Abstract
tRNA anticodon damage inflicted by the Kluyveromyces lactis γ-toxin underlies an RNA-based innate immune system that distinguishes self from nonself species. γ-toxin arrests the growth of Saccharomyces cerevisiae by incising a single phosphodiester 3′ of the wobble base of tRNAGlu(UUC) to generate a break with 2′,3′-cyclic phosphate and 5′-OH ends. Recombinant γ-toxin cleaves oligonucleotide substrates in vitro that mimic the anticodon stem–loop of tRNAGlu. A single 2′-deoxy sugar substitution at the wobble nucleoside abolishes anticodon nuclease activity. To gain further insights to γ-toxin's substrate specificity, we tested deoxynucleoside effects at positions other than the site of transesterification. The results attest to a stringent requirement for a ribonucleoside at the uridine 5′ of the wobble base. In contrast, every other nonwobble ribonucleoside in the anticodon loop can be replaced by a deoxy without significantly affecting γ-toxin's cleavage activity. Whereas either the 5′ half or the 3′ half of the anticodon stem can be replaced en bloc with DNA without a major effect, simultaneously replacing both strands with DNA interfered strongly, signifying that γ-toxin requires an A-form helical conformation of the anticodon stem. We purified γ-toxin mutants identified previously as nontoxic in vivo and gauged their anticodon nuclease activities in vitro. The results highlight Glu9 and Arg151 as candidate catalytic residues, along with His209 implicated previously. By analogy to other endoribonucleases, we speculate that γ-toxin drives transesterification by general acid-base catalysis (via His209 and Glu9) and transition-state stabilization (via Arg151).
Keywords: ribotoxin, tRNA anticodon stem–loop, transesterification
INTRODUCTION
Target-specific endoribonuclease toxins (“ribotoxins”) are elaborated by bacteria and fungi as a defense against nonself species. Transfer RNAs figure prominently as the specific targets of microbial ribotoxins, exemplified by the bacterial tRNA anticodon nucleases PrrC, colicin D, and colicin E5 (Ogawa et al. 1999; Tomita et al. 2000; Blanga-Kanfi et al. 2006; Meineke et al. 2011) and the fungal tRNA anticodon nucleases Kluyveromyces lactis γ-toxin and Pichia acaciae toxin PaT (Lu et al. 2005; Klassen et al. 2008). These ribotoxins incise specific tRNAs via a transesterification reaction that yields 2′,3′ cyclic phosphate and 5′-OH ends. Their target preferences are influenced by base modifications within the anticodon loop. For example, K. lactis γ-toxin cleaves tRNAGlu(UUC) at a single phosphodiester 3′ of the modified wobble base mcm5s2U (5-methoxycarbonylmethyl-2-thiouridine) of the UUC anticodon. γ-toxin can cleave two other yeast tRNAs (Gln and Lys) that have an mcm5s2U wobble base, albeit less efficiently than it incises tRNAGlu, which is γ-toxin's principal target in vivo (Butler et al. 1994; Lu et al. 2005). K. lactis γ-toxin has no apparent primary structure similarity to other known tRNA ribotoxins or to any known ribonucleases or phosphotransferases. Indeed, K. lactis γ-toxin has no credible similarity to any protein in public databases. These features make γ-toxin an attractive model system to elucidate novel principles of tRNA recognition, wobble base discrimination, and transesterification chemistry.
The Byström laboratory has pioneered biochemical studies of the γ-toxin nuclease activity. The investigators found that cleavage of native mcm5s2U-modified tRNAGlu in vitro was about 100-fold more efficient than incision of hypo-modified tRNAGlu (Lu et al. 2005). The wobble uridine modification requirement could be evaded by including high concentrations of trimethylamine oxide (TMAO) in the reaction mixtures (Lu et al. 2008). In effect, TMAO conferred a gain of function, enabling γ-toxin to efficiently cleave either (1) unmodified tRNAGlu prepared by transcription in vitro or (2) a minimized synthetic 17-mer RNA oligonucleotide substrate corresponding to the anticodon stem–loop of tRNAGlu (illustrated in Fig. 1; Lu et al. 2008). The unmodified 17-mer RNA (R17), when 5′ 32P-labeled, is incised by γ-toxin at the correct phosphodiester 3′ of the first uridine of the UUC anticodon to generate a single 32P-labeled 8-mer product (Lu et al. 2008; Keppetipola et al. 2009). The minimized R17 substrate has been extremely useful in dissecting the influence of nucleobase sequence on substrate utilization. For example, Lu et al. (2008) reported that cleavage by γ-toxin is stringently dependent on the anticodon loop sequence U34U35C36A37C38 derived from wild-type tRNAGlu. Indeed, any single base change within the U34U35C36A37 element abolished tRNase activity. Nucleobase changes that disrupted or destabilized the anticodon stem duplex also abolished activity, although changes in stem primary structure that preserved the duplex RNA secondary structure did not (Lu et al. 2008).
FIGURE 1.
