Abstract
Global species extinction typically represents the endpoint in a long sequence of population declines and local extinctions. In comparative studies of extinction risk of contemporary mammalian species, there appear to be some universal traits that may predispose taxa to an elevated risk of extinction. In local population-level studies, there are limited insights into the process of population decline and extinction. Moreover, there is still little appreciation of how local processes scale up to global patterns. Advancing the understanding of factors which predispose populations to rapid declines will benefit proactive conservation and may allow us to target at-risk populations as well as at-risk species. Here, we take mammalian population trend data from the largest repository of population abundance trends, and combine it with the PanTHERIA database on mammal traits to answer the question: what factors can be used to predict decline in mammalian abundance? We find in general that environmental variables are better determinants of cross-species population-level decline than intrinsic biological traits. For effective conservation, we must not only describe which species are at risk and why, but also prescribe ways to counteract this.
Keywords: anthropogenic threat, biodiversity decline, Living Planet Index, phylogenetic generalized linear model, population time series, species abundance
1. Introduction
More than a decade and a half on from May's [1] discussion of the issue of scale in ecology, a gap still exists in understanding how population processes scale up from local populations to the global species level. This line of research could be particularly illuminating for understanding extinction risk. There is currently a disconnection between discernment of what factors correlate with extinction risk at a species level, and those that predict risk at the population level where the majority of conservation action is focused. The link is important as the disappearance of a population is a prelude to species-level extinction, which makes population decline a far more sensitive indicator of the loss of biodiversity than species extinction [2,3]. Understanding how species-level extinction risk and population-level declines interrelate could help in the design of more proactive conservation actions. One of the first steps along this road is attempting to understand what predisposes populations to elevated risk of extinction.
Over the past decade and a half, phylogenetic comparative methods have provided researchers with a rigorous tool to explore and understand the underlying processes and patterns of contemporary species extinction [4]. Identifying the underlying causes of species decline and searching for correlates of extinction risk has proved useful in revealing general patterns across groups. Specifically, comparative analyses on mammalian species have shown some consistent predictors across mammalian orders [5–8], but mixed results under changing extinction pressures [9,10] (table 1).
Table 1.
Trait–risk relationship hypotheses tested in previous mammalian studies.
| trait | hypothesis | references | |
|---|---|---|---|
| intrinsic traits | body mass | larger bodied mammals are more likely to have an elevated risk of extinction as they are targeted for hunting and persecuted. In addition, larger species usually have lower reproductive rates and thus slower population growth, making them more sensitive to exploitation, and low population density | [6,7,11–16] |
| head body length | head body length has not been tested as a predictor of extinction in previous studies; however, it is strongly correlated with body mass | not tested previously | |
| inter-birth interval | species with slow life histories are less able to compensate for increased mortality through increased fecundity and are therefore more vulnerable to population extinction; a longer inter-birth interval is thus associated with an elevated extinction risk | [12,17] | |
| litter size | species with a low reproductive output, mediated by life-history attributes such as litter size, are more likely to be at greater risk of extinction | [5,7,11,18] | |
| age at first birth | the predictions regarding inter-birth interval above are also applicable here, as both are important components of reproductive rate; some studies have looked at the closely related age at sexual maturity | [7,11,14,19] | |
| geographical range size | small geographical range size is associated with elevated extinction risk because of its relationship with low population size and the concomitant risks from demographic and environmental stochasticity. A small geographical range indicates habitat specificity, i.e. small habitat breadth, which is likely to affect a species' ability to withstand habitat modification, and increases the risk that the entire species range is in the firing line | [6,7,11,12,14,17,19–21] | |
| home range size | larger home range size, which is a necessary product of larger body size, is also associated with higher extinction risk; this is because species where individuals have large home ranges, reflecting high resource demands, are particularly vulnerable to habitat loss and degradation and, in particular, to reserve edge effects in fragmented habitats; by contrast, small home ranges suggest lower individual energetic requirements and higher population densities, and thus a lower risk of extinction in low-productivity environments or remnant habitat patches | [5,12,14,22–24] | |
| extrinsic factors | average human population density | higher human population density within a species' range means more competition for resources, and more opportunity for conflict and exploitation, and thus a higher risk of extinction; in addition, habitat degradation, fragmentation and destruction are more likely to occur in densely populated locations | [6,14,20] |
| precipitation | precipitation, along with temperature, plays a complex role in its effect on mammal population sizes; in times of drought and in regions of low productivity or resource availability, populations may be highly variable, leaving them more vulnerable to extinction; precipitation is also related to latitude, and is likely to become an important factor in light of continuing climate change | [14,15] | |
| temperature | mean annual temperature is used as a measure of available ambient energy; temperature and precipitation are closely related, and predictions are as above | [25] | |
| AET | actual evapotranspiration (AET) reflects the joint availability of energy and water, and is regarded as an index of primary productivity; a low mean AET is therefore associated with an elevated risk of extinction | [14,19] | |
| PET | mean potential evapotranspiration rate (PET) is strongly influenced by temperature and reflects the potential availability of energy and water; a low PET is therefore associated with an elevated risk of extinction | not tested previously | |
| habitat breadth | narrow habitat breadth suggests specialization and therefore poor ecological adaptability and flexibility; specialists are more susceptible to habitat modification and loss, and thus at an increased risk of extinction; in addition, narrow habitat breadth is associated with a small geographical range, which can put an entire species at risk | [15,22,26,27] |
There are two non-mutually exclusive types of variables that may account for variation in risk among species and populations: intrinsic biological factors and extrinsic abiotic or geographical factors. One null model offered to describe extinction is that of a field of bullets [28], whereby in a metaphor drawn from trench warfare, extinction is a random process. Many studies have established that extinction is likely to be taxonomically non-random (e.g. [29–31]). What predisposes species and populations to extinction should therefore be explained by intrinsic biology (the ability of a species to withstand a threatening process), the severity of impact (mediated through geography) and the interaction between the two [32,33].
