spermatogenic cells by sedimentation velocity. A morphological characterization. Dev Biol 49: 119–131 [DOI] [PubMed] [Google Scholar]
- Rybanska I, Gursky J, Faskova M, Salazar EP, Kimlickova-Polakovicova E, Kleibl K, Thompson LH, Pirsel M 2010. Newly identified CHO ERCC3/XPB mutations and phenotype characterization. Mutagenesis 25: 179–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Siomi H, Siomi MC 2007. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev 21: 1603–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC 2009. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461: 1296–1299 [DOI] [PubMed] [Google Scholar]
- Saldanha AJ 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20: 3246–3248 [DOI] [PubMed] [Google Scholar]
- Shima JE, McLean DJ, McCarrey JR, Griswold MD 2004. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod 71: 319–330 [DOI] [PubMed] [Google Scholar]
- Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, Kondoh G, Okawa K, Chujo T, Suzuki T, et al. 2009. The TDRD9–MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell 17: 775–787 [DOI] [PubMed] [Google Scholar]
- Siddiqui AS, Khattra J, Delaney AD, Zhao Y, Astell C, Asano J, Babakaiff R, Barber S, Beland J, Bohacec S, et al. 2005. A mouse atlas of gene expression: Large-scale digital gene-expression profiles from precisely defined developing C57BL/6J mouse tissues and cells. Proc Natl Acad Sci 102: 18485–18490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci 101: 6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas K, Sung DY, Yang J, Johnson K, Thompson W, Millette C, McCarrey J, Breitberg A, Gibbs R, Walker W 2005. Identification, characterization, and functional analysis of sp1 transcript variants expressed in germ cells during mouse spermatogenesis. Biol Reprod 72: 898–907 [DOI] [PubMed] [Google Scholar]
- Thomson T, Lin H 2009. The biogenesis and function of PIWI proteins and piRNAs: Progress and prospect. Annu Rev Cell Dev Biol 25: 355–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unhavaithaya Y, Hao Y, Beyret E, Yin H, Kuramochi-Miyagawa S, Nakano T, Lin H 2009. MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J Biol Chem 284: 6507–6519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oers JM, Roa S, Werling U, Liu Y, Genschel J, Hou H Jr, Sellers RS, Modrich P, Scharff MD, Edelmann W 2010. PMS2 endonuclease activity has distinct biological functions and is essential for genome maintenance. Proc Natl Acad Sci 107: 13384–13389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW 1995. Serial analysis of gene expression. Science 270: 484–487 [DOI] [PubMed] [Google Scholar]
- Werner A, Carlile M, Swan D 2009. What do natural antisense transcripts regulate? RNA Biol 6: 43–48 [DOI] [PubMed] [Google Scholar]
- Yang G, Zhang YL, Buchold GM, Jetten AM, O'Brien DA 2003. Analysis of germ cell nuclear factor transcripts and protein expression during spermatogenesis. Biol Reprod 68: 1620–1630 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li J, Kong L, Gao G, Liu QR, Wei L 2007. NATsDB: Natural Antisense Transcripts DataBase. Nucleic Acids Res 35: D156–D161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, Xiol J, Reuter M, Eckardt S, Leu NA, McLaughlin KJ, Stark A, Sachidanandam R, Pillai RS, Wang PJ 2010. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc Natl Acad Sci 107: 11841–11846 [DOI] [PMC free article] [PubMed] [Google Scholar]