Abstract
The 5′ UTR of HIV-2 genomic RNA contains signaling motifs that regulate specific steps of the replication cycle. Two motifs of interest are the C-box and the G-box. The C-box is found in the 5′ untranslated region upstream of the primer binding site, while the G-box is found downstream from the major splice donor site, encompassing the gag start codon and flanking nucleotides. Together the C-box and the G-box form a long-range base-pairing interaction called the CGI. We and others have previously shown that formation of the CGI affects RNA dimerization in vitro and the positions of the C-box and the G-box are suggestive of potential roles of the CGI in other steps of HIV-2 replication. Therefore, we attempted to elucidate the role of the CGI using a viral SELEX approach. We constructed proviral DNA libraries containing randomized regions of the C-box or G-box paired with wild-type or mutant base-pairing partners. These proviral DNA libraries were transfected into COS-7 cells to produce viral libraries that were then used to infect permissive C8166 cells. The “winner” viruses were sequenced and further characterized. Our results demonstrate that there is strong selective pressure favoring viruses that can form a branched CGI. In addition, we show that the mutation of the C-box alone can enhance RNA encapsidation, and mutation of the G-box can alter the levels of Gag protein isoforms. These results suggest coordinated regulation of RNA translation, dimerization, and encapsidation during HIV-2 replication.
Keywords: C-box, G-box, CGI, HIV-2, viral SELEX
INTRODUCTION
HIV replication is highly orchestrated, requiring tight regulation of each step of the viral cycle to ensure successful virus synthesis and propagation. The 5′ untranslated region (5′ UTR) of the HIV RNA genome contains several regulatory motifs including TAR (trans-activation responsive element), the polyadenylation signal, the C-box, the primer binding site (PBS), stem–loop 1 (SL1), and the G-box (Fig. 1A; Berkhout 1996; Lanchy et al. 2003b). The C-box, a pyrimidine-rich region, has been implicated in the regulation of genomic RNA dimerization and translation through a base-pairing interaction with the G-box, a purine-rich region encompassing the gag start codon and flanking nucleotides (Fig. 1A; Lanchy et al. 2003b; Strong et al. 2009).
FIGURE 1.
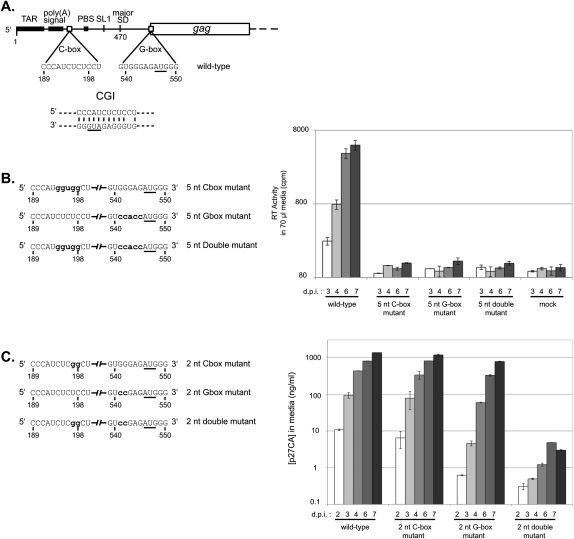
Compensatory mutation analysis of the CGI. (A) Schematic of HIV-2 genomic RNA. (TAR) Trans-activation region; (poly(A) signal) poly(A) signal domain; (C-box) the core of the C-box; (PBS) the tRNA primer binding site; (SL1) the stem–loop one dimerization signal; (major SD) the major splice donor site; (G-box) the core of the G-box; and (gag) the Gag protein-coding region. A proposed C-box/G-box interaction (CGI) is illustrated below. The AUG initiation codon of Gag, found in the G-box, is underlined. (B, left panel) Schematic of the 5-nt mutation series. Only the C-box and G-box regions of the unspliced genomic RNAs are represented. Nucleotide positions that are mutated are in bold lowercase letters. (Right panel) Replication kinetics of the wild-type, the 5-nt C-box mutant, the 5-nt G-box mutant, and the 5-nt double mutant viruses in C8166 cells. Equal amounts of viral particles (10 ng of p27 capsid, as determined by ELISA) produced during COS-7 transfection were used to infect C8166 cells. Viral replication was measured using reverse transcriptase quantification. Numbers below the x-axis represent days post-infection. The y-axis error bars represent the standard error of duplicate experiments. (C, left panel) Schematic of the 2-nt mutation series. Only the C-box and G-box regions of the unspliced genomic RNAs are represented. Nucleotide positions that are mutated are in bold lowercase letters. (Right panel) Replication kinetics of wild-type, the 2-nt C-box mutant, the 2-nt G-box mutant, and the 2-nt double mutant viruses in C8166 cells. Equal amounts of viral particles (10 ng of p27 capsid, as determined by ELISA) produced during COS-7 transfection were used to infect C8166 cells. Viral replication was measured using p27 capsid quantification. Numbers below the x-axis represent days post-infection. The y-axis error bars represent the standard error of duplicate experiments.
Several previous studies suggested that both the presence of the C-box and the G-box and their ability to base-pair affected dimerization in HIV-2 (Dirac et al. 2002; Lanchy et al. 2003a,b, 2004; Baig et al. 2009) and in HIV-1 (Abbink and Berkhout 2003; Damgaard et al. 2004; Abbink et al. 2005; Song et al. 2008). It was postulated that the C-box–G-box interaction, termed “CGI” in HIV-2 (Baig et al. 2008) or “U5-AUG duplex” in HIV-1 (Abbink and Berkhout 2003), could modulate translation through occlusion of the gag start codon encompassed by the G-box (Abbink and Berkhout 2003; Lanchy et al. 2003a,b; Damgaard et al. 2004). Mutational studies designed to test two proposed structural conformations of the HIV-1 leader, namely, the branched multiple-hairpin (BMH) and the long-distance interaction (LDI) conformations, did not appear to support a role for the U5-AUG duplex, which would be disrupted in a large-scale LDI-BMH conversion, in translational control (Abbink et al. 2005). However, the main focus of that study was the BMH-LDI interconversion, and it did not directly probe the U5-AUG duplex. In HIV-2, directed mutagenesis of the leader suggested that the C-box does play a role in translational regulation, as deletion or mutation of the C-box to disrupt the CGI led to enhanced translation of HIV-2 chimeric reporter constructs (Strong et al. 2009).
HIV-2 and SIVmac are unique among the primate lentiviruses in that they have a conserved intron contained entirely within the 5′ untranslated leader region (Viglianti et al. 1990, 1992; Chatterjee et al. 1993; Strong et al. 2009). The C-box is located at the 3′ end of the intron and is completely removed upon 5′ UTR splicing. Previously we have shown that removal of this intron results in increased translation of gag mRNA, apparently through the combined effects of overall shortening of the leader and removal of secondary structure that could impede ribosome scanning (Strong et al. 2009). Detailed in vitro translation studies demonstrated that mutations or deletions of the C-box that were designed to disrupt the proposed CGI resulted in increased translational efficiency. However, attempts to directly test the CGI by making compensatory mutations in the G-box were confounded by the exquisite sensitivity of the G-box to changes, apparently because the G-box encompasses the Kozak context for the gag start codon (Strong et al. 2009). Thus, the importance and the role(s) of the CGI have been difficult to address experimentally.
Here we used a viral SELEX technique (Berkhout and Klaver 1993; Baig et al. 2009) to generate viable virus variants at the C-box or the G-box. When either of these sequences was randomized alone, wild-type-like sequences rapidly emerged as the dominant species. This led us to question whether this was the result of local sequence constraints or the requirement to maintain base-pairing compatibility with another viral sequence. To address this, we built two additional proviral DNA libraries, one that was simultaneously randomized at both the C-box and the G-box, and one that contained a lethal mutated G-box sequence combined with a randomized C-box. Each of these libraries was designed to test, with the use of non-native sequence in the C-box and/or G-box, whether base-pairing between these sites was important for virus viability. These co-randomization and forced evolution experiments reproducibly demonstrated that the requirement for base-pairing in the CGI is finely tuned, with lack of base-pairing being as deleterious as overly stable extended base-pairing arrangements between these elements. Sequence analysis of winning viruses suggested that the CGI is directly involved in a branched three-way RNA junction, consistent with a previously proposed model (Lanchy et al. 2003b).