The vast majority of previous analyses of correlates of extinction in mammals have been conducted using global-level metrics that classify species into categorical classes of extinction risk, such as IUCN (International Union for Conservation of Nature) Red List status (e.g. [6,7]). Others have used national level equivalent classifications (e.g. National Red List Status [34]), or site-level measures of persistence (e.g. [22]). Species-level classifications such as the IUCN Red List are designed to capture multiple symptoms of threat (e.g. population size, trend, geographical range size; [35]) and integrate them into one level of risk [36]. However, the biological correlates and phylogenetic distribution of these symptoms of risk need not necessarily be the same [37]. To date, differences in the correlates of different measures of risk have been largely ignored (but see [37]); examining different metrics of extinction risk may be important.
A composite risk measure such as IUCN Red List category is uninformative about trends in status. To examine global correlates of decline, it is possible to examine a subset of species within a group that are categorically classed as declining (e.g. rapidly declining amphibians [38]), though in most groups measured to date, the number of species undergoing a category transition is rather few. Therefore, common species that are declining in abundance at a sub-threshold rate (and are therefore likely to be classed as Least Concern) would not be picked up in such analyses, a concern as decline in common species is becoming more prevalent (e.g. [39,40]). A more detailed pattern of trends in abundance allows the study of symptoms of risk across the Red List categories. Here, we combine two large datasets, one of mammalian biological trait information [41] and one of population trend data [3,42]; www.livingplanetindex.org) in a phylogenetic framework [43] to examine whether correlates of elevated species-level extinction risk in mammals are the same as those that predispose mammal populations to decline.
2. Methods
(a). Population data
Time-series trends for mammal species were collated from the Living Planet database ([3,42]; www.livingplanetindex.org). These data were collected from published scientific literature, online databases (e.g. [44,45]) and grey literature. Following a modified version of Collen et al. [3], data were included only if:
— a measure of change in population size was available for at least 10 years;
— information was available on how the data were collected and what the units of measurement were;
— the geographical location of the population was provided;
— the data were collected using the same method on the same population throughout the time series;
— the data source was referenced and traceable; and
— at least 50 per cent of data points for the species lie within the time period 1970 to 2005.
Ancillary information to the time-series data was collated at the population level. The specific location of each population was used to identify the region and biome which it occupies, and information from the original data source used to record the primary threat to that population.
(b). Mammalian life history and extrinsic data
We collated mammalian biological trait information from the PanTHERIA database [41]. From the 30 traits in this database, we refined our analyses to a set of 13 correlates of extinction risk (table 2) taken from a review of the published literature on mammalian extinction risk (table 1). Additionally, using the habitat classification scheme in the IUCN Red List of Threatened Species [46], a measure of habitat breadth was calculated as the sum of all the major habitat types occupied by each species. The complete dataset was partitioned on the basis of threatening process (habitat loss or over-exploitation). We recorded the threatening process from the original source of the population abundance data (see electronic supplementary material).
Table 2.
Number of species each correlative trait analysed.
| predictor variable | number of species |
|---|---|
| body mass | 279 |
| head–body length | 226 |
| inter-birth interval | 179 |
| litter size | 265 |
| geographical range area | 222 |
| age at first birth | 131 |
| home range | 147 |
| human population density | 222 |
| precipitation | 220 |
| temperature | 220 |
| AET | 219 |
| PET | 219 |
| habitat breadth | 291 |
(c). Spatial congruence of risk measures
We compiled species from the IUCN mammal species dataset ([47,48]; n = 5490 species) and filtered it to obtain species that had a geographical range map and non-extinct Red List status. During IUCN Red List assessments, a species-level population trend is recorded under the categories increasing, stable, decreasing and unknown. We subdivided the mammal ranges into these categories: increasing (n = 80), stable (n = 1330), decreasing (n = 1630) and unknown (n = 2356). We also extracted species categorized as threatened (n = 1143, i.e. all species classified as Vulnerable, VU; Endangered, EN and Critically Endangered, CR). We compared this global proxy of species population trend with data collated at a site-level from the population abundance database.
From the measure of change in population abundance collected from the Living Planet Index database, we classified a decline of −2 per cent or more as a decreasing trend (n = 72), an increase of +2 per cent or more as an increasing trend (n = 74) and the remainder as stable (n = 138). To obtain representative global species richness maps, a hexagonal grid was overlaid onto these species distributions (see [47,49]). The grid was defined on an icosahedron and projected to the sphere using the inverse Icosahedral Snyder Equal Area (ISEA) Projection, thus taking into account the Earth's spherical nature, and consisting of cells of approximately 23 300 km2. Species richness was calculated as the number of species polygons intersecting each hexagonal grid cell using the extension ‘Counting overlapping polygons’ [50] in ArcView GIS v. 3.3 [51]. In addition, measures of population change from terrestrial species in the population dataset (see below) were averaged for each terrestrial biome [52]. All maps were plotted in ArcMap v. 9.2 [53]. We evaluated spatial congruence of risk measures by comparing global patterns for decline at two different scales.
(d). Analyses
All analyses were carried out in R v. 2.11.1 [54]. We computed three measures of population change using raw population trend time-series data from 292 mammal species of 1384 populations. These were:
— slope of a linear regression of year against population size (LRS);
— mean annual change (MAC) in population size calculated using a Generalized Additive Modelling framework; and
— total change (TC) in population size over time.
We compiled three measures of population change for each species by calculating the mean of the logged trend values from its constituent populations. Previous analyses of population trend have used total measures of population change over a time period, or categorical estimates of proportional change (e.g. [55]). Any predictive trait which might correlate with change in abundance will have greater force when lineages vary markedly in that trait, and where there is greatest variation in abundance [56]. As the LRS and MAC techniques resulted in a range of values with a small amount of variance, we also analysed TC as this yielded a greater variance across species.