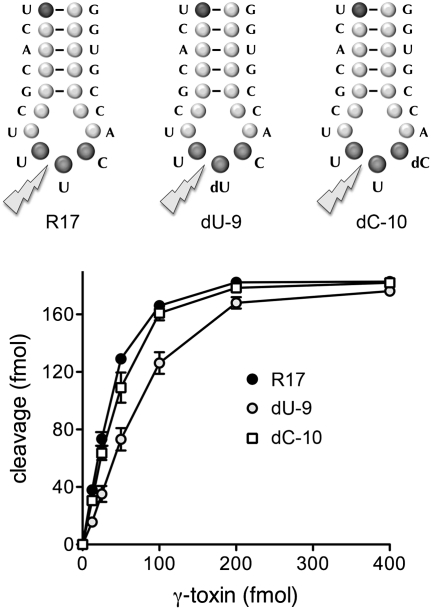
Effect of nonwobble anticodon deoxynucleosides on γ-toxin activity in vitro. The 17-mer oligonucleotide substrates corresponding to the anticodon stem–loop of tRNAGlu are shown. The R17 substrate is all-RNA. The dU-9 and dC-10 substrates contain single deoxynucleosides in the UUC anticodon triplet, as indicated. Nuclease reaction mixtures (10 μL) contained 200 fmol 5′ 32P-labeled 17-mer substrates and γ-toxin as specified. The products were analyzed by denaturing PAGE. Reaction of γ-toxin with the R17, dU9, and dC-10 substrate yielded single 32P-labeled 8-mer cleavage products of identical electrophoretic mobility (data not shown). The extents of 17-mer cleavage are plotted as a function of input γ-toxin.
Our interest has been to (1) delineate structure–activity relations for γ-toxin that might illuminate its active site and catalytic mechanism and (2) further define the determinants of substrate recognition. With respect to the former aim, we conducted an extensive mutational analysis of K. lactis γ-toxin, focusing on the functional groups likely to be involved in catalysis or RNA recognition (e.g., histidine, arginine, lysine, glutamate). The biological activity of γ-toxin was assayed using a Saccharomyces cerevisiae strain containing a galactose-inducible γ-toxin expression cassette. We tested the effects of 62 single alanine mutations and thereby identified 22 residues essential for galactose-induced toxicity in vivo (Supplemental Fig. S1). We proceeded to determine their relevant properties by testing the effects of conservative substitutions for each of the essential amino acids (Keppetipola et al. 2009). Biochemical characterization of recombinant γ-toxin highlighted its RNase A–like chemical mechanism and a putative role of the essential His209 residue (the only histidine present in γ-toxin) as a general acid or general base in catalysis of transesterification. Incision of the 17-mer stem–loop by γ-toxin was abolished by ribose 2′-H or 2′-F modifications of the wobble uridine (Keppetipola et al. 2009), as expected for a site-specific transesterifying endonuclease that recognizes only one scissile phosphodiester in the tRNA substrate.
Here we extend our studies of γ-toxin by querying its specificity for RNA nucleosides at positions other than the site of transesterification. The results of this analysis attest to stringent requirements for a ribonucleoside at the uridine 5′ of the wobble base in the anticodon loop and for an A-form helical conformation of the anticodon stem. We also characterize biochemically a collection of γ-toxin mutants that we identified previously as inactive in vivo in arresting yeast cell growth. Survey of their anticodon nuclease activities in vitro highlights Glu9 and Arg151 as candidate catalytic residues.
RESULTS AND DISCUSSION
Effect of anticodon loop deoxynucleosides on γ-toxin cleavage activity
The UUC anticodon nucleobases and the wobble uridine ribose are critical for incision of an anticodon stem–loop substrate (Lu et al. 2008; Keppetipola et al. 2009). Here we tested whether replacing the flanking anticodon nucleoside sugars with deoxyribose had any impact on anticodon nuclease activity. The unmodified RNA substrate used in these experiments was a 5′ 32P-labeled 17-mer oligonucleotide (R17; 5′-pUCACGCUUUCACCGUGG) corresponding to the anticodon stem–loop of tRNAGlu (Fig. 1). Variant 17-mers dU-9 and dC-10 contained single deoxynucleosides in the U8U9C10 anticodon triplet (underlined above). The presence of the deoxynucleosides in the dU-9 and dU-10 substrates was verified by partial alkaline hydrolysis as follows. The 5′ 32P-labeled R17, dU-9, and dC-10 oligonucleotides were incubated in a solution of 40 mM NaHCO3, 60 mM Na2CO3 (pH 10.2) for 12 min at 95°C. The partially cleaved RNAs were then resolved by denaturing PAGE in parallel with the product of R17 cleavage by γ-toxin (the 5′ 32P-labeled 8-mer, 5′-pUCACGCUU>p). Whereas the ladder of R17 alkaline hydrolysis products was evenly distributed in 1-nucleotide (nt) electrophoretic increments, the alkaline hydrolysis ladders derived from dU-9 and dU-10 “skipped” the 2′-H sugars, leaving single gaps whereby the 9-mer and 10-mer hydrolysis products were missing from the dU-9 and dU-10 ladders, respectively (data not shown).
The R17, dU-9, and dC10 stem–loops (20 nM) were reacted with recombinant γ-toxin for 30 min. The extents of cleavage were proportional to input γ-toxin (Fig. 1). The endpoint cleavage values at the saturating enzyme were similar for the three substrates tested, i.e., 88%–91% of input stem–loop was incised (Fig. 1). We calculated specific activities from the initial slopes of the titration curves (by linear regression in Prism) and then normalized the values to R17 (defined as 100%). The specific activities of γ-toxin with dU-9 and dC-10 stem–loops were 54% and 85% of the R17 control, respectively.
