Abstract
The phylum Crenarchaeota includes hyperthermophilic micro-organisms subjected to dynamic thermal conditions. Previous transcriptomic studies of Sulfolobus solfataricus identified vapBC6 as a heat-shock (HS)-inducible member of the Vap toxin–antitoxin gene family. In this study, the inactivation of the vapBC6 operon by targeted gene disruption produced two recessive phenotypes related to fitness, HS sensitivity and a heat-dependent reduction in the rate of growth. In-frame vapBC6 deletion mutants were analyzed to examine the respective roles of each protein. Since vapB6 transcript abundance was elevated in the vapC6 deletion, the VapC6 toxin appears to regulate abundance of its cognate antitoxin. In contrast, vapC6 transcript abundance was reduced in the vapB6 deletion. A putative intergenic terminator may underlie these observations by coordinating vapBC6 expression. As predicted by structural modeling, recombinant VapC6 produced using chaperone cosynthesis exhibited heat-dependent ribonucleolytic activity toward S. solfataricus total RNA. This activity could be blocked by addition of preheated recombinant VapB6. In vivo transcript targets were identified by assessing the relative expression of genes that naturally respond to thermal stress in VapBC6-deficient cells. Preferential increases were observed for dppB-1 and tetR, and preferential decreases were observed for rpoD and eIF2 gamma. Specific VapC6 ribonucleolytic action could also be demonstrated in vitro toward RNAs whose expression increased in the VapBC6-deficient strain during heat shock. These findings provide a biochemical mechanism and identify cellular targets underlying VapBC6-mediated control over microbial growth and survival at temperature extremes.
Keywords: antitoxin, RNA degradation, heat shock, archaea, transcription
INTRODUCTION
Despite the intrinsically high temperatures found in hydrogeothermal niches, thermotolerance and the heat-shock response are probably essential for endogenous microbial survival and proliferation. Frequent shifts in geological activity foster steep thermal gradients, necessitating the capacity to respond rapidly to this type of challenge, particularly by planktonic organisms. Diverse taxa belonging to the order Sulfolobales within the archaeal phylum Crenarchaeota are common inhabitants of acidic geothermal pools. The heat-shock response of the thermoacidophile, Sulfolobus shibatae, was found to involve a generalized reduction in protein synthesis with continued production of a eukaryotic-like molecular chaperone, termed the thermosome (Trent et al. 1991). Additional components of the heat-shock (HS) response in this organism include adaptive heat resistance by pre-treatment (Trent et al. 1990) and heat-induced shifts in the proteome (Cerchia et al. 2000). In a related species, Sulfolobus solfataricus, the HS response transcriptome associated with a nonlethal temperature shift involved approximately one-third of the genome (Tachdjian and Kelly 2006). However, transcription factors that control thermotolerance are missing in crenarchaeotal genomes such as those characterized in the other major archaeal phylum, the Euryarchaeota (Thompson et al. 1999; Vierke et al. 2003; Liu et al. 2007). Eukaryotic factors (Vuister et al. 1994; Yamamoto et al. 2008; Westerheide et al. 2009) and bacterial sigma factors (Rodriguez et al. 2008; Wagner et al. 2009) also are not evident in crenarchaeotal genomes, and an underlying mechanism governing the heat-shock response in these organism has not been identified.
Type II Toxin–antitoxin (TA) loci are found in nearly all prokaryotic genomes and are thought to promote survival during environmental stress (Gerdes et al. 2005; Makarova et al. 2009). Type II TA systems consist of an intracellular protein toxin coexpressed with a cognate protein antitoxin. These two components are usually contiguous and encoded on the genome or plasmids. Antitoxins have been shown to bind and inactivate cognate toxins, while the antitoxin can be susceptible to protease cleavage due to its natively unfolded C-terminal domain (Gerdes et al. 2005; Pandey and Gerdes 2005). When not bound to the antitoxin, some toxins act as ribonucleases and cleave mRNA, thus modulating protein synthesis (Jensen and Gerdes 1995). Consequently, imbalanced concentrations of stable toxin and unstable antitoxin may control protein expression in the cell (Jensen and Gerdes 1995).
Currently, there are eight major TA families encoded on plasmids and chromosomes (Gerdes 2000; Pandey and Gerdes 2005), though this number continues to increase as more genomes are examined (Makarova et al. 2009). In archaea, there are three toxin–antitoxin families that have been identified: RelBE, Phd/Doc, and VapBC (Gerdes et al. 2005). The Vap (Virulence associated protein) family make up ∼40% of all TAs in prokaryotes (Gerdes et al. 2005), with at least 26 identified in S. solfataricus (Tachdjian and Kelly 2006; Jorgensen et al. 2009). While the target and cognate protease have been identified for E. coli RelBE and Phd/Doc families, the VapBC family, although the most abundant of all the TAs, has no known targets nor proteases (Gerdes et al. 2005).
In the Vap family, VapB is the unstable antitoxin and VapC is the stable toxin. The VapC toxin is identified by the presence of a PIN (PilT N-terminal) domain, which is indicative of RNase activity (Anantharaman and Aravind 2003). The crystal structure of an archaeal PIN domain toxin, PAE2754, shows homology with RNase H, but exhibits DNA not RNA nuclease activity (Arcus et al. 2004; Bunker et al. 2008). Bacterial VapC's have been studied in some detail. VapC-1 of nontypeable Haemophilus influenzae has ribonuclease activity that could be inhibited by the VapB-1 antitoxin (Daines et al. 2007). FitB of Neisseria gonorrhoeae modulates virulence through transcellular trafficking and structurally resembles PAE2754, although insolubility of the recombinantly produced protein precluded assessment of its activity (Hopper et al. 2000; Mattison et al. 2006). NtrR of Sinorhizobium meliloti controls metabolism under symbiosis and stress (Bodogai et al. 2006). Finally, while VapC5 from several species of Mycobacteria have undergone structural (Miallau et al. 2009) and in vivo characterization, a biologic function has yet to be identified (Robson et al. 2009). In S. solfataricus, the strong and rapid heat-shock induction of certain vapBC loci (Tachdjian and Kelly 2006), combined with preliminary information on an uncomplemented vapBC mutant (Cooper et al. 2009), provided the impetus to investigate the function of the toxin and antitoxin belonging to a single family member, vapBC6, and a role in thermotolerance.
RESULTS
The effect of vapB6 disruption on thermotolerance and fitness
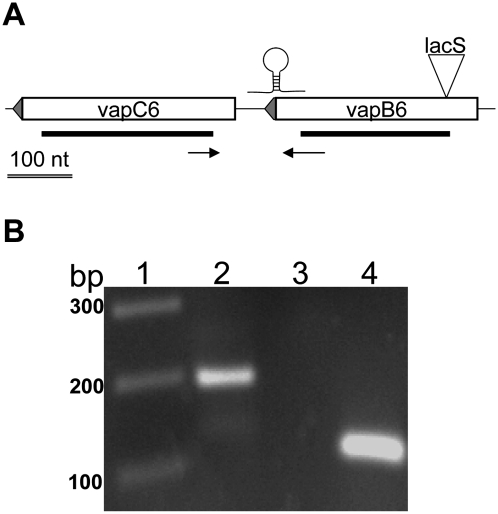
Unlike most vapBC loci in S. solfataricus (Sso) strain 98/2, the vapB6 and vapC6 genes are separated by a 61-nt intergenic region (Fig. 1A). RT–PCR analysis was used to test whether these genes were cotranscribed (Fig. 1B). A cDNA product spanning the intergenic region was evident using total RNA extracted from cells growing at 80°C and at a pH of 3. As this intergenic RNA was only evident at a high PCR cycle number, efforts to verify heat-shock induction of vapBC6 transcription were conducted. Five minutes after a 10°C heat shock (80°–90°C), the abundance of the intergenic RNA increased dramatically and remained at this level for at least an additional 45 min (data not shown). These results demonstrate that the vapBC6 genes of Sso strain 98/2 are cotranscribed. They also demonstrate that vapBC6 heat-shock induction is analogous to that observed previously in strain P2 (Tachdjian and Kelly 2006).
FIGURE 1.
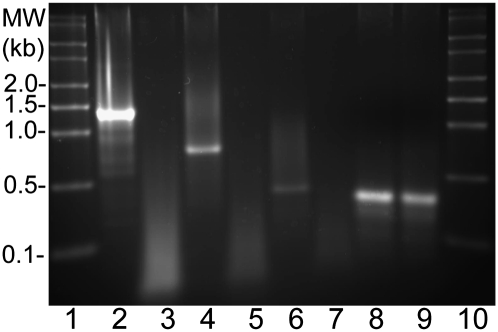
RT–PCR analysis of vapBC6. (A) Schematic of vapBC6 locus. Both vapB6 (SSO1494) and vapC6 (SSO1493) are transcribed from the antisense strand (leftward). Sites are shown for: the lacS disruption (inverted triangle); putative intragenic terminator; in frame deletions (heavy lines beneath boxed ORFs) and primers (horizontal arrows) that verify cotranscription. A 100-nt scale bar is indicated. (B) Cotranscription of vapB6 and vapC6. Detection of transcript complementary to the intergenic region used primers VapBC6RT-F, located 51 nt from the 3′ end of vapB6 and VapBC6RT-R located 51 nt from the 5′ end of vapC6. Samples: (lane 1) MW markers; (lane 2) complete reaction; (lane 3) no addition of reverse transcriptase; (lane 4) 7S RNA positive control using primers 7S-F and 7S-R.