Variables were log-transformed to normalize distributions and equalize error variance. We first carried out single predictor regressions of our three measures of population change against each of the 13 predictor variables. We used two methods: firstly, non-phylogenetic ordinary least squares regressions, which assume all species values are independent. Secondly, we built phylogenetic generalized linear models (PGLMs; [57] using the R package CAIC (Comparative Analysis using Independent Contrasts; available at http://r-forge.r-project.org/projects/caic), which enabled us to control for the effect of phylogeny, accounting for the fact that species are more similar than you would expect by chance [58,59]. The advantage of the PGLM approach over those that assume complete phylogenetic dependence (as in independent contrasts), or complete independence (as in non-phylogenetic regressions) is that the phylogenetic dependence of the data is incorporated into the structure of the model error term [57,60,61]. Therefore, in each regression, a maximum-likelihood estimate of λ (a multiplier of the off-diagonal elements of a phylogenetic variance–covariance matrix that best fits the data; [57]) is calculated, and used to control the degree of phylogenetic non-independence in the data variable. Following the work of Jones & Purvis [62], we reduced heteroscedasticity by repeating our PGLMs after the removal of influential points (those with a studentized residual exceeding ±3).
We built a multivariate model following the heuristic procedure described by Purvis et al. [7] to build a minimum adequate model from the full set of predictor variables. We then carried out non-phylogenetic analyses, partitioning the dataset into species threatened by different threatening processes. We looked for differences between traits that predispose populations to decline in abundance owing to over-exploitation and habitat loss. PGLMs were not used as these data were only available at the population level, where no phylogeny exists.
3. Results
(a). Global model of population decline
We analysed the change in population size of 292 species of mammals, compiled from 1384 estimates of population trend. As has been apparent in previous analyses of mammalian extinction risk, a non-phylogenetic framework resulted in slightly greater significance of predictive traits (see electronic supplementary material, table S1). Hereafter, we only discuss the results of the PGLM analysis (table 3). We found that species with high mean annual temperature within their range (t = −2.78, d.f. = 188, p = 0.01) and high potential evapotranspiration (PET) rate within their range (t = −2.44, d.f. = 187, p = 0.02) declined in abundance more rapidly (as measured by total population change: table 3). There was broad agreement of significant predictive correlates of change in abundance between the three different measures (table 3). For mean regression slope of change in population abundance, we also found that geographical range area was significantly positively associated with population decline, such that species with smaller ranges were found to be declining in abundance more rapidly (t = 2.32, d.f. = 190, p = 0.02). We found no significant multivariate model of any of the three measures of population change.
Table 3.
Single predictor phylogenetic generalized linear models for: total population change; mean annual change; and mean linear regression slope. GR, geographical range.
| predictor | slope | s.e. | t | n | adjusted r2 | Λ | log-likelihood |
|---|---|---|---|---|---|---|---|
| total population change | |||||||
| body mass | 0.01 | 0.01 | 1.18 | 248 | |||
| head–body length | 0.06 | 0.04 | 1.67* | 204 | 0.01 | 0.09 | −186.92 |
| inter-birth interval | −0.01 | 0.04 | −0.21 | 166 | |||
| litter size | −0.07 | 0.08 | −0.82 | 239 | |||
| GR area | 0.01 | 0.02 | 0.52 | 191 | |||
| age at first birth | 0.01 | 0.06 | 0.09 | 121 | |||
| home range | 0.00 | 0.01 | 0.12 | 137 | |||
| average human population density | 0.02 | 0.03 | 0.66 | 191 | |||
| precipitation | −0.04 | 0.06 | −0.62 | 189 | |||
| temperature | 0.00 | 0.00 | −2.78*** | 189 | 0.04 | 0.0001 | −162.89 |
| AET | −0.10 | 0.07 | −1.37 | 188 | |||
| PET | −0.17 | 0.07 | −2.44** | 188 | 0.03 | 0.0001 | −163.06 |
| habitat breadth | 0.03 | 0.05 | 0.54 | 252 | |||
| mean annual change | |||||||
| body mass | 0.000 | 0.001 | 0.531 | 248 | |||
| head–body length | 0.002 | 0.003 | 0.610 | 204 | |||
| inter-birth interval | −0.002 | 0.004 | −0.385 | 166 | |||
| litter size | −0.008 | 0.007 | −1.218 | 239 | |||
| GR area | 0.001 | 0.002 | 0.860 | 191 | |||
| age at first birth | −0.002 | 0.005 | −0.398 | 121 | |||
| home range | 0.000 | 0.001 | 0.034 | 137 | |||
| average human population density | 0.001 | 0.003 | 0.287 | 191 | |||
| precipitation | −0.005 | 0.005 | −0.912 | 189 | |||
| temperature | 0.000 | 0.000 | −1.839* | 189 | 0.01 | 0.0001 | 302.07 |
| AET | −0.009 | 0.006 | −1.432 | 188 | |||
| PET | −0.011 | 0.006 | −1.833* | 188 | 0.01 | 0.0001 | 300.01 |
| habitat breadth | 0.000 | 0.004 | 0.060 | 252 | |||
| mean linear regression slope | |||||||
| body mass | 0.000 | 0.001 | 0.559 | 248 | |||
| head–body length | 0.002 | 0.003 | 0.931 | 204 | |||
| inter-birth interval | 0.001 | 0.004 | 0.174 | 166 | |||
| litter size | −0.003 | 0.006 | −0.511 | 239 | |||
| GR area | 0.004 | 0.002 | 2.322** | 191 | 0.02 | 0.73 | 317.09 |
| age at first birth | −0.002 | 0.008 | −0.201 | 121 | |||
| home range | 0.001 | 0.001 | 0.555 | 137 | |||
| average human population density | 0.004 | 0.003 | 1.429 | 191 | |||
| precipitation | 0.000 | 0.006 | 0.037 | 189 | |||
| temperature | 0.000 | 0.000 | −1.839* | 189 | 0.01 | 0.59 | 313.33 |
| AET | −0.005 | 0.007 | −0.736 | 188 | |||
| PET | −0.014 | 0.007 | −1.899* | 188 | 0.02 | 0.73 | 311.73 |
| habitat breadth | 0.005 | 0.004 | 1.270 | 252 | |||
*p ≤ 0.1, **p < 0.05, ***p < 0.01.