RESULTS
Mutations in the C-box and G-box are deleterious to HIV-2 in cell culture
We first addressed the role of the CGI in HIV-2 replication by investigating the effects of mutations and presumed compensatory mutations of the C-box and G-box. Mutant viruses containing five altered nucleotides in either the C-box (positions 194–198) or the G-box (positions 541–545) were replication-defective in cell culture (Fig. 1B). The compensatory mutant, containing simultaneously mutated C-box and G-box, failed to rescue replication (Fig. 1B). Because 5-nt mutations of the C-box or G-box were lethal, we attempted more subtle 2-nt mutations in the C-box (positions 197–198), G-box (position 541–542), and compensatory mutations in both (Fig. 1C). The 2-nt C-box mutant reached close to wild-type replication levels (Fig. 1C). The 2-nt G-box mutant showed reduced levels of replication. Interestingly, combining the C-box and G-box mutations resulted in viruses that were replication-defective (Fig. 1C). Thus, our first attempts to create a non-wild-type CGI using compensatory mutations resulted in replication-defective viruses, hampering analysis of sequence requirements using cell culture studies. To circumvent the limitations of building and testing substitution mutants one at a time, we adopted a viral SELEX approach.
Viral SELEX of C-box and G-box randomized libraries
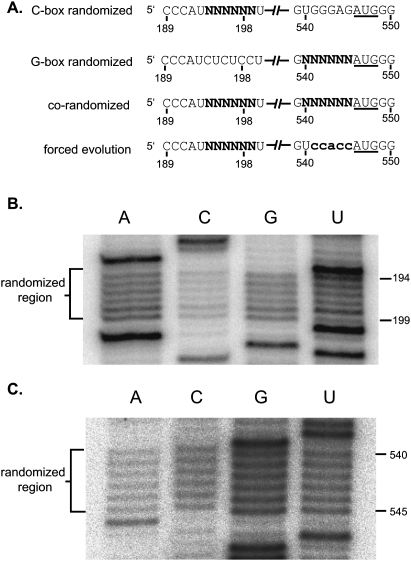
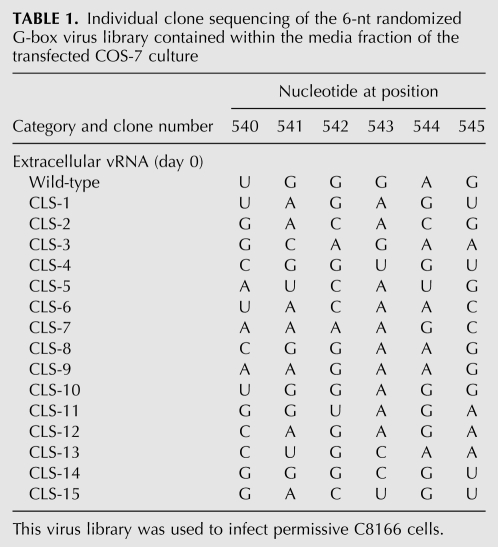
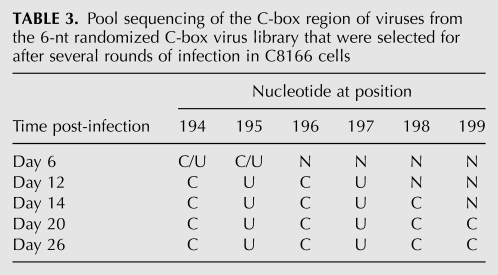
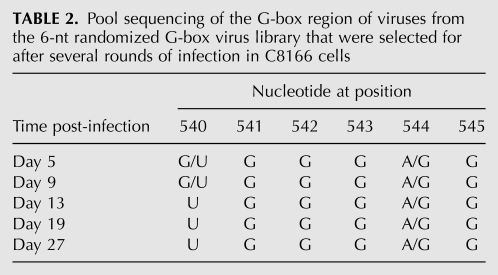
Six nucleotides of the C-box (nucleotides 194–199) or 6 nt of the G-box (nucleotides 540–545) were randomized using degenerate primers to make PCR libraries containing up to 4096 possible sequence combinations of either the C-box or the G-box region of the genome. Each PCR library was used to construct a library of full-length infectious HIV-2 proviral DNA clones using a ligation-independent protocol (see Materials and Methods). The degeneracy of each randomized proviral DNA library was verified through pool sequencing. The C-box and G-box proviral DNA libraries were independently transfected into COS-7 cells to generate virus. COS-7 cytoplasmic and media fractions for both libraries were harvested. Viral RNAs purified from the media fraction were examined for degeneracy of the randomized region (Fig. 2). Virus harvested from the media fraction was used to infect permissive C8166 cells. Emergence of dominant virus sequences was quite rapid, with the two winning sequences for the G-box library (5′ 540-UGGGRG, where R is G or wt A) dominating by day 13 (cf. Tables 1 and 2) and the wild-type C-box sequence (5′ 194-CUCUCC) dominating the C-box library by day 20 (Table 3). The more rapid emergence of dominant sequences suggested that there was stronger selective pressure on the G-box sequence than the C-box. By day 5, four out of the six positions in the randomized G-box were wild type, with the two remaining positions 540 and 544 being U or G and A or G, respectively (Table 2). The C-box, on the other hand, contained pyrimidines at positions 194 and 195 by day 6, with the remaining four positions (196–199) still relatively diverse (Table 3). However, only the wild-type C-box sequence was found by day 20.
FIGURE 2.
C-box and G-box randomized libraries used in this study. (A) Schematic of the C-box and G-box elements in the four randomized viral libraries. Only the C-box and G-box regions of the unspliced genomic RNAs are represented. Nucleotide positions containing an “N” have an equal probability of being A, C, G, or U. The G-box mutation used in the forced selection is in bold lowercase letters. (B,C) Viral RNAs purified from the media fraction of transfected COS-7 cells were reverse-transcribed into cDNA. Primer extension of the different cDNA showed the degeneracy of the C-box randomized region in the C-box randomized viral library (B) and the G-box randomized region in the G-box randomized viral library (C).
TABLE 1.
Individual clone sequencing of the 6-nt randomized G-box virus library contained within the media fraction of the transfected COS-7 culture
TABLE 3.
Pool sequencing of the C-box region of viruses from the 6-nt randomized C-box virus library that were selected for after several rounds of infection in C8166 cells
TABLE 2.
Pool sequencing of the G-box region of viruses from the 6-nt randomized G-box virus library that were selected for after several rounds of infection in C8166 cells
The rapid emergence of wild-type sequence seen in both the C-box and G-box randomized libraries made us ask whether this was because of local sequence requirements, mutual base-pairing requirements (i.e., CGI), or both. To address this question, we built a new library (Fig. 2A, co-randomized library) with both the C-box (positions 194–199) and G-box (positions 540–545) randomized on the same proviral DNA.