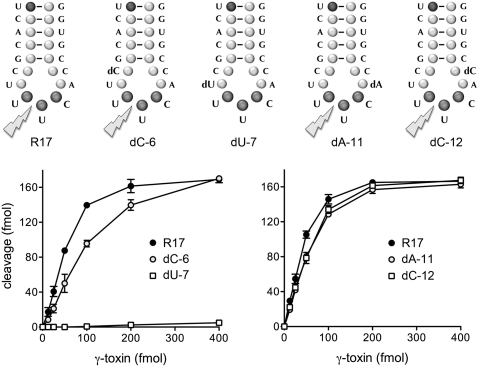
We extended the deoxyribose replacement experiment to the other 4 nt of the 7-mer anticodon loop, C6U7UUCA11C12, numbered according to their position in the 17-mer stem–loop (Fig. 2). Focusing first on the 5′ side of the loop, we found that the dU-7 substrate was virtually uncleavable by γ-toxin at concentrations saturating for cleavage of the R17 control and the modified dC-6 substrate (Fig. 2, left panel). The specific activities of the dU-7 and dC-6 stem–loops were 0.7% and 56% of the R17 control, respectively. Shifting to the 3′ side of the loop, we found that the dA-11 and dC-12 stem–loops were cleaved quite well by γ-toxin (Fig. 2, right panel). The specific activities of the dA-11 and dC-12 stem–loops were both 76% of the R17 control. We conclude that γ-toxin stringently requires a ribonucleoside at the uridine 5′ of the wobble base of the UUC anticodon but is relatively indifferent to deoxy sugar substitutions elsewhere in the anticodon loop (exclusive of the scissile wobble nucleotide).
FIGURE 2.
γ-toxin cleavage requires a ribose 5′ of the scissile wobble nucleotide. The 17-mer oligonucleotide substrates are shown. The dC-6, dU-7, dA-11, and dC-12 substrates contain single deoxynucleosides in the anticodon loop, as indicated. Nuclease reaction mixtures (10 μL) contained 200 fmol 5′ 32P-labeled 17-mer substrates and γ-toxin as specified. The extents of 17-mer cleavage are plotted as a function of input γ-toxin.
Effect of installing DNA in the stem segments of the anticodon stem–loop
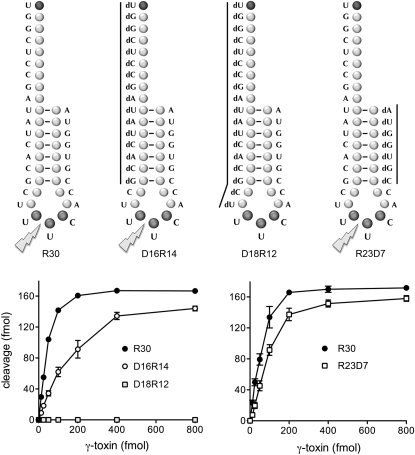
We tested the impact of replacing the proximal or distal RNA stem segments en bloc with DNA segments, without altering the nucleobase sequence. In these experiments, the control RNA substrate was a 5′ 32P-labeled 30-mer oligonucleotide (R30) consisting of the anticodon stem–loop of tRNAGlu(UUC) plus a 9-nt 5′ single-strand leader (Fig. 3). The R30 substrate was incised by γ-toxin to yield a single radiolabeled 19-mer product as a consequence of transesterification 3′ of the wobble uridine (Tanaka and Shuman 2011; data not shown). The extent of R30 cleavage was proportional to input γ-toxin (Fig. 3). The specific activity of R30 as a substrate and the reaction endpoint (in the experiment in Fig. 3, left panel) were virtually identical to that observed for the R17 substrate (in Fig. 2, right panel), signifying that the 5′ single-stranded RNA leader sequence did not adversely affect γ-toxin activity.
FIGURE 3.
Effect of 5′ or 3′ stem deoxynucleosides on γ-toxin activity. The 30-mer oligonucleotide substrates are shown. The R30 substrate is all-RNA. D16R14 is a chimera in which the proximal (5′) 16 nucleotides (nt) are deoxys and the distal (3′) 14 nt are ribos. D18R12 is composed of a proximal 18-nt DNA segment and a distal 12-nt RNA segment. R23D7 is composed of a proximal 23-nt RNA segment and a distal 7-nt DNA segment. Nuclease reaction mixtures contained 200 fmol 5′ 32P-labeled 30-mer substrates, and γ-toxin as specified. The extents of 30-mer cleavage are plotted as a function of input γ-toxin.
We initially replaced the leader and the 5′ stem segment with a DNA segment, thereby yielding a chimeric substrate, D16R14, composed of 16 deoxynucleotides linked to 14 ribonucleotides (Fig. 3). The D16R14 substrate was cleaved by γ-toxin with 33% the specific activity of the R30 control (Fig. 3, left panel). Extending the 5′-DNA segment 2 nt into the anticodon loop, in the case of the chimeric substrate D18R12 (Fig. 3), rendered the stem–loop inert for cleavage by γ-toxin at an enzyme concentration in fourfold excess of the level that sufficed for saturating incision of the R30 control substrate (Fig. 3, left panel). This result confirms the essentiality of the ribonucleoside at the uridine proximal to the wobble base that was demonstrated above in Figure 2.
We proceeded to replace the 3′ RNA stem segment with DNA, yielding the R23D7 chimera composed of 23 ribonucleotides linked to seven deoxynucleotides (Fig. 3). The specific activity of γ-toxin with the R23D7 substrate was 54% of the R30 control (Fig. 3, right panel). These results attest to the lack of a stringent requirement by γ-toxin for RNA in either the proximal or distal segments that anneal to form the stem of the anticodon stem–loop.
