Abstract
PIWI-interacting RNAs (piRNAs) are 23–30-nucleotide-long small RNAs that act as sequence-specific silencers of transposable elements in animal gonads. In flies, genetics and deep sequencing data have led to a hypothesis for piRNA biogenesis called the ping-pong cycle, where antisense primary piRNAs initiate an amplification loop to generate sense secondary piRNAs. However, to date, the process of the ping-pong cycle has never been monitored at work. Here, by large-scale profiling of piRNAs from silkworm ovary and embryos of different developmental stages, we demonstrate that maternally inherited antisense-biased piRNAs trigger acute amplification of secondary sense piRNA production in zygotes, at a time coinciding with zygotic transcription of sense transposon mRNAs. These results provide on-site evidence for the ping-pong cycle.
Keywords: silkworm, piRNA, transposon, ping-pong cycle
INTRODUCTION
Controlling transposon activity is essential for host genomes especially in germ line cells, as an unfavorable mutation caused by transposition can be transmitted to the next generation (Malone and Hannon 2009). To prevent this, organisms have evolved an elegant defense system against transposons (Malone and Hannon 2009). In animal germ lines, PIWI proteins and associated PIWI-interacting RNAs (piRNAs) are at the center of this defense system. piRNAs are 23–30-nucleotide-long small RNAs that act as guides for PIWI proteins which exhibit slicer activity (Saito et al. 2006; Gunawardane et al. 2007; Nishida et al. 2007; Thomson and Lin 2009). Mutations in the piRNA pathway genes cause transposon derepression and defects in germ line development (Klattenhoff and Theurkauf 2008).
In contrast to small interfering RNAs (siRNAs) and microRNAs (miRNAs), piRNA production does not require Dicer proteins, suggesting that yet-unidentified piRNA precursors may not be stable double-stranded (Vagin et al. 2006; Houwing et al. 2007). Instead, piRNA biogenesis seems to be dependent on slicer activity of PIWI proteins (Brennecke et al. 2007; Gunawardane et al. 2007). The fly genome encodes three PIWI proteins, Piwi, Aubergine (Aub), and Argonaute3 (Ago3). Brennecke et al. and Gunawardane et al. discovered that Aub- and Piwi-bound antisense piRNAs often begin with uridine at their 5′ ends (1U), while Ago3-bound sense piRNAs are enriched for adenosine at position 10 (10A). Importantly, Aub-bound antisense and Ago3-bound sense piRNAs often overlap precisely by 10 nucleotides from their 5′ ends. Moreover, all fly PIWI proteins showed slicer activity (Saito et al. 2006; Gunawardane et al. 2007; Nishida et al. 2007). As PIWIs cleave their targets between position 10 and 11 (Saito et al. 2006; Gunawardane et al. 2007), the 10-nt overlaps between sense and antisense piRNAs could be explained by PIWI protein-mediated cleavage. Based on these data, the current model called the ping-pong cycle describes that Aub-bound antisense “primary” piRNAs define 5′ ends of Ago3-bound sense “secondary” piRNAs, which in turn create 5′ ends of Aub-bound antisense “secondary” antisense piRNAs (Brennecke et al. 2007; Gunawardane et al. 2007). Supporting this model, loss of ago3 dramatically reduced not only sense piRNAs but also Aub-bound antisense piRNAs (Li et al. 2009a). The 10-nt overlaps, often called ping-pong signatures, are conserved across phyla, including mice, zebrafish, and silkworm (Aravin et al. 2008; Houwing et al. 2008; Kawaoka et al. 2009). However, to date, biochemical evidence for the ping-pong model has been lacking, and the process of piRNA amplification has never been monitored at work.
Silkworm is an emerging model to study piRNAs. Silkworm ovary and ovary-derived cell line BmN4 express two PIWI proteins and piRNAs with significant ping-pong signatures (Kawaoka et al. 2008a,b, 2009). In addition, silkworm embryos provide an ideal system to monitor developmental piRNA profiles. After mating, a female moth continuously lays approximately three hundred eggs within 3–4 hours. Fertilization occurs just before laying. This allowed us to collect highly developmentally synchronized egg samples. Here, taking advantage of these points, we analyzed piRNAs prepared from pharate ovary and developing embryos. We, for the first time, visualized developmental, ping-pong-dependent piRNA amplification that was coupled with zygotic transcription of sense transposon transcripts. Our study provides on-site evidence for the ping-pong model.