(b). Predictors under different threats
We found that populations threatened by habitat loss with a higher age at first birth were significantly declining (table 4). However, for populations threatened by over-exploitation, we found populations which had small litter sizes were significantly declining (table 4: t = 2.20, d.f. = 60, p = 0.03), in addition to the same results for PET and mean annual temperature found above.
Table 4.
Single predictor ordinary least squares regressions for total population change for populations declining owing to habitat loss and over-exploitation. GR, geographical range.
| predictor | slope | s.e. | t | d.f. |
|---|---|---|---|---|
| habitat loss | ||||
| body mass | 0.0000 | 0.0000 | −1.14 | 56 |
| head–body length | 0.0001 | 0.0001 | 0.76 | 47 |
| inter-birth interval | −0.0001 | 0.0004 | −0.14 | 37 |
| litter size | −0.0680 | 0.0732 | −0.93 | 54 |
| GR area | 0.0000 | 0.0000 | −0.96 | 52 |
| age at first birth | −0.0002 | 0.0001 | −1.77* | 35 |
| home range | 0.0003 | 0.0003 | 1.34 | 37 |
| average human population density | 0.0004 | 0.0014 | 0.28 | 52 |
| precipitation | −0.0012 | 0.0018 | −0.68 | 51 |
| temperature | −0.0009 | 0.0011 | −0.82 | 51 |
| AET | −0.0001 | 0.0003 | −0.43 | 51 |
| PET | −0.0002 | 0.0002 | −0.71 | 51 |
| habitat breadth | −0.0108 | 0.0113 | −0.95 | 57 |
| overexploitation | ||||
| body mass | 0.0000 | 0.0000 | 0.41 | 62 |
| head–body length | 0.0000 | 0.0000 | 0.13 | 54 |
| inter-birth interval | −0.0001 | 0.0002 | −0.40 | 46 |
| litter size | 0.1412 | 0.0641 | 2.20** | 60 |
| GR area | 0.0000 | 0.0000 | 0.60 | 39 |
| age at first birth | 0.0001 | 0.0002 | 0.29 | 29 |
| home range | 0.0000 | 0.0000 | 0.24 | 29 |
| average human population density | 0.0014 | 0.0019 | 0.72 | 39 |
| precipitation | −0.0008 | 0.0023 | −0.35 | 39 |
| temperature | −0.0022 | 0.0010 | −2.27** | 39 |
| AET | −0.0005 | 0.0003 | −1.41 | 39 |
| PET | −0.0005 | 0.0002 | −2.08** | 39 |
| habitat breadth | 0.0198 | 0.0154 | 1.28 | 65 |
*p ≤ 0.1, **p < 0.05, ***p < 0.01.
(c). Spatial congruence of decline across scales
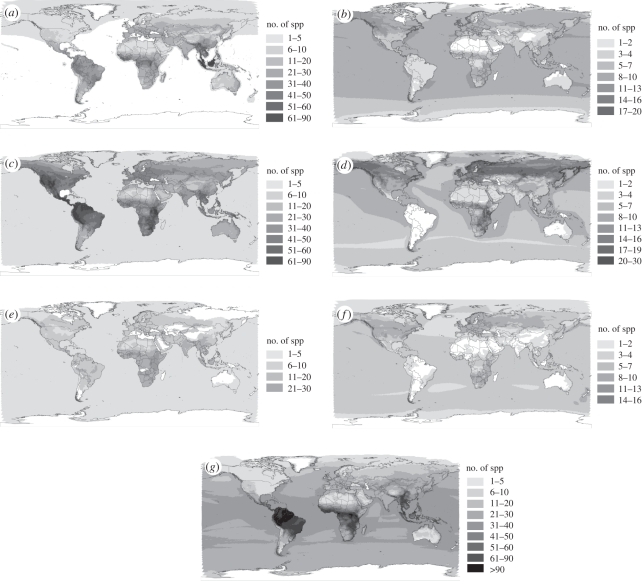
Figure 1 shows the patterns of population trend at two scales, for species-level population trends (figure 1a,c,e,g) and site-level change in population abundance (figure 1b,d,f). We found the greatest differences between the two scales for species declining in abundance (figure 1a,b). Incomplete species coverage at a site scale results in large differences for South America and southeast Asia in particular, which are most likely explained by poor data coverage. However, there is also an aggregation of species with declining population trends in the Arctic region of North America, which is apparent at a site scale but not at a species level. Species with stable abundance trends (figure 1c,d) and increasing abundance trends (figure 1e,f) show broadly similar results, though with a large data gap in South America once again for stable populations measured at a site scale (figure 1d). Species categorized by IUCN as having unknown population trends show particularly large aggregations of species in central and South America, and areas of Eastern and Southern Africa (figure 1g).
Figure 1.
Global patterns of species and population-level decline in mammals for (a) species flagged as declining (IUCN), (b) species that have declining total population trend, (c) species flagged as stable (IUCN), (d) species that have stable total population trend, (e) species flagged as increasing (IUCN), (f) species that have increasing total population trend, (g) species flagged as unknown (IUCN). Dark colours are accumulations of high numbers of species and light colours denote low numbers of species.