Viral SELEX of the co-randomized library
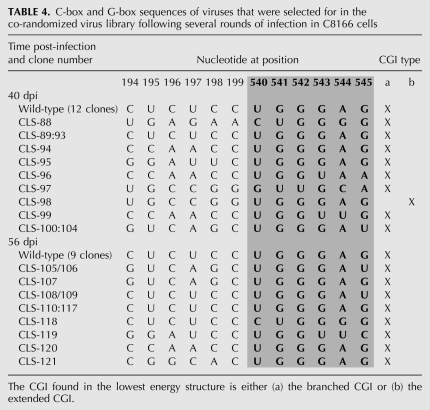
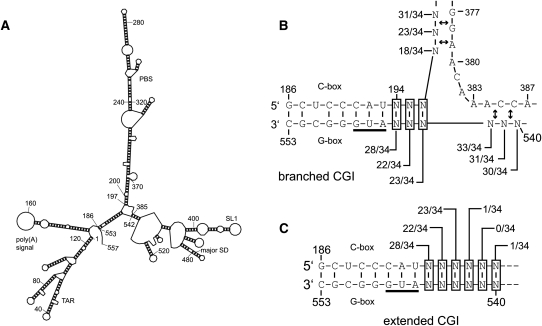
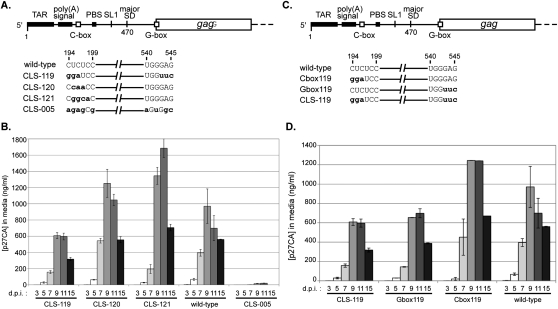
The co-randomized viral library (Fig. 2A) was constructed and tested using the same techniques as described for the C-box and G-box libraries. Mfold (Zuker 2003) prediction of secondary structure of all winner viral sequences revealed several key characteristics. First, non-wild-type viruses represented a large proportion of selected viruses on day 40 (12 out of 17) and day 56 (nine out of 17) (Table 4). Second, all of the selected viruses were predicted by Mfold to form two nearly isoenergetic conformations of CGI, an extended form and a branched form (Fig. 3). The extended CGI consisted of continuous base-pairing through the entirety of the C-box and G-box (Fig. 3C). The branched CGI consisted of a three-way RNA junction with base-pairing of the 5′ proximal C-box with the 3′ proximal G-box, while the 5′ proximal G-box was base-paired with the nucleotide 385 region, similar to a structure previously proposed (Fig. 3B; Lanchy et al. 2003b). The branched CGI was found in lowest energy structures for all but one of the viral sequences (Table 4). Selection for wild-type sequence at the 5′ side of the proximal G-box (positions 540–542) was readily apparent, represented by 15 out 17 viruses on day 40 and 16 out of 17 viruses by day 56 (Table 4). Although a significant percentage of viruses with wild-type sequence emerged from our co-randomized library, it also produced CGI-forming viruses with non-wild-type sequences (12 out of 17 sequences by day 40). To determine whether these non-wild-type viruses were viable, three late-round viruses (CLS-119, CLS-120, and CLS-121) and one early-round virus (CLS-005) were selected for individual testing to compare their viral replication to that of wild type (Fig. 4A,B). To show that the C-box and G-box mutations are solely responsible for the observed viral phenotypes, we cloned these mutations into the original wild-type HIV-2 backbone. All of the late-round viruses were predicted to form the branched CGI, while the CLS-005 was not. As shown in Figure 4B, the early round CLS-005 displayed defective viral replication, while late-round CLS-119, CLS-120, and CLS-121 were replication-competent when compared to wild type. These results showed first that the CGI is important for viral replication and second that it can be studied outside of a wild-type context.
TABLE 4.
C-box and G-box sequences of viruses that were selected for in the co-randomized virus library following several rounds of infection in C8166 cells
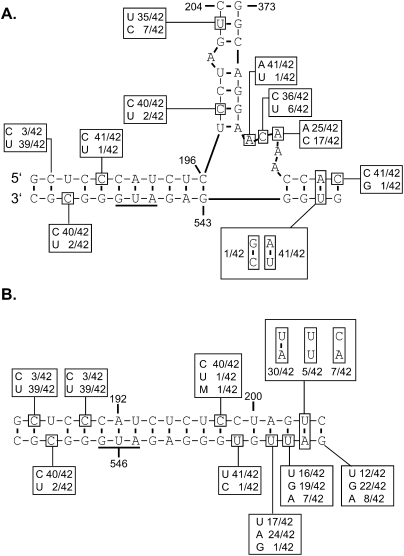
FIGURE 3.
Summary of the base-pairing interactions involving the C-box and G-box found in the predicted optimal RNA structures from the co-randomized viral selection. (A) The most stable secondary structure predicted by Mfold for nucleotides 1–557 of the wild-type HIV-2 genomic RNA. (B,C) When analyzed similarly, the 34 viral sequences from the evolved co-randomized library (Table 4) grouped into two CGI conformations, the branched CGI (B) and the extended CGI (C). Each potential base-pairing interaction of the randomized nucleotides is shown. Each ratio indicates the number of times this base-pairing is found in the most stable CGI-containing structure for each of the 34 viral sequences. (Double arrows) A level of flexibility exists with regard to the base-pairing partner for the randomized nucleotide. Depending on the identity of the randomized nucleotide, the base-pairing partner in the designated region may shift by one or two positions.
FIGURE 4.
Replication kinetics of mutant HIV-2 viruses. (A) A schematic of the five HIV-2 genomic RNAs produced in this study. The CLS-119 RNA contains a mutated C-box and a mutated G-box. The CLS-120 RNA contains a mutated C-box and a wild-type G-box. The CLS-121 RNA contains a mutated C-box and a wild-type G-box. The CLS-005 RNA contains a mutated C-box and a mutated G-box. The mutations are in bold lowercase letters. (B) Replication kinetics of wild-type, CLS-119, CLS-120, CLS-121, and CLS-005 viruses in C8166 cells. Equal amounts of viral particles (10 ng of p27 capsid, as determined by ELISA) produced during COS-7 transfection were used to infect C8166 cells. Viral replication was measured using p27 capsid quantification. The bars indicate standard error for two experiments. (C,D) Mutant HIV-2 viruses harboring G-box mutations display attenuated replication. (C) A schematic of the four HIV-2 genomic RNAs produced in this study. The C-box119 RNA contains a mutated C-box and a wild-type G-box. The G-box119 RNA contains a wild-type C-box and a mutated G-box. The CLS-119 RNA contains a mutated C-box and a mutated G-box. The mutations are in bold lowercase letters. (D) Replication kinetics of wild-type, C-box119, G-box119, and CLS-119 viruses in C8166 cells. Equal amounts of viral particles (10 ng of p27 capsid, as determined by ELISA), produced during COS-7 transfection were used to infect C8166 cells. Viral replication was measured using p27 capsid quantification. The y-axis error bars indicate standard error for the two experiments.
Forced evolution of C-box randomized viruses with a defective G-box
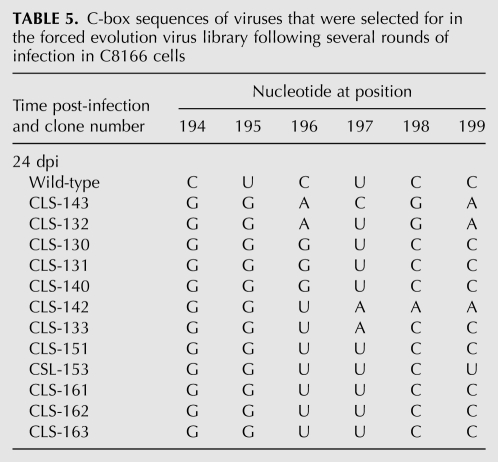
To further evaluate the relationship of C-box and G-box sequences, we built a new randomized viral library that forced the viruses to compensate for a defective mutant G-box sequence with novel sequence combinations at the C-box. Six nucleotides of the C-box (nucleotides 194–199) were randomized using degenerate primers in an HIV-2 background containing a mutated G-box that is lethal by itself (Figs. 1B, 2A). The proviral DNA library was transfected into COS-7 cells, and the virions produced were used to infect C8166 cells. Emergence of viable virus occurred after 3 wk. The mutated G-box sequence (5′ 541-CCACC) was conserved in all sequenced clones (data not shown). All selected viruses contained guanosines at positions 194 and 195 of the C-box (Table 5). There was more variability seen in position 196, although the wild-type cytosine was completely absent in all selected viruses. Wild-type sequence (UCC) was found at positions 197–199 for most of the selected viruses, which would not be expected to base-pair with the mutated G-box nucleotides 540–552, located upstream of the gag AUG start codon. This experiment showed that mutations in the C-box rescued a lethal mutant G-box mutation. Moreover, these results corroborated results from the co-randomized library, demonstrating selective pressure for base-pairing between the 5′ proximal C-box and 3′ proximal G-box.
TABLE 5.