RESULTS
Maternal deposition of PIWI/piRNA complexes in silkworm embryos
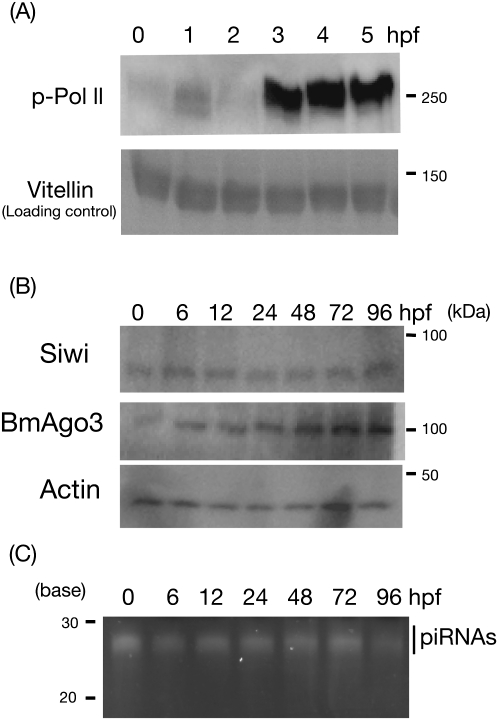
After fertilization, an animal embryo at the one-cell stage is transcriptionally inactive, and early embryogenesis requires maternal factors (Schier 2007). At a distinct point of embryogenesis, the embryo initiates zygotic transcription (Schier 2007). This transition is called maternal to zygote transition (MZT). Transcriptional states of embryos can be monitored by phosphorylation of RNA polymerase II (Vastenhouw et al. 2010). To characterize the silkworm embryogenesis, we performed Western blots with anti-phosphorylated RNA polymerase II (pol II) antibody (Vastenhouw et al. 2010). Phosphorylated pol II was detected from 3–4 h post-fertilization (hpf), suggesting that MZT occurs at this stage in the silkworm (Fig. 1A).
FIGURE 1.
Maternal to zygote transition and maternal deposition of PIWI/piRNA complexes in the silkworm embryos. (A) Proteins from eggs of 0, 1, 2, 3, 4, and 5 hpf were analyzed with anti-phosphorylated RNA polymerase II antibody. Vitellin, a major egg protein served as a loading control. (B) Proteins from eggs of 0, 6, 12, 24, 48, 72, and 96 h post-fertilization (hpf) were analyzed by Western blotting with anti-Siwi, anti-BmAgo3, and anti-actin antibodies. Actin served as a loading control. (C) 10 μg of total RNAs from 0-, 6-, 12-, 24-, 48-, 72-, and 96-hpf eggs were analyzed by urea-containing acrylamide gel. Signals were visualized by SYBRGold staining.
Next, we asked whether PIWI/piRNA complexes are maternally inherited in silkworm embryos. We revealed by Western blotting that Siwi and BmAgo3, two silkworm PIWI proteins (Kawaoka et al. 2008b) were detected before zygotic activation (Fig. 1B). Moreover, piRNAs were abundantly detected in 0–1-hpf embryos (Fig. 1C). These results proved maternal deposition of PIWI/piRNA complexes in the silkworm. Expressions of PIWI/piRNA complexes lasted as long as we surveyed during embryogenesis (Fig. 1C).
Features of maternally inherited piRNAs
Fly adult ovaries contain gonadal germ line cells and somatic support cells (Malone et al. 2009). Somatic follicle epithelium is shed from the laid egg. By comparing ovarian and early embryonic piRNA libraries, Malone et al. identified two distinct pools of piRNAs—soma-dominant piRNAs, and germ line-enriched, maternally inherited piRNAs (Malone et al. 2009). In the silkworm, ovarian development occurs exclusively during pupation and pharate adult development before hatching (Yamauchi and Yoshitake 1984). The structure of pharate adult ovary is nearly identical to that of flies (Yamauchi and Yoshitake 1984). In contrast, the nature of the silkworm adult ovary is different from the fly adult ovary, as the silkworm adult ovary contains only matured eggs, which are mainly composed of germ line cells.