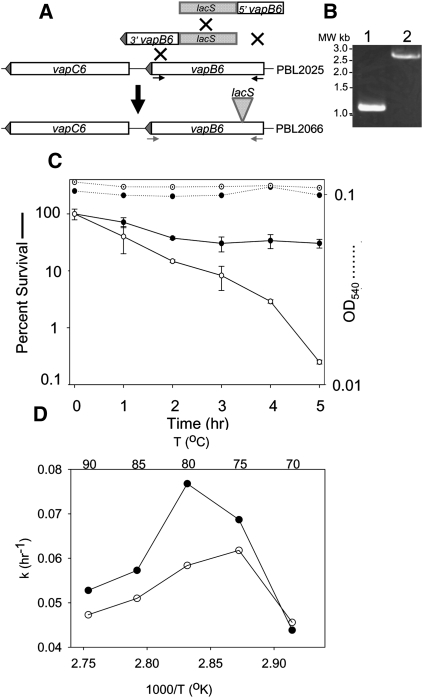
The functional role played by vapBC6 in thermotolerance was examined initially by gene disruption using an expressed copy of Sso lacS (β-glycosidase). To simplify construction of this type of mutation, a new strategy based on multiplex linear DNA recombination was used (Maaty et al. 2009). In this strategy, two DNA fragments, one joining the 5′ end of vapB6 to lacS and the other joining the 3′ end of vapB6 to lacS, were generated by overlap extension PCR and then cotransformed into lacS-deleted strain PBL2025 (Fig. 2A). Lactose utilizing recombinants arose as a result of a simultaneous triple recombination event. These were shown to encode a lacS-disrupted copy of vapB6 by PCR (Fig. 2B) and by DNA sequence analysis. The insertion was placed 51 nt 3′ of the vapB6 start codon, resulting in strain PBL2066 (Fig. 1A).
FIGURE 2.
Construction and characterization of a vapB6::lacS disruption. (A) Multiplex linear recombination for vapB6 disruption. Two overlap extension PCR products containing lacS and either the 5′ or 3′ end of vapB6 were cotransformed into PBL2025. Recombinants arose by three crossover events (X's) between the two incoming fragments and the corresponding chromosomal allele. Primers used to verify identities of vapB6 alleles are indicated. (B) Mutant genotype. PCR amplification of the wild-type (lane 1) and lacS disrupted alleles of vapB6. (C) Thermotolerance of the vapB6 mutant. Wild-type (•) and vapB6::lacS disruption mutant (○) were cultivated in rich medium and shifted from 80°C to 95°C for the times indicated. Culturable cells were enumerated on solid medium with incubation at 80°C. (D) Arrhenius plot. The growth rate constant (k), which is related inversely to generation time, is portrayed on the y-axis. The temperature in °K is indicated on the x-axis, and the temperature in °C is indicated across the top of the figure. Strains were: wild-type (•) and vapB6::lacS disruption mutant (○). Values for the average of two independent experiments are shown.
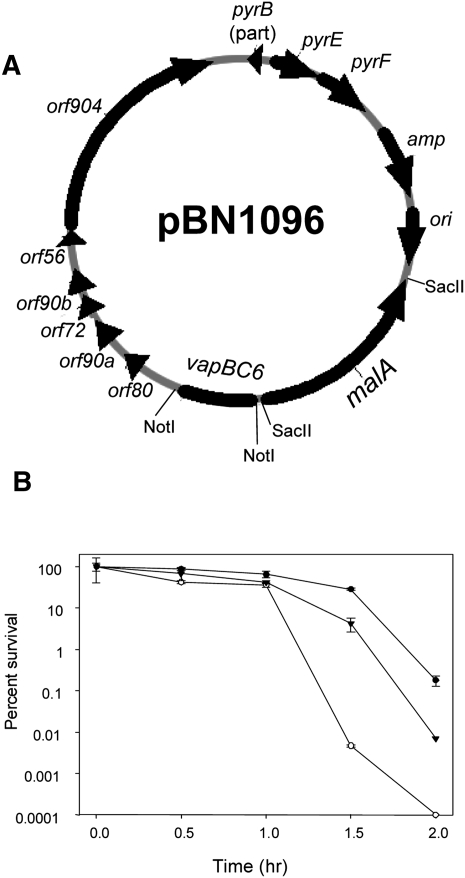
Thermotolerance was measured as the percentage of culturable cells surviving exposure to lethal heat shock (Fig. 2C). After exposure, cells were transferred to solid medium and incubated at 80°C until colonies formed. A normally nonlethal temperature shift from 80°C to 95°C resulted in preferential killing of the vapB6 disruption strain PBL2066. Killing was evident after 1 h at 95°C and became more pronounced with continued exposure. Thermotolerance of the mutant was reduced 100-fold relative to the wild type after 5 h of heat shock. Under the conditions used, the wild-type strain exhibited a 50% reduction in thermotolerance after 2 h of exposure, but then stabilized at this level for the duration of the exposure period. No significant cell lysis was apparent for either strain during the entire exposure period, indicating that cells retained their integrity despite a loss of viability.
Since vapBC6 is expressed during growth at normal temperatures (Fig. 1B), an effect resulting from mutation of this locus could also have an impact at other temperatures. To assess the role of VapBC6 on biological fitness, the rate of growth at temperatures ranging from 70°C to 90°C was evaluated for the vapB6 disruption mutant and wild-type strain (Fig. 2D). Generation times were determined across the temperature growth range of the organism and the results are depicted in an Arrhenius plot, where growth rate is plotted as a function of the reciprocal of absolute temperature. Cells were cultivated in a glucose minimal medium to maximize anabolic requirements, and thereby enhance detection of differences in fitness. Reduced rates of growth were observed for the vapB6 disruption mutant at all temperatures above 70°C relative to the wild-type control strain.
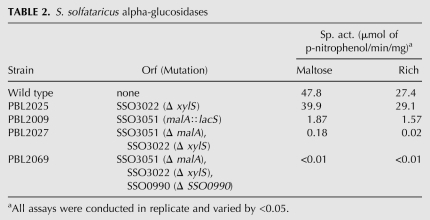
Complementation analysis was conducted to confirm that reduced thermotolerance and growth rate of the vapB6 disruption mutant arose from vapBC6 deficiency. Two genetic markers were required to conduct this test; lacS was used for vapB6 disruption, and a new genetic marker, Sso malA (Rolfsmeier et al. 1998), was used as the genetic marker gene to select for a vapBC6 recombinant shuttle plasmid (Table 1). Simultaneous use of these genetic markers required construction of a strain (PBL2069) that was deficient in utilization of both lactose and maltose as sole carbon and energy sources (Table 2). Although Sso encodes six annotated α-glucosidases (She et al. 2001; Kim et al. 2004), deletion of only three was sufficient to create an absolute catabolic defect for maltose utilization. α-glucosidase-specific activities for several strains were measured to assess the relative importance of each of these genes. Simultaneous inactivation of malA (SSO3051), xylS (SSO3022), and SSO0990, resulting in strain PBL2069, reduced α-glucosidase activity over 100-fold (Table 2) and blocked growth on maltose as the sole carbon and energy source. A vapB6 disruption mutant derivative of PBL2069 was then constructed by linear DNA recombination using the PCR-amplified vapB6 mutation as the recombination template, creating strain PBL2081. Plasmid pBN1080 encoding the wild-type vapBC6 operon, and malA as a selectable marker (Fig. 3A), was then transformed into PBL2069 by selection for maltose utilization, creating strain PBL2082. Thermotolerance of PBL2082, relative to the wild-type strain (PBL2025) and the vapB6 disruption mutant (PBL2066), was then examined (Fig. 3B). After 90 min of exposure, the complemented mutant was over 100-fold more thermotolerant than the uncomplemented mutant and remained so until a complete loss of culturability of the mutant was reached. In all samples, the parental wild-type strain was still about 10-fold more resistant to heat killing than the complemented mutant. This difference may arise from increased vapBC6 gene dosage due to use of the low, but not single-copy vector, pRN1 (Berkner et al. 2007), for construction of the complementation plasmid (pBN1096). Overproduction of toxins are well known to be toxic (Christensen-Dalsgaard and Gerdes 2006; Daines et al. 2007), therefore increased vapBC6 gene dosage may also be deleterious and compromise survival during heat shock. Complementation of the vapB6 disruption mutant defect in growth rate at normal growth temperatures could also be demonstrated; in minimal glucose medium, the complemented vapB6 disruption mutant grew better than the otherwise isogenic uncomplemented vapB6::lacS disruption mutant that carried the malA plasmid (pBN1080), but lacking vapBC6.