4. Discussion
In line with studies of species-level extinction risk, our analyses show that population decline is determined by a number of interacting aspects of biology, geography and threat. Specifically, we find that mammals living in areas with a high rate of mean PET and high mean temperature in their range are more likely to be experiencing population decline. The likely explanation for these findings is that as a measure of primary productivity, species with high values of PET are likely to be found in the tropics, which show much greater levels of population decline than other regions [42]. In an analysis of extinction risk in ungulates [19], actual evapotranspiration was included as a measure of primary productivity, which is thought to be a confounding factor when using mean human population density [63]. We cannot yet rule out this effect though the correlation of PET with more rapidly declining tropical species seems to be the more likely explanation.
Overall, support for biological correlates across the species set is weak, with low r2 values indicating low explanatory power. These relatively small effects suggest that we should look into extrinsic processes to explain the variation in population trend that we observe. It could be that species experiencing rapid changes in population size could be those with traits that cause them to be especially prone to certain extrinsic pressures, or that when pressures are of very high intensity, species biology ceases to matter. Partitioning our dataset into different rates of decline could be revealing. The correlates of rapid change in abundance could, for example, be associated with novel threats, and concomitantly rapid increase in abundance with cessation of threats [64], whereas persistent but low level threats may result in greater association with measures of intrinsic species biology.
To our knowledge, there have been very few studies which have looked for correlates of population decline. Thomas [37] found that British birds with declining populations tended to be phylogenetically clustered, suggesting that there are potential biological and geographical characteristics that unite them. In a study of mammals in Ghanaian National Parks [22], Brashares found that population persistence was correlated with degree of isolation of the population and the type of mating system, which was attributed to monogamous species being particularly vulnerable to demographic stochasticity. We find that our results are broadly comparable to those found at a species level in mammals [6,11], though with fewer significant determinants of risk identified.
At a global species level, many analyses have found small geographical range correlates strongly with higher extinction risk [6,7,12,17,19], though its correlation can be confounded, owing to the presence of geographical range in processes such as Red List classification (though see [7]). At a population level, our study did not find a consistent significant correlation of geographical range with population decline.
The associations of correlates of extinction risk are believed to vary under different threatening processes [65]. The fact that we found few significant predictors of population decline for populations impacted by habitat loss could be owing to small sample sizes (our largest predictor had 58 populations), or lack of variance across predictor trait or dependent variable (i.e. too little variation in abundance to explain). There is accumulating evidence that rather than rapid declines in abundance, slow declines in common species under the effects of habitat loss are becoming more prevalent [39]. Such slow declines are difficult to pick up in this type of analysis, though may have major ramifications for processes such as ecosystem services, as by definition the common more abundant species are likely to shape the ecosystems that they inhabit, contributing much of the structure, biomass and energy turnover [66]. Over-exploitation and disease outbreaks, on the other hand perhaps, cause local populations to decline more quickly [35]. We find the mean rate of change in population abundance between the two threat types is more rapid in over-exploited populations. Within over-exploited species, our results suggest that species with larger litter size are less subject to declining populations. Larger litters contribute to fast population growth, and are probably part of the reason that smaller species are in general less extinction-prone [5].
As monitoring of population level change in abundance improves in breadth and scope [67], the picture observed here will become more complete. Unfortunately, some parts of the world have clear gaps in population monitoring, for example tropical regions of South America and southeast Asia [68]. The latter in particular is known to be one of the most rapidly changing regions, in terms of land cover, with higher rate of forest loss than any other region [69]. Site-level data on change in population abundance show some distinct advantages over broad scale but coarse classifications of trend, such as those used in IUCN Red Listing. For example, site-level information has recently revealed ‘hotspots’ of decline in abundance within the Arctic, a trend which is not yet apparent using global species information ([70]; though see [71]). This type of fine scale information is at the cost of breadth of coverage though, and is currently taxonomically limited.
Finally, population trends categorized as ‘unknown’ [47,48] appear to be congregated in tropical regions in particular, specifically in areas that are undergoing rapid change in land cover such as Central America and southeast Asia. While it is apparent that such areas are globally the most depauperate in biodiversity information at all scales [68], there is clear indication that inroads must be made towards enhancing data collection in these regions, which potentially have the most biodiversity to lose.
5. Conclusion
Rates of decline in abundance documented in cross-species datasets provide some of the most compelling evidence of changing rates of biodiversity loss. To address and reverse these declines, we must understand the interactions between human impact, species and their environment. In mammals, environmental variables are generally better determinants of cross-species population-level decline rates than intrinsic biological traits. However, with threats changing in intensity, spatial scale and type, more targeted analyses across a broader range of species are required to truly understand this variation, and provide a basis for proactive conservation management.
Acknowledgements
We are grateful to Rachel Burrows, Jenny Beschizza, Olivia Daniel, Annemarie Greenwood, Nicola Harrison, Gayle Kothari, Julia Latham, Robyn Manley, Jenny Martin, Fiona Pamplin, Sandra Tranquilli and Sarah Whitmee for data collection; Mike Gill and Thomas Galewski for project support; and to Danny Richman and his team for the new online database. WWF International provided funding (L.M. and J.L.). B.C. is supported by the Rufford Foundation.