To compare the characters of both germ line (maternal) and somatic piRNAs with those in flies, we sequenced and analyzed piRNAs from pharate ovary and early embryos. Strong maternal deposition indicates that a piRNA is enriched in germ line, whereas soma-dominant piRNAs show weak maternal deposition.
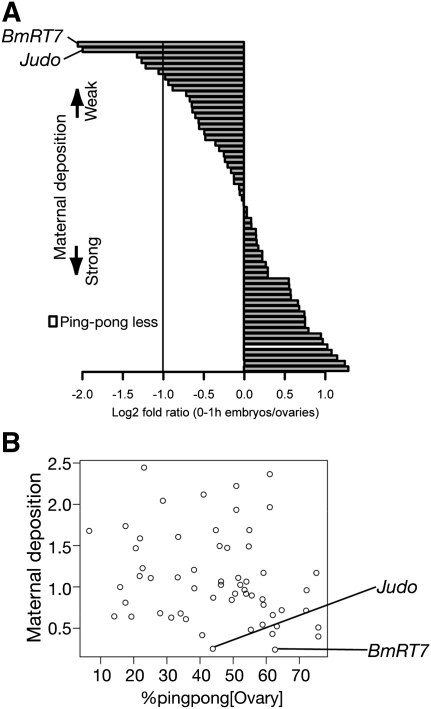
We used 60 elements among 121 annotated transposons that are most heavily targeted by ovarian piRNAs (>500 reads per million [RPM]) (Fig. 2A). We calculated the maternal deposition and found that 54 of 60 transposon-derived piRNAs were effectively maternally deposited, indicating that these piRNAs were enriched in germ line. On the other hand, six elements exhibited weak maternal deposition, suggesting that these are expressed predominantly in gonadal soma.
FIGURE 2.
Maternal piRNAs and ping-pong signatures in silkworm embryos. (A) Log2 fold ratio between normalized ovarian and 0 hpf-derived piRNA reads against 60 well annotated transposons. An element lacking statistically significant ping-pong signature is represented by white bar. (B) Relationships between maternal deposition and % ping-pong in pharate ovary are displayed as a scatter plot. Maternal deposition is defined as normalized early (0 hpf) embryonic reads/normalized ovarian piRNA reads. % ping-pong is expressed as the likelihood for the average piRNA mapping to an individual transposon with ping-pong partners.
The fly somatic piRNA pool lacks ping-pong signatures, suggesting that the ping-pong cycle may operate mainly in gonadal germ line cells (Lau et al. 2009; Li et al. 2009a; Malone et al. 2009; Saito et al. 2009; Haase et al. 2010; Olivieri et al. 2010; Saito et al. 2010). To investigate relationships between maternal deposition and the ping-pong cycle in the silkworm, we calculated the statistical significance of a ping-pong signature for individual transposons (see Materials and Methods) (Fig. 2A; Supplemental Table S1). Within the selected 60 transposons, only R2Bm lacks a statistically significant ping-pong signature, but R2Bm-derived piRNAs were efficiently inherited to early embryos via germ line cells. On the other hand, although Judo-derived piRNAs harbored significant ping-pong signature in the ovary, they were soma-dominant. In fact, we observed no correlation between % ping-pong (Brennecke et al. 2008; Malone et al. 2009) (see Materials and Methods) and maternal deposition among 60 transposons (Fig. 2B). We also confirmed soma-dominant expressions of Judo and BmRT7 mRNAs by qRT-PCR (Supplemental Fig. S1). Collectively, as long as we surveyed based on two independent methods, the ping-pong cycle may operate both in gonadal soma and germ line cells in the silkworm.