TABLE 1.
Archaeal strains
TABLE 2.
S. solfataricus alpha-glucosidases
FIGURE 3.
Complementation of vapB6::lacS disruption using malA-based selection. (A) Complementation plasmid pBN1096. Plasmid pBN1096 was derived from plasmid pBN1080 by insertion of vapBC6 into the NotI site. Plasmid pBN1080 was derived from pBN1074 (Berkner et al. 2007) by replacment of lacS with malA at the SacII site. (B) Percent survival of the vapB6 disruption mutant and complemented strain during heat shock. Cultures were shifted to 95°C and samples were taken thereafter every 0.5 h. Strains were: wild-type (PBL2025, •); vapB6::lacS disruption (PBL2081, ○), vapB6::lacS complemented mutant (PBL2082, ▾).
Coupling of vapB6 and vapC6 expression
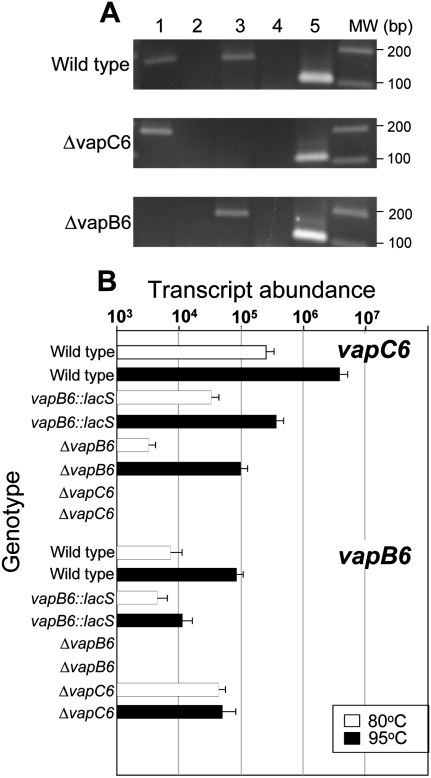
Though the vapB6 disruption mutant phenotype suggested that a deficiency of VapB6 was responsible for HS sensitivity, alternative mechanisms such as VapC6 toxin deficiency or VapB6 and C6 deficiency remained plausible. To discriminate between these possibilities, in-frame deletion mutants lacking either vapB6 or vapC6 were constructed and evaluated. In-frame deletions were created by markerless exchange using internally truncated reading frames (Fig. 1A; Schelert 2006). RT–PCR using reverse primers complementary to sequences within the deleted region confirmed loss of each respective transcript (Fig. 4A). Total RNA derived from cells growing at normal growth temperatures (80°C) was used as the template in these reactions. The ΔvapC6 mutant (PBL2080) lacked detectable vapC6 mRNA and the ΔvapB6 mutant (PBL2078) lacked detectable vapB6 mRNA. Thermal conditions producing rates of killing of the wild type similar to those used to test complementation (Fig. 3B) indicated that both vapB6 and vapC6 in-frame deletions mutants were thermally sensitive after at least 1 h of heat shock relative to the wild type (data not shown). To clarify the basis for this result, levels of vapB6 and vapC6 transcript abundance were determined by qRT–PCR in each of the respective deletion mutants, as well as both the vapB::lacS disruption and wild-type strains, during normal growth and in response to heat shock (Fig. 4B). Abundance of vapBC6 mRNA was normalized to the transcript abundance of tbp as an internal control for each RNA sample tested, variation was <10% in all cases. In general, both vapB6 and vapC6 mRNA levels were heat-shock inducible in all strains examined; however, their abundance under steady-state growth conditions was differentially affected by the various mutations. At 80°C, the abundance of vapB6 mRNA increased eightfold in the vapC6 in-frame deletion strain relative to the wild type. In contrast, the abundance of vapC6 mRNA decreased over 10-fold in both the vapB6 disruption mutant and the in-frame vapB6 deletion mutant during growth at 80°C. This latter observation is consistent with the heat-shock sensitivity of both mutant strains. No transcript was evident for vapB6 in the vapB6 in-frame deletion strain or for vapC6 in the vapC6 in-frame deletion strain. Examination of the vapBC6 locus identified a single putative intergenic terminator located between vapB6 and vapC6 (Fig. 1A). This stem–loop would sequester the C-terminal 33 nt of the vapB6 coding region and 23 nt of the flanking vapB6–vapC6 intergenic region. Analysis using MFOLD (ver. 2.3) (Zuker 2003) indicates that this stem–loop has a ΔG of −2.45 kcal/mol and Tm 86.3°C at standard conditions (80°C, 1 M NaCl, 0 Mg) and, therefore, could play a role in the expression of vapC6 in a manner that would depend on vapB6 translation. These data indicate that expression of these genes is coupled, and suggest that RNA degradation, perhaps mediated by VapC6, could be involved in modulating transcript abundance in response to heat shock.
FIGURE 4.
Expression and coupling of vapB6 and vapC6. (A) Detection of vapBC6 transcript. RT–PCR was used to detect transcripts present in wild-type (PBL2025), the vapC6 deletion strain (PBL2080), and the vapB6 deletion strain (PBL2078). (Lanes 1,2) vapB6 transcript with reverse transcriptase (RTase) added or omitted, respectively; (lanes 3,4) vapC6 transcript with RTase added or omitted, respectively; (lane 5) 7S RNA. Location of vapBC6 deleted regions is shown in Figure 1A. (B) Abundance of vapB6 and vapC6 mRNA. qRT–PCR primers used to assess transcript abundance were complementary to regions that remained present in all strains tested. RNA was extracted from cells growing exponentially at the temperatures indicated: (open bars) 80°C; (closed bars) 95°C. Strain genotypes for vapB6 and vapC6, respectively, are indicated across the top of the figure. Strain and relevant vapBC6 genotypes were: + +, wild-type; d +, vapB6::lacS disruption; − +, vapB6 deletion strain; + −, vapC6 deletion. The tbp mRNA was used as an internal control in each RNA sample to normalize vapBC6 values; tbp mRNA varied in abundance by <15% in these experiments.
Structure and activity of VapC6
VapC homologs from bacteria have been reported to have ribonuclease activity (Daines et al. 2007; Miallau et al. 2009), while VapC (PAE0151) from the archaeon Pyrobaculum aerophilum has DNA exonuclease activity (Arcus et al. 2004). To clarify the situation for the Sso VapC6, the amino acid sequence for SSO1493 was analyzed with the 3D-JIGSAW prediction tool (Bates and Sternberg 1999; Bates et al. 2001) using PDB values for the PIN-domain protein and putative toxin from Pyrobaculum aerophilum PAE0151 (PDB:2FE1) (Arcus et al. 2004). The model showed >90% secondary structure homology with the template, and the confidence level of structure predictions (or “accuracy”) was >95%, indicative of a high-quality match. VapC6 is missing about 30 residues at the beginning of the template when compared with PAE0151, resulting in a more open conformation at the active site. When comparing this predicted stucture to a known ribonuclease, T4 RNAse H (PDB: 1TFR), using an in silico superimposition tool LOCK 2 (Shapiro and Brutlag 2004), all of the predicted catalytic residues in VapC-6 matched known T4 RNAse catalytic residues (Bhagwat et al. 1997), and the overall RNAse H active site configuration was conserved in the toxin (data not shown).
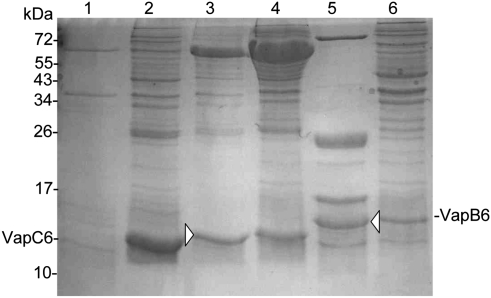
To test the prediction that the S. solfataricus VapC6 was a ribonuclease, efforts to produce this and its cognate antitoxin, VapB6, as recombinant proteins in E. coli were undertaken. VapC6 or hexahistidine N- or C-terminal-tagged derivatives expressed in E. coli formed insoluble products. Cosynthesis with the E. coli molecular chaperone, DnaK (HSP70), was evaluated as a means of overcoming protein insolubility (Blum et al. 1992b). The expression of dnaK was induced concomitantly with vapC6 using the lacI-repressible Ptac promoter, as described previously (Blum et al. 1992a). Compatible replication of the dnaK expression plasmid with the vapC6 expression plasmid required construction of a P15A replicon for the dnaK vector. This was accomplished by exchanging the origin and genetic marker of plasmid pBN13 (Blum et al. 1992a) with the origin (P15A) and genetic marker (tet) from plasmid pACYC184 (Chang and Cohen 1978). Comparison of soluble levels of VapC6 encoding a C-terminal polyhistidine tag (VapC63′his) were then evaluated either with or without chaperone coexpression (Fig. 5). DnaK coexpression shifted VapC63′his from an insoluble product present in centrifugal pellets to a soluble form present in centrifugal supernatants. VapB63′his was produced in E. coli in a soluble form and purified by nickel affinity chromatography. The identities of both proteins were confirmed by tandem mass spectrometry of excised material (Fig. 5, arrowheads).