γ-toxin senses an A-form helical conformation of the anticodon stem
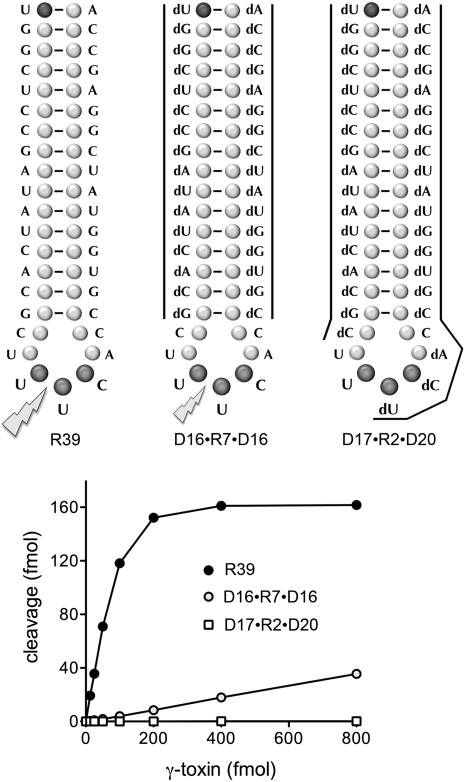
The ability of γ-toxin to incise model tRNAGlu(UUC) substrates wherein the stems of the stem–loop are RNA:DNA or DNA:RNA hybrids raised the question of whether the entire stem can be replaced by DNA. To address this issue, we tested a set of 5′ 32P-labeled 39-mer substrates modeled on the R39 stem–loop illustrated in Figure 4. R39 is composed of a 16-bp stem and a 7-nt loop. Like R30 (from which it derives by “filling-in” the nucleotides complementary to the leader sequence), the R39 substrate was incised by γ-toxin 3′ of the wobble uridine to yield a single radiolabeled 19-mer product (Supplemental Fig. S2; Tanaka and Shuman 2011). The extent of R39 cleavage was proportional to input γ-toxin (Fig. 4). (By way of comparison, the specific activity of γ-toxin with the R39 substrate in the experiment in Fig. 4 was 85% of the R30 specific activity seen in the experiment in Fig. 3, right panel.) Thus, the longer stem is not an impediment to γ-toxin activity. We intentionally incorporated the longer stem so that, when simultaneously replacing both stem segments with DNA, as in the D16•R7•D16 chimera shown in Figure 4, we would maintain a stable duplex structure of the 16-bp all-DNA stem. (We thought it potentially problematic to replace the 7-bp RNA stem structure in the “minimized” R17 substrate with a less stable DNA-DNA hybrid.)
FIGURE 4.
Effect of an all-DNA stem on γ-toxin activity. The 39-mer oligonucleotide substrates are shown. The R39 substrate is all-RNA. D16•R7•D16 is a chimera in which the 16-bp stem is all-DNA and the anticodon loop is all-RNA. In D17•R2•D20 only the wobble U and the 5′ flanking U are ribonucleosides. Nuclease reaction mixtures contained 200 fmol 5′ 32P-labeled 30-mer substrates and γ-toxin as specified. The extents of 39-mer cleavage are plotted as a function of input γ-toxin.
The D16•R7•D16 substrate contains DNA in both stem segments but maintains an all-RNA anticodon loop (Fig. 4). Partial alkaline hydrolysis confirmed that D16•R7•D16 had seven sequential alkali-labile ribonucleoside phosphodiester linkages flanked by alkali-resistant deoxynucleotides (Supplemental Fig. S2). D16•R7•D16 was a very feeble substrate for γ-toxin; its specific activity was just 3% of the R39 control (Fig. 4). This result is highly instructive in light of the relatively benign effects of installing DNA in just one of the strands of the duplex stem (Fig. 3). A likely explanation is that γ-toxin requires that the stem duplex adopt an A-form helical conformation, which is generally the case for RNA:RNA duplexes and RNA:DNA hybrids. In contrast, DNA:DNA duplexes adopt a B-form helical conformation, which we surmise is inimical to stem–loop substrate recognition and incision.
The D17•R2•D20 substrate is all-DNA except for the wobble nucleotide and the 5′-flanking uridine (Fig. 4), which we have shown are the only two individual positions at which a ribose sugar is strictly essential. Partial alkaline hydrolysis verified that the D17•R2•D20 substrate contained only two sequential alkali-labile ribonucleoside phosphodiester linkages flanked by alkaline-resistant deoxynucleotides (Supplemental Fig. S2). We detected no cleavage of the D17•R2•D20 substrate by γ-toxin at an enzyme concentration in fourfold excess of the level that sufficed for saturating incision of the R39 control substrate (Fig. 4).