Developmental profiling of the silkworm embryonic piRNAs
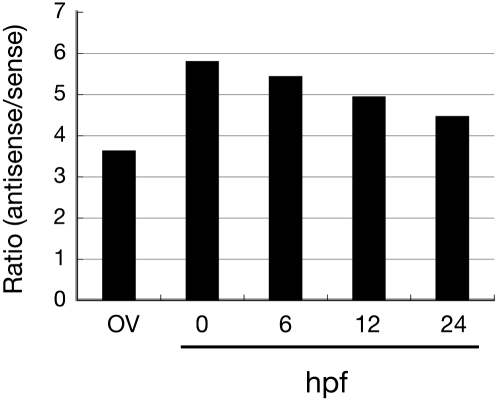
In the developmental context, how the ping-pong cycle initiates is currently unknown. The maternal piRNA pool can be defined as an input for ping-pong amplification. To obtain evidence for the initiation of zygotic piRNA biogenesis by maternal piRNAs, we analyzed piRNAs from developing embryos (0, 6, 12, and 24 hpf). We mapped cloned reads to 121 transposons and investigated strand bias (Fig. 3). As shown in Figure 3, early embryonic piRNAs were much more antisense-biased than ovarian piRNAs (sixfold vs. 3.5-fold), demonstrating that antisense piRNAs are more enriched in germ lines than in gonadal soma. Strong antisense bias slightly and gradually declined according to embryonic development (Fig. 3). Decrease of antisense piRNAs could account for strand bias changes. Alternatively, accumulation of sense piRNAs may explain why antisense strand bias reduced.
FIGURE 3.
Strand bias of ovarian and embryonic piRNAs. Strand bias of 121 transposon-derived piRNAs from pharate ovary, 0-, 6-, 12-, and 24-hpf embryos.
To test these possibilities, we constructed a heat map showing relative abundance of sense and antisense piRNAs on an element (piRNA expression in 0-hpf embryos = 1) (Fig. 4). Although MZT occurs at 3–4 hpf in the silkworm (Fig. 1A), overall piRNA profiles between 0 and 6 hpf were remarkably similar, indicating that MZT does not affect the piRNA biogenesis. Throughout embryonic development, expression levels of antisense piRNAs were comparable to 0-hpf embryos (Fig. 4A). In contrast, after 12 hpf, we found a significant increase of specific sets of sense piRNAs (Figs. 4B and 5).
FIGURE 4.
Developmental profiling of embryonic piRNAs. Relative expressions of antisense (A) and sense (B) piRNAs derived from 60 elements (0-hpf embryo = 1). Blue indicates relative expression (RE) meets 0.5 ≤ RE ≤ 2.0. Light blue corresponds to RE < 0.5, and red indicates RE > 2.
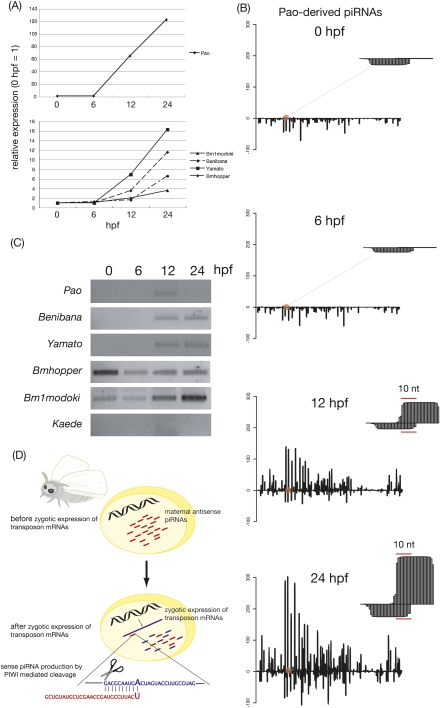
FIGURE 5.
Zygotic amplification of sense piRNAs by maternal antisense piRNAs. (A) Relative expressions of a set of piRNAs whose sense piRNA increase was most drastic. (B) Density plot of 0-, 6-, 12-, and 24-hpf piRNAs derived from Pao. Representative ping-pong site was enlarged. Plots were generated by base calling method. (C) Sense strand specific RT-PCR for Pao, Yamato, Bm1modoki, Benibana, and Kaede. (D) Model depicting zygotic piRNA amplification by maternal piRNAs in the silkworm.
Amplification of secondary piRNAs during embryogenesis
The most prominent piRNA change we observed during embryogenesis was the accumulation of a set of sense piRNAs at 12–24 hpf (Figs. 4B and 5A). We plotted expression levels of piRNAs derived from five transposons whose sense piRNA accumulations were most drastic (Fig. 5A). At 12 hpf, Pao-derived sense piRNAs rapidly increased (60-fold). The abundance of Pao-derived sense piRNAs peaked at 24 hpf by 120-fold. The same pattern was obtained for Yamato, Bmhopper, Benibana, and Bm1modoki.