References
- 1.May R. M. 1994. The effects of spatial scale on ecological questions and answers. In Large-scale ecology and conservation biology (eds Edwards P. J., May R. M., Webb N. R.), pp. 1–18 Oxford, UK: Blackwell Scientific [Google Scholar]
- 2.Ceballos G., Ehrlich P. R. 2002. Mammal population losses and the extinction crisis. Science 296, 904–907 10.1126/science.1069349 (doi:10.1126/science.1069349) [DOI] [PubMed] [Google Scholar]
- 3.Collen B., Loh J., Holbrook S., McRae L., Amin R., Baillie J. E. M. 2009. Monitoring change in vertebrate abundance: the Living Planet Index. Conserv. Biol. 23, 317–327 10.1111/j.1523-1739.2008.01117.x (doi:10.1111/j.1523-1739.2008.01117.x) [DOI] [PubMed] [Google Scholar]
- 4.Purvis A. 2008. Phylogenetic approaches to the study of extinction. Annu. Rev. Ecol. Syst. 39, 301–319 10.1146/annurev.ecolsys.063008.102010 (doi:10.1146/annurev.ecolsys.063008.102010) [DOI] [Google Scholar]
- 5.Cardillo M. 2003. Biological determinants of extinction risk: why are smaller species less vulnerable? Anim. Conserv. 6, 1–7 10.1017/S1367943003003093 (doi:10.1017/S1367943003003093) [DOI] [Google Scholar]
- 6.Cardillo M., Mace G. M., Jones K. E., Bielby J., Bininda-Emonds O. R. P., Sechrest W., Orme C. D. L., Purvis A. 2005. Multiple causes of high extinction risk in large mammal species. Science 309, 1239–1241 10.1126/science.1116030 (doi:10.1126/science.1116030) [DOI] [PubMed] [Google Scholar]
- 7.Purvis A., Gittlemann J. L., Cowlishaw G., Mace G. M. 2000. Predicting extinction risk in declining species. Proc. R. Soc. Lond. B 267, 1947–1952 10.1098/rspb.2000.1234 (doi:10.1098/rspb.2000.1234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purvis A., Cardillo M., Grenyer R., Collen B. 2005. Correlates of extinction risk: phylogeny, biology, threat and scale. In Phylogeny and conservation (eds Purvis A., Brooks T. M., Gittleman J. L.), pp. 295–316 Cambridge, UK: Cambridge University Press [Google Scholar]
- 9.Fisher D. O., Owens I. P. F. 2004. The comparative method in conservation biology. Trends Ecol. Evol. 19, 391–398 10.1016/j.tree.2004.05.004 (doi:10.1016/j.tree.2004.05.004) [DOI] [PubMed] [Google Scholar]
- 10.Isaac N. J. B., Cowlishaw G. 2004. How species respond to multiple extinction threats: evidence from primates. Proc. R. Soc. Lond. B 271, 1135–1141 10.1098/rspb.2004.2724 (doi:10.1098/rspb.2004.2724) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cardillo M., Mace G. M., Gittleman J. L., Purvis A. 2006. Latent extinction risk and the future battlegrounds of mammal conservation. Proc. Natl Acad. Sci. USA 103, 4157–4161 10.1073/pnas.0510541103 (doi:10.1073/pnas.0510541103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harcourt A. H., Schwartz M. W. 2001. Primate evolution: a biology of holocene extinction and survival on the southeast Asian Sunda shelf islands. Am. J. Phys. Anthropol. 114, 4–17 (doi:10.1002/1096-8644(200101)114:1<4::AID-AJPA1001>3.0.CO;2-6) [DOI] [PubMed] [Google Scholar]
- 13.Cardillo M., Bromham L. 2001. Body size and risk of extinction in Australian mammals. Conserv. Biol. 15, 1435–1440 10.1046/j.1523-1739.2001.00286.x (doi:10.1046/j.1523-1739.2001.00286.x) [DOI] [Google Scholar]
- 14.Cardillo M., Mace G. M., Gittleman J. L., Jones K. E., Bielby J., Purvis A. 2008. The predictability of extinction: biological and external correlates of decline in mammals. Proc. R. Soc. B 275, 1441–1448 10.1098/rspb.2008.0179 (doi:10.1098/rspb.2008.0179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher D. O., Blomberg S. P., Owens I. P. F. 2003. Extrinsic versus intrinsic factors in the decline and extinction of Australian marsupials. Proc. R. Soc. Lond. B 270, 1801–1808 10.1098/rspb.2003.2447 (doi:10.1098/rspb.2003.2447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson C. N. 2002. Determinants of loss of mammal species during the Late Quaternary ‘megafauna’ extinctions: life history and ecology, but not body size. Proc. R. Soc. Lond. B 269, 2221–2227 10.1098/rspb.2002.2130 (doi:10.1098/rspb.2002.2130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones K. E., Purvis A., Gittleman J. L. 2003. Biological correlates of extinction risk in bats. Am. Nat. 161, 601–614 10.1086/368289 (doi:10.1086/368289) [DOI] [PubMed] [Google Scholar]
- 18.Smith A. P., Quin D. G. 1996. Patterns and causes of extinction and decline in Australian conilurine rodents. Biol. Conserv. 77, 243–267 10.1016/0006-3207(96)00002-X (doi:10.1016/0006-3207(96)00002-X) [DOI] [Google Scholar]
- 19.Price S. A., Gittleman J. L. 2007. Bushmeat hunting, habitat loss and global extinction in the Artiodactyla. Proc. R. Soc. B 274, 1845–1851 10.1098/rspb.2007.0505 (doi:10.1098/rspb.2007.0505) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cardillo M., Purvis A., Sechrest W., Gittleman J. L., Bielby J., Mace G. M. 2004. Human population density and extinction risk in the world's carnivores. PLoS Biol. 2, 909–914 10.1371/journal.pbio.0020197 (doi:10.1371/journal.pbio.0020197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ceballos G., Ehrlich P. E., Sobero J., Salazar I., Fay J. P. 2005. Global mammal conservation: what must we manage? Science 309, 603–607 10.1126/science.1114015 (doi:10.1126/science.1114015) [DOI] [PubMed] [Google Scholar]