C-box sequences of viruses that were selected for in the forced evolution virus library following several rounds of infection in C8166 cells
Effects of artificially evolved C-box and G-box sequences on viral replication
Although the co-randomized and forced evolution library results supported a role in viral replication for the C-box and G-box through their base-pairing interaction, our 5-nt and 2-nt mutational studies suggested that the sequence of each region may also have a CGI-independent role in viral replication. To probe the individual roles of the C-box and G-box, a viral sequence (CLS-119) from the co-randomized library was selected for testing. We chose CLS-119 for three reasons. First, CLS-119 was a selected (viable) virus, predicted to form the branched CGI (Fig. 3A,B). Second, the CLS-119 C-box and G-box were non-native, composed equally of both wild-type and non-wild-type nucleotides (Table 4). The CLS-119 C-box contained both purines (positions 194–196, GGA) and pyrimidines (positions 197–199, UCC), as did the G-box with mostly purines (positions 540–542, UGG) and pyrimidines (positions 543–545, UCC). Third, although represented in the winning pool, CLS-119 displayed delayed viral replication compared to the other selected viruses as well as wild type (Fig. 4B). We therefore hypothesized that this delay may be a result of the individual contributions of the C-box and the G-box.
To determine the individual effects of the non-native CLS-119 C-box and G-box sequences, we built two additional clones and compared viral replication to that of wild type. All of the clones were predicted to form the branched CGI, which allowed us to concentrate on individual effects of the C-box and G-box regions instead of effects of the CGI. The CLS-119 clone contained both the mutated C-box and G-box. The C-box119 and G-box119 clones contained either the CLS-119 C-box or the CLS-119 G-box, respectively. COS-7 cells were transfected with the wild-type, C-box119, G-box119, and CLS-119 full-length HIV-2 plasmids (Fig. 4C). Virions from the COS-7 supernatant were harvested and used to infect permissive C8166 cells. Viral replication of the C-box119 mutant, as determined by ELISA, reached wild-type-like levels, whereas the G-box119 and CLS-119 mutants exhibited attenuated replication (Fig. 4D). The shared feature of the G-box119 and CLS-119 mutants is the presence of the mutated G-box. To determine whether the decrease in replication was a result of a packaging deficiency linked to the presence of the G-box, intracellular and virion RNA levels were examined.
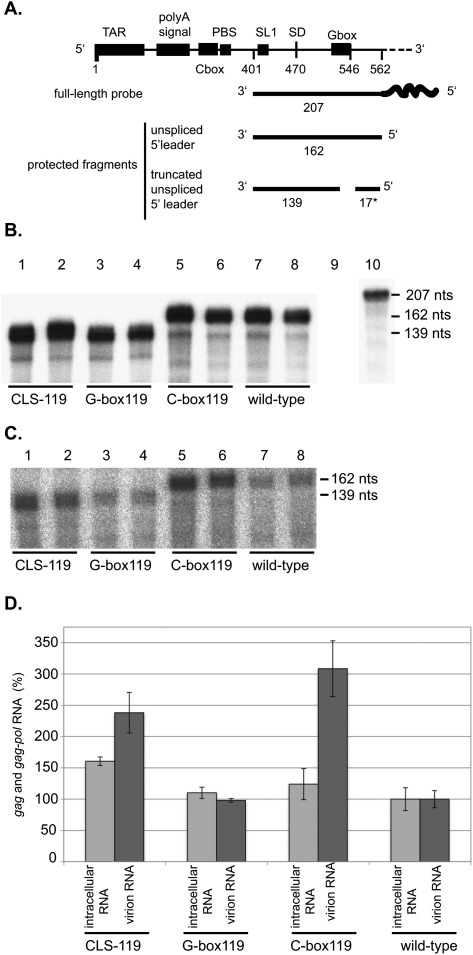
We measured intracellular viral RNA levels and virion RNA content using an RNase protection assay (RPA) (Fig. 5A). Intracellular gag RNA levels were very similar, while RNA levels found in the virion fractions differed (Fig. 5). Surprisingly, the C-box119 virus had higher levels of genomic RNA encapsidated (normalized to extracellular capsid protein p27) than the other viruses. This suggested that in addition to affecting dimerization, translation, and 5′ UTR splicing, the C-box also plays a role in encapsidation. However, the RPA results could not explain why the presence of the G-box resulted in decreased replication. Because the mutated G-box encompasses the Kozak sequence of the gag initiation site, we hypothesized that perhaps this mutation altered the translation of gag mRNA. Therefore, we then sought to determine if the attenuated viral replication seen with the G-box119 and CLS-119 viruses was the result of decreased translational efficiency. To that end, we examined both intracellular and virion fractions of transfected COS-7 cells for total Gag levels (Fig. 6). The intracellular and virion protein fractions were examined via Western blotting, using a primary antibody against p27 capsid. Both the CLS-119 and G-box119 viruses showed a marked decrease in total Gag levels both inside and outside the cell compared to the C-box119 and wild-type viruses (Fig. 6C,D).
FIGURE 5.
C-box mutation results in increased RNA encapsidation. (A) A schematic of the radioactive probe and predicted protected fragments based on the presence of a wild-type or mutant G-box. (B) RNase protection assay of intracellular RNAs obtained from duplicate experiments. DNA templates used for each transfection are listed below each lane (lanes 1–8). Lanes 9 and 10 contain yeast RNA used as a nonspecific target control, incubated with or without RNases, respectively. (C) RNase protection assay of virion RNAs produced from duplicate HIV-2 DNA transfected COS-7 cell experiments. DNA templates used for each transfection are listed below each lane (lanes 1–8). (D) Quantification of B and C. RNA levels were normalized to wild-type levels, set at 100% for both intracellular and virion samples. Identities of RNA source, intracellular or virion, and DNA templates used for each transfection are listed on the x-axis. The y-axis represents gag and gag-pol RNA levels in percent. The y-axis error bars indicate standard error for the two experiments.
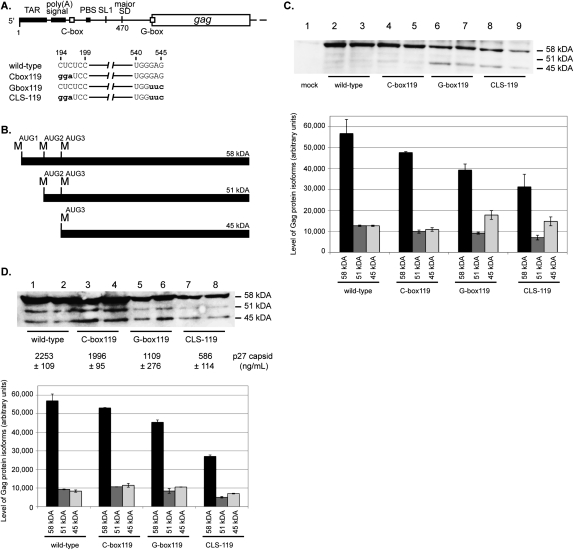
FIGURE 6.
Effects of a mutated G-box on Gag production in cell culture. (A) A schematic of the four HIV-2 genomic RNAs produced in this study. The wild-type genomic RNA contains a wild-type C-box and a wild-type G-box. The C-box119 genomic RNA contains a mutated C-box and a wild-type G-box. The G-box119 genomic RNA contains a wild-type C-box and a mutated G-box. The double 119 genomic RNA contains a mutated C-box and a mutated G-box. The mutations are in bold lowercase letters. (B) A schematic of the three HIV-2 Gag protein isoforms that are synthesized in cell culture. The 58-, 51-, and 45-kDa isoforms are synthesized when translation initiates from methionine codons at positions 546, 744, and 897, respectively. (C) Western blot analysis of intracellular proteins from duplicate experiments where COS-7 cells were transfected with equimolar amounts of the wild-type, C-box119, G-box119, or CLS-119 proviral DNAs. (Lane 1) The mock-transfected sample. DNA templates used in each transfection are designated below each lane. The relative proportion of each isoform band was quantified and is graphically represented. The y-axis error bars represent the standard error of duplicate experiments. (D) Western blot analysis of virion proteins produced from duplicate experiments where COS-7 cells were transfected with equimolar amounts of the wild-type, C-box119, G-box119, or CLS-119 proviral DNAs. DNA templates used in each transfection are designated below each lane. The averaged level of capsid p27 protein in the COS-7 media was quantified (ELISA) and is indicated below the Western blot. The relative proportion of each isoform band was quantified and is graphically represented. The y-axis error bars represent the standard error of the duplicate experiments.