Effect of γ-toxin glutamate mutations on anticodon nuclease activity in vitro
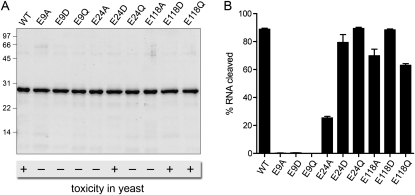
We previously conducted an extensive survey of mutational effects on γ-toxin's ability to arrest growth of S. cerevisiae, which identified three essential glutamates (Glu9, Glu24, Glu118) out of 21 glutamates subjected to alanine scanning. Structure–activity relations at Glu9, Glu24, and Glu118 were clarified by conservative substitutions with aspartate and glutamine. The mutational effects on cytotoxicity are summarized in Figure 5A. To gain further insights to which, if any, of the essential glutamate residues might be implicated in catalysis of RNA transesterification, we produced the E9A, E9D, E9Q, E24A, E24D, E24Q, E118A, E118D, and E118Q proteins in Escherichia coli and purified them from soluble bacterial extracts, in parallel with the active “wild-type” γ-toxin (Fig. 5A). The recombinant proteins were tested in parallel for their ability to cleave the R17 stem–loop substrate (Fig. 5B), which revealed striking differences in mutational effects on transesterification. For example, among the alanine changes that eliminated toxicity in vivo, only E9A abolished RNA cleavage in vitro.
FIGURE 5.
Effects of glutamate mutations on anticodon nuclease activity in vitro. (A) Aliquots (2 μg) of the recombinant wild-type (WT) and mutant γ-toxin polypeptides as specified were analyzed by SDS-PAGE (15% acrylamide). The Coomassie blue-stained gel is shown. The positions and sizes (in kDa) of marker polypeptides are indicated on the left. Below the figure is a synopsis of the toxicity (+) or lack thereof (−) of the WT and mutant proteins when expressed in vivo in yeast (Keppetipola et al. 2009). (B) Anticodon nuclease reaction mixtures contained 200 fmol 5′ 32P-labeled R17 stem–loop and 100 fmol of the indicated γ-toxin preparations. The products were analyzed by denaturing PAGE, and the extents of RNA cleavage were quantified.
The yield of cleaved stem–loop by E118A (70%) was only slightly less than wild-type (89%), which we take to mean that the requirement for Glu118 for cytotoxicity in vivo is not at the level of catalysis. It is worth emphasizing that the in vitro cleavage reactions with stem–loop substrates are performed under conditions that elide the in vivo requirement for the mcm5s2U wobble base for efficient cleavage of tRNAGlu(UUC) (Lu et al. 2008). Thus, the in vitro cleavage assay may not be sensitive to mutations that affect the recognition of the modified wobble uridine base. Glu118 is a candidate for such a role. The conservative E118D and E118Q mutants regained toxicity in vivo, suggesting that hydrogen bonding is the key property at this position. In this light, it was not surprising that the E118D and E118Q proteins were effective anticodon nucleases in vitro (88% and 63% cleavage, respectively) (Fig. 5B).
Whereas changing Glu24 to Ala reduced, but did not ablate, cleavage activity in vitro (to 25% yield), the conservative E24D and E24Q changes restored the extents of R17 cleavage to 79% and 89%, respectively (Fig. 5B). Taking into account that the E24D mutant was cytotoxic in vivo, whereas E24Q was not (Fig. 5A), we infer that Glu24 is also not a direct catalyst of transesterification. The requirement for a carboxylate at this position for cytotoxicity, but not incision of a synthetic stem–loop substrate, raises the prospect that Glu24 aids in recognition of the mcm5 modification of the wobble uridine.
The E9A, E9D, and E9Q mutants were uniformly defective in cleaving the R17 substrate, in agreement with their lack of toxicity in yeast. These results implicate Glu9 either as a catalyst of transesterification (e.g., as a general acid or general base) or as essential for proper γ-toxin folding (e.g., as a component of a salt-bridge). We are attracted to the idea that Glu9 and His209, both of which are strictly essential for toxicity in vivo and stem–loop cleavage in vitro, are the acid-base catalysts in the γ-toxin transesterification reaction. There is precedent for such an arrangement in transesterifying endoribonucleases RNase T1 and barnase. The active site of RNase T1 is composed of two histidines, one arginine, and one glutamate that contact the scissile phosphate and are critical for activity (Steyaert 1997). The prevailing model for the RNase T1 reaction posits that one of the histidines acts as a general acid to expel the leaving group. The role of general base is imputed to the glutamate, assisted by the second histidine residue (Steyaert 1997; Loverix and Steyaert 2001). (Note that there is only one histidine side-chain in the entire γ-toxin polypeptide.) Barnase relies on a single histidine general acid; a single glutamate is proposed to act as the general base (Buckle and Fersht 1994).