For detailed analysis, we visualized developmental piRNA expression as a density plot (Fig. 5B). Pao-derived piRNAs showed a marked antisense bias at 0 hpf. The piRNA population did not change after MZT (6 hpf). At 12 hpf, we observed an increase in sense piRNAs, followed by a slight antisense piRNA increase. Strikingly, we noted that almost all amplification occurred at the ping-pong site (represented as enlarged pictures in Fig. 5B). Indeed, >80% of increasing sense piRNAs had their partner. Consistent with this, we observed an overall increase on 10A bias of sense piRNAs (Supplemental Fig. S2). These data indicated that slicer activity of PIWI proteins mediate selective amplifications of sense secondary piRNAs.
We hypothesized that abundance of sense piRNAs depends on zygotic expression of sense transposon mRNA, a theoretical target of maternal antisense piRNAs. To test this idea, we investigated the expression profiles of sense transposon mRNA by using strand- specific RT-PCR (Fig. 5C). In this experiment, we reverse-transcribed total RNAs with a specific primer for each transposon, rather than the oligo(dT) primer or random primer; therefore, RT efficiency is different among transposons. Thus, even though we used the same amount of total RNAs for input, we cannot directly compare expression levels among transposons. This is why we did not see a correlation between the amount of sense piRNAs produced from a specific transposon (Fig. 5A) and the expression level of corresponding transposon (Fig. 5C). Sense mRNAs of Pao, Yamato, and Benibana were not maternally deposited (Fig. 5C). These mRNAs were immediately transcribed at 12 hpf. Sense Pao mRNA then rapidly became undetected. Importantly, the timing of their zygotic expression was coupled with the timing of sense piRNA accumulation (Fig. 5C).
In contrast to Pao, Yamato, and Benibana, sense mRNAs of Bmhopper and Bm1modoki were detected as early as 0-hpf embryos and slightly decreased at 6 hpf, suggesting that these transposon mRNAs were maternally deposited and being slowly degraded. Although sense mRNAs of these two elements were present during this period, corresponding sense piRNAs remained unamplified in zygotes. At 12 hpf, Bmhopper and Bm1modoki increased again, most likely reflecting their zygotic transcription. In response to this, sense piRNAs started to accumulate (Fig. 5A). Sense transcription of Kaede, whose piRNA profile did not change during embryogenesis (Fig. 4), was not detected (Fig. 5C). Taken together, we envision that antisense maternal piRNAs kick-start ping-pong amplification of secondary sense piRNAs and that it is coupled with zygotic transcription of sense transposon mRNAs (Fig. 5D). Our data do not exclude the possibility that sense transposon mRNAs could be processed into zygotic primary sense piRNAs via a ping-pong-independent primary processing pathway, partially contributing zygotic sense piRNA accumulation.
DISCUSSION
Here, by using silkworm developing embryos as a model system, we monitored the developmental process of piRNA profiles. We found that many ovarian piRNAs are actually maternally inherited via germ line cells in early embryos (Figs. 1 and 2). In flies, piRNAs deriving from the flamenco locus are not effectively maternally deposited (Malone et al. 2009). Importantly, flamenco-derived piRNAs are depleted of ping-pong signatures (Li et al. 2009a; Malone et al. 2009). Thus there is a clear correlation between maternal deposition and ping-pong significance in flies. In contrast, in the silkworm, we observed no correlation between maternal deposition and ping-pong significance (Fig. 2; Supplemental Table S1). For instance, Judo-derived piRNAs are predominant in soma but retain the ping-pong signature, indicating that the ping-pong cycle may not be specific for germ line in the silkworm. Although we could not exclude the possibility that still unannotated transposable elements show features of flamenco-derived piRNAs, we suggest that the silkworm genome does not harbor a soma-specific piRNA (transposon) cluster such as flamenco.