In addition to differences in total Gag levels, there were also differences in the relative expression of three isoforms of Gag between viruses with a mutant G-box and those with a wild-type G-box. Ohlmann and coworkers have shown that these three isoforms of Gag are expressed during HIV-2 infection, which they attributed to alternative initiation events (Herbreteau et al. 2005; Weill et al. 2010). The first isoform, p58, initiates synthesis from a start codon located at nucleotides 546–548 (AUG1). The second isoform, p51, initiates synthesis from a start codon located at positions 744–746 (AUG2). The third isoform, p45, initiates synthesis from a start codon located at positions 897–899 (AUG3). Interestingly, clones containing the mutant G-box, i.e., G-box119 and CLS-119, exhibited an increased level of the p45 Gag isoform initiated from AUG3 compared to wild type and to the C-box119 mutant (Fig. 6C, cf. lanes 2–5 and lanes 6–9).
In vitro translation of luciferase RNAs containing mutated C-box, G-box, or both
To test whether the different levels of Gag isoforms seen in clones containing the mutant G-box were caused by altered start codon usage, we assayed the translation of the HIV-2 C-box and G-box mutant constructs in an in vitro rabbit reticulocyte translation assay. As shown in Figure 7, constructs containing a mutated G-box demonstrated differences in gag start codon usage compared to constructs with a wild-type G-box. Both CLS-119 and G-box119 gag-luciferase constructs demonstrated a slight increase in usage of the AUG3 start codon, similar to what was seen in cell culture. These data suggest that the different levels of the isoforms are caused by altered start codon usage, and that this difference is directly influenced by the mutation in the G-box.
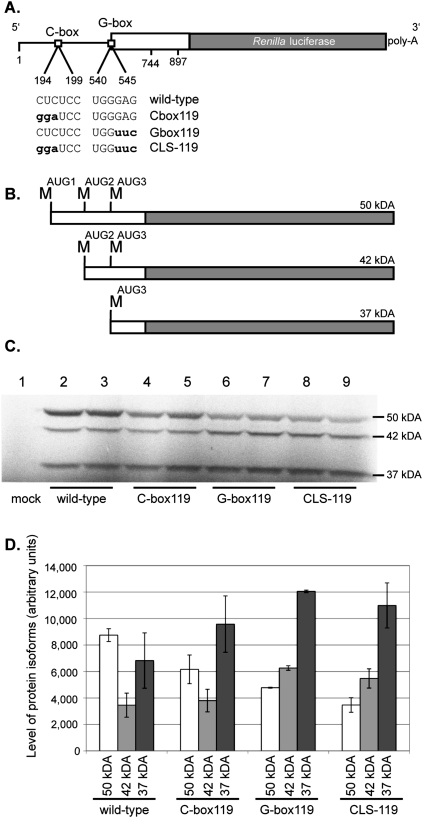
FIGURE 7.
In vitro translation of luciferase mRNAs harboring mutated HIV-2 5′ leaders. (A) A schematic of the four mRNA constructs used in the S35 in vitro translation assays. The mutated nucleotides are in bold lowercase letters. (B) A schematic of the three HIV-2 Gag-luciferase fusion protein isoforms that are synthesized in vitro. The 50-, 42-, and 37-kDa isoforms are synthesized when translation initiates from methionine codons located at positions 546, 744, and 897, respectively. (C) Duplicate S35 in vitro translation assays of the wild-type, C-box119, G-box119, and CLS-119 mRNAs. (Lane 1) A mock where no mRNA was added to the translation reaction. RNAs used in each translation reaction are indicated below each lane. (D) Quantification of protein isoform expression levels. The RNAs used are indicated on the x-axis, along with the protein isoforms produced and quantified. The level of protein isoforms in arbitrary units is indicated on the y-axis. The y-axis error bars represent the standard error of the duplicate experiments.
DISCUSSION
In this study, we used viral SELEX to determine the role of the CGI in viral replication. Generating a collection of unique viruses that differed only in their C-box and G-box regions allowed us to circumvent the experimental limitations of building and testing one unique mutant at a time. Testing these viruses in a competitive cell culture setting allowed us to examine in detail viable viruses with CGI sequence combinations not found in nature. Our results using this viral evolution technique emphasize the importance of the CGI for viral replication and fine-tune our understanding of base-pairing arrangements in this region. Furthermore, we show that in addition to a CGI-dependent role, the C-box and G-box have CGI-independent roles in viral replication.
We demonstrate here that mutation of the C-box affects HIV-2 RNA encapsidation levels (Fig. 5), suggesting a role for the C-box in the packaging process. Mutation of the C-box may contribute to encapsidation through structural rearrangements of the 5′ UTR, exposing previously hidden motifs while trapping others (D'Souza and Summers 2004). Indeed, in HIV-2, destabilization of the CGI was shown to free RNA elements essential for dimerization (SL1) and encapsidation (ψ) from structural entrapment (Baig et al. 2008). Another possibility is that the enhanced encapsidation phenotype may be due to enhanced binding of NC to the mutated C-box. It is known that nucleocapsid protein (NC) mediates genomic RNA encapsidation by binding RNA elements found in the 5′ leader region (Rein et al. 1998; D'Souza and Summers 2005). Studies of HIV-1 leader RNA have suggested a role for the C-box as a binding partner of NC, wherein the binding of NC disrupts the CGI (Spriggs et al. 2008; Wilkinson et al. 2008). Additionally, in vitro HIV-1 mutant RNAs harboring guanosines in the C-box displayed enhanced NC binding affinity when compared to wild type (Spriggs et al. 2008). Here, the mutated CLS-119 C-box differs from the wild-type C-box at the 5′ proximal positions 194–196, with the sequence GGA instead of CUC. It is possible that the presence of two non-wild-type guanosines in the mutant C-box may influence NC binding and encapsidation in HIV-2. It is conceivable that wild-type C-box functions as a good but not ideal encapsidation enhancer, similar to what we have shown for the G-box and its modulation of translation. However, we cannot rule out an indirect effect of the CLS-119 C-box mutation on the presentation of RNA signals such as encapsidation elements.
Our studies on the G-box demonstrated the sensitivity of this region to mutation. Besides its CGI-dependent context, the G-box sequence is also constrained by the basic requirements for a start codon and an adjacent glycine codon to serve as the myristoylation signal (Gottlinger et al. 1989). In addition, G-box nucleotides are involved in translation initiation as part of the Kozak sequence. It is notable that according to the Kozak consensus rules, the context of the wild-type start codon AUG1 is good, but not ideal (Kozak 1986). Interestingly, when we mutated the G-box sequence closer to an ideal consensus sequence, resulting mutant viruses were not viable. This suggested that enhancement of AUG1 translational efficiency is not necessarily beneficial in the overall scheme of HIV-2 replication. Conversely, we show that viruses displaying decreased AUG1 translational efficiency compared to wild type, are viable (Fig. 4D) but display altered ratios of the three isoforms of Gag. The decrease in p58 Gag production is expected due to the mutation of the AUG1 Kozak consensus sequence; however, the slight increase in p45 Gag production (AUG3 initiation) is notable. The p45 Gag isoform was previously shown to be necessary for efficient viral replication, but its role remains obscure (Herbreteau et al. 2005).
Previous studies on HIV-2 translation have suggested that the genomic RNA contains an IRES located downstream from the gag AUG1 initiation codon (Herbreteau et al. 2005; Weill et al. 2010). According to this model, HIV-2 IRES recruits ribosomes for translational initiation at AUG1 and farther downstream at AUG2 and AUG3 to generate the three Gag isoforms. IRES initiation at each of the three AUGs occurs independently of one another (Ricci et al. 2008). Interestingly, we observed that mutation of nucleotides immediately upstream of AUG1 resulted in altered Gag isoform ratios. Although it is possible that the 3-nt G-box mutation affected IRES-dependent gag initiations, a more likely scenario is leaky scanning through AUG1. It has been shown recently that initiation at the HIV-1 AUG1 gag initiation site primarily occurs through ribosomal scanning (Berkhout et al. 2011). Since our HIV-2 G-box mutation results in a weaker Kozak context for AUG1, leaky scanning at this initiation site could enhance the use of at least one of the downstream start codons. Several laboratories have shown that in HIV-1 the strength of upstream AUGs can influence initiation at downstream AUGs (Schwartz et al. 1992; Anderson et al. 2007; Krummheuer et al. 2007). Mutation of a weak upstream HIV-1 rev Kozak sequence to a strong initiating sequence resulted in decreased leaky scanning and poor initiation at the downstream vpu AUG (Anderson et al. 2007). Here the CLS-119 AUG1 is embedded in a relatively poor Kozak context compared to the wild-type AUG1, resulting in increased initiation at downstream AUG3. In this case, of the three potential gag initiation codons in CLS-119, AUG3 is predicted to have the best Kozak context; thus, a scanning ribosome could scan past AUG1 and 2 and initiate translation at AUG3 more often than when the Kozak context of AUG1 is wild type (Kozak 1986, 1987). Taken together, we propose that the strength of the Kozak context sequence for the HIV-2 gag AUG1 start codon is evolutionarily fine-tuned to allow balanced synthesis of Gag isoforms and that, in combination with the C-box, there could be a higher-order structure component to the use of the AUG1 start codon as well.