FIGURE 5.
In vivo solubilization of VapC63′His and VapB63′His. SDS-PAGE analysis of E. coli crude extracts (lanes 2,4,6) or Ni-NTA column eluants (lanes 1,3,5). Extracts were prepared from strains containing plasmids expressing vapC63′His (lanes 1,2), both vapC63′His and dnaK (lanes 3,4), and vapB63′His (lanes 5,6). Protein identities that were determined by tandem mass spectrometry are indicated (arrowheads).
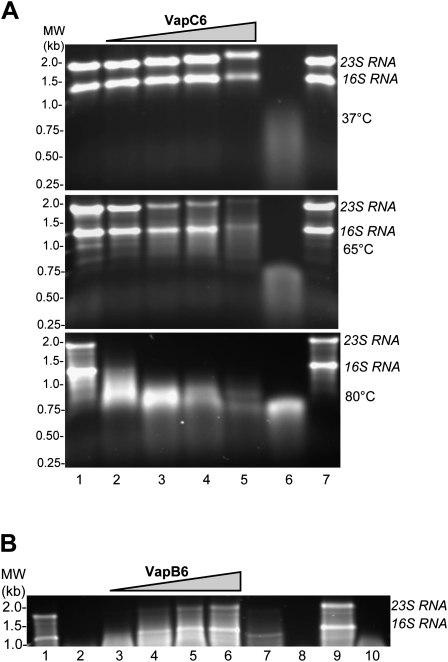
Nickel-affinity column-purified VapC63′his was then tested for activity toward homologous total RNA extracted from S. solfataricus. RNase activity was monitored by the extent of 16 and 23S rRNA degradation on agarose gels following incubation under various conditions (Fig. 6A). To exclude any contribution in these assays from contaminating RNAse activity that might copurify from E. coli extracts, mock extracts were also assayed that had been prepared from an otherwise identical strain lacking VapC63′his (Fig. 6A, lane 7). At 80°C, S. solfataricus total RNA was rapidly degraded by VapC63′his in a manner that was proportional to the quantity of protein added. Residual activity was still apparent following incubation at 65°C, while no activity was evident at 37°C. In contrast, VapC63′his did not exhibit any exo- or endonucleolytic activity toward DNA at 80°C. DNAase activity was tested using double-stranded circular DNA, double-stranded linear DNA, as well as a single-stranded linear DNA (data not shown, see Materials and Methods). To evaluate the potential interaction between VapC63′his and the antitoxin VapB6, the effect of addition of affinity-purified VapB63′his was tested on ribonuclease activity (Fig. 6B). VapB63′his and VapC63′his were preincubated separately at 95°C for 10 min, then combined and incubated at 90°C for an additional 10 min, followed by RNA addition and incubation for 30 min at 80°C. VapB63′his addition blocked the action of VapC63′His and prevented RNA degradation with increasing potency as its relative amount increased. Addition of similar amounts of a VapB63′his mock affinity-purifed extract had a negligible effect on VapC63′His ribonuclease activity even at the highest ratio tested (20:1). Modulation of VapC63′His RNAse activity could arise from other proteins present in the heat fractionated affinity-purifed recombinant E. coli extracts. However, since VapC63′His RNAse activity was both strongly heat dependent and specifically inhibited by addition of the cognate VapB63′his antitoxin, a mechanism whereby copurifying the E. coli protein would confer these properties seems unlikely. In summary, these data suggest that recombinant VapC6 and VapB6 form a heat-activated inactive complex that has lost ribonucleolytic activity, thereby providing a mechanism to regulate the activity of the VapC6 toxin in vivo.
FIGURE 6.
Characterization of VapC63′His activity and interaction with VapB63′His. (A) VapC63′His activity. S. solfataricus total RNA (4 μg) was treated at various temperatures with increasing amounts of VapC63′His. (Lane 1) No VapC63′His; (lanes 2–5) 0.75 μg, 2.25 μg, 3.75 μg, 7.5 μg VapC63′His; (lane 6) Ribonuclease A; (lane 7) VapC6 mock extract. (B) VapB63′His effect. VapB63′His effect on VapC63′His ribonuclease activity toward total Sso RNA was examined at 80°C . (Lane 1) Sso RNA (4 μg); (lane 2) VapC63′His (0.75μg); (lanes 3–6) VapB63′His and VapC63′His at ratios of 1:1, 5:1, 10:1 and 20:1; (lane 7) VapB6 mock extract and VapC63′His at 20:1; (lane 8) Ribonuclease A; (lane 9) VapB63′His; (lane 10) VapC63′His pretreated at 95°C.
Transcriptomic effects of VapBC6 deficiency
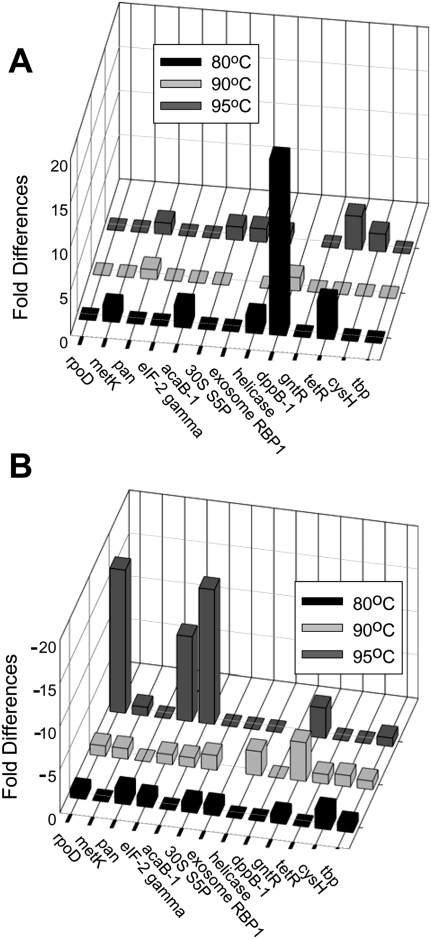
To identify the in vivo targets of VapBC6, the impact of VapBC6 deficiency was examined on the heat-shock transcriptome. A qRT–PCR analysis was conducted surveying 13 different mRNAs identified previously as the strongest responders to heat-shock stimulus in this organism (Tachdjian and Kelly 2006). Levels of the target mRNAs were evaluated in the wild-type and vapB6 disruption strains during growth under normal conditions at 80°C, and 5 min after culture temperatures had increased to either 90°C or 95°C. The level of the tbp mRNA was evaluated as an internal control in all reactions. Several transcripts were more abundant in the mutant relative to the wild type (Fig. 7A). The most apparent was the Oligo/dipeptide transport permease (dppB-1, SSO1274), followed by TetR a transcriptional regulator (SSO2506). Transcripts whose abundance was less in the mutant relative to the wild type (Fig. 7B) included RNA polymerase subunit RpoD (SSO0271), translation initiation factor subunit eIF-gamma (SSO0412), Acetyl-CoA c-acetyltransferase (acetoacetyl-CoA thiolase; AcaB1, SSO0534), and transcription factor GntR (SSO1589). All transcript abundance values for both strains are indicated (Supplemental data; Supplemental Table 3). The variations between replicate determinations are provided for all determinations along with internal values for tbp. While the mechanism underlying the relative reduction in abundance of some transcripts in the vapB6 disruption mutant is as yet unclear, VapC6 may act in a direct manner to modulate the abundance of those transcripts whose levels increased preferentially in cells lacking the ribonuclease. This hypothesis was tested by measuring the action of VapC6 at 80°C toward several RNAs. Runoff RNAs were prepared as described (Hoang et al. 2004) for four genes; SSO1274 (dppB-1), SSO2506 (tetR), SSO1494 (vapB6), and SSOr01 (7SRNA). While three of these (dppB-1, tetR, vapB6) underwent an increase in abundance in the vapC6 disruption mutant upon heat shock (Fig. 7), the 7SRNA did not vary appreciably under these conditions or, as demonstrated previously, in response to heat shock in wild-type cells (Tachdjian and Kelly 2006). Treatment of purified runoff RNAs with recombinant VapC6 at 80°C resulted in preferential degradation of dppB-1, tetR, and vapB6, while no degradation was apparent for 7SRNA (Fig. 8).
