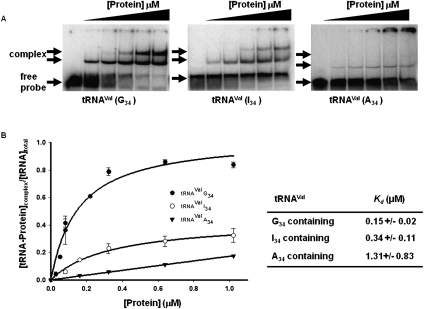
FIGURE 1.
Wild-type TbADAT2/3 stably binds tRNA in vitro. Radioactively labeled tRNAVal (8 nM) from T. brucei was incubated with increasing concentrations of recombinant TbADAT2/3 expressed in E. coli and purified by Ni2+-chelate chromatography. (A) Representative electrophoretic mobility shift assay (EMSA) to determine the extent of tRNA binding. Right panel shows TbADAT2/3 binding to an A34-containing tRNA (natural substrate). Middle and left panels show a similar experiment but with either an I34- or G34-containing tRNA. In all panels, lane 1 shows a mock reaction in which the probe was incubated in binding buffer in the absence of enzyme. Lanes 2–6 show tRNA incubated with increasing concentrations of the enzyme (0.052, 0.18, 0.35, 0.65, and 1.18 μM, respectively). “Free probe” denotes the migration of the unbound tRNA, and “complex” denotes the migration of the TbADAT2/3 bound tRNA. (B) The reaction products from A were used to calculate the fraction of tRNA bound by calculating the percent of the probe shifted divided by the total (bound and unbound) probe in each reaction. These values were plotted against TbADAT2/3 concentration in μM and fitted to a single exponential curve, and the dissociation constant (Kd) was calculated by nonlinear regression using SigmaPlot software.