Effect of γ-toxin arginine mutations on anticodon nuclease activity in vitro
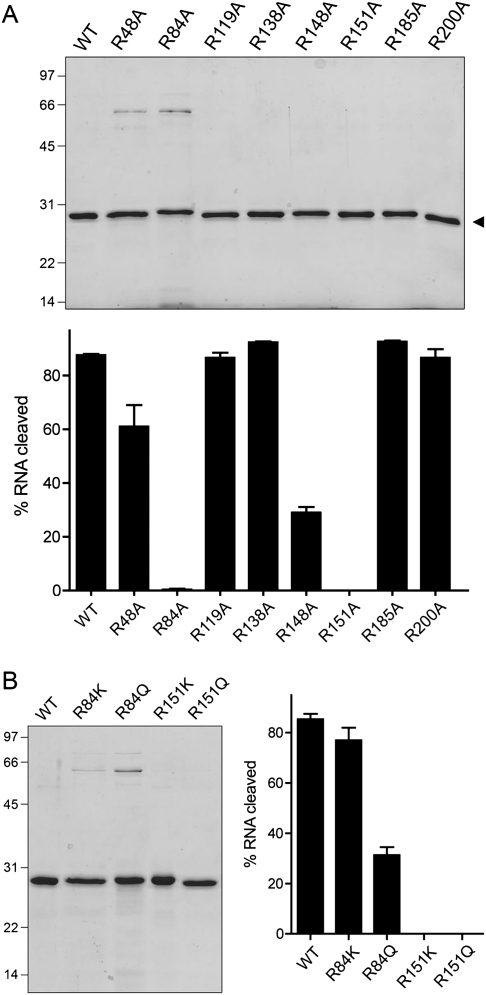
We previously identified seven arginines as essential for cytotoxicity among the 13 γ-toxin arginines subjected to alanine scanning (Keppetipola et al. 2009). Here we produced and purified the nontoxic R48A, R84A, R119A, R148A, R151A, R185A, and R200A proteins, in parallel with wild-type γ-toxin and one of arginine mutants (R138A) that retained cytotoxicity in yeast (Fig. 6A, top panel). The recombinant proteins were tested in parallel for their ability to cleave the R17 stem–loop substrate (Fig. 6A, bottom panel). The toxic wild-type and R138A proteins incised the RNA to extents of 88% and 92%, respectively. The in vitro cleavage activities of five of the nontoxic mutants—R119A (87% yield), R185A (93%), R200A (87%), R48A (61%), and R148A (29%)—militated against direct catalytic roles for Arg119, Arg185, Arg200, Arg48, or Arg148. In contrast, the R84A and R151A mutations ablated stem–loop cleavage in vitro (Fig. 6A, bottom panel). Structure–activity relations at Arg84 and Arg151 were determined previously via conservative substitutions with lysine and glutamine. Mutants R84A, R84Q, R151A, and R151Q were nontoxic in yeast (Keppetipola et al. 2009). We proceeded to purify the four conservative mutants and test the recombinant proteins for anticodon nuclease activity in vitro (Fig. 6B). Replacing the essential Arg84 with lysine and glutamine restored nuclease activity (to yields of 77% and 33%, respectively, vs. 85% for wild-type γ-toxin). This lack of concordance between positive nuclease activity and absence of toxicity in the conservative mutants weighs against a direct and singular role for Arg84 in transesterification. In contrast, the nontoxic R151K and R151Q mutants were inactive as anticodon nucleases (Fig. 6B). The inability of lysine to replace Arg151 suggests that bidentate electrostatic or hydrogen-bonding interactions are the key feature of this side-chain. Arginines classically play a role in ground-state binding and transition-state stabilization during phosphoryl transfer reactions by making bidentate contacts to the phosphate oxygens. Several of the transesterifying ribonucleases with known structures have a catalytic arginine in their active sites. The active site of colicin E5 (a tRNA ribotoxin) includes a single arginine that coordinates both nonbridging oxygens of the scissile phosphodiester (Yajima et al. 2006). The active site of RNase T1 has a single arginine that contacts the scissile phosphodiester (Zegers et al. 1998). The active site of barnase includes two arginines that contact the scissile phosphodiester (Buckle and Fersht 1994). In this light, we speculate that γ-toxin Arg151 might coordinate the scissile wobble phosphodiester (while acknowledging an alternative explanation whereby Arg151 is essential for protein folding, e.g., as part of a salt-bridge).
FIGURE 6.
Effects of arginine mutations on anticodon nuclease activity in vitro. (A) Aliquots (1 μg) of the recombinant wild-type (WT) and Arg-to-Ala mutant γ-toxin polypeptides as specified were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The positions and sizes (in kDa) of marker polypeptides are indicated on the left. Anticodon nuclease reaction mixtures contained 200 fmol 5′ 32P-labeled 17-mer stem–loop RNA and 200 fmol of the indicated γ-toxin preparations. The extents of RNA cleavage are plotted. (B) Conservative mutations of Arg84 and Arg151. Aliquots (1 μg) of the WT and mutant γ-toxin polypeptides as specified were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown in the left panel. Anticodon nuclease reaction mixtures contained 200 fmol 5′ 32P-labeled 17-mer stem–loop RNA and 500 fmol of the indicated γ-toxin preparations. The extents of RNA cleavage are shown.
Identification of tyrosines required for toxicity in vivo
We extended the structure–function analysis by introducing single-alanine mutations in lieu of the 11 tyrosine residues in γ-toxin and then surveying the mutants for cytotoxicity in yeast. We chose to target the full set of tyrosines in light of insights from the structure of tRNA splicing endonuclease, which has a tyrosine in the active site that likely serves as a general acid catalyst of RNA transesterification (Xue et al. 2006). The activity of the Tyr-to-Ala γ-toxin mutants was assayed using a S. cerevisiae strain containing a galactose-inducible γ-toxin expression cassette on a centromeric plasmid. In the presence of glucose, γ-toxin expression is repressed and yeast cells grow normally. In the presence of galactose, γ-toxin is produced and, if the toxin is active, yeast growth is arrested. The results are compiled in Supplemental Table S1. Mutants Y19A and Y128A retained toxicity in yeast, signifying that Tyr19 and Tyr128 are inessential. In contrast, mutants Y14A, Y23A, Y50A, Y55A, Y72A, Y101A, Y113A, Y152A, and Y165A were nontoxic. We proceeded to introduced phenylalanine, leucine, and serine in lieu of the eight essential tyrosines. In no instance was a Tyr-to-Leu or Tyr-to-Ser mutant active in yeast (Supplemental Table S1). The instructive findings were as follows. First, toxicity was restored when phenylalanine was installed in place of Tyr23, Tyr55, Tyr72, and Tyr152, signifying that the phenyl rings were necessary and sufficient for function at these four tyrosine positions. Thus, Tyr23, Tyr55, Tyr72, and Tyr152 are not implicated as proton donors or acceptors in catalysis or as hydrogen-bond donors in RNA recognition or protein folding. Second, toxicity was not restored when Tyr14, Tyr50, Tyr101, Tyr113, or Tyr165 were replaced by phenylalanine, signifying that the tyrosine hydroxyl groups are critical at these five positions, presumably via hydrogen-bonding interactions or, possibly, proton transfer.