FIGURE 7.
Transcriptomic effects of VapBC6 deficiency. Relative transcript abundance of genes were determined by qRT–PCR in total RNA samples from the vapB6 disruption mutant and wild-type strain. (A) Transcripts whose abundance increased in the vapB6::lacS mutant relative to the wild type during heat shock. (B) Transcripts whose abundance decreased in the vapB6::lacS mutant relative to the wild type during heat shock. RNA levels were determined from cells growing exponentially at 80°C (filled bars), or 5 min after temperatures of the cultures reached 90°C (light gray bars), or 95°C (dark gray bars).
FIGURE 8.
Treatment of runoff RNAs at 80°C using VapC6. RNA standards (lanes 1,10); dppB-1 RNA (lanes 2,3); tetR RNA (lanes 4,5); vapB6 RNA (lanes 6,7); 7SRNA (lanes 8,9). RNAs treated with VapC6 (lanes 3,5,7,9). Amounts of RNA and VapC6 added to reactions were identical to those described in the legend to Figure 6.
DISCUSSION
The S. solfataricus VapBC6 toxin–antitoxin (TA) is the first example of a cellular component required for heat-shock (HS) survival in the archaeal phylum, Crenarchaeota. The toxin, VapC6, appears to play an essential ribonucleolytic role in this process. The vapBC6 genes are also the first example of a gene pair that are required for normal rates of growth in this organism. This finding, together with the apparent absence of heat-shock transcription factors found in Crenarchaeota, suggest that TA pairs like VapBC6 contribute strongly to heat-shock gene regulation in this phylum. Since VapC6 was found to be a ribonuclease, RNA depletion mediated by an increase in ribonuclease abundance is a requirement for cell survival. This mechanism could explain the basis for prior reports of a strong repression in protein synthesis during the heat-shock response of the related species, S. shibatae (Trent et al. 1990).
VapC6 rapidly degraded total S. solfataricus RNA in vitro; therefore, this capacity in vivo could provide a rapid response mechanism for modulating protein synthesis. An initial survey of 13 genes selected because of their response to heat shock (HS) (Tachdjian and Kelly 2006) revealed strong changes in the abundance of several transcripts during HS of the vapB6::lacS disruption mutant relative to the wild type. Those undergoing large preferential increases included the oligopeptide permase, dppB-1, as well as the transcription factor, tetR, suggesting that these transcripts are direct targets of the VapC6 ribonuclease. Genes whose abundance preferentially decreased during HS in the vapB6::lacS disruption mutant included the RNA polymerase subunit, rpoD, and the translation initiation factor, eIF-2 gamma. Since depletion of the corresponding two proteins would block both transcription and translation, their modulation could underly the HS sensitive phenotype of the vapB6::lacS disruption mutant. While the mechanism for this effect remains to be elucidated, it demonstrates that the VapBC6 proteins are required to maintain normal levels of essential cellular components.
Unexpectedly, deletion of vapC6 resulted in the elevated abundance of the vapB6 transcript during growth at 80°C. This finding could mean that VapC6 normally degrades the mRNA of its cognate antitoxin, VapB6, suggesting that there is another level of regulatory control operating to coordinate the relative abundance of the toxin and antitoxin proteins. The HS-sensitive phentoype of the vapB6 disruption reflects reduced vapC6 mRNA abundance due to premature transcription termination mediated by the lacS terminator (transcriptional polarity). Despite this effect, levels of vapC6 mRNA retained HS inducibility, albeit at reduced levels. This regulatory pattern most likely reflects readthrough of the lacS terminator from transcripts initiating at the vapBC6 promoter and not an internal initation site, since RNAseq data of S. solfataricus fail to detect any 5′ primary transcript reads from within this region (Wurtzel et al. 2010). In addition, vapB6 deficiency (by in-frame deletion) resulted in a reduction of vapC6 mRNA. This effect could be mediated through premature transcription termination at a putative intergenic terminator identified at the 3′ end of vapB6. Translation of vapB6 would normally compete with formation of this structure, preventing premature transcription termination and reduction in vapC6 expression. Inhibition of vapB6 translation resulting from either vapB6 mutation, might compromise stem–loop formation. This type of coupling mechanism could coordinate expression of both Vap products in the wild-type strain. Alternatively, VapB6 deficiency may allow VapC6 to degrade itself. As the VapC6 protein is likely to be longer lived than vapC6 mRNA, an autodegradatory mechanism is plausible. Such a strategy resembles that used by other TA families like ccd, which regulate plasmid replication where the toxin is long lived and antitoxin is short lived; if cells lose the TA genes, they die because the long-lived toxin inactivates plasmid replication (Madl et al. 2006). Since recombinant VapB6 was found to be a potent inhibitor in vitro of VapC6 ribonucleolytic acitivity, coordinate increases in both gene products may not elevate ribonuclease activity without a corresponding reduction in antitoxin abundance. While this could arise by elevated rates of antitoxin proteolysis, in vivo measurements of toxin and antitoxin abundance are necessary to resolve this process.
VapBC6 is only one of 22 copies of this two-gene family (and one solitary toxin) in Sso (Tachdjian and Kelly 2006; Jorgensen et al. 2009). While several other vap loci are heat-shock inducible, including, for example, vapBC8 and vapBC22 (Tachdjian and Kelly 2006), most others are not. The finding that a single vapBC locus was required for thermotolerance leaves open the possibility that other heat-shock-inducible vap loci are also involved in the heat-shock response. In addition, it also suggests that other vap loci may play distinct and important physiologic roles. The occurrence in S. solfataricus of multiple vapBC loci that are closely related in sequence composition raises the possibility that interactions occur between noncognate pairs. Such noncognate interactions or “cross talk” could increase the functional role of these proteins by expanding their capacity to respond to diverse stimuli and control diverse genes. However, while vapC6 inactivation by in-frame deletion had a strong impact on long-term heat-shock survival, the absence of vapB6 may be more important for the initial HS response. This may indicate that VapB6 plays a specific regulatory role toward VapC6, but has little control over other VapC proteins. Experiments are underway to determine the affinity of VapB6 toward other recombinant VapCs.
Initial efforts to prepare recombinant soluble VapC6 were not successful because of protein insolubility. As described previously for other proteins (Blum et al. 1992b), molecular chaperones can be used to overcome this constraint. DnaK is a highly characterized molecular chaperone and a heat-shock protein that plays a critical role in protein folding impacting thermotolerance, stress, and other physiologic traits. Simultaneous overproduction of DnaK in E. coli greatly improved the solubility of VapC6. This effect may have general applicability for promoting recovery of soluble forms of other S. solfataricus VapC proteins. The results presented here also introduce a genetic advance for S. solfataricus. Complementation of the vapB6::lacS disruption mutant depended on the development of a new genetic marker encoded by malA and a host strain that had lost the catabolic ability to grow on maltose. The S. solfataricus genome is thought to encode several α-glucosidases (Kim et al. 2004). Deletion of three of these genes was sufficient to disable maltose catabolism and create a strong selection for restoration of this trait by malA introduction. The malA marker expands the list of genes that can be used to conduct studies with S. solfataricus (Martusewitsch et al. 2000; Worthington et al. 2003b; Sowers et al. 2006; Berkner et al. 2007).
This study reinforces the importance of TA loci such as the Vaps for post-transcriptional regulation in microorganisms. Given the apparent lack of several bacterial post-transcriptional regulation mechanisms in the archaea, the prospect that TA loci play an even more critical role in stress response cannot be ruled out and is currently under investigation.
MATERIALS AND METHODS
Archaeal cultivation and strains
All S. solfataricus cell lines were derivatives of the wild-type strain 98/2 (Rolfsmeier and Blum 1995) and are listed with plasmids in Table 1 (primers are listed in the Supplemental Materials). S. solfataricus strains were cultivated in a basal salts medium (Allen 1959) with added carbon sources and adjusted to a pH of 3.0 at 80°C in screw-top flasks, with aeration in orbital baths (Rolfsmeier et al. 1998; Worthington et al. 2003a). Sugars were prepared as 10% (w/v) stocks, filter sterilized, and added at a final concentration of 0.2% (w/v). Rich medium contained tryptone 0.2% (w/v). Growth in liquid medium was followed spectrophotometrically at a wavelength of 540 nanometers. Solid medium contained 0.6% (w/v) gelrite (Kelco) and 8.0 mM magnesium chloride added to rich liquid medium.