The main focus of this study was to determine the necessity of the CGI in HIV-2 replication. Our viral SELEX work supports a role for the association of the C-box with the G-box, with the caveat that both local and long-distance constraints govern the CGI. Local sequence constraints limit nucleotide identity in both the C-box and G-box. Sequence identities in these regions appear to affect 5′ UTR splicing, Gag or NC binding, and the Kozak consensus sequence. However, our co-randomization and forced evolution experiments demonstrated reproducibly that partial base-pairing across this region is necessary, as seen in the branched CGI conformation (Fig. 3A,B). Indeed, the forced evolution experiment clearly illustrates that a relationship between the C-box and G-box exists in HIV-2 replication. A lethal viral replication phenotype associated with the presence of the mutant G-box (CCACC) was rescued by the presence of the selected C-boxes. The selected C-boxes in this library all contain guanosines at positions 194 and 195, which are compatible for base-pairing with the mutant G-box cytosines at positions 544 and 545. Interestingly, extension of the CGI through additional base-pairing did not emerge significantly among non-wild-type surviving viruses from both the forced evolution and co-randomized libraries. Instead, it seems that selective pressure may be exerted at these remaining positions (197–199) to keep the CGI from becoming too stable and to allow the 5′ proximal G-box to form a long-distance base-pairing interaction with the nucleotide 385 region. In all sequences forming the branched CGI, nucleotides 200–204 (just downstream from the randomized C-box) were base-paired with nucleotides 379–376, irrespective of the identity of randomized nucleotides 197–199. It is notable that the base-pairing interaction of the 5′ proximal G-box with the nucleotide 385 region could vary through shifting of 1 or 2 nt depending on the identity of the randomized G-box nucleotides.
Our 2-nt and 5-nt mutations experiments support the pressure for CGI flexibility, as we saw that high-stability base-pairing across the entire mutated CGI results in a lethal phenotype. The replication-defective 2-nt double mutant is predicted to be unable to form two of the three branches of the branched CGI. Similarly, the 5-nt double mutant is also predicted to be unable to form two of the three branches of the branched CGI. Only when the 5-nt G-box was paired with a randomized C-box that did not extend base-pairing through the CGI, were viable viruses obtained. Furthermore, analysis of published HIV-2/SIV sequences (Kuiken et al. 2010) shows several examples of nucleotide variations that maintain base-pairing that is consistent with our “artificial phylogeny” and supports a branched three-way RNA junction (Fig. 8). In particular, a key element in the branching CGI structure is the base-pairing of some of G-box nucleotides with the 385 region. These four base pairs are highly conserved, which is further illustrated by a conservative compensatory mutation in the isolate SIVsmmSL92.
FIGURE 8.
Nucleotide identities of nonconserved position among natural isolates of HIV-2 and SIV superimposed on the two potential CGI conformations. The consensus branched (A) and extended (B) CGI structures from the analysis of 42 HIV-2 and SIV sequences from the HIV Sequence Compendium 2010 are indicated, with the proportions of alternative base pairs shown (Kuiken et al. 2010). Those nucleotides not absolutely conserved are boxed.
Our data emphasize the importance of the CGI in vivo and suggest a model in which some degree of CGI association is necessary, as seen in the branched CGI model (Fig. 3A,B; Lanchy et al. 2003b), but because of roles in multiple steps of the HIV-2 replication cycle, the CGI needs to be dynamic. The dynamic nature of base-pairing in the 5′ leader has previously been supported in the context of the LDI/BMH riboswitch affecting the BMH and LDI conformational equilibrium in HIV-1 RNA (Abbink et al. 2005). Although it has been shown that the base-pairing between the C-box and G-box in mature HIV-1 particles is maintained in the open conformation (Wilkinson et al. 2008), its behavior in HIV-2 still needs to be investigated. Mapping the dynamic range of the CGI during select replication steps would provide further insight into the CGI's mode of action.
MATERIALS AND METHODS
Construction of plasmids for generation of randomized C-box, G-box, co-randomized, and forced evolution proviral libraries
To prevent contamination by wild-type proviral DNA plasmid in the C-box, G-box, co-randomized, and forced evolution libraries, we used parent plasmids called pSCR2, pAUG1, pCboxg, and pGboxc, respectively. The pSCR2, pAUG1, pCboxg, and pGboxc plasmids harbor deleterious mutations in the encapsidation signal (Baig et al. 2009), the gag initiation codon, the C-box region (194-GGUGG-198), and the G-box region (541-CCACC-545), respectively.
To create a vector for generating the C-box, co-randomized, and forced evolution libraries, a derivative of pSCR2 plasmid called pSCR2AfeIΔ(173–2030) was constructed, in which a fragment encompassing nucleotides 173 to 2030 of the viral genome, including part of the noncoding region, and most of the Gag-coding region up to the XhoI site was deleted and substituted with eight nucleotides to introduce a unique AfeI restriction site and a spacer region. All nucleotide numbering in the present study is referenced to the RNA sequence of HIV-2 (ROD isolate, GenBank no. M15390). The cloning region in the pSCR2AfeIΔ(173–2030) vector is as follows: 5′-160GCCAGTTAGAAGCgctaagtc180//2031CTCGAG-3′, where AfeI and XhoI sites are underlined and the lowercase letters indicate the changed nucleotides. pSCR2AfeIΔ(173–2030) was constructed as follows: A fragment containing the long terminal repeat (LTR) sequence up to nucleotide 172, followed by a 14-nt sequence containing the remaining half of the AfeI site, the spacer region, and the XhoI site, was amplified using a sense primer (M13 forward [−41] binding upstream of a unique AatII site) and an antisense primer (asXhoCbox, containing the XhoI site, spacer region, and AfeI site). The amplified product was digested with AatII and XhoI and ligated into the pSCR2 plasmid vector missing the original AatII–XhoI fragment. The ligated product was transformed in Escherichia coli DH5α cells, and ampicillin-resistant colonies were selected, followed by plasmid purification and sequencing.
To create a vector for generating the G-box randomized library, a derivative of pAUG1 plasmid called pAUG1AfeIΔ was constructed, in which a fragment encompassing nucleotides 498 to 2030 was deleted and substituted with eight nucleotides to introduce a unique AfeI restriction site and a spacer region. The cloning region of the pAUG1AfeIΔ(498–2030) vector is as follows: 5′-481ACACCAAAAACTGTAGCgctaagtc505//2031CTCGAG-3′, where AfeI and XhoI sites are underlined and the lowercase letters indicate the changed nucleotides. pAUG1AfeIΔ(498–2030) was constructed as follows: A fragment containing the LTR sequence up to nucleotide 498, followed by a 14-nt sequence containing the remaining half of the AfeI site, the spacer region, and the XhoI site, was amplified using a sense primer (M13 forward [−41] binding upstream of a unique AatII site) and an antisense primer (asXhoGbox, containing the XhoI site, spacer region, and AfeI site). The amplified product was digested with AatII and XhoI and ligated into the pAUG1 plasmid vector missing the original AatII–XhoI fragment. The ligated product was transformed in E. coli DH5α cells, and ampicillin-resistant colonies were selected, followed by plasmid purification and sequencing.