Identification of aspartates required for toxicity in vivo
Here we introduced single-alanine mutations in lieu of 17 aspartate residues in γ-toxin and surveyed the mutants for galactose-induced toxicity in yeast. We envisioned that aspartates might play a role in recognizing the anticodon loop nucleobases that dictate the tRNAGlu(UUC) target specificity of γ-toxin in vivo. The results are compiled in Supplemental Table S2. Six of the mutants were fully toxic in yeast (D74A, D135A, D139A, D210A, D211A, and D221A), signifying that these six aspartate side-chains are inessential. The D190A mutant was partially active (scored as ± for toxicity in Supplemental Table S1), i.e., galactose-induced D190A expression merely slowed yeast growth (as evinced by smaller colony size compared to the vector control) rather than arresting growth and preventing colony formation (as seen with wild-type γ-toxin). Ten of the Asp-to-Ala changes resulted in loss of toxicity. We then tested the effects of conservative glutamate and asparagine changes at the 10 essential aspartates (Supplemental Table S2). The structure–activity relations fell into three categories. First, aspartate was strictly essential at positions 180, 202, and 206; i.e., neither glutamate nor asparagine restored toxicity. Apparently, the Asp180, Asp202, and Asp206 carboxylates are critical, and there are constraints on the distance from the main-chain to the carboxylates that cannot accommodate the longer glutamates. Second, replacing aspartate with glutamate restored toxicity fully (at Asp123, Asp173, and Asp175) or partially (at Asp15 and Asp76), whereas asparagine had no salutary effect. In these cases, the carboxylate is critical, and the longer glutamate can be accommodated either easily or with some loss of function. Third, introduction of either glutamate or asparagine restored toxicity fully (at Asp44) or partially (at Asp188), implying that hydrogen bonding is the relevant parameter at these positions.
CONCLUDING REMARKS
Here we conducted a systematic analysis of the RNA specificity of a eukaryal tRNA anticodon nuclease. We find that the wobble uridine and the 5′ flanking uridine are the only ribose-specific constituents of the tRNAGlu(UUC) stem–loop incised by K. lactis γ-toxin in vitro. Whereas deoxy substitutions are tolerated at other nucleosides, there is strong interference with anticodon cleavage when both strands of the anticodon stem are replaced by DNA, signifying that γ-toxin requires A-form conformation of the stem duplex. This work, together with previous studies of nucleobase specificity (Lu et al. 2008), highlights two factors that govern site-specific transesterification by γ-toxin: the nucleobase identity and ribose sugar content of the tRNAGlu U33U34U35C36A37 anticodon loop, and the secondary structure (but not the sequence or duplex length) of the anticodon stem. Our analysis of the effects of γ-toxin mutations on RNA incision in vitro hints at catalytic roles for Glu9, His209, and Arg151. New mutational studies with an in vivo readout of toxicity extend the functional map of an enzyme that has no homologs. Crystal structures of γ-toxin, alone and in complex with RNA, will be needed to definitively interpret the mutational and deoxy-interference results, assign the active site, and understand the basis for tRNA recognition.
MATERIALS AND METHODS
Recombinant γ-toxin from bacteria
The intracellular γ-toxin C13A-C177A-C231A protein was produced in E. coli as an N-terminal His10Smt3 fusion, isolated from a soluble bacterial lysate by Ni-agarose chromatography, digested with Smt3-protease Ulp1 to remove the tag, and recovered as tag-free γ-toxin by a second round of Ni-agarose chromatography as described previously (Keppetipola et al. 2009). Missense mutations were introduced into γ-toxin-(C13A-C177A-C231A) gene by two-stage overlap extension PCR. The pET28 plasmid inserts were sequenced completely to verify the intended coding sequences. The mutated γ-toxin proteins were purified in parallel with the “wild-type” C13A-C177A-C231A protein. Protein concentrations were determined by using the Biorad dye reagent with bovine serum albumin as the standard.
Anticodon nuclease assay
Reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 7.5), 2 M TMAO, 20 nM (200 fmol) 5′ 32P-labeled stem–loop RNA (or RNA-DNA chimera), and γ-toxin as specified were incubated for 30 min at 4°C. The reactions were quenched by adding 10 μL of 90% formamide, 50 mM EDTA. The samples were heated at 95°C for 1 min and then analyzed by electrophoresis through a 15-cm 24% polyacrylamide gel containing 8 M urea in 0.5× TBE (45 mM Tris-borate, 1.2 mM EDTA) at 12 W for ∼90 min. The radiolabeled products were visualized by autoradiography, and the extents of RNA cleavage were quantified by scanning the gel with a Fujix BAS2500 imager. Each datum plotted in the graphs of wild-type γ-toxin cleavage of different stem–loop substrates is an average of three independent enzyme titration experiments ±SEM. Each datum in the bar graphs comparing single-point assays of wild-type and mutant γ-toxins is an average of three independent experiments ±SEM.