The vapB6::lacS disruption strain, PBL2066, was created by simultaneous transformation of multiple overlap extension PCR products, and recombinants were enriched in a lactose minimal medium, as described previously (Sowers et al. 2006). The lacS gene including promoter and terminator was inserted 51-bp downstream from the vapB6 start codon. Primers Vap6B-LF and VapB6-LR-lacS were used to amplify 527-nt upstream of the lacS insertion, including an overlap portion that complements the 5′ end of lacS. Primers Vap6B-RF-lacS and Vap6B-RR amplified 557-bp downstream from the lacS insertion site, including a portion that complements the 3′ end of lacS. A total of 1630 bp of lacS, including flanking regions was amplified using primers Vap6B-lacS-F and Vap6B-lacS-R. OLE PCR was used to join the upstream PCR product with lacS, producing a 2157-bp amplicon to join the downstream PCR product with lacS, producing a 2189-bp amplicon. The two chimeric PCR products were cotransformed into cells, and recombinants arising from three crossover events were recovered. Purified isolates were recovered as described (Schelert et al. 2006). The genotype of strain PBL2066 was confirmed by colony PCR using primers Vap6B-LF and Vap6B-RR to amplify the wild-type (1073 bp) or mutant alleles (2706 bp).
An in-frame vapB6 deletion strain (PBL2078) was constructed using plasmid pPB1093 by markerless exchange as described (Schelert et al. 2006). Overlap extension PCR (OLE PCR) was used to create a 228-bp in-frame deletion that begins 39 bp downstream from the vapB6 start codon and ends 267 bp downstream from the start codon, leaving a 72-bp gene product. A 487-nt amplicon upstream of the deletion was created using primers XmaVapB6out-F and VapB6delOR-R. A 530-bp amplicon 3′ to the deletion site was made using forward primers VapB6delOR-F and SbfVapB6out-R. An OLE PCR amplicon was cloned at XmaI and SbfI sites in plasmid pPB1035, creating plasmid pPB1093, used subsequently to make strain PBL2078. Presence of the vapB6 in-frame deletion was confirmed by DNA sequence of PCR amplicons using primers XmaVapB6out-F and SbfVapB6out-F for the wild-type (1244 bp) and mutant alleles (1016 bp). An in-frame vapC6 deletion strain (PBL2080) was constructed following the strategy used for the vapB6 in-frame deletion strain. A 507-bp OLE PCR product used to create the deletion was generated using primers XmaIVapC6-F and VapC6delOL-R. A 427-bp amplicon located 3′ of the deletion was produced using primers VapC6delOL-F and SbfIVapC6-R. OLE PCR produced a 934-bp product that was digested with XmaI and SbfI and cloned into plasmid pPB1035, resulting in plasmid pPB1094, used subsequently to produce strain PBL2078. The 255-bp deletion in vapC6 begins 36 bp downstream from the vapC6 start codon and ends 30 bp upstream of the vapC6 stop codon, leaving a 66-nt vapC6 gene. Presence of the vapC6 in-frame deletion was confirmed by DNA sequence of PCR amplicons using forward primer XmaIVapC6-F and reverse primer SbfIVapC6-R for the wild-type (1189 bp) and mutant alleles (934 bp).
Maltose-based selections using malA
PBL2069 was derived from strain PBL2027 (Schelert et al. 2006), which carried a spontaneous chromosomal deletion from SSO3017 through SSO3052 that removed lacS (SSO3019), xylS (SSO3022), and malA (SSO3051). While a malA-deficient strain (malA::lacS, PBL2009) exhibited normal growth rates on maltose as the sole carbon and energy source, simultaneous loss of SSO3022 reduced the growth rate by only one-third. Therefore, SSO0990 was inactivated, creating strain PBL2069 by in-frame deletion, resulting in loss of positions 691–1854 of the open reading frame. This region was removed by markerless exchange (Schelert et al. 2006) following recombination with an overlap PCR amplicon consisting of two paired segments, generated using primers 0990-BamHI-F and 0990-Del-R and primers 0990-Del-F and 0990-BamHI-R. PBL2069 could not grow on maltose as the sole carbon and energy source.
Strain PBL2081, a vapB6::lacS-disrupted derivative of PBL2069, was created by linear DNA transformation of the vapB6::lacS disruption mutation amplified using genomic DNA from strain PBL2066 and primers Vap6B-LF and Vap6B-RR. PCR and DNA sequencing were used to confirm the vapB6 genotype of strain PBL2081. Complementation of PBL2081 was performed using a derivative of pJlacS (Berkner et al. 2007), in which lacS had been replaced with malA, including its promoter and terminator at the SacII site; vapBC6 was inserted at the NotI site using primers NotIVapB6out-F and NotIVapC6out-R to make plasmid pPB1096. To maintain the plasmid in strain PBL2081, cells were grown in 0.2% maltose minimal media prior to heat-shock analysis. α-Glucosidase assays were conducted as described (Rolfsmeier and Blum 1995) using the fluorescent maltose analog, p-nitrophenylglucoside (10 mM) as substrate in 100 mM sodium acetate (pH 4.5) buffer at 80°C. All samples were assayed in duplicate, and the averages of the sample results are shown.
Heat-shock analysis
Heat-shock experiments designed to determine heat inactivation rates for S. solfataricus used 50-mL cultures of rich medium grown to an optical density at λ540 of 0.1 at 80°C, which is equivalent to 1.0 × 108 cells/mL. To minimize gas exchange, flask caps were lined with neoprene seals (0.66-mm thick). Heat shock was initiated by transferring flasks to a second orbital-shaking glycerol bath equilibrated at 95°C; flask contents required 20 min to reach the new temperature. Samples were removed periodically to determine cell density and viability. Viable counts were determined before and after heat shift by plating 10-fold serial dilutions of heated cells on rich medium plates, followed by incubation at 80°C. Heat inactivation rates were determined in replicate by comparing the number of surviving colonies normalized to those present in pre-heat-shock samples. Cell lysis was evaluated by determining relative total cell counts over the course of the experiments.
qRT–PCR analysis
Quantitative reverse transcription-PCR (RT–PCR) using SYBR-I Green and a real-time PCR instrument (Eppendorf Mastercycler) was performed as described by the manufacturer or, as described previously (Bradford et al. 2005), using a variable range of PCR cycles (Nakayama et al. 1992; Marone et al. 2001) with RNA prepared according to methods described previously (Haseltine et al. 1999; Bini et al. 2002). The exponential range of PCR product abundance (Noonan et al. 1990) was determined for all targets, and product qualities were verified by examination of melting curves. Parallel RT–PCR amplifications were used to evaluate RNA levels from experimental genes relative to those of the reference gene, 7S RNA, as described elsewhere (Lesur and Campbell 2004). RNA was treated to remove DNA by addition of 1 U of DNAse I (Fermentas) per microgram of total RNA at RT for 15 min and then neutralized with 2 μL of 25 mM EDTA and incubated at 70°C for 10 min. cDNA synthesis used 20 pmoles of PCR antisense primer, 20 mM dNTPs mix (Invitrogen), and 200 U of M-MuLV RT (Fermentas) for 60 min at 37°C. Synthesized cDNA was subjected to standard PCR and analyzed using 2% (w/v) TBE agarose gels.
Recombinant Vap protein synthesis and purification
Recombinant plasmids were constructed using E. coli DH5α, and characterized by restriction analysis and DNA sequencing, then transferred into BL21 (DE3) for expression analysis. VapC6 and VapB6 C-terminal polyhistidine-tagged expression plasmids were constructed by cloning vapC6 or vapB6 at the NdeI and XhoI sites of pET21b (Novagen), using primers SSO1493-F and SSO1493-R or SSO1494-F and SSO1494-R, respectively. Expression of these plasmids was evaluated in E. coli strain BL21(DE3), with or without plasmid pBN19. Plasmid pBN19 was a derivative of pBN13 (Blum et al. 1992a) in which a 2.4-Kb MscI–ScaI fragment encoding ColEI and part of bla was replaced with a 2.3-Kb XmnI–MscI fragment from pACYC184 encoding p15A and tet, thereby conferring compatibility with pET21b. To obtain the Vap proteins, recombinant E. coli strains pregrown in Luria Broth medium with ampicillin (100 μg/mL) and tetracycline (10 μg/mL) were stored at 4°C overnight and used as inocula for large-scale batch cultures. Induction of recombinant proteins used 1-L LB cultures with antibiotics grown to an OD600 of 0.1 and treated with 0.1 mM IPTG, followed by incubation at room temperature overnight as indicated previously (Blum et al. 1992a,b). Cells were recovered by centrifugation at 4000g, resuspended in Buffer A (50 mM Tris-Hcl at pH 7.8, 500 mM NaCl, 10% glycerol, 20 mM imidazole) at 50 OD600 U/mL, sonicated (80W x 30 sec × 10 times), and clarified by centrifugation (34,220g for 20 min). The supernatant was then fractionated by Ni-NTA chromatography. The sample was concentrated sixfold using centrifugal columns (Millipore, 3500 MWCO) and examined by SDS PAGE using 16% resolving gel and 5% stacking gel. Protein identities were determined by tandem mass spectrometry electrospray analysis using material excised from SDS–polyacrylamide gels (Worthington et al. 2003b; Schelert 2006). Gel slices were infused with trypsin to digest the protein, and the resulting peptides were recovered and separated by capillary electrophoresis prior to analysis. To assess the extent of contamination by E. coli ribonuclease, mock samples were prepared from induced cultures of a strain lacking vap expression plasmids, and these were processed exactly as for recombinant protein samples, including heat fractionation and Ni-NTA chromatography followed by centrifugal concentration.