Generation of the C-box, G-box, co-randomized, and forced evolution proviral DNA libraries
To generate the proviral DNA libraries, we used a single-step ligation-independent DNA cloning protocol (In-Fusion; Clontech) as described previously (Baig et al. 2009). Both the pSCR2AfeIΔ and pAUG1AfeIΔ vectors were prepared by an AfeI and XhoI digestion, followed by gel electrophoresis and gel extraction (5PRIME).
To construct the C-box randomized insert, a PCR product (nucleotides 157 to 2050) was generated using a mutagenic sense primer (sCboxrandom) with degenerate nucleotides at positions 194 to 199, an antisense primer (asROD2050), and the pCboxG plasmid as a template. To construct the G-box randomized insert, a PCR product (nucleotides 483 to 2050) was generated using a mutagenic sense primer (sGboxrandom) with degenerate nucleotides at positions 540 to 545, an antisense primer (asROD2050), and the pSCR2 plasmid as a template. To construct the co-randomized insert, overlap extension of two PCR products was performed, followed by a PCR using a sense primer (sROD157) and an antisense primer (asROD2050). The first PCR product (157–571) was generated using a mutagenic sense primer (sCboxrandom) with degenerate nucleotides at positions 194 to 199, a mutagenic antisense primer (asGboxrandom) with degenerate nucleotides at positions 540 to 545, and the pCboxg plasmid as a template. The second PCR product (551–2050) was generated using a sense primer (sROD551), an antisense primer (asROD2050), and the pCboxg plasmid as a template. To construct the forced evolution randomized insert, a PCR product (nucleotides 157–2050) was generated using a mutagenic sense primer (sCboxrandom) with degenerate nucleotides at positions 194 to 199, an antisense primer (asROD2050), and the pGboxc plasmid as a template. All PCR products (randomized inserts) were purified by agarose gel electrophoresis.
The C-box randomized, co-randomized, or forced evolution randomized insert was combined with the digested pSCR2AfeIΔ(173–2030) vector at a 2:1 ratio in an infusion reaction. The G-box randomized insert was cloned into the digested pAUG1AfeIΔ vector at a 2:1 ratio using the Infusion Clontech kit. Each of the resulting recombined products was transformed into E. coli DH5α cells, and DNA was extracted from bulk transformation reactions to obtain each of the randomized proviral DNA libraries. Degeneracy of the proviral DNA libraries was checked by sequencing.
Construction of individual plasmids with C-box and G-box mutations
To construct full-length plasmids with mutated and nonmutated C-box and G-box regions, we used the In-Fusion strategy described previously with the pSCR2AfeIΔ(173–2030) vector but with inserts that contained the desired C-box and G-box sequences. Each insert was produced using overlap extension with two PCR products as previously described. All overlap extensions contained a PCR product (551–2050) generated using a sense primer (sROD551), an antisense primer (asROD2050), and the pCboxg plasmid as a template. The C-box119 PCR product was generated with a sense primer (sCboxCLS119), an antisense primer (asGboxwt), and the pCboxg plasmid as a template. The G-box119 PCR product was generated with a sense primer (sCboxwt), an antisense primer (asGboxCLS119), and the pGboxc plasmid as a template. The CLS-119 PCR product was generated with a sense primer (sCboxCLS119), an antisense primer (asGboxCLS119), and the pGboxc plasmid as a template. The CLS-120, CLS-121, and CLS-005 proviral DNAs were constructed as described for CLS-119, using their designated primers.
Cell culture and transfection
COS-7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum, glutamine, penicillin, and streptomycin (Invitrogen). COS-7 cells were transiently transfected using the Trans-IT-COS transfection kit (Mirus). Cells and media were harvested 24–48 h post-transfection. The amount of HIV-2 particles in the media was determined by measurement of the viral capsid (CAp27) protein levels (Retro-Tek SIV p27 Antigen ELISA kit; Zeptometrix).
Cell culture and infection
C8166 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% fetal calf serum, glutamine, penicillin, and streptomycin (Invitrogen). C8166 cells (2 × 104 cells) were infected with media from COS-7 transfected cells (10 ng of HIV-2 capsid as determined by ELISA). The C8166 cells were washed twice 12 h post-infection, followed by resuspension in RPMI/fetal bovine serum. A media aliquot (day 0) was taken to serve as a reference. A daily aliquot of media was harvested and spun to remove cells, and then the supernatant was frozen at −80°C. Viral replication was followed by quantification of the CAp27 protein (Retro-Tek SIV p27 Antigen ELISA kit; Zeptometrix) or reverse transcriptase (Amersham Biosciences Quan-T-RT assay system) levels in the supernatant sample.
Protein isolation and analysis
Intracellular COS-7 RNA and proteins were harvested 48 h after transfection by washing and scraping cells in phosphate-buffered saline. To prevent Gag cleavages, 10 μM (final concentration) saquinavir (an HIV protease inhibitor) was added to the media of the transfected COS-7 cells 24 h before harvesting. One-half of the cells were harvested in RNA lysis buffer for genomic RNA quantitation (see below). The remaining half of the cells were lysed in radioimmunoprecipitation assay buffer (Santa Cruz Biotechnology Inc.; Tris-HCl, 50 mM at pH 7.4, NaCl 150 mM, SDS 0.1%, Nonidet P-40 0.1%, 0.5% sodium deoxycholate with protease inhibitors). Following lysis on ice, samples were fractionated on SDS–8% polyacrylamide gels. Electrotransfer onto polyvinylidene difluoride (PVDF) membranes was then performed. To visualize Gag proteins, the Western blot was probed using the primary antibody biotin-labeled anti-capsid antibody (p27 ELISA kit from Zeptometrix) followed by incubation with streptavidin–horseradish peroxidase (p27 ELISA kit from Zeptometrix). Detection was performed by electrochemiluminescence (ECL Plus Western Blotting Detection Reagents from GE Healthcare) using a Fujifilm LAS-3000.
Supernatants from transfected cells were filtrated through 0.45-μm filters, and virions were collected by a 1-h centrifugation at 21,000g and 4°C. Pelleted virions were resuspended in radioimmunoprecipitation assay buffer, and their Gag proteins were visualized as described above.
RNA isolation and analysis
Total cellular RNA and extracellular viral RNA fractions from transfected COS-7 cultures were purified with the Absolutely RNA mini-prep kit (Stratagene). Virions were pelleted by centrifuging an aliquot of the media for 1 h at 4°C and 21,000g. Purified RNAs from the cells and viruses were used in RNase protection assays (RPA III kit; Ambion). The RNase protection assay was performed using an antisense RNA probe complementary to the 401–562 region of HIV-2 ROD isolate. The antisense region was cloned into the pGEM7Zf(+) vector (Novagen) allowing for 41 nt of vector origin at the 5′ end of the T7 transcript. This non-HIV-2 tail was used to confirm RNase's digestion efficiency during the RPA experiment. Three protected bands are expected: one 162-nt band corresponding to the 401–562 region (wild-type G-box), one 139-nt band corresponding to the 401–539 region (mutant G-box), and one 17-nt band corresponding to the 546–562 region (mutant G-box).
Luciferase template construction for in vitro transcription
Luciferase chimeric constructs were built containing the first 920 nt of HIV-2 fused to a Renilla luciferase reporter gene. The HIV-2 inserts (1–920 nt) were generated using PCR. A sense primer (sNHErod) containing an NheI site and an antisense primer (asBsoBI920) containing a BsoBI site were used to amplify the first 1–920 nt of the C-box119, G-box119, and CLS-119 constructs as well as wild-type HIV-2 genomic RNA, ROD isolate (GenBank M15390; genomic RNA sequence starts at 1). All constructs were checked by DNA sequencing.
In vitro transcription
The HIV-2-luciferase plasmids were linearized via ClaI digestion. These linearized DNA templates were then used in transcription reactions to synthesize RNA (mScript mRNA Production System; Epicentre). Following completion of the incubation, the reactions were treated with RNase-free DNase, followed by ammonium acetate precipitation. The RNAs were then 5′-capped and polyadenylated (mScript mRNA Production System; Epicentre), followed by ammonium acetate precipitation.