Materials
RNA oligonucleotides and RNA–DNA chimaeras used as substrates in anticodon nuclease assays were purchased from Dharmacon. The RNAs were deprotected according to the vendor's instructions and 5′ 32P-labeled by reaction with T4 polynucleotide kinase and [γ32P]ATP. The labeled RNA was purified by preparative gel electrophoresis and stored at −20°C. TMAO was purchased from Sigma.
Test of γ-toxin activity in budding yeast
A haploid S. cerevisiae strain was transformed with plasmid pLF16 (CEN LEU2 UASGAL-γ-toxin), which allows for galactose-inducible, glucose-repressible expression of an intracellular 232-aa version of K. lactis γ-toxin that lacks the N-terminal 17-aa signal peptide (Supplemental Fig. S1; Jablonowski et al. 2006; Keppetipola et al. 2009). Missense mutations were introduced into the γ-toxin gene of pLF16 via two-stage overlap extension PCR. The mutated pLF16 plasmids were transformed into yeast in parallel with the wild-type pLF16. Leu+ transformants were selected on glucose-containing medium lacking leucine. Cultures were grown in selective liquid media at 30°C, adjusted to A600 of 0.1, and then diluted serially in 10-fold increments. Aliquots (3 μL) of each dilution were spotted on glucose-containing and galactose-containing agar plates, which were incubated at 30°C.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM46330. S.S. is an American Cancer Society Research Professor.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2722711.
REFERENCES
- Blanga-Kanfi S, Amitsur M, Azem A, Kaufmann G 2006. PrrC-anticodon nuclease: functional organization of a prototypal bacterial restriction RNase. Nucleic Acids Res 34: 3209–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckle AM, Fersht AR 1994. Subsite binding in an RNase: structure of a barnase-tetranucleotide complex at 1.76-Å resolution. Biochemistry 33: 1644–1653 [DOI] [PubMed] [Google Scholar]
- Butler AR, White JH, Folawiyo Y, Edlin A, Gardiner D, Stark MJR 1994. Intracellular expression of Kluyveromyces lactis toxin gamma subunit mimics treatment with exogenous toxin and distinguishes two classes of toxin-resistant mutant. Mol Cell Biol 14: 6306–63168065362 [Google Scholar]
- Jablonowski D, Zink S, Mehlgarten C, Daum G, Schaffrath R 2006. tRNAGlu wobble uridine methylation by Trm9 identifies elongator's key role for zymocin-induced cell death in yeast. Mol Microbiol 59: 677–688 [DOI] [PubMed] [Google Scholar]
- Keppetipola N, Jain R, Meineke B, Diver M, Shuman S 2009. Structure-activity relationships in Kluyveromyces lactis γ-toxin, a eukaryal tRNA anticodon nuclease. RNA 15: 1036–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen R, Paluszynski JP, Emhoff S, Pfeiffer A, Fricke J, Meinhardt F 2008. The primary target of the killer toxin from Pichia acaciae is tRNAGln. Mol Microbiol 69: 681–697 [DOI] [PubMed] [Google Scholar]
- Loverix S, Steyaert J 2001. Deciphering the mechanism of RNase T1. Methods Enzymol 341: 305–323 [DOI] [PubMed] [Google Scholar]
- Lu J, Huang B, Esberg A, Johanson M, Byström AS 2005. The Kluyveromyces lactis gamma-toxin targets tRNA anticodons. RNA 11: 1648–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Esberg A, Huang B, Byström AS 2008. The Kluyveromyces lactis γ-toxin, a ribonuclease that recognizes the anticodon stem loop of tRNA. Nucleic Acids Res 36: 1072–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meineke B, Schwer B, Schaffrath R, Shuman S 2011. Determinants of eukaryal cell killing by the bacterial ribotoxin PrrC. Nucleic Acids Res 39: 687–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Tomita K, Ueda T, Watanabe K, Uozumi T, Masaki H 1999. A cytotoxic ribonuclease targeting specific tRNA anticodons. Science 283: 2097–2100 [DOI] [PubMed] [Google Scholar]
- Steyaert J 1997. A decade of protein engineering on ribonuclease T1: atomic dissection of the enzyme-substrate interactions. Eur J Biochem 247: 1–11 [DOI] [PubMed] [Google Scholar]
- Tanaka N, Shuman S 2011. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J Biol Chem 286: 7727–7731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Ogawa T, Uozumi T, Watanabe K, Masaki H 2000. A cytotoxic ribonuclease which specifically cleaves four isoaccepting arginine tRNAs at their anticodon loops. Proc Natl Acad Sci 97: 8278–8283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Calvin K, Li H 2006. RNA recognition and cleavage by a splicing endonuclease. Science 312: 906–910 [DOI] [PubMed] [Google Scholar]
- Yajima S, Inoue S, Ogawa T, Nonaka T, Ohsawa K, Masaki H 2006. Structural basis for sequence-dependent recognition of colicin E5 tRNase by mimicking the mRNA–tRNA interaction. Nucleic Acids Res 34: 6074–6082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegers I, Loris R, Dehollander G, Haikal AF, Poortmans F, Steyaert J, Wyns L 1998. hydrolysis of a slow cyclic thiophosphate substrate of RNase T1 analyzed by time-resolved crystallography. Nat Struct Biol 5: 280–283 [DOI] [PubMed] [Google Scholar]