Ribonuclease and deoxyribonuclease assays
For VapC6 assays using total cellular RNA as a substrate, S. solfataricus RNA was prepared as described previously (Bini et al. 2002). Four micrograms of S. solfataricus total RNA or runoff RNAs was used as substrate for all ribonuclease assays unless otherwise specified. Ribonuclease A (Sigma) was used as a positive control for the assay in amounts of 0.0246 U. The reactions were carried out in 10 mM MgCl2, 150 mM NaCl, 50 mM Tris-HCl at pH 6.0 at 80°C for 1 h. A total of 250 mM KCl was also added to each reaction to ensure stability of the RNA substrate at high temperature as described (Hethke et al. 1999; Bini et al. 2002). All recombinant and mock samples were dialyzed against 50 mM Tris-HCl (pH 6.0), 100 mM NaCl before assay. Reactions were initiated by addition of RNA substrate to preincubated components and then incubated for 1 h at the temperatures indicated. Protein concentrations of Ni-column-purified and dialyzed samples were VapC6 (0.75 mg/mL), VapB6 (0.61mg/mL). VapB6 and C6 proteins were incubated first at 95°C for 10 min separately, then various volumetric ratios as indicated were combined and incubated at 80°C for 2 h prior to RNA substrate addition. Reactions were fractionated by gel electrophoresis using 0.8% TBE agarose submerged gels and visualized by Ethidium Bromide staining with a UVP EC3 4210 Imaging System.
DNAse activity was assessed using VapC6 and three different DNA substrates, including (1) a circular dsDNA comprised of plasmid pUC19 and a 4.0-kb S. solfataricus insert consisting of a disruption of SSO2651 by SSO0615–0616; (2) linear dsDNA consisting of a PCR amplicon of the pUC19 insert, and (3) ssDNA generated from the PCR amplicon denatured by heating at 95°C followed by transfer to the 80°C reaction mixture. S. solfataricus total RNA was used as a positive control for enzyme activity. Nuclease assays were carried out in 10 mM MgCl2, 150 mM NaCl, 250 mM KCl, 50 mM Tris-HCl (pH 6.0) at 80°C for 1 h. Reactions were initiated by addition of 1 μL of VapC6 (0.75mg/mL) to reactions containing substrates (1 μg of DNA or 2 μg of RNA) and then incubated for 1 h. Reactions were fractionated by gel electrophoresis using 0.8% TBE submerged gels and visualized by Ethidium Bromide staining with a UVP EC3 4210 Imaging System.
Runoff RNAs for VapC6 assays used specific DNA templates prepared as described (Hoang et al. 2004). The selected genes were cloned into plasmid pT7T3/18U at HindIII and BamHI sites. Runoff RNA lengths (indicated in parentheses) and the following primers were used: SSO1274F-HindIII and SSO1274R-BamHI (1000 nt), SSO2506F-HindIII and SSO2506R-BamHI (597 nt), SSO1494F-HindIII and SSO1494R-BamHI (303 nt), and SSO7Sr-F-HindIII and SSO7Sr-R-BamHI (366 nt). Plasmids were linearized by digestion with EcoRI and gel extracted following electrophoresis using (Qiagen Gel Extraction Kit). In-vitro transcription used phage T3 RNA polymerase (Promega) and DNA templates were removed using DNase I without subsequent inactivation followed by RNA purification.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article (Table 3: Transcriptomic effects of VapBC6 deficiency; Table 4: List of Primers).
ACKNOWLEDGMENTS
We thank Georg Lipps for plasmid pJlacS. This research was supported in part by grants from NIH (R01GM090209) and DOD (HDTRA1-09-1-0030) to P.B. and from NSF (CBET-0730091) to R.M.K. C.C. acknowledges support from a T32 NIH Biotechnology Traineeship.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2679911.
REFERENCES
- Allen MB 1959. Studies with Cyanidium caldarium, an anomalously pigmented chlorophyte. Arch Mikrobiol 32: 270–277 [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L 2003. New connections in the prokaryotic toxin-antitoxin network: relationship with the eukaryotic nonsense-mediated RNA decay system. Genome Biol 4: R81 doi: 10.1186/gb-2003-4-12-r81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcus VL, Backbro K, Roos A, Daniel EL, Baker EN 2004. Distant structural homology leads to the functional characterization of an archaeal PIN domain as an exonuclease. J Biol Chem 279: 16471–16478 [DOI] [PubMed] [Google Scholar]
- Bates PA, Sternberg MJ 1999. Model building by comparison at CASP3: using expert knowledge and computer automation. Proteins 37 Suppl 3: 47–54 [DOI] [PubMed] [Google Scholar]
- Bates PA, Kelley LA, MacCallum RM, Sternberg MJE 2001. Enhancement of protein modelling by human intervention in applying the automatic programs 3D-JIGSAW and 3D-PSSM. Protein Struct Funct Genet (Suppl) 5: 39–46 [DOI] [PubMed] [Google Scholar]
- Berkner S, Grogan D, Albers SV, Lipps G 2007. Small multicopy, non-integrative shuttle vectors based on the plasmid pRN1 for Sulfolobus acidocaldarius and Sulfolobus solfataricus, model organisms of the (cren-)archaea. Nucleic Acids Res 35: e88 doi: 10.1093/nar/gkm449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagwat M, Meara D, Nossal NG 1997. Identification of residues of T4 RNase H required for catalysis and DNA binding. J Biol Chem 272: 28531–28538 [DOI] [PubMed] [Google Scholar]
- Bini E, Dikshit V, Dirksen K, Drozda M, Blum P 2002. Stability of mRNA in the hyperthermophilic archaeon Sulfolobus solfataricus. RNA 8: 1129–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum P, Ory J, Bauernfeind J, Krska J 1992a. Physiological consequences of DnaK and DnaJ overproduction in Escherichia coli. J Bacteriol 174: 7436–7444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum P, Velligan M, Lin N, Matin A 1992b. DnaK-mediated alterations in human growth hormone protein inclusion bodies. Biotechnology 10: 301–304 [DOI] [PubMed] [Google Scholar]
- Bodogai M, Ferenczi S, Bashtovyy D, Miclea P, Papp P, Dusha I 2006. The ntrPR operon of Sinorhizobium meliloti is organized and functions as a toxin-antitoxin module. Mol Plant Microbe Interact 19: 811–822 [DOI] [PubMed] [Google Scholar]
- Bradford WD, Cahoon L, Freel SR, Hoopes LL, Eckdahl TT 2005. An inexpensive gel electrophoresis-based polymerase chain reaction method for quantifying mRNA levels. Cell Biol Educ 4: 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunker RD, McKenzie JL, Baker EN, Arcus VL 2008. Crystal structure of PAE0151 from Pyrobaculum aerophilum, a PIN-domain (VapC) protein from a toxin-antitoxin operon. Proteins 72: 510–518 [DOI] [PubMed] [Google Scholar]
- Cerchia L, Rossi M, Guagliardi A 2000. An archaeal chaperonin-based reactor for renaturation of denatured proteins. Extremophiles 4: 1–7 [DOI] [PubMed] [Google Scholar]
- Chang AC, Cohen SN 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol 134: 1141–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen-Dalsgaard M, Gerdes K 2006. Two higBA loci in the Vibrio cholerae superintegron encode mRNA cleaving enzymes and can stabilize plasmids. Mol Microbiol 62: 397–411 [DOI] [PubMed] [Google Scholar]
- Cooper CR, Daugherty AJ, Tachdjian S, Blum PH, Kelly RM 2009. Role of vapBC toxin-antitoxin loci in the thermal stress response of Sulfolobus solfataricus. Biochem Soc Trans 37: 123–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daines DA, Wu MH, Yuan SY 2007. VapC-1 of nontypeable Haemophilus influenzae is a ribonuclease. J Bacteriol 189: 5041–5048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K 2000. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J Bacteriol 182: 561–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Christensen SK, Lobner-Olesen A 2005. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol 3: 371–382 [DOI] [PubMed] [Google Scholar]
- Haseltine C, Montalvo-Rodriguez R, Bini E, Carl A, Blum P 1999. Coordinate transcriptional control in the hyperthermophilic archaeon Sulfolobus solfataricus. J Bacteriol 181: 3920–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hethke C, Bergerat A, Hausner W, Forterre P, Thomm M 1999. Cell-free transcription at 95 degrees: thermostability of transcriptional components and DNA topology requirements of Pyrococcus transcription. Genetics 152: 1325–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang V, Bini E, Dixit V, Drozda M, Blum P 2004. The role of cis-acting sequences governing catabolite repression control of lacS expression in the archaeon Sulfolobus solfataricus. Genetics 167: 1563–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper S, Wilbur JS, Vasquez BL, Larson J, Clary S, Mehr IJ, Seifert HS, So M 2000. Isolation of Neisseria gonorrhoeae mutants that show enhanced trafficking across polarized T84 epithelial monolayers. Infect Immun 68: 896–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Gerdes K 1995. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol Microbiol 17: 205–210 [DOI] [PubMed] [Google Scholar]