In vitro translation and protein analysis
Comparison translation reactions were performed using equimolar amounts (46 nmol) of the tested RNAs. The RNAs were initially denatured in water (final volume of 25.2 μL) at 65°C, then snap-cooled for 3 min on ice. The translation mix (70 μL of rabbit reticulocyte lystate, 1 μL of amino acid mixture minus leucine, 1 μL of [35S]methionine [Perkin Elmer], and 2.8 μL of 2.5 M KCl) (Flexi Rabbit Reticulocyte Lysate System; Promega) was added to each mRNA sample to initiate the reaction. The translation reactions were then incubated at 30°C. Five minutes into the 45-min incubation, each reaction was spiked with 1 mM nonradioactive methionine. The samples were again placed at 30°C. Upon completion of the incubation, reactions were placed on ice. A 10-μL aliquot of each reaction was combined with 20 μL of 4 M urea protein loading dye. The samples were denatured for 2 min at 90°C and run on an SDS-polyacrylamide gel. The radioactive protein isoforms were visualized using a Fuji FLA-3000.
Prediction of RNA secondary structures
Mfold version 3 (Zuker 2003) was used to predict the secondary structures for the wild-type HIV-2 ROD and the 34 individual clonal RNA sequences isolated from the co-randomized viral library selected at days 40 and 56. The analyzed RNA fragments represent the first 580 nt of the genomic RNA sequences. The software used is found on the mfold server (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form) (Zuker 2003). The 10 most stable secondary structures for each RNA were visually analyzed, and the base-pairing partners of the C-box and G-box, presence or absence of the CGI, and type of CGI were recorded.
ACKNOWLEDGMENTS
We thank Michalee Moen for critical reading of the manuscript. This work was funded by the National Institutes of Health grant AI45388 to J.S.L. and a Grant-In-Aid of Research from Sigma Xi, the Scientific Research Society to C.L.S. The plasmid pROD10 was provided by the EU Programme EVA/MRC Centralised Facility for AIDS Reagents, NIBSC, UK (Grant Number QLK2-CT-1999-00609 and GP828102). The following reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAD, NIH: C8166-145 (Cat # 404) from Dr. Robert Gallo.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2564311.
REFERENCES
- Abbink TE, Berkhout B 2003. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J Biol Chem 278: 11601–11611 [DOI] [PubMed] [Google Scholar]
- Abbink TE, Ooms M, Haasnoot PC, Berkhout B 2005. The HIV-1 leader RNA conformational switch regulates RNA dimerization but does not regulate mRNA translation. Biochemistry 44: 9058–9066 [DOI] [PubMed] [Google Scholar]
- Anderson JL, Johnson AT, Howard JL, Purcell DF 2007. Both linear and discontinuous ribosome scanning are used for translation initiation from bicistronic human immunodeficiency virus type 1 env mRNAs. J Virol 81: 4664–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baig TT, Strong CL, Lodmell JS, Lanchy JM 2008. Regulation of primate lentiviral RNA dimerization by structural entrapment. Retrovirology 5: 65 doi: 10.1186/1742-4690-5-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baig TT, Lanchy JM, Lodmell JS 2009. Randomization and in vivo selection reveal a GGRG motif essential for packaging human immunodeficiency virus type 2 RNA. J Virol 83: 802–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B 1996. Structure and function of the human immunodeficiency virus leader RNA. Prog Nucleic Acid Res Mol Biol 54: 1–34 [DOI] [PubMed] [Google Scholar]
- Berkhout B, Klaver B 1993. In vivo selection of randomly mutated retroviral genomes. Nucleic Acids Res 21: 5020–5024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B, Arts K, Abbink TE 2011. Ribosomal scanning on the 5′-untranslated region of the human immunodeficiency virus RNA genome. Nucleic Acids Res doi: 10.1093/nar/gkr113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee P, Garzino-Demo A, Swinney P, Arya SK 1993. Human immunodeficiency virus type 2 multiply spliced transcripts. AIDS Res Hum Retroviruses 9: 331–335 [DOI] [PubMed] [Google Scholar]
- Damgaard CK, Andersen ES, Knudsen B, Gorodkin J, Kjems J 2004. RNA interactions in the 5′ region of the HIV-1 genome. J Mol Biol 336: 369–379 [DOI] [PubMed] [Google Scholar]
- Dirac AM, Huthoff H, Kjems J, Berkhout B 2002. Regulated HIV-2 RNA dimerization by means of alternative RNA conformations. Nucleic Acids Res 30: 2647–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza V, Summers MF 2004. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature 431: 586–590 [DOI] [PubMed] [Google Scholar]
- D'Souza V, Summers MF 2005. How retroviruses select their genomes. Nat Rev Microbiol 3: 643–655 [DOI] [PubMed] [Google Scholar]
- Gottlinger HG, Sodroski JG, Haseltine WA 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci 86: 5781–5785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbreteau CH, Weill L, Decimo D, Prevot D, Darlix JL, Sargueil B, Ohlmann T 2005. HIV-2 genomic RNA contains a novel type of IRES located downstream of its initiation codon. Nat Struct Mol Biol 12: 1001–1007 [DOI] [PubMed] [Google Scholar]
- Kozak M 1986. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44: 283–292 [DOI] [PubMed] [Google Scholar]
- Kozak M 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15: 8125–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krummheuer J, Johnson AT, Hauber I, Kammler S, Anderson JL, Hauber J, Purcell DF, Schaal H 2007. A minimal uORF within the HIV-1 vpu leader allows efficient translation initiation at the downstream env AUG. Virology 363: 261–271 [DOI] [PubMed] [Google Scholar]
- Kuiken C, Foley B, Leitner T, Apetrei C, Hahn B, Mizrachi I, Mullins J, Rambaut A, Wolinsky S, Korber B 2010. HIV sequence compendium 2010. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM [Google Scholar]
- Lanchy JM, Ivanovitch JD, Lodmell JS 2003a. A structural linkage between the dimerization and encapsidation signals in HIV-2 leader RNA. RNA 9: 1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanchy JM, Rentz CA, Ivanovitch JD, Lodmell JS 2003b. Elements located upstream and downstream of the major splice donor site influence the ability of HIV-2 leader RNA to dimerize in vitro. Biochemistry 42: 2634–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanchy JM, Szafran QN, Lodmell JS 2004. Splicing affects presentation of RNA dimerization signals in HIV-2 in vitro. Nucleic Acids Res 32: 4585–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein A, Henderson LE, Levin JG 1998. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem Sci 23: 297–301 [DOI] [PubMed] [Google Scholar]
- Ricci EP, Herbreteau CH, Decimo D, Schaupp A, Datta SA, Rein A, Darlix JL, Ohlmann T 2008. In vitro expression of the HIV-2 genomic RNA is controlled by three distinct internal ribosome entry segments that are regulated by the HIV protease and the Gag polyprotein. RNA 14: 1443–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Felber BK, Pavlakis GN 1992. Mechanism of translation of monocistronic and multicistronic human immunodeficiency virus type 1 mRNAs. Mol Cell Biol 12: 207–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R, Kafaie J, Laughrea M 2008. Role of the 5′ TAR stem–loop and the U5-AUG duplex in dimerization of HIV-1 genomic RNA. Biochemistry 47: 3283–3293 [DOI] [PubMed] [Google Scholar]
- Spriggs S, Garyu L, Connor R, Summers MF 2008. Potential intra- and intermolecular interactions involving the unique-5′ region of the HIV-1 5′-UTR. Biochemistry 47: 13064–13073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong CL, Lanchy JM, Dieng-Sarr A, Kanki PJ, Lodmell JS 2009. A 5′UTR-spliced mRNA isoform is specialized for enhanced HIV-2 gag translation. J Mol Biol 391: 426–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viglianti GA, Sharma PL, Mullins JI 1990. Simian immunodeficiency virus displays complex patterns of RNA splicing. J Virol 64: 4207–4216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viglianti GA, Rubinstein EP, Graves KL 1992. Role of TAR RNA splicing in translational regulation of simian immunodeficiency virus from rhesus macaques. J Virol 66: 4824–4833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill L, James L, Ulryck N, Chamond N, Herbreteau CH, Ohlmann T, Sargueil B 2010. A new type of IRES within gag coding region recruits three initiation complexes on HIV-2 genomic RNA. Nucleic Acids Res 38: 1367–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM 2008. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol 6: e96 doi: 10.1371/journal.pbio.0060096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]