- 22.Brashares J. S. 2003. Ecological, behavioural and life-history correlates of mammal extinctions in West Africa. Conserv. Biol. 17, 733–743 10.1046/j.1523-1739.2003.01592.x (doi:10.1046/j.1523-1739.2003.01592.x) [DOI] [Google Scholar]
- 23.Harcourt S. A. 1998. Ecological indicators of risk for primates, as judged by species' susceptibility to logging. In Behavioural ecology and conservation ecology (ed. Caro T.), pp. 56–79 New York, USA: Oxford University Press [Google Scholar]
- 24.Woodroffe R., Ginsberg J. R. 1998. Edge effects and the extinction of populations inside protected areas. Science 280, 2126–2128 10.1126/science.280.5372.2126 (doi:10.1126/science.280.5372.2126) [DOI] [PubMed] [Google Scholar]
- 25.Mayhew P. J., Jenkins G. B., Benton T. G. 2008. A long-term association between global temperature and biodiversity, origination and extinction in the fossil record. Proc. R. Soc. B 275, 47–53 10.1098/rspb.2007.1302 (doi:10.1098/rspb.2007.1302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harcourt A. H., Coppeto S. A., Parks S. A. 2002. Rarity, specialization and extinction in primates. J. Biogeogr. 29, 445–456 10.1046/j.1365-2699.2002.00685.x (doi:10.1046/j.1365-2699.2002.00685.x) [DOI] [Google Scholar]
- 27.Jernvall J., Wright P. C. 1998. Diversity components of impending primate extinctions. Proc. Natl Acad. Sci. USA 95, 11 279–11 283 10.1073/pnas.95.19.11279 (doi:10.1073/pnas.95.19.11279) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raup D. M. 1991. Extinction: bad genes or bad luck? New York, NY: Norton; [PubMed] [Google Scholar]
- 29.Purvis A., Agapow P.-M., Gittleman J. L., Mace G. M. 2000. Nonrandom extinction and the loss of evolutionary history. Science 288, 328–330 10.1126/science.288.5464.328 (doi:10.1126/science.288.5464.328) [DOI] [PubMed] [Google Scholar]
- 30.Bielby J., Cunningham A. A., Purvis A. 2006. Taxonomic selectivity in amphibians: ignorance, geography or biology? Anim. Conserv. 9, 135–143 10.1111/j.1469-1795.2005.00013.x (doi:10.1111/j.1469-1795.2005.00013.x) [DOI] [Google Scholar]
- 31.Cardillo M. 2011. Phylogenetic structure of mammal assemblages at large geographic scales: linking phylogenetic community ecology with macroecology. Phil. Trans. R. Soc. B 366, 2545–2553 10.1098/rstb.2011.0021 (doi:10.1098/rstb.2011.0021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purvis A., Jones K. E., Mace G. M. 2000. Extinction. BioEssays 22, 1123–1133 (doi:10.1002/1521-1878(200012)22:12<1123::AID-BIES10>3.0.CO;2-C) [DOI] [PubMed] [Google Scholar]
- 33.Reynolds J. D. 2003. Life histories and extinction risk. In Macroecology: concepts and consequences (eds Blackburn T. M., Gaston K. J.), pp. 195–217 Oxford, UK: Blackwell [Google Scholar]
- 34.Collen B., Bykova E., Ling S., Milner-Gulland E. J., Purvis A. 2006. Extinction risk: a comparative analysis of Central Asian vertebrates. Biodivers. Conserv. 15, 1859–1871 10.1007/s10531-005-4303-6 (doi:10.1007/s10531-005-4303-6) [DOI] [Google Scholar]
- 35.Mace G. M., Collar N. J., Gaston K. J., Hilton-Taylor C., Akcakaya H. R., Leader-Williams N., Milner-Gulland E. J., Stuart S. N. 2008. Quantification of extinction risk: IUCN's system for classifying threatened species. Conserv. Biol. 22, 1424–1442 10.1111/j.1523-1739.2008.01044.x (doi:10.1111/j.1523-1739.2008.01044.x) [DOI] [PubMed] [Google Scholar]
- 36.IUCN 2001. IUCN Red List Categories and Criteria version 3.1. Gland, Switzerland: IUCN [Google Scholar]
- 37.Thomas G. H. 2008. Phylogenetic distributions of British birds of conservation concern. Proc. R. Soc. B 275, 2077–2083 10.1098/rspb.2008.0549 (doi:10.1098/rspb.2008.0549) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bielby J., Cooper N., Cunningham A. A., Garner T. W. J., Purvis A. 2008. Predicting declines in the world's frogs. Conserv. Lett. 1, 82–90 10.1111/j.1755-263X.2008.00015.x (doi:10.1111/j.1755-263X.2008.00015.x) [DOI] [Google Scholar]
- 39.Gaston K. J. 2010. Valuing common species. Science 327, 154–155 10.1126/science.1182818 (doi:10.1126/science.1182818) [DOI] [PubMed] [Google Scholar]
- 40.Robinson R. A., et al. 2010. Emerging infectious disease leads to rapid population declines of common British birds. PLoS ONE 5, e12215. 10.1371/journal.pone.0012215 (doi:10.1371/journal.pone.0012215) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones K. E., et al. 2009. PanTHERIA: a species-level database of life-history, ecology and geography of extant and recently extinct mammals. Ecology 90, 2648. 10.1890/08-1494.1 (doi:10.1890/08-1494.1) [DOI] [Google Scholar]
- 42.Loh J., Green R. E., Ricketts T., Lamoreux J. F., Jenkins M., Kapos V., Randers J. 2005. The Living Planet Index: using species population time series to track trends in biodiversity. Phil. Trans. R. Soc. B 360, 289–295 10.1098/rstb.2004.1584 (doi:10.1098/rstb.2004.1584) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bininda Emonds O. R. P., et al. 2007. The delayed rise of present-day mammals. Nature 446, 507–512 10.1038/nature05634 (doi:10.1038/nature05634) [DOI] [PubMed] [Google Scholar]
- 44.NERC Centre for Population Biology, Imperial College. 2010. The Global Population Dynamics database, version 2. See http://www.sw.ic.ac.uk/cpb/cpb/gpdd.html