- Jorgensen MG, Pandey DP, Jaskolska M, Gerdes K 2009. HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J Bacteriol 191: 1191–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Park JT, Kim YW, Lee HS, Nyawira R, Shin HS, Park CS, Yoo SH, Kim YR, Moon TW, et al. 2004. Properties of a novel thermostable glucoamylase from the hyperthermophilic archaeon Sulfolobus solfataricus in relation to starch processing. Appl Environ Microbiol 70: 3933–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesur I, Campbell JL 2004. The transcriptome of prematurely aging yeast cells is similar to that of telomerase-deficient cells. Mol Biol Cell 15: 1297–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Vierke G, Wenke AK, Thomm M, Ladenstein R 2007. Crystal structure of the archaeal heat shock regulator from Pyrococcus furiosus: a molecular chimera representing eukaryal and bacterial features. J Mol Biol 369: 474–488 [DOI] [PubMed] [Google Scholar]
- Maaty WS, Wiedenheft B, Tarlykov P, Schaff N, Heinemann J, Robison-Cox J, Valenzuela J, Dougherty A, Blum P, Lawrence CM, et al. 2009. Something old, something new, something borrowed; how the thermoacidophilic archaeon Sulfolobus solfataricus responds to oxidative stress. PLoS ONE 4: e6964 doi: 10.1371/journal.pone.0006964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madl T, Van Melderen L, Mine N, Respondek M, Oberer M, Keller W, Khatai L, Zangger K 2006. Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA. J Mol Biol 364: 170–185 [DOI] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Koonin EV 2009. Comprehensive comparative-genomic analysis of Type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol Direct 4: 19 doi: 10.1186/1745-6150-4-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marone M, Mozzetti S, De Ritis D, Pierelli L, Scambia G 2001. Semiquantitative RT-PCR analysis to assess the expression levels of multiple transcripts from the same sample. Biol Proced Online 3: 19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martusewitsch E, Sensen CW, Schleper C 2000. High spontaneous mutation rate in the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by transposable elements. J Bacteriol 182: 2574–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison K, Wilbur JS, So M, Brennan RG 2006. Structure of FitAB from Neisseria gonorrhoeae bound to DNA reveals a tetramer of toxin-antitoxin heterodimers containing pin domains and ribbon-helix-helix motifs. J Biol Chem 281: 37942–37951 [DOI] [PubMed] [Google Scholar]
- Miallau L, Faller M, Chiang J, Arbing M, Guo F, Cascio D, Eisenberg D 2009. Structure and proposed activity of a member of the VapBC family of toxin-antitoxin systems. VapBC-5 from Mycobacterium tuberculosis. J Biol Chem 284: 276–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama H, Yokoi H, Fujita J 1992. Quantification of mRNA by non-radioactive RT-PCR and CCD imaging system. Nucleic Acids Res 20: 4939 doi: 10.1093/nar/20.18.4939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan KE, Beck C, Holzmayer TA, Chin JE, Wunder JS, Andrulis IL, Gazdar AF, Willman CL, Griffith B, Von Hoff DD, et al. 1990. Quantitative analysis of MDR1 (multidrug resistance) gene expression in human tumors by polymerase chain reaction. Proc Natl Acad Sci 87: 7160–7164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey DP, Gerdes K 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res 33: 966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson J, McKenzie JL, Cursons R, Cook GM, Arcus VL 2009. The vapBC operon from Mycobacterium smegmatis is an autoregulated toxin-antitoxin module that controls growth via inhibition of translation. J Mol Biol 390: 353–367 [DOI] [PubMed] [Google Scholar]
- Rodriguez F, Arsene-Ploetze F, Rist W, Rudiger S, Schneider-Mergener J, Mayer MP, Bukau B 2008. Molecular basis for regulation of the heat shock transcription factor σ32 by the DnaK and DnaJ chaperones. Mol Cell 32: 347–358 [DOI] [PubMed] [Google Scholar]
- Rolfsmeier M, Blum P 1995. Purification and characterization of a maltase from the extremely thermophilic crenarchaeote Sulfolobus solfataricus. J Bacteriol 177: 482–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfsmeier M, Haseltine C, Bini E, Clark A, Blum P 1998. Molecular characterization of the α-glucosidase gene (malA) from the hyperthermophilic archaeon Sulfolobus solfataricus. J Bacteriol 180: 1287–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelert J 2006. Mercury resistance in Sulfolobus solfataricus. In Microbiology. University of Nebraska-Lincoln, Lincoln, NE [Google Scholar]
- Schelert J, Dixit V, Hoang V, Simbahan J, Drozda M, Blum P 2004. Occurrence and characterization of mercury resistance in the hyperthermophilic archaeon Sulfolobus solfataricus by use of gene disruption. J Bacteriol 186: 427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelert J, Drozda M, Dixit V, Dillman A, Blum P 2006. Regulation of mercury resistance in the crenarchaeote Sulfolobus solfataricus. J Bacteriol 188: 7141–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J, Brutlag D 2004. FoldMiner and LOCK 2: protein structure comparison and motif discovery on the web. Nucleic Acids Res 32: W536–W541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CC-Y, Clausen Ib G, Curtis BA, De Moors A, et al. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci 98: 7835–7840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers KR, Blum PH, DasSharma S 2006. Gene transfer in archaea. In Methods for general and molecular microbiology (ed. Reddy C). ASM Press, Washington, DC [Google Scholar]
- Tachdjian S, Kelly RM 2006. Dynamic metabolic adjustments and genome plasticity are implicated in the heat shock response of the extremely thermoacidophilic archaeon Sulfolobus solfataricus. J Bacteriol 188: 4553–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DK, Palmer JR, Daniels CJ 1999. Expression and heat-responsive regulation of a TFIIB homologue from the archaeon Haloferax volcanii. Mol Microbiol 33: 1081–1092 [DOI] [PubMed] [Google Scholar]
- Trent JD, Osipiuk J, Pinkau T 1990. Acquired thermotolerance and heat shock in the extremely thermophilic archaebacterium Sulfolobus sp. strain B12. J Bacteriol 172: 1478–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent JD, Nimmesgern E, Wall JS, Hartl FU, Horwich AL 1991. A molecular chaperone from a thermophilic archaebacterium is related to the eukaryotic protein t-complex polypeptide-1. Nature 354: 490–493 [DOI] [PubMed] [Google Scholar]
- Vierke G, Engelmann A, Hebbeln C, Thomm M 2003. A novel archaeal transcriptional regulator of heat shock response. J Biol Chem 278: 18–26 [DOI] [PubMed] [Google Scholar]
- Vuister GW, Kim SJ, Orosz A, Marquardt J, Wu C, Bax A 1994. Solution structure of the DNA-binding domain of Drosophila heat shock transcription factor. Nat Struct Biol 1: 605–614 [PubMed] [Google Scholar]
- Wagner MA, Zahrl D, Rieser G, Koraimann G 2009. Growth phase- and cell division-dependent activation and inactivation of the {σ}32 regulon in Escherichia coli. J Bacteriol 191: 1695–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerheide SD, Anckar J, Stevens SM Jr, Sistonen L, Morimoto RI 2009. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science 323: 1063–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington P, Blum P, Perez-Pomares F, Elthon T 2003a. Large-scale cultivation of acidophilic hyperthermophiles for recovery of secreted proteins. Appl Environ Microbiol 69: 252–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington P, Hoang V, Perez-Pomares F, Blum P 2003b. Targeted disruption of the α-amylase gene in the hyperthermophilic archaeon Sulfolobus solfataricus. J Bacteriol 185: 482–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel O, Sapra R, Chen F, Zhu Y, Simmons BA, Sorek R 2010. A single-base resolution map of an archaeal transcriptome. Genome Res 20: 133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N, Maeda Y, Ikeda A, Sakurai H 2008. Regulation of thermotolerance by stress-induced transcription factors in Saccharomyces cerevisiae. Eukaryot Cell 7: 783–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]