As shown in Figures 3 and 4, maternally deposited piRNAs are mainly antisense, and their population is not affected by MZT and subsequent embryonic development (Figs. 3 and 4). The majority of maternally deposited piRNAs continued to express at almost the same level throughout embryogenesis (Fig. 4). We have previously found a rapid decrease of PIWI mRNAs at 20 hpf (Kawaoka et al. 2008b), whereas protein expressions did not alter until 96 hpf during embryogensis. Collectively, we suggest that PIWI/piRNA complexes are highly stable during embryogenesis.
In contrast to antisense piRNAs, most importantly, our analyses revealed an acute amplification of a specific set of secondary piRNAs, which is coupled with zygotic transcription of sense transposon mRNAs. The efficiency of sense piRNA accumulation is not simply dependent on the availability of sense transposon mRNAs; for particular transposons, although we detected maternally transmitted sense mRNAs at 0 to 6 hpf, corresponding sense piRNAs were not amplified until their zygotic transcription occurred at 12 hpf. Therefore, zygotes may detect de novo transcription of transposon mRNAs and trigger piRNA-mediated combat against them. Alternatively, a factor(s) required for sense piRNA accumulation (i.e., the ping-pong amplification) in zygotes may be simply missing in 0- to 6-hpf embryos.
Accumulating sense piRNAs overlapped by 10 nucleotides with maternal antisense primary piRNAs, suggesting that PIWI/antisense piRNA complexes cleaved sense transposon mRNAs to generate sense piRNAs (Fig. 5). Taken altogether, our current data provide direct evidence for ping-pong cycle-dependent “amplification” of piRNAs.
MATERIALS AND METHODS
Insect
Wild type (WT) strain p50T was reared on fresh mulberry leaves in an insect rearing chamber under short-day conditions (12L: 12D).
Western blotting
Lysate preparation and Western blotting were performed as described previously (Kawaoka et al. 2009). Antibodies used were anti-Siwi (1:10,000) and anti-BmAgo3 (1:5000) (13), anti-actin (1:1000; Santa Cruz), and anti-phosphorylated polymerase II (1:250; Santa Cruz).
piRNA library construction
Total RNA was prepared using Trizol reagent (Invitrogen) according to the manufacturer's protocol. The total RNA (10 μg) was loaded onto a 15% denaturing polyacrylamide gel containing 8 M urea, electrophoresed, and then stained with SYBRGold (Invitrogen). Signals were visualized using LAS-1000 film (Fujifilm). As silkworm piRNAs are visible as a distinct band by SYBRGold staining (see Fig.1B), we could easily gel-purify the piRNA-containing fraction. Small RNA libraries were constructed using a small RNA cloning kit (Takara). DNA sequencing was performed using the Solexa genetic analysis system (Illumina) (Kawaoka et al. 2009). One nanogram of the prepared cDNA was used for the sequencing reactions with the Illumina GA. 10,000–15,000 clusters were generated per “tile,” and 36 cycles of the sequencing reactions were performed. The protocols of the cluster generation and sequence reactions were according to the manufacturer's instructions.
Sequence analysis
Solexa sequencing generated reads of up to 36 nucleotides in length. The 3′ adaptor sequences were identified and removed, allowing for up to two mismatches. Reads without adaptor sequences were discarded. Reads shorter than 23 nucleotides or longer than 30 nucleotides were excluded, resulting in reads of 23–30 nucleotides. Alignment to the B. mori genome, 121 annotated transposons, and 1668 ReAS clones were performed with SOAP2 (ver. 2.20) allowing no mismatch (Li et al. 2009b). In this manuscript, we refer to piRNA as 23–30-nt RNAs matching transposable elements. The total number of perfect genome-mapping reads reflects the sequencing depth. To compare the reads among different data sets, reads were expressed in reads per million (RPM) by normalizing to the total number of perfect genome-mapping in each library.
Ping-pong signature
Ping-pong pairs were defined as precise 10-nt overlaps between sense and antisense piRNAs matching each of the 121 transposons. The observed abundance of ping-pong pairs (O) was defined as the sum of all the piRNA reads that joined in ping-pong pairs. For estimating the expectation (E), we counted the number of piRNA reads that form ping-pong pairs when we randomly map the n number of antisense piRNAs and the m number of sense piRNAs to each transposon. By computing this process 100,000 times for each transposon, we calculated the score X = (O – Ei) (i = 1, 2, …, 100,000). The P-value represents k/100,000, where k shows how many times X meets (O – Ei) < 0. The R-code for this analysis will be provided upon request. In addition, we evaluated ping-pong significance by calculating the likelihood for average piRNA mapping to an element to have a complementary ping-pong partner (% ping-pong) (Brennecke et al. 2008; Malone et al. 2009).