- 45.EBCC—European Bird Census Council 2006. European common bird index: population trends of European common birds 2005 update. See http://www.ebcc.info/ (accessed 02 June 2006)
- 46.IUCN 2007. IUCN habitat classification scheme. See http://www.iucn.org/themes/ssc/sis/authority.htm (accessed 28th February 2007)
- 47.Schipper J., et al. 2008. The biogeography of diversity, threat, and knowledge in the world's terrestrial and aquatic mammals. Science 322, 225–230 10.1126/science.1165115 (doi:10.1126/science.1165115) [DOI] [PubMed] [Google Scholar]
- 48.IUCN 2010. IUCN Red List of Threatened Species. Version 2010.3. See www.iucnredlist.org
- 49.Clausnitzer V., et al. 2009. Odonata enter the biodiversity crisis debate: the first global assessment of an insect group. Biol. Conserv. 142, 1864–1869 10.1016/j.biocon.2009.03.028 (doi:10.1016/j.biocon.2009.03.028) [DOI] [Google Scholar]
- 50.Smith R. J. 2004. Count overlapping polygons: an ArcView GIS extension. See http://arcscripts.esri.com/details.asp?dbid=13345
- 51.Environmental Systems Research Institute 2002. ArcView® GIS 3.3. See http://www.esri.com
- 52.Olson D. M., et al. 2001. Terrestrial ecoregions of the world: a new map of life on Earth. Bioscience 51, 933–938 10.1641/0006-3568(2001)051[0933:TEOTWA]2.0.CO;2 (doi:10.1641/0006-3568(2001)051[0933:TEOTWA]2.0.CO;2) [DOI] [Google Scholar]
- 53.Environmental Systems Research Institute 2006. ArcMap™ 9.2. See http://www.esri.com
- 54.R Development Core Team 2010. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org [Google Scholar]
- 55.Sanderson F. J., Donald P. F., Pain D. J., Burfield I. J., van Bommel F. P. J. 2006. Long-term population declines in Afro-Palearctic migrant birds. Biol. Conserv. 131, 93–105 10.1016/j.biocon.2006.02.008 (doi:10.1016/j.biocon.2006.02.008) [DOI] [Google Scholar]
- 56.Gittleman J. L., Purvis A. 1998. Body size and species-richness in carnivores and primates. Proc. R. Soc. Lond. B 265, 113–119 10.1098/rspb.1998.0271 (doi:10.1098/rspb.1998.0271) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Freckleton R. P., Harvey P. H., Pagel M. 2002. Phylogenetic analysis and comparative data: a test and review of evidence. Am. Nat. 160, 712–726 10.1086/343873 (doi:10.1086/343873) [DOI] [PubMed] [Google Scholar]
- 58.Garland T., Harvey P. H., Ives A. R. 1992. Procedures for analysis of comparative data using phylogenetically independent contrasts. Syst. Biol. 41, 18–32 [Google Scholar]
- 59.Purvis A., Rambaut A. 1995. Comparative analysis by independent contrasts (CAIC): an Apple Macintosh application for analysing comparative data. Comput. Appl. Biosci. 11, 247–251 [DOI] [PubMed] [Google Scholar]
- 60.Pagel M. 1999. Inferring the historical patterns of biological evolution. Nature 401, 877–884 10.1038/44766 (doi:10.1038/44766) [DOI] [PubMed] [Google Scholar]
- 61.Rohlf F. J. 2001. Comparative methods for the analysis of continuous variables: geometric interpretations. Evolution 55, 2143–2160 [DOI] [PubMed] [Google Scholar]
- 62.Jones K. E., Purvis A. 1997. An optimum body size for mammals? Comparative evidence from bats. Funct. Ecol. 11, 751–756 10.1046/j.1365-2435.1997.00149.x (doi:10.1046/j.1365-2435.1997.00149.x) [DOI] [Google Scholar]
- 63.Balmford A., Moore J. L., Brooks T., Burgess N., Hansen L. A., Williams P., Rahbek C. 2001. Conservation conflicts across Africa. Science 291, 2616–2619 10.1126/science.291.5513.2616 (doi:10.1126/science.291.5513.2616) [DOI] [PubMed] [Google Scholar]
- 64.Mace G. M., Collen B., Fuller R. A., Boakes E. H. 2010. Population and geographic range dynamics: implications for conservation planning. Phil. Trans. R. Soc. B 365, 3743–3751 10.1098/rstb.2010.0264 (doi:10.1098/rstb.2010.0264) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Owens I. P. F., Bennett P. M. 2000. Ecological basis of extinction risk in birds: habitat loss versus human persecution and introduced predators. Proc. Natl Acad. Sci. USA 97, 12 144–12 148 10.1073/pnas.200223397 (doi:10.1073/pnas.200223397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gaston K. J., Fuller R. A. 2008. Commonness, population depletion and conservation biology. Trends Ecol. Evol. 23, 14–19 10.1016/j.tree.2007.11.001 (doi:10.1016/j.tree.2007.11.001) [DOI] [PubMed] [Google Scholar]
- 67.Pereira H. M., et al. 2010. Global biodiversity monitoring: filling the gap where it counts the most. Front. Ecol. Environ. 8, 459–460 10.1890/10.WB.23 (doi:10.1890/10.WB.23) [DOI] [Google Scholar]
- 68.Collen B., Ram M., Zamin T., McRae L. 2008. The tropical biodiversity data gap: addressing disparity in global monitoring. Tropic. Conserv. Sci. 1, 75–88 See tropicalconservationscience.org [Google Scholar]
- 69.Millennium Ecosystem Assessment 2005. Ecosystems and human well-being: wetlands and water synthesis. Washington, DC: World Resources Institute [Google Scholar]
- 70.McRae L., et al. Arctic Species Trend Index 2010: tracking trends in Arctic wildlife. CAFF CBMP Report no. 20. Akureyri, Iceland: CAFF International Secretariat
- 71.Foden W., et al. 2009. Species susceptibility to climate change impacts. In Wildlife in a changing world: an analysis of the 2008 IUCN Red List of Threatened Species (eds Vié J.-C., Hilton-Taylor C., Stuart S. N.), pp. 77–87 Gland, Switzerland: IUCN [Google Scholar]