Strand specific RT-PCR
Total RNAs extracted from embryos were reverse transcribed using avian myeloblastosis virus (AMV) reverse transcriptase (TaKaRa) with strand-specific primers listed in Supplemental Table S2, and PCR was performed using KOD-plus polymerase (TOYOBO).
Data deposition
piRNAs sequenced in this study are deposited in DRA000173 and DRA000317 [DNA Data Bank of Japan (DDBJ)].
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank P.B. Kwak for critical comments on the manuscript, and M. Kawamoto and E. Sekimori for their technical assistance. Sh.K. is a recipient of fellowships from the Japan Society for the Promotion of Science. This work was supported in part by Special Coordination Funds for Promoting Science and Technology from the Ministry of Education, Culture, Sports, Science, and Technology of the Japanese Government (MEXT) (No. 22115502 to Su.K., No. 21710208 to K.K., No. 17018007 to T.S., and the Professional Program for Agricultural Bioinformatics), by the National Bio-Resource Project “Silkworm” of MEXT, and Grant-in-Aid for Scientific Research on Innovative Areas (“Functional machinery for non-coding RNAs”) to Su.K. and Y.T.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2709411.
REFERENCES
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ 2008. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322: 1387–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590 [DOI] [PubMed] [Google Scholar]
- Haase AD, Fenoglio S, Muerdter F, Guzzardo PM, Czech B, Pappin DJ, Chen C, Gordon A, Hannon GJ 2010. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev 24: 2499–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. 2007. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell 129: 69–82 [DOI] [PubMed] [Google Scholar]
- Houwing S, Berezikov E, Ketting RF 2008. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J 27: 2702–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka S, Hayashi N, Katsuma S, Kishino H, Kohara Y, Mita K, Shimada T 2008a. Bombyx small RNAs: genomic defense system against transposons in the silkworm, Bombyx mori. Insect Biochem Mol Biol 38: 1058–1065 [DOI] [PubMed] [Google Scholar]
- Kawaoka S, Minami K, Katsuma S, Mita K, Shimada T 2008b. Developmentally synchronized expression of two Bombyx mori Piwi subfamily genes, SIWI and BmAGO3 in germ-line cells. Biochem Biophys Res Commun 367: 755–760 [DOI] [PubMed] [Google Scholar]
- Kawaoka S, Hayashi N, Suzuki Y, Abe H, Sugano S, Tomari Y, Shimada T, Katsuma S 2009. The Bombyx ovary-derived cell line endogenously expresses PIWI/PIWI-interacting RNA complexes. RNA 15: 1258–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Theurkauf W 2008. Biogenesis and germline functions of piRNAs. Development 135: 3–9 [DOI] [PubMed] [Google Scholar]
- Lau NC, Robine N, Martin R, Chung WJ, Niki Y, Berezikov E, Lai EC 2009. Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res 19: 1776–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. 2009a. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137: 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J 2009b. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967 [DOI] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ 2009. Molecular evolution of piRNA and transposon control pathways in Drosophila. Cold Spring Harb Symp Quant Biol 74: 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, Saito K, Mori T, Kawamura Y, Nagami-Okada T, Inagaki S, Siomi H, Siomi MC 2007. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA 13: 1911–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J 2010. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J 29: 3301–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC 2006. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev 20: 2214–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC 2009. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461: 1296–1299 [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC 2010. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev 24: 2493–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier AF 2007. The maternal-zygotic transition: death and birth of RNAs. Science 316: 406–407 [DOI] [PubMed] [Google Scholar]
- Thomson T, Lin H 2009. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol 25: 355–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324 [DOI] [PubMed] [Google Scholar]
- Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF 2010. Chromatin signature of embryonic pluripotency is established during genome activation. Nature 464: 922–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi H, Yoshitake N 1984. Developmental stages of ovarian follicles of the silkworm, Bombyx mori L. J Morphol 179: 21–31 [DOI] [PubMed] [Google